Abstract
The herpes simplex virus type 1 (HSV-1) mutant KUL25NS, containing a null mutation within the UL25 gene, was isolated and characterized by McNab and coworkers (A. R. McNab, P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa, J. Virol. 72:1060–1070, 1998). This mutant was able to cleave the concatemeric products of viral DNA replication into monomeric units, but in contrast to wild-type (wt) HSV-1, they were degraded by DNase treatment, indicating that they were not stably packaged into virus capsids. I have examined the packaging of the KUL25NS genome and an HSV-1 amplicon in cells infected with the mutant virus. In contrast to the previous results, a low level of KUL25NS DNA was resistant to DNase digestion, indicating that it was retained in capsids. The proportion of this packaged DNA present as full-length genomes was much lower than in cells infected by wt HSV-1, and there was a significant overrepresentation of the long terminus and underrepresentation of the short terminus. KUL25NS was less impaired in stably packaging amplicon DNA than in packaging its own genome, and the packaged molecules contained approximately equimolar amounts of the two terminal fragments. Below about 100 kbp, the packaged amplicon molecules exhibited an abundance and size distribution similar to those generated using wt HSV-1 as a helper, but the mutant was relatively impaired in packaging longer amplicon molecules. Both packaged genomic and amplicon DNAs were retained in the nuclei of KUL25NS-infected cells. These results suggest that the UL25 protein may play an important role during the later stages of the head-filling process, prior to release of capsids into the cytoplasm.
The pathways of herpes simplex virus type 1 (HSV-1) genome replication and virion morphogenesis converge at the stage of DNA maturation and encapsidation. The substrates for DNA packaging are the concatemeric products of DNA synthesis and a preassembled icosahedral structure, designated the procapsid, in which the capsid shell proteins surround an internal proteinaceous scaffold (20, 21, 26, 41). Encapsidation proceeds by a complex mechanism in which the cleavage of concatemers into monomeric units is tightly coupled with their insertion into the procapsid (for a review, see reference 10).
An integral component of the scaffold is the virus-encoded protease, activation of which is necessary for the conversion of the procapsid into a more angularized and stable capsid form (6, 8, 23, 24, 26). Three types of angularized capsid are seen in the nuclei of infected cells (9, 10, 25). C capsids result from insertion of the viral genome and the concomitant loss of the scaffold and represent the precursors of infectious virus particles. B capsids retain an internal scaffold, while A capsids contain neither DNA nor scaffold. The A and B capsid forms are considered to be dead-end products, and it is thought likely that the former are generated as a consequence of abortive DNA-packaging events (2, 30, 32).
In addition to the major components of the procapsid, six other viral proteins, encoded by the genes UL6, UL15, UL17, UL28, UL32, and UL33, are necessary for cleavage and packaging of concatemeric HSV-1 DNA. Mutants with lesions in these genes exhibit a characteristic phenotype in which viral DNA accumulates in the form of concatemers, genomic terminal fragments are absent, the nucleus contains B but not C capsids, and the replicated viral DNA is susceptible to the action of exogenously added DNase (10, 12, 28).
Studies of mutants affected in a seventh viral protein, the UL25 product, have suggested that this product also plays an important role in the generation of DNA-containing capsids. Addison et al. (3) characterized two temperature-sensitive (ts) mutants with lesions in UL25. One of these, ts1204, had a defect in initiating infection at the nonpermissive temperature which could be overcome by an initial brief incubation at the permissive temperature. In addition, both ts1204 and the second mutant, ts1208, were defective at a later stage of infection. Although the question was not directly examined, it was concluded that efficient viral DNA synthesis occurred, since viral polypeptide production was very similar to that of wild-type (wt) HSV-1. Both mutants assembled capsids in the nuclei of cells incubated at nonpermissive temperature, albeit in smaller numbers than wt virus, and the majority of these had the appearance of B capsids in the published electron micrographs. The most striking feature, however, was the absence of detectable DNA-containing C capsids, suggesting that the mutants were defective at the stage of DNA encapsidation.
A UL25-null mutant (KUL25NS) was subsequently isolated in a complementing cell line, and its behavior was characterized in the parental untransformed Vero cells (15). The virus synthesized viral DNA, but only A and B capsids were present in infected cell nuclei. Like the mutants defective in DNA packaging, replicated KUL25NS DNA was completely susceptible to added DNase. However, a crucial difference was that both unit-length KUL25NS genomes and terminal genomic DNA fragments were detected in infected cells. These data reveal a novel phenotype for an HSV-1 mutant in which cleavage of concatemeric DNA occurs in the absence of stable DNA packaging. Based largely upon the accumulation of A capsids, McNab et al. (15) concluded that rather than uncoupling cleavage and packaging, the absence of the UL25 protein may have resulted in an abortive packaging process in which viral DNA was only transiently associated with the capsid. The essential function of the UL25 protein during DNA packaging was therefore deduced to be in retaining DNA in capsids.
UL25 protein has been detected in preparations of procapsids; A, B, and C capsids; and virions. Consistent with a role in DNA retention, C capsids contain more UL25 protein than B capsids, which in turn contain more than procapsids (4, 11, 15, 22, 31, 39, 43). In addition, UL25 protein has recently been reported to interact with both capsid shell proteins and viral DNA, suggesting a possible role in anchoring the genome to the capsid (22).
A convenient approach to investigating the cis-acting elements and trans-acting functions that participate in the HSV-1 DNA encapsidation process employs amplicons: bacterial plasmid vectors into which have been inserted functional copies of a viral DNA replication origin and a packaging signal (1, 7, 19, 34, 35). When cells transfected with the amplicon are superinfected with wt HSV-1, the presence of the two viral cis-acting signals allows the plasmid to be replicated as a concatemer and packaged in virus particles. During the course of experiments in which baby hamster kidney (BHK) cells were transfected with an amplicon and superinfected with the UL25-null mutant, KUL25NS, I unexpectedly observed that significant amounts of the replicated amplicon were stably packaged. This finding prompted a more detailed examination of the packaging of mutant genomes and amplicon DNA in Vero and BHK cells.
MATERIALS AND METHODS
Cells and viruses.
BHK 21 clone 13 (hereafter referred to as BHK) cells were grown in Glasgow minimal essential medium (MEM) supplemented with 10% tryptose phosphate broth, 10% newborn calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. After transfection or infection, the cells were maintained in Glasgow MEM supplemented with 5% newborn calf serum and the same antibiotics (EC5). Vero cells were maintained throughout in Dulbecco's MEM containing 5% fetal calf serum and the same antibiotics (EFC5). The HSV-1 UL25- and UL28-null mutants (KUL25NS and gCB) were propagated on Vero cell-derived complementing cell lines, 8-1 and C1, respectively (15, 40). Stocks of wt HSV-1 (strain 17 syn+ [14]) were prepared and titrated in BHK or Vero cells. Wt HSV-1 strain KOS, the parent of KUL25NS (15), and a phenotypically wt virus, 25R22, generated by marker rescue of KUL25NS with a cloned HSV-1 fragment containing the UL25 gene of HSV-1 strain 17 syn+ (a kind gift of P. Targett-Adams), were also used in some experiments.
