Summary
The vacuolar H+-ATPase (V-ATPase) is a functionally conserved multimeric complex localized at the membranes of many organelles where its proton-pumping action is required for proper lumen acidification. The V-ATPase complex is composed of several subunits, some of which have been linked to human disease. We and others previously reported pathogenic dominantly acting variants in ATP6V1B2, the gene encoding the V1B2 subunit, as underlying a clinically variable phenotypic spectrum including dominant deafness-onychodystrophy (DDOD) syndrome, Zimmermann-Laband syndrome (ZLS), and deafness, onychodystrophy, osteodystrophy, intellectual disability, and seizures (DOORS) syndrome. Here, we report on an individual with features fitting DOORS syndrome caused by dysregulated ATP6V1C1 function, expand the clinical features associated with ATP6V1B2 pathogenic variants, and provide evidence that these ATP6V1C1/ATP6V1B2 amino acid substitutions result in a gain-of-function mechanism upregulating V-ATPase function that drives increased lysosomal acidification. We demonstrate a disruptive effect of these ATP6V1B2/ATP6V1C1 variants on lysosomal morphology, localization, and function, resulting in a defective autophagic flux and accumulation of lysosomal substrates. We also show that the upregulated V-ATPase function affects cilium biogenesis, further documenting pleiotropy. This work identifies ATP6V1C1 as a new gene associated with a neurodevelopmental phenotype resembling DOORS syndrome, documents the occurrence of a phenotypic continuum between ZLS, and DDOD and DOORS syndromes, and classify these conditions as lysosomal disorders.
Keywords: vacuolar ATPase, ATP6V1B2, ATP6V1C1, Zimmermann-Laband syndrome, DOORS syndrome, DDOD syndrome, neurodevelopmental disorder, pleiotropy, lysosome, autophagy
V-ATPase is a multimeric complex localized at the membranes of organelles where its proton-pumping action is required for lumen acidification. We report that increased ATP6V1C1 function causes DOORS syndrome and show a phenotypic continuum between ZLS, DDOD syndrome, and DOORS syndrome due to activating ATP6V1B2 variants, classifying them as lysosomal disorders.
Introduction
The vacuolar H+-ATPase (V-ATPase) is an electrogenic proton pump located in the membranes of intracellular organelles, including secretory granules, endosomes, lysosomes, and Golgi cisternae. By its proton-pumping action, V-ATPase is required for proper lumen acidification1 and is involved in several physiological processes, including membrane trafficking, protein degradation, and pH homeostasis.2,3 This multimeric complex is highly conserved through evolution, and its presence in yeast indicates that the enzyme is required for basic cellular physiology. Consistently, functional dysregulation of the pump underlies a number of clinically heterogeneous human diseases.4 Loss-of-function (LoF) variants in genes encoding V-ATPase components (ATP6V1B1 [MIM: 192132] and ATP6V0A4 [MIM: 605239]) underlie autosomal recessive distal renal tubular acidosis with or without sensorineural hearing loss (MIM: PS179800),5 biallelic variants in ATP6V1A (MIM: 607027) cause a recessive form of cutis laxa (MIM: 617403),3 and LoF variants in ATP6V0A3 (MIM: 604592) cause osteopetrosis, a recessive disorder characterized by increased bone density due to impaired bone resorption (MIM: PS259700).6 Dysregulated function of these subunits have also been implicated in neurological diseases, including dominant developmental encephalopathy with epilepsy (MIM: 618012), which is caused by variants in ATP6V1A (MIM: 607027),7 a dominant early-onset form of epilepsy, with or without developmental delay (DD) (MIM: 620465),8 associated with variants in ATP6V0C (MIM: 108745), and X-linked parkinsonism with spasticity (MIM: 300911), congenital disorder of glycosylation (MIM: 301045), and intellectual developmental disorder, Hedera type (MIM: 300423), which are due to hemizygous variants in ATP6AP2 (MIM: 300556).9,10,11
V-ATPase is composed of a catalytic cytosolic V1 region comprising eight subunits (stoichiometry A3, B3, C, D, E3, F, G3, and H), catalyzing ATP hydrolysis, and a membrane-embedded VO region made up of eight subunits (stoichiometry a, c9, c″, d, e, f, ATP6AP1, and ATP6AP2) that is responsible for the proton transport from the cytoplasm to the lumen or extracellular space.4,12 The ATP6V1B2 and ATP6V1C1 subunits belong to the V1 complex and are ubiquitously expressed. We and others reported that dominantly acting variants in ATP6V1B2 are associated with a wide clinical spectrum, including dominant deafness-onychodystrophy (DDOD) syndrome (MIM: 124480), deafness, onychodystrophy, osteodystrophy, intellectual disability, and seizures (DOORS) syndrome (MIM: 220500), and Zimmermann-Laband syndrome (ZLS [MIM: PS135500]).13,14,15,16,17,18 Individuals with DDOD syndrome usually show normal development and cognitive function, while those with DOORS syndrome and ZLS present with intellectual disability (ID), with or without seizures. These disorders share hypoplasia/aplasia of nails and terminal phalanges; a recognizable craniofacial appearance, gingival overgrowth, and hypertrichosis also characterize ZLS. Of note, a recurrent truncating variant in ATP6V1B2 (c.1516C>T, p.Arg506∗) has been reported in both DDOD and DOORS syndromes,16,17,18 suggesting that these conditions are within a spectrum of a single disorder caused by altered V-ATPase function. No clinical phenotype has been reported to be caused by altered function of ATP6V1C1, to date.
Here, we causally link a de novo missense variant in ATP6V1C1 to a neurodevelopmental phenotype with features resembling DOORS syndrome, and more accurately define the clinical spectrum of dominantly acting ATP6V1B2 variants, which is consistent with a phenotypic continuum having DDOD syndrome and ZLS as extreme entities. We also provide evidence that all tested variants result in a functional increase in the V-ATPase pump activity, which has pleiotropic effects in cells, affecting lysosomal morphology and function, autophagy, and cilium biogenesis, defining a neurodevelopmental spectrum primed by generalized defective lysosome function.
Subjects and methods
Subjects
Affected subjects were identified through networking, including GeneMatcher and other professional links (e.g., European Reference Networks). Clinical data and DNA samples were collected, stored, and used under institutional review board-approved protocols. All procedures were conducted in accordance with the ethical standards for use of human participants in medical experimentation, after written informed consent was obtained from the parents or legal guardians of all subjects. The study was approved by the Ospedale Pediatrico Bambino Gesù Institutional Review Board (ref. 1702_OPBG_2018). DNA from leukocytes, hair bulb cells, and skin fibroblasts were extracted using standard protocols. Explicit permission was obtained to publish the photographs of the subjects shown in Figure 1.
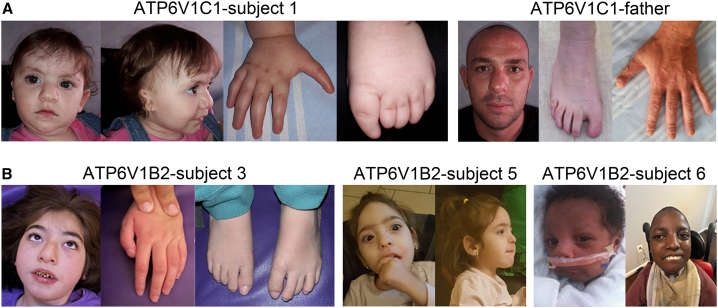
Figure 1.
Craniofacial features of the individuals carrying dominant acting ATP6V1C1/ATP6V1B2 variants
(A) Subject 1 at 2 years (left). Note the occurrence of bushy eyebrow, up-slanted palpebral fissures, epicanthal folds, microtia, broad nasal bridge, bifid nasal tip, deep philtrum, thin upper lip, prominent lower vermillion, nail aplasia of hands and feet, hypoplastic terminal phalanges and triphalangeal thumbs/big toes. Subject 1’s father (right). Note nail aplasia of feet and hypoplastic terminal phalanges of hands and feet. Minor dysmorphism includes broad nasal bridge, bulbus nose, deep philtrum, and prominent upper and lower vermillion.
(B) Subject 3 at 8 years (left). Note facial hypotonia, straight eyebrow, hypertelorism, ptosis, long eyelashes, tubular nose with broad nasal base, short and deep philtrum and everted thin upper lip with prominent upper vermilion. Subject 5 at 4 years 7 months (middle panels). Note the high forehead, bitemporal narrowing, coarse face, brushy and straight eyebrows, long eyelashes, prominent upper and lower vermilion and large mouth. Subject 6 at 13 years (right). Note the bitemporal narrowing, coarse face with full-cheeks, thin upper lip, micrognathia/retrognathia, and short neck. A detailed clinical characterization of affected subjects is reported in Tables 1 and S3 and Supplemental Material (Clinical Reports).
Exome sequencing analyses
DNA of the affected subject and her parents was extracted from circulating leukocytes and sequenced using Illumina paired end technology coupled with the SureSelect AllExon V.5 (Aligent) enrichment kit. Exome sequencing (ES) raw data were processed and analyzed using an in-house implemented pipeline previously described,19,20 according to the GATK’s Best Practices.21 The UCSC GRCh37/hg19 version of genome assembly was used as a reference for reads alignment by means of BWA-MEM tool,22 and the subsequent variant calling with HaplotypeCaller (GATK v3.7).21 Variants’ functional annotation was made by SnpEff v.5.0 and dbNSFP v.4.2 tools.23,24 The most relevant in silico impact predictions were also evaluated, such as Combined Annotation Dependent Depletion v.1.6,25 Mendelian Clinically Applicable Pathogenicity v.1.3, and InterVar v.2.0.1.26,27 By filtering against our population-matched database (approximately 3,000 whole ES [WES]) and public databases (dbSNP150 and gnomAD V.2.0.1), the analysis was focused on high-quality rare variants that affect coding sequences and splice site regions.
