Abstract
Abstract
The growing use of artificial neural network (ANN) tools for computed tomography angiography (CTA) data analysis underscores the necessity for elevated data protection measures. We aimed to establish an automated defacing pipeline for CTA data. In this retrospective study, CTA data from multi-institutional cohorts were utilized to annotate facemasks (n = 100) and train an ANN model, subsequently tested on an external institution’s dataset (n = 50) and compared to a publicly available defacing algorithm. Face detection (MTCNN) and verification (FaceNet) networks were applied to measure the similarity between the original and defaced CTA images. Dice similarity coefficient (DSC), face detection probability, and face similarity measures were calculated to evaluate model performance. The CTA-DEFACE model effectively segmented soft face tissue in CTA data achieving a DSC of 0.94 ± 0.02 (mean ± standard deviation) on the test set. Our model was benchmarked against a publicly available defacing algorithm. After applying face detection and verification networks, our model showed substantially reduced face detection probability (p < 0.001) and similarity to the original CTA image (p < 0.001). The CTA-DEFACE model enabled robust and precise defacing of CTA data. The trained network is publicly accessible at www.github.com/neuroAI-HD/CTA-DEFACE.
Relevance statement
The ANN model CTA-DEFACE, developed for automatic defacing of CT angiography images, achieves significantly lower face detection probabilities and greater dissimilarity from the original images compared to a publicly available model. The algorithm has been externally validated and is publicly accessible.
Key Points
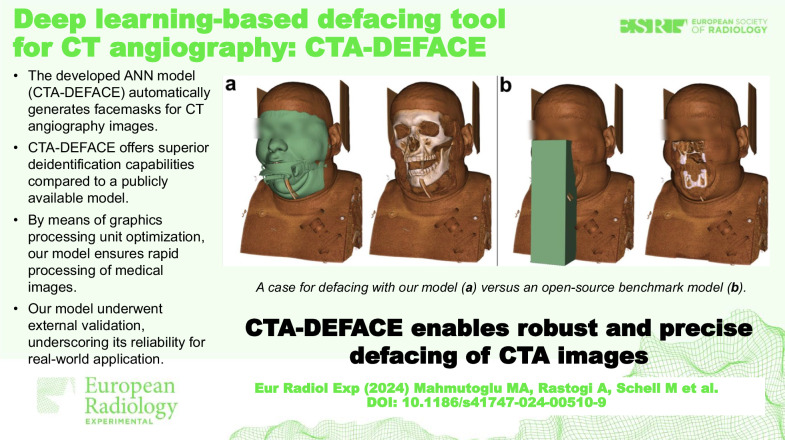
The developed ANN model (CTA-DEFACE) automatically generates facemasks for CT angiography images.
CTA-DEFACE offers superior deidentification capabilities compared to a publicly available model.
By means of graphics processing unit optimization, our model ensures rapid processing of medical images.
Our model underwent external validation, underscoring its reliability for real-world application.
Graphical Abstract
Keywords: Artificial intelligence, Computed tomography angiography, Data anonymization, Image processing (computer-assisted), Neural network (computer)
Background
Computed tomography angiography (CTA) plays a crucial role in evaluating patients with vascular malformations, aneurysms, and tumors, and notably in diagnosing vessel occlusion in acute ischemic stroke. Machine/deep learning technology is increasingly applied to CTA to assess cervical artery anatomy [1] and stenosis [2], radiomics signatures of carotid plaques [3] and to develop automated tools for detecting vessel occlusions [4–7]. This last task often relies on large datasets acquired through data-sharing initiatives. Additionally, the clinical use of such tools may necessitate the sharing of patient images, emphasizing the need for robust data protection protocols.
A critical aspect of deidentifying medical images is the removal or distortion of identifiable facial features [8–10]. While numerous defacing tools are available for magnetic resonance imaging [11–13], they are less available for computed tomography (CT) or positron emission tomography [14–16].
We developed a neural network-based approach for defacing CTA images (CTA-DEFACE), based on the nnU-Net framework [17] and compared our CTA-DEFACE model against the publicly available facemask generator function from the ICHSEG library [18].
