Abstract
The mammalian imprinted Dlk1-Dio3 domain contains multiple lncRNAs, mRNAs, the largest miRNA cluster in the genome and four differentially methylated regions (DMRs), and deletion of maternally expressed RNA within this locus results in embryonic lethality, but the mechanism by which this occurs is not clear. Here, we optimized the model of maternally expressed RNAs transcription termination in the domain and found that the cause of embryonic death was apoptosis in the embryo, particularly in the liver. We generated a mouse model of maternally expressed RNAs silencing in the Dlk1-Dio3 domain by inserting a 3 × polyA termination sequence into the Gtl2 locus. By analyzing RNA-seq data of mouse embryos combined with histological analysis, we found that silencing of maternally expressed RNAs in the domain activated apoptosis, causing vascular rupture of the fetal liver, resulting in hemorrhage and injury. Mechanistically, termination of Gtl2 transcription results in the silencing of maternally expressed RNAs and activation of paternally expressed genes in the interval, and it is the gene itself rather than the IG-DMR and Gtl2-DMR that causes the aforementioned phenotypes. In conclusion, these findings illuminate a novel mechanism by which the silencing of maternally expressed RNAs within Dlk1-Dio3 domain leads to hepatic hemorrhage and embryonic death through the activation of apoptosis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-024-05462-2.
Keywords: Long non-coding RNAs, Easi-CRISPR, Maternally expressed RNAs, Embryonic lethality, Mouse
Introduction
Genomic imprinting is a unique epigenetic regulation that results in parent-specific gene expression and is essential for normal development and growth in mammals [1, 2]. As an important genomic region, the genes and elements in the Dlk1-Dio3 imprinting region play a key role in embryonic development, cell differentiation and disease occurrence [3, 4]. The study of this region provides insights into the mechanisms of genomic imprinting and the functions of imprinted genes in organism development and disease.
The long non-coding RNA (lncRNA) gene trap locus 2 (Gtl2), which is known as maternally expressed gene 3 (MEG3) in humans and has an imprinting pattern of maternal expression and paternal imprinting, is an imprinting gene located in the Dlk1-Dio3 imprinting gene cluster [5]. Downstream maternally expressed intergenic transcripts are transcribed in the same direction as those of Gtl2, and canonical promoter sequences are apparently absent from this region, with all lncRNAs regulated by common cis-elements and epigenetic control to produce a large polycistronic transcription unit [6]. Furthermore, the tissue-specific features of Gtl2 and these microRNAs (miRNAs) are strikingly similar, suggesting coordinated regulation of the expression of these genes [7–9]. Three studies have shown that Gtl2 knockout causes death of mouse. But curiously, the lethal genotypes and phenotypes of mice in these studies were different, and some were even contradictory.
When the 9.8 kb region of Gtl2-DMR and exon 1–5 was knocked out, the maternal knockout mice were born normally, but all died within 4 weeks, while the paternal knockout mice showed growth retardation and perinatal mortality, meanwhile, the homozygous knockout mice could survive normally [10]. Another study also knocked out a 5.9 kb fragment in the similar region of Gtl2, but the results were completely different: maternal and homozygous knockout mice resulted in perinatal death, while paternal knockout mice were born and developed normally [11]. By constructing mouse models with deletion of Meg3 1–4 exon (Meg3Δ1−4) and deleton of Meg3 2–4 exon deletion (Meg3Δ2−4), it was found that maternal Meg3Δ1−4 caused the death of embryos at embryonic day 13.5 (E13.5), and the maternal Meg3Δ2−4 mice were able to be born normally, however, the data information of homozygous double deletion in parents was missing [12]. Although these three reports all focused on Gtl2 gene knockout, the conclusions obtained were not mutually corroborative and even contradictory. After comparing the three reported gene knockout regions, it was found that the large knockout gene may lead to the deletion of miRNAs, Gtl2-DMR and genomic regulatory elements within the Gtl2 gene, or interfere with the active open chromatin region in the Gtl2 locus, so the obtained experimental results did not reflect the true role of Gtl2. In addition, several articles have pointed out that gene knockout is not suitable for the study of lncRNA function, which may cause false positive results [13, 14]. Therefore, a new method that can accurately describe the function of lncRNA is urgently needed.
The mammalian fetal liver can not only participate in glucose and lipid metabolism, but also synthesize substances such as hemoglobin, coagulation factors and bilirubin [15–19]. Meanwhile, as a major hematopoietic organ, the fetal liver provides the embryo with essential erythrocytes and other blood components [20, 21]. In addition, when the blood vessels of fetal liver are damaged and hemorrhage occurs, it will cause serious consequences such as liver damage, abdominal hemorrhage, embryonic development restriction and even death [22, 23]. Previous studies have mainly explored the role of fetal liver in metabolism, hematopoiesis and regeneration, but lack of exploration of liver vascular development. There is a close relationship between Dlk1-Dio3 domain and the fetal liver. Previous studies have described that the gene expression in this region has an important impact on the metabolism and hematopoietic functions of the fetal liver [20, 21, 24], but the description of the development of the liver vascular system is lacking.
In this study, the Easi-CRISPR strategy was used to successfully establish Gtl2 transcription termination mice model by inserting 3 × polyA termination sequence [25]. Our previous studies have reported the importance of this domain for placental vasculature [25]. Subsequently, we observed that silencing of maternally expressed RNAs in Dlk1-Dio3 domain resulted in embryonic lethality due to liver vascular damage, fatal hepatic hemorrhage in mice. Precise gene expression analysis showed that after Gtl2 transcription termination, maternally expressed RNAs in the Dlk1-Dio3 domain were almost completely silenced, while paternally expressed genes were activated. Together, this study explained that silencing of maternally expressed RNAs in Dlk1-Dio3 domain caused apoptosis in embryo, especially in liver, leading to vascular damage, causing large area of liver hemorrhage and embryonic death.
Materials and methods
Animals
We improved and optimized the Easi-CRISPR gene knock-in method [26]. According to the prediction analysis of the websites ((http://crispr.wustl.edu/ and http://www.rgenome.net/cas-designer/), we found an efficient sgRNA after Gtl2 promoter, formed RNP complex with Cas9 protein and gRNA, and then microinjected the fertilized male pronucleus together with ssDNA, and efficiently established the mouse model of transcription termination of maternally expressed RNA in Dlk1-Dio3 domain. Gtl2+/+ female mice were mated with Gtl2+/polyA male mice for propagation and expansion of the system. Male and female mice were mated at 6:00p.m. and vaginal suppositories were tested at 9:00a.m., the next day, and the presence of vaginal plugs was marked as E0.5. B6D2F1 (BDF1) female mice were induced to superovulate, and the superovulated females were mated with BDF1 male mice for zygote collection. The initial pronuclear injection was performed in these zygotes. The SNP site on the Dlk1 locus could distinguish whether the newborn mice were of C57BL/6N or DBA/2J background, and finally one male mouse of C57BL/6N and DBA/2J background was selected as founder mice of the two strains for subsequent experiments. The mice used for biallelic expression and methylation analysis were derived from the hybrid offspring of DBA/2J and C57BL/6N strains, while the remaining experimental strains in this study were all from DBA/2J mice. Wide-type DBA/2J and C57BL/6N mice were purchased from Beijing, Weitonglihua.
RNA-seq analysis
We selected one sample from each of the four genotypes and extracted 1 μg of total RNA, and the cDNA library was generated and sequenced on Illumina sequencing platform provided by Biomarker technologies (Wuhan, China) (www.biomarker.com.cn). We used FASTQ to assess the quality of sequencing data, and poor-quality reads were removed. We selected mouse GRCm38/mm10 as the reference genome, and relied on several comprehensive databases such as Ensembl (https://asia.ensembl.org/index.html) and GENCODE (https://www.gencodegenes.org/) to identify and annotate mRNAs and lncRNAs. Then, we compared high-quality reads to the reference genome by HISAT2 [27], quantified reads by StringTie [28], and finally calculated the expression level of each mRNA and lncRNA (FPKM). Differentially expressed lncRNAs, miRNAs and mRNAs were screened were screened by EdgeR [29], with fold change more than 2.0, and p-value less than 0.05. Target genes of lncRNAs were predicted by its cis-interaction and trans-interaction [30, 31]. Ensembl and UCSC Genome Browser (https://www.genome.ucsc.edu/) were used to identify neighboring genes within 1Mbps of lncRNAs as their cis-target genes. And ENCORI (https://rnasysu.com/encori/index.php) were used to predict the trans-target genes of lncRNAs. And based on the complementary pairing principle of mRNA 3′UTR [32], we predicted the target genes of miRNAs by miRbase (http://mirbase.org) and Targetscan (https://www.targetscan.org/). Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) and Metascape (https://metascape.org/) were used to analyze the potential functions of target genes of lncRNAs and miRNAs, and mRNA in KEGG pathway and Go terms.
qRT-PCR analysis
Total RNA was extracted from brain, heart, liver, lung, muscle and embryo at E12.5. cDNA of mRNA and lncRNA were synthesized by using PrimeScript™ RT reagent Kit (TakaRa, RR047Q) in a 10 μl reaction solution containing 500 ng total RNA. cDNA of miRNA was synthesized by using Mir-X™ miRNA First-Strand Synthesis (TakaRa) in a 10 μl reaction solution containing 500 ng total RNA. The gene expression levels were measured by real-time PCR with the SYBR Green PCR Master Mix. β-Actin was used as control for mRNA and lncRNA. U6 was used as control for miRNA. Values from wild-type were designated as 1. Values from Gtl2+/polyA, Gtl2polyA/+ and Gtl2polyA/polyA were normalized against those from Gtl2+/+. Primers' sequences were shown in Tab. S1.
