Abstract
The VP22 protein of herpes simplex virus type 2 (HSV-2) is a major component of the virion tegument. Previous work with HSV-1 indicated that VP22 is phosphorylated during infection, and phosphorylation may play a role in modulating VP22 localization in infected cells. It is not clear, however, when phosphorylation occurs in infected cells or how it is regulated. Less is known about the synthesis and phosphorylation of HSV-2 VP22. To study the complete biosynthetic history of HSV-2 VP22, we generated a monoclonal antibody to the carboxy terminus of VP22. Using immunoprecipitation and Western blot analyses, we show that HSV-2 VP22 can be found in three distinct isoforms in infected cells, two of which are phosphorylated. Like HSV-1 VP22, HSV-2 VP22 is synthesized ca. 4 h after infection, and the isoform later incorporated into virions is hypophosphorylated. In addition, we demonstrate for the first time (i) that newly synthesized VP22 is phosphorylated rapidly after synthesis, (ii) that this phosphorylation occurs in a virus-dependent manner, (iii) that the HSV-2 kinase UL13 is capable of inducing phosphorylation of VP22 in the absence of other viral proteins, (iv) that phosphorylated VP22 is very stable in infected cells, (v) that phosphorylated isoforms of VP22 are gradually dephosphorylated late in infection to produce the virion tegument form, and (vi) that this dephosphorylation occurs independently of viral DNA replication or virion assembly. These results indicate that HSV-2 VP22 is a stable protein that undergoes highly regulated, virus-dependent phosphorylation events in infected cells.
The tegument of the herpesvirus virion is an amorphous structure that lies between the capsid and the envelope. The function of this subvirion structure appears to be twofold: to link the capsid of the newly forming viral particle with the envelope and to introduce regulatory proteins into a newly infected cell. The capacity of herpes simplex virus type 1 (HSV-1) and HSV-2 tegument proteins to regulate viral gene expression and to subvert cellular functions has been the focus of considerable study. For example, it is known that the tegument proteins VP16 and vhs transactivate viral α genes and degrade host and viral mRNAs, respectively (14, 15, 22). These and other tegument proteins help to create a cellular environment that is advantageous to virus replication. VP22 is an abundant HSV tegument protein that possesses the remarkable property of intercellular spread (5). VP22 also can associate with and stabilize microtubules (7), interact with the transactivating protein VP16 (6), and associate with chromatin (13, 23). Although the function(s) of VP22 has not been established, its properties suggest that VP22 must play an important role in the HSV life cycle.
Posttranslational modification is a common mechanism to regulate protein localization and function. VP22 is a 300-amino-acid protein that has been shown in both HSV-1 and HSV-2 to be posttranslationally modified by phosphorylation (1, 8, 13, 23). Phosphorylation of HSV-1 VP22 has been implicated in directing VP22 to distinct subcellular regions, with the hypophosphorylated form localizing to the cytoplasm and phosphorylated forms localizing to the nucleus (24). HSV-1 VP22 appears to be phosphorylated early in infection, whereas at later times during infection the majority of VP22 is hypophosphorylated (24). Considerable evidence from in vitro kinase assays has implicated casein kinase II (CKII) in the phosphorylation of HSV-1 VP22 (8), and other data suggest that a virus-associated kinase may also be involved (9). A viral kinase, UL13, has been implicated in inducing phosphorylation of HSV-1 VP22 based on observations that a protein of ca. 38 kDa is phosphorylated in in vitro kinase assays using lysates of cells infected with wild-type virus, but not when the lysates derive from cells infected with UL13− virus (2, 20). However, lack of an observable mobility difference in VP22 from transfected and infected cells led to the conclusion that UL13 does not play a major role in phosphorylation of VP22 (8). Whether UL13 is itself capable of phosphorylating VP22 is unknown. It is also not clear whether phosphorylation of VP22 is regulated, although this seems likely since the fastest-mobility form accumulates late in infection (24) and hypophosphorylated VP22 is preferentially incorporated into extracellular virions (8, 17).
HSV-2 VP22 is highly homologous to HSV-1 VP22, with 65% identity at the amino acid level. The homology shared by these two proteins suggests that they may act in similar ways. Despite the potential importance of VP22 in human disease caused by HSV-2, information about HSV-2 VP22 is limited. It is known to exist in one phosphorylated and one hypophosphorylated form and to be ADP-ribosylated (1). We generated a monoclonal antibody (MAb) to VP22 that is not sensitive to phosphorylation to more fully characterize the phosphorylation pattern of HSV-2 VP22 in the context of the infected cell. Better separation of the HSV-2 VP22 isoforms on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) compared to those of HSV-1 facilitated examination of their phosphorylation pattern. Based on its similarity to HSV-1 VP22, we used HSV-2 VP22 as a model to investigate whether VP22 is differentially phosphorylated at various stages of the HSV-2 infectious cycle and whether this phosphorylation is regulated, possibly to control VP22 localization or function.
MATERIALS AND METHODS
Viruses and cells.
Vero cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). The HSV-2 186 strain was used for most viral infections. HSV-2 strain 5BlacZ contains a lacZ insertion into the UL29 open reading frame of strain 186 that results in an ICP8-null phenotype (3). hr259, an ICP4-deficient mutant of strain 186 (28), was kindly provided by Priscilla Schaffer.
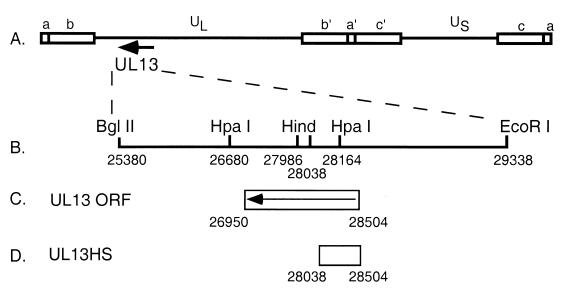
A UL13-null mutant of HSV-1 KOS was constructed as shown in Fig. 1. A 4-kb BglII-EcoRI fragment of the EcoRI D genomic segment that contains the entire UL13 open reading frame was cloned into pGem7 (Promega) to yield plasmid pUL13. This plasmid was cut with HindIII to remove a 52-bp segment and then blunted; a linker was then introduced which contains stop codons in all three reading frames plus a unique HpaI site (29). This mutation was predicted to truncate the carboxy-terminal 70% of the protein. This plasmid, pUL13HS, was then cotransfected into Vero cells with full-length KOS DNA, and the progeny were screened for the presence of a new HpaI site in UL13. Marker rescue of the mutant virus was performed with pUL13. The genotypes of the UL13 mutant virus, UL13HS, and of its marker rescuant, UL13HSMR, were confirmed by Southern blot when restricted with HpaI and probed with pUL13 (data not shown).