Methods
Dataset
This retrospective multicenter study was approved by the local ethics committee. CTA data from two cohorts for model training and a third distinct cohort for testing was used. Fifty patients previously treated at Heidelberg University Hospital (cohort 1) and 50 patients from three primary/secondary care hospitals of the regional stroke consortium Rhein-Neckar with acute teleneurology/teleradiology coverage through the Heidelberg University Hospital (cohort 2), were used for model training. These patients were diagnosed with acute ischemic stroke and CTA-confirmed vessel occlusion. Testing involved 50 patients who underwent CTA for suspected acute ischemic stroke at Bonn University Hospital’s Department of Neuroradiology (cohort 3).
Cohorts 1 and 2 were partitioned equally for a balanced representation of each cohort in both training processes. Scanner and acquisition parameters are depicted in Supplementary Table 1, and patient demographics are in Supplementary Tables 2 and 3.
Study design
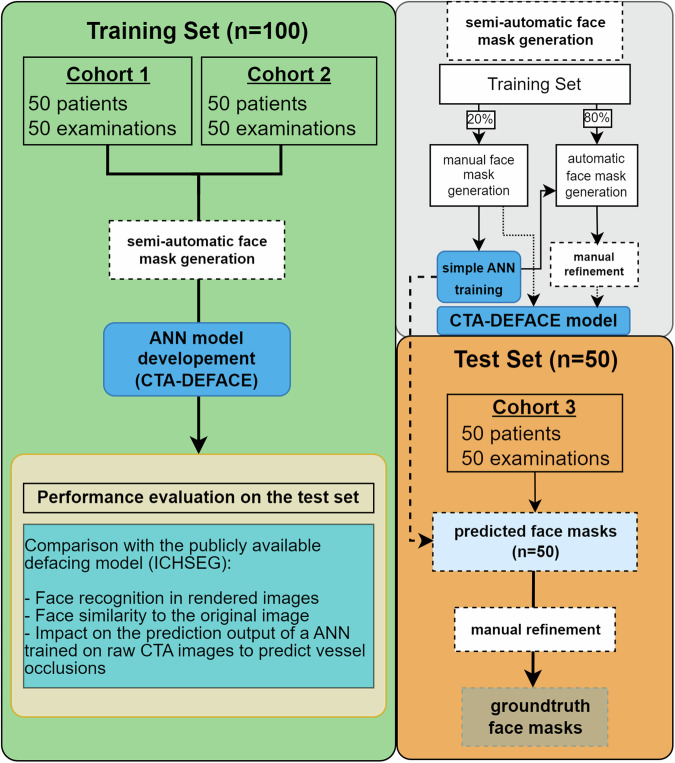
Figure 1 shows the flowchart of model training and testing. The state-of-the-art nnU-Net was used for training, automatically configuring hyperparameters based on the dataset characteristics [17]. The model was trained on NVIDIA DGX A100 (NVIDIA, Santa Clara, CA, USA) using 2 A100-SXM4 graphics processing units (GPUs) of 40GB each with AMD EPYC 7742 64-Core Processor and 1 TB RAM. The inference runtimes for predictions were measured on a local workstation with an Intel Xeon E5/Core i7 3.1 GHz CPU and NVIDIA TITAN RTX GPU.
Fig. 1.
Flowchart of the procedures for CTA-DEFACE and model training and external testing
Model development
To train the CTA-DEFACE model, the entire training dataset (n = 100) was divided into two equal and disjoint sets from both HD and FAST cohorts (n = 50 each). Face mask generation for CTA data was done in Slicer 3D (version 5.4.0), annotating soft face tissue from forehead to chin, including the nose, lips, and masseter muscles. An initial nnU-Net was trained on 20% of the training set (ten patients each from cohorts 1 and 2). Subsequently, this model predicted facemasks from the remaining 80% of the training set and the complete test set. These predictions were manually refined to generate ground truth facemasks. The final nnU-Net training was done using the entire CTA dataset (n = 100). The resulting predicted facemasks on the external test set (cohort 3), were compared with the ground truth.
Face detection, recognition, and validation
To validate our model, we utilized the publicly available CT defacing tool “ct_face_mask” from the “ICHSEG” package in R [18] for comparative analysis. Predicted facemasks from ICHSEG and our model on the test set were replaced with the 10th percentile value of the original image to represent void space. The processing time for face segmentation was measured for both methods.
