Abstract
Previously we have shown that the establishment of an oriP replicon is dependent on its epigenetic modification, which occurs in only 1 to 10% of proliferating cells (E. R. Leight and B. Sugden, Mol. Cell. Biol. 21:4149–4161, 2001). To gain insights into the cis-acting requirements for the establishment of oriP replicons, we monitored the replication of oriP plasmid derivatives for several weeks following their introduction into cells. In EBNA-1-positive 143B and H1299 cells, plasmids containing only the region of dyad symmetry (DS) of oriP replicated but were lost more rapidly from cells than were oriP plasmids, demonstrating that the family of repeats (FR) of oriP acts in cis to stimulate replication in these cells. Unexpectedly, we found that the DS plasmid was established efficiently in 293/EBNA-1 cells, being lost at a rate of only 8% per cell generation over 24 days posttransfection. However, plasmids containing the FR in addition to the DS of oriP replicated but were lost at a rate of approximately 30% per cell generation in 293/EBNA-1 cells, indicating that the FR inhibits oriP's establishment in this cell line. FR's enhancement of transcription of a promoter in cis and FR's ability to inhibit replication fork movement do not account solely for oriP's inefficient establishment. In addition, DNA looping between FR and DS neither stimulates nor inhibits replication. Deletion of 11 EBNA-1 binding sites in the FR or replacement of the FR with DS sequences, however, does overcome the inhibitory activity of the FR, thereby allowing efficient establishment of the oriP derivative in 293/EBNA-1 cells.
Epstein-Barr virus (EBV) is a gammaherpesvirus that causes infectious mononucleosis (12, 42) and is associated with several malignancies, including Hodgkin's disease, gastric carcinoma, and B-cell lymphomas in immunocompromised individuals (9, 18, 23, 26, 51). EBV also has been found to contribute causally to Burkitt's lymphoma and nasopharyngeal carcinoma in prospective epidemiological surveys (10, 17, 31). EBV's ability to induce and maintain proliferation of the B cells it infects likely underlies its contribution to these malignancies. Within these latently infected cells, the viral genome is present as a circular plasmid that is synthesized only once per cell cycle during S-phase (1, 61) and is efficiently partitioned to daughter cells (29, 36, 55, 56), akin to the genome of its host cell. We use the term replication to encompass these DNA synthesis and partitioning events. Only two viral components are required for the replication of the EBV genome, the latent origin oriP and its binding protein EBNA-1; all else is contributed by the cell (40, 59, 62).
oriP is composed of two cis-acting elements, the family of repeats (FR) and the region of dyad symmetry (DS), which are separated by approximately 1 kbp of DNA. The FR contains 20 imperfect copies of a 30-bp repeat to which EBNA-1 binds site specifically and with high affinity (5, 6, 44). The DS, which contains four low-affinity EBNA-1-binding sites, is the site at or near which DNA synthesis initiates (15). The FR, in conjunction with EBNA-1, is thought to ensure faithful partitioning (or maintenance) of oriP replicons. This supposition is based partially on the findings that (i) plasmids lacking 14 or more EBNA-1 binding sites in the FR of oriP fail to support long-term replication in D98/Raji and Raji cells (8, 47, 57) and (ii) addition of the FR to a plasmid containing mammalian or viral autonomously replicating sequences allows long-term replication in EBNA-1-positive cells (30, 34).
Plasmids containing oriP support efficient replication in EBNA-1-positive cells selected to retain them, being lost at a rate of 2 to 4% per cell generation after removal of selection (29, 56), a rate of loss which resembles that of ARS/CEN plasmids in Saccharomyces cerevisiae (33). We refer to these plasmids as “established” replicons in that they support efficient DNA synthesis and partitioning each cell cycle. Unexpectedly we have found that upon introduction of oriP plasmids into a population of EBNA-1-positive cells, oriP plasmids replicate but are lost precipitously from cells during 2 weeks posttransfection (>25% rate of loss per cell generation) (37). Upon investigation of these disparate observations, we have found that oriP replicons must be modified epigenetically for establishment, and this modification occurs in only 1 to 10% of transfected cells (37). Our observations of oriP plasmids are consistent with studies on EBV that demonstrate that EBV DNA is lost rapidly from proliferating cells following infection (45) and is established in approximately 1% of infected cells (27, 58).
We wished to gain insights into the cis-acting requirements for the establishment of an oriP replicon in different cell lines. We therefore monitored the replication levels of oriP plasmid derivatives in multiple EBNA-1-positive cell lines over several weeks following their introduction into cells. Here we report that cell lines differ in their ability to support the replication of a plasmid containing only the DS of oriP. In 143B and H1299 cells that express EBNA-1, DS plasmids support replication, albeit inefficiently, and are lost more rapidly from cells than are oriP plasmids. That is, EBNA-1-binding sites within the FR clearly stimulate replication in 143B and H1299 cells. Unexpectedly, however, plasmids containing only the DS of oriP were established efficiently in 293/EBNA-1 cells, providing efficient DNA synthetic and partitioning functions over 24 days posttransfection. Plasmids containing the FR in addition to the DS supported replication but were lost precipitously in 293/EBNA-1 cells and all other cell lines analyzed (27, 37, 45, 58), indicating that the FR inhibits establishment of oriP replicons in 293/EBNA-1 cells and likely inhibits establishment of oriP replicons in all other cell lines. We show that this inefficient establishment of oriP is not due to FR's enhancement of transcription from the oriP replicon or to DNA looping between the FR and DS.
In addition, the FR's ability to act as a replication fork barrier does not solely underlie the inefficient establishment of oriP replicons. Deletion of EBNA-1 binding sites within the FR or substitution of the FR with DS sequences, however, overcame the inhibitory activity of the FR, allowing efficient establishment of the oriP plasmid derivative in 293/EBNA-1 cells. Our findings indicate that the FR of oriP possesses inhibitory as well as stimulatory activities with respect to replication. By defining the mechanism by which an epigenetic event overcomes the inhibitory activity of the FR, therapeutic agents may be developed to prevent the establishment of EBV's plasmid replicon.
MATERIALS AND METHODS
Plasmids.
2278, the oriP test plasmid, and 2276, the prokaryotic backbone plasmid, are derived from 2275, a plasmid containing a ColE1 origin and supF marker for propagation in Escherichia coli and a neomycin phosphotransferase gene driven by the thymidine kinase promoter of herpes simplex virus. 2278 was constructed by inserting oriP between the HpaI and NsiI sites of 2275. 2276 was constructed by replacing the neomycin phosphotransferase gene with the firefly luciferase gene, generating a length polymorphism upon amplification with primers 1588 and 86, as described below. 2402.7, the test plasmid containing only the DS of oriP, was constructed by inserting the DS between the EcoRV and SalI sites of 2275. The test plasmid lacking the DS of oriP, 2331, was constructed by deleting DS from 2278 by digestion with EcoRV and religation.
The ΔTKp plasmid 2619 was constructed by addition of a BglII linker at the EcoRI site of 2278, followed by destruction of the herpes simplex virus thymidine kinase promoter by digestion with BglII and religation. 2278 was digested with BstXI and religated to remove the right-hand 11 EBNA-1 binding sites in the FR and 353 nucleotides between the FR and DS (nucleotides 7708 to 8378 of the B95-8 strain of EBV), generating plasmid 2617 (9FR+DS). Three EBNA-1-binding sites from the FR (nucleotides 7458 to 7548 of the B95-8 strain of EBV) were introduced between the Bpu10I and XbaI sites of 2278 to construct plasmid 2777 (3FR+DS). The DS was introduced between the Bpu10I and SalI sites of 2278 in order to replace the FR of oriP, generating plasmid 2776 (DS+DS).
Gerber et al. have defined a minimal cis-acting sequence within the murine ribosomal DNA (rDNA) locus (+642 to +747 of this locus) that is sufficient for inhibiting replication fork movement in vitro when HeLa protein extract is provided (16). This cis-acting sequence was introduced between the Bpu10I and XbaI sites of 2278 to construct plasmid 2785 (RFB→DS), in which RFB→ refers to the replication fork barrier (RFB) in the +642 to +747 orientation. Given that the rDNA RFB is orientation dependent, whereas the FR is not (11, 16), an additional rDNA RFB was introduced between the BamHI and HindIII sites of 2785 to generate plasmid 2789 (RFB→DS←RFB).
Plasmid 2775 (FR∗DS) was constructed by inserting the DS between the Bpu10I and EcoRV sites of 2331. The DS was inserted between the Bpu10I and EcoRV sites of 2777 to generate plasmid 2784 (3FR∗DS).