Biallelic expression of Dlk1
Chromosome-specific gene expression was determined by SNP analysis. In Dlk1 cDNA (NM_010052), SNP is a G in C57BL/6N and an A in DBA/2J, respectively. Total RNA was isolated from E12.5 embryos obtained by crossing Gtl2+/+ DBA/2J with Gtl2+/polyA C57BL/6N mice and crossing Gtl2+/polyA DBA/2J with Gtl2+/+ C57BL/6N mice. The region containing the polymorphism was amplified by RT-PCR. The allelic expression of Dlk1 was determined by sequencing PCR products. Primers' sequences were shown in Tab. S1.
DNA methylation analysis
Genomic DNAs were isolated from E12.5 embryos (DBA/2J x C57BL/6N). After bisulfite treatment by EZ DNA Methylation-Gold™ kit (Zymo Research), the converted genomic DNAs were amplified by PCR for Gtl2-DMR and IG-DMR, which primers' sequences were shown in Tab. S1 Results were read by DNAMAN Version 6 after sequencing (Comate Bioscience, China). Primers' sequences were shown in Tab. S1.
Histologyical analysis
Fetal livers of E12.5 were fixed with 4% paraformaldehyde of 24 h, dehydrated with ethanol and transparent with xylene, then paraffin blocks were prepared, sliced 5 μm and stained with hematoxylin/eosin. Images were captured with Zeiss optical microscope (Germany).
Vascular area analysis
We regarded the closed interval surrounded by CD31 signal as one vessel, and quantified the area of the enclosed vessel by imageJ. Within a view, the average vessel area per unit area is obtained by (total vessel area / total number) /view area.
Preparation of blood smears
Peripheral blood of mouse embryos at E12.5 were taken to make blood pictures and stained with Wright-Giemsa Stain kit (Solarbio, G1021). Images were captured with Zeiss optical microscope (Germany).
Immunohistochemistry
Fetal livers of E12.5 were fixed with 4% paraformaldehyde of 24 h. After blocking with 2% BSA/PBS, sections were incubated with specific primary antibodies at 4 ℃ overnight, and sections were treated with HRP-conjugated secondary antibodies. DAB kit (ZSGB-BIO, China) was used for chemical staining.
Assay of Caspase 9 activity
The Caspase 9 spectrophotometric assay kit is based on the combination of a Caspase 9 sequence-specific peptide (acetyl-Asp-Glu-Val-Asp p-nitroanilide, Ac-LEHD-pNA) coupled to the yellow luminescent group p-nitroaniline (pNA). When the substrate was cleaved by Caspase 9, the yellow luminescent group pNA was free, and its absorbance value could be measured by a microplate reader or a spectrophotometer (λ = 405 nm). Embryo or liver tissue was taken in a petri dish, cut into pieces, added with lysate (400ul/100 mg), homogenized with a glass homogenizer on ice, left in an ice bath for 5 min, and centrifuged. The supernatant was sucked into a new EP tube. After Ac-LEHD-pNA was added and mixed, the mixture was incubated at 37 °C for 2 to 4 h, and OD405 could be determined when the color change was obvious. Finally, the activation degree of Caspase 9 is determined by calculating the multiple of (ODsample value-ODblank control)/(ODnegative value-ODblank control) (Elabscience, E-CK-A313).
TUNEL apoptosis assay
TUNEL apoptosis assay was detected using frozen sections of fetal liver at E12.5. After fixing with paraformaldehyde, and washing with PBS solution, the assay was used by one step TUNEL Apoptosis Assay kit (Beyotime, C1088). Images were captured with Olympus fluorescence inverted microscope (Japan).
In situ hybridization
Paraffin sections were made as described above. Sections were treated with 2 mg/ml proteinase K in PBS and terminated wit 1 mg/mL glycine. Antisense riboprobes were used for hybridization overnight at 65 °C, alkaline phosphatase-conjugated anti-DIG antibody (Roche, 54,732,420, dilution 1:5000) was incubated overnight at 4 °C, and NBT/BCIP Stock Solution (Roche, 11,681,451,001) was used for chemical staining. Images were captured with Zeiss optical microscopy. Primers’ sequences of probe were shown in Tab. S1.
Statistics
Values are shown as the mean ± SEM as indicated. All statistical analyses were generated using GraphPad Prism 6. Student’s t test was used for comparisons between two groups. Statistical significance was defined as p value of < 0.05 (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Results
Establishment and genetic analysis of Gtl2 transcription termination mouse
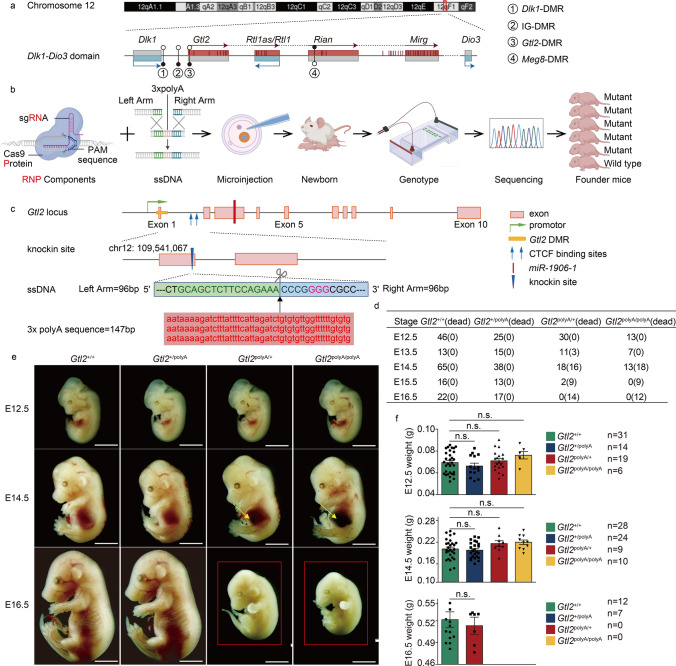
Gtl2 is located within the Dlk1-Dio3 imprinting domain at the end of mouse chromosome 12 (Fig. 1a). Previous analysis of the Gtl2 locus has revealed a differentially methylated region (Gtl2-DMR), a microRNA (miR-1906), two CTCF binding sites and some active open chromatin regions between exons 1–5 [12, 33]. In order to silence the Gtl2 gene without affecting the action and activity of other transcription factors, we identified a highly efficient sgRNA site at the end of the first exon, and used 96 bp of each upstream and downstream sgRNA as the left and right homology arms to form ssDNA with the inserted 3 × polyA (Fig. 1b and c). The combined ssDNA and RNP complex (sgRNA and Cas9 protein) were microinjected into the male pronucleus of the zygote (Fig. S1a), cultured in vitro until the blastocyst stage (Fig. S1b), and the embryos were transferred into two surrogate mice [25]. Among the six newborn mice (Fig. S1c), five founder mice with successful 3 × polyA knock-in were identified by PCR (Fig. 1b and S1d). Taken, we successfully generated a mutant mouse model that only silenced the expression of ncRNA in Dlk1-Dio3 domain without deleting other active elements.
Fig. 1.