Constructs
The human ATP6V1B2 (RefSeq: NM_001693.4) and ATP6V1C1 (RefSeq: NM_001695.5) coding sequences were cloned in pCMV-Myc/HisA (Invitrogen) and in pFLAG-CMV (SIGMA) tagged expression vectors, respectively. The missense variants identified in two different subunits of ATP6V1 complex (p.Ala332Val, p.Gln376Lys, p.Tyr328His, p.Arg485Pro, ATP6V1B2; p.Glu289Lys, and ATP6V1C1) were introduced by site-directed mutagenesis using the QuikChange XL kit (Agilent Technologies) in accordance with the manufacturer’s protocol. All generated constructs were checked by direct sequencing.
Cell cultures
Functional characterization of ATP6V1B2 and ATP6V1C1 mutants was carried out on cultured skin fibroblasts obtained from subcutaneous biopsies of enrolled individuals, as well as in transiently transfected cell lines. Primary human fibroblasts and 293T cell lines were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (EuroClone) and 1% penicillin-streptomycin, at 37°C with 5% CO2. Specifically, 293T cells were seeded in six-well plates the day before transfection. Monolayer were transfected at 70%–80% confluency with wild-type (WT) or mutants C1-Flag-tagged or B2-Myc-tagged expression plasmids, using Fugene 6 transfection reagent (Promega). Forty-eight hours after transfection, cells were lysed and the level of ATP6V1B2 and ATP6V1C1 was assessed by immunoblotting with an anti-Myc (Cell Signaling) or anti Flag (SIGMA) antibodies respectively. ATP6V1B2 endogenous level was evaluated on fibroblasts lysates using a rabbit polyclonal anti-ATP6V1B2 antibody (Abcam). Membranes were probed with an anti-GAPDH antibody (Santa Cruz Biotechnology) for protein normalization.
Confocal analysis
Confocal analysis was performed on a Zeiss LSM 980 with Airyscan2, using the 63× oil objective and excitation spectral laser lines at 405, 488, 546, 594, and 633 nm. Quantitative analyses were performed using the Zen 3.3 software and mean fluorescence intensity (MFI) ± SEM of signals were determined in at least 3 independent experiment per marker (≥20 cells per experimental condition, in each repeat), and plotted following the formula: Final MFI = MFI (region of interest) − MFI (background). Signal colocalization was performed using the Pearson correlation coefficient.
Primary cilium staining
Cells were plated onto cover slips, maintained 24 h in low serum medium to promote emission of cilia and then fixed in 4% paraformaldehyde (PFA). Primary cilium was stained with a rabbit polyclonal anti-ARL13B antibody (Abcam) followed by goat anti-rabbit Alexa Fluor 594 (red), while basal bodies were stained with mouse monoclonal anti-pericentrin (Abcam) followed by goat anti-mouse Alex Fluor 488 (green) and nuclei with DAPI (blue).
Subcellular co-localization of ATP6V1B2 and lysosomes
For immunofluorescence, patients’ and controls fibroblasts were seeded at the density of 20 × 103, in 24-well cluster plates onto 12-μm cover glasses. After 24 h of culture in complete medium, cells were fixed with 3% PFA (30 min at 4°C). After permeabilization with 0.5% Triton X-100 (10 min at room temperature [RT]), fibroblasts were stained with mouse monoclonal anti-Lamp1 antibody (Cell Signaling), and rabbit polyclonal ATP6V1B2 antibody (Abcam), followed by the appropriate secondary antibody (Life Technologies) and DAPI.
Labeling lysosomes with DQ-BSA
To assess the autolysosome function, a red BODIPY dye-conjugated bovine serum albumin (DQBSA, Invitrogen) based assay was used. This BSA derivative is so heavily labeled that the fluorophore is self-quenched. Proteolysis of this compound results in dequenching and release of brightly fluorescent fragments. Thus, the use of DQ-BSA is useful for the visualization of intracellular proteolytic activity. Control and patient’s fibroblasts were seeded in 24-well cluster plates onto 12-mm cover glasses and maintained in culture complete medium for 24 h. After 24 h, cells were incubated with the BSA derivative (10 μg/mL) for 1 h at 37°C in complete culture medium, and then in starvation medium (Earle’s balanced salt solution [EBSS]) for 16 h to induce the autophagic flux. After incubation, the coverslips were counterstained with DAPI and observed under confocal microscopy. Proteolysis of this BSA derivative results in the release of fluorescence (590 nm).
LysoSensor staining
For CLSM analyses patients’ and controls fibroblasts were seeded on coverslips at a density of 300 × 103. After 24 h of culture in complete medium, cells were rinsed with an isotonic buffer pH 7.4 (105 mM NaCl, 5 mM KCl, 6 mM HEPES-acid, 4 mM HEPES-Na, 5 mM NaHCO3, 60 mM mannitol, 5 mM glucose, 0.5 mM MgCl2, 1.3 mM CaCl2) and then incubated with 2 μM LysoSensor Yellow/Blue DND-160 (Thermo Fisher Scientific) for 3′ at RT. Then cells were extensively rinsed with the isotonic buffer and observed on a Zeiss LSM 980. Images were then acquired using a 63× oil objective, laser and filter settings were adjusted according to the fluorescence excitation and emission requirements of the reagent: excitation 365 nm and dual emission at 440 nm (neutral pH 5–7, blue) and 540 nm (acidic pH 3–5, yellow).
Endo-lysosomal pH analysis by LysoSensor yellow/blue dextran
To measure the pH of endo-lysosomal compartments, 2 × 105 fibroblasts were plated on a 35-mm imaging dish with a glass bottom (Ibidi), after 48 h of culture in complete medium were incubated overnight with 0.1 mg/mL LysoSensor Yellow/Blue dextran (#L22460, Thermo Fisher Scientific) in complete medium at 37°C in a humidified atmosphere with 5% CO2. The loading solution was then removed, cells were washed and live imaged in complete medium (without phenol red) with excitation at 360 nm and double emission at 450–470 nm (blue) and 520–540 nm (yellow) on a Zeiss LSM980 confocal microscope. The pH calibration curve was generated using an isotonic buffer (105 mM NaCl, 5 mM KCl, 6 mM HEPES-acid, 4 mM HEPES-Na, 5 mM NaHCO3, 60 mM mannitol, 5 mM glucose, 0.5 mM MgCl2, 1.3 mM CaCl2) supplemented with 20 μM nigericin. Each buffer solution was adjusted to the appropriate final pH, ranging from 2.5 to 6.5, using 1 N NaOH or 1 N HCl.
Cells (>50) were analyzed both in fluorescence and in bright field with the Zeiss Zen 3.3 software. ROIs within cells that were reflective of LysoSensor Yellow/Blue dextran-labeled organelles were selected and the fluorescence intensity values of such regions were recorded for emissions at both wavelengths (450 and 540 nm). Additionally, ROIs outside of cells (background) were selected and the background fluorescence intensity values were recorded for each emission wavelength.
After subtracting the background fluorescence intensity values for each emission wavelength from the corresponding fluorescence intensity values relating to LysoSensor Yellow/Blue dextran-labeled organelles, the ratio of intensity 450 nm:540 nm was calculated for each region of interest and for each pH calibration curve buffer. Next, the data for the pH calibration curve were fitted to a linear regression using GraphPad Prism5 software and the data of experimentally measured ratios in samples were converted into absolute pH values by interpolation in the pH calibration curve.
Filipin fluorescence staining of free cholesterol
Filipin dye is used as a pro-fluorescence probe to stain the cholesterol in cells or tissues. Filipin binds to model crystalline phase lipid bilayers containing cholesterol, and to liquid phase bilayers with or without cholesterol, in an orientation that forms membrane pores. Due to its fluorescence shift upon binding to cholesterol, filipin is visible under simple UV trans-illumination. Briefly, control and patient fibroblasts were seeded in 24-well cluster plates onto 12-mm cover glasses and maintained in culture complete medium for 24 h. After 24 h, cells were rinsed twice with PBS, fixed in with 3% PFA, washed twice with PBS, and incubated with 1 mL of 1.5 mg glycine/mL PBS for 10 min at RT to quench the PFA. Fixed cells were washed three times and labeled with the Filipin staining solution for 2 h at RT, followed by three washes before imaging. Cells were observed in PBS by fluorescence microscopy using a UV filter set (340–380 nm excitation, 40 nm dichroic, 430-nm-long pass filter).
Evaluation of LC3I/II levels by western blot analysis
To monitor the function of the autophagic pathway, primary control and patients’ fibroblasts were incubated in EBSS (Gibco) medium for 2, 4, and 8 h with and without addition of the inhibitor bafilomycin A1 (200 nM for 4 h, Sigma). LC3I/II expression level was assessed by western blot analysis and immunofluorescence microscopy. For western blot analyses, cell pellets were washed twice in PBS and lysed in RIPA buffer (20 min, 4°C). Lysates were centrifuged at 10,000×g (15 min, 4°C), and 20 μg cell extracts were separated by 15% SDS-PAGE, and transferred to PVDF membranes (Bio-Rad). Blots were incubated with rabbit polyclonal anti-LC3 antibody (Cell Signaling). Membranes were re-probed with an anti-GAPDH antibody (Santa Cruz Biotechnology) to normalize protein content. For immunofluorescence, patients’ and controls fibroblasts were seeded at the density of 20 × 103 in 24-well cluster plates onto 12-μm cover glasses. After 24 h of culture in complete medium, fibroblasts were treated with EBSS medium to promote the autophagosomes formation, for 4 and 8 h with or without bafilomycin (4 h). Then were fixed with absolute chilled methanol for 10 min at −20°C. Cells were stained with a rabbit polyclonal anti-LC3I/II antibody (Cell Signaling) only or co-stained with mouse monoclonal anti-Lamp1 antibody (Cell Signaling) followed by the appropriate secondary antibodies (Life Technologies) and DAPI.