1728 contains oriP and encodes hygromycin B phosphotransferase and a derivative of EBNA-1 containing only five copies of the Gly-Gly-Ala repeat (2). 2048 is pcDNA3 (Invitrogen) in which the neomycin phosphotransferase gene was deleted by digestion with EcoRV and Bst1107I and religated. 2145 encodes enhanced green fluorescent protein (EGFP), whose expression is driven by the cytomegalovirus (CMV) promoter and which lacks the neomycin phosphotransferase cassette. 2264, the competitor DNA, contains a fragment of DNA composed of pBR322 and LEU2 flanked by primer 1588 and primer 86 binding sites, so that amplification by PCR yields a 656-bp product.
Plasmid constructions were confirmed by sequencing and/or restriction enzyme digestions. Our plasmid database is accessible at http://mcardle.oncology.wisc.edu/sugden/.
Cell lines and transfections.
The cell lines used for the replication assays include 143B, a human osteosarcoma cell line (ATCC CRL-8303); C33A/EBNA-1, a human cervical carcinoma cell line (ATCC HTB-31) into which plasmid 1553, which expresses EBNA-1 and hygromycin B phosphotransferase, was integrated (2); H1299/1728#3, a p53-null human lung carcinoma cell line (ATCC CRL-5803) that maintains the 1728 oriP/EBNA-1 expression plasmid (37); 293/EBNA-1, a human embryonic kidney cell line that stably expresses EBNA-1 and neomycin phosphotransferase (ATCC CRL 10852); and the 293/1728#5 cell clone, which was selected to stably replicate the 1728 oriP/EBNA-1 expression plasmid (37). Cell lines were grown in Dulbecco's modified Eagle's medium with high glucose and supplemented with 10% fetal bovine serum and 200 U of penicillin and 200 μg of streptomycin sulfate per ml. 143B cells were grown in medium containing calf serum instead of fetal bovine serum. 293/EBNA-1, C33A/EBNA-1, H1299/1728#3, and 293/1728#5 cell lines were also grown in the presence of G418 sulfate (200 μg/ml) and 100, 300, and 200 μg of hygromycin B/ml, respectively. Cells were grown at 37°C in a humidified 5% CO2 atmosphere. The doubling time of these cell lines is 22 to 24 h.
For the time course experiments, calcium phosphate precipitates containing equimolar amounts of test plasmid (10 μg of oriP plasmid) and prokaryotic backbone plasmid (6.5 μg), 5 μg of 2145, an expression vector for EGFP, and 10 μg of 1728, an oriP-based EBNA-1 expression plasmid, or 10 μg of 2048, an empty expression plasmid, were prepared and placed onto 15-cm dishes containing adherent cell lines (48). The 1728 and 2048 plasmids were not introduced into C33A/EBNA-1, H1299/1728#3, and 293/EBNA-1 cell lines. H1299/1728#3 cells were suspended by trypsinization, mixed with the precipitate, and plated onto a 15-cm dish. Medium was changed 5 to 8 h after addition of the precipitate.
DNAs were introduced into 143B cells by electroporation (32). At 2 days posttransfection, cells were harvested, and the percent EGFP-positive cells was enumerated as a measure of the transfection efficiency. (The transfection efficiency of oriP plasmids should be equivalent to or greater than that of the EGFP expression vector, as the FR of oriP promotes plasmid retention [34, 41].) Cells were expanded on 15-cm dishes and harvested at 4 to 6 days posttransfection, at which time a dilution of cells was replated such that the dishes were near confluence at the next time point, and the remaining cells were prepared by Hirt extraction, as described below. This assay allows us to track the fate of replicated plasmids in a population of cells in the absence of selection. We have shown previously that the growth rates of the untransfected and transfected cells are indistinguishable (37).
Quantitative competitive PCR assay.
At the indicated time points posttransfection, cells were harvested, and low-molecular-weight DNA was extracted by the method of Hirt (25) and prepared as described (37).
A modified quantitative competitive PCR assay (28) was used to measure the amount of replicated, DpnI-resistant plasmid DNA present at various times posttransfection. Five PCRs were performed per sample using decreasing amounts of competitor DNA: 9 pg (corresponding to approximately 3.12 × 106 molecules), 3 pg, 600 fg, 120 fg, and 24 fg. (The competitor DNA was linearized and quantified as described [37].) A total of 105 cell equivalents of digested, Hirt-extracted DNA were added to a tube containing competitor DNA, 1 × Taq buffer (Roche), 0.2 mM each of a mix of deoxynucleoside triphosphates, 10 pmol of each primer (0.17 μM each), and 1.5 U of Taq DNA polymerase (Roche) in a total volume of 60 μl. DNA was amplified by a touchdown protocol in a Hybaid Omn-E thermocycler for 22 cycles using the following conditions: 94°C for 60 s, 60°C for 30 s, and 72°C for 75 s for two cycles; in the remaining cycles, DNA was denatured at 94°C for only 30 s and the annealing temperature decreased by 1°C every second cycle until reaching 55°C. The primers used included 5′-GATCAAGAGACAGGATGAGGATCG-3′ (primer 1588), which lies in the herpes simplex virus thymidine kinase promoter region, and the previously described primer 5′-ACGATTCCGAAGCCCAACCTTTCA-3′ (primer 86) (28). The sizes of the amplified products generated from the primers are 656 bp for the competitor DNA, 931 bp for the oriP test plasmid, and 1,164 bp for the prokaryotic backbone plasmid.
One-third of the PCR was electrophoresed through a 1.2% agarose gel using 0.5 × TBE (Tris-borate-EDTA) buffer containing ethidium bromide (100 ng/ml). Signals were captured with a charge-coupled device camera (IS-1000 digital imaging system; Alpha Innotech Corporation) and analyzed with ImageQuant software (Molecular Dynamics). The number of molecules of replicated oriP test plasmid and prokaryotic backbone plasmid was determined by interpolation between known quantities of competitor DNA. This number was divided by the number of transfected cells analyzed to give the average number of molecules per transfected cell.
Cesium chloride density gradient analysis.
Equimolar amounts of an oriP plasmid (10 μg of oriP) and a plasmid containing only the DS of oriP (6.5 μg of DS) were introduced independently into 293/EBNA-1 cells together with 5 μg of 2145 (an expression vector for EGFP). At 2 days posttransfection, cells were harvested and the percent EGFP-positive cells was enumerated as a measure of the transfection efficiency. For each transfection, approximately 50% of the cells were EGFP positive. The cells were expanded on 15-cm dishes and labeled with 100 μM bromodeoxyuridine and 200 μM deoxycytidine for 17 h at 5 days posttransfection (61). Plasmid DNA was isolated by Hirt extraction of approximately 5 × 107 cells (as described above), linearized with XhoI, and mixed with CsCl to a density of 1.74 g/ml in a 5-ml volume (61). These samples were spun at 60,000 rpm for 19 h in a Beckman type NVT 65.2 rotor and fractionated into 200-μl aliquots, and the refractive index of every fourth fraction was determined. One fourth of each fraction was denatured, transferred to Gene Screen Plus hybridization membrane (NEN Life Sciences) via dot-blotting (Schleicher and Schuell Minifold I), and hybridized to labeled DNAs encompassing the neomycin phosphotransferase gene. Signals were captured with a phosphorimager and analyzed with ImageQuant software (Molecular Dynamics).
RESULTS
Plasmids containing only DS of oriP support replication with various efficiencies in different cell lines.
To gain insight into the cis-acting requirements for the establishment of an oriP replicon in different cell lines, we monitored the replication levels of oriP plasmid derivatives containing wild-type oriP (oriP), only the DS of oriP (DS), and only the FR of oriP (FR) in multiple EBNA-1-positive cell lines at early times posttransfection. The fate of these replicated plasmids was then monitored over 2 weeks following their introduction into cells. To do this, equimolar amounts of a test plasmid (oriP, DS, or FR) and prokaryotic backbone plasmid (which serves as an internal negative control) were introduced into 143B cells with or without an expression plasmid for EBNA-1 or into C33A/EBNA-1, H1299/1728#3, and 293/EBNA-1 cells that stably express EBNA-1. Cells were grown in the absence of selection. At different times following transfection, plasmid DNA was isolated by Hirt extraction and digested exhaustively with DpnI, and the level of replicated, DpnI-resistant DNA was determined by quantitative competitive PCR (28).