Generation and phenotypic analysis of founder mice. a Schematic representation of Dlk1-Dio3 domain in chromosome12. Maternally expressed genes are in red, paternally expressed genes are in blue and silenced genes are in gray, miRNAs are shown as short vertical lines, differentially methylated regions are shown as circles. Filled circle, methylated; open circle, unmethylated. b Schematic representation of mutant mouse production by Easi-CRISPR: preparation of RNP components and ssDNA, microinjection, generation of founder offspring, PCR genotyping and off-target detection and obtaining of founder mice. c Schematic representation of 3 × polyA termination sequence insertion sites. d Survival and death number of embryos from E12.5 to E16.5. e Representative pictures of embryo at E12.5, E14.5 and E16.5, abdominal hemorrhage are indicated by yellow arrows, dead embryos are in red boxes. Scale bar, 2 mm. f Bar chat showing embryo weight of E12.5, E14.5 and E16.5, each dot represents an embryo. Error bars, ± SEM, n.s., p > 0.05
Since Gtl2 is a maternally expressed gene, we maintained the generated mice by crossing wild type (Gtl2+/+) or Gtl2 transcription termination occurs on the paternal alleles (Gtl2+/polyA) of male mice with Gtl2+/+ or Gtl2+/polyA females (Fig. S1e). We selected 3–4 generations of mice for the parents of embryos obtained. Mice developed normally in Gtl2+/polyA mice, while embryonic death began at E13.5 and all mice died at E16.5 (Fig. 1e) when Gtl2 transcription termination occurred on the maternal allele (Gtl2polyA/+) and the homozygous biallelic (Gtl2polyA/polyA) (Fig. S1f). The mortality rate of Gtl2polyA/+ was 0%, 21.43%, 47.06%, 77.78% and 100% from E12.5 to E16.5, respectively, and then the mortality rate of Gtl2polyA/polyA was 0%, 0%, 58.06%, 100% and 100% form E12.5 to E16.5, respectively (Fig. 1d and S1g). So far, we have found that there is a slight difference in the time of death between Gtl2polyA/+ and Gtl2polyA/polyA. According to the representative images of embryos, we observed that there was no significant difference in the four-genotype embryos at E12.5. There was no obvious change in Gtl2+/polyA, however, the Gtl2polyA/+ and Gtl2polyA/polyA mice showed abdominal hemorrhage at E14.5 (Fig. 1e). There was no obvious change in Gtl2+/polyA at E16.5, whereas the Gtl2polyA/+ and Gtl2polyA/polyA mice were all dead at this stage (Fig. 1e). Gtl2polyA/+ and Gtl2polyA/polyA were slightly but not significantly heavier than Gtl2+/+, and Gtl2+/polyA was almost the same compared to Gtl2+/+, and the trends of embryo weight change were the same at E12.5, E14.5 and E16.5 (Fig. 1f). Together, these data show that the silencing of ncRNAs on Dlk1-Dio3 domain results in little effect on Gtl2+/polyA, and a slight increase in embryo weight, abdominal hemorrhage and embryonic death on Gtl2polyA/+ and Gtl2polyA/polyA.
Termination of Gtl2 transcription resulted in silencing of the maternally expressed RNAs in the Dlk1-Dio3 domain
We next attempt to elucidate the molecular mechanisms responsible for the above results. To this end, we performed whole transcriptome sequencing in four-genotype embryos at E12.5 and detected 29,158 lncRNAs and 2395 miRNAs (Tab. S2). After excluding genes with that FPKM is 0 in all the four genotypes, we screened out differential expression lncRNAs (DElncRNAs) (Tab. S3) and differential expression miRNAs (DEmiRNAs) (Tab. S4), fold change more than 2.0, and p-value less than 0.05.
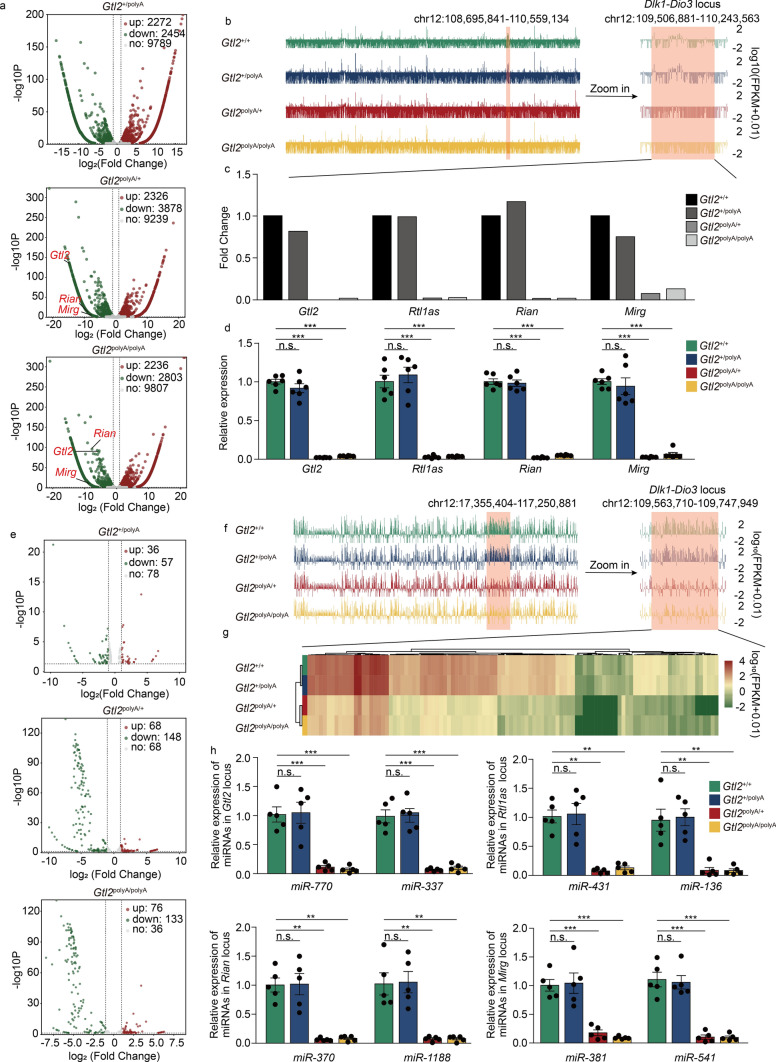
Firstly, we identified 2272 upregulated and 2454 downregulated lncRNAs in Gtl2+/polyA, 2326 upregulated and 3878 downregulated lncRNAs in Gtl2polyA/+ and 2236 upregulated and 2803 downregulated lncRNAs in Gtl2polyA/polyA (Fig. 2a) (Tab. S3). RNA-seq data of all lncRNAs showed that Gtl2polyA/+and Gtl2polyA/polyA had a significant downward trend in Dlk1-Dio3 domain (Fig. 2b), mainly in Gtl2, Rtl1as (miR-127), Rian and Mirg (Fig. 2c). The results of qRT-PCR were consistent with the trend of RNA-seq (Fig. 2d), indicating that the termination of Gtl2 transcription led to the silencing of lncRNA in the domain.
Fig. 2.
Termination of Gtl2 transcription resulted in silencing of the maternally expressed RNA in the Dlk1-Dio3 domain. a Volcano plot of DElncRNAs in Gtl2polyA/+ and Gtl2polyA/polyA embryos at E12.5. b lncRNA-seq of embryos at E12.5. c The expression of Gtl2, Rtl1as, Rian and Mirg in RNA-seq data. d qRT-PCR analysis of Gtl2, Rtl1as, Rian and Mirg, β-actin was used as an internal control, n = 6 Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001. e Volcano plot of DEmiRNAs in Gtl2polyA/+ and Gtl2polyA/polyA. f miRNA-seq of embryos at E12.5. g The expression of miRNAs in Dlk1-Dio3 domain in RNA-seq data. h qRT-PCR analysis of miRNAs in Gtl2, Rtl1as, Rian and Mirg locus, U6 was used as an internal control, n = 5. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001
The lncRNAs and miRNAs in the Dlk1-Dio3 domain constitute a large polycistronic transcription unit, and maternally expressed intergenic transcripts downstream of Gtl2 are transcribed in the same direction as Gtl2, and there is no typical promoter sequence present in this region [5]. Therefore, we guessed that miRNAs in the domain would also be severely affected by the termination of Gtl2 transcription. We identified 36 upregulated and 57 downregulated miRNAs in Gtl2+/polyA, 68 upregulated and 148 downregulated miRNAs in Gtl2polyA/+ and 76 upregulated and 133 downregulated miRNAs in Gtl2polyA/polyA (Fig. 2e) (Tab. S4). As most of the downregulated miRNAs were located in the Dlk1-Dio3 domain in Gtl2polyA/+ and Gtl2polyA/polyA (Fig. 2f), we clustered the miRNAs in the domain and found that the four genotypes could be clearly divided into two groups: the survival group including Gtl2+/+ and Gtl2+/polyA, and the death group including Gtl2polyA/+ and Gtl2polyA/polyA, among them, the expression of miRNAs in Gtl2+/polyA were almost unchanged, while the expression of miRNAs in Gtl2polyA/+ and Gtl2polyA/polyA decreased significantly (Fig. 2g). In Gtl2, Rtl1as, Rian and Mirg loci, one miRNA with high expression and one with low expression were selected for qRT-PCR verification, the results were identical to RNA-seq data, indicating that the termination of Gtl2 transcription led to the silencing of miRNAs in the domain (Fig. 2h). Taken together, these data indicated that our model successfully terminated the transcription of the maternally expressed RNAs in the Dlk1-Dio3 domain and the termination of embryonic development might be closely related to the silencing of these genes.