Plasmids.
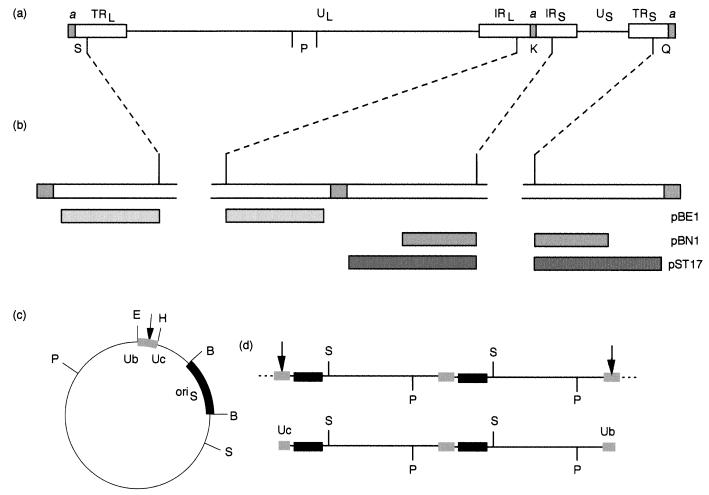
Amplicon pSA1 (Fig. 1c) contains a copy of the HSV-1 oriS DNA replication origin and a 200-bp packaging signal spanning the junction between two tandem a sequences inserted into the vector pAT153 (1). Plasmids pGX2 and pGX153 contain the HSV-1 BamHI K and P fragments (Fig. 1a), respectively, inserted into pAT153. Since BamHI K contains sequences from both the long repeat (RL) and the short repeat (RS), pGX2 can hybridize to fragments originating from the joint region and both termini. Plasmids suitable for specific detection of the joint region and either the L or S terminus of HSV-1 DNA were derived by introducing deletions that removed the a sequence and all of one terminus from pGX2. Thus, plasmid pBE1 contains sequences corresponding to HSV-1 nucleotides 596 to 2905 of TRL, and pBN1 and pST17 contain sequences corresponding to nucleotides 148825 to 150562 and 148825 to 151857, respectively, of TRS (Fig. 1b; nucleotide numbering taken from reference 14).
FIG. 1.
HSV-1 and amplicon DNAs. (a) Structure of the HSV-1 genome showing the positions of the unique (UL and US) and repeated (TRL, IRL, IRS, and TRS) regions of the genome. The a sequence is shaded, and for simplicity only single copies are shown at the L terminus and joint. The locations of BamHI fragments K, P, Q, and S are indicated. (b) Expanded representation of the BamHI joint fragment, K, and terminal fragments, S and Q. The regions within these fragments corresponding to the inserts in plasmids pBE1, pBN1, and pST17 are shown as shaded boxes. (c) Structure of the amplicon, pSA1 (4.4 kbp). The fragments containing oriS and the packaging signal are shown as thickened lines. The relative positions of the Ub and Uc regions (which contain the pac1 and pac2 signals, respectively) are indicated. The site of cleavage for packaging (arrow) occurs within the DR1 element between Ub and Uc. E, H, B, S, and P indicate the positions of EcoRI, HindIII, BamHI, SalI, and PstI restriction endonuclease sites. (d) The upper line represents the structure of a concatemer generated by pSA1 replication (only two complete copies of the monomeric plasmid are depicted). Cleavage and packaging of concatemeric DNA generates molecules with Uc and Ub at opposite ends (lower line). Digestion of packaged DNA with PstI (P) or SalI (S) yields fragments corresponding to pSA1 monomers plus specific terminal fragments. The terminal SalI fragments containing Uc and Ub (equivalent to the L and S termini of the viral genome) are 1.3 and 3.1 kbp, respectively. The sizes of the corresponding PstI terminal fragments are 3.5 and 0.9 kbp.
Virus infection and DNA analysis.
Monolayers of BHK or Vero cells in 35-mm-diameter petri dishes were infected with 5 PFU of wt HSV-1 or KUL25NS/cell in a volume of 200 μl. One hour after the addition of virus, the inoculum was removed; the cells were washed with 0.14 M NaCl, exposed to 0.1 M glycine–0.14 M NaCl (pH 3.0) for 1 min to inactivate residual virus, and washed with maintenance medium; and incubation continued for 18 h at 37°C in 2 ml of EC5 or EFC5, as appropriate. The medium was removed, and the cells were resuspended in Tris-buffered saline (137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 5.5 mM glucose, 25 mM Tris-HCl, pH 7.4) and divided into two equal samples, which were used to prepare total cellular and DNase-resistant (encapsidated) DNAs (36). The cells from both samples were pelleted and resuspended in 184 μl of reticulocyte standard buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2) containing 0.5% NP-40. An equal volume of 2× CLB (20 mM Tris-HCl [pH 7.5], 2 mM EDTA, 1.2% sodium dodecyl sulfate, 1 mg of protease [Sigma grade XIV]/ml) was either added immediately (total cellular DNA) or after incubation in the presence of 200 μg of DNase I/ml, with occasional mixing, for 20 min at 37°C (encapsidated DNA). After the addition of protease, all samples were incubated for 1 h at 37°C, extracted sequentially with phenol and chloroform, and precipitated with ethanol, and the nucleic acids were redissolved in 10 mM Tris-HCl (pH 7.5)–1 mM EDTA containing 5 μg of RNase A/ml and 50 U of RNase T1/ml. Control experiments (data not shown) indicated that incubation of cells infected with wt HSV-1 or KUL25NS in the presence of DNase I for times between 5 and 300 min did not affect the recovery of DNase-resistant DNA.
The preparation of nuclear and cytoplasmic DNAs was similar except that after initial resuspension in reticulocyte standard buffer containing 0.5% NP-40, the cells were first incubated for 10 min on ice and centrifuged for 10 s at 2,000 × g to pellet the nuclei. Gel analysis of the DNAs was performed as previously described (37). Samples of DNA corresponding to the yield from 4 × 105 cells were cleaved with various enzymes, and the resulting fragments were separated by agarose gel electrophoresis, transferred to a Hybond-N membrane (Amersham), and detected by hybridization to appropriate 32P-labeled probes. In most experiments with DNA from cells that received the amplicon pSA1, DpnI was included in the enzyme reaction to specifically digest unreplicated input plasmid DNA. Phosphorimages of Southern blots were acquired using the Personal Molecular Imager and analyzed with Quantity One software (Bio-Rad).
DNA transfections.
Monolayers of cells in 35-mm-diameter petri dishes were transfected by the calcium phosphate procedure followed by treatment with dimethyl sulfoxide at 4 h (38). Each monolayer received 0.5 ml of precipitate containing 0.5 μg of pSA1 and 12 μg of calf thymus carrier DNA. Two hours after treatment with dimethyl sulfoxide, the transfected cells were infected with 5 PFU of wt HSV-1 or KUL25NS/cell in a volume of 200 μl and treated as described above prior to the preparation of DNA.