Electron microscopy
Control and patients-derived fibroblast were fixed in 1% glutaraldehyde and 4% PFA in 0.1 M sodium cacodylate buffer (pH 7.2), overnight, at 4°C, and processed as described previously.28 Cells were washed in cacodylate buffer and post-fixed with 1% OsO4 in 0.1 M sodium cacodylate buffer for 1 h at RT, treated with 1% tannic acid in 0.05 M cacodylate buffer for 30 min, and rinsed in 1% sodium sulfate in 0.05 M cacodylate buffer for 10 min. Post-fixed specimens were washed, dehydrated through a graded series of ethanol solutions (30%–100% ethanol) and embedded in Agar 100 (Agar Scientific). Ultrathin sections, obtained by an UC6 ultramicrotome (Leica Microsystems), were stained with uranyl zerosolution (Agar Scientific) and Reynolds’ lead citrate and examined at 100 kV with a FEI/Philips EM 208S Transmission Electron Microscope equipped with acquisition system/Megaview SIS camera (Olympus).
Structural analyses
Structures with pdb codes 6wm2 and 3vr6 were used in our analysis. In 6wm2, the A and B subunits are present as trimers with chain identifiers A, B, and C for the subunits A, while D, E, and F identify the subunits B. To avoid confusion with the letters identifying the subunits, we refer to these chains as α, β, γ, δ, ε, and ζ, respectively. The β/ε and the α/δ interfaces represent the open and semi-open configurations, while the γ/ζ is the only one bound to ADP and is representative of the closed state. The C subunit is present as a monomer (with chain identifier O). All the figures were obtained by using the software Chimera.29 The same software package was used to carry out the structural analysis.
Results
Genetic and clinical findings
In the context of an intramural research program dedicated to individuals affected by undiagnosed diseases, trio-based ES was performed to reach diagnosis in an individual with a phenotype partially overlapping DOORS syndrome and ZLS, who was negative for mutations in the previously identified disease genes. The main clinical features of the individual included DD/ID, electroencephalogram abnormalities, a bulbous nose with a bifid nasal tip and additional minor dysmorphism, microtia with malformation of inner and outer ear, profound sensorineural deafness, short stature, nail aplasia of hands and feet, hypoplastic terminal phalanges, and triphalangeal thumbs/big toes (Figure 1A; Supplemental Material, Clinical Reports). Of note, occurrence of minor digit anomalies (i.e., hypoplastic nails and terminal phalanges of hands and feet) was also observed in her father, who however did not show any additional feature reported in his daughter (Figure 1A). Sequencing statistics and data output are reported in Table S1. ES data analysis allowed us to identify a previously unreported, apparently de novo, missense variant, c.865G>A (p.Glu289Lys [C1E289K]), affecting ATP6V1C1. No functionally relevant variants compatible with known Mendelian disorders based on the inheritance model and clinical presentation were identified. ATP6V1C1 was considered as an excellent candidate based on its functional link with ATP6V1B2, the high degree of conservation of the affected residue, and the predicted damaging consequences of the amino acid substitution on protein function (Figure S1A; Table S2). The heterozygous variant was validated via Sanger sequencing, which confirmed its occurrence in all tested tissues (Figure S2). The variant was not present in the healthy brother and mother, while testing multiple paternal tissues (i.e., leukocytes, fibroblasts, hair bulb cells, and oral mucosal epithelial cells) documented a low level of mosaicism for the variant in hair bulb and buccal cells, supporting a post-zygotic onset of the variant in the father, in line with the clinical findings (Figure S2). Based on these findings, manual inspection of the WES data in the father confirmed a low-level mosaicism in the sample (estimated as 7%, 6 reads out of 87).
Networking failed to identify additional cases with putative pathogenic ATP6V1C1 variants. Based on the clinical overlap and functional link, phenotypic and molecular data on six unrelated subjects with bona fide pathogenic heterozygous ATP6V1B2 variants were collected to more precisely characterize the clinical and molecular spectra of the disorder, and investigate the mechanism of disease. All variants were missense (NM_001693.4:c.995C>T, p.Ala332Val [B2A332V]; c.982T>C, p.Tyr328His [B2Y328H]; c.983A>G, p.Tyr328Cys; c.1120G>C, p.Glu374Gln; c.1126C>A, p.Gln376Lys [B2Q376K]; and c.1127A>G, p.Gln376Arg), occurred de novo, had not previously been reported in public databases (e.g., gnomAD), involved conserved residues located within regions of the protein that were intolerant to variation (Figure S1B), and were predicted to have a disruptive impact on protein function (Table S2). The clinical phenotype of the subjects was variable but invariably characterized by dysmorphic features, DD with impact both on cognitive and motor functions, ID, and seizures. A subset of individuals showed horizontal nystagmus/visual impairment, growth delay, brain abnormalities (e.g., brain atrophy and several degrees of myelination delay), deafness, gingival overgrowth, and minor skeletal abnormalities (e.g., scoliosis, pes planus, joint hyperextensibility). No individuals had onychodystrophy, osteodystrophy, or skeletal dysplasia. The severity of the condition was fatal in one individual (subject 2) (Supplemental Material, Clinical Reports). The observed clinical spectrum was tentatively associated with DOORS, DDOD, and/or ZLS without reaching the diagnostic criteria for none of these disorders (Table 1; Figure 1B).
Table 1.
Major clinical features of the identified subjects with ATP6V1B2/ATP6V1C1 variants, and comparison with DDOD syndrome, DOORS syndrome, and ZLS
| DD | ID | Seizures | Hypoplasia/aplasia terminal phalanges and/or onychodystrophy | Hypertrichosis | Gingival overgrowth | Facial dysmorphisms | Deafness | Osteodystrophy | |
|---|---|---|---|---|---|---|---|---|---|
| DDOD | +/− | + | – | + | – | – | +/− | + | – |
| DOORS | + | + | + | + | +/− | – | + | + | + |
| ZLS | + | + | +/− | + | +/− | + | + | +/− | – |
| ATP6V1B2 (p.Arg506∗) | 8/20 (40%) | 8/20 (40%) | 7/20 (35%) | 20/20 (100%) | 1/20 (5%) | 1/20 (5%) | 18/18 (100%) | 20/20 (100%) | 0/20 (0%) |
| ATPV1B2 (missense changes) | 5/11 (45%) | 9/11 (81%) | 9/11 (81%) | 3/10 (30%) | 4/11 (36%) | 5/11 (45%) | 11/11 (100%) | 1/10 (10%) | 0/11 (0%) |
| S1 (ATP6V1C1) | + | + | + | + | – | – | bushy eyebrow, up-slanted palpebral fissures, epicanthal folds, microtia, broad nasal bridge, "bifid” nasal tip, deep philtrum, thin upper lip, prominent lower vermillion | + | – |
| S2 (ATP6V1B2, p.Ala332Val) | + | + | + | – | – | + | bitemporal narrowing, long eyelashes, deep philtrum, everted upper lip, micrognathia, full cheeks, high-arched palate, blue sclerae | NA | – |
| S3 (ATP6V1B2, p.Tyr328His) | + | + | + | – | – | – | facial hypotonia, straight eyebrow, eyes appear wide set, ptosis, long eyelashes, tubular nose with broad nasal base, decreased facial movement/wrinkles, short and deep philtrum, everted thin upper lip with prominent upper vermilion, mouth open at rest | – | – |
| S4 (ATP6V1B2, p.Tyr328Cys) | + | + | + | – | – | – | high forehead, dolichocephaly, bitemporal narrowing, straight eyebrows, long eyelashes, short philtrum, slight micrognathia, posteriorly rotated ears | – | – |
| S5 (ATP6V1B2, p.Gln376Lys) | + | + | + | – | – | – | high forehead, dolichocephaly, bitemporal narrowing, coarse face, brushy and straight eyebrows, long eyelashes, prominent upper and lower vermilion, large mouth | – | – |
| S6 (ATP6V1B2, p.Gln376Arg) | + | + | + | – | – | – | dolichocephaly, bitemporal narrowing, coarse face with full-cheeks, thin upper lip, micrognathia/retrognathia, glossoptosis, high arched palate, and short neck | – | – |
| S7 (ATP6V1B2, p.Glu374Gln) | + | + | + | – | – | – | dolichocephaly, bitemporal narrowing, mildly coarse face, short philtrum, everted upper lip, thin upper lip, prominent upper and lower vermilions, large mouth | – | – |
NA, not available.
Functional and structural analyses
We investigated the functional consequences of the identified ATP6V1B2 and ATP6V1C1 variants, together with the p.Arg485Pro change in ATP6V1B2 [B2R485P], which had previously been associated with ZLS.13 To assess whether the identified variants impact protein stability, the protein levels of all ATP6V1B2/ATP6V1C1 mutants were evaluated in transiently transfected 293T cells, documenting a negligible effect of each of the tested variants (Figure S3). Consistently, similar levels of endogenous ATP6V1B2 were also found in primary fibroblasts obtained from affected (B2A332V, B2Q376K, and B2R485P) and control individuals (Figure S3). Assessment of the endogenous level of the ATP6V1C1 protein was not possible due to the poor sensitivity of all tested commercially available antibodies.
