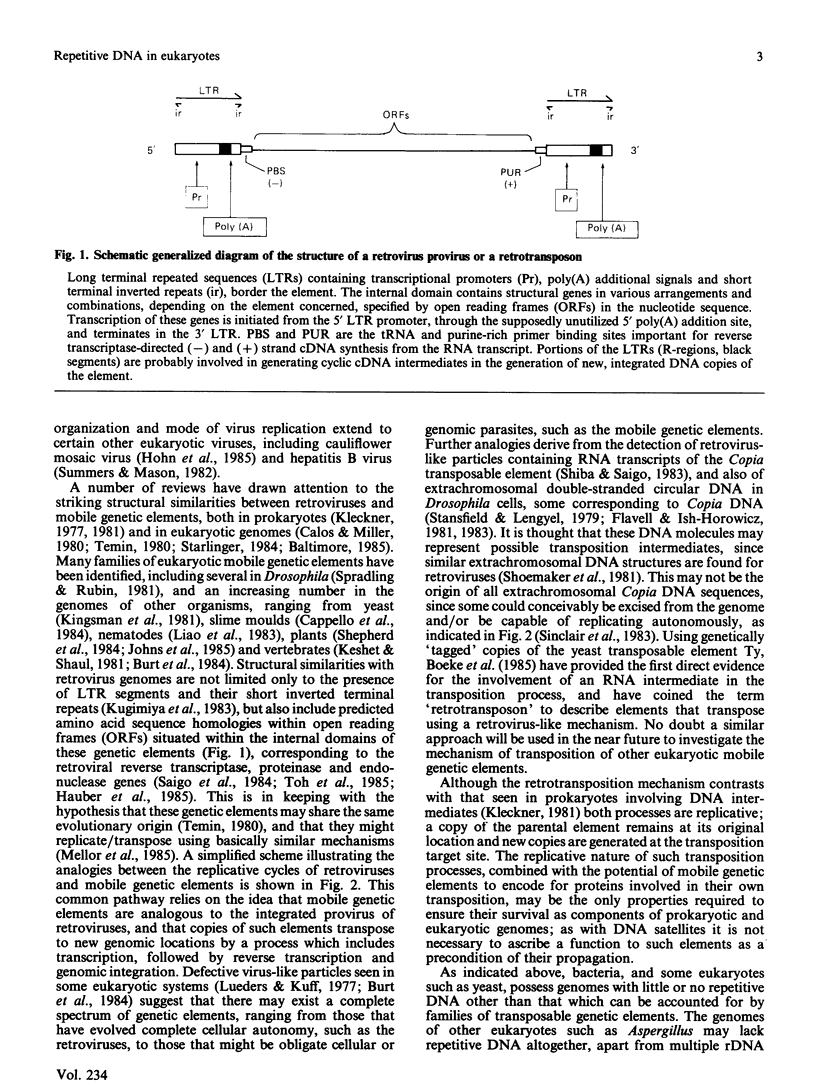
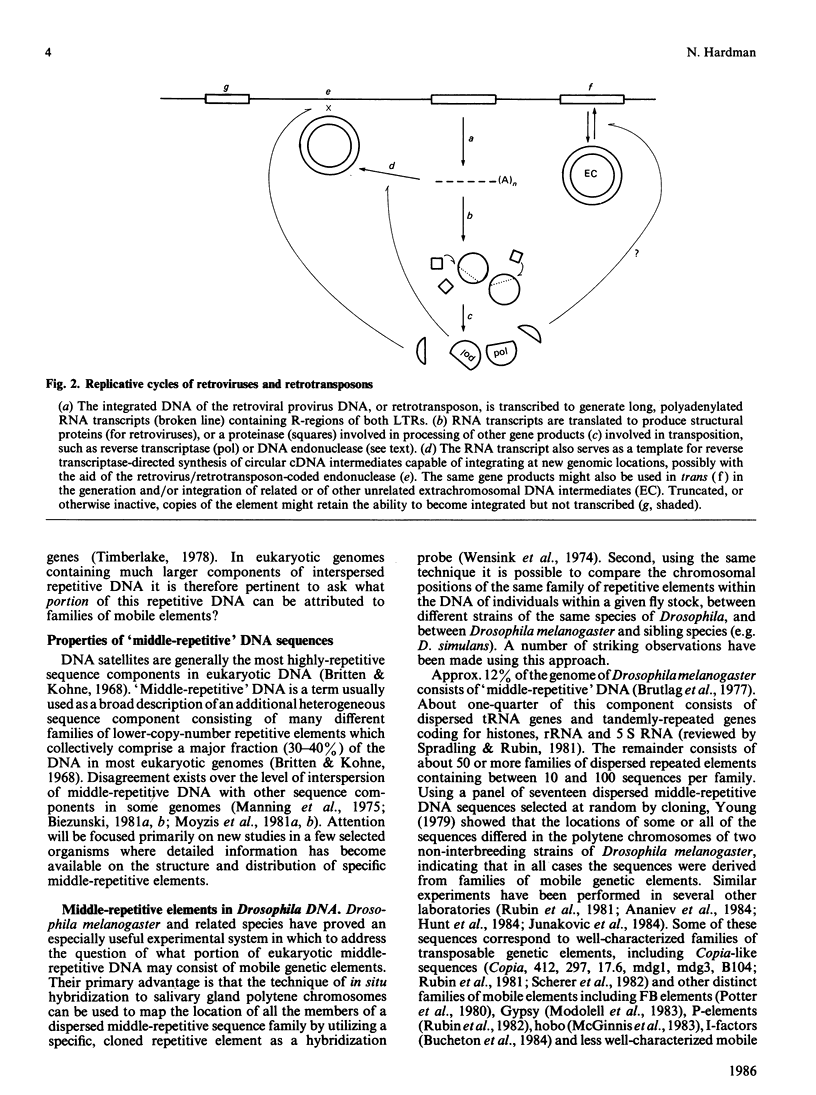
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell. 1985 Mar;40(3):481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- Bennett K. L., Hastie N. D. Looking for relationships between the most repeated dispersed DNA sequences in the mouse: small R elements are found associated consistently with long MIF repeats. EMBO J. 1984 Feb;3(2):467–472. doi: 10.1002/j.1460-2075.1984.tb01829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M., Richmond M. H., Petrocheilou V. The inactivation of tet genes on a plasmid by the duplication of one inverted repeat of a transposon-like structure which itself mediates tetracycline resistance. Plasmid. 1980 Mar;3(2):135–149. doi: 10.1016/0147-619x(80)90105-5. [DOI] [PubMed] [Google Scholar]
- Biezunski N. Structure and distribution of inverted repeats (Palindromes). I. Analysis of DNA of Drosophila melanogaster. Chromosoma. 1981;84(1):87–109. doi: 10.1007/BF00293365. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Judd B. H. A copy of the copia transposable element is very tightly linked to the Wa allele at the white locus of D. melanogaster. Cell. 1981 Sep;25(3):705–711. doi: 10.1016/0092-8674(81)90177-x. [DOI] [PubMed] [Google Scholar]
- Bishop J. M. Cellular oncogenes and retroviruses. Annu Rev Biochem. 1983;52:301–354. doi: 10.1146/annurev.bi.52.070183.001505. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bostock C. J., Clark E. M., Harding N. G., Mounts P. M., Tyler-Smith C., van Heyningen V., Walker P. M. The development of resistance to methotrexate in a mouse melanoma cell line. I. Characterisation of the dihydrofolate reductases and chromosomes in sensitive and resistant cells. Chromosoma. 1979;74(2):153–177. doi: 10.1007/BF00292270. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Gene regulation for higher cells: a theory. Science. 1969 Jul 25;165(3891):349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Appels R., Dennis E. S., Peacock W. J. Highly repeated DNA in Drosophila melanogaster. J Mol Biol. 1977 May 5;112(1):31–47. doi: 10.1016/s0022-2836(77)80154-x. [DOI] [PubMed] [Google Scholar]
- Brûlet P., Kaghad M., Xu Y. S., Croissant O., Jacob F. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5641–5645. doi: 10.1073/pnas.80.18.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A., Paro R., Sang H. M., Pelisson A., Finnegan D. J. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984 Aug;38(1):153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- Burt D. W., Reith A. D., Brammar W. J. A retroviral provirus closely associated with the Ren-2 gene of DBA/2 mice. Nucleic Acids Res. 1984 Nov 26;12(22):8579–8593. doi: 10.1093/nar/12.22.8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan H. G. The organization of genetic units in chromosomes. J Cell Sci. 1967 Mar;2(1):1–7. doi: 10.1242/jcs.2.1.1. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Cappello J., Cohen S. M., Lodish H. F. Dictyostelium transposable element DIRS-1 preferentially inserts into DIRS-1 sequences. Mol Cell Biol. 1984 Oct;4(10):2207–2213. doi: 10.1128/mcb.4.10.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Selfish DNA and the origin of introns. Nature. 1985 May 23;315(6017):283–284. doi: 10.1038/315283b0. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Hearst J. E. An electron microscopic study of mouse foldback DNA. Cell. 1975 Aug;5(4):429–446. doi: 10.1016/0092-8674(75)90062-8. [DOI] [PubMed] [Google Scholar]