PFGE.
Nuclei and cytoplasm were prepared and treated with DNase as described above. Digestion was terminated by the addition of one-third volume 4× CLB plus gel loading buffer, and the samples were mixed gently to minimize shearing of DNA. Electrophoresis was carried out on a Bio-Rad DR-II apparatus. Samples were loaded onto a 1% agarose (Bio-Rad; pulsed-field gel electrophoresis [PFGE]-certified) gel in 0.5× Tris-borate-EDTA using a pipette with a wide tip, and the DNAs were resolved with a voltage gradient of 6 V/cm for 18 h at 14°C and a linear switch time gradient from 1 to 15 s. After electrophoresis, the gels were stained with ethidium bromide, photographed, and blotted. A 45-min treatment of the gel with 0.25 M HCl was included prior to the alkali denaturation step.
Total DNA from infected cells was analyzed by embedding the cells in 1% agarose (CleanCut; Bio-Rad) blocks and performing lysis and proteinase K digestion in situ as recommended by the manufacturer. The blocks were washed, and pieces containing one-third of the cells from a 35-mm-diameter petri dish were inserted into the wells of a precast 1% agarose gel. Electrophoresis and subsequent treatment of the gel were as described above.
RESULTS
Packaging of KUL25NS DNA in Vero cells.
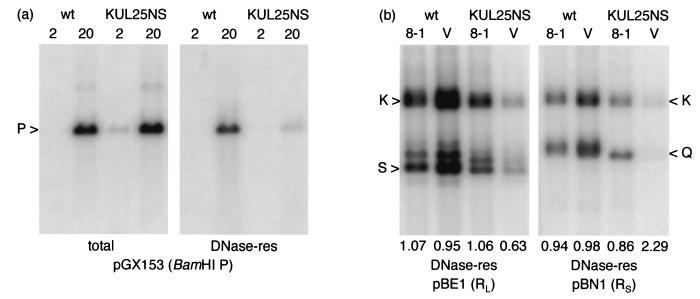
Vero cell monolayers were infected with wt HSV-1 or the UL25-null mutant KUL25NS, and total cellular and DNase-resistant DNAs were prepared at 2 h postinfection (p.i.) (before viral DNA synthesis had commenced) and 20 h p.i. Samples were cleaved with BamHI and analyzed by Southern blot hybridization to a labeled pGX153 probe that detects the BamHI P fragment (Fig. 2a). The signals obtained for the 2-h-p.i. total-DNA samples represent input DNA. It was frequently observed that a more intense signal was obtained with stocks of KUL25NS than with wt HSV-1, presumably because of the presence of larger numbers of noninfectious particles. Examination of the lanes containing DNA prepared 20 h p.i. reveals that the two viruses accumulated similar amounts of replicated DNA. In each case, a proportion of the viral DNA was resistant to digestion with DNase I (i.e., it had been stably packaged in virus capsids), but this was much greater in the case of wt HSV-1. Approximately 30 and 0.8% of the replicated wt HSV-1 and KUL25NS DNAs, respectively, were recovered in the DNase-resistant samples.
FIG. 2.
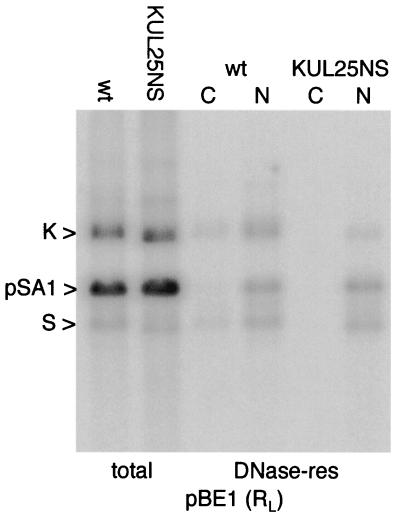
Packaging of KUL25NS DNA in Vero and 8-1 cells. (a) Monolayers of Vero cells were infected with wt HSV-1 or KUL25NS, and total and DNase-resistant (res) DNAs were prepared at either 2 or 20 h p.i. as indicated above the lanes. Samples were cleaved with BamHI, and the fragments were resolved by electrophoresis through a 0.8% agarose gel, transferred to a nylon membrane, and hybridized to 32P-labeled pGX153 (containing BamHI P) DNA. (b) Monolayers of Vero (V) or 8-1 cells were infected with the indicated virus, and DNase-resistant DNA was prepared 20 h p.i. Duplicate samples, cleaved with BamHI, were hybridized to either pBE1 or pBN1 as shown. The numbers below the lanes indicate the ratio of radioactivity present in the joint (K) to that in terminal (S or Q) fragments. The measurement for the L-terminal fragment included the bands containing one (labeled S), two, or three (bands above S) copies of the a sequence. It should be noted that BamHI fragments K, Q, and S of KUL25NS (strain KOS) are slightly smaller than the corresponding fragments of wt HSV-1 (strain 17 syn+). The panels were obtained from single phosphorimager exposures of the washed membrane (i.e., all lanes were processed identically).
In order to confirm that the lesion in UL25 was responsible for the phenotype, parental Vero cells and the Vero-derived complementing line 8-1, which express only UL25 (15), were infected with wt HSV-1 and KUL25NS. DNase-resistant DNAs were prepared 20 h p.i., and BamHI-digested samples were analyzed using pBE1 and pBN1 as probes (Fig. 2b). In 8-1 cells, each of the probes detected similar amounts of packaged wt HSV-1 and KUL25NS DNAs. In contrast, in Vero cells, the amounts of packaged wt HSV-1 and KUL25NS DNAs were approximately 2-fold higher and 10-fold lower, respectively, than in 8-1 cells. Thus, the lesion in UL25 of KUL25NS is responsible for a reduction in DNA packaging efficiency of at least 20-fold in Vero cells.
The amounts of radioactivity in the bands corresponding to the joint fragment, the long terminus, and the short terminus (BamHI fragments K, S, and Q, respectively) were measured, and the ratio of joint to terminal fragment was calculated for each lane in Fig. 2b. In the case of fragment S from the long terminus, the measurement included the major fragment containing a single copy of the a sequence plus the fragments corresponding to the presence of two and three copies. For wt HSV-1 in both cell lines, and KUL25NS in 8-1 cells, the ratios were, as expected, close to unity. However, for KUL25NS in Vero cells, there was a significant overrepresentation of the L terminus (1.6-fold) and an underrepresentation of the S terminus (2.3-fold) compared to the joint fragment.
Packaging of KUL25NS DNA in BHK cells.