FIG. 1.
Construction of a UL13-null mutant of HSV-1 KOS. (A) Linear representation of the HSV-1 genome with the location of the UL13 locus indicated by an arrow. (B) Expanded view of the BglII-EcoRI fragment of the D genomic segment containing the UL13 open reading frame, which was inserted into pGem7. HindIII sites used to remove a 52-bp fragment and replace it with a stop codon linker are indicated. (C) Location of the UL13 open reading frame within the BglII-EcoRI fragment. (D) Coding sequences remaining in UL13HS.
For Western blot analyses, virus infections were carried out in six-well tissue culture plates. Cell monolayers were preincubated on ice for 15 min, and then virus was added in a volume of 300 μl of phosphate-buffered saline (PBS) (supplemented with 2% newborn calf serum and 1% glucose) and incubated on ice for 1 h. The inoculum was removed, and DMEM at 37°C containing 10% FCS was added. This was considered time zero in our experiments. In experiments involving transfection, six-well Vero cell monolayers were transfected with 2 μg of DNA and Lipofectamine reagent (Gibco) according to the procedure recommended by the manufacturer. For metabolic labeling experiments, six-well cell monolayers were preincubated for 30 min in a methionine-cysteine-free medium, and then 50 μCi of 35S-labeled methionine and cysteine ([35S]methionine-cysteine; NEN) was added in a 200-μl volume per well. For phosphate labeling experiments, 500 μCi of [32P]orthophosphate (NEN) was added in a 200-μl volume per well.
Plasmids.
We engineered a bacterial expression plasmid, pRSet-VP22(c150), that expresses the carboxy-terminal 150 amino acids of VP22 from HSV-1 strain KOS with an amino-terminal His (6) tag, by cloning nucleotides 105,944 to 105,485 of HSV-1 strain KOS (based on the sequence of HSV-2 strain 17 [accession no. NC001806]) containing the carboxy-terminal 345 bp of VP22 into the HindIII-NcoI sites of pRSetC (Invitrogen). To generate the plasmids pcDNA-VP22(2) (HSV-2 VP22 under the control of the human cytomegalovirus immediate-early [HCMV IE] promoter), pcDNA-VP22(2)-3′-HA (VP22 fused to the influenza virus hemagglutinin [HA] epitope), and pcDNA-VP22(2)-MKO [pcDNA-VP22(2) with the initial methionine mutated to alanine], VP22 of HSV-2 strain 186 (12) was amplified by PCR with Taq DNA polymerase (Promega), and the PCR product was cloned into the BamHI/EcoRI sites of pcDNA3.1-Zeo+ (Invitrogen). To generate the plasmids pSG5-UL13 and pSG5-US3, the UL13 and US3 open reading frames of HSV-2 strain 186 were amplified by PCR, and the products were cloned into the EcoRI/BamHI sites of pSG5 (Stratagene). PCR-amplified sequences were verified by sequencing.
Antibodies.
His-tagged VP22(c150) for use as an antigen was expressed from the pRSet-VP22(c150) plasmid in Escherichia coli BL21(DE3) and was purified as a denatured protein on nickel-nitrilotriacetic acid-agarose (Qiagen) as recommended by the manufacturer. Purified protein was dialyzed against PBS containing 0.01% SDS and was used to immunize BALB/c mice. Hybridomas were developed by the Saint Louis University Hybridoma Development Service and were screened by Western blot against the immunizing antigen. One clone, designated 22-3, was selected and subcloned based on high specific reactivity and low background; its isotype is immunoglobulin G1 (IgG1). MAb H1119, used to detect HSV ICP27 (16), was the generous gift of Steve Rice. MAbs 12CA5 and 3F10 (Roche) recognize an influenza virus hemagglutinin epitope. The IgG1 mouse MAb specific for duck hepatitis B virus polymerase, MAb 6 (31), was used as an isotype control in immunoprecipitations.
Immunoprecipitation.
Vero cell monolayers were washed twice with sterile PBS, scraped in PBS, and solubilized in radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 1% sodium deoxycholate, 1% SDS, 150 mM NaCl, 20 mM Tris-HCl [pH 7.4]). RIPA buffer was supplemented with 2 μg of aprotinin/ml, 3 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride before use. Cells were incubated in RIPA buffer on ice for 15 min and then clarified at 12,000 × g at 4°C for 15 min. Cleared lysates were incubated with antibody-protein A/G-agarose bead (Oncogene Research) complexes for 3 h at 4°C, followed by three washes with RIPA buffer. Protein was released from the beads by boiling in 2× Laemmli buffer, and eluted protein was analyzed by SDS-PAGE and Western blot.
Western blot analyses.
Whole-cell lysates were solubilized in 2× Laemmli buffer and resolved by SDS-PAGE. Electrophoresed protein was transferred to polyvinylidene difluoride membranes, which were then blocked in Tris-buffered saline–Tween 20 containing 5% nonfat dry milk. 22-3 antibody and secondary antibody incubations were also carried out in this buffer. Anti-mouse antibody conjugated to alkaline phosphatase (Promega) was used as the secondary antibody, and bands were visualized by using NBT/BCIP (Promega) according to the manufacturer's instructions.
Virus purification.
Stocks of partially purified HSV-2 virions were prepared from the supernatant of infected cells as previously described (19). HSV-2 virions were then gradient purified (30). Briefly, virus was resuspended in 1 ml of PBS and layered onto a 12-ml 5 to 15% Ficoll 400 gradient (Sigma). The gradient was centrifuged at 26,000 × g for 2 h at 4°C. Twelve 1-ml fractions were collected, and virus in the fractions was pelleted at 80,000 × g for 2 h at 4°C and resuspended in 200 μl of PBS.
RESULTS
Generation of MAb 22-3 and immunological detection of VP22.