Silencing of maternally expressed RNAs in Dlk1-Dio3 domain results in hemorrhage in embryos
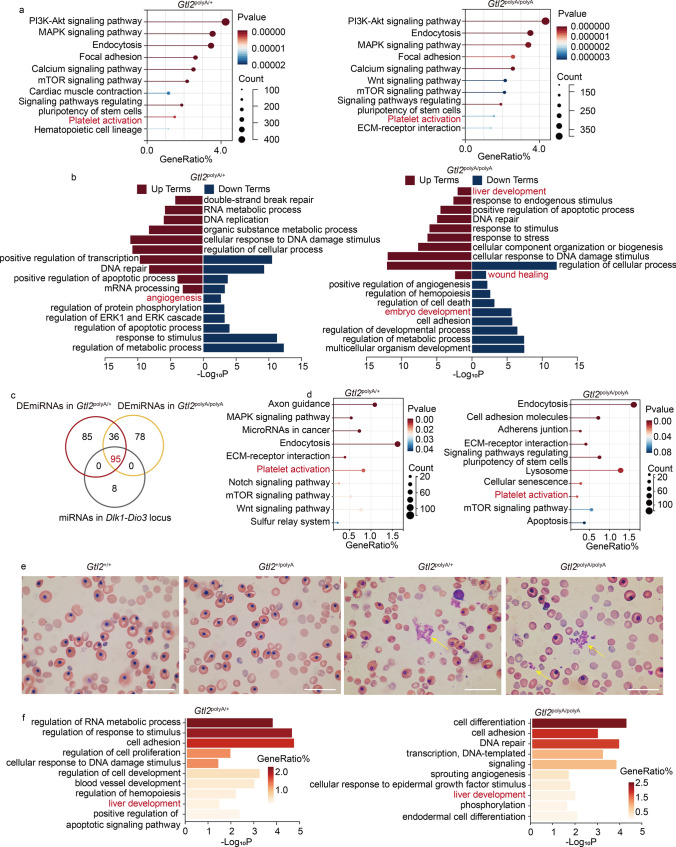
Because platelets undergo activities such as activation and aggregation when injured or bleeding [34], the platelet activation in Kyoto Encyclopedia of Genes and Genomes (KEGG) that appeared in Gtl2polyA/+ and Gtl2polyA/polyA, but not in Gtl2+/polyA, caught our attention (Fig. 3a and S2a) (Tab. S5). Gene Ontology (GO) enrichment analysis revealed that GO terms enriched in both Gtl2polyA/+ and Gtl2polyA/polyA, but not in Gtl2+/polyA, were mainly associated with positive regulation of apoptotic process, embryo development, angiogenesis and wound healing (Fig. 3b and S2b) (Tab. S6). These features are associated with the phenotype of bleeding.
Fig. 3.
Termination of maternally expressed RNAs in Dlk1-Dio3 domain transcription leads to hemorrhage. a KEGG analysis of target genes of DElncRNAs in Gtl2polyA/+ and Gtl2polyA/polyA embryos at E12.5. b GO analysis of target genes of DElncRNAs in Gtl2polyA/+ and Gtl2polyA/polyA embryos at E12.5. c Venn diagram shows the intersection of DEmiRNAs in Gtl2polyA/+, DEmiRNAs in Gtl2polyA/polyA, and all miRNAs in Dlk1-Dio3 domain. d KEGG analysis of target genes of DEmiRNAs in Dlk1-Dio3 domain in Gtl2polyA/+ and Gtl2polyA/polyA embryos at E12.5. e Blood smear of peripheral blood at E12.5, yellow arrows point to avtivatee and aggregated platelets, Scale bar, 40 μm. f GO analysis of target genes of DEmiRNAs in Gtl2polyA/+ and Gtl2polyA/polyA embryos at E12.5
We hypothesized that miRNA changes in the death group may play more roles. Therefore, we found the common DEmiRNAs of Gtl2polyA/+ and Gtl2polyA/polyA, and overlapped with the miRNAs in the Dlk1-Dio3 domain to obtain 95 DEmiRNAs (Fig. 3c). We further analyzed the KEGG and GO of target genes of these 95 DEmiRNAs in Gtl2polyA/+ and Gtl2polyA/polyA (Tab. S4). KEGG enrichment analysis revealed platelet aggregation in both Gtl2polyA/+ and Gtl2polyA/polyA (Fig. 3d and S2c) (Tab. S7). In parallel, activated and aggregated platelets were also identified from cord blood smears of Gtl2polyA/+ and Gtl2polyA/polyA embryos (Fig. 3e). This suggests that bleeding or even injury did occur in Gtl2polyA/+ and Gtl2polyA/polyA embryos. In addition, VEGF signaling pathway also appeared in Gtl2polyA/+ (Fig. 3e), and combined with the related terms such as angiogenesis that appeared in GO of DElncRNAs (Fig. 3b), we speculated that the termination of maternally expressed RNA transcription may cause some effects on blood vessels. GO enrichment analysis revealed liver development in both Gtl2polyA/+ and Gtl2polyA/polyA, but not in Gtl2+/polyA (Fig. 3f and S2d) (Tab. S8), and the same term was also appeared in the GO enrichment of lncRNAs (Fig. 3b). Taken together, the DEmiRNAs suggested that the organ affected by ncRNA silencing was the liver.
Silencing of maternally expressed RNAs in Dlk1-Dio3 domain results in apoptosis
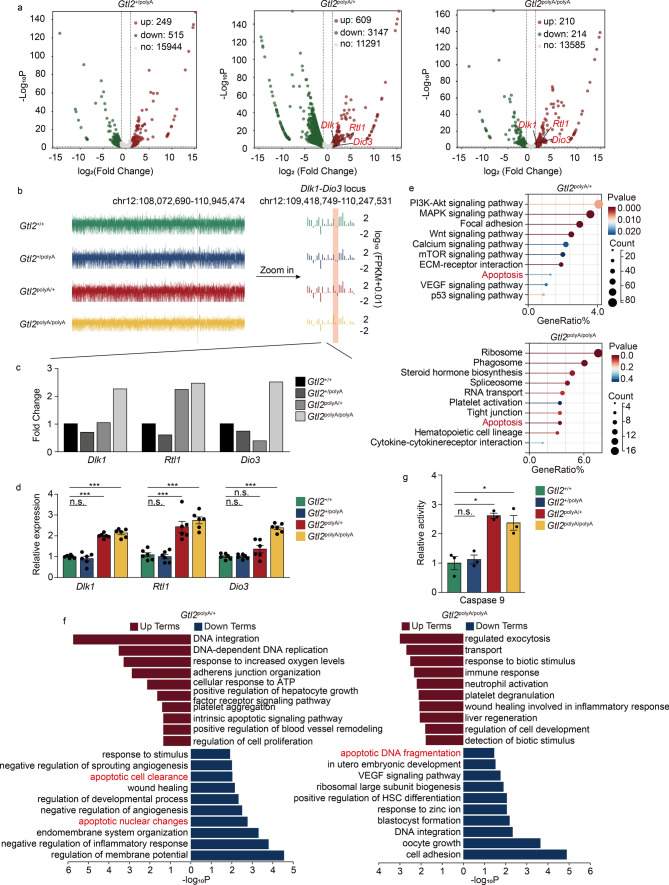
Finally, we identified 249 upregulated and 515 downregulated mRNAs in Gtl2+/polyA, 609 upregulated and 3147 downregulated mRNAs in Gtl2polyA/+ and 220 upregulated and 214 downregulated mRNAs in Gtl2polyA/polyA (Fig. 4a) (Tab. S9). We found the Dlk1-Dio3 region in the mRNA profile (Fig. 4b), RNA-seq showed that the expression levels of Dlk1 and Rtl1 in Gtl2polyA/+and Gtl2polyA/polyA were 2 times higher than those in Gtl2+/+, but there was no significant difference between Gtl2+/polyA and Gtl2+/+. The expression levels of Dio3 in Gtl2polyA/polyA were 2 times higher than those in Gtl2+/+, while the expression level in Gtl2+/polyA and Gtl2polyA/+ was slightly lower than that in Gtl2+/+, which may account for the lower expression of Dio3. (Fig. 4c). The results of qRT-PCR were consistent with the trend of RNA-seq (Fig. 4d), indicating that the termination of transcription of the maternally expressed RNAs in Dlk1-Dio3 domain activates the expression of the paternally expressed genes.
Fig. 4.