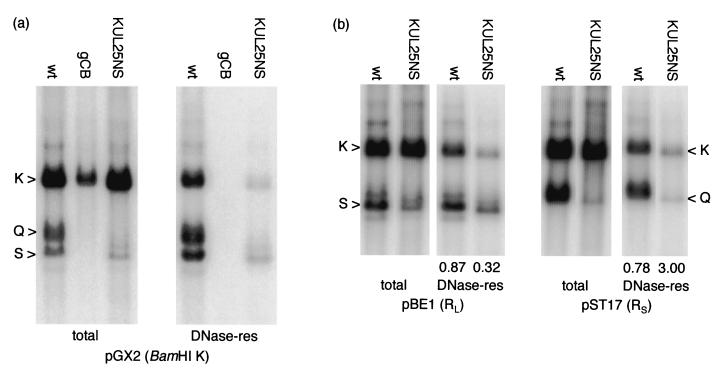
The behavior of KUL25NS was similarly investigated in BHK cells. Total and DNase-resistant DNAs were prepared 20 h p.i. with wt HSV-1, KUL25NS, or the UL28-null mutant gCB. BamHI-digested DNA was analyzed by hybridization to pGX2, which detects the joint fragment and both termini. The total-cellular-DNA samples (Fig. 3a) show that KUL25NS and wt HSV-1 replicated to similar levels (as judged by fragment K), but greatly reduced levels of terminal fragments were present in the KUL25NS DNA. In contrast, the proportion of KUL25NS DNA packaged was much lower than for wt HSV-1. For both viruses, the amounts of terminal fragment detected in the total- and DNase-resistant-DNA samples were similar, suggesting that most of the terminal fragments detected in the total-DNA samples originate from molecules that have already been packaged. DNA from the mutant gCB was detected in the total-DNA samples but not the DNase-resistant-DNA samples, in agreement with previous results (40) and confirming that the low levels of DNase-resistant KUL25NS DNA detected are not a result of incomplete nuclease digestion.
FIG. 3.
Packaging of KUL25NS DNA in BHK cells. (a) Monolayers of BHK cells were infected with the viruses indicated, and total and DNase-resistant (res) DNAs were prepared 18 h p.i. Samples, cleaved with BamHI, were analyzed by hybridization to 32P-labeled pGX2 (containing BamHI K) DNA. (b) Monolayers of BHK cells were infected with wt HSV-1 or KUL25NS as indicated, and DNase-resistant DNA was prepared 20 h p.i. Duplicate samples, cleaved with BamHI, were hybridized to labeled pBE1 or pST17 as indicated, and the numbers below the lanes containing DNase-resistant DNA indicate the ratio of radioactivity present in the joint (K) to that in terminal (S or Q) fragments, measured as described in the legend to Fig. 2. All lanes were processed identically.
The above-mentioned data indicate that KUL25NS displays similar phenotypes in Vero and BHK cells: namely, that DNA was stably packaged but at a greatly reduced level compared to that in wt HSV-1. However, the data differ in two important aspects from the results obtained with the same virus by McNab et al. (15). Those workers found first, that similar amounts of KUL25NS and wt HSV-1 genomic termini were present in total cellular DNA and, second, that the replicated DNA of the mutant was completely susceptible to DNase digestion.
An independent experiment was performed with BHK cells to examine separately the L- and S-terminal fragments of wt HSV-1 and KUL25NS in total cellular and DNase-resistant DNAs (Fig. 3b). Comparison of the total-cellular-DNA samples harvested 18 h p.i. again revealed similar replication of the two viruses but lower levels of KUL25NS terminal fragments. Quantification of the blot indicated that the amounts of the L (BamHI S)- and S (BamHI Q)-terminal fragments in total cellular DNA from KUL25NS-infected cells were reduced at least 2- and 10-fold, respectively, relative to those in wt HSV-1. The amount of terminal fragment detected in total DNA was, in each instance, similar to the amount present in the corresponding DNase-resistant-DNA sample, indicating that total cellular DNA contains relatively few free termini of either wt HSV-1 or KUL25NS DNA. It should be noted, however, that precise quantification of the amounts of terminal fragments in these experiments is difficult, particularly in the total-cellular-DNA samples, because of the presence of a background smear of hybridization.
The ratios of joint fragment to L terminus for packaged wt HSV-1 and KUL25NS DNAs were 0.87 and 0.32, respectively, and the corresponding values for the S terminus were 0.78 and 3.00. Thus, as noted in Vero cells, there is relative overrepresentation of the L terminus and underrepresentation of the S terminus in packaged KUL25NS DNA. The proportions of wt HSV-1 and KUL25NS DNAs packaged in the experiments shown in Fig. 3 were assessed from measurements of the radioactivity present in the BamHI K fragments. For wt HSV-1, 41 and 26% of the total DNA was recovered in the DNase-resistant fractions, with the amounts of packaged KUL25NS DNA in the corresponding experiments being 0.9 and 3.9%, respectively. Thus, packaging of the mutant was reduced approximately 6- to 40-fold compared to wt HSV-1. These values are representative of the variation noted in a large number of independent experiments, but they need to be interpreted with caution, since it is clear that relative packaging efficiency is not constant throughout the KUL25NS genome.
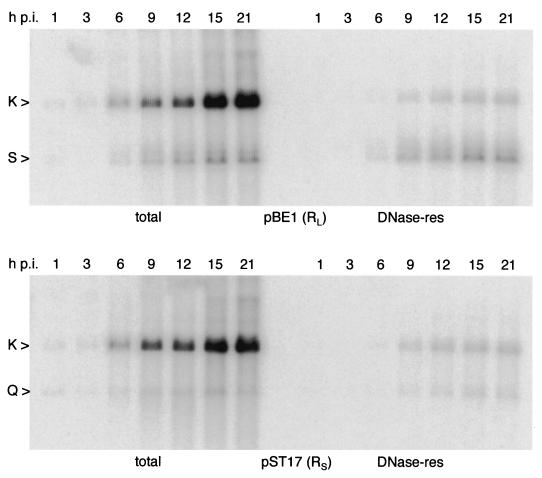
Figure 4 shows a time course for KUL25NS DNA replication and packaging in BHK cells. Each of the terminal fragments (BamHI Q and S) accumulates to similar levels in the total and DNase-resistant fractions throughout the course of the infection, with no evidence for an excess of unpackaged termini at any stage. Visual inspection also reveals that at all times, relative to the joint fragment BamHI K, the L terminus is overrepresented and the S terminus is underrepresented in packaged DNA.
FIG. 4.
Time course for KUL25NS DNA replication and packaging in BHK cells. Monolayers of BHK cells were infected with KUL25NS, and total and DNase-resistant (res) DNAs were prepared at the indicated times (h p.i.). BamHI-cleaved samples were analyzed by hybridization to 32P-labeled pBE1 (top) or pST17 (bottom).
PFGE of packaged KUL25NS DNA.