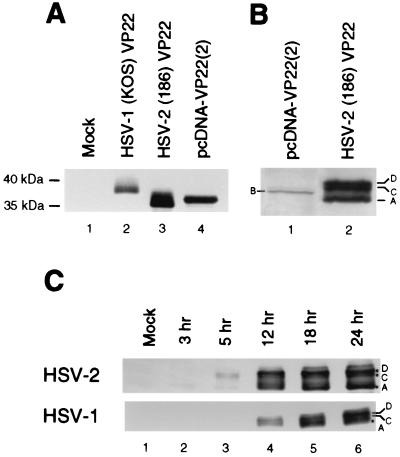
We chose to generate MAbs to the carboxy terminus of VP22 due to the high degree of homology in this region between HSV-1 and HSV-2. A His tag (6) was added to the carboxy-terminal 150 amino acids of VP22 from HSV-1, strain KOS, and the protein was expressed in E. coli, isolated by denaturing nickel affinity chromatography, and used to immunize BALB/c mice. A panel of hybridomas was generated from these mice. One MAb that exhibited high specific reactivity to the immunizing antigen and low background binding (designated 22-3) was tested for cross-reactivity by using Western blots of VP22 derived from HSV-1- or HSV-2-infected cells (Fig. 2). 22-3 exhibited no reactivity to an uninfected Vero cell lysate (Fig. 2A, lane 1) but did recognize VP22 in lysates from either HSV-1- or HSV-2-infected cells (Fig. 2A, lanes 2 and 3, respectively). 22-3 also recognized VP22 from cells transfected with pcDNA-VP22(2), a plasmid containing the HSV-2 VP22 open reading frame under the control of the HCMV IE promoter (Fig. 2A, lane 4). An isotype-matched MAb directed against the duck hepatitis B virus polymerase (MAb 6; IgG1) did not react with VP22 (data not shown).
FIG. 2.
Detection of VP22 with MAb 22-3. (A) 22-3 specifically recognizes HSV-1 and HSV-2 VP22. Vero cells were mock infected (lane 1), infected with HSV-1 strain KOS (lane 2) or HSV-2 strain 186 (lane 3) for 18 h, or transfected with pcDNA-VP22(2) (lane 4). Cells were disrupted in Laemmli buffer, and protein was resolved by SDS-PAGE. VP22 was detected by Western blot with 22-3. (B) Magnified view of HSV-2 VP22 Western analysis. The fastest-migrating form in lane 6 was designated A, and the slower-mobility forms were designated C and D. Transfected cells (lane 5) exhibit a distinct isoform designated B. (C) Vero cells were mock infected (lane 1) or infected with HSV-2 186 (lanes 2 to 6, upper panel) or HSV-1 KOS (lanes 2 to 6, lower panel) at an MOI of 5 for the indicated times. Cells were then lysed by addition of 1× Laemmli buffer, and proteins were resolved by SDS-PAGE and detected by Western blotting with 22-3.
Figure 2B shows a magnified view of a Western blot of pcDNA-VP22(2)-transfected or HSV-2-infected cell lysates probed with 22-3. Bands of four distinct mobilities were evident; these 38-, 37-, 36-, and 35-kDa isoforms were designated D, C, B, and A, respectively. The B isoform was found predominantly in transfected cells (Fig. 2B, lane 1), and the D, C, and A isoforms were present predominantly in HSV-2-infected cells (Fig. 2B, lane 2). A similar isoform pattern was found in HSV-1-infected cells, although the mobility of the entire HSV-1 VP22 cluster was slower (37 to 39 kDa) and more condensed (Fig. 2A, lane 2). The distribution of the various isoforms over time in infected cells did not differ significantly between HSV-1 and HSV-2 (Fig. 2C), although the appearance of the A isoform in HSV-1-infected cells was delayed compared to HSV-2-infected cells (Fig. 2C, lane 4).
HSV-2 VP22 expression begins ca. 4 to 6 h after infection.
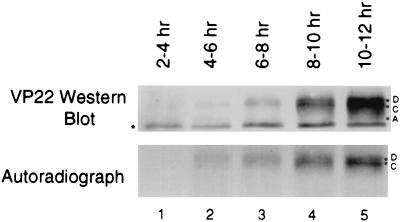
To examine the biosynthesis of HSV-2 VP22, we infected cells at a multiplicity of infection (MOI) of 5 and metabolically labeled the cultures with [35S]methionine-cysteine for 2-h intervals until 12 h postinfection. At the end of each labeling period the cells were washed, lysed, and immunoprecipitated with MAb 22-3. Autoradiographs of the samples demonstrated that 35S-labeled VP22 was detectable starting when cells were labeled from 4 to 6 h after infection (Fig. 3, lane 2), with a gradual increase in labeling intensity over the following 6 h. Additional Western blots verified the time of first expression as 4 h (data not shown). We observed 35S labeling of only the D and C isoforms by autoradiography (Fig. 3), although a small amount of the A isoform was detected by Western blot in the 10- to 12-h samples (Fig. 3, lane 5). This suggested that the A isoform was not produced until later in infection or that the A isoform was derived from the D and/or C isoforms that had been made earlier in infection. The fastest-migrating band on the Western blot (Fig. 3, asterisk) is an immunoprecipitation artifact also present in control lanes (Fig. 4A).
FIG. 3.
Expression of HSV-2 VP22 begins 4 to 6 h after infection. Vero cells were infected with HSV-2 at an MOI of 5. Infected cells were incubated with [35S]methionine-cysteine for the times indicated. After incubation with the label, cells were lysed in RIPA buffer, and equal volumes were immunoprecipitated with 22-3. VP22 was detected by Western analysis by using 22-3, and 35S was detected by autoradiography. The band labeled with an asterisk is an artifact observed during immunoprecipitation.
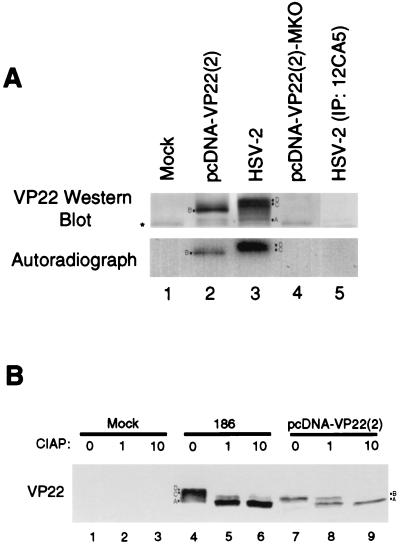
FIG. 4.
HSV-2 VP22 is a phosphoprotein. (A) HSV-2 can be labeled with 32P. Vero cells were mock infected, infected with HSV-2 at an MOI of 5, or transfected with pcDNA-VP22(2) or pcDNA-VP22(2)-MKO. Cells were incubated with [32P]orthophosphate and lysed in RIPA buffer. Equal volumes were immunoprecipitated with the MAbs 22-3 (lanes 1 to 4) or 12CA5 (lane 5), and precipitated proteins were resolved by SDS-PAGE. VP22 was detected by Western analysis by using MAb 22-3, and 32P was detected by autoradiography. (B) HSV-2 VP22 is sensitive to CIAP. Cells were mock infected (lanes 1 to 3), infected with HSV-2 (lanes 4 to 6), or transfected with pcDNA-VP22(2) (lanes 7 to 9). Cells were lysed in RIPA buffer, and equal volumes were mock treated or treated with 1 or 10 U of CIAP for 30 min at 37°C. Proteins were resolved by SDS-PAGE, and VP22 was detected by Western analysis with MAb 22-3.