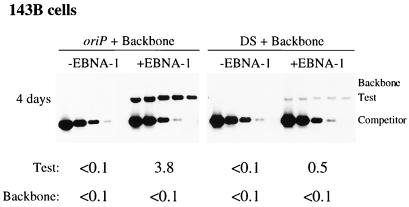
Under our conditions DpnI cleaves input methylated plasmid prepared from dam+ E. coli and hemimethylated plasmid which has undergone one round of DNA synthesis in mammalian cells (3). In a representative experiment in 143B cells, the level of replicated oriP test plasmid detected at 4 days posttransfection in the presence of EBNA-1 was greater than 38 times the level detected in the absence of EBNA-1 (Fig. 1). The plasmid containing only the DS of oriP supported EBNA-1-dependent replication, albeit at 13% of the efficiency of the oriP plasmid. This replicated DS plasmid was undetectable (<0.1 molecule per cell) by 7 days posttransfection (data not shown). These experiments demonstrate that the DS of oriP is not sufficient for efficient replication in 143B cells and indicate that the EBNA-1-binding sites within the FR stimulate replication by promoting DNA synthesis at the DS and/or by maintaining the newly synthesized plasmid. Even though the FR is present in cis, replicated oriP plasmids are lost precipitously from the population of transiently transfected 143B cells and are established in only 6 to 10% of these successfully transfected cells (37).
FIG. 1.
Plasmid containing only the DS of oriP supports EBNA-1-dependent replication, albeit inefficienty, at 4 days posttransfection in 143B cells. A test plasmid containing oriP (oriP) or only the DS of oriP (DS) was introduced into 143B cells together with an equimolar amount of prokaryotic backbone plasmid (Backbone) either with (+EBNA-1) or without (−EBNA-1) an expression plasmid for EBNA-1. At 4 days posttransfection, plasmid DNA was isolated by Hirt extraction and digested with XhoI and DpnI, and the level of replicated, DpnI-resistant DNA was determined by quantitative competitive PCR. PCRs were performed using 6 × 105 cell equivalents and a competitor DNA standard curve (9 pg, 3 pg, 600 fg, 120 fg, and 24 fg). Numbers below each gel refer to the average number of molecules present per transfected cell.
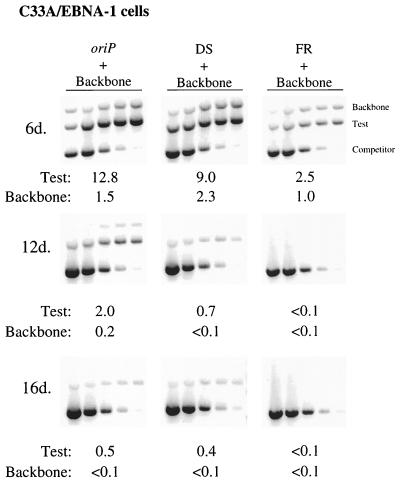
To determine if the DS plasmid supports inefficient replication in other cell lines, we monitored its replication in parallel to the oriP plasmid in three cell lines that stably express EBNA-1 (C33A/EBNA-1, H1299/1728#3, and 293/EBNA-1 cell lines). In C33A/EBNA-1 cells, as shown in a representative experiment (Fig. 2), the DS plasmid supports replication as efficiently as the oriP plasmid at 6 days posttransfection (Wilcoxon rank sum test: P [two-sided] = 0.28; n = 3), whereas a plasmid containing only the FR of oriP supports replication with a decreased efficiency compared to the oriP plasmid (Wilcoxon rank sum test: p [one-sided] = 0.04; n = 2). The replicated DS plasmid pool was lost precipitously from the cell population, so that the level of replicated DNA detected at 16 days posttransfection was 4% of the level detected at 6 days posttransfection. This rapid loss of replicated DS plasmids is analogous to the loss of oriP plasmids from the majority of transiently transfected cells (37), and is indicative of the inefficient establishment of these replicons.
FIG. 2.
Plasmid containing only the DS of oriP replicates within C33A/EBNA-1 cells but is lost precipitously during 2 weeks posttransfection, as is the oriP plasmid. Equimolar amounts of an oriP test plasmid (oriP), a test plasmid containing only the DS of oriP (DS), and a test plasmid containing only the FR of oriP (FR) were introduced independently into C33A/EBNA-1 cells together with a prokaryotic backbone plasmid (Backbone). At the indicated times posttransfection, plasmid DNA was isolated by Hirt extraction and digested with XhoI and DpnI, and the level of replicated, DpnI-resistant DNA was determined by quantitative competitive PCR. PCRs were performed using 105 cell equivalents and a competitor DNA standard curve (9 pg, 3 pg, 600 fg, 120 fg, and 24 fg). Numbers below each gel refer to the average number of molecules present per transfected cell.6d., day 6 posttransfection.
In the H1299/1728#3 cell line, the DS plasmid likewise supported replication with a similar efficiency as the oriP plasmid at 5 days posttransfection. However, the level of replicated DS plasmid decreased by a factor of greater than 80 between 5 and 15 days posttransfection, whereas the level of replicated oriP plasmid decreased by a factor of only 25 during this time (data not shown). That is, in both the C33A/EBNA-1 and H1299/1728#3 cell lines, plasmids containing only the DS of oriP replicate but are lost precipitously from transfected cells, as are oriP plasmids. This precipitous loss of the replicated DS plasmid population could result from inefficient synthesis, inefficient maintenance, or both.
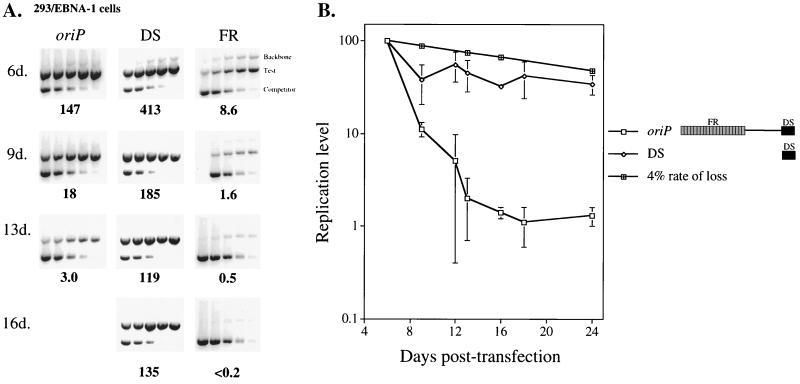
We and others have observed that plasmids containing only the DS of oriP support replication as efficiently as oriP plasmids at 2 to 4 days posttransfection in 293 cells which harbor EBNA-1 (37, 60). We therefore monitored the fate of replicated DS plasmids over 3 weeks following their introduction into 293 cells that express EBNA-1 stably (293/EBNA-1). Surprisingly, we found that while oriP plasmids were lost precipitously from the majority of transfected cells during this time (approximately a 33% rate of loss per cell generation between 6 and 18 days posttransfection; n = 7 [37]), the DS plasmid was established efficiently in transfected cells, being lost at a rate of only 8% per cell generation (Fig. 3B) (n = 7). That is, in one representative experiment (Fig. 3A), the level of replicated oriP plasmid detected at 13 days posttransfection was 2% of the level detected at 6 days posttransfection. However, the DS plasmid supported replication more efficiently than the oriP plasmid at 6 days posttransfection (Wilcoxon rank sum test: P [two-sided] = 0.009; n = 7), and this replicated DS plasmid pool decreased by a factor of only 3 between 6 and 16 days posttransfection.
FIG. 3.
