Abstract
The hepatitis B virus posttranscriptional regulatory element (PRE) is an RNA element that increases the expression of unspliced mRNAs, apparently by facilitating their export from the nucleus. We have identified a cellular protein that binds to the PRE as the polypyrimidine tract binding protein (PTB), which shuttles rapidly between the nucleus and the cytoplasm. Mutants of the PRE with mutations in PTB binding sites show markedly decreased activity, while cells that stably overexpress PTB show increased PRE-dependent gene expression. Export of PTB from the nucleus, like PRE function, is blocked by a mutant form of Ran binding protein 1 but not by leptomycin B. Therefore, PTB is important for PRE activity and appears to function as an export factor for PRE-containing mRNAs.
Eucaryotic mRNA transcription takes place in the nucleus, but translation occurs in the cytoplasm, necessitating the export of mRNA through nuclear pores. In mammalian cells, this export is strictly controlled, in that only fully spliced and processed mRNA is exported (22, 27). Part of the control is at the level of retention of incompletely spliced mRNA, probably by splicing factors binding to splice sites. However, this retention mechanism cannot explain all of the available data. First, some genes give rise to alternatively spliced transcripts, in which some of the mature mRNAs still contain splice sites. Second, for at least some cellular genes (e.g., the β-globin gene), removal of all introns leads to a defect in the export of mRNA to the cytoplasm (4). Therefore, the presence of splice sites does not always preclude RNA export, while the absence of splice sites does not always lead to export. These data imply that at least some mRNAs contain cis-acting elements that can effect export independently of splicing.
In recent years, the existence of such RNA export elements has been confirmed. The best-studied element is the Rev response element (RRE) of human immunodeficiency virus (HIV) and related lentiviruses (6, 14). Like almost all retroviruses, HIV contains only one promoter that gives rise to a transcript that is alternatively spliced, resulting in the export of completely spliced, partially spliced, and unspliced mRNAs into the cytoplasm. HIV codes for a trans-acting protein product that modulates the relative amounts of completely spliced versus incompletely spliced and unspliced messages. This protein, called Rev, binds to the RRE in the nucleus and strongly enhances the export of RRE-containing, unspliced or incompletely spliced transcripts. It has become clear that Rev contains a leucine-rich nuclear export signal (NES) that allows it to form a trimolecular complex with two cellular proteins, Crm1 (exportin) and Ran (8, 9, 29, 34). This complex, together with its RNA cargo, interacts with components of the nuclear pore in order to migrate into the cytoplasm. Rev is then stripped off the mRNA in the cytoplasm and is recycled back into the nucleus by virtue of a nuclear localization signal.
Other retroviruses also contain RNA export elements which bind to cellular rather than viral trans-acting proteins. The simian retrovirus element is called the constitutive transport element (CTE) (3, 43) and is exported by binding to TAP, the human homolog of the yeast RNA export protein Mex67p (1, 2, 23). RNA helicase A also plays a role in this export pathway (36).
Hepatitis B virus (HBV) is not a retrovirus but also undergoes reverse transcription during its replication and utilizes only unspliced mRNA for expressing its gene products (10). Recently, HBV transcripts were found to contain an element that facilitates the export of HBV transcripts to the cytoplasm (7, 16, 19, 39). This posttranscriptional regulatory element (PRE) can restore export to an unspliced, HIV-derived transcript in the absence of Rev and RRE (7, 20). In addition, the PRE can increase the export of RNA transcribed from an intronless version of the β-globin gene (20). Therefore, the PRE has the attributes of an RNA export element, although effects on other posttranscriptional processing events in the nucleus have also been found and may further contribute to PRE function (18).