- Chen H. R., Barker W. C. Nucleotide sequences of the retroviral long terminal repeats and their adjacent regions. Nucleic Acids Res. 1984 Feb 24;12(4):1767–1778. doi: 10.1093/nar/12.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke H. J., Schmidtke J., Gosden J. R. Characterisation of a human Y chromosome repeated sequence and related sequences in higher primates. Chromosoma. 1982;87(5):491–502. doi: 10.1007/BF00333470. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Are introns structural elements or evolutionary debris? Nature. 1985 Feb 7;313(6002):434–435. doi: 10.1038/313434b0. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Jolly D. J., Rubin C. M., Friedmann T., Schmid C. W. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol. 1981 Sep 5;151(1):17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Schmid C. W. An electron microscope study of the DNA sequence organization of the human genome. J Mol Biol. 1976 Sep 25;106(3):773–790. doi: 10.1016/0022-2836(76)90264-3. [DOI] [PubMed] [Google Scholar]
- DiGiovanni L., Haynes S. R., Misra R., Jelinek W. R. Kpn I family of long-dispersed repeated DNA sequences of man: evidence for entry into genomic DNA of DNA copies of poly(A)-terminated Kpn I RNAs. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6533–6537. doi: 10.1073/pnas.80.21.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980 Apr 17;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Dover G. A., Flavell R. B. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984 Oct;38(3):622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Dowsett A. P., Young M. W. Differing levels of dispersed repetitive DNA among closely related species of Drosophila. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4570–4574. doi: 10.1073/pnas.79.15.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk G., Leis J., Longiaru M., Skalka A. M. Selective cleavage in the avian retroviral long terminal repeat sequence by the endonuclease associated with the alpha beta form of avian reverse transcriptase. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6745–6749. doi: 10.1073/pnas.80.22.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden F. C., Musti A. M., Sobieski D. A. Clusters of repeated sequences of chicken DNA are extensively methylated but contain specific undermethylated regions. J Mol Biol. 1981 May 15;148(2):129–151. doi: 10.1016/0022-2836(81)90509-x. [DOI] [PubMed] [Google Scholar]
- Fanning T. G. Characterization of a highly repetitive family of DNA sequences in the mouse. Nucleic Acids Res. 1982 Aug 25;10(16):5003–5013. doi: 10.1093/nar/10.16.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Size and structure of the highly repetitive BAM HI element in mice. Nucleic Acids Res. 1983 Aug 11;11(15):5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. Extrachromosomal circular copies of the eukaryotic transposable element copia in cultured Drosophila cells. Nature. 1981 Aug 13;292(5824):591–595. doi: 10.1038/292591a0. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. The origin of extrachromosomal circular copia elements. Cell. 1983 Sep;34(2):415–419. doi: 10.1016/0092-8674(83)90375-6. [DOI] [PubMed] [Google Scholar]
- Fry K., Salser W. Nucleotide sequences of HS-alpha satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell. 1977 Dec;12(4):1069–1084. doi: 10.1016/0092-8674(77)90170-2. [DOI] [PubMed] [Google Scholar]
- Gall J. G., Atherton D. D. Satellite DNA sequences in Drosophila virilis. J Mol Biol. 1974 Jan 5;85(4):633–664. doi: 10.1016/0022-2836(74)90321-0. [DOI] [PubMed] [Google Scholar]