The genetic changes within the Dlk1-Dio3 domain leading to apoptosis. a Volcano plot of DEmRNAs in Gtl2polyA/+ and Gtl2polyA/polyA embryos at E12.5. b mRNA-seq of embryos at E12.5. c The expression of Dlk1, Rtl1 and Dio3 in RNA-seq data. d qRT-PCR analysis of Dlk1, Rtl1, and Dio3, β-actin was used as an internal control, n = 6. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001. e KEGG analysis of DEmRNAs in Gtl2polyA/+ and Gtl2polyA/polyA. f GO analysis of target genes of DEmRNAs in Gtl2polyA/+ and Gtl2polyA/polyA. g Relative activity of Caspase 9, n = 3. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *, p < 0.05
KEGG enrichment analysis revealed that apoptosis was enriched in DEmRNAs of both Gtl2polyA/+ and Gtl2polyA/polyA, but not of Gtl2+/polyA (Fig. 4e and S3a) (Tab. S10). GO terms enriched in DEmRNAs of both Gtl2polyA/+ and Gtl2polyA/polyA were mainly associated with apoptosis, wound healing, VEGF signaling pathway and platelet aggregation (Fig. 4f and S3b) (Tab. S11). Among them, platelet aggregation was the same as the experimental results of ncRNA enrichment, wound healing and VEGF signaling pathway appeared again, which had to attract our attention and needed further verification.
In addition, apoptosis occurs simultaneously with the above pathways, we speculated that the termination of maternally expressed RNA transcription may cause some effects on blood vessels and hemorrhage, and the relative activity of Caspase 9 in Gtl2polyA/+ and Gtl2polyA/polyA was significantly higher than that in Gtl2+/+, but there was no significant change in Gtl2+/polyA (Fig. 4g). Taken together, these data suggest that silencing of maternally expressed RNAs in Dlk1-Dio3 domain cause apoptosis, which may be responsible for vascular damage and hemorrhage.
Silencing of maternally expressed RNAs in Dlk1-Dio3 domain results in vascular damaged and hemorrhage in fetal livers
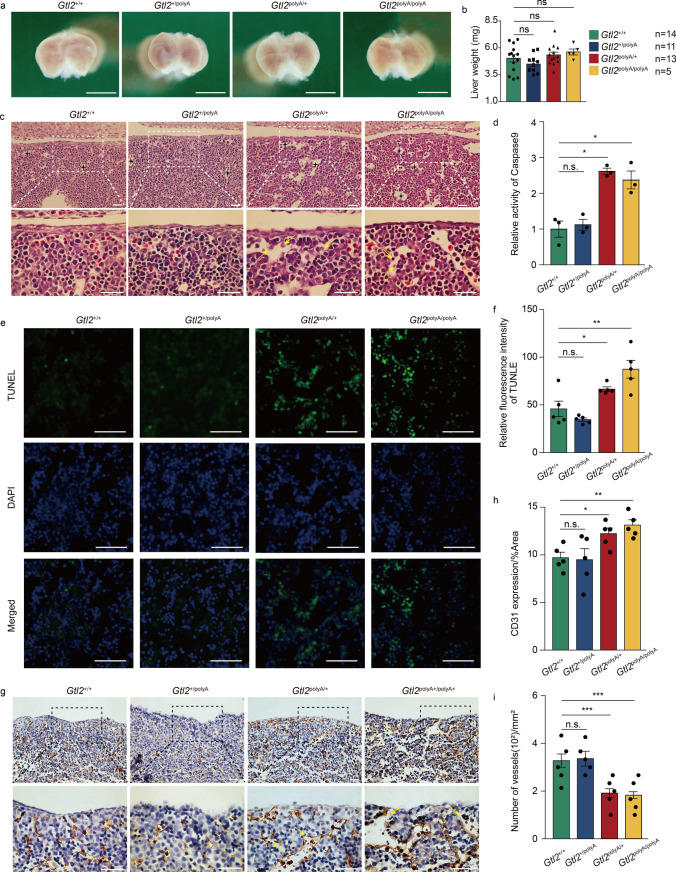
The GO functional enrichment of the maternally expressed RNA in the interval was concentrated in the liver (Fig. 3b and f), so we speculated that the above phenomenon may be the most obvious in the liver. To determine the effect of silencing the maternally expressed RNAs in the Dlk1-Dio3 domain on fetal liver, we obtained fetal livers from all four-genotypes at E12.5. It was found that there was no significant change in appearance of the three mutants compared with Gtl2+/+ (Fig. 5a). Immunofluorescence of Ki67 at E14.5 fetal livers showing that significant cell proliferation occurred in Gtl2polyA/+ and Gtl2polyA/polyA, but not in Gtl2+/polyA fetal livers (Fig. S4). The above results have shown that the increase in body and fetal liver weight may be due to cell proliferation which was consistent with the results of KEGG and GO enrichment by the ncRNAs. The trend of weight in four-genotype fetal livers was consistent with that of embryos (Fig. 5b). To further observe the changes at the cellular level in the fetal livers, histological analysis showed that there was no significant change between Gtl2+/polyA and Gtl2+/+ (Fig. 5C). The area of blood vessels in the fetal livers of Gtl2polyA/+ and Gtl2polyA/polyA increased, and some blood vessels were damaged (Fig. 5c). At the same time, a variety of cells, such as hepatocytes and endothelial cells, showed suspected apoptosis phenomena such as cell volume reduction, membrane rupture, karyopyknosis and cell fragmentation (Fig. 5c). Single cell suspension of intact liver was prepared for caspase 9 activity detection. The results showed that the relative activities of Gtl2polyA/+ and Gtl2polyA/polyA were more than 2 times higher than that of Gtl2+/+ (Fig. 5d). TUNEL enabled us to locate apoptosis in fetal livers, which also confirmed the previous speculation that cells in fetal liver did indeed undergo apoptosis (Fig. 5e). Relative fluorescence intensity of TUNEL showed that the apoptosis degree of Gtl2polyA/+ and Gtl2polyA/polyA were significantly higher than that of Gtl2+/+, which was consistent with the previous results (Fig. 5f). These results suggest that termination transcription of maternally expressed RNA in Dlk1-Dio3 domain leads to apoptosis in fetal livers.
Fig. 5.
Termination of maternally expressed RNAs in Dlk1-Dio3 domain transcription leads to apoptosis and vascular damage in fetal liver. a Representative pictures of fetal liver at E12.5. Scale bar, 1 mm. b Bar chat showing weight of fetal livers at E12.5, each dot represents a fetal liver. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05. c Representative images of HE staining of fetal livers of each genotype at E12.5. Scale bar, 40 μm. d Relative activity of Caspase 9 in fetal liver, n = 3. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *p < 0.05. e TUNEL fluorescence staining of apoptotic cells in fetal livers, Scale bar, 40 μm. f Quantitative analysis of the immunohistochemical staining for TUNEL, n = 5. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *, p < 0.05; **, p < 0.01. g Representative images of immunohistochemistry staining for CD31, apoptosis of endothelial cells are shown as yellow arrows. Scale bar, 40 μm. h Quantitative analysis of the immunohistochemical staining for CD31, n = 5. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *, p < 0.05; **, p < 0.01. i Number of blood vessels per square millimeter in each genotype fetal livers, n = 5. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001
Next, combined with the previous histological observation of vascular rupture, we further analyzed the blood vessels of the fetal liver. Immunohistochemistry of CD31 was localized to liver endothelial cells and the results showed fragments of endothelial cell apoptosis, which damaged blood vessels and caused erythrocytes to flow out of the damaged blood vessels, resulting in liver hemorrhage, which was consistent with previous histological analysis (Fig. 5g). Quantitative analysis of vessel area showed that Gtl2+/polyA, Gtl2polyA/+ and Gtl2polyA/polyA were 0.98-fold, 1.26-fold and 1.35-fold larger than that of Gtl2+/+ (Fig. 5h). The number of blood vessels per square millimeter showed that Gtl2+/polyA, Gtl2polyA/+ and Gtl2polyA/polyA were 1.03-fold, 0.58-fold and 0.56-fold of Gtl2+/+, respectively (Fig. 5i). That is, the average vessel area of Gtl2polyA/+ and Gtl2polyA/polyA was significantly larger than that of Gtl2+/+, but the number of vessels was significantly less than that of Gtl2+/+, and the cause of vascular injury and hepatic hemorrhage was apoptosis of endothelial cells.