The PRE, like the CTE, functions in the absence of any viral proteins and thus presumably binds to one or more cellular trans-acting export factors. These factors apparently utilize an export pathway different from that of either Rev or TAP, since a mutant form of Ran binding protein 1 (RBP1) blocks export mediated by RRE and PRE but not by CTE (41), while leptomycin B blocks export mediated by RRE but not by PRE or CTE (30, 41). Two cellular proteins, approximately 35 and 55 kDa in mass, that bind specifically to functionally important regions of the PRE were previously identified (21). The smaller protein was previously shown to be glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (40), whose functional role in PRE export is still unclear. In this report, we present evidence that the larger protein is the polypyrimidine tract binding protein (PTB), or hnRNP I, an RNA binding protein previously postulated to play a role in mRNA export. Mutants of the PRE that show decreased binding to PTB also show decreased export activity. Overexpression of PTB leads to an increase in PRE-mediated gene expression. PTB shuttles rapidly between the nucleus and the cytoplasm, and its export from the nucleus is blocked by mutant RBP1 but not by leptomycin B. Therefore, PTB has the properties of a cellular effector of PRE function.
MATERIALS AND METHODS
Plasmids.
The chloramphenicol acetyltransferase (CAT) expression plasmid pDM138 was obtained from T. Parslow and contains the simian virus 40 (SV40) enhancer-early promoter driving the expression of a chimeric CAT–HIV type 1 (HIV-1) mRNA that is not highly expressed unless an export element is present within the transcribed region (15). Plasmid pDM138-PRE contains the entire HBV PRE inserted downstream of the CAT gene in the transcribed region of pDM138 (20). Clustered point mutants of pDM138-PRE were obtained by two-stage PCR (11). Plasmid pDM121, which expresses HIV-1 Rev, was obtained from T. Parslow (15). The plasmid that expresses NES-mutated RBP1 was obtained from B. Felber (42). Plasmid pCMV-PTB-myc/cyto expresses human PTB with a myc epitope tag at the C terminus. It was constructed by using PCR to generate the entire human PTB cDNA and inserting this fragment into the XhoI and NotI sites of plasmid pCMV-myc/cyto (Invitrogen). Plasmid pGEM-FIII1, which contains the fragment III region of PRE under the transcriptional control of a T7 promoter, has been previously described (40). Mutants of pGEM-FIII1 were made by two-stage PCR (11).
RNA-protein interaction.
Northwestern blotting, UV cross-linking, and RNA electrophoretic mobility shift assays were performed as described previously (21, 40). The probes were synthesized by transcription in vitro from pGEM-FIII1 or mutants thereof in the presence of [α-32P]UTP. Oligoribonucleotides were purchased from IDT. Western blotting following Northwestern blotting was performed by using purified rabbit antibodies to human PTB (obtained from A. Gil and P. Sharp) at a final concentration of 500 ng/ml, followed by alkaline phosphatase-labeled secondary antibody and chemiluminescence detection using an Amersham Pharmacia ECL kit. Immunoprecipitation following UV cross-linking was performed as described previously (40) using 10 μg of rabbit antibodies to PTB or 10 μg of rabbit antibodies to integrin α1 (Chemicon International) as a negative control.
Selection of PTB-expressing cell lines.
Plasmid PTB-Flag/pcDNA3.1, expressing full-length PTB with a 3′-end Flag tag (Sigma) under the control of the cytomegalovirus immediate-early promoter, was transfected into DBT cells, a murine astrocytoma cell line (13), using the lipid reagent DOTAP (Roche). At 12 h after transfection, the medium was removed from the cells and replaced by selection medium, which consists of modified Eagle's medium with 1× Earle's salts and supplemented with 7% newborn calf serum, 1% glutamine-penicillin-streptomycin solution, 10% tryptose phosphate broth (Difco), and 500 ng of G418 sulfate (Omega Scientific)/ml. The transfected cells were cultured for 4 weeks with refreshment of the selection medium every 3 days. Colonies were picked and examined for the expression of Flag-tagged PTB with both Western blotting and immunofluorescence using an antibody against the Flag tag (Sigma), and one representative clone was picked for the experiments described here. Exogenous PTB is predominantly localized in the nucleus, similar to endogenous PTB (data not shown). Cells transfected with the empty vector plasmid were similarly selected.