CTA images from the test set were visualized in Slicer 3D (version 5.4.0) with “CT-Muscle” preset. Rendered images were captured from an anterior viewpoint for the original image and the defaced versions with ICHSEG and CTA-DEFACE. Examples of each algorithm are illustrated in Supplementary Fig. 1.
Unlike classical segmentation tasks, defacing algorithms cannot be compared with traditional metrics like dice similarity coefficient (DSC) due to the lack of actual ground truth. Therefore, to compare ICHSEG and CTA-DEFACE, we employed two neural networks from previous studies [16, 19]. Face detection in rendered images was conducted using a multitask cascaded convolutional neural network (MTCNN) [20], which integrates three convolutional neural network structures for face recognition, bounding box regression, and facial landmark localization. Further validation involved quantifying the face identifiability of CTA images before and after defacing. If a face was detected by MTCNN, FaceNet [21] was used to extract the face embedding vector to verify whether the face matched the rendered original image. Face embedding vectors were calculated by FaceNet on the rendered original CTA image and after defacing with ICHSEG and CTA-DEFACE models. We calculated the Euclidean distance, a measure of similarity, between the embeddings of the original CTA image and the images produced by two defacing strategies (less distance representing a greater similarity).
Statistical analysis
Statistical analyses were performed using Python (version 3.8.13) and R (version 4.0.5). The performance of each model was assessed on the cohort 3 test set. Face detection probabilities, calculated on the captions of rendered images, underwent analysis using a nonparametric Friedman rank sum test with a post-hoc pairwise Wilcoxon signed-rang exact test with Bonferroni correction. Euclidian distance for face embedding vectors was computed to examine similarities. Paired t-tests compared the defacing algorithms. If normal distributions were met, descriptive statistics were provided in terms of mean and standard deviation.
Results
CTA-DEFACE model characteristics
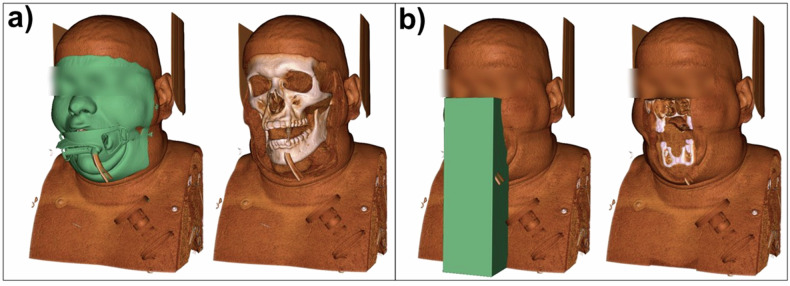
The facemask generated by CTA-DEFACE covers soft tissue and skin from the forehead (above the frontal sinus) to the chin (at the level of hyoid bone), which includes soft tissue around the eyes, nose, masseter muscles, and lips. In cases where external devices are present, such as protective glass or an intubation tube, they are also depicted by the segmentation mask. In contrast, the ICHSEG library’s “ct_face_mask” function predicts a rectangular prism covering the mouth and nose (Fig. 2).
Fig. 2.
An illustrative case from the test set for CTA-DEFACE (a) and the “ct_face_mask” function from the ICHSEG library (b). Illustrations are rendered images of CT volumes in Slicer 3D software with the “CT-Muscle” preset. Left: predicted segmentation masks. Right: volume rendering after subtracting the facemask and replacing it with the 10th percentile of the HU value of the image to represent empty air. Eyes are blurred for anonymization purposes
Segmentation and face recognition metrics
The CTA-DEFACE model achieved a DSC of 0.94 ± 0.02 (mean ± standard deviation) on ground truth facemasks. For each test case, the average face segmentation time was 0.2 ± 0.1 min (mean ± standard deviation) with the CTA-DEFACE model and 36.3 ± 9.2 min with ICHSEG.