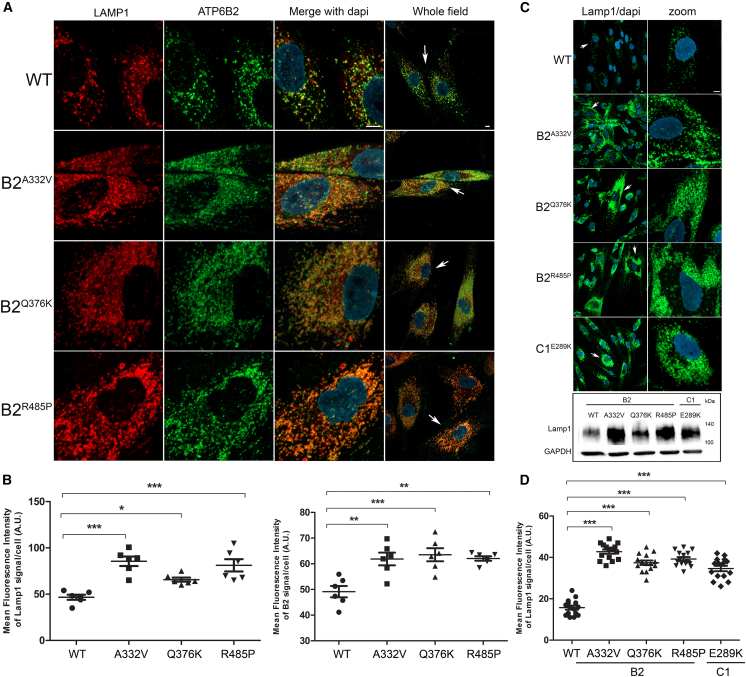
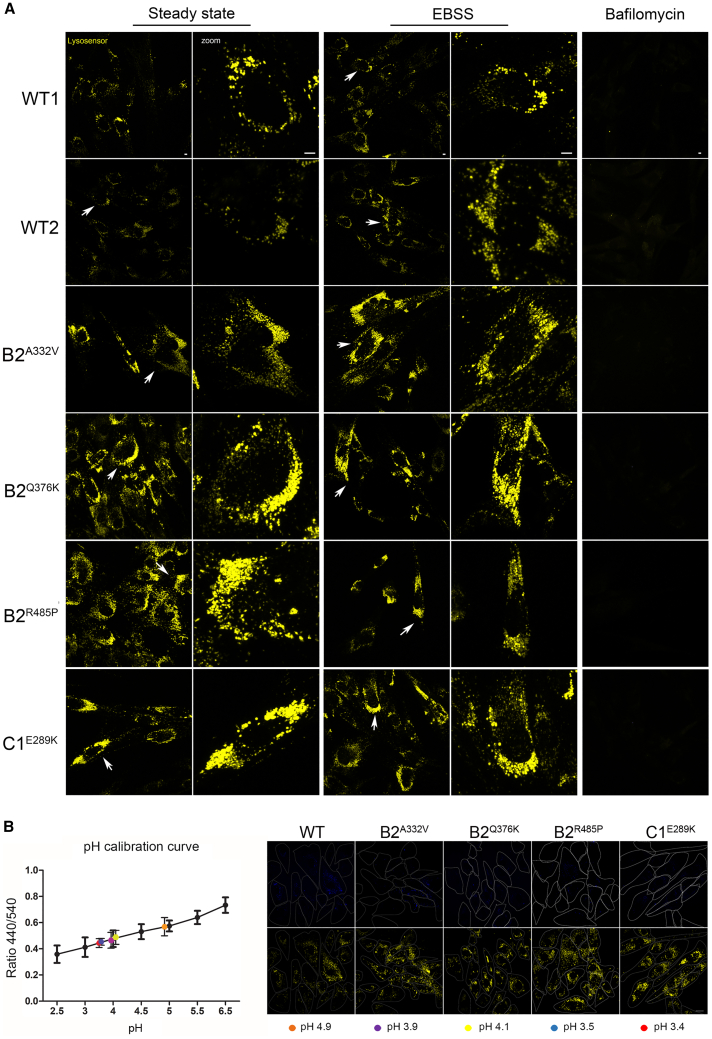
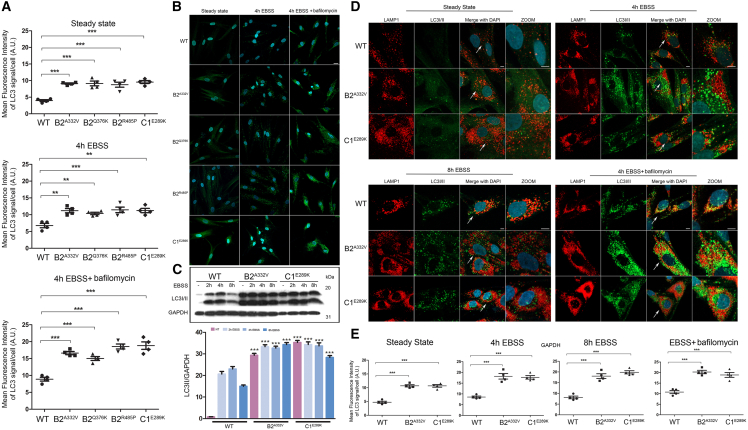
The vacuolar V-ATPase is present in the membranes of many organelles, where its proton-pumping action is required to maintain their acidic luminal pH, especially in lysosomes. Using patient and control fibroblasts, we first analyzed the subcellular localization of B2 subunits. By immunofluorescence confocal microscopy analysis, we observed proper localization of WT and all tested mutant B2 subunits (B2A332V, B2Q376K, and B2R485P) to lysosomes, as indicated by their co-localization with LAMP1 (Figures 2A, 2B, and S4). Notably, a marked increase in the size and number of lysosomes in cells endogenously expressing each of the tested B2 and C1 mutants was observed by immunofluorescence analysis and western blot analyses (Figures 2C and 2D), as observed in lysosomal storage disorders. Then, we evaluated a possible impact of altered ATPase pump function on lysosome function by assessing the accumulation of ceramide and cholesterol as representative of lysosomes substrates. Analyses performed using specific probes (i.e., BTR ceramide and Filipin) showed a significantly increased storage of both substrates in all tested patients’ cell lines compared with controls cells (Figures 3A-3D), suggesting a generalized defective hydrolytic activity of lysosomal enzymes possibly as a result of an altered vacuolar ATPase proton function. To evaluate the lysosomal acidification status, we first used LysoSensor Yellow/Blue DND-160 as a dye (Figure 4A). Cells were treated with this probe at steady state condition, after 4 h of EBSS treatment to induce autophagy, and after 4 h of treatment with EBSS together with bafilomycin, the latter used to inhibit the V-ATPase function.30 We observed a significant higher acidity of lysosomes in all patients’ cell lines compared with control cells, indicating a hyperactive behavior of the proton pumps assembling the mutant B2/C1 subunits. As expected, bafilomycin treatment promoted the complete loss of staining indicating the efficient inhibition of both mutant and WT proton-pumping function. To further validate this finding, we performed experiments to measure the intraorganellar pH in patient-derived fibroblasts by treating cells with the ratiometric LysoSensor Yellow/Blue dextran (Figure 4B). These assays revealed a substantial decrease of intra-organellar pH in patients’ cells compared with control cells, further supporting an increase of proton-pumping activity in the former.
Figure 2.
Disease-causing ATP6V1B2 and ATP6V1C1 variants affect lysosome morphology but maintain a proper lysosomal localization
(A and B) Confocal laser scanning microscopy analysis shows a proper ATP6V1B2 subcellular localization with lysosomes. Cells were stained with Lamp1 (lysosome marker, red) and ATP6V1B2 (B2 subunit marker, green) antibodies, and DAPI (DNA marker, blue). Representative images are shown (A) together with the quantitative data and statistical analysis (B). Arrows indicate the selected cells reported in the enlarged images. Scale bar, 5 μm. MFI ± SEM of B2 and Lamp1 signal per cell was quantified (≥20 cells per experimental condition, in six independent experiments per marker), and plotted in the corresponding graphs, defining final MFI as MFI (region of interest [ROI]) − MFI (background). Bars indicate mean ± SEM, p values were calculated by one-way ANOVA with Tukey’s correction for multiple testing.
(C and D) Representative images reporting the altered morphology, distribution, and number of lysosomes in cells from individuals carrying heterozygous ATP6V1B2 missense variants (p.Ala332Val [A332V], p.Gln376Lys [Q376K], and p.Arg485Pro [R485P]) compared with control cells are shown (C). Arrows indicate the selected cells reported in the enlarged images. Scale bar is 5 μm. In the same panel, western blot analysis shows the higher expression level of lysosomes marker in patients’ fibroblasts than control cells. GAPDH was used to normalize the experiments. Quantitative data and statistical analysis are also shown (D). MFI and SEM were calculated as above.
Figure 3.
Effects of autophagic flux dysregulation promoted by ATP6V1B2 and ATP6V1C1 mutants
Confocal laser scanning microscopy analyses show a significant storage of cholesterol (A and B) and ceramide (C and D) on patients’ fibroblasts compared with control cells. Cells were stained with filipin (cholesterol) (A) and BTR-ceramide (C) probes, respectively. Scale bars are 10 μm (A), 20 μm (C, left), and 2 μm (C, right). Arrows indicate the selected cells reported in the enlarged images. MFI ± SEM of filipin (B) and ceramide (D) signal per cell, was quantified (3 independent experiments, ≥20 cells per condition, in each repeat), and plotted in the corresponding graphs, defining Final MFI as MFI (region of interest) – MFI (background). WT1 and WT2 were pooled together vs. patient’s samples. Bars indicate mean ± SEM, p values were calculated by One way ANOVA with Tukey’s correction for multiple testing.