HSV-2 VP22 is rapidly phosphorylated after expression.
The slower-migrating isoforms of HSV-1 VP22 are phosphorylated, but the fastest-migrating form is not phosphorylated (8, 24). A single phosphorylated HSV-2 VP22 isoform has been previously described (1). In our metabolic labeling experiment, the D and C isoforms of HSV-2 VP22 were immediately detected. If their slower migration was due to phosphorylation, this would suggest that HSV-2 VP22 was phosphorylated very rapidly after synthesis.
To determine whether newly expressed HSV-2 VP22 was phosphorylated, we tested the capacity of the D and C isoforms of HSV-2 VP22 to incorporate [32P]orthophosphate (Fig. 4A). Vero cell monolayers were mock transfected or transfected with pcDNA-VP22(2) (wild-type VP22) or with pcDNA-VP22(2)-MKO (VP22 start codon mutant). Alternatively, cells were infected with HSV-2. Cultures were labeled with 32P from 36 to 38 h posttransfection or from 6 to 8 h postinfection. Cells transfected with pcDNA-VP22(2) (Fig. 4A, lane 2) showed 32P incorporation into the B isoform. Cells infected with HSV-2 (Fig. 4A, autoradiograph, lane 3) incorporated 32P into the D and C isoforms. The A isoform was present in the Western blot (Fig. 4A, lanes 2 and 3), but it did not contain 32P, indicating that it was not phosphorylated during the labeling period. Because we frequently detect a low level of the A isoform (in cells infected at an MOI of ≥5) prior to the time when it is detected by metabolic labeling (10 to 12 h postinfection; Fig. 3, lane 5, bottom band), the A isoform detected here most likely was derived from the input virus. To confirm phosphorylation of VP22, we used calf intestinal alkaline phosphatase (CIAP) to dephosphorylate the protein. Treatment of infected cell lysates with increasing concentrations of CIAP resulted in loss of the D and C isoforms and accumulation of the A isoform (Fig. 4B, lanes 4 to 6). Similarly, CIAP treatment of transfected cell lysates resulted in loss of the B isoform and accumulation of the A isoform (Fig. 4B, lanes 7 to 9), indicating that the A isoform is either not phosphorylated or is hypophosphorylated. Thus, the VP22 D and C isoforms in HSV-2-infected cells and the B isoform in transfected cells are phosphorylated isoforms of VP22 and, because the D and C isoforms are the earliest detected isoforms of VP22 in HSV-2-infected cells, we conclude that VP22 is phosphorylated very rapidly after translation.
Phosphorylation to generate the D and C isoforms is dependent on infection.
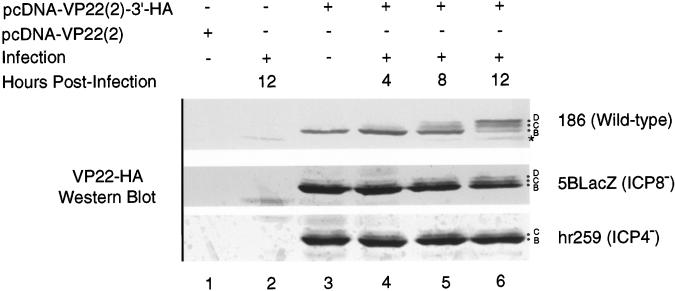
To examine the contribution of viral infection to phosphorylation of VP22, we tagged the carboxy terminus of VP22 with an influenza virus HA epitope to permit detection of transfected VP22 during viral infection (Fig. 5). Cells were mock transfected, transfected with pcDNA-VP22(2), or transfected with pcDNA-VP22(2)-3′-HA. Cells were then either mock infected or infected with HSV-2 186 strain for the indicated times. Cell lysates were prepared and Western blotted with MAb 12CA5 to detect the HA-tagged VP22. 12CA5 did not react with nontagged VP22 (Fig. 5, lane 1) and had minimal cross-reactivity to a protein in HSV-2-infected cells of 34 kDa (Fig. 5, lane 2). Cells transfected with HA-tagged VP22 exhibited a single band at 36 kDa (Fig. 5, lane 3). In cells infected with wild-type HSV-2, there was a decrease in the mobility of the HA-tagged VP22 that began at 4 h after infection and continued until at least 12 h after infection (Fig. 5, lanes 4 to 6). The two additional isoforms of VP22(2)–3′-HA apparent after infection are reminiscent of the D and C isoforms in infected cells. Treatment of the samples from lanes 3 and 6 with CIAP yielded a single band that migrated faster than the HA-tagged VP22 produced in transfected cells in the absence of HSV-2 infection (data not shown), suggesting that the HA-tagged bands in Fig. 5, lane 6, are equivalent to the phosphorylated D, C, and B isoforms of VP22. These data further suggest that during infection either a viral kinase or cellular kinase activated by HSV-2 phosphorylates VP22 differently than VP22 is phosphorylated in transfected cells.
FIG. 5.
Phosphorylation to the D and C isoforms is dependent on infection. Vero cells were mock transfected or transfected with pcDNA-VP22(2)–3′-HA. At 24 h posttransfection, cells were either mock infected or infected with 186, hr259, or 5BlacZ at an MOI of 1 for the indicated times. Cells were lysed in 2× Laemmli buffer, and proteins were resolved by SDS-PAGE. HA-tagged VP22 was detected by Western analysis using MAb 12CA5. The fastest-migrating band (✽) is an irrelevant protein that reacts with 12CA5.
To determine the kinetic class of viral gene products that may activate phosphorylation to the D and C isoforms, we transfected cells with the HA-tagged VP22 and then infected them with wild-type HSV-2 186, hr259 or 5BlacZ viruses. hr259 is an ICP4−/− virus derived from 186 that arrests viral gene expression after the α gene class (28). 5BlacZ is a replication-defective virus derived from strain 186 that produces all HSV-2 gene products except ICP8 and the γ2 proteins (3). In transfected cells subsequently infected with hr259, the D isoform was not observed, and the relative proportions of the B and C isoforms of HA-tagged VP22 were not changed by infection (Fig. 5, lanes 4 to 6). In contrast, transfected cells infected for 8 h with 5BlacZ showed a shift in the phosphorylation pattern to a greater proportion of the C isoform (and faint D isoform) relative to the amount of B isoform (Fig. 5, lane 5), similar to that seen in cells infected with wild-type virus. By 12 h after infection, the D isoform had become apparent and the amounts of the D and C isoforms relative to the B isoform had further increased (Fig. 5, lane 6). The C and D isoforms were, however, less prominent in cells infected with 5BlacZ compared to cells infected with wild-type virus. These results suggest that either ICP4 or a β or γ1 gene product is required for efficient phosphorylation of VP22 to the D and C isoforms.