(A) Plasmid containing only the DS of oriP supports efficient replication over 16 days posttransfection in 293/EBNA-1 cells, while replicated oriP plasmids are lost precipitously. Equimolar amounts of an oriP test plasmid (oriP), a test plasmid containing only the DS of oriP (DS), and a test plasmid lacking the DS of oriP (FR) were introduced independently into 293/EBNA-1 cells together with a prokaryotic backbone plasmid (Backbone). At the indicated times posttransfection, plasmid DNA was isolated by Hirt extraction and digested with XhoI and DpnI, and the level of replicated, DpnI-resistant DNA was determined by quantitative competitive PCR. PCRs were performed using 105 cell equivalents and a competitor DNA standard curve (9 pg, 3 pg, 600 fg, 120 fg, and 24 fg), except for the day 9 time point (9d.) for the FR experiment, in which 9 pg of competitor was not used. Numbers below each gel refer to the average number of molecules present per transfected cell for each test plasmid (oriP, DS, and FR). The amount of replicated oriP plasmid present at 6 days posttransfection and the amount of replicated DS plasmid present at each time point were quantified from separate gels in which 3.3 × 103 cell equivalents were assayed. The amount of replicated backbone plasmid detected at 6 days posttransfection in the oriP, DS, and FR experiments was 3.0, 3.6, and 0.8 molecules per transfected cell, respectively, and was undetectable at the latter times (<0.2 molecule/transfected cell). The 16-day time point for the oriP test plasmid is not shown because the cells were lost to contamination after the day 13 harvest. (B) DS plasmids are stable during 24 days posttransfection in 293/EBNA-1 cells, while oriP plasmids are lost precipitously. Shown is a graphic representation of experiments conducted as described above. The replication level of each plasmid was plotted versus the days posttransfection. For each independent experiment, the level of replicated test plasmid detected at 6 days posttransfection was set to 100% and the replication level at later time points was set relative to this point (100% = 439 ± 251 oriP and 754 ± 324 DS molecules per transfected cell). The data points are representative of the number of independent experiments listed: 6 days, n = 7; 9 days, n = 3; 12 days, n = 4; 13 days, n = 3; 16 days, n = 2; 18 days, n = 4; 24 days, n = 4. Three experiments in which the replication of the oriP test plasmid was monitored at 6, 9, 13, and 16 days posttransfection were presented previously (37). Note that the level of replicated oriP plasmid decreased approximately 100-fold from the cell population between 6 and 18 days posttransfection, then remained stable between 18 and 24 days. This finding is consistent with our previous observation that oriP plasmids are established in only 1% of transfected 293 cells (37). The 4% rate-of-loss curve, depicted by black cross-hatched boxes, is theoretical and is based on previous studies of established, drug-resistant cell clones (29, 56).
A plasmid containing only the FR of oriP supported replication with only 6% of the efficiency of an oriP plasmid at 6 days posttransfection, and this replicated DNA was lost precipitously, as was the oriP plasmid (Fig. 3A). These experiments clearly demonstrate that plasmids containing only the DS of oriP can be efficiently synthesized and maintained each cell cycle in 293/EBNA-1 cells. The FR, whose stimulatory function is not required in 293/EBNA-1 cells, acts in cis to promote the rapid loss of oriP plasmids in these cells.
To determine if the stability of the DS plasmid was restricted to the 293/EBNA-1 cell clone analyzed, we monitored the fate of the oriP and DS plasmids in the 293/1728#5 cell clone that stably maintains the 1728 oriP/EBNA-1 expression plasmid (37). The replicated DS plasmid pool remained stable between 5 and 16 days posttransfection (Fig. 4). However, addition of the FR in cis resulted in the precipitous loss of the replicated oriP plasmid population in the 293/1728#5 cell clone (37), as seen in 293/EBNA-1 cells. Given that oriP plasmids and EBV itself are established inefficiently in all proliferating cell lines analyzed to date (27, 37, 45, 58), the inhibitory activity of the FR observed in 293/EBNA-1 cells and 293/1728#5 cells likely underlies the inefficient establishment detected in all of these cell lines. We therefore pursued experiments to characterize the inhibitory activity of the FR in 293/EBNA-1 cells.
FIG. 4.
Replicated DS plasmids are stable during 16 days posttransfection in the 293/1728#5 cell clone, as seen in the 293/EBNA-1 cell clone. The 293/1728#5 cell clone was isolated as described (37). This cell line was selected to stably replicate the 1728 plasmid, which contains oriP and an EBNA-1 expression cassette. The levels of replicated DS plasmid (DS) and prokaryotic backbone plasmid (Backbone) were monitored over 16 days posttransfection as described in the legend to Fig. 3A. Numbers below each gel refer to the average number of replicated DS plasmids present per transfected cell. The replicated backbone plasmid was present at less than one copy per transfected cell for each time point. These data were confirmed by Southern blot analysis (data not shown).
FR of oriP does not limit DNA synthesis at the DS.
Why does a plasmid containing only the DS of oriP support stable replication within 293/EBNA-1 cells, yet addition of FR to this plasmid promotes the replicon's precipitious loss from the majority of transfected cells? This observation is counterintuitive, because the FR has been shown to act in cis to promote the maintenance of plasmids containing a putative viral or mammalian origin in 293 and 143B cell lines (30, 34). Given that the DS plasmid supported replication more efficiently than the oriP plasmid at 6 days posttransfection (Wilcoxon rank sum test: P [two-sided] = 0.009; n = 7), we postulated that the FR may impose licensing on the DS of oriP so that it is synthesized only once per cell cycle. In the absence of the FR, the DS plasmid would be amplified each cell cycle, as is the replicon of bovine papillomavirus (BPV), and these DS replicons would be maintained if four EBNA-1-binding sites within the DS are sufficient for plasmid maintenance in 293/EBNA-1 cells.
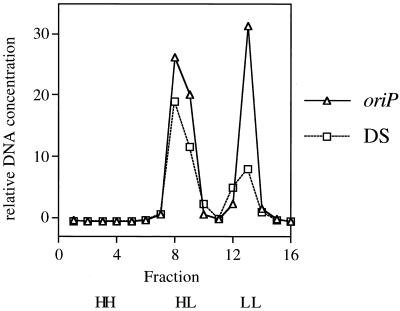
To test this hypothesis, we monitored the amount of plasmid that underwent one or additional rounds of DNA synthesis during one cell division cycle. Equimolar amounts of an oriP plasmid and a plasmid containing only the DS of oriP were introduced independently into 293/EBNA-1 cells, and the cells were labeled with bromodeoxyuridine for 17 h at 5 days posttransfection. Plasmid DNA was isolated by Hirt extraction, linearized, and separated on a CsCl gradient. Southern blotting was then used to quantify the amount of plasmid DNA that underwent zero (light-light [LL]), one (heavy-light [HL]), or multiple (heavy-heavy [HH]) rounds of DNA synthesis. Contrary to our hypothesis, the DS plasmid supported DNA synthesis only once per cell cycle, as did the oriP plasmid (Fig. 5). Namely, the FR of oriP does not limit DNA synthesis at the DS; rather, the DS alone possesses the cis-acting sequences required for licensing. This finding is consistent with the observation of Shirakata et al. that DS plasmids must traverse early G1 phase prior to their synthesis in S-phase (50).
FIG. 5.
Plasmid containing only the DS of oriP is synthesized once per cell cycle, as is an oriP replicon (1, 61). Equimolar amounts of an oriP plasmid (oriP) and a plasmid containing only the DS of oriP (DS) were introduced independently into 293/EBNA-1 cells, and the cells were labeled with bromodeoxyuridine for 17 h at 5 days posttransfection. Plasmid DNA was isolated by Hirt extraction of approximately 5 × 107 cells, linearized with XhoI, and separated on a CsCl density gradient. The refractive index of every fourth fraction was determined, one-fourth of each fraction was transferred to a nylon membrane, and plasmid DNA present in the LL, HL, and HH peaks was determined by Southern blotting (48). The relative DNA concentration was plotted versus fraction number. No plasmid DNA was detected at the density at which HH DNAs should migrate. The limit of detection was approximately 1 molecule per transfected cell (25 pg).
In addition to its synthetic function, the four EBNA-1-binding sites within the DS of oriP must also allow efficient plasmid maintenance in 293/EBNA-1 cells, given that other replicons that are efficiently synthesized once per cell cycle do not support stable replication in 293 cells (21, 34). That is, Krysan et al. have shown that plasmids containing putative mammalian origins support efficient replication at 4 days posttransfection in 293 cells; however, these plasmids were lost unless EBNA-1 and its binding sites in the FR were provided (34). Likewise, we have observed that a plasmid containing an efficient DNA synthetic element comprised of eight copies of the viral Rep* element (30) is lost at twice the rate of a DS plasmid in 293/EBNA-1 cells (Leight and Sugden, unpublished observations), indicating that the DS must possess efficient DNA synthetic and maintenance functions in these cells.
Efficient transcriptional activation from an oriP replicon does not promote its precipitous loss.
The FR clearly inhibits the establishment of an oriP replicon in 293/EBNA-1 cells, and an epigenetic event is required to overcome this inhibitory activity (37). But how does the FR inhibit establishment? The FR possesses two features that are not shared by the DS. First, the FR acts as a potent transcriptional enhancer, activating transcription of a promoter present in cis by 10- to 100-fold when EBNA-1 is provided in trans (4, 35, 46, 57). Second, when EBNA-1 dimers are bound to the 20 binding sites in the FR, the DNA synthesis machinery cannot proceed efficiently through this region (11, 15). The FR therefore scores as a replication fork barrier (RFB) in two-dimensional gel electrophoresis studies (15). We examined whether either of these attributes of the FR underlies its inhibitory activity.