Figure 4.
Expression of ATP6V1B2 and ATP6V1C1 mutants is associated with increased acidification of lysosomes
(A) Patients’ and control cells were incubated with 2 μM LysoSensor Yellow/Blue DND-160 (Thermo Fisher Scientific) and observed on a Zeiss LSM 980 confocal microscope. Images were acquired using a 63× oil objective, laser and filter settings were adjusted according to the fluorescence excitation and emission requirements of the reagent. Scale bar, 5 μm.
(B) Evaluation of lysosomal pH values in patient-derived fibroblasts. The pH calibration curve performed using WT1 cells, and obtained by plotting the fluorescence intensity 450/540 ratios as a function of pH, was fitted with linear regression using GraphPad Prism5 software (left). Data are means ± SEM from >50 cells analyzed for each pH value. Colored filled circled represent the experimentally measured ratios converted into absolute pH values by interpolation in the pH calibration curve. Representative images of fibroblasts incubated with LysoSensor Yellow/Blue dextran and visualized in live at 450 nm (top) and 540 nm (bottom) emission wavelengths (right). The cell contours recognition was performed in bright field images and used to effectively quantify fluorescence intensity. Scale bar, 20 μm. All panels are identical in scale.
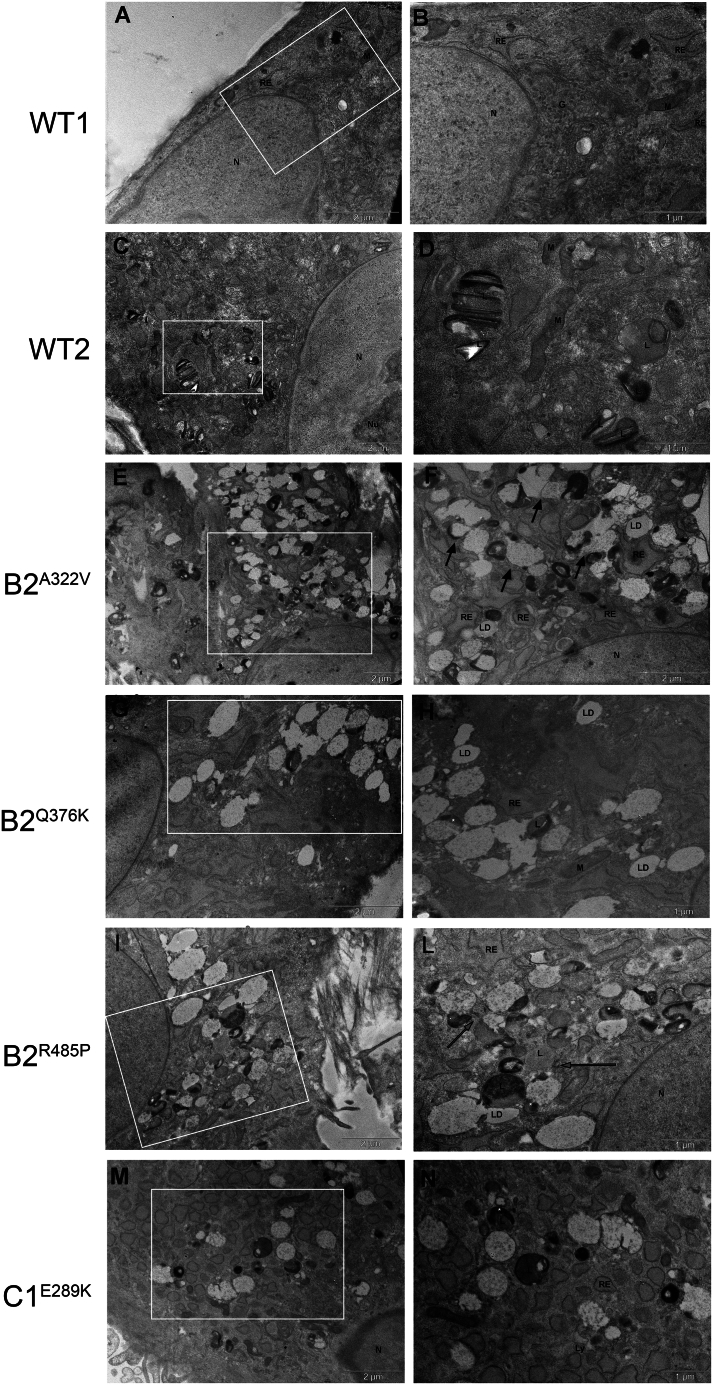
Ultrastructural analyses were performed to better characterize the effects of aberrant V-ATPase functioning. Control fibroblasts showed identifiable cytoplasmic components, including well-organized mitochondria, slightly dilated endoplasmic reticulum, and lysosomes that were filled with typical multilamellar and electron-dense material (Figures 5A–5D). Remarkably, compared with control cells, patients’ fibroblasts showed a high number of enlarged vesicles containing lighter granular material, which resembled early lysosomes, and vacuoles containing osmiophilic material with different electron densities, reminiscent of lysosomes. In particular, in fibroblasts expressing B2A322V and B2R485P many of these vesicles seemed to be partially fused (Figures 5E and 5F) or in close proximity to each other (Figures 5I–5L). Cells endogenously expressing B2Q376K showed accumulation of large heterogeneous vacuoles filled with osmiophilic material and substances with different electron densities (Figures 5G and 5H), while cells expressing the C1E289K variant showed only vesicles uniform in size (Figures 5M and 5N). Dilated endoplasmic reticulum and lipid droplets were also visible in the cytoplasm of the cells. Overall, these findings were suggestive of aberrant lysosome maturation and functioning, as commonly observed in lysosomal storage diseases,31,32 in line with the data collected by immunofluorescence analysis.
Figure 5.
Electron microscopy analysis
Electron micrographs showing the ultrastructure of control (A–D) and patient-derived fibroblasts (E–N).
(E and F) Fibroblasts from subjects B2A322V and (I–L) B2R485P showed vesicles partially fused to each other (black arrows) and in close proximity (gray arrows).
(G–H) B2Q376K patients’ cells showed accumulation of large heterogeneous vacuoles whereas the cells carrying the C1E289K mutant presented vesicles uniform in size.
(M and N) (white asterisks) Heterogeneous substances and osmiophilic material. G, Golgi apparatus; N, nucleus; n, nucleolus; L, lysosome; LD, lipid droplets; M, mitochondria; RE, rough endoplasmic reticulum.
Since proper acidification of lysosomes is required for autophagosome-lysosome fusion, we hypothesized that the altered lysosomal function might also result in an impairment of the autophagic flux. Immunofluorescence and western blot analysis using an anti LC3I/II antibody as probe showed a significant accumulation of autophagosomes at steady state condition as well as after autophagy flux induction with EBSS in all tested patients’ fibroblasts compared with control cells (Figures 6A-6C, S5, and S6). Remarkably, these organelles, which appeared as enlarged vesicles mainly accumulated in the perinuclear region, did not colocalize with lysosomes after autophagic flux induction and bafilomycin treatment (Figures 6B and S6). To further assay the extent of dysregulation of the autophagic flux and/or lysosomal function, we evaluated the fluorescence emission of a BSA derivative (DQ-BSA) conjugated to a self-quenched fluorophore in patients’ fibroblasts and control cells after starvation with EBSS for 16 h to induce autophagy. We did not observe dequenched DQ-BSA in patients’ cells over the time period, further supporting a defective autophagosome-lysosomal function (Figure S7).
Figure 6.
An impaired autophagic flux in patients’ fibroblasts promotes LC3I/II proteins accumulation and affects autophagosomes number and subcellular localization
(A–C) Confocal laser scanning microscopy (CSLM) analysis performed on patients’ fibroblasts show a significant high level of LC3I/II proteins already in the steady state condition, which remains constant after treatment with EBSS for 4 h without bafylomicin, to indicate an autophagic flux impairment. Graphs reporting MFI ± SEM of LC3I/II (A) and representative images (B) are shown. Fixed cells were stained with rabbit monoclonal anti-LC3I/II antibody followed by rabbit anti-goat Alexa Fluor 488 (green). Nuclei are visualized by DAPI staining (blue). Scale bar, 20 μm. Western blot analyses performed on patients’ fibroblasts (C) confirm the data obtained by immunofluorescence, showing an anomalous accumulation of LC3I/II in the steady state condition and after autophagic flux induction with EBSS for 2, 4, and 8 h. Equal amounts of cell lysates were resolved by 15% polyacrylamide gel electrophoresis. Membranes were probed with an anti-LC3I/II antibody and then re-probed with an anti-GAPDH antibody for data normalization. Non-treated cells (i.e., cells cultured in steady state conditions) were reported as internal controls (−). Bars indicate mean ± SEM, p values were calculated by one-way ANOVA with Tukey’s correction for multiple testing. MFI ± SEM signals per cell were quantified (4 independent experiments per marker, ≥20 cells per experimental condition, in each repeat), and plotted in the corresponding graphs, defining Final MFI as MFI (region of interest [ROI]) − MFI (background). Bars indicate mean ± SEM, p values were calculated by one-way ANOVA with Tukey’s correction for multiple testing.
(D and E) CSLM analysis shows an unusual subcellular localization of autophagosomes in patients’ cells compared with control cells (D). In particular, these organelles localized near the nucleus and a few number colocalized with lysosomes residing in the peripheral region of the cell in the steady state condition as well as after treatment with EBSS without bafilomycin. Fixed cells were stained with rabbit monoclonal anti-LC3I/II antibody followed by anti-rabbit Alexa Fluor 488 (green). The lysosomes were stained with an anti-mouse Lamp1 followed by anti-mouse Alexa Fluyor-546 (red). Nuclei are visualized by DAPI staining (blue). Scale bar, 5 μm. Quantitative data assessing the LC3I/II levels and statistical analysis are also shown (D). MFI and SEM were calculated as above.