Cell culturing, transfection, and protein and RNA analyses.
HuH-7 human hepatoma cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Stably transfected DBT cells were grown in G418 selection medium (see above). Transient transfection was done with the calcium phosphate method and 60-mm dishes (5 Prime→3 Prime, Inc.). Two days after transfection, the cells were harvested for either CAT assays or nuclear and cytoplasmic RNA extraction as described previously (41) or nuclear export assays as described below. RNA samples were electrophoresed on a denaturing 1% agarose gel, transferred to a nitrocellulose membrane, and probed for CAT and GAPDH RNAs as described previously (41).
Nuclear export assays.
To assay the nuclear export of endogenous PTB, the human-mouse heterokaryon assay was used (25). HuH-7 human hepatoma cells and mouse NIH 3T3 cells were separately cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. At 30 h posttransfection, the transfected HuH-7 cells were mixed with an equal number of NIH 3T3 cells, and the mixture was seeded onto a two-well chamber slide. After overnight incubation, 50 μg of cycloheximide/ml was added to the culture medium to inhibit protein synthesis for 45 min. The cocultured cells were then fused with 50% polyethylene glycol 8000 in serum-free medium for 1 min, washed once with medium containing 50 μg of cycloheximide/ml, and incubated in medium containing 50 μg of cycloheximide/ml for 15 min before fixation and immunostaining. In some experiments, 2.5 ng of leptomycin B/ml (courtesy of M. Yoshida) (28) was added to the cocultured cells 1 h before the addition of cycloheximide. To assay the export of exogenously expressed PTB, HuH-7 cells were transfected with pCMV-PTB-myc/cyto, which expresses human PTB with a myc epitope tag at the C terminus. In some experiments, HuH-7 cells were cotransfected with a plasmid that expresses RBP1 with a mutated NES, fused to green fluorescent protein (GFP) (42).
Because a small but significant portion of Rev can be present in the cytoplasm at steady state (33), the heterokaryon assay is not suitable for examining the nuclear export of Rev. Instead, we examined the subcellular localization of Rev following actinomycin D treatment, which decreases the relative rate of Rev import versus export and hence causes Rev to localize to the cytoplasm at steady state (33, 35). Briefly, HuH-7 cells were transfected with pDM121, which expresses HIV-1 Rev. After 2 days, the cells were treated with 50 μg of cycloheximide/ml for 45 min followed by 1 μg of actinomycin D/ml for 1 h and processed for immunostaining. In some experiments, 2.5 ng of leptomycin B/ml was added 1 h before the addition of cycloheximide.
Immunofluorescence microscopy.
Cells were fixed and permeabilized with 100% methanol for 5 min. Immunostaining with primary antibody was done by incubating the cells with a mouse antibody specific for human PTB (courtesy of S. Huang and D. Spector) (17), a mouse anti-myc antibody (Invitrogen), or a mouse anti-Rev antibody (Intracel Corporation) at a 1:100 dilution in phosphate-buffered saline (PBS) for 1 h. After washing of the cells with PBS, secondary antibody staining was done by incubating the cells with PBS containing fluorescein isothiocyanate-labeled goat anti-mouse antibody (Sigma) at a 1:25 dilution and saturated 4′,6′-diamidino-2-phenylindole (DAPI) at a 1:100 dilution for 1 h. The stained cells were washed, placed on coverslips, and visualized with a Zeiss epifluorescence microscope.
RESULTS
Binding of PTB to PRE.