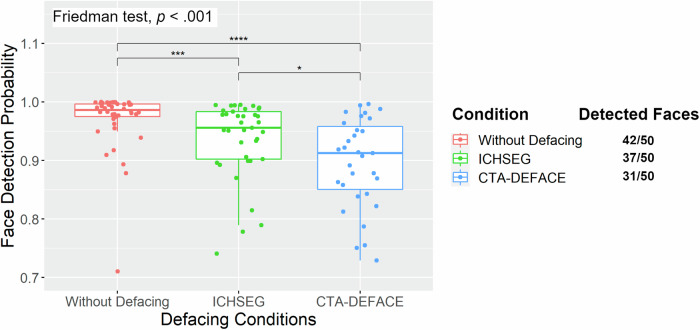
Faces were detected by the MTCNN network on 42/50 (84%, 95% confidence interval: 71–93%) of original images without defacing, 37/50 (74%, 95% confidence interval: 60–85%) of images defaced with ICHSEG, and 31/50 (62%, 95% confidence interval: 47–75%) of images defaced with CTA-DEFACE (Fig. 3).
Fig. 3.
Face detection probabilities by multitask cascaded convolutional neural network (MTCNN) on the rendered CTA images are illustrated in boxplots for the original CTA volume image (left), defaced image with ICHSEG (center) and defaced image with CTA-DEFACE (right). Undetected faces were excluded. The number of detected faces is indicated next to each method (on the right). Nonparametric Friedman rank sum test with post-hoc pairwise comparison using Wilcoxon signed rand exact were calculated. CTA, Computed tomography angiography, *p = 0.04, ***p < 0.001, ****p < 0.0001
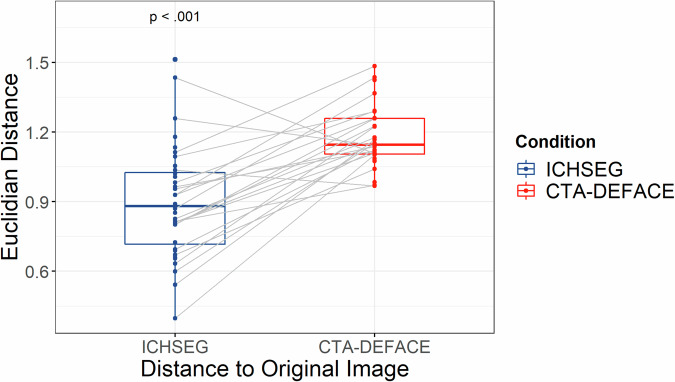
Face embedding vectors after defacing with our model showed a significantly increased distance from the original image (Fig. 4).
Fig. 4.
Euclidian distance of face embedding vectors between the original CTA images and defaced images (ICHSEG versus CTA-DEFACE) are shown as boxplots. Lower values indicate greater similarity to the original image. The paired t-test p-value between groups is displayed in the top left. CTA, Computed tomography angiography
Discussion
Reidentification of individuals in brain imaging is highly accurate using face-recognition software [8–10], posing challenges for head CT data in both research and clinical applications. We developed a defacing model (CTA-DEFACE) for CTA data, based on the state-of-the-art segmentation algorithm nnU-Net [17], which automatically generates an anatomical facemask from the forehead to the chin covering the facial soft tissue. We compared our model to a publicly available defacing function “ct_face_mask” from the ICHSEG library [18], which uses a rectangular prism to cover the mouth and nose. The CTA-DEFACE model resulted in significantly lower face detection probabilities and higher dissimilarity to the original image compared to ICHSEG. Furthermore, our model deidentified patient faces faster by leveraging GPU parallel computation.
The performance of defacing algorithms is difficult to compare with traditional metrics such as DSC since an actual ground truth does not exist. Recent algorithms for CT data have already adopted an “anatomical” approach by segmenting facial soft tissue [14], but comparing the correctness of these segmentations of those algorithms is not meaningful. We chose ICHSEG for comparison due to its different defacing strategy [18]. When segmentation masks were replaced by void space, our model revealed significantly lower face detection probabilities and higher dissimilarity to the original image, suggesting that removing more anatomical facial features than only mouth and nose is necessary for robust de-identification.
The application of machine learning models in the medical field, particularly for the head and neck, remains limited. Examples include automated detection of brain hemorrhage [22, 23] and intracranial thrombus in vessels [22–25]. Similar limitations affect defacing tools, as the inclusion of facial features depends on various technical factors such as: (i) scanned area, (ii) slice thickness, (iii) presence of foreign materials like eye protection, (iv) motion artifacts, (v) beam-hardening, and (vi) patient positioning. These factors need to be addressed when applying automated defacing tools.