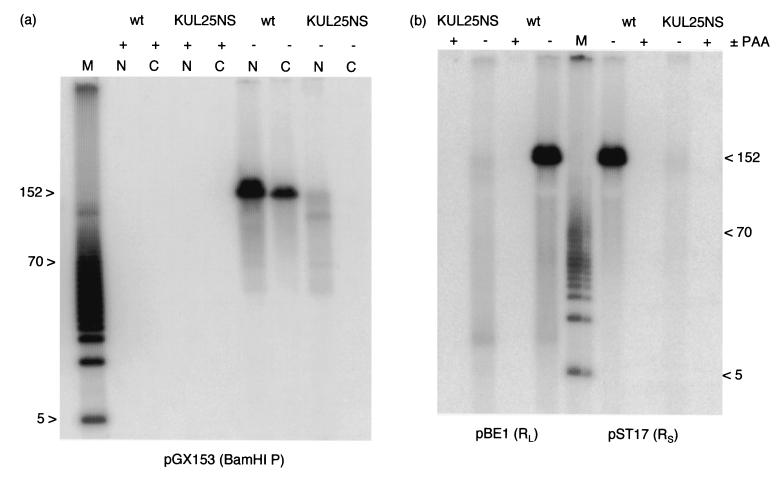
Monolayers of BHK cells infected with wt HSV-1 or KUL25NS were incubated in the absence or presence of phosphonoacetic acid (PAA) to block viral DNA synthesis. The cells were fractionated into nuclei and cytoplasm, and DNase-resistant DNA was prepared so as to minimize the degree of shearing. Samples were resolved by PFGE, and the gel was blotted and hybridized to labeled pGX153 (which detects BamHI P). Figure 5a shows that no viral DNA was detectable in the samples obtained from cells treated with PAA, and hence any present in the untreated samples must represent newly synthesized, and not input, DNA. Wt HSV-1 DNA was detected in both the nucleus and cytoplasm, and major bands corresponding to full-length genomes were present in each instance. In contrast, KUL25NS DNA was present only in the nuclear fraction. The abundance and distribution of KUL25NS fragments of less than about 125 kbp was very similar to that of wt HSV-1 in the nucleus, but the amount of full-length genomes was reduced over 20-fold.
FIG. 5.
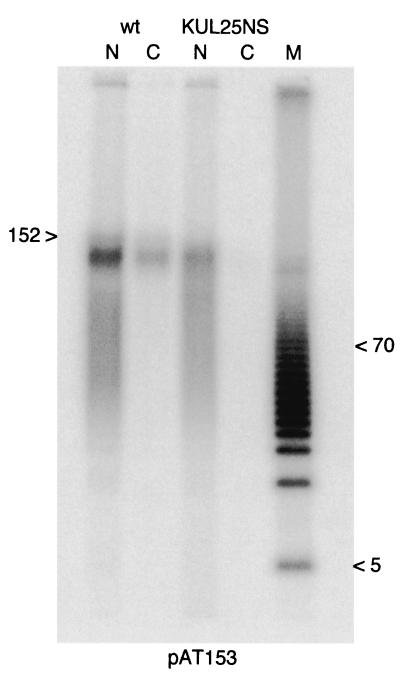
PFGE analysis of packaged genomic DNAs. (a) Monolayers of BHK cells were infected with wt HSV-1 or KUL25NS as indicated and incubated in either the presence (+) or absence (−) of 200 μg of PAA/ml. At 18 h p.i., the cells were fractionated into nuclei (N) and cytoplasm (C), and DNase-resistant DNA was prepared by a gentle method without phenol extraction. Samples were subjected to PFGE, and the gel was blotted and hybridized to 32P-labeled pGX153 (which contains BamHI P). (b) Monolayers of BHK cells were infected with wt HSV-1 or KUL25NS and incubated for 17 h in either the presence (+) or absence (−) of 200 μg of PAA/ml. Duplicate samples of DNase-resistant nuclear DNA were resolved on a pulsed-field gel and blotted. The membrane was divided in two and hybridized to pBE1 or pST17, as indicated. The markers (M) are 5-kbp ladders (Bio-Rad), and the positions of the 5- and 70-kbp fragments are shown. The 152-kbp band corresponds to full-length HSV-1 DNA. (c) Monolayers of BHK cells were infected with KUL25NS, 25R22, wt HSV-1 strain 17 syn+ (wt 17), or wt HSV-1 strain KOS (wt KOS) as indicated and incubated for 17 h. The cells were harvested, embedded in agarose, lysed, and digested with proteinase K in situ. Samples were resolved on a pulsed-field gel, blotted, and hybridized to pGX2. The positions of the wells and of 152-kbp linear genomes are indicated.
A similar experiment was performed in which duplicate samples of nuclear DNase-resistant DNA were resolved by PFGE. The two halves of the blot were hybridized to probes specific for RL and RS (pBE1 and pST17, respectively) (Fig. 5b). Again, both probes detected abundant full-length wt HSV-1 genomes but very small amounts of similarly sized KUL25NS molecules. The distributions of molecules smaller than approximately 125 kbp were similar for both viruses. Interestingly, the probe specific for the S terminus hybridized primarily to molecules greater than 50 kbp in size, whereas fragments between about 5 and 50 kbp were more strongly detected with the L-terminal probe.
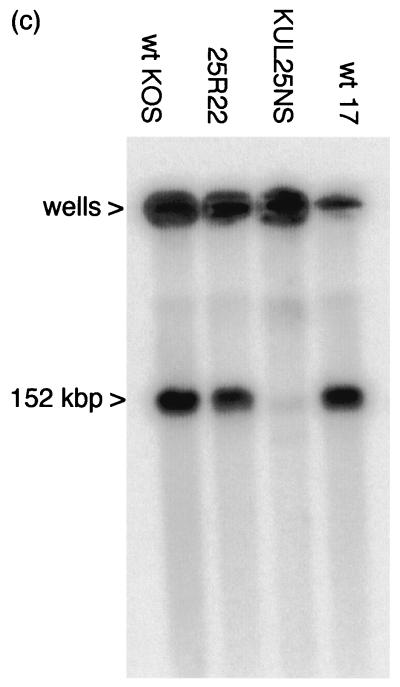
Total DNAs from cells infected with wt HSV-1 (strain 17 syn+) and KUL25NS were also examined by PFGE. In this experiment, two other phenotypically wt viruses were included: HSV-1 strains KOS (the parent of KUL25NS) and 25R22, which was generated by marker rescue of the KUL25NS lesion. Viral DNA was detected by hybridization to labeled pGX2. Figure 5c shows that each of the viruses generated concatemeric DNA that was retained in the wells of the gel. Similar amounts of linear genomes were present in the cells infected with 25R22 and the two wt strains, but the amount of linear KUL25NS DNA was over 30-fold lower. This result is consistent with the observations that the terminal fragments of KUL25NS are less abundant than those of wt HSV-1 in total cellular DNA. In addition, the data indicate that HSV-1 strain 17 syn+ cleaves and packages DNA with efficiency similar to that of the true parent (strain KOS) and the rescuant virus (25R22).
Packaging of amplicon DNAs in cells infected with KUL25NS.
BHK cells were transfected with the amplicon pSA1 and subsequently superinfected with wt HSV-1 or KUL25NS. Total DNA and DNase-resistant nuclear and cytoplasmic DNAs were prepared, and samples cleaved with a combination of BamHI and DpnI were analyzed by hybridization to labeled pBE1 DNA (Fig. 6). The use of this probe allows detection of the amplicon at the same time as the viral joint and L-terminal fragments. It can be seen that the two viruses replicate pSA1 to similar extents and that similar amounts of packaged pSA1 are present in the nuclear DNase-resistant DNA fractions. In contrast, approximately sixfold less of the viral BamHI K fragment was present in the packaged nuclear fraction from KUL25NS-infected cells, in agreement with the earlier results obtained in untransfected cells. Both packaged amplicon and viral genomes were present in the cytoplasm of wt HSV-1- but not KUL25NS-infected cells.