- Gebhard W., Meitinger T., Höchtl J., Zachau H. G. A new family of interspersed repetitive DNA sequences in the mouse genome. J Mol Biol. 1982 May 25;157(3):453–471. doi: 10.1016/0022-2836(82)90471-5. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Saedler H. DNA sequence of the mini-insertion IS2--6 and its relation to the sequence of IS2. Nature. 1978 Oct 19;275(5681):611–617. doi: 10.1038/275611a0. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Green M. M. Transposable elements in Drosophila and other Diptera. Annu Rev Genet. 1980;14:109–120. doi: 10.1146/annurev.ge.14.120180.000545. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Queen C., Singer M. F. Interspersed repeated sequences in the African green monkey genome that are homologous to the human Alu family. Nucleic Acids Res. 1981 Nov 11;9(21):5553–5568. doi: 10.1093/nar/9.21.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Skowronski J., Singer M. F. Defining the beginning and end of KpnI family segments. EMBO J. 1984 Aug;3(8):1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D., Di Carlo M., Zopf D., Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984 Oct;3(10):2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D., Krause E., Melli M., Dobberstein B. The organization of the 7SL RNA in the signal recognition particle. Nucleic Acids Res. 1983 Nov 11;11(21):7363–7374. doi: 10.1093/nar/11.21.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman N., Bell A. J., McLachlan A. Organisation of inverted repeat sequences in hamster cell nuclear DNA. Biochim Biophys Acta. 1979 Oct 25;564(3):372–389. doi: 10.1016/0005-2787(79)90029-7. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L., Brown A. J., McLachlan A. Distribution of inverted repeat sequences in nuclear DNA from Physarum polycephalum. Eur J Biochem. 1979 Feb 15;94(1):179–187. doi: 10.1111/j.1432-1033.1979.tb12884.x. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L. Characterization of foldback sequences in Physarum polycephalum nuclear DNA using the electron microscope. Eur J Biochem. 1977 Apr 1;74(2):275–283. doi: 10.1111/j.1432-1033.1977.tb11391.x. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L., Fergie R. C., Gerrie L. M. Sequence organisation in nuclear DNA from Physarum polycephalum. Interspersion of repetitive and single-copy sequences. Eur J Biochem. 1980 Jan;103(2):247–257. doi: 10.1111/j.1432-1033.1980.tb04309.x. [DOI] [PubMed] [Google Scholar]
- Hauber J., Nelböck-Hochstetter P., Feldmann H. Nucleotide sequence and characteristics of a Ty element from yeast. Nucleic Acids Res. 1985 Apr 25;13(8):2745–2758. doi: 10.1093/nar/13.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Toomey T. P., Leinwand L., Jelinek W. R. The Chinese hamster Alu-equivalent sequence: a conserved highly repetitious, interspersed deoxyribonucleic acid sequence in mammals has a structure suggestive of a transposable element. Mol Cell Biol. 1981 Jul;1(7):573–583. doi: 10.1128/mcb.1.7.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist G. Organisation and evolution of Drosophila virilis heterochromatin. Nature. 1975 Oct 9;257(5526):503–506. doi: 10.1038/257503a0. [DOI] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Hunt J. A., Bishop J. G., 3rd, Carson H. L. Chromosomal mapping of a middle-repetitive DNA sequence in a cluster of five species of Hawaiian Drosophila. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7146–7150. doi: 10.1073/pnas.81.22.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran P., Forget B. G., Weissman S. M. Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981 Oct;26(2 Pt 2):141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- John B., Miklos G. L. Functional aspects of satellite DNA and heterochromatin. Int Rev Cytol. 1979;58:1–114. doi: 10.1016/s0074-7696(08)61473-4. [DOI] [PubMed] [Google Scholar]