With respect to data protection regulations, the integration of automated defacing tools into workflows or preprocessing pipelines of commercially available stroke detection programs should consider the fact that defacing medical images removes a substantial portion of facial tissue, altering image characteristics (Supplementary Fig. 2). Therefore, to integrate the CTA-DEFACE model or other defacing protocols into automated stroke detection pipelines, it may be necessary to either retrain the stroke detection models on defaced volumes or perform defacing in the postprocessing.
This study has limitations. First, the number of patients is limited, albeit from different cohorts. The effective de-identification capability of CTA-DEFACE was demonstrated on an external dataset obtained from a different CT vendor. However, a larger dataset is necessary to further evaluate the reproducibility of our results. Second, only CTA data was addressed. Future studies should validate our model including unenhanced and other contrast-enhanced head/neck CT protocols.
In conclusion, our results show that the CTA-DEFACE model effectively segments and removes facial soft tissue from CTA images faster than a publicly available CT defacing method, resulting in significantly lower face detection probabilities and higher dissimilarity to the original image. Future research should evaluate the potential of training algorithms (e.g., stroke algorithms) on defaced data.
Supplementary information
Additional file 1: Supplementary Table 1: Computed tomography angiography imaging features are depicted. Supplementary Table 2: Patient demographics in Cohort 1, 2 and 3 for CTA-DEFACE model training and testing. Pearson’s chi-squared test was used for comparing categorical variables and Kruskal Wallis test was used for comparing continuous variables between the training and test set. Supplementary Table 3: Patient demographics in Cohort 1, 2 and 3 for CTA-BET model training and testing. Pearson’s chi-squared test was used for comparing categorical variables and Kruskal Wallis test was used for comparing continuous variables between the training and test set. Supplementary Fig. 1: Representative cases for face detection are illustrated for rendered CTA images (from top to bottom: original CTA image, ICHSEG defacing, CTA-DEFACE defacing). The anterior point-of-view for image acquisition was maintained in each image, without considering head rotations. The probabilities of face detection by multitask cascaded convolutional neural network (MTCNN) are provided below the images, with “N/A” indicating that no face was detected. Eyes are blurred for anonymization purposes. Supplementary Fig. 2: Two cases demonstrating false-positive predictions by the automated vessel occlusion network (referred to as VO-ANN in this study) are displayed. In these cases, VO-ANN generated a bounding box (depicted in green) indicating a left internal carotid artery (ICA) occlusion. Both cases underwent visual examination by a radiologist with 5 years of experience. In the first case, the bounding box captured the low-contrast enhancement of the left internal jugular vein (IJV), with no occlusion observed in the left internal (ICA) or external carotid artery (ECA). In the second case, the bounding box depicted an area with calcified carotid plaque (no occlusion, no high-grade stenosis). These false-positive predictions were not observed after defacing with our CTA-DEFACE model, while reappearing after defacing with the ICHSEG model.
Acknowledgements
We obtained limited support from large language models (LLMs) during the revision of this article. Specifically, ChatGPT-3.5 and ChatGPT-4o (developed by OpenAI) were used during the review phase to improve language and condense text due to word count constraints.
Abbreviations
- ANN
Artificial neural network
- CT
Computed tomography
- CTA
Computed tomography angiography
- DSC
Dice similarity coefficient
- GPU
Graphics processing unit
- MTCNN
Multitask convolutional neural network
Authors contributions
MAM, GB, and PV designed the study. MAM, AR, MS, GB, and PV performed quality control of CTA data. MAM and GB performed pre-processing of CTA data. MAM and AR performed development, training, and application of artificial neural networks (ANN). MAM, AR, MS, M Baumgartner, and PV performed post-processing of the data generated by the ANN. MAM, AR, MS, and PV performed the statistical analyses. MAM, AR, MS, MF-D, MB, GB, M Bendszus, and PV interpreted the findings with essential input from all co-authors. MAM, AR, MS, and GB prepared the first draft of the manuscript. All authors critically revised and approved the final version of the manuscript. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication. All authors had access to all the data reported in the study. MAM, AR, MS, GB, and PV accessed and verified all the data reported in the study.