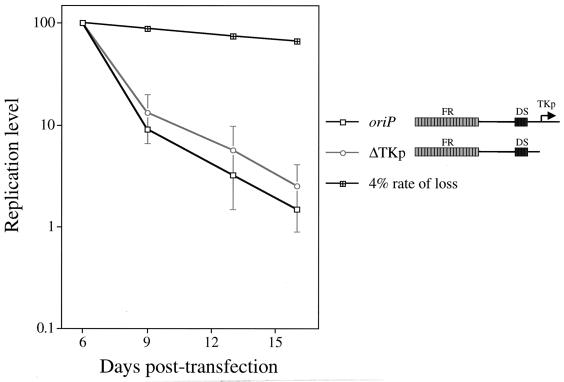
The Calos laboratory has shown an inverse correlation between the transcription and replication of a plasmid containing autonomously replicating sequences (22). We therefore postulated that an inverse relationship between replication and transcription of an oriP plasmid exists. The oriP plasmid analyzed in our studies contains both replicative (oriP) and transcriptional (FR enhancer and the thymidine kinase promoter) elements. We therefore addressed whether elimination of transcriptional activation from the thymidine kinase promoter by its deletion could rescue the replication defect of the oriP plasmid. The levels of replicated oriP plasmid (oriP) and oriP plasmid lacking the thymidine kinase promoter (ΔTKp) were monitored over 16 days following their introduction into 293/EBNA-1 cells. The ΔTKp plasmid supported replication but was lost precipitously, as was the oriP plasmid (Fig. 6), demonstrating that FR's potent transcriptional activation of the thymidine kinase promoter does not underlie the replicon's precipitous loss. There is not an inverse relationship between transcription and replication of an oriP plasmid as is seen with plasmids containing autonomously replicating human sequences (22).
FIG. 6.
Efficient transcriptional activation from an oriP plasmid does not promote its precipitous loss. The level of replicated oriP test plasmid (oriP) and oriP test plasmid lacking the thymidine kinase promoter (ΔTKp) was monitored over 16 days posttransfection in 293/EBNA-1 cells as described in the legend to Fig. 3A. The replication level of these plasmids was plotted versus the days posttransfection. For each independent experiment, the level of replicated test plasmid detected at 6 days posttransfection was set to 100%, and the replication level at later time points was set relative to this point (100% = 403 oriP and 318 ± 175 ΔTKp molecules per transfected cell). The loss of the ΔTKp plasmid is representative of three independent experiments conducted in parallel to the oriP test plasmid. The 4% rate-of-loss curve, depicted by black cross-hatched boxes, is theoretical and is based on previous studies of established, drug-resistant cell clones (29, 56).
Multiple EBNA-1 binding sites in FR of oriP promote inefficient establishment of plasmid replicon.
Given that the FR's enhancement of transcription from the oriP replicon does not underlie the replicon's precipitous loss, we addressed whether the FR, by acting as a potent RFB, promotes this inefficient establishment. oriP plasmids that initiate DNA synthesis during the last 3 h of S-phase may not be completely synthesized by the end of S-phase, as 3 or more hours are devoted to DNA synthesis through the FR (given that the FR is detected as an RFB in EBV and assuming that DNA elongation occurs at a rate of 1 kbp/min) (15). These plasmids may be eliminated from the cell if a checkpoint is not imposed to ensure duplication of the oriP plasmid prior to entry into G2 phase. The FR, by acting as an RFB, may thereby promote the loss of plasmids whose origin fires late during S-phase.
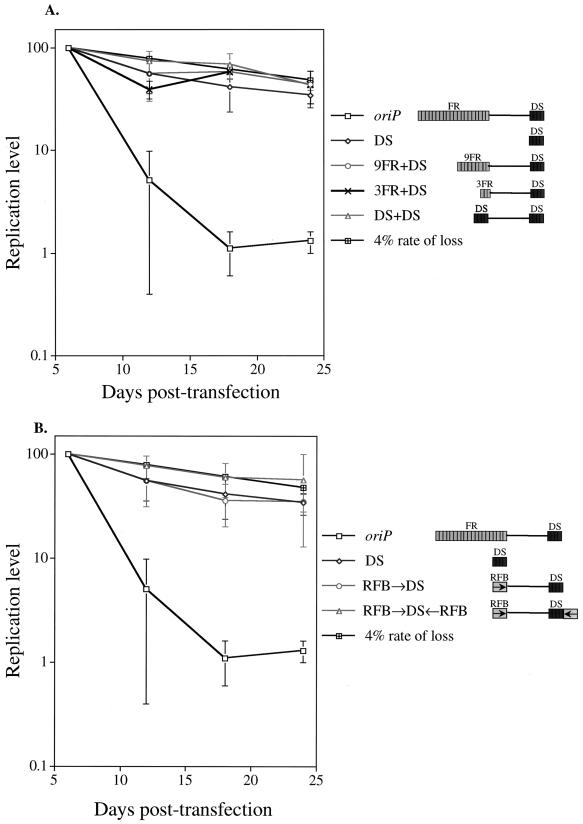
Platt et al. have shown that the number of EBNA-1-binding sites is critical for RFB formation (40). In their studies, the DS is not detected as an RFB, yet when it is trimerized and introduced in place of the FR, it scores as an RFB (43). To test our hypothesis, we therefore determined whether deletion of EBNA-1-binding sites within the FR or substitution of the FR with low-affinity binding sites from the DS could rescue the replication defect. oriP plasmid derivatives containing only nine and three EBNA-1 binding sites from the FR (9FR+DS and 3FR+DS, respectively) and a derivative in which the FR was replaced by the DS (DS+DS) were introduced independently into 293/EBNA-1 cells, and the level of replicated plasmid DNA was monitored over 24 days posttransfection in parallel to the oriP and DS plasmids. The 9FR+DS, 3FR+DS, and DS+DS plasmids supported stable replication over the time course, as did the plasmid containing only the DS of oriP (Fig. 7A). That is, deletion of EBNA-1-binding sites within the FR or substitution of the FR with DS sequences overcomes the inhibitory activity of the FR, allowing efficient establishment of the replicon.
FIG. 7.
(A) Multiple EBNA-1-binding sites within the FR of oriP inhibit the replicon's establishment. oriP plasmid derivatives containing permutations in the number and type (high versus low affinity) of EBNA-1-binding sites were introduced into 293/EBNA-1 cells, and the level of replicated plasmid DNA was monitored over 24 days posttransfection as described in the legend to Fig. 3A. The oriP and DS plasmids were analyzed in four independent experiments conducted in parallel to the oriP plasmid derivatives shown. (These data are also presented in Fig. 7B and 8, as all oriP plasmid derivatives were analyzed simultaneously.) The replication level of these plasmids was plotted versus the days posttransfection. For each of four independent experiments, the level of replicated test plasmid detected at 6 days posttransfection was set to 100%, and the replication level at later time points was set relative to this point (100% = 625 ± 121 oriP, 967 ± 258 DS, 876 ± 175 9FR+DS, 1149 ± 405 3FR+DS, and 1,498 ± 487 DS+DS molecules per transfected cell). The 3FR+DS plasmid data are representative of three independent experiments. The 4% rate-of-loss curve, depicted by black cross-hatched boxes, is theoretical and is based on previous studies of established, drug-resistant cell clones (29, 56). (B) An RFB from the rDNA locus, when introduced in place of the FR of oriP, does not inhibit establishment of the replicon. oriP plasmid derivatives containing one (RFB→DS) or two (RFB→DS←RFB) copies of the rDNA RFB (16) in place of the FR of oriP were introduced into 293/EBNA-1 cells, and the level of replicated plasmid DNA was monitored over 24 days posttransfection as described in the legend to Fig. 3A. The oriP and DS plasmids were analyzed in four independent experiments conducted in parallel to the oriP plasmid derivatives shown. (The data are also presented in Fig. 7A and 8, as all oriP plasmid derivatives were analyzed simultaneously.) The replication level of these plasmids was plotted versus the days posttransfection. For each of four independent experiments, the level of replicated test plasmid detected at 6 days posttransfection was set to 100%, and the replication level at later times was set relative to this point (100% = 625 ± 121 oriP, 967 ± 258 DS, 1,043 ± 462 RFB→DS, and 843 ± 215 RFB→DS←RFB molecules per transfected cell). The 4% rate-of-loss curve, depicted by black cross-hatched boxes, is theoretical and is based on previous studies of established, drug-resistant cell clones (29, 56).