- Johns M. A., Mottinger J., Freeling M. A low copy number, copia-like transposon in maize. EMBO J. 1985 May;4(5):1093–1101. doi: 10.1002/j.1460-2075.1985.tb03745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb V. F., Glasser S., King D., Lingrel J. B. A cluster of repetitive elements within a 700 base pair region in the mouse genome. Nucleic Acids Res. 1983 Apr 11;11(7):2177–2184. doi: 10.1093/nar/11.7.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J., Brown P. C., Schimke R. T. Amplified dihydrofolate reductase genes in unstably methotrexate-resistant cells are associated with double minute chromosomes. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5669–5673. doi: 10.1073/pnas.76.11.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet E., Shaul Y. Terminal direct repeats in a retrovirus-like repeated mouse gene family. Nature. 1981 Jan 1;289(5793):83–85. doi: 10.1038/289083a0. [DOI] [PubMed] [Google Scholar]
- Keyl H. G. A demonstrable local and geometric increase in the chromosomal DNA of Chironomus. Experientia. 1965 Apr 15;21(4):191–193. doi: 10.1007/BF02141878. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T. Estimate of the genome size of various microorganisms. J Bacteriol. 1969 Jun;98(3):1400–1401. doi: 10.1128/jb.98.3.1400-1401.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsman A. J., Gimlich R. L., Clarke L., Chinault A. C., Carbon J. Sequence variation in dispersed repetitive sequences in Saccharomyces cerevisiae. J Mol Biol. 1981 Feb 5;145(4):619–632. doi: 10.1016/0022-2836(81)90306-5. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Koch A. L. Enzyme evolution. I. The importance of untranslatable intermediates. Genetics. 1972 Oct;72(2):297–316. doi: 10.1093/genetics/72.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kugimiya W., Ikenaga H., Saigo K. Close relationship between the long terminal repeats of avian leukosis-sarcoma virus and copia-like movable genetic elements of Drosophila. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3193–3197. doi: 10.1073/pnas.80.11.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird C. D. Chromatid structure: relationship between DNA content and nucleotide sequence diversity. Chromosoma. 1971 Mar 16;32(4):378–406. doi: 10.1007/BF00285251. [DOI] [PubMed] [Google Scholar]
- Lauth M. R., Spear B. B., Heumann J., Prescott D. M. DNA of ciliated protozoa: DNA sequence diminution during macronuclear development of Oxytricha. Cell. 1976 Jan;7(1):67–74. doi: 10.1016/0092-8674(76)90256-7. [DOI] [PubMed] [Google Scholar]
- Leach D. R., Stahl F. W. Viability of lambda phages carrying a perfect palindrome in the absence of recombination nucleases. 1983 Sep 29-Oct 5Nature. 305(5933):448–451. doi: 10.1038/305448a0. [DOI] [PubMed] [Google Scholar]
- Lerman M. I., Thayer R. E., Singer M. F. Kpn I family of long interspersed repeated DNA sequences in primates: polymorphism of family members and evidence for transcription. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3966–3970. doi: 10.1073/pnas.80.13.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Rubin G. M. The unstable wDZL mutation of Drosophila is caused by a 13 kilobase insertion that is imprecisely excised in phenotypic revertants. Cell. 1982 Sep;30(2):543–550. doi: 10.1016/0092-8674(82)90251-3. [DOI] [PubMed] [Google Scholar]