Funding
PV is funded through an Else Kröner Clinician Scientist Endowed Professorship by the Else Kröner Fresenius Foundation (reference number: 2022_EKCS.17). MAM is founded through an Else Kröner Research College for young physicians (reference number: 2023_EKFK.02). GB and MS are funded by the Physician-Scientist Program of Heidelberg University, Faculty of Medicine. M Baumgartner is partially funded through Helmholtz Imaging. Open Access funding enabled and organized by Projekt DEAL.
Data availability
The CTA-DEFACE model has been released as an open-source tool accompanied by thorough documentation at www.github.com/neuroAI-HD/CTA-DEFACE.
Declarations
Ethics approval and consent to participate
This retrospective study of the imaging data was approved by the local ethics committee of the University of Heidelberg and the requirement to obtain informed consent was waived (S-247/2009, approval 10/2010). External anonymized data was received through a data-sharing agreement between both hospitals (approval 01/2023).
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1186/s41747-024-00510-9.
References
- 1.Nageler G, Gergel I, Fangerau M et al (2023) Deep learning-based assessment of internal carotid artery anatomy to predict difficult intracranial access in endovascular recanalization of acute ischemic stroke. Clin Neuroradiol 33:783–792. 10.1007/s00062-023-01276-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucek RA, Puchner S, Kanitsar A, Rand T, Lammer J (2007) Automated CTA quantification of internal carotid artery stenosis: a pilot trial. J Endovasc Ther 14:70–76. 10.1583/06-1905.1 [DOI] [PubMed] [Google Scholar]
- 3.Shi J, Sun Y, Hou J et al (2023) Radiomics signatures of carotid plaque on computed tomography angiography: an approach to identify symptomatic plaques. Clin Neuroradiol 33:931–941. 10.1007/s00062-023-01289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brugnara G, Baumgartner M, Scholze ED et al (2023) Deep-learning based detection of vessel occlusions on CT-angiography in patients with suspected acute ischemic stroke. Nat Commun 14:4938. 10.1038/s41467-023-40564-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meijs M, Meijer FJA, Prokop M, van Ginneken B, Manniesing R (2020) Image-level detection of arterial occlusions in 4D-CTA of acute stroke patients using deep learning. Med Image Anal 66:101810. 10.1016/j.media.2020.101810 [DOI] [PubMed] [Google Scholar]
- 6.Stib MT, Vasquez J, Dong MP et al (2020) Detecting large vessel occlusion at multiphase CT angiography by using a deep convolutional neural network. Radiology 297:640–649. 10.1148/radiol.2020200334 [DOI] [PubMed] [Google Scholar]
- 7.Thamm F, Taubmann O, Jürgens M, Ditt H, Maier A (2022) Detection of large vessel occlusions using deep learning by deforming vessel tree segmentations. In: Maier-Hein KH, Deserno TM, Handels H, Maier A, Palm C, Tolxdorff T (eds) Bildverarbeitung für die Medizin 2022. Proceedings of the German Workshop on medical image computing, Heidelberg, vol 1. Springer Fachmedien Wiesbaden; Imprint Springer Vieweg, Wiesbaden, pp 444–498
- 8.Schwarz CG, Kremers WK, Therneau TM et al (2019) Identification of anonymous MRI research participants with face-recognition software. N Engl J Med 381:1684–1686. 10.1056/NEJMc1908881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz CG, Kremers WK, Wiste HJ et al (2021) Changing the face of neuroimaging research: comparing a new MRI de-facing technique with popular alternatives. Neuroimage 231:117845. 10.1016/j.neuroimage.2021.117845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz CG, Kremers WK, Lowe VJ et al (2022) Face recognition from research brain PET: an unexpected PET problem. Neuroimage 258:119357. 10.1016/j.neuroimage.2022.119357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischoff-Grethe A, Ozyurt IB, Busa E et al (2007) A technique for the deidentification of structural brain MR images. Hum Brain Mapp 28:892–903. 10.1002/hbm.20312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schimke N, Hale J (2011) Quickshear defacing for neuroimages. HealthSec 11:11