FIG. 6.
Replication and packaging of pSA1 in BHK cells. Monolayers of BHK cells were transfected with pSA1 and superinfected with either wt HSV-1 or KUL25NS. At 17 h p.i., total cellular, cytoplasmic DNase-resistant (res; C), and nuclear DNase-resistant (N) DNAs were prepared. Samples, cleaved with BamHI plus DpnI, were hybridized to 32P-labeled pBE1 DNA. The positions of the BamHI K and S fragments of the viral DNA and of pSA1 monomers are indicated.
Quantification of several independent repeat experiments indicated that stable packaging of replicated pSA1 molecules in KUL25NS-infected cells occurred with between 30 and 100% of wt efficiency. Thus, KUL25NS appears significantly less impaired in packaging replicated amplicons than in packaging its own genome.
Terminal fragments of packaged amplicons.
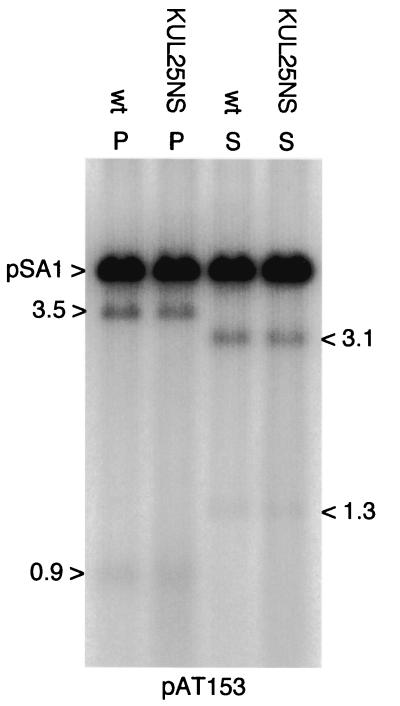
As shown in Fig. 1d, packaged amplicons consist of tandem head-to-tail repeats of the input plasmid. The packaging signal in pSA1 corresponds to the fragment containing Uc-DR1-Ub that spans two tandem copies of the a sequence. The Uc and Ub regions of the a sequence lie adjacent to the L and S termini, respectively, of the HSV-1 genome, and the site for cleavage of concatemeric DNA lies within DR1 (for a review, see reference 27). Digestion of replicated and packaged pSA1 molecules with PstI or SalI generates fragments the size of linearized pSA1 plus specific smaller fragments from each terminus (Fig. 1d). The 1.3- and 3.1-kbp fragments generated by SalI digestion of packaged pSA1 terminate in Uc and Ub, respectively, and are therefore equivalent to the L and S termini of the viral genome. The sizes of the corresponding PstI terminal fragments are 3.5 and 0.9 kbp, respectively.
To examine the termini of packaged amplicons, DNase-resistant nuclear DNA from cells transfected with pSA1 and superinfected with either wt HSV-1 or KUL25NS was digested with DpnI plus either SalI or PstI. The fragments were detected by hybridization of the blot to labeled pAT153 DNA. As shown in Fig. 7, the digest patterns and relative abundance of the terminal fragments were essentially indistinguishable for the two helper viruses. Therefore, in contrast to genomic KUL25NS DNA, there is no evidence for one terminus being present in significantly greater amounts than the other.
FIG. 7.
Terminal fragments of packaged pSA1 DNA. Monolayers of BHK cells were transfected with pSA1 and superinfected with either wt HSV-1 or KUL25NS. DNase-resistant DNA was prepared from the nuclei of infected cells 18 h p.i., and samples were cleaved with either PstI plus DpnI (P) or SalI plus DpnI (S). The fragments generated were detected by hybridization to 32P-labeled pAT153 DNA. The positions and sizes (in kilobase pairs) of the four terminal fragments and the position of unit-length pSA1 molecules are indicated.
PFGE analysis of packaged amplicon DNA.
DNase-resistant DNA from the nuclei and cytoplasm of cells transfected with pSA1 and superinfected with either wt HSV-1 or KUL25NS was prepared and analyzed by PFGE as described above, except that the probe was labeled pAT153 DNA. Figure 8 shows that with a wt helper, packaged DNA was present in both the nuclear and cytoplasmic fractions, although molecules smaller than about 100 kbp were in relatively lower abundance in the cytoplasm. This is in agreement with the findings of Vlazny et al. (42), who reported that although capsids in the nucleus contain defective genomes comprising integral numbers of repeats up to the length of a standard HSV-1 genome, only the largest species are present in cytoplasmic capsids. Although not clearly resolved in this gel, the characteristic ladder of packaged nuclear molecules described by Vlazny et al. (42) was apparent in other experiments.
FIG. 8.
PFGE analysis of packaged amplicon DNA. BHK cell monolayers were transfected with pSA1 and superinfected with either wt HSV-1 or KUL25NS. Nuclear and cytoplasmic DNase-resistant DNAs were prepared and analyzed by PFGE as described in the legend to Fig. 5, except that the probe was 32P-labeled pAT153. The markers (M) are a 5 kbp ladder (Bio-Rad), and the positions of the 5- and 70-kbp fragments are shown. The blot was reprobed with labeled pGX153, and the position of unit-length HSV-1 genomes (152 kbp) is indicated.
Packaged amplicons were found only in the nuclei of KUL25NS-infected cells. The size distribution was very similar to that of the corresponding fraction from cells infected with wt HSV-1 with the exception that the largest molecules were present in lower amounts. Interestingly, the bands corresponding to the largest packaged amplicons in the nuclear and cytoplasmic fractions of cells infected with wt HSV-1 exhibited a slightly faster mobility than genome-length HSV-1 DNA (detected by reprobing of the blot with labeled pGX153). It is not clear whether these molecules are actually shorter than standard genomes or if the difference in migration arises because of some other property, for example, their tandemly reiterated structure or lower G+C content.
In control experiments, cells transfected with pSA1 and superinfected with wt HSV-1 or KUL25NS were incubated for 18 h in the absence or presence of 200 μg of PAA/ml prior to the preparation and analysis of DNase-resistant DNA. Hybridization to labeled pAT153 DNA failed to detect any pSA1 DNA in the samples incubated with PAA (data not shown), demonstrating that the signals observed in Fig. 8 are not due to the presence of unreplicated input pSA1 molecules.