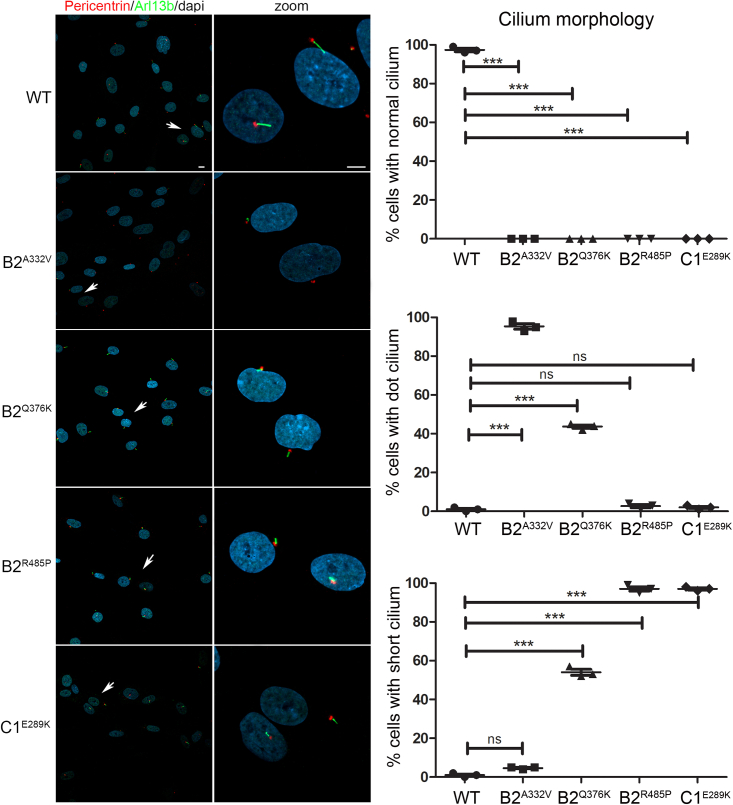
Increasing evidence testifies the interplay between cilia and autophagy.33,34,35 On one hand, the autophagy core machinery is known to localize at ciliary compartments; on the other, the primary cilium, representing a sensorial antenna of cells, promotes the activation of the autophagic flux in response to diverse stimuli. Autophagy can also control the level of positive or negative regulators of cilium biogenesis, influencing cilia formation and elongation. Based on these considerations, we investigated the effect of altered B2 and C1 subunit function on primary cilium biogenesis and morphology in primary fibroblasts from affected subjects. The analysis performed in starved fibroblasts revealed the presence of aberrant primary cilium formation in all cell lines (Figure 7). Specifically, the cilium was either shorter compared with control cells or absent (dot cilium), documenting a pleiotropic effect of each of the tested variants also involving cilium biogenesis.
Figure 7.
ATP6V1B2 and ATP6V1C1 variants are associated with an aberrant primary cilium morphology
Representative confocal images showing altered primary cilium morphology in patients’ fibroblasts compared with control cells. Specifically, cilia with altered morphology (either short or characterized by a basal body in absence of any visible cilium [dot cilium], zoomed images) were invariably observed in fibroblasts heterozygous for the all variants studied. Primary cilia are labeled with ARL13B (green), basal bodies and nuclei are labeled with pericentrin (red) and DAPI (blue), respectively. Scale bars, 10 μm (left) and 2 μm (right). Cells were analyzed for each line over three independent experiments (50 cells/line each) for a total of 150 cells/line scored. p values were calculated by one-way ANOVA with Tukey’s correction for multiple testing. Graph bars show mean ± SEM.
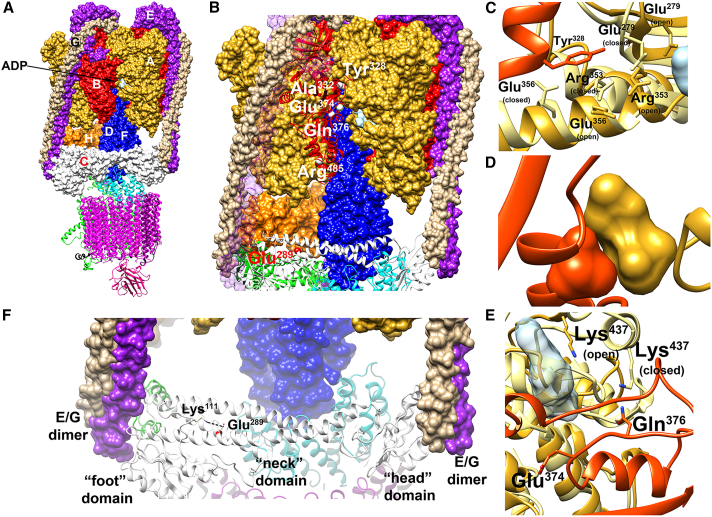
Recently, the cryoelectron microscopy structures of the human V-ATPase in three rotational states representing the sequential conformational changes after ATP hydrolysis and release were solved.12 We used these structures to explore the possible structural and dynamics effects of the identified amino acid substitutions. In general, ATP hydrolysis in the V1 catalytic subcomplex drives a general rearrangement of the pump, leading to proton translocation (Figure 8A). The ATP binding site is located at the A/B interface, in the hexamer formed by subunits A and B, though only the A subunits are directly involved in ATP hydrolysis. The C subunit interacts with two E/G heterodimers and is part of the “stator” ensuring the reciprocal stationary configuration of the VO and V1 subcomplexes (Figure 8A).36 The mutated Tyr328, Ala332, and Gln376 residues of the B2 subunit are located at the interface with the A subunit, where the ATP/ADP binds to the complex (Figure 8B). In particular, Tyr328 is close to multiple residues of the A subunit that undergo significant conformational transformations in the transition between the open and closed states (Figure 8C), suggesting that the p.Tyr328Cys and p.Tyr328His substitutions could affect the ADP/ATP exchanges and, in turn, the transition between the open and closed states. Ala332 interacts very tightly with Ser316 and Asn317 in the A subunit. The p.Ala332Val substitution would introduce steric clashes (Figure 8D). The proximity to the ATP/ADP binding region suggests a possible effect of the substitution on the ATP/ADP binding and exchange; however, in this case, a non-specific perturbation of the A/B interface cannot be ruled out. Gln376 is quite close to ADP (minimum distance below 8 Å from the phosphate groups). Similar to Tyr328, the residues in the A subunit surrounding Gln376, including Lys437, display significant variations when examining the different states of the complex (Figure 8E). The p.Gln376Arg substitution, introducing a positive net charge, very likely perturbs the correct dynamics of the A/B interface, and the interactions with the ATP/ADP phosphate. It is worth mentioning that variants introducing a change in the net charge of the side chain of Glu374, which is only two residues apart from Gln376, have already been associated with ZLS (p.Glu374Gln),14 and an epileptic neurodevelopmental phenotype (p.Glu374Gly).37 The possible effects of the p.Arg485Pro substitution have been discussed previously,13 based on a homology model. Overall, the recently reported structures confirm the previously reported analyses. Arg485 is far from the ADP/ATP-binding site, making a significant impact of its substitution with proline on the ATP binding unlikely. In contrast, the substitution is predicted to destabilize the helical tract encompassing residue 485 and weakens a possible salt bridge with Asp507, likely perturbing the interactions within the V1 subcomplex. Finally, Glu289 is located within the so-called neck domain of boot-shaped subunit C. It does not interact with residues of other subunits but is involved in an ion pair interaction with Lys111 (Figure 8F). This electrostatic interaction, stabilizing the peculiar topology of the C subunit, is strongly perturbed by the charge inversion caused by the p.Glu289Lys substitution, likely destabilizing the correct interaction with the E/G heterodimers, and in turn perturbing the correct transduction of the signal after ATP release. Overall, coherently with the observed effects on the pumping activity, all the identified substitutions were expected to perturb the transduction of the signal caused by ATP hydrolysis. However, the predicted mechanism of perturbation seems to be different for the various substitutions. Specifically, those involving the B subunit are predicted to affect the dynamics of the A3B3 hexamer, whose conformational change leads to hydrogen pumping and/or exert a direct effect on the ATP/ADP binding site, while the p.Glu289Lys substitution in the C subunit is predicted to perturb the correct orientation of the E/G heterodimers.
Figure 8.
Location of the mutated residues of the B2 and C1 subunits in the cryo-EM structures of human V-ATPase
(A) Side view of the of human V-ATPase complex (PDB: 6wm2). The subunits belonging to the V1 and VO regions are reported as surface and ribbon, respectively. The B and C subunits in the V1 region are colored red and white, respectively. ADP is reported as a light blue surface in the V1 region at the interface between the A and B subunits.
(B) The V-ATPase complex in the region containing the residues discussed in the present study. The C subunit and one of the three B subunits are reported as ribbons. The Cα atoms of the muted residues are reported as white and red spheres for the B and C subunits, respectively. For the sake of clarity, one of the E subunits is not reported.
(C) p.Tyr328Cys and p.Tyr328His substitutions (B2 subunit). The helix tract in the B subunit comprising Tyr328 is reported as a red ribbon (chain ζ), Tyr328 is represented as red sticks. Two different conformations of the A subunit are reported as ribbons, corresponding to the closed (chain β, light yellow) and open (chain γ, gold) states, respectively. The residues in the A subunit in contact with Tyr328 (minimum distance less than 0.5 nm) and populating different conformations in the two states are indicated and reported as sticks.
