ABSTRACT
We present a metagenome assembled genome (MAG) of an anaerobic bacterium from a nitrate-reducing, benzene-degrading enrichment culture (NRBC). The draft Thermincolales genome consists of 20 contigs with a total length of 4.09 Mbp and includes putative carboxylase genes likely involved in benzene activation.
KEYWORDS: anaerobic, benzene, nitrate reduction, metagenome assembled genome, Thermincolales, carboxylase
ANNOUNCEMENT
NRBC [previously known as Cartwright-NO3 (1)] is a nitrate-reducing, benzene-degrading microbial enrichment culture established in 1995 from a gasoline-contaminated site (Latitude: 43.722647, Longitude: −79.463022), and is grown in a defined anaerobic mineral medium repeatedly amended with benzene (300–400 µM) and ~2 mM nitrate (2, 3). A benzene-degrading bacterium first classified as a Peptococacceae by 16S rRNA gene sequence analysis (NCBI accession: KJ522755.1) was identified in the mixed culture (1); it was later grouped within the Thermincola genus (4) using the SILVA SSU 138 database (5). Here, we report a near-complete genome of this uncultured benzene-degrading bacterium to better understand its phylogeny and metabolism.
The assembly of the Thermincola MAG used multiple previously reported data sets (6). Illumina paired-end (NCBI accession: SRR24043423) and mate-pair (NCBI accession: SRR24043417) reads were obtained in 2013 from an NRBC subculture called CartCons19. Paired-end reads were sequenced using the TrueSeq DNA Library Prep kit LT, and mate-pair reads were sequenced using the Nextera Mate Pair Library Preparation Kit, according to the manufacturer’s (Illumina) instructions with no additional quality assurance measures. All raw reads were processed using Trimmomatic v. 0.32 (7) before using Abyss v. 1.3.7 (8) to create unitigs that were merged with scaffolds generated in ALL-PATHS-LG v. 4.7.0 (9) using a gap-filling Perl script (10) based on the script in Text S1 of Tang et al. (11). Due to the high number of undefined nucleotides in this metagenome assembly (JARXNP010000000), further steps were taken. In 2018, a NRBC subculture (FeS-Dialysis) was sequenced using the HiSeq PE Cluster Kit v4 cBot (Illumina) with no additional quality control measures (NCBI accession: SRR24043422) and was assembled using IDBA v. 1.1.1 (12) and binned using metaBAT v. 2.12 (13). A bin assigned to Thermincola in 157 contigs (NCBI accession: JAVSMV000000000.1) was retrieved as reported previously (6). These 157 contigs were incorporated into the above Abyss/ALL-PATHS-LG gap-filling workflow generating a 26-contig assembly that was curated by read mapping using BBMap v. 38.94 (14) to resolve ambiguities. Finally, long reads from a 2020 NRBC subculture called 10L-NRBC, sequenced according to the manufacturer’s instructions using PacBio RSII with the SMRTbell Express Template Prep Kit 2.0 (SRR24043419) without shearing or size selection (Pacific Biosciences), were used to join adjacent contigs using the de novo assembly tool in Geneious v. 8.1.8 (15), resulting in a 20-contig assembly. Read mapping visualization was done using Geneious v. 8.1.8, and genome annotation was performed using NCBI’s Prokaryotic Genomic Annotation Pipeline, PGAP v. 1.2 (16). A more detailed description with all data sets and scripts is provided in Figshare (10).