- 13.Theyers AE, Zamyadi M, O’Reilly M et al (2021) Multisite comparison of MRI defacing software across multiple cohorts. Front Psychiatry 12:617997. 10.3389/fpsyt.2021.617997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasserthal J, Breit H-C, Meyer MT et al (2023) TotalSegmentator: robust segmentation of 104 anatomic structures in CT images. Radiol Artif Intell 5:e230024. 10.1148/ryai.230024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muschelli J (2019) Recommendations for processing head CT data. Front Neuroinform 13:61. 10.3389/fninf.2019.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selfridge AR, Spencer BA, Abdelhafez YG, Nakagawa K, Tupin JD, Badawi RD (2023) Facial anonymization and privacy concerns in total-body PET/CT. J Nucl Med 64:1304–1309. 10.2967/jnumed.122.265280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:203–211. 10.1038/s41592-020-01008-z [DOI] [PubMed] [Google Scholar]
- 18.Muschelli J, Sweeney EM, Ullman NL, Vespa P, Hanley DF, Crainiceanu CM (2017) PItcHPERFeCT: primary intracranial hemorrhage probability estimation using random forests on CT. Neuroimage Clin 14:379–390. 10.1016/j.nicl.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Zhang Y (2021) MTCNN and FACENET based access control system for face detection and recognition. Aut Control Comp Sci 55:102–112. 10.3103/S0146411621010090 [Google Scholar]
- 20.Zhang K, Zhang Z, Li Z, Qiao Y (2016) Joint face detection and alignment using multitask cascaded convolutional networks. IEEE Signal Process Lett 23:1499–1503. 10.1109/LSP.2016.2603342 [Google Scholar]
- 21.Schroff F, Kalenichenko D, Philbin J (2015) FaceNet: a unified embedding for face recognition and clustering. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). IEEE, Boston, pp 815–823
- 22.Schmitt N, Mokli Y, Weyland CS et al (2022) Automated detection and segmentation of intracranial hemorrhage suspect hyperdensities in non-contrast-enhanced CT scans of acute stroke patients. Eur Radiol 32:2246–2254. 10.1007/s00330-021-08352-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seyam M, Weikert T, Sauter A, Brehm A, Psychogios M-N, Blackham KA (2022) Utilization of artificial intelligence-based intracranial hemorrhage detection on emergent noncontrast CT images in clinical workflow. Radiol Artif Intell 4:e210168. 10.1148/ryai.210168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1: Computed tomography angiography imaging features are depicted. Supplementary Table 2: Patient demographics in Cohort 1, 2 and 3 for CTA-DEFACE model training and testing. Pearson’s chi-squared test was used for comparing categorical variables and Kruskal Wallis test was used for comparing continuous variables between the training and test set. Supplementary Table 3: Patient demographics in Cohort 1, 2 and 3 for CTA-BET model training and testing. Pearson’s chi-squared test was used for comparing categorical variables and Kruskal Wallis test was used for comparing continuous variables between the training and test set. Supplementary Fig. 1: Representative cases for face detection are illustrated for rendered CTA images (from top to bottom: original CTA image, ICHSEG defacing, CTA-DEFACE defacing). The anterior point-of-view for image acquisition was maintained in each image, without considering head rotations. The probabilities of face detection by multitask cascaded convolutional neural network (MTCNN) are provided below the images, with “N/A” indicating that no face was detected. Eyes are blurred for anonymization purposes. Supplementary Fig. 2: Two cases demonstrating false-positive predictions by the automated vessel occlusion network (referred to as VO-ANN in this study) are displayed. In these cases, VO-ANN generated a bounding box (depicted in green) indicating a left internal carotid artery (ICA) occlusion. Both cases underwent visual examination by a radiologist with 5 years of experience. In the first case, the bounding box captured the low-contrast enhancement of the left internal jugular vein (IJV), with no occlusion observed in the left internal (ICA) or external carotid artery (ECA). In the second case, the bounding box depicted an area with calcified carotid plaque (no occlusion, no high-grade stenosis). These false-positive predictions were not observed after defacing with our CTA-DEFACE model, while reappearing after defacing with the ICHSEG model.
Data Availability Statement
The CTA-DEFACE model has been released as an open-source tool accompanied by thorough documentation at www.github.com/neuroAI-HD/CTA-DEFACE.