While these findings are consistent with a model in which disruption of RFB formation overcomes the inhibitory activity of the FR, they do not address whether an RFB per se can act in cis to promote the loss of a plasmid containing only the DS of oriP in 293/EBNA-1 cells. We therefore asked whether an RFB, when introduced into a plasmid containing only the DS of oriP (RFB→DS), could promote the replicon's inefficient establishment. We chose the rDNA RFB for this purpose because this RFB has been conserved in organisms ranging from S. cerevisiae to humans (7, 39) and can function efficiently even in the presence of EBNA-1 (39). In addition, this conserved site of RFB formation can also function in the context of a plasmid (16). Given that the rDNA RFB is a directional (or polar) barrier while the FR is not (11, 16), a plasmid containing two copies of the rDNA RFB in a tail-to-tail orientation (RFB→DS←RFB) was also analyzed (Fig. 7B). These plasmids were introduced independently into 293/EBNA-1 cells, and the level of replicated plasmid DNA was monitored over 24 days posttransfection in parallel to the oriP and DS plasmids.
The RFB→DS and RFB→DS←RFB plasmids were efficiently established, supporting efficient DNA synthesis and maintenance each cell generation over the 24-day period (Fig. 7B). This experiment demonstrates that an RFB alone does not impose instability on a DS plasmid. Rather, multiple EBNA-1-binding sites in the FR of oriP possess an independent inhibitory function that promotes the inefficient establishment of plasmid replicons.
Spacing between FR and DS does not affect establishment of an oriP replicon.
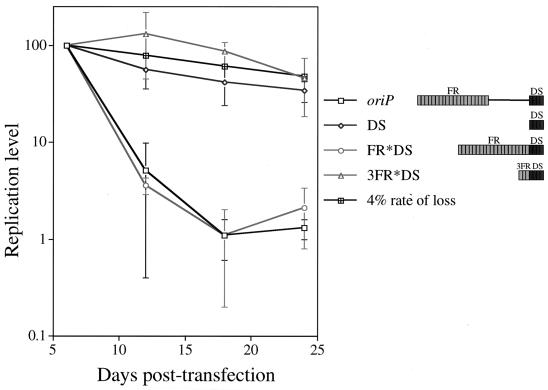
EBNA-1 dimers bound to the FR can associate with EBNA-1 dimers bound to the DS, resulting in the formation of intramolecular “looped” DNA complexes and intermolecular “linked” DNA complexes, as visualized by electron microscopy (14, 54). DNA looping and linking between the FR and DS are not required for replication in that, in 293/EBNA-1 cells, plasmids containing only the DS of oriP are established efficiently. We wished to address whether intramolecular DNA looping between the FR and DS could contribute to the inefficient establishment of oriP replicons. Given that it is less energetically favorable for a trans-acting factor to loop two cis elements when the distance between those elements is decreased (20), we constructed an oriP plasmid derivative in which the 963 bp separating FR and DS were deleted (FR*DS) (Fig. 8). DNA looping is unlikely to occur in the FR*DS plasmid, as Su et al. did not detect looped DNA complexes in electron microscopy studies with a DNA fragment containing the FR, and the length of the loops with an oriP DNA fragment detected ranged from 1,000 to 1,780 bp (54). DNA loops between the juxtaposed FR and DS would be less than 800 bp.
FIG. 8.
Spacing between the FR and DS of oriP has no effect on establishment of the replicon. The 963 nucleotides separating the FR and DS of oriP were deleted in the context of the oriP test plasmid and the 3FR+DS plasmid, the resulting plasmids (FR*DS and 3FR*DS, respectively) were introduced into 293/EBNA-1 cells, and the level of replicated DNA was monitored over 24 days posttransfection as described in the legend to Fig. 3A. The oriP and DS plasmids were analyzed in four independent experiments conducted in parallel to the oriP plasmid derivatives shown. (The data are also presented in Fig. 7A and 7B, as all oriP plasmid derivatives were analyzed simultaneously.) The replication level of these plasmids was plotted versus the days posttransfection. For each of four independent experiments, the level of replicated test plasmid detected at 6 days posttransfection was set to 100%, and the replication level at later times was set relative to this point (100% = 625 ± 121 oriP, 967 ± 258 DS, 439 ± 222 FR*DS, and 817 ± 576 3FR*DS molecules per transfected cell). The 3FR*DS data are representative of three independent experiments except for the day 24 time point, which is representative of two experiments. The 4% rate-of-loss curve, depicted by black cross-hatched boxes, is theoretical and is based on previous studies of established, drug-resistant cell clones (29, 56).
This FR*DS plasmid was introduced into 293/EBNA-1 cells in parallel to the oriP and DS plasmids, and the level of replicated DNA was monitored over 24 days posttransfection. The FR*DS plasmid was lost precipitously from cells between 6 and 18 days posttransfection (Fig. 8), as was the oriP plasmid, demonstrating that DNA looping between the FR and DS does not underlie the inefficient establishment of an oriP replicon. To ensure that juxtaposition of the FR and DS does not impose a novel replication defect, we monitored the replication of an oriP plasmid derivative containing three EBNA-1 binding sites from the FR juxtaposed to the DS (3FR*DS). This plasmid supported efficient replication over 24 days posttransfection, as did the 3FR+DS plasmid (Fig. 7A and 8), confirming that juxtaposition of the FR and DS does not impose a novel replication defect.
These experiments demonstrate that the spacing between FR and DS does not affect the establishment of an oriP replicon. This conclusion is also supported by the studies of the DS+DS, 9FR+DS, and 3FR+DS plasmids. DNA looping is expected to occur efficiently between distant EBNA-1-binding sites in these plasmids, given that single EBNA-1-binding sites that are separated by 930 nucleotides loop with the same frequency as an oriP DNA fragment when EBNA-1 is present in trans (19). These plasmids were established efficiently, however, demonstrating that DNA looping between distal EBNA-1 binding sites does not underline the inefficient establishment of oriP replicons.
DISCUSSION
Cell lines vary in their ability to support replication of oriP plasmid derivatives.
We have monitored the fate of plasmids containing oriP or only the DS of oriP for 24 days following their introduction into various EBNA-1-positive cell lines. While oriP plasmids are lost precipitously and established inefficiently in all 22 cell lines analyzed to date (27, 37, 45, 58), surprisingly we have found that cell lines vary dramatically in their ability to support the replication of a DS plasmid. DS plasmids were eliminated rapidly from 143B cells, so that the level of replicated DS plasmid detected at 4 days posttransfection was only 10% of the level of wild-type oriP and dropped below the level of detection (<0.1 molecule/cell) by 7 days posttransfection. Replicated DS plasmids were lost at an intermediate rate in C33A/EBNA-1 cells. That is, the DS plasmid supported replication as efficiently as the oriP plasmid at 6 days posttransfection (Wilcoxon rank sum test: P [two-sided] = 0.28; n = 3) and was lost at a rate of approximately 28% per cell generation between 6 and 16 days following transfection, as was the oriP plasmid. In 293/EBNA-1 cells, however, the DS plasmid was established efficiently, being lost at a rate of only 8% per cell generation, while the oriP plasmid was lost at a rate of 33% per cell generation between 6 and 18 days posttransfection. This replicated DS plasmid was likewise stable in the 293/1728#5 cell clone.
Variations between cell lines were also observed in historical studies in which the level of replicated oriP and DS DNA was monitored at 2 to 5 days posttransfection. In these experiments, the DS plasmid supported replication with less than 6% of the efficiency of the oriP plasmid in Raji cells and EBNA-1-expressing 143B cells (30, 57), whereas the DS plasmid supported replication as efficiently as the oriP plasmid in D98/Raji cells and EBNA-1-positive HeLa and 293 cells (24, 49, 60). Interestingly, cell line differences are not limited to the replication of DS plasmids. Stillman and Gluzman have shown that 293 cell extracts support efficient DNA synthesis of simian virus 40 DNA while HeLa cell extracts support only 12 to 25% of that level (53). In addition, while human papillomavirus replicates efficiently in 293 cells, it replicates poorly in HeLa cells due to limiting levels of cyclin E/CDK2 (38).