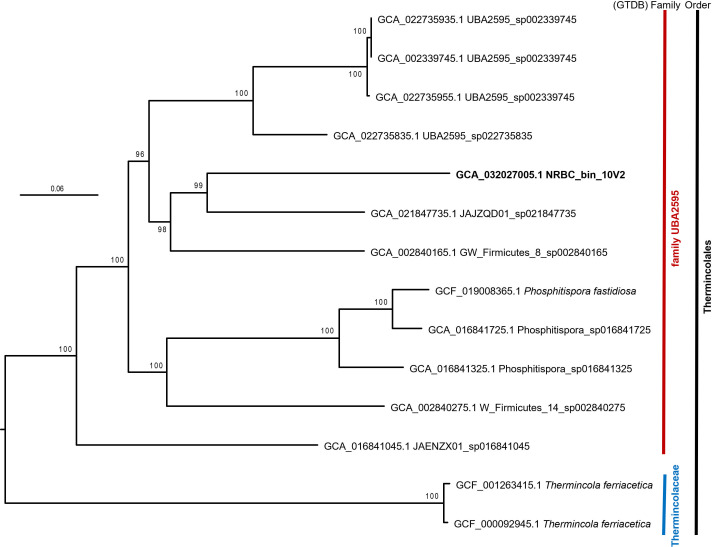
The 20-contig MAG is 4,085,792 bp with an average GC content of 44.5%, N50 of 277,970 bp, 100% completeness, and 0% contamination scores, as determined by CheckM v. 1.2.2 using the lineage-wf command (17). At least four distinct 16S rRNA amplicon sequence variants classified to the genus Thermincola have been identified in previous sequencing analyses of NRBC subcultures (4); however, no complete 16S rRNA genes were incorporated into the 20-contig MAG. Phylogenomic analysis using GTDB-tk (Release 214.1) classifies this MAG at the order level as a member of the Thermincolales (18, 19) in the placeholder family UBA2595 and likely represents a novel genus within the Thermincolales (Fig. 1).
Fig 1.
Maximum likelihood phylogenetic tree of Thermincolales genomes. The MAG reported is highlighted in bold. The remaining genomes are other members from the order Thermincolales from GTDB R214.1 obtained using the GToTree v1.8.1 pipline (20). Single-copy conserved genes were extracted using the Firmicutes.hmm profile and aligned by MUSCLE and concatenated by GToTree. The 23,147 amino acid alignments were imported to Geneious Prime v2023.2.1 (15), sites with >90% gaps were removed, and a maximum likelihood tree was constructed using RAxML v8 (21) with the WAG+GAMMA substitution model and 100 bootstrap replicates. Bootstrap support values are reported as numbers on branches. The tree was rooted by midpoint rooting. The GTDB order and family assignments are shown on the right.
ACKNOWLEDGMENTS
This study was funded by Genomic Application Partnership Program (GAPP) grants OGI-102 and OGI-107 which were supported by Genome Canada, Genome Ontario Genomics, the Government of Ontario, SiREM, Alberta Innovates, Mitacs, Federated Co-Operatives Limited, and Imperial Oil. We acknowledge the contributions of Roya Gitiafroz, Shen Guo, Elisse Magnusson, and Fei Luo for their previous work maintaining and analyzing NRBC cultures.
J.Z.X.: Formal Analysis, Conceptualization, Investigation, Methodology, Writing - Original draft, Writing – Review and Editing C.L.N.: Formal Analysis, Conceptualization, Investigation, Methodology, Writing - Original draft, Writing – Review and Editing O.M.: Conceptualization, Formal Analysis, methodology C.R.A.T.: Formal Analysis, Writing – Review and Editing E.A.E.: Conceptualization, Investigation, Methodology, Writing – Review and Editing, Funding
Contributor Information
Elizabeth A. Edwards, Email: elizabeth.edwards@utoronto.ca.
Elinne Becket, California State University San Marcos, San Marcos, California, USA.
DATA AVAILABILITY
The 20-contig version of the Thermincolales MAG has been deposited to NCBI under the accession number JAVSMW000000000.1. The original 157-contig MAG was previously deposited under accession number JAVSMV000000000.1. Illumina reads can be found under accession numbers SRR24043417, SRR24043423, and SRR24043422, whereas PacBio reads are found under accession number SRR24043419. Assemblies and bins are available at NCBI under Project PRJNA951427 and on FigShare (https://doi.org/10.6084/m9.figshare.22637596.v4). The latter also includes MAG statistics and FASTA files as well as detailed assembly steps and scripts. An older version of this draft genome is available in the US DOE Joint Genome Institute’s Integrated Microbial Genomes (IMG) system (ID: 2835707023).