- Liao L. W., Rosenzweig B., Hirsh D. Analysis of a transposable element in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3585–3589. doi: 10.1073/pnas.80.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonberg N., Gilbert W. Intron/exon structure of the chicken pyruvate kinase gene. Cell. 1985 Jan;40(1):81–90. doi: 10.1016/0092-8674(85)90311-3. [DOI] [PubMed] [Google Scholar]
- Lueders K. K., Kuff E. L. Sequences associated with intracisternal A particles are reiterated in the mouse genome. Cell. 1977 Dec;12(4):963–972. doi: 10.1016/0092-8674(77)90161-1. [DOI] [PubMed] [Google Scholar]
- Macreadie I. G., Scott R. M., Zinn A. R., Butow R. A. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell. 1985 Jun;41(2):395–402. doi: 10.1016/s0092-8674(85)80012-x. [DOI] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Manning J. E., Schmid C. W., Davidson N. Interspersion of repetitive and nonrepetitive DNA sequences in the Drosophila melanogaster genome. Cell. 1975 Feb;4(2):141–155. doi: 10.1016/0092-8674(75)90121-x. [DOI] [PubMed] [Google Scholar]
- Martin S. L., Voliva C. F., Burton F. H., Edgell M. H., Hutchison C. A., 3rd A large interspersed repeat found in mouse DNA contains a long open reading frame that evolves as if it encodes a protein. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2308–2312. doi: 10.1073/pnas.81.8.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazrimas J. A., Hatch F. T. A possible relationship between satellite DNA and the evolution of kangaroo rat species (genus Dipodomys). Nat New Biol. 1972 Nov 22;240(99):102–105. doi: 10.1038/newbio240102a0. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Shermoen A. W., Beckendorf S. K. A transposable element inserted just 5' to a Drosophila glue protein gene alters gene expression and chromatin structure. Cell. 1983 Aug;34(1):75–84. doi: 10.1016/0092-8674(83)90137-x. [DOI] [PubMed] [Google Scholar]
- Mellor J., Fulton S. M., Dobson M. J., Wilson W., Kingsman S. M., Kingsman A. J. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature. 1985 Jan 17;313(5999):243–246. doi: 10.1038/313243a0. [DOI] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis R. K., Bonnet J., Li D. W., Ts'o P. O. An alternative view of mammalian DNA sequence organization. II. Short repetitive sequences are organized into scrambled tandem clusters in Syrian hamster DNA. J Mol Biol. 1981 Dec 25;153(4):871–896. doi: 10.1016/0022-2836(81)90457-5. [DOI] [PubMed] [Google Scholar]
- Nader W. F., Edlind T. D., Huettermann A., Sauer H. W. Cloning of Physarum actin sequences in an exonuclease-deficient bacterial host. Proc Natl Acad Sci U S A. 1985 May;82(9):2698–2702. doi: 10.1073/pnas.82.9.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevers P., Saedler H. Transposable genetic elements as agents of gene instability and chromosomal rearrangements. Nature. 1977 Jul 14;268(5616):109–115. doi: 10.1038/268109a0. [DOI] [PubMed] [Google Scholar]