Slower mobility isoforms of transfected HSV-2 VP22 are not produced upon infection with UL13− virus.
The increased phosphorylation of VP22 during infection compared with transfection may be attributed to either a virus-encoded kinase or to activation of a cellular kinase upon virus infection. Previous reports concerning the activity of the UL13 kinase indicate that a protein of ca. 38 kDa in HSV-1-infected cell extracts becomes heavily phosphorylated during in vitro kinase assays and that this protein is hypophosphorylated in extracts of cells infected with UL13-deficient virus (2, 20). We therefore sought direct evidence that UL13 could phosphorylate VP22 in vivo.
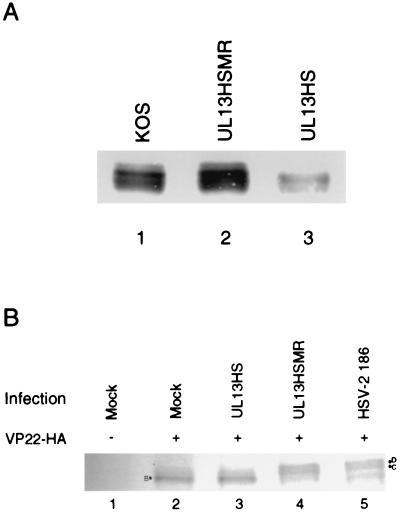
First, we examined the capacity of a UL13 kinase-deficient virus to phosphorylate VP22 in infected cells. No HSV-2 strains deficient in UL13 activity have been reported but, due to the similarity between HSV-1 and HSV-2 VP22, we used the UL13− HSV-1 strain, UL13HS, and its marker rescuant, UL13HSMR. Western blot of lysates from Vero cells infected for 12 h with wild-type HSV-1 KOS or UL13HSMR revealed the expected cluster of VP22 isoforms (Fig. 6A, lanes 1 and 2). Cells infected with UL13HS, however, did not contain the slowest-mobility isoform(s) of VP22 (Fig. 6A, lane 3). Prolonged infection of cells with UL13HS did not lead to the appearance of the slowest-mobility isoform(s) (data not shown). UL13− viruses have a general defect in late gene expression (26). Therefore, to confirm these observations, we transfected cells with pcDNA-VP22(2)–3′-HA and then infected the cells with UL13HS, UL13HSMR, or wild-type HSV-2 strain 186 (Fig. 6B) and used an HA-specific MAb to detect transfected VP22. Cells transfected with pcDNA-VP22(2)–3′-HA only or transfected and then infected with the UL13− mutant produce only the B isoform (Fig. 6B, lanes 2 and 3). Cells transfected with pcDNA-VP22(2)–3′-HA and infected with either UL13HSMR or HSV-2 produce both the D and C isoforms (Fig. 6B, lanes 4 and 5). These data indicate that UL13, in combination with a cellular kinase (that generates the B isoform in transfected cells), is necessary for the phosphorylation of VP22 in infected cells through either direct or indirect mechanisms.
FIG. 6.
(A) UL13− virus does not generate the slowest-mobility isoform(s) of VP22 in infected cells. Vero cell monolayers were infected at an MOI of 5 with wild-type HSV-1 KOS (lane 1), UL13HSMR (lane 2), or UL13HS (lane 3). At 12 h postinfection, cells were disrupted in Laemmli buffer, and proteins were resolved by SDS-PAGE. Transferred protein was detected by Western blot with 22-3. (B) UL13− virus does not phosphorylate transfected VP22 to the D and C isoforms. Vero cells were mock transfected (lane 1) or transfected with pcDNA-VP22(2)–3′-HA (lanes 2 to 5). At 16 h after transfection, the cells were mock infected (lanes 1 and 2) or infected with UL13HS (lane 3), UL13HSMR (lane 4), or HSV-2 (lane 5) at an MOI of 5. Cells were infected for 16 h and disrupted in 1× Laemmli buffer, and proteins were resolved by SDS-PAGE. HA-tagged VP22 was detected by Western analysis by using MAb 3F10.
Cotransfection of HSV-2 UL13 and VP22 expression plasmids increases VP22 phosphorylation.
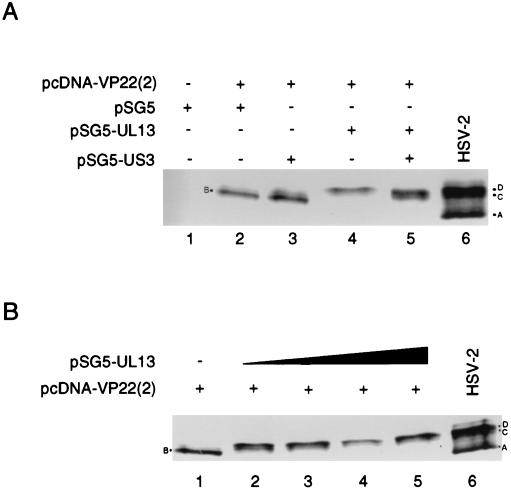
We next determined whether either UL13 or a second viral kinase, US3, would be sufficient for generation of the D and C isoforms of VP22. Vero cell monolayers were transfected with pcDNA-VP22(2) alone or in combination with plasmids expressing the HSV-2 UL13 or US3 kinases. Transfection of pcDNA-VP22(2) or pcDNA-VP22(2) and pSG5-US3 resulted in the characteristic B isoform (Fig. 7A, lanes 2 and 3), whereas cells cotransfected with pcDNA-VP22(2) and pSG5-UL13 contained the D isoform (Fig. 7A, lane 4). Transfection of cells with pcDNA-VP22(2), pSG5-UL13, and pSG5-US3 resulted in the total loss of the B isoform and greatly increased accumulation of the D and C isoforms. These data indicate that UL13 but not US3 is able to induce VP22 phosphorylation in the absence of all other viral proteins.
FIG. 7.
(A) UL13 can induce the phosphorylation of VP22. Vero cells were mock transfected (lane 1) or transfected with pcDNA-VP22(2) in combination with pSG5 (empty vector, lane 2), pSG5-UL13 (lane 3), pSG5-US3 (lane 4), or pSG5-UL13 plus pSG5-US3 (lane 5). Transfections were incubated for 24 h and lysed with 1× Laemmli buffer, and proteins were resolved by SDS-PAGE. Lysates from HSV-2-infected cells are shown in lane 6. VP22 was detected by Western analysis with MAb 22-3. (B) VP22 phosphorylation is responsive to the concentration of cotransfected UL13-expressing plasmid. Vero cells were transfected with pcDNA-VP22(2) alone (lane 1) or with increasing amounts of pSG5-UL13 (125 ng, lane 2; 250 ng, lane 3; 500 ng, lane 4; 1,000 ng, lane 5). Transfected cells were incubated for 24 h and lysed with 1× Laemmli buffer, and proteins were resolved by SDS-PAGE. VP22 was detected by Western analysis by using MAb 22-3.