REFERENCES
- 1. Luo F, Gitiafroz R, Devine CE, Gong Y, Hug LA, Raskin L, Edwards EA. 2014. Metatranscriptome of an anaerobic benzene-degrading, nitrate-reducing enrichment culture reveals involvement of carboxylation in benzene ring activation. Appl Environ Microbiol 80:4095–4107. doi: 10.1128/AEM.00717-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burland SM, Edwards EA. 1999. Anaerobic benzene biodegradation linked to nitrate reduction. Appl Environ Microbiol 65:529–533. doi: 10.1128/AEM.65.2.529-533.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ulrich AC, Beller HR, Edwards EA. 2005. Metabolites detected during biodegradation of 13C6-benzene in nitrate-reducing and methanogenic enrichment cultures. Environ Sci Technol 39:6681–6691. doi: 10.1021/es050294u [DOI] [PubMed] [Google Scholar]
- 4. Toth CRA, Luo F, Bawa N, Webb J, Guo S, Dworatzek S, Edwards EA. 2021. Anaerobic benzene biodegradation linked to the growth of highly specific bacterial clades. Environ Sci Technol 55:7970–7980. doi: 10.1021/acs.est.1c00508 [DOI] [PubMed] [Google Scholar]
- 5. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiao JNC, Molenda O, Toth C, Edwards EA. 2024. Metagenomic and genomic sequences from a nitrate-reducing benzene-degrading enrichment culture. Submitted to Microbiology Resource Announcements MRA00294-24 [DOI] [PMC free article] [PubMed]
- 7. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Butler J, MacCallum I, Kleber M, Shlyakhter IA, Belmonte MK, Lander ES, Nusbaum C, Jaffe DB. 2008. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res 18:810–820. doi: 10.1101/gr.7337908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao J, Nesbo C, Molenda O, Toth C, Edwards E. 2024. FASTA files and relevant statistics for draft MAGs generated from a nitrate-reducing benzene-degrading enrichment culture. doi: 10.6084/m9.figshare.22637596.v3 [DOI]
- 11. Tang S, Gong Y, Edwards EA. 2012. Semi-automatic in silico gap closure enabled de novo assembly of two Dehalobacter genomes from metagenomic data. PLoS One 7:e52038. doi: 10.1371/journal.pone.0052038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng Y, Leung HCM, Yiu SM, Chin FYL. 2011. Meta-IDBA: a de novo assembler for metagenomic data. Bioinformatics 27:i94–i101. doi: 10.1093/bioinformatics/btr216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner, abstr 9th annual genomics of energy & environment meeting, Walnut Creek, CA, 2014-03-17. Available from: https://www.osti.gov/biblio/1241166
- 15. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, Hugenholtz P. 2022. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res 50:D785–D794. doi: 10.1093/nar/gkab776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee MD. 2019. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35:4162–4164. doi: 10.1093/bioinformatics/btz188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 20-contig version of the Thermincolales MAG has been deposited to NCBI under the accession number JAVSMW000000000.1. The original 157-contig MAG was previously deposited under accession number JAVSMV000000000.1. Illumina reads can be found under accession numbers SRR24043417, SRR24043423, and SRR24043422, whereas PacBio reads are found under accession number SRR24043419. Assemblies and bins are available at NCBI under Project PRJNA951427 and on FigShare (https://doi.org/10.6084/m9.figshare.22637596.v4). The latter also includes MAG statistics and FASTA files as well as detailed assembly steps and scripts. An older version of this draft genome is available in the US DOE Joint Genome Institute’s Integrated Microbial Genomes (IMG) system (ID: 2835707023).