(D) p.Ala332Val substitution (B2 subunit). Sketch of the interface comprising Ala332 in one of the three interfaces (chains γ/ζ in the PDB file); in the sketch, Ala332 (in the B subunit) and Ser316 and Asn317 (in the A subunit) are reported as red and gold surfaces, respectively.
(E) p.Glu374Gln, p.Gln376Arg and p.Gln376Lys substitutions (B2 subunit). Interfaces between the A and B subunits in the region surrounding Gln376 and Glu374 in the B subunits from the 6wm2 structure. The backbone of the strand comprising Glu374 and Gln376 is reported as red ribbon; Glu374 and Gln376 are represented as sticks. Two different conformations of the A subunit are reported as ribbons, corresponding to the closed (chain β, light yellow) and open (chain γ, gold) states, respectively. Lys437 in chain A is reported as sticks and colored by atoms, with the carbons colored as the corresponding ribbon. The ADP molecule in the γ chain is reported as a semi-transparent light blue surface.
(F) p.Glu289Lys substitution (C1 subunit). The C subunit is reported as a white ribbon. Glu289 is reported as sticks, and the backbone is colored red. Lys111 is also reported as white sticks.
Discussion
Here we provide evidence that ATP6V1C1 variants can underlie a neurodevelopmental phenotype with features resembling DOORS syndrome, and report that dominantly acting ATP6V1B2 variants are causally associated with a phenotypic continuum having DDOD syndrome and ZLS as extreme phenotypes. We show that variants involving these two subunits of the V-ATPase pump promote enhanced function of the complex, which affects lysosomal morphology and function, autophagy and cilium biogenesis.
By mediating proton transport in intracellular compartments, V-ATPases play a central role in regulating electrochemical gradients between membranes, which is crucial in a variety of cellular processes. In addition, V-ATPases might also localize at the plasma membrane of a number of cells, mediating their specialized functions, including renal acidification, bone resorption, homeostasis of cytoplasmic pH and sperm maturation.4 V-ATPase functional pleiotropy, the tissue/cell-specific expression pattern of the different subunits, as well as the gain-of-function (GoF) vs. LoF effect of individual disease-causing variants explain the striking phenotypic heterogeneity associated with dysregulated V-ATPase function. Exemplarily, biallelic LoF variants in ATP6V0A4 cause a form of renal tubular acidosis,38 which is characterized by a derangement of acid-base balance due to an inability to secrete acid in the distal nephron. The physiological impact of these LoF variants is related to the specific expression of the encoded accessory subunits at the level of the proton-secreting cells in the distal nephron.38 A strikingly different clinical phenotype (neurodevelopmental disorder with epilepsy and brain atrophy [MIM: 619971]) is associated with biallelic LoF variants in ATP6V0A1,39 which encodes a different “a” subunit of the VO subcomplex that is highly expressed in the central nervous system. Remarkably, dominantly acting variants in the same gene causing LoF of the pump have been reported to underlie a severe form of developmental and epileptic encephalopathy (MIM: 619970).39 In the present series, the multisystemic involvement of the phenotypic spectrum resulting from the dysregulated function of the B2 and C1 subunits is in line with the wide expression pattern of both subunits. Of note, two distinct classes of dominant ATP6V1A mutations were recently identified to cause a developmental encephalopathy with distinct pathomechanisms implicating both GoF and LoF converging to an altered lysosomal homeostasis.40 Our findings provide a new example of clinical phenotypes resulting from GoF mutations enhancing V-ATPase function.
Pathogenic dominantly acting variants in ATP6V1B2 have been associated with different phenotypes clinically fitting DOORS and DDOD syndromes, and ZLS, indicating that these conditions represent a variable expression of a single disorder caused by altered V-ATPase function (Table 1). Specifically, individuals with DDOD syndrome primarily share sensorineural deafness and onychodystrophy with digital anomalies characterized by bulbous fingertips of digits, and finger-like thumbs/triphalangeal thumb. Some affected individuals may also show tooth abnormalities (e.g., oligodontia, conical teeth, and late dentition). DOORS syndrome shares sensorineural hearing loss, onychodystrophy, and digital abnormalities (i.e., triphalangeal thumb and hypoplastic/absent distal phalanges) with DDOD syndrome. Osteodystrophy is reported in most cases as well as a neurodevelopmental/neurological involvement, which includes seizures. Facial dysmorphisms are also reported (e.g., bitemporal narrowing, coarse facies, ear abnormalities, broad nasal bridge, large nose, bulbous nasal tip, anteverted nares, long philtrum, thick everted lower lip, downturned corners of the mouth, and high-arched palate). Facial features of DOORS syndrome have a significant overlap with ZLS, a condition also associated with organomegaly, gingival hypertrophy, and hypertrichosis, which are not usually reported in DDOD and DOORS syndromes. ZLS is more frequently caused by GoF variants in the potassium transporters KCNH1 and KCNN3,13,41 which are not related to lysosomal acidification; the different pathways involved might explain phenotypic differences. The present findings further support these observations, documenting the presence of a phenotypic continuum among these disorders. Notwithstanding the relatively small number of affected individuals reported thus far, first genotype-phenotype correlations are emerging. Specifically, sensorineural hearing loss and onychodystrophy appear to invariably occur in individuals carrying the recurrent truncating variant (p.Arg506∗) in ATP6V1B2, with or without neurodevelopmental involvement. Moreover, the mutation cluster affecting residues located close to the ADP/ATP binding site (Gly325, Tyr328, Ala332, Glu374, Gln376, and Leu398) seems to be associated with a phenotype principally characterized by DD affecting both cognitive and motor functions, ID, seizures and distinctive facial features (high forehead, bitemporal narrowing, coarse face, bushy and straight highbrows, deep and short philtrum, prominent lower vermillion, and large mouth), with onychodystrophy, gingival overgrowth, and hypertrichosis affecting just a minority of affected individuals. A subset of individuals shows brain abnormalities (e.g., brain atrophy and several degree of myelination delay). Consistent with these findings, the clinical features observed in the subject with the p.Glu289Lys substitution in ATP6V1C1 show a substantial overlap with DOORS syndrome, further supporting the occurrence of a phenotypic continuum characterizing DOORS syndrome, DDOD syndrome, and ZLS, which should be considered as a single clinically variable phenotypic spectrum/nosologic entity.
In conclusion, we show a continuum in the clinical spectrum associated with dominant variants affecting the ATP6V1C1 and ATP6V1B2 subunits causing upregulation of the V-ATPase function. Notwithstanding the multiple processes that are altered in cells expressing these variants, increased lysosomal acidification seems to be the driver event of such pleiotropy, indicating that these disorders can be considered as lysosomal diseases. These findings provide a rationale for the use of molecules targeting the upregulated V-ATPase function to ameliorate evolutive features in these subjects.
Data and code availability
WES datasets have not been deposited in a public repository due to privacy and ethical restrictions but will be made available on request. The ATP6V1C1 c.865G>A (p.Glu289Lys; NM_001695.5) has been submitted to ClinVar (SCV004814160).
Acknowledgments
The authors thank the participating families, Kerstin Kutsche (University Medical Center Hamburg-Eppendorf, Hamburg, Germany) for providing primary fibroblasts carrying the heterozygous ATP6V1B2 Arg485Pro amino acid substitution, and Serenella Venanzi (Istituto Superiore di Sanità, Rome) for technical assistance. This work was supported by the Italian Ministry of Health (RCR-2022-23682289 and PNRR-MR1-2022-12376811 to M.T.), Italian Ministry of University and Research (PNRR_M4C2I1.3-HEAL ITALIA PE 00000019 to L.S., FOE_2020 to M.T.), and Fondazione Bambino Gesù (Vite Coraggiose), and the Telethon Undiagnosed Diseases Program (GSP15001). J.S.C., A.H., and C.S. receive support from the NIH National Institute of Child Health and Human Development (P50 HD103538). J.S.C. additionally receives support from the NIH National Institute of Neurologic Disorders and Stroke (1U24NS131172-01) and U.S. Department of Health and Human Services, Health Resources and Services Administration (MCH T7317245). This work has been carried out in the frame of the ERN-ITHACA research activities.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100349.
Contributor Information
Marco Tartaglia, Email: marco.tartaglia@opbg.net.
Elisabetta Flex, Email: elisabetta.flex@iss.it.
Web Resources
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/;
gnomAD, http://gnomad.broadinstitute.org;
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/;
MetaDome, https://stuart.radboudumc.nl/metadome/;
GeneMatcher, https://genematcher.org/
CADD, https://cadd.gs.washington.edu/
M-CAP, http://bejerano.stanford.edu/mcap/
InterVar, http://wintervar.wglab.org.