Silencing of maternally expressed RNAs in Dlk1-Dio3 domain had no significant effect on other organs
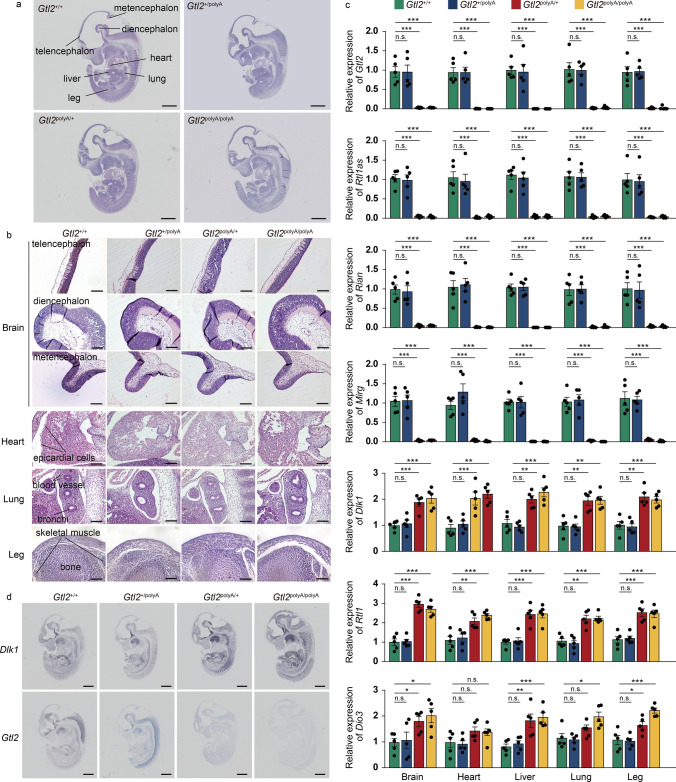
In order to investigate whether silencing of maternally expressed RNAs in Dlk1-Dio3 domain has an effect on tissues other than liver, we conducted a further investigation in embryos. Firstly, histological morphology showed that no significant difference, except liver, was observed in the mutant embryo as a whole (Fig. 6a). Secondly, as previous studies have confirmed the effect of maternally expressed RNA silencing in this domain on brain, lung and skeletal muscle [10, 11, 35], we mainly observed these organs (Fig. 6b). We divided the brain into three parts, telencephalon, diencephalon and metencephalon and the results showed that the brain of E12.5 was not fully developed, and there was no significant difference in telencephalon, diencephalon and metencephalon in the mutant (Fig. 6b). The tracheas of all mutants and Gtl2+/+ in lung were well developed, and there were some small blood vessels, and the alveoli were not yet formed at this stage (Fig. 6b). There were no significant differences in the bones of the legs, nor any defects in the skeletal muscles (Fig. 6b). It is worth mentioning that our currently published study reported that Gtl2 transcription termination caused developmental abnormalities in the heart caused by slow proliferation of epicardial cells (Fig. 6b) [36]. These results suggest that silencing of maternally expressed RNAs transcription in Dlk1-Dio3 at E12.5 does not have a significant effect on major organs except the liver, and the exploration of the effect on the heart needs to be further improved.
Fig. 6.
Termination of maternally expressed RNAs in Dlk1-Dio3 domain transcription had no significant effect on other embryonic organs. a Representative images of HE staining of embryos at E12.5. Scale bar, 1 mm. b Representative images of HE staining of major organs. Scale bar, 80 μm. c qRT-PCR analysis of lncRNAs and mRNAs in Dlk1-Dio3 domain of major organs, β-actin was used as an internal control, n = 5. Error bars, ± SEM, Student’s t-test, n.s., p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001. d In situ hybridization of Dlk1 and Gtl2. Scale bar, 1 mm
qRT-PCR detected the expression of genes in each organ within the range, and the expression trend of maternally expressed RNA in each organ was basically the same, all of them were significantly down-regulated to almost silence, and the expression of paternally expressed genes, Dlk1 and Rtl1, in brain, heart, liver, lung and leg increased about twice, which is basically the same as that of embryo (Fig. 6c). The changes of Dio3 in heart and lung were not very obvious, which may be the reason for the low expression of Dio3 within the two organs (Fig. 6c). We selected Dlk1 and Gtl2 as high expression genes from maternally expressed RNA and paternally expressed RNA, respectively, and in situ hybridization showed the same expression as qRT-PCR, and both Dlk1 and Gtl2 were highly expressed in liver, but not in other organs (Fig. 6d and S5a, b). Thus, the above results indicate that the changes of genes in major organs in the interval are basically the same as those in the whole embryo, and the effect on the liver is greater than that on the other organs, which needs further observation.
Silencing of Gtl2, but not Gtl2-DMR and IG-DMR, leads to Dlk1 activation
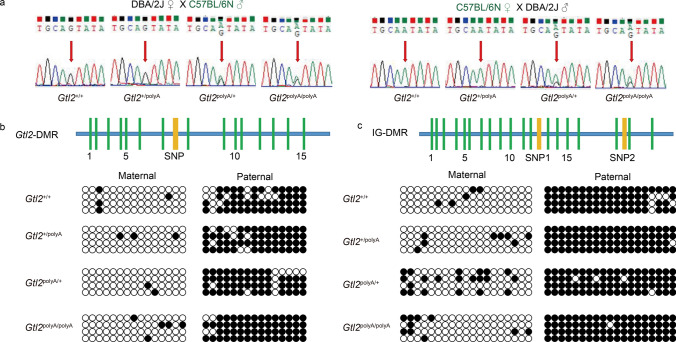
To explore whether the cause of the phenotype was caused by changes in the gene itself or by the methylation status of the Gtl2-DMR and IG-DMR, we found a SNP(A/G) site on Dlk1 that can distinguish between C57BL/6N(G) and DBA/2J(A). After obtaining the cDNA of the embryo of the female mouse of DBA/2J and the male mouse of C57BL/6N after mating, the cDNA fragment containing the SNP site was sequenced. The expression in Dlk1 of Gtl2+/+ and Gtl2+/polyA was mono-allelic, while the expression of Gtl2polyA/+ and Gtl2polyA/polyA was bi-allelic. At the same time, we also tested the offspring obtained by mating male DBA/2J mice with female C57BL/6N mice, and the same conclusion was obtained (Fig. 7a). This means that the silencing of maternally expressed RNAs in Dlk1-Dio3 cause the imprinting loss of Dlk1, although, paternally expressed genes were not edited in the domain, Dlk1, Rtl1 and Dio3 were still affected, which was also likely to have some impact on embryonic development.
Fig. 7.
Loss of imprinting occurs in Dlk1, which was not caused by Gtl2-DMR and IG-DMR. a Biallelic expression of Dlk1 in embryos at E12.5, red arrow points to SNPs. b, c Methylation analysis of Gtl2-DMR (b) and IG-DMR(c). Methylated CpG dinucleotides are represented by filled circles, while unmethylated CpG dinucleotides are represented by open circles
Subsequently, we examined Gtl2-DMR and IG-DMR. The lengths of IG-DMR and Gtl2-DMR detected in this study were 497 bp and 385 bp, correspondingly. These sequences contained 19 CpG and 2 SNP sites, and 15 CpG and 1 SNP sites in C57BL/6N and DBA/2J hybrids, respectively [11, 34] (Fig. 7b and c). The results shown that both of two DMRs maintained the parental origin specific methylation status in E12.5 embryos (Fig. 7b and c). Taken together, the above data indicated that the variation of paternally expressed genes in the domain were caused by silencing of Gtl2 rather than by changes in methylation status of Gtl2-DMR and IG-DMR.
Discussion
In this study, we used the improved Easi-CRISPR technique to knock 3 × polyA powerful transcriptional termination sequence into exon1 of Gtl2, and finally obtained a mouse model that silenced only the Gtl2 gene itself without altering other active elements. By using RNA sequencing analysis, we identified that the termination of maternally expressed RNA transcription in the Dlk1-Dio3 would cause apoptosis of cells in the embryos, especially in the liver, resulting in vascular damage which further caused liver hemorrhage, injury, and finally led to embryonic death.
Previous functional studies of the maternally expressed RNA in the Dlk1-Dio3 imprinted region have all adopted the method of large deletions of the Gtl2 gene sequence, which result in the deletion of regulatory elements at the edited locus together [10–12]. Although there was a phenomenon of perinatal death, the cause of death was alveolar dysplasia or skeletal muscle deficiency, which was different from the present study [10, 11]. In addition, other studies have shown that the method of large fragment deletion to study the function of lncRNA will result in false positive results [13, 14]. It has been shown that the insertion of small fragments does not delete any genomic native sequence and does not change staining mass conformation has been proved to be a more effective method for lncRNA study [14, 37]. By inserting the sequence of 3 × poyA to block the expression of maternally expressed RNA in the Dlk1-Dio3 domain, without deleting other components, the results obtained are more persuasive. Here, we provide a new method to study lncRNA, which can more accurately elucidate the function and regulatory mechanism of the gene itself.