- Nunberg J. H., Kaufman R. J., Chang A. C., Cohen S. N., Schimke R. T. Structure and genomic organization of the mouse dihydrofolate reductase gene. Cell. 1980 Feb;19(2):355–364. doi: 10.1016/0092-8674(80)90510-3. [DOI] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Olmo E., Morescalchi A. Evolution of the genome and cell sizes in salamanders. Experientia. 1975 Jul 15;31(7):804–806. doi: 10.1007/BF01938475. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Owens G. P., Chaudhari N., Hahn W. E. Brain "identifier sequence" is not restricted to brain: similar abundance in nuclear RNA of other organs. Science. 1985 Sep 20;229(4719):1263–1265. doi: 10.1126/science.2412293. [DOI] [PubMed] [Google Scholar]
- Palm D., Goerl R., Burger K. J. Evolution of catalytic and regulatory sites in phosphorylases. Nature. 1985 Feb 7;313(6002):500–502. doi: 10.1038/313500a0. [DOI] [PubMed] [Google Scholar]
- Pearston D. H., Gordon M., Hardman N. Transposon-like properties of the major, long repetitive sequence family in the genome of Physarum polycephalum. EMBO J. 1985 Dec 16;4(13A):3557–3562. doi: 10.1002/j.1460-2075.1985.tb04117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples O. P., Hardman N. An abundant family of methylated repetitive sequences dominates the genome of Physarum polycephalum. Nucleic Acids Res. 1983 Nov 25;11(22):7777–7788. doi: 10.1093/nar/11.22.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples O. P., Robinson A. C., Whittaker P. A., Hardman N. Sequence organisation in nuclear DNA from Physarum polycephalum. Genomic organisation of DNA segments containing foldback sequences. Biochim Biophys Acta. 1983 Nov 17;741(2):204–213. doi: 10.1016/0167-4781(83)90060-x. [DOI] [PubMed] [Google Scholar]
- Peoples O. P., Whittaker P. A., Pearston D., Hardman N. Structural organization of a hypermethylated nuclear DNA component in Physarum polycephalum. J Gen Microbiol. 1985 May;131(5):1157–1165. doi: 10.1099/00221287-131-5-1157. [DOI] [PubMed] [Google Scholar]
- Perlman J. One side of a deletion breakpoint from the Drosophila melanogaster genome contains a transposable element. Gene. 1983 Jan-Feb;21(1-2):87–94. doi: 10.1016/0378-1119(83)90150-6. [DOI] [PubMed] [Google Scholar]
- Perlman S., Phillips C., Bishop J. O. A study of foldback DNA. Cell. 1976 May;8(1):33–42. doi: 10.1016/0092-8674(76)90182-3. [DOI] [PubMed] [Google Scholar]
- Potter S., Truett M., Phillips M., Maher A. Eucaryotic transposable genetic elements with inverted terminal repeats. Cell. 1980 Jul;20(3):639–647. doi: 10.1016/0092-8674(80)90310-4. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G. The mechanism of nucleolar dominance in Xenopus hybrids. Cell. 1984 Aug;38(1):38–44. doi: 10.1016/0092-8674(84)90524-5. [DOI] [PubMed] [Google Scholar]
- Rees H., Jones R. N. Structural basis of quantitative variation in nuclear DNA. Nature. 1967 Nov 25;216(5117):825–826. doi: 10.1038/216825b0. [DOI] [PubMed] [Google Scholar]
- Robertson A. D., Moyzis R. K., Bonnet J., Ts'o P. O. A probabilistic analysis of DNA sequence organization. J Mol Biol. 1981 Dec 25;153(4):864–870. doi: 10.1016/0022-2836(81)90456-3. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. Long interspersed sequences in mammalian DNA. Properties of newly identified specimens. Biochim Biophys Acta. 1985 Feb 20;824(2):113–120. doi: 10.1016/0167-4781(85)90087-9. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Brorein W. J., Jr, Dunsmuir P., Flavell A. J., Levis R., Strobel E., Toole J. J., Young E. Copia-like transposable elements in the Drosophila genome. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):619–628. doi: 10.1101/sqb.1981.045.01.080. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Kidwell M. G., Bingham P. M. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982 Jul;29(3):987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Saigo K., Kugimiya W., Matsuo Y., Inouye S., Yoshioka K., Yuki S. Identification of the coding sequence for a reverse transcriptase-like enzyme in a transposable genetic element in Drosophila melanogaster. Nature. 1984 Dec 13;312(5995):659–661. doi: 10.1038/312659a0. [DOI] [PubMed] [Google Scholar]