Supplemental information
References
- 1.Sun-Wada G.H., Wada Y. Role of vacuolar-type proton ATPase in signal transduction. Biochim. Biophys. Acta. 2015;1847:1166–1172. doi: 10.1016/j.bbabio.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Cotter K., Capecci J., Sennoune S., Huss M., Maier M., Martinez-Zaguilan R., Forgac M. Activity of plasma membrane V-ATPases is critical for the invasion of MDA-MB231 breast cancer cells. J. Biol. Chem. 2015;290:3680–3692. doi: 10.1074/jbc.M114.611210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Damme T., Gardeitchik T., Mohamed M., Guerrero-Castillo S., Freisinger P., Guillemyn B., Kariminejad A., Dalloyaux D., van Kraaij S., Lefeber D.J., et al. Mutations in ATP6V1E1 or ATP6V1A Cause Autosomal-Recessive Cutis Laxa. Am. J. Hum. Genet. 2020;107:374. doi: 10.1016/j.ajhg.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 5.Bruce L.J., Cope D.L., Jones G.K., Schofield A.E., Burley M., Povey S., Unwin R.J., Wrong O., Tanner M.J. Familial distal renal tubular acidosis is associated with mutations in the red cell anion exchanger (band 3, AE1) gene. J. Clin. Invest. 1997;100:1693–1707. doi: 10.1172/JCI119694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frattini A., Orchard P.J., Sobacchi C., Giliani S., Abinun M., Mattsson J.P., Keeling D.J., Andersson A.-K., Wallbrandt P., Zecca L., et al. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat. Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 7.Fassio A., Esposito A., Kato M., Saitsu H., Mei D., Marini C., Conti V., Nakashima M., Okamoto N., Olmez Turker A., et al. De novo mutations of the ATP6V1A gene cause developmental encephalopathy with epilepsy. Brain. 2018;141:1703–1718. doi: 10.1093/brain/awy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattison K.A., Tossing G., Mulroe F., Simmons C., Butler K.M., Schreiber A., Alsadah A., Neilson D.E., Naess K., Wedell A., et al. ATP6V0C variants impair V-ATPase function causing a neurodevelopmental disorder often associated with epilepsy. Brain. 2023;146:1357–1372. doi: 10.1093/brain/awac330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poorkaj P., Raskind W.H., Leverenz J.B., Matsushita M., Zabetian C.P., Samii A., Kim S., Gazi N., Nutt J.G., Wolff J., et al. A novel X-linked four-repeat tauopathy with parkinsonism and spasticity. Mov. Disord. 2010;25:1409–1417. doi: 10.1002/mds.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rujano M.A., Cannata Serio M., Panasyuk G., Péanne R., Reunert J., Rymen D., Hauser V., Park J.H., Freisinger P., Souche E., et al. Mutations in the X-linked ATP6AP2 cause a glycosylation disorder with autophagic defects. J. Exp. Med. 2017;214:3707–3729. doi: 10.1084/jem.20170453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedera P., Alvarado D., Beydoun A., Fink J.K. Novel mental retardation-epilepsy syndrome linked to Xp21.1-p11.4. Ann. Neurol. 2002;51:45–50. doi: 10.1002/ana.10051. [DOI] [PubMed] [Google Scholar]
- 12.Wang L., Wu D., Robinson C.V., Wu H., Fu T.M. Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly. Mol. Cell. 2020;80:501–511.e3. doi: 10.1016/j.molcel.2020.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kortüm F., Caputo V., Bauer C.K., Stella L., Ciolfi A., Alawi M., Bocchinfuso G., Flex E., Paolacci S., Dentici M.L., et al. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat. Genet. 2015;47:661–667. doi: 10.1038/ng.3282. [DOI] [PubMed] [Google Scholar]
- 14.Popp B., Ekici A.B., Thiel C.T., Hoyer J., Wiesener A., Kraus C., Reis A., Zweier C. Exome Pool-Seq in neurodevelopmental disorders. Eur. J. Hum. Genet. 2017;25:1364–1376. doi: 10.1038/s41431-017-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw M., Winczewska-Wiktor A., Badura-Stronka M., Koirala S., Gardner A., Kuszel Ł., Kowal P., Steinborn B., Starczewska M., Garry S., et al. EXOME REPORT: Novel mutation in ATP6V1B2 segregating with autosomal dominant epilepsy, intellectual disability and mild gingival and nail abnormalities. Eur. J. Med. Genet. 2020;63 doi: 10.1016/j.ejmg.2019.103799. [DOI] [PubMed] [Google Scholar]
- 16.Beauregard-Lacroix E., Pacheco-Cuellar G., Ajeawung N.F., Tardif J., Dieterich K., Dabir T., Vind-Kezunovic D., White S.M., Zadori D., Castiglioni C., et al. DOORS syndrome and a recurrent truncating ATP6V1B2 variant. Genet. Med. 2021;23:149–154. doi: 10.1038/s41436-020-00950-9. [DOI] [PubMed] [Google Scholar]
- 17.Yuan Y., Zhang J., Chang Q., Zeng J., Xin F., Wang J., Zhu Q., Wu J., Lu J., Guo W., et al. De novo mutation in ATP6V1B2 impairs lysosome acidification and causes dominant deafness-onychodystrophy syndrome. Cell Res. 2014;24:1370–1373. doi: 10.1038/cr.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menendez I., Carranza C., Herrera M., Marroquin N., Foster J., Cengiz F.B., Bademci G., Tekin M. Dominant deafness-onychodystrophy syndrome caused by an ATP6V1B2 mutation. Clin. Case Rep. 2017;5:376–379. doi: 10.1002/ccr3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radio F.C., Pang K., Ciolfi A., Levy M.A., Hernández-García A., Pedace L., Pantaleoni F., Liu Z., de Boer E., Jackson A., et al. SPEN haploinsufficiency causes a neurodevelopmental disorder overlapping proximal 1p36 deletion syndrome with an episignature of X chromosomes in females. Am. J. Hum. Genet. 2021;108:502–516. doi: 10.1016/j.ajhg.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motta M., Fasano G., Gredy S., Brinkmann J., Bonnard A.A., Simsek-Kiper P.O., Gulec E.Y., Essaddam L., Utine G.E., Guarnetti Prandi I., et al. SPRED2 loss-of-function causes a recessive Noonan syndrome-like phenotype. Am. J. Hum. Genet. 2021;108:2112–2129. doi: 10.1016/j.ajhg.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Li C., Mou C., Dong Y., Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12:103. doi: 10.1186/s13073-020-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G. M-CAP eliminates a majority of variants with uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 27.Li Q., Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marucci G., Zullino I., Bertuccini L., Camerini S., Cecchetti S., Pietrantoni A., Casella M., Vatta P., Greenwood A.D., Fiorillo A., Lalle M. Re-Discovery of Giardiavirus: Genomic and Functional Analysis of Viruses from Giardia duodenalis Isolates. Biomedicines. 2021;9:654. doi: 10.3390/biomedicines9060654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., Abeliovich H., Abildgaard M.H., Abudu Y.P., Acevedo-Arozena A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yam G.H.F., Bosshard N., Zuber C., Steinmann B., Roth J. Pharmacological chaperone corrects lysosomal storage in Fabry disease caused by trafficking-incompetent variants. Am. J. Physiol. Cell Physiol. 2006;290:C1076–C1082. doi: 10.1152/ajpcell.00426.2005. [DOI] [PubMed] [Google Scholar]
- 32.Neikirk K., Vue Z., Katti P., Rodriguez B.I., Omer S., Shao J., Christensen T., Garza Lopez E., Marshall A., Palavicino-Maggio C.B., et al. Systematic Transmission Electron Microscopy-Based Identification and 3D Reconstruction of Cellular Degradation Machinery. Adv. Biol. 2023;7 doi: 10.1002/adbi.202200221. [DOI] [PubMed] [Google Scholar]
- 33.Orhon I., Dupont N., Pampliega O., Cuervo A.M., Codogno P. Autophagy and regulation of cilia function and assembly. Cell Death Differ. 2015;22:389–397. doi: 10.1038/cdd.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pampliega O., Orhon I., Patel B., Sridhar S., Díaz-Carretero A., Beau I., Codogno P., Satir B.H., Satir P., Cuervo A.M. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Z., Lin M.G., Stowe T.R., Chen S., Zhu M., Stearns T., Franco B., Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502:254–257. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasanthakumar T., Rubinstein J.L. Structure and Roles of V-type ATPases. Trends Biochem. Sci. 2020;45:295–307. doi: 10.1016/j.tibs.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Veltra D., Kosma K., Papavasiliou A., Tilemis F.N., Traeger-Synodinos J., Sofocleous C. A novel pathogenic ATP6V1B2 variant: Widening the genotypic spectrum of the epileptic neurodevelopmental phenotype. Am. J. Med. Genet. 2022;188:3563–3566. doi: 10.1002/ajmg.a.62971. [DOI] [PubMed] [Google Scholar]
- 38.Smith A.N., Skaug J., Choate K.A., Nayir A., Bakkaloglu A., Ozen S., Hulton S.A., Sanjad S.A., Al-Sabban E.A., Lifton R.P., et al. Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat. Genet. 2000;26:71–75. doi: 10.1038/79208. [DOI] [PubMed] [Google Scholar]
- 39.Aoto K., Kato M., Akita T., Nakashima M., Mutoh H., Akasaka N., Tohyama J., Nomura Y., Hoshino K., Ago Y., et al. ATP6V0A1 encoding the a1-subunit of the V0 domain of vacuolar H+-ATPases is essential for brain development in humans and mice. Nat. Commun. 2021;12:2107. doi: 10.1038/s41467-021-22389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fassio A., Esposito A., Kato M., Saitsu H., Mei D., Marini C., Conti V., Nakashima M., Okamoto N., Olmez Turker A., et al. De novo mutations of the ATP6V1A gene cause developmental encephalopathy with epilepsy. Brain. 2018;141:1703–1718. doi: 10.1093/brain/awy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauer C.K., Schneeberger P.E., Kortüm F., Altmüller J., Santos-Simarro F., Baker L., Keller-Ramey J., White S.M., Campeau P.M., Gripp K.W., Kutsche K. Gain-of-Function Mutations in KCNN3 Encoding the Small-Conductance Ca2+-Activated K+ Channel SK3 Cause Zimmermann-Laband Syndrome. Am. J. Hum. Genet. 2019;104:1139–1157. doi: 10.1016/j.ajhg.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WES datasets have not been deposited in a public repository due to privacy and ethical restrictions but will be made available on request. The ATP6V1C1 c.865G>A (p.Glu289Lys; NM_001695.5) has been submitted to ClinVar (SCV004814160).