Gtl2-induced apoptosis is a type of programmed cell death that occurs after many stimuli, infections, or injuries and plays a key role in physiological processes such as embryogenesis [38]. Gtl2 down-regulation can regulate FOXO1 and FOXO4 to promote apoptosis through competitive binding of miR-361-5P and miR-23b-3p, respectively [39–41]. Another study found that knockout of Gtl2 promotes apoptosis of vascular endothelial cells and adipose-derived stem cells by activating the Bcl-2/Bax pathway through the p53 signaling pathway [42, 43]. Deletion Gtl2 resulted in the down-regulation of lncRNAs and miRNAs in Dlk1-Dio3 domain, activated the PI3K-mTOR pathway, enhanced metabolic activity, ultimately leading to HSC apoptosis and embryonic death [24]. MEG3 (or Gtl2) inhibited the expression of RASA1 by mediating the histone methylation of the promoter of RASA1 gene by EZH2, thereby activating the RAS-MAPK pathway and enhancing the proliferative and invasive capacities of trophoblasts, and the study suggested that the silent of MEG3 (or Gtl2) maybe a risk fact for unexplained recurrent spontaneous abortion (URSA) [44]. Meanwhile, Dlk1 acts as an inhibitor of vascular differentiation and inhibits vessel sprouting by antagonizing the NOTCH pathway [45], and it can also play an important role in hematopoiesis and hepatocyte development [46–48]. That means increased dose of Dlk1 promoted the proliferation of endothelial cells, but inhibited the differentiation of blood vessels, causing the blood vessels in fetal liver to become thicker and fewer (Fig. 5e–g). In addition, it has been reported that Rtl1 is essential for the maintenance of fetal capillaries, and both its loss and overproduction lead to late fetal and/or neonatal lethality in mice [49]. In addition, our previous study has also shown that maternally expressed RNAs silencing can cause defective narrowing of blood vessels in the placental labyrinth, leading to fetal vascular obstruction and ultimately affecting the exchange function of fetal capillaries [25].
In the process of obtaining embryos, Gtl2polyA/+ embryos, but not Gtl2polyA/polyA ones, were found to die at E13.5 (mortality rate was 21.43%), which possibly due to the fact that the Gtl2polyA/+ phenotype appeared earlier than Gtl2polyA/polyA embryos. At E12.5, Gtl2polyA/+ embryos were in the agonal state, leading to a large change in gene expression. At the same time, although the number of DEGs was smaller in Gtl2polyA/polyA embryos, the changes of gene expression were more significant in Dlk1-Dio3 region, which may account for the severe mortality after E13.5. We therefore hypothesized that Gtl2 transcriptional termination may function by regulating different mechanisms in Gtl2polyA/+ and Gtl2polyA/polyA embryos. However, this requires a lot of work to confirm. Up to now, there is little information on Gtl2 transcriptional termination homozygous mice, and most studies have been limited to termination of paternal or maternal transcription [12, 24]. In the future, we will devote ourselves to studying the genetic changes of Gtl2polyA/+ and Gtl2polyA/polyA embryos after E12.5, and give a more comprehensive explanation of this phenomenon through RNA-seq, single-cell RNA-seq, spatial transcriptomics and some other ways.
The Dlk1-Dio3 imprinted region contains the largest miRNA cluster in the mammalian genome. In this study, our RNA-seq and qRT-PCR data showed that these miRNAs and lncRNAs within the region have the same expression pattern as Gtl2, supporting the hypothesis that these ncRNAs act as a large polycistronic transcription unit [6]. Silencing of the maternally expressed ncRNAs in this region re-activate the normally repressed protein-coding genes on the maternal allele, however, the methylation status of IG-DMR and Gtl2-DMR remained unchanged, which was consistent with previous reported results [12, 33]. As an enhancer that activates Gtl2-Rian-Mirg polycistron, IG-DMR regulates the Dlk1-Dio3 imprinting domain through Gtl2-DMR [12, 50, 51]. Gtl2-DMR methylation level are associated with CTCF binding, Gtl2 expression, and inhibition of paternally expressed genes [33]. However, we detected no change in Gtl2-DMR, which may be caused by premature transcriptional termination of Gtl2. This also indicates that the Gtl2 itself plays a role in the inhibition of Dlk1. Recent studies have shown that the expression level of imprinted lncRNA was related to the degree of in-cis repression, which was related to the formation of lncRNA-protein aggregates [52]. Gtl2 interact with components of polycomb repressive complexes (PRC) and was thought to be part of this aggregate. However, whether such aggregates form within the Dlk1-Dio3 interval and how Gtl2 itself inhibits Dlk1 in cis needs to be further investigated [52–54]. Furthermore, studies have shown that Gtl2 expression, but not other ncRNAs in the domain, controls inhibition of Dlk1 [33].
In summary, our study further revealed that transcription termination of the maternally expressed RNAs in the Dlk1-Dio3 imprinted domain activated the apoptotic and caused severe consequences such as vascular rupture, hepatic hemorrhage and embryonic death, indicating that maternally expressed RNAs in the domain play a crucial role in the regulation of apoptosis and embryogenesis. Although the study has not yet identified the key targets of silencing of maternally expressed RNA in Dlk1-Dio3 domain in inducing apoptosis, it links the region with vascular and hepatic development, which will be of great significance for the study of embryonic development and even disease occurrence. At the same time, we used Easi-CRISPR method to study the function of lncRNA, which may provide a new way to study the function of lncRNA.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank BMKCloud for the analysis of the RNA-seq data. We thank members of our teams for helpful discussion.
Author contributions
Conceptualization, HRY and QW; Methodology, HRY, XH and ZBH; Validation, ZBH; Formal Analysis, RC and YZ; Resources, XMJZ, XQT, ZYW and BJL; Investigation, MYW and RC; Writing-Original Draft, HRY; Writing-Review & Editing, MYW, HJH, ZBH, XH, YZ and QW; Visualization, HRY and MYW; Funding Acquisition, YZ and QW.
Funding
This work was supported by grants from the Key Research and Development Program of Heilongjiang (2023ZX06C09), the State Key Laboratory of Urban Water Resource and Environment of Harbin Institute of Technology (2022TS27), the National Natural Science Foundation of China (U20A20376).
Data availability
All data supporting this study are available within the article and the Supplementary Materials. The RNA-seq data have been deposited in Gene Expression Omnibus database (GEO) under accession number GSE274171.
Declarations
Conflict of interests
The authors declare no competing financial interests.
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Harbin Institute of Technology (Date: 19 June 2023 /No: IACUC-2023066).
Consent for publication
All authors discussed the results and approved the final manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yan Zhang, Email: zhangtyo@hit.edu.cn.
Qiong Wu, Email: kigo@hit.edu.cn.
References
- 1.Bartolomei MS (2009) Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev 23:2124–2133. 10.1101/gad.1841409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferguson-Smith AC (2011) Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 12:565–575. 10.1038/nrg3032 [DOI] [PubMed] [Google Scholar]
- 3.Enterina JR, Enfield KSS, Anderson C et al (2017) DLK1-DIO3 imprinted locus deregulation in development, respiratory disease, and cancer. Expert Rev Respir Med 11:749–761. 10.1080/17476348.2017.1355241 [DOI] [PubMed] [Google Scholar]
- 4.Sanceau J, Poupel L, Joubel C et al (2024) DLK1/DIO3 locus upregulation by a β-catenin-dependent enhancer drives cell proliferation and liver tumorigenesis. Mol Ther 32:1125–1143. 10.1016/j.ymthe.2024.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocha STD, Edwards CA, Ito M et al (2008) Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet 24:306–316. 10.1016/j.tig.2008.03.011 [DOI] [PubMed] [Google Scholar]
- 6.Tevendale M, Watkins M, Rasberry C et al (2006) Analysis of mouse conceptuses with uniparental duplication/deficiency for distal chromosome 12: comparison with chromosome 12 uniparental disomy and implications for genomic imprinting. Cytogenet Genome Res 113:215–222. 10.1159/000090835 [DOI] [PubMed] [Google Scholar]
- 7.Labialle S, Marty V, Bortolin‐Cavaillé M, et al (2014) The miR‐379/miR‐410 cluster at the imprinted Dlk1‐Dio3 domain controls neonatal metabolic adaptation. The EMBO Journal 33:2216–2230. 10.15252/embj.201387038 [DOI] [PMC free article] [PubMed]
- 8.Royo H, Basyuk E, Marty V et al (2007) Bsr, a Nuclear-retained RNA with Monoallelic Expression. MBoC 18:2817–2827. 10.1091/mbc.e06-10-0920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Runte M (2001) The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet 10:2687–2700. 10.1093/hmg/10.23.2687 [DOI] [PubMed] [Google Scholar]