- Scherer G., Tschudi C., Perera J., Delius H., Pirrotta V. B104, a new dispersed repeated gene family in Drosophila melanogaster and its analogies with retroviruses. J Mol Biol. 1982 May 25;157(3):435–451. doi: 10.1016/0022-2836(82)90470-3. [DOI] [PubMed] [Google Scholar]
- Schindler C. W., Rush M. G. The KpnI family of long interspersed nucleotide sequences is present on discrete sizes of circular DNA in monkey (BSC-1) cells. J Mol Biol. 1985 Jan 20;181(2):161–173. doi: 10.1016/0022-2836(85)90082-8. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Deininger P. L. Sequence organization of the human genome. Cell. 1975 Nov;6(3):345–358. doi: 10.1016/0092-8674(75)90184-1. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Manning J. E., Davidson N. Inverted repeat sequences in the Drosophila genome. Cell. 1975 Jun;5(2):159–172. doi: 10.1016/0092-8674(75)90024-0. [DOI] [PubMed] [Google Scholar]
- Shafit-Zagardo B., Brown F. L., Zavodny P. J., Maio J. J. Transcription of the KpnI families of long interspersed DNAs in human cells. Nature. 1983 Jul 21;304(5923):277–280. doi: 10.1038/304277a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Conversion of RNA to DNA in mammals: Alu-like elements and pseudogenes. Nature. 1983 Feb 10;301(5900):471–472. doi: 10.1038/301471a0. [DOI] [PubMed] [Google Scholar]
- Shepherd N. S., Schwarz-Sommer Z., Blumberg vel Spalve J., Gupta M., Wienand U., Saedler H. Similarity of the Cin1 repetitive family of Zea mays to eukaryotic transposable elements. Nature. 1984 Jan 12;307(5947):185–187. doi: 10.1038/307185a0. [DOI] [PubMed] [Google Scholar]
- Sherratt D., Arthur A., Burke M. Transposon-specified, site-specific recombination systems. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):275–281. doi: 10.1101/sqb.1981.045.01.040. [DOI] [PubMed] [Google Scholar]
- Shiba T., Saigo K. Retrovirus-like particles containing RNA homologous to the transposable element copia in Drosophila melanogaster. Nature. 1983 Mar 10;302(5904):119–124. doi: 10.1038/302119a0. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Hoffman J., Goff S. P., Baltimore D. Intramolecular integration within Moloney murine leukemia virus DNA. J Virol. 1981 Oct;40(1):164–172. doi: 10.1128/jvi.40.1.164-172.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Skowronski J., Singer M. F. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Unequal crossover and the evolution of multigene families. Cold Spring Harb Symp Quant Biol. 1974;38:507–513. doi: 10.1101/sqb.1974.038.01.055. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Kunes S. M., Schultz D. W., Taylor A., Triman K. L. Structure of chi hotspots of generalized recombination. Cell. 1981 May;24(2):429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- Sobieski D. A., Eden F. C. Clustering and methylation of repeated DNA: persistence in avian development and evolution. Nucleic Acids Res. 1981 Nov 25;9(22):6001–6015. doi: 10.1093/nar/9.22.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Stanfield S. W., Lengyel J. A. Small circular DNA of Drosophila melanogaster: chromosomal homology and kinetic complexity. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6142–6146. doi: 10.1073/pnas.76.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. M., Rothblum K. N., Schwartz R. J. Intron-dependent evolution of chicken glyceraldehyde phosphate dehydrogenase gene. Nature. 1985 Feb 7;313(6002):498–500. doi: 10.1038/313498a0. [DOI] [PubMed] [Google Scholar]