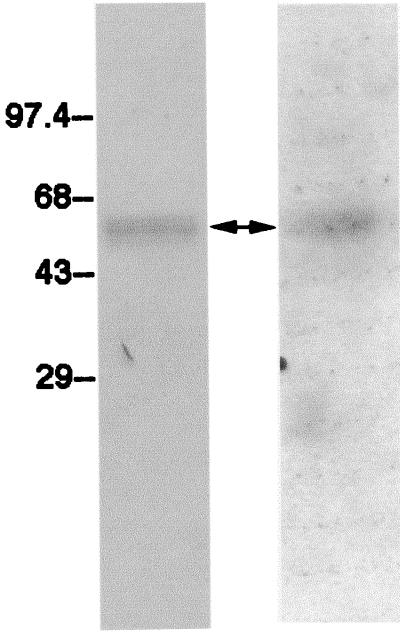
A functionally important subelement of the PRE was previously localized between map positions 1487 and 1582 of HBV strain adw2 (21). This subelement, named fragment III, binds strongly to an ∼55-kDa cellular protein, as revealed by Northwestern blotting of nuclear proteins separated by gel electrophoresis and probed with 32P-labeled fragment III (Fig. 1, right panel). In order to identify this protein, antibodies to known RNA binding proteins of similar masses were used in Western blotting of the nuclear extracts. As shown in Fig. 1, left panel, blotting with antibodies to PTB revealed a pattern similar to that obtained in Northwestern blotting with fragment III. This result raised the possibility that the PRE binding protein is PTB.
FIG. 1.
Serial Northwestern and Western blotting of nuclear proteins. Nuclear proteins were isolated from HuH-7 cells and electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel. The separated proteins were transferred to a polyvinylidene difluoride membrane. PRE binding proteins (arrow) were revealed by Northwestern blotting with a 32P-labeled PRE fragment III probe and autoradiography (right panel). Northwestern blotting is relatively inefficient in detecting the binding of GAPDH to fragment III; hence, the GAPDH band is not visible in the autoradiogram. The blot was washed, and PTB was detected by Western blotting with rabbit antibodies against human PTB (left panel). Numbers at left are kilodaltons.
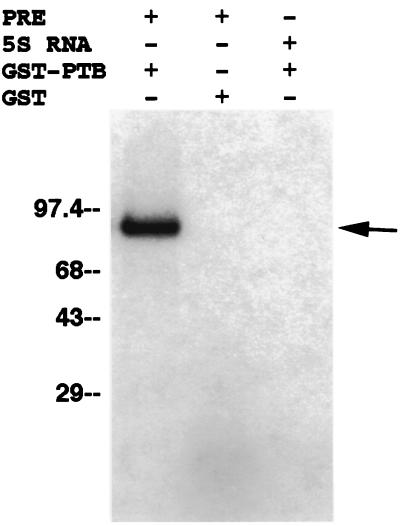
To confirm that PTB can bind to the PRE, we used two different methods to detect the binding of fragment III by a fusion protein containing PTB and glutathione S-transferase (GST) purified from bacteria (courtesy of A. Gil and P. Sharp) (12). The first method was UV cross-linking to 32P-labeled probes, which revealed that fragment III indeed bound to GST-PTB (Fig. 2, left lane). This binding was specific, since fragment III did not bind to GST present at more than eight times the concentration of GST-PTB (Fig. 2, middle lane) and since 5S RNA from Xenopus did not bind GST-PTB (Fig. 2, right lane). In the second experiment, we used an electrophoretic mobility shift assay (EMSA). Again, GST-PTB bound labeled fragment III, as assessed by the retarded mobility of the probe, while GST did not (data not shown; see also Fig. 5). Therefore, purified PTB expressed as a fusion protein in bacteria can bind fragment III in vitro.
FIG. 2.
UV cross-linking of a recombinant GST-PTB fusion or GST to PRE fragment III or 5S RNA. GST-PTB (3 μg) (right and left lanes) and GST (25 μg) (middle lane) purified from bacteria were separately incubated with 32P-labeled PRE fragment III (left and middle lanes) or 5S RNA (right lane) and cross-linked with UV irradiation. After RNase digestion, the mixtures were electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel and labeled proteins (arrow) were detected by autoradiography. Numbers at left are kilodaltons.