- 10.Takahashi N, Okamoto A, Kobayashi R et al (2009) Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum Mol Genet 18:1879–1888. 10.1093/hmg/ddp108 [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Cheunsuchon P, Nakayama Y et al (2010) Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development 137:2643–2652. 10.1242/dev.045724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu W, Botticelli EM, Kery RE et al (2019) Meg3-DMR, not the Meg3 gene, regulates imprinting of the Dlk1-Dio3 locus. Dev Biol 455:10–18. 10.1016/j.ydbio.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyal A, Myacheva K, Groß M, et al (2016) Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res gkw883. 10.1093/nar/gkw883
- 14.Paralkar VR, Taborda CC, Huang P et al (2016) Unlinking an lncRNA from Its Associated cis Element. Mol Cell 62:104–110. 10.1016/j.molcel.2016.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swenson KS, Wang D, Jones AK et al (2023) Metformin Disrupts Signaling and Metabolism in Fetal Hepatocytes. Diabetes 72:1214–1227. 10.2337/db23-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Ramirez CA, Nakano H, Law RC et al (2024) Atlas of fetal metabolism during mid-to-late gestation and diabetic pregnancy. Cell 187:204-215.e14. 10.1016/j.cell.2023.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin K, Lan X, Huang P, et al (2023) Molecular basis of polycomb group protein-mediated fetal hemoglobin repression. Blood blood.2022019578. 10.1182/blood.2022019578 [DOI] [PMC free article] [PubMed]
- 18.Saffarzadeh M, Grunz K, Nguyen TS et al (2020) Macrophage protease-activated receptor 2 regulates fetal liver erythropoiesis in mice. Blood Adv 4:5810–5824. 10.1182/bloodadvances.2020003299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh S, Okada H, Koyano K et al (2023) Fetal and neonatal bilirubin metabolism. Front Pediatr 10:1002408. 10.3389/fped.2022.1002408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao L, Paik NY, Sanborn MA et al (2023) Hematopoietic Jagged1 is a fetal liver niche factor required for functional maturation and engraftment of fetal hematopoietic stem cells. Proc Natl Acad Sci USA 120:e2210058120. 10.1073/pnas.2210058120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokomizo T, Ideue T, Morino-Koga S et al (2022) Independent origins of fetal liver haematopoietic stem and progenitor cells. Nature 609:779–784. 10.1038/s41586-022-05203-0 [DOI] [PubMed] [Google Scholar]
- 22.De Jong IEM, Hunt ML, Chen D et al (2023) A fetal wound healing program after intrauterine bile duct injury may contribute to biliary atresia. J Hepatol 79:1396–1407. 10.1016/j.jhep.2023.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anwar I, Ashfaq UA, Shokat Z (2020) Therapeutic Potential of Umbilical Cord Stem Cells for Liver Regeneration. CSCR 15:219–232. 10.2174/1568026620666200220122536 [DOI] [PubMed] [Google Scholar]
- 24.Qian P, He XC, Paulson A et al (2016) The Dlk1-Gtl2 Locus Preserves LT-HSC Function by Inhibiting the PI3K-mTOR Pathway to Restrict Mitochondrial Metabolism. Cell Stem Cell 18:214–228. 10.1016/j.stem.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, He H, Yu H et al (2024) Maternal RNA transcription in Dlk1-Dio3 domain is critical for proper development of the mouse placental vasculature. Commun Biol 7:363. 10.1038/s42003-024-06038-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M (2018) Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat Protoc 13:195–215. 10.1038/nprot.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pertea M, Kim D, Pertea GM et al (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11:1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR : a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Zhu H, Li L et al (2024) New mechanism of LncRNA: In addition to act as a ceRNA. Non-coding RNA Research 9:1050–1060. 10.1016/j.ncrna.2024.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnepper AP, Marques LF, Wolf IR et al (2024) Potential global cis and trans regulation of lncRNAs in Saccharomyces cerevisiae subjected to ethanol stress. Gene 920:148521. 10.1016/j.gene.2024.148521 [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP (2009) MicroRNAs: Target Recognition and Regulatory Functions. Cell 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farhadova S, Ghousein A, Charon F, et al (2024) The long non-coding RNA Meg3 mediates imprinted gene expression during stem cell differentiation. Nucleic Acids Research gkae247. 10.1093/nar/gkae247 [DOI] [PMC free article] [PubMed]
- 34.Wilner GD, Nossel HL, LeRoy EC (1968) Aggregation of platelets by collagen. J Clin Invest 47:2616–2621. 10.1172/JCI105944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez ME, Karaczyn A, Wu Z et al (2024) Transgenerational epigenetic self-memory of Dio3 dosage is associated with Meg3 methylation and altered growth trajectories and neonatal hormones. Epigenetics 19:2376948. 10.1080/15592294.2024.2376948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teng X, He H, Yu H et al (2024) LncRNAs in the Dlk1-Dio3 Domain Are Essential for Mid-Embryonic Heart Development. IJMS 25:8184. 10.3390/ijms25158184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson KM, Anderson DM, McAnally JR et al (2016) Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539:433–436. 10.1038/nature20128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morana O, Wood W, Gregory CD (2022) The Apoptosis Paradox in Cancer IJMS 23:1328. 10.3390/ijms23031328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsao CW, Aday AW, Almarzooq ZI, et al (2022) Heart Disease and Stroke Statistics—2022 Update: A Report From the American Heart Association. Circulation 145:. 10.1161/CIR.0000000000001052 [DOI] [PubMed]
- 40.Wang A, Hu N, Zhang Y et al (2019) MEG3 promotes proliferation and inhibits apoptosis in osteoarthritis chondrocytes by miR-361-5p/FOXO1 axis. BMC Med Genomics 12:201. 10.1186/s12920-019-0649-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Li X, Wang Z (2021) lncRNA MEG3 inhibits pituitary tumor development by participating in cell proliferation, apoptosis and EMT processes. Oncol Rep 45:40. 10.3892/or.2021.7991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y (2020) MEG3 regulates apoptosis of adipose-derived stem cells. Mol Med Report. 10.3892/mmr.2020.11059 [Google Scholar]
- 43.Yu Y, Kou D, Liu B et al (2020) LncRNA MEG3 contributes to drug resistance in acute myeloid leukemia by positively regulating ALG9 through sponging miR-155. Int J Lab Hematology 42:464–472. 10.1111/ijlh.13225 [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Liu X, Gao Y (2021) The long noncoding RNA MEG3 regulates Ras-MAPK pathway through RASA1 in trophoblast and is associated with unexplained recurrent spontaneous abortion. Mol Med 27:70. 10.1186/s10020-021-00337-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang C-C, Kuo H-M, Wu P-C et al (2018) Soluble delta-like 1 homolog (DLK1) stimulates angiogenesis through Notch1/Akt/eNOS signaling in endothelial cells. Angiogenesis 21:299–312. 10.1007/s10456-018-9596-7 [DOI] [PubMed] [Google Scholar]
- 46.Li L, Forman SJ, Bhatia R (2005) Expression of DLK1 in hematopoietic cells results in inhibition of differentiation and proliferation. Oncogene 24:4472–4476. 10.1038/sj.onc.1208637 [DOI] [PubMed] [Google Scholar]
- 47.Persson-Augner D, Lee Y-W, Tovar S et al (2014) Delta-Like 1 Homologue (DLK1) Protein in Neurons of the Arcuate Nucleus That Control Weight Homeostasis and Effect of Fasting on Hypothalamic DLK1 mRNA. Neuroendocrinology 100:209–220. 10.1159/000369069 [DOI] [PubMed] [Google Scholar]
- 48.Tanimizu N, Tsujimura T, Takahide K et al (2004) Expression of Dlk/Pref-1 defines a subpopulation in the oval cell compartment of rat liver. Gene Expr Patterns 5:209–218. 10.1016/j.modgep.2004.08.003 [DOI] [PubMed] [Google Scholar]
- 49.Balusu S, Horré K, Thrupp N et al (2023) MEG3 activates necroptosis in human neuron xenografts modeling Alzheimer’s disease. Science 381:1176–1182. 10.1126/science.abp9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aronson BE, Scourzic L, Shah V et al (2021) A bipartite element with allele-specific functions safeguards DNA methylation imprints at the Dlk1-Dio3 locus. Dev Cell 56:3052-3065.e5. 10.1016/j.devcel.2021.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiura H, Komiyama J, Shirai M et al (2007) DNA methylation imprints on the IG-DMR of the Dlk1 – Gtl2 domain in mouse male germline. FEBS Lett 581:1255–1260. 10.1016/j.febslet.2007.02.034 [DOI] [PubMed] [Google Scholar]
- 52.Schertzer MD, Braceros KCA, Starmer J et al (2019) lncRNA-Induced Spread of Polycomb Controlled by Genome Architecture, RNA Abundance, and CpG Island DNA. Mol Cell 75:523-537.e10. 10.1016/j.molcel.2019.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanli I, Lalevée S, Cammisa M et al (2018) Meg3 Non-coding RNA Expression Controls Imprinting by Preventing Transcriptional Upregulation in cis. Cell Rep 23:337–348. 10.1016/j.celrep.2018.03.044 [DOI] [PubMed] [Google Scholar]
- 54.Kaneko S, Bonasio R, Saldaña-Meyer R et al (2014) Interactions between JARID2 and Noncoding RNAs Regulate PRC2 Recruitment to Chromatin. Mol Cell 53:290–300. 10.1016/j.molcel.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting this study are available within the article and the Supplementary Materials. The RNA-seq data have been deposited in Gene Expression Omnibus database (GEO) under accession number GSE274171.