Clearly, cell lines vary in their support of replication of plasmids containing only four EBNA-1 binding sites from the DS. But what underlies this differential replication of DS plasmids? Previously, Aiyar et al. have shown that cell lines vary in their ability to eliminate replicated DNAs and transcribed DNAs (3). These DNAs were lost at a rate of 50% per cell generation in 293 cells, but were lost more rapidly in C33A and 143B cells (70 and 80% rate of loss per cell generation, respectively) (3). These differences in the maintenance of newly synthesized plasmids, however, cannot solely account for our observations in 293/EBNA-1 cells. That is, if the efficient replication of DS plasmids in 293/EBNA-1 cells is due to efficient DNA synthesis at the DS coupled with a maintenance function provided by the cell, all plasmids containing an efficient origin should replicate efficiently over 24 days posttransfection. On the contrary, we have observed that a plasmid containing an efficient DNA synthesis element comprised of eight copies of the viral Rep* element (30) is lost at twice the rate of a DS plasmid in 293/EBNA-1 cells (Leight and Sugden, unpublished observations). Likewise, plasmids containing putative mammalian origins are lost from 293 cells unless EBNA-1 and its binding sites in the FR are provided (34). These findings demonstrate that the DS alone possesses efficient DNA synthesis and maintenance functions in 293/EBNA-1 cells but not in C33A/EBNA-1, H1299/1728#3, or EBNA-1-positive 143B cells. This feature of 293/EBNA-1 cells allowed us to unveil the inhibitory activity of the FR—an inhibitory activity that likely underlies the inefficient establishment of oriP replicons shared by 293/EBNA-1 cells and the other 21 cell lines analyzed to date (27, 37, 45, 58).
We propose that the differences observed in cell lines are due to variations in the levels of cellular trans-acting factors involved in EBNA-1-mediated DNA synthesis and/or maintenance functions. These factors may be abundant in 293 cells, so that four DNA-bound EBNA-1 dimers are sufficient for their recruitment. In 143B cells, these factors would be limiting, and additional DNA-bound EBNA-1 dimers (and therefore additional EBNA-1-binding sites from the FR) must be employed for their recruitment. A prediction for this model is that a cDNA(s) from 293 cells should complement the replication defect of the DS plasmid in EBNA-1-positive 143B cells.
FR can stimulate and inhibit replication of oriP replicons.
EBNA-1 binding sites from the FR clearly contribute to DNA synthesis and/or maintenance of oriP plasmids in some cell lines. That is, we and others have shown that oriP plasmid derivatives containing fewer than seven EBNA-1 binding sites from the FR fail to support short-term replication in Raji, D98/Raji, and EBNA-1-positive 143B cells (8, 30, 47, 57). However, even with the intact FR, oriP plasmids are established in only 1 to 10% of transfected cells (37). By monitoring the replication of oriP plasmid derivatives in multiple cell lines, we unexpectedly found that the FR also acts in cis to promote the rapid loss of replicated oriP plasmids from the population of transfected 293/EBNA-1 cells and 293/1728#5 cells. This inhibitory activity of the FR likely underlies the inefficient establishment of oriP replicons in all cells, as oriP replicons are established inefficiently in all 21 cell lines analyzed to date (27, 37, 45, 58). Previously we have shown that oriP plasmids must be modified epigenetically for establishment (37). (Note that in Fig. 3B the level of replicated oriP plasmid decreased approximately 100-fold from the cell population between 6 and 18 days posttransfection, then remained stable between 18 and 24 days. This is consistent with our previous observation that oriP plasmids are established in only 1% of transfected 293 cells [37].) Our findings indicate that this epigenetic event is required to overcome an inhibitory activity of the FR.
How does the FR inhibit the establishment of oriP replicons? This still remains an enigma. Our studies, however, have defined parameters for FR's inhibitory activity in 293/EBNA-1 cells. We have shown that DNA looping between FR and DS neither stimulates nor inhibits replication (Fig. 8). Additionally, the FR does not limit DNA synthesis at the DS, as plasmids lacking the FR of oriP are licensed analogously to oriP plasmids (Fig. 5) (1, 61). Likewise, the FR does not inhibit activities of the DS by competing for EBNA-1. Namely, only 10% of EBNA-1 molecules are bound to the FR at 6 days posttransfection (given that there are approximately 3 × 104 EBNA-1 molecules per cell [52; Wang and Sugden, unpublished observations], 100 replicated oriP plasmids per cell, and 20 binding sites for EBNA-1 dimers within the FR), and previous studies have shown that the FR enhances EBNA-1's binding to the DS (13, 54). Furthermore, two attributes of the FR that are not shared by the DS, FR's enhancement of transcription of a promoter in cis and FR's ability to inhibit replication fork movement, do not solely account for oriP's inefficient establishment. Deletion of 11 EBNA-1-binding sites in the FR or replacement of the FR with DS sequences, however, does overcome the inhibitory activity of the FR, allowing efficient establishment of the oriP derivative in 293/EBNA-1 cells (Fig. 7A).
Why do 20 EBNA-1-binding sites from FR inhibit replication whereas 9 EBNA-1-binding sites do not? There are two likely answers. First, an inhibitory factor may bind the 11 right-hand binding sites from FR, and deletion of these sites relieves inhibition. Alternatively, the 20 binding sites in the FR may act cooperatively to allow binding of an inhibitory factor. Such an inhibitory factor may be involved in modulation of chromatin structure or covalent modification of the FR. We find the first possibility unlikely, as the right-hand and left-hand EBNA-1-binding sites in the FR behave similarly in supporting transcription and replication (8, 57), and the nine left-hand EBNA-1-binding sites from the FR show >95% sequence identity with the 11 right-hand EBNA-1-binding sites (based on the Smith-Waterman algorithm). Identification of cellular factors that bind to the FR of oriP in transiently transfected cells, in which oriP plasmids are lost at ∼30% per cell generation, and in established cell clones, in which oriP plasmids are lost at ∼2 to 4% per cell generation (29, 56), should provide insights into the epigenetic event that underlies the establishment of oriP replicons and EBV's plasmid replicon.
Clearly the FR has both inhibitory and stimulatory functions with respect to oriP's replication, but given that seven EBNA-1 binding sites from the FR are sufficient for plasmid maintenance in Raji and D98/Raji cells (47, 57) and are not even required in 293 cells, why has EBV evolved to retain all 20 EBNA-1-binding sites in the FR? We can only speculate that these 20 binding sites contribute to an essential function of the virus, such as efficient transcription of latent gene promoters within the established host cell. Even though EBV has kept these 20 binding sites in the FR at the expense of its establishment in proliferating cells, it has accomplished an astonishing feat—assimilation by the host cell as an extrachromosomal replicon that is synthesized only once per cell cycle and faithfully partitioned to daughter cells (1, 29, 56, 61).
ACKNOWLEDGMENTS
We are grateful to Paul Ahlquist, Paul Lambert, and our colleagues for helpful comments on the manuscript. We thank Ping Hua for construction of several plasmids.
This work was supported by Public Health Service grants CA-22443, CA-07175, and T32-CA-09135. Bill Sugden is an American Cancer Society Research Professor.