Comparison of the isoform patterns in lanes 4 and 5 from Fig. 7A suggested that phosphorylation of VP22 was sensitive to UL13 levels because only half as much UL13-expressing plasmid was used during cotransfection with US3-expressing plasmid. Alternatively, generation of the C isoform was dependent on the presence of both kinases. We therefore examined VP22 mobility in the presence of increasing amounts of pSG5-UL13. Vero cells were transfected with pcDNA-VP22(2) alone (Fig. 7B, lane 1) or were cotransfected with increasing amounts of pSG5-UL13 (Fig. 7B, lanes 2 to 5). At a low concentration of the UL13-expressing plasmid, VP22 was found in both D and C isoforms (Fig. 7B, lane 2), but increasing concentrations of pSG5-UL13 drove VP22 phosphorylation into the D isoform. Increasing concentrations of pSG5-US3 did not alter VP22 phosphorylation (data not shown). These results suggest that UL13 concentration is critical to the accumulation of the D and C isoforms in HSV-infected cells and that US3 apparently is not necessary for VP22 phosphorylation.
VP22 is a stable protein that is gradually dephosphorylated.
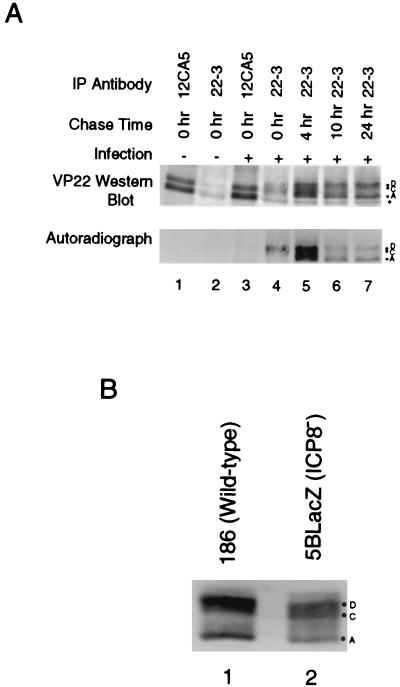
Pulse-chase experiments were performed on infected cell monolayers to examine the stability of VP22 during infection. HSV-2-infected cells were pulsed with [35S]methionine-cysteine from 6 to 8 h postinfection and were chased with unlabeled medium for various times up to 32 h postinfection. Cells were then lysed and immunoprecipitated with 22-3 or with 12CA5 (a control antibody). The label was exclusively incorporated into the D and C isoforms immediately after the labeling period (Fig. 8A, lane 4), as seen in Fig. 3. At later times, however, some of the label was chased into a band migrating as the hypophosphorylated A isoform of VP22 (Fig. 8A, lanes 5, 6, and 7). This result indicates that VP22 is very stable because it could be detected for at least 24 h after it was synthesized. Furthermore, because the pulse-chase experiments first detected labeled VP22 in the D and C isoforms, and only after prolonged chase was the A isoform detected, we conclude that at least some the A isoform is derived from the D and/or the C isoforms during the course of infection. In light of the results in Fig. 4, the A isoform is most likely generated by gradual dephosphorylation of the D and/or C isoforms beginning ca. 10 to 12 h postinfection.
FIG. 8.
(A) HSV-2 VP22 is a stable protein that is gradually dephosphorylated. Vero cells were mock infected or infected with HSV-2 and then labeled with [35S]methionine-cysteine from 6 to 8 h postinfection. Labeling medium was replaced with label-free medium, and cells were incubated for the indicated times. Cells were lysed in RIPA buffer, equal volumes of the lysates were immunoprecipitated with 22-3 (lanes 2, 4, 5, 6, and 7) or 12CA5 (lanes 1 and 3), and proteins were resolved by SDS-PAGE. VP22 was detected by Western analysis with 22-3, and 35S was detected by autoradiography. (B) Dephosphorylation of VP22 occurs independently of viral DNA replication and virion assembly. Vero cells were infected at an MOI of 5 for 12 h with HSV-2 186 (lane 1) or 5BlacZ (lane 2). Cells were then lysed in 1× Laemmli buffer, and proteins were resolved by SDS-PAGE. VP22 was detected by Western analysis with MAb 22-3.
To determine whether dephosphorylation of VP22 to the A isoform in infected cells is dependent on a virus-encoded product, we infected Vero cells for 12 or 24 h with wild-type HSV-2 or the 5BlacZ mutant virus and then analyzed the lysates by Western blot by using 22-3. At 12 h postinfection, the D and C isoforms predominated in wild-type-virus-infected cells and in cells infected with 5BlacZ, but the A isoform was also present (Fig. 8B). By 24 h after infection, significant accumulation of the A isoform had occurred in cells infected with wild-type virus and also with the ICP8 mutant virus (data not shown). Therefore, dephosphorylation of VP22 late in infection occurs independently of γ2 protein synthesis, viral DNA synthesis, and assembly of viral particles.
The A isoform is associated with infectious virions.
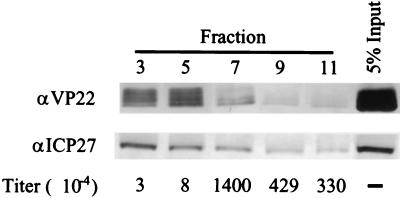
A hypophosphorylated form of HSV-1 VP22 is incorporated into virions (8, 17). Since the A isoform of HSV-2 VP22 is found late in infection and is hypophosphorylated, it seemed likely that the HSV-2 A isoform would also be incorporated into virions. To test this possibility, we purified HSV-2 virions derived from infected cell supernatants on a 5 to 15% Ficoll gradient and assayed the resulting fractions for infectivity, for the presence of VP22, and for the presence of ICP27, a viral protein not associated with virions (16) (Fig. 9). Infectivity was 100- to 1,000-fold greater in the denser half of the gradient (Fig. 9, fractions 7 to 11) with comparatively little present in the lighter fractions (Fig. 9, fractions 3 and 5), indicating that mature virions sedimented in the denser portion of the gradient. All fractions of the gradient contained VP22; however, the D, C, and A isoforms were not evenly distributed throughout the gradient. The D and C isoforms were predominant in the lighter fractions (Fig. 9, fractions 3 and 5), and the A isoform was predominant in the denser fractions (Fig. 9, fractions 7 to 11). This indicated that the greatest infectivity was associated with the A isoform in fractions that did not contain significant amounts of the D and C isoforms, whereas less-dense structures associated with the D and C isoforms contained very little infectious virus. Western blot analysis showed the majority of ICP27 to be in the lighter fractions (Fig. 9, fractions 3 and 5), although some was also found in the denser fractions, most likely bound to cellular structures. These results suggested that nonparticulate proteins remained predominantly in the upper region of the gradient, separate from the infectious virions found predominantly in the bottom half of the gradient. Therefore, we conclude that HSV-2 virions preferentially incorporate the A isoform of VP22.