DISCUSSION
The experiments presented in this paper extend the characterization of the HSV-1 UL25-null mutant KUL25NS. The principal findings are that (i) stable packaging of the KUL25NS genome into a DNase-resistant form occurs at low efficiency, (ii) the L terminus is overrepresented and the S terminus is underrepresented in packaged KUL25NS DNA molecules, (iii) the null mutant packages amplicon DNA relatively more efficiently than it packages its own genome, (iv) the two termini of packaged amplicons are present in similar amounts, and (v) neither packaged viral genomes nor amplicons are transported into the cytoplasm in KUL25NS-infected cells. In contrast to the results of McNab and coworkers (15), there was little evidence that cleavage of concatemers into genome monomers was nearly as efficient as in cells infected with wt HSV-1 or to support the suggestion that in the absence of the UL25 protein significant amounts of cleaved DNA failed to be retained in capsids. The reason why large amounts of DNase-sensitive unit-length KUL25NS genomes were observed by McNab et al. (15) but not in this study is not clear, but the use of different cell lines is unlikely to be the explanation, since I observed similar behavior of KUL25NS in Vero and BHK cells (Fig. 2). One possible explanation is that in my experiments full-length KUL25NS genomes were indeed efficiently packaged into capsids and subsequently released as previously proposed (15), but they were then rapidly degraded. Although this possibility cannot be fully excluded, such a marked difference in cellular behavior seems unlikely. As described in detail below, my analysis of the terminal fragments of KUL25NS in total and DNase-resistant DNAs can best be reconciled with a model in which packaging of KUL25NS DNA is initiated but fails to go to completion. Despite this important difference, the data presented in this paper nevertheless support two important conclusions of McNab et al. (15), namely, that the UL25 protein is not absolutely required for DNA cleavage and that it does play an essential role at some stage between the initiation of encapsidation and the release of DNA-containing capsids into the cytoplasm.
The mechanism of HSV-1 DNA encapsidation remains relatively poorly understood, and many fundamental issues, such as the polarity of packaging, remain to be resolved. It has previously been noted for several herpesviruses that during DNA packaging only one terminus, that containing the conserved pac2 signal, is associated with high-molecular-weight concatemeric DNA (13, 16, 17, 29, 33, 44). This observation provides tentative support for a model in which the directionality of packaging of herpesvirus DNA is controlled by the initial insertion of the pac2-containing terminus into the capsid (16, 17). Since the pac2 signal of HSV-1 lies within the Uc region at the L terminus, the model predicts that this end is inserted into the capsid and packaging proceeds towards the S terminus. The DNA being packaged remains associated with the concatemer until a terminating cleavage event occurs that generates the S terminus of the packaged genome and exposes a free L terminus on the concatemer, which is available to reinitiate the packaging process. Two different cleavage events are therefore envisaged to be associated with HSV-1 DNA packaging. The first would occur to initiate the packaging of a DNA concatemer and would generate free L- and S-terminal fragments, with the L terminus being inserted into a capsid and the S terminus remaining unencapsidated. The second type would generate the S terminus of a packaged genome and a free L terminus, allowing packaging to progress along the concatemer.
Consideration of the composition of the DNA samples allows several of the results presented here to be evaluated in light of the above proposals. It is reasonable to assume that when infected cells are harvested for the preparation of total or DNase-resistant DNA, capsids that have either completed or are still in the process of packaging DNA are present in the nucleus. Additionally, although some release of DNA from partially filled capsids may occur, it seems probable that DNase-resistant molecules shorter than unit length correspond closely to the portion of an incompletely packaged genome that is first inserted into the capsid.
Figures 3b and 4 indicate that not only were terminal fragments less abundant in total DNA from KUL25NS-infected cells than in that from cells infected with wt HSV-1, but also that the KUL25NS S terminus was present in significantly lower abundance than the L terminus. Since the initial cleavage of concatemeric DNA to begin packaging would be expected to generate equimolar amounts of the two terminal fragments in total DNA, this suggests that the S terminus is less stable than the L terminus, in agreement with the suggestion that, following the first cleavage event on a concatemer, the L terminus is inserted into a capsid while the S terminus remains unprotected.
The relative impairment of KUL25NS DNA packaging was least when assessed by the amount of L-terminal fragment (BamHI S) present in the DNase-resistant fraction or by hybridization to a probe from RL (Fig. 3b and 5b). In some experiments, only an approximately twofold reduction in BamHI S was observed in KUL25NS-infected cells, demonstrating that the initiation of DNA packaging is not greatly impaired in the mutant. The frequent aborting of packaging at an early stage (i.e., prior to the second cleavage event, which would generate a full-length genome), rather than the failure to retain a cleaved full-length genome in the capsid, could explain the presence of the empty A capsids seen in KUL25NS-infected cells (15). It is pertinent to note that if such early abortive packaging events were to occur at sites along a concatemer separated by the length of a single viral genome, they might possibly result in the generation of the unencapsidated unit-length molecules seen by McNab et al. (15). These ideas, however, remain to be tested experimentally.
Support for a model in which the direction of packaging of the HSV-1 genome is from L to S is provided by the observed overrepresentation of the L terminus and underrepresentation of the S terminus of KUL25NS in DNase-resistant DNA (Fig. 2 and 3) and also by the data obtained upon hybridization of samples resolved by PFGE to probes from RL (pBE1), UL (pGX153), and RS (pST17). The RL probe was able to hybridize to much smaller fragments of wt and KUL25NS DNase-resistant DNAs than the RS probe (Fig. 5). If packaging occurred from L to S, the RL, UL, and RS probes would be expected to hybridize efficiently to fragments larger than approximately 3, 48, and 128 kbp, respectively. The detection of fragments smaller than 128 kbp by the RS probe is probably because of limited shearing of DNA occurring during extraction. Evidence for such shearing is provided by the presence of a smear of DNA extending down to about 50 kbp in the wt HSV-1 cytoplasmic fraction, in which only full-length packaged genomes would be expected to be present (Fig. 5a). Ideally, DNase treatment could be performed on agarose-embedded cells prior to PFGE. However, in such experiments, we have encountered difficulty in inactivating the enzyme prior to capsid lysis and protease digestion, resulting in poor recovery of packaged DNA.
The PFGE analysis of packaged KUL25NS and wt HSV-1 DNAs also indicated that although the mutant was impaired in the overall level of DNA it packaged, the difference was much smaller for molecules up to about 100 kbp in size than for those closer in size to full-length genomes (Fig. 5). This observation suggests that the UL25 protein may play a role in the final stages of inserting a genome length of DNA into the capsid. Several possible modes of action can be envisaged, some of which are analogous to well-characterized events occurring during DNA encapsidation by double-stranded DNA bacteriophages (for reviews, see references 5 and 18). For example, the UL25 protein could be involved in overcoming some rate-limiting step or energy barrier as the DNA content of the capsid increases, promoting a change in capsid structure to allow insertion of more DNA, stabilization of the interaction between the packaging complex and capsid, or enhancing the activity of the terminase. The presence of larger amounts of UL25 protein in C capsids than in procapsids or B capsids (11, 15, 31, 43) suggests that, like the bacteriophage lambda protein gpD (5, 18), its role in DNA packaging may be related to its incorporation into capsids actively packaging DNA. The low level of S-terminal fragment and the weak band of unit-length DNA seen in the pulsed-field gels nevertheless indicate that, even in the absence of UL25 protein, a small proportion of apparently full-length genomes are packaged.