REFERENCES
- 1.Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987;61:1743–176. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Sugden B. Fusions between Epstein-Barr viral nuclear antigen-1 of Epstein-Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J Biol Chem. 1998;273:33073–33081. doi: 10.1074/jbc.273.49.33073. [DOI] [PubMed] [Google Scholar]
- 3.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambinder R F, Mullen M A, Chang Y N, Hayward G S, Hayward S D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambinder R F, Shah W A, Rawlins D R, Hayward G S, Hayward S D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. DNA sequence and expression of the B95–8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 7.Brewer B J, Fangman W L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 8.Chittenden T, Lupton S, Levine A J. Functional limits of oriP, the Epstein-Barr virus plasmid origin of replication. J Virol. 1989;63:3016–3025. doi: 10.1128/jvi.63.7.3016-3025.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford D H, Thomas J A, Janossy G, Sweny P, Fernando O N, Moorhead J F, Thompson J H. Epstein Barr virus nuclear antigen positive lymphoma after cyclosporin A treatment in patient with renal allograft. Lancet. 1980;i:1355–1356. doi: 10.1016/s0140-6736(80)91800-0. [DOI] [PubMed] [Google Scholar]
- 10.de-The G, Geser A, Day N E, Tukei P M, Williams E H, Beri D P, Smith P G, Dean A G, Bronkamm G W, Feorino P, Henle W. Epidemiological evidence for causal relationship between Epstein-Barr virus and Burkitt's lymphoma from Ugandan prospective study. Nature. 1978;274:756–761. doi: 10.1038/274756a0. [DOI] [PubMed] [Google Scholar]
- 11.Ermakova O V, Frappier L, Schildkraut C L. Role of the EBNA-1 protein in pausing of replication forks in the Epstein-Barr virus genome. J Biol Chem. 1996;271:33009–33017. doi: 10.1074/jbc.271.51.33009. [DOI] [PubMed] [Google Scholar]
- 12.Evans A. Viral infections in oral medicine. New York, N.Y: Elsevier North Holland; 1982. [Google Scholar]
- 13.Frappier L, Goldsmith K, Bendell L. Stabilization of the EBNA1 protein on the Epstein-Barr virus latent origin of DNA replication by a DNA looping mechanism. J Biol Chem. 1994;269:1057–1062. [PubMed] [Google Scholar]
- 14.Frappier L, O'Donnell M. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc Natl Acad Sci USA. 1991;88:10875–10879. doi: 10.1073/pnas.88.23.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 16.Gerber J K, Gogel E, Berger C, Wallisch M, Muller F, Grummt I, Grummt F. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell. 1997;90:559–567. doi: 10.1016/s0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 17.Geser A, de The G, Lenoir G, Day N E, Williams E H. Final case reporting from the Ugandan prospective study of the relationship between EBV and Burkitt's lymphoma. Int J Cancer. 1982;29:397–400. doi: 10.1002/ijc.2910290406. [DOI] [PubMed] [Google Scholar]
- 18.Glaser S L, Lin R J, Stewart S L, Ambinder R F, Jarrett R F, Brousset P, Pallesen G, Gulley M L, Khan G, O'Grady J, Hummel M, Preciado M V, Knecht H, Chan J K, Claviez A. Epstein-Barr virus-associated Hodgkin's disease: epidemiologic characteristics in international data. Int J Cancer. 1997;70:375–382. doi: 10.1002/(sici)1097-0215(19970207)70:4<375::aid-ijc1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 19.Goldsmith K, Bendell L, Frappier L. Identification of EBNA1 amino acid sequences required for the interaction of the functional elements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1993;67:3418–3426. doi: 10.1128/jvi.67.6.3418-3426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goryshin I, Kil Y V, Reznikoff W S. DNA length, bending, and twisting constraints on IS50 transposition. Proc Natl Acad Sci USA. 1994;91:10834–10838. doi: 10.1073/pnas.91.23.10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haase S B, Calos M P. Replication control of autonomously replicating human sequences. Nucleic Acids Res. 1991;19:5053–5058. doi: 10.1093/nar/19.18.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haase S B, Heinzel S S, Calos M P. Transcription inhibits the replication of autonomously replicating plasmids in human cells. Mol Cell Biol. 1994;14:2516–2524. doi: 10.1128/mcb.14.4.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanto D W, Frizzera G, Purtilo D T, Sakamoto K, Sullivan J L, Saemundsen A K, Klein G, Simmons R L, Najarian J S. Clinical spectrum of lymphoproliferative disorders in renal transplant recipients and evidence for the role of Epstein-Barr virus. Cancer Res. 1981;41:4253–4261. [PubMed] [Google Scholar]
- 24.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 26.Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, Tanaka S, Sato E, Osato T. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci USA. 1994;91:9131–9135. doi: 10.1073/pnas.91.19.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai S, Nishikawa J, Takada K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol. 1998;72:4371–4378. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchmaier A L, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirchmaier A L, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirchmaier A L, Sugden B. Rep*: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein-Barr virus. J Virol. 1998;72:4657–4666. doi: 10.1128/jvi.72.6.4657-4666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein G, Glovanella B C, Lindahl T, Fialkow P J, Singh S, Stehlin J S. Direct evidence for the presence of Epstein-Barr virus DNA and nuclear antigen in malignant epithelial cells from patients with poorly differentiated carcinoma of the nasopharynx. Proc Natl Acad Sci USA. 1974;71:4737–4741. doi: 10.1073/pnas.71.12.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutson J C, Yee D. Electroporation: parameters affecting transfer of DNA into mammalian cells. Anal Biochem. 1987;164:44–52. doi: 10.1016/0003-2697(87)90365-4. [DOI] [PubMed] [Google Scholar]
- 33.Koshland D, Kent J C, Hartwell L H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 34.Krysan P J, Haase S B, Calos M P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langle-Rouault F, Patzel V, Benavente A, Taillez M, Silvestre N, Bompard A, Sczakiel G, Jacobs E, Rittner K. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J Virol. 1998;72:6181–6185. doi: 10.1128/jvi.72.7.6181-6185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leight E R, Sugden B. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 37.Leight E R, Sugden B. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol Cell Biol. 2001;21:4149–4161. doi: 10.1128/MCB.21.13.4149-4161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin B Y, Ma T, Liu J S, Kuo S R, Jin G, Broker T R, Harper J W, Chow L T. HeLa cells are phenotypically limiting in cyclin E/CDK2 for efficient human papillomavirus DNA replication. J Biol Chem. 2000;275:6167–6174. doi: 10.1074/jbc.275.9.6167. [DOI] [PubMed] [Google Scholar]
- 39.Little R D, Platt T H, Schildkraut C L. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupton S, Levine A J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niederman J C, McCollum R W, Henle G, Henle W. Infectious mononucleosis: clinical manifestations in relation to EB virus antibodies. JAMA. 1968;203:205–209. doi: 10.1001/jama.203.3.205. [DOI] [PubMed] [Google Scholar]
- 43.Platt T H, Tcherepanova I Y, Schildkraut C L. Effect of number and position of EBNA-1 binding sites in Epstein-Barr virus oriP on the sites of initiation, barrier formation, and termination of replication. J Virol. 1993;67:1739–1745. doi: 10.1128/jvi.67.3.1739-1745.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 45.Reisman D, Sugden B. An EBNA-negative, EBV-genome-positive human lymphoblast cell line in which superinfecting EBV DNA is not maintained. Virology. 1984;137:113–126. doi: 10.1016/0042-6822(84)90014-x. [DOI] [PubMed] [Google Scholar]
- 46.Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Shirakata M, Hirai K. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J Biochem (Tokyo) 1998;123:175–181. doi: 10.1093/oxfordjournals.jbchem.a021907. [DOI] [PubMed] [Google Scholar]
- 50.Shirakata M, Imadome K I, Hirai K. Requirement of replication licensing for the dyad symmetry element-dependent replication of the Epstein-Barr virus oriP minichromosome. Virology. 1999;263:42–54. doi: 10.1006/viro.1999.9965. [DOI] [PubMed] [Google Scholar]
- 51.Siebert J D, Ambinder R F, Napoli V M, Quintanilla M L, Banks P M, Gulley M L. Human immunodeficiency virus-associated Hodgkin's disease contains latent, not replicative, Epstein-Barr virus. Hum Pathol. 1995;26:1191–1195. doi: 10.1016/0046-8177(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 52.Sternas L, Middleton T, Sugden B. The average number of molecules of Epstein-Barr nuclear antigen 1 per cell does not correlate with the average number of Epstein-Barr virus (EBV) DNA molecules per cell among different clones of EBV-immortalized cells. J Virol. 1990;64:2407–2410. doi: 10.1128/jvi.64.5.2407-2410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stillman B W, Gluzman Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol Cell Biol. 1985;5:2051–2060. doi: 10.1128/mcb.5.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su W, Middleton T, Sugden B, Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci USA. 1991;88:10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugden B, Leight E R. EBV's plasmid replicon: an enigma in cis and trans. Curr Top Microbiol Immunol. 2001;258:3–11. doi: 10.1007/978-3-642-56515-1_1. [DOI] [PubMed] [Google Scholar]
- 56.Sugden B, Warren N. Plasmid origin of replication of Epstein-Barr virus, oriP, does not limit replication in cis. Mol Biol Med. 1988;5:85–94. [PubMed] [Google Scholar]
- 57.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang L, Maruo S, Takada K. CD21-mediated entry and stable infection by Epstein-Barr virus in canine and rat cells. J Virol. 2000;74:10745–10751. doi: 10.1128/jvi.74.22.10745-10751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yates J L, Camiolo S M, Bashaw J M. The minimal replicator of Epstein-Barr virus oriP. J Virol. 2000;74:4512–4522. doi: 10.1128/jvi.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]