- Straus N. A. Comparative DNA renaturation kinetics in amphibians. Proc Natl Acad Sci U S A. 1971 Apr;68(4):799–802. doi: 10.1073/pnas.68.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Milner R. J., Gottesfeld J. M., Lerner R. A. Identifier sequences are transcribed specifically in brain. Nature. 1984 Mar 15;308(5956):237–241. doi: 10.1038/308237a0. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Function of the retrovirus long terminal repeat. Cell. 1982 Jan;28(1):3–5. doi: 10.1016/0092-8674(82)90367-1. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Origin of retroviruses from cellular moveable genetic elements. Cell. 1980 Oct;21(3):599–600. doi: 10.1016/0092-8674(80)90420-1. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981 Nov;27(1 Pt 2):1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Thomas C. A., Jr The genetic organization of chromosomes. Annu Rev Genet. 1971;5:237–256. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- Timberlake W. E. Low repetitive DNA content in Aspergillus nidulans. Science. 1978 Dec 1;202(4371):973–975. doi: 10.1126/science.362530. [DOI] [PubMed] [Google Scholar]
- Toh H., Kikuno R., Hayashida H., Miyata T., Kugimiya W., Inouye S., Yuki S., Saigo K. Close structural resemblance between putative polymerase of a Drosophila transposable genetic element 17.6 and pol gene product of Moloney murine leukaemia virus. EMBO J. 1985 May;4(5):1267–1272. doi: 10.1002/j.1460-2075.1985.tb03771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Van Arsdell S. W., Denison R. A., Bernstein L. B., Weiner A. M., Manser T., Gesteland R. F. Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell. 1981 Oct;26(1 Pt 1):11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Voliva C. F., Jahn C. L., Comer M. B., Hutchison C. A., 3rd, Edgell M. H. The L1Md long interspersed repeat family in the mouse: almost all examples are truncated at one end. Nucleic Acids Res. 1983 Dec 20;11(24):8847–8859. doi: 10.1093/nar/11.24.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Tabata S., Pachl C. The clustered and scrambled arrangement of moderately repetitive elements in Drosophila DNA. Cell. 1979 Dec;18(4):1231–1246. doi: 10.1016/0092-8674(79)90235-6. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Palindromes in chromosomes. J Mol Biol. 1974 Mar 25;84(1):115–138. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]
- Wyman A. R., Wolfe L. B., Botstein D. Propagation of some human DNA sequences in bacteriophage lambda vectors requires mutant Escherichia coli hosts. Proc Natl Acad Sci U S A. 1985 May;82(9):2880–2884. doi: 10.1073/pnas.82.9.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. W. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z., Bingham P. M. Regulation of white locus expression: the structure of mutant alleles at the white locus of Drosophila melanogaster. Cell. 1982 Sep;30(2):529–541. doi: 10.1016/0092-8674(82)90250-1. [DOI] [PubMed] [Google Scholar]
- von der Helm K. Cleavage of Rous sarcoma viral polypeptide precursor into internal structural proteins in vitro involves viral protein p15. Proc Natl Acad Sci U S A. 1977 Mar;74(3):911–915. doi: 10.1073/pnas.74.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]