Experiments using plasmid pSA1 demonstrated that KUL25NS stably packaged replicated amplicon DNA relatively more efficiently than it packaged its own genome and in amounts approaching that encapsidated in the presence of wt HSV-1 (Fig. 6 and 8). As noted with the genomic DNAs, the difference in packaging efficiency between cells infected with wt HSV-1 and KUL25NS was most apparent in the highest-molecular-weight species. In contrast to KUL25NS genomic DNA, however, the Uc-containing termini (equivalent to the L terminus of viral DNA) were not represented at significantly higher levels in packaged DNA than those containing Ub (equivalent to the S terminus) (Fig. 7). These results are again compatible with the mutant being relatively unimpaired in cleavage to initiate packaging and the early stages of capsid filling. Unlike concatemeric genomes, in which directly repeated copies of the packaging signals are separated by a full genome length, replicated pSA1 molecules contain direct repeats of the Uc-DR1-Ub fragment at approximately 4.5-kbp intervals. Thus, even though the later stages of head filling may proceed inefficiently in KUL25NS-infected cells, the presence of regularly spaced packaging signals gives ample opportunity for the occurrence of cleavage events to terminate packaging, as was previously observed to occur with tandemly repeated defective HSV-1 DNA (42).
In agreement with previous work (42), in cells infected with wt-HSV-1 only the largest packaged amplicon molecules were transported from the nucleus into the cytoplasm. As noted above for genomic DNA, a proportion of packaged amplicon molecules in KUL25NS-infected cells appeared to be of sizes similar to these species (Fig. 8) but were nevertheless retained in the nucleus. These observations suggest that, although not absolutely necessary for DNA cleavage and packaging, the presence of UL25 protein in the capsid may be an important trigger for translocation into the cytoplasm. If the acquisition of a full complement of UL25 protein by the C capsid is one of the requirements for recognition by the transport machinery and incorporation of the protein occurs during the late stages of capsid filling, this may provide an explanation for why capsids containing only short amplicon molecules or defective genomes are selectively retained in the nucleus.
In summary, the results presented in this paper identify possible roles for the UL25 protein during the late stages of DNA encapsidation and in the maturation of the virus particle. Although other possibilities cannot be conclusively excluded, the imbalance between the abundance of L and S termini in stably packaged KUL25NS DNA is most simply explained by a model in which the directionality of packaging of the HSV-1 genome is from L to S.
ACKNOWLEDGMENTS
I am very grateful to Fred Homa for the provision of mutants gCB and KUL25NS and complementing cell lines 8-1 and C1. My thanks also go to Valerie Preston, Duncan McGeoch, and Andrew Davison for helpful discussions and comments on the manuscript.
REFERENCES
- 1.Abbotts A P, Preston V G, Hughes M, Patel A H, Stow N D. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J Gen Virol. 2000;81:2999–3009. doi: 10.1099/0022-1317-81-12-2999. [DOI] [PubMed] [Google Scholar]
- 2.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 3.Addison C, Rixon F J, Palfreyman J W, O'Hara M, Preston V G. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 4.Ali M A, Forghani B, Cantin E M. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology. 1996;216:278–283. doi: 10.1006/viro.1996.0061. [DOI] [PubMed] [Google Scholar]
- 5.Catalano C E. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol Life Sci. 2000;57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church G A, Wilson D W. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deiss L P, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao M, Matusick-Kumar L, Hurlburt W, DiTusa S F, Newcomb W W, Brown J C, McCann III P J, Deckman I, Colonno R J. The protease of herpes simplex virus type 1 is essential for functional capsid formation and virus growth. J Virol. 1994;68:3702–3712. doi: 10.1128/jvi.68.6.3702-3712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson W, Roizman B. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 11.Kaelin K, Dezélée S, Masse M J, Bras F, Flamand A. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J Virol. 2000;74:474–482. doi: 10.1128/jvi.74.1.474-482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamberti C, Weller S K. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–2473. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 15.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McVoy M A, Adler S P. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McVoy M A, Nixon D E, Hur J K, Adler S P. The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J Virol. 2000;74:1587–1592. doi: 10.1128/jvi.74.3.1587-1592.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murialdo H. Bacteriophage lambda DNA maturation and packaging. Annu Rev Biochem. 1991;60:125–153. doi: 10.1146/annurev.bi.60.070191.001013. [DOI] [PubMed] [Google Scholar]
- 19.Nasseri M, Mocarski E S. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology. 1988;167:25–30. doi: 10.1016/0042-6822(88)90050-5. [DOI] [PubMed] [Google Scholar]
- 20.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 21.Newcomb W W, Homa F L, Thomsen D R, Trus B L, Cheng N, Steven A, Booy F, Brown J C. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogasawara M, Suzutani T, Yoshida I, Azuma M. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J Virol. 2001;75:1427–1436. doi: 10.1128/JVI.75.3.1427-1436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preston V G, Coates J A V, Rixon F J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983;45:1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Register R B, Shafer J A. Alterations in catalytic activity and virus maturation produced by mutation of the conserved histidine residues of herpes simplex virus type 1 protease. J Virol. 1997;71:8572–8581. doi: 10.1128/jvi.71.11.8572-8581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 26.Rixon F J, McNab D. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J Virol. 1999;73:5714–5721. doi: 10.1128/jvi.73.7.5714-5721.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 28.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Severini A, Morgan A R, Tovell D R, Tyrrell D L J. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 30.Shao L, Rapp L M, Weller S K. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology. 1993;196:146–162. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- 31.Sheaffer A K, Newcomb W W, Gao M, Yu D, Weller S K, Brown J C, Tenney D J. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation J. Virol. 2001;75:687–698. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman G, Bachenheimer S. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 33.Slobedman B, Simmons A. Concatemeric intermediates of equine herpesvirus type 1 DNA replication contain frequent inversions of adjacent long segments of the viral genome. Virology. 1997;229:415–420. doi: 10.1006/viro.1997.8447. [DOI] [PubMed] [Google Scholar]
- 34.Smiley J R, Duncan J, Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J Virol. 1990;64:5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spaete R R, Frenkel N. The herpes simplex virus amplicon: a new eukaryotic defective-virus cloning-amplifying vector. Cell. 1982;82:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- 36.Stow N D. Transient assays for HSV origin and replication protein function. In: Brown S M, MacLean A R, editors. Methods in molecular medicine. 10. Herpes simplex virus protocols. Totowa, N.J: Humana Press Inc.; 1998. pp. 215–226. [DOI] [PubMed] [Google Scholar]
- 37.Stow N D, McMonagle E C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983;130:427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- 38.Stow N D, Wilkie N M. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J Gen Virol. 1976;33:447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- 39.Taus N S, Baines J D. Herpes simplex virus DNA cleavage/packaging: the UL28 gene encodes a minor component of B capsids. Virology. 1998;252:443–449. doi: 10.1006/viro.1998.9475. [DOI] [PubMed] [Google Scholar]
- 40.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 42.Vlazny D A, Kwong A, Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full length viral DNA. Proc Natl Acad Sci USA. 1982;79:1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]