FIG. 9.
The VP22 A isoform is associated with infectivity. Supernatants from infected cell cultures were centrifuged to concentrate virus, and the mixture was fractionated by sedimentation through a Ficoll gradient. Titers of fractions were determined to assess infectivity, and the fractions were analyzed by Western blot for the presence of VP22 and ICP27.
DISCUSSION
VP22 of HSV-1 and HSV-2 has been previously described as a phosphoprotein that exists in three and two mobility forms, respectively, in SDS-PAGE of infected cell lysates (1, 24). The phosphorylation of HSV-1 VP22 can occur early after synthesis, within 5 to 7 h of infection (24). At this time postinfection, HSV-1 VP22 exists in its slowest-mobility isoform, but it is later found in faster-migrating forms (24). The form incorporated into the extracellular virion is a hypophosphorylated form of rapid mobility (8, 17). These observations have suggested regulated phosphorylation of HSV-1 VP22. We have demonstrated here that phosphorylation and dephosphorylation of HSV-2 VP22 indeed occur in a highly regulated fashion. We have now completed the picture of the biosynthetic history of VP22 by demonstrating that phosphorylation of VP22 occurs essentially coincident with its synthesis, beginning 4 to 5 h postinfection, and that phosphorylation of VP22 to the slowest-migrating isoforms found in infected cells occurs in a virus-dependent manner. In addition, we have demonstrated that the phosphorylated, slowest-mobility isoforms of VP22 are gradually dephosphorylated to a hypophosphorylated isoform, which is the form eventually incorporated into virions, and that dephosphorylation of the D and C isoforms occurs independent of virion assembly. Both the phosphorylated and hypophosphorylated isoforms of VP22 are very stable in infected cells. Finally, we have shown that the UL13 viral kinase can stimulate phosphorylation of VP22 in transfected cells in the absence of other viral proteins. Because HSV-1 and HSV-2 VP22 share many similar properties, this complete view of the life cycle of HSV-2 VP22 may also be largely applicable to VP22 of HSV-1.
Only two distinct VP22 isoforms were previously detected in HSV-2-infected cells, one of which is phosphorylated (1). We demonstrate that at least three isoforms of HSV-2 VP22 exist in infected cells, two of which are phosphorylated. The number and pattern of VP22 isoforms in HSV-2 infected cells is similar to the pattern reported in HSV-1-infected cells (24). The kinase activity mediating phosphorylation of VP22 is a matter of conjecture. HSV-2 VP22 expressed via transfection is phosphorylated and migrates more slowly than the hypophosphorylated form incorporated into virions, indicating that a cellular kinase acts upon newly synthesized VP22. Elliott et al. have provided strong evidence that the cellular kinase CKII is involved in phosphorylation of VP22 because phosphorylation occurs at CKII consensus sites (9) and CKII can phosphorylate VP22 in vitro (8). The conclusion that phosphorylation of VP22 depends predominantly on a cellular kinase is based on the interpretation that the phosphorylated form of HSV-1 VP22 found in transfected cells is indistinguishable from that found in infected cells (8). We have now shown with HSV-2 VP22 that the mobility form found in transfected cells is different from the two phosphorylated isoforms found in infected cells and that phosphorylation to the D and C isoforms is dependent on a viral gene product. Our observation that VP22 is hypophosphorylated in UL13− virus-infected cells relative to cells infected with wild-type virus argues that the presence of the UL13 viral kinase affects phosphorylation of VP22 in vivo. This effect could be a direct phosphorylation of VP22 by UL13 or an indirect effect such as UL13-mediated activation and/or redirection of CKII.
Our observation that UL13 induces phosphorylation of VP22 in the absence of other viral proteins suggests that direct phosphorylation to the D and C isoforms in vivo by UL13 is a viable possibility. UL13 is synthesized as a γ1 gene product and enters newly infected cells as a component of the virion tegument (21). The observation that phosphorylation of tegument proteins, including VP22, leads to dissociation of the tegument complex (18) is consistent with a role for UL13 in VP22 phosphorylation. In addition, our observation that infection of VP22-transfected cells with ICP8− but not ICP4− HSV-2 activates efficient phosphorylation to the D and C isoforms is consistent with a requirement for a γ1 (or a β) gene product. Finally, we have demonstrated in cotransfection experiments that phosphorylation of VP22 by UL13 to the D and C isoforms is sensitive to the amount of UL13-expressing plasmid in the cell. Because some γ1 gene products are less abundant in cells infected with ICP8 mutant viruses (11), involvement of UL13 in in vivo phosphorylation of VP22 suggests a possible explanation for our observation that the levels of the D and C isoforms are reduced relative to the B isoform in transfected cells subsequently infected with 5BlacZ (Fig. 5, lane 6). It must be emphasized, however, that whether UL13 directly phosphorylates VP22 to the D and C isoforms or indirectly activates or causes relocation of a cellular kinase remains unresolved. Finally, we found no effect of the US3 viral kinase on transfected, HA-tagged VP22, alone or in combination with UL13, in agreement with previous observations that the phosphorylation of a ca. 38-kDa protein is unchanged in US3-deficient HSV-1- or HSV-2-infected cells (4, 27).
VP22 is a component of the virion tegument, and we demonstrated that, as in HSV-1 (8), the hypophosphorylated A isoform of HSV-2 VP22 is incorporated into virus particles. This suggests that VP22 should be relatively stable in the infected cell and, indeed, we can detect VP22 at least 24 h after its synthesis. Interestingly, at least a portion of the D and C isoforms, as well as the A isoform, is stable for long periods after synthesis in HSV-2-infected cells. Pulse-chase experiments revealed that the D and C isoforms of VP22 are gradually dephosphorylated, resulting in the accumulation of the A isoform late in infection. The A isoform accumulates in cells infected with wild-type virus or ICP8-deficient virus, but not in VP22-transfected cells. Thus, the phosphatase activity most likely is provided by the host cell but is activated by virus infection independently of viral DNA replication or initiation of virion assembly, neither of which occurs in cells infected with ICP8-deficient virus (10). Curiously, we have not observed significant accumulation of an A isoform of transfected HA-tagged VP22 in cells subsequently infected with HSV-2. Further experiments will be necessary to determine the reason for this difference.
The regulation of HSV-2 VP22 phosphorylation is intricate, since two separate phosphorylated isoforms and one hypophosphorylated isoform of the protein can be identified in infected cells, and a unique phosphorylated isoform is found after transfection. The scheme is further complicated by dephosphorylation of the D and C isoforms to produce the A isoform that is incorporated into virions. The complexity of these phosphorylation events and the fact that, after its synthesis, VP22 is found in phosphorylated forms throughout infection imply that VP22 may play more than just a structural role in the HSV-2 replication cycle. Although all attempts to completely ablate VP22 synthesis have failed to produce virus, it has been recently reported that infectious HSV-1 can be produced without incorporating full-length VP22 (25). Incorporation of modest changes into the UL49 gene may thus be a feasible means of determining the role of VP22 in HSV biology. Studies to further dissect how VP22 phosphorylation is regulated and how this affects function of the protein are in progress.
ACKNOWLEDGMENTS
We thank Priscilla Schaffer for providing the hr259 virus and Steve Rice for the H1119 antibody. The technical assistance of Jon Halling, John Patton, Amy Ruff, and Rob Reass is greatly appreciated. Helpful discussions with Sam Speck, Skip Virgin, and members of their laboratories are gratefully acknowledged, as are useful discussions with members of the Morrison and Leib laboratories.
This work was supported by funds from Saint Louis University to J.E.T. and L.A.M. and by PHS awards EY10707 to D.A.L. and P30-EY02689 to the Department of Ophthalmology at Washington University.
REFERENCES
- 1.Blaho J, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 α regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL49 gene products. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]
- 2.Coulter L J, Moss H W, Lang J, McGeoch D J. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 3.Da Costa X J, Bourne N, Stanberry L R, Knipe D M. Construction and characterization of a replication-defective HSV-2. Virology. 1997;232:1–12. doi: 10.1006/viro.1997.8564. [DOI] [PubMed] [Google Scholar]
- 4.Daikoku T, Kurachi R, Tsurumi T, Nishiyama Y. Identification of a target protein of US3 protein kinase of herpes simplex virus type 2. J Gen Virol. 1994;75:2065–2068. doi: 10.1099/0022-1317-75-8-2065. [DOI] [PubMed] [Google Scholar]
- 5.Elliot G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1998;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 6.Elliott G, Mouzakitis G, O'Hare P. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol. 1995;69:7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott G, O'Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott G, O'Reilly D, O'Hare P. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology. 1996;226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 9.Elliott G, O'Reilly D, O'Hare P. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J Virol. 1999;73:6203–6206. doi: 10.1128/jvi.73.7.6203-6206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao M, Knipe D M. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J Virol. 1989;63:5258–5267. doi: 10.1128/jvi.63.12.5258-5267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao M, Knipe D M. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J Virol. 1991;65:2666–2675. doi: 10.1128/jvi.65.5.2666-2675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall L M, Draper K G, Frink R J, Costa R H, Wagner E K. Herpes simplex virus mRNA species mapping in EcoRI fragment I. J Virol. 1982;43:594–607. doi: 10.1128/jvi.43.2.594-607.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knopf K, Kaerner H. Virus-specific basic phosphoproteins associated with herpes simplex virus type 1 (HSV-1) particles and the chromatin of HSV-1-infected cells. J Virol. 1980;46:405–414. doi: 10.1099/0022-1317-46-2-405. [DOI] [PubMed] [Google Scholar]
- 14.Kwong A D, Frenkel N. The herpes simplex virus virion host shutoff function. J Virol. 1989;63:4834–4839. doi: 10.1128/jvi.63.11.4834-4839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong A D, Frenkel N, Kruper J A. Herpes simplex virus virion host shutoff function. Proc Natl Acad Sci USA. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mears W E, Lam V, Rice S A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J Virol. 1995;69:935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meredith D M, Lindsay J A, Halliburton I W, Whittaker G R. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J Gen Virol. 1991;72:2771–2775. doi: 10.1099/0022-1317-72-11-2771. [DOI] [PubMed] [Google Scholar]
- 18.Morrison E E, Wang Y F, Meredith D M. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison L A, Da Costa X J, Knipe D M. Influence of mucosal and parenteral immunization with a replication-defective mutant of HSV-2 on immune responses and protection from genital challenge. Virology. 1998;243:178–187. doi: 10.1006/viro.1998.9047. [DOI] [PubMed] [Google Scholar]
- 20.Overton H, McMillan D, Hope L, Wong-Kai-In P. Production of host shutoff-defective mutants of herpes simplex virus type 1 by inactivation of the UL13 gene. Virology. 1994;202:97–106. doi: 10.1006/viro.1994.1326. [DOI] [PubMed] [Google Scholar]
- 21.Overton H, McMillan D J, Klavinskis L S, Hope L, Ritchie A J, Wong-Kai-In P. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology. 1992;190:184–192. doi: 10.1016/0042-6822(92)91204-8. [DOI] [PubMed] [Google Scholar]
- 22.Pellett P E, McKnight J L C, Jenkins F J, Roizman B. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing α genes. Proc Natl Acad Sci USA. 1985;82:5870–5874. doi: 10.1073/pnas.82.17.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinard M F, Simard R, Bibor-Hardy V. DNA-binding proteins of herpes simplex virus type 1-infected BHK cell nuclear matrices. J Gen Virol. 1987;68:727–735. doi: 10.1099/0022-1317-68-3-727. [DOI] [PubMed] [Google Scholar]
- 24.Pomeranz L E, Blaho J A. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pomeranz L E, Blaho J A. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J Virol. 2000;74:10041–10054. doi: 10.1128/jvi.74.21.10041-10054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purves F C, Ogle W O, Roizman B. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purves F C, Spector D, Roizman B. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J Virol. 1991;65:5757–5764. doi: 10.1128/jvi.65.11.5757-5764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith C A, Schaffer P A. Mutants defective in herpes simplex virus type 2 ICP4: isolation and preliminary characterization. J Virol. 1987;61:1092–1097. doi: 10.1128/jvi.61.4.1092-1097.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strelow L I, Leib D A. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J Virol. 1995;69:6779–6786. doi: 10.1128/jvi.69.11.6779-6786.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szilagyi J F, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 31.Yao E, Gong Y, Chen N, Tavis J E. The majority of duck hepatitis B virus reverse transcriptase in cells is nonencapsidated and is bound to a cytoplasmic structure. J Virol. 2000;74:8648–8657. doi: 10.1128/jvi.74.18.8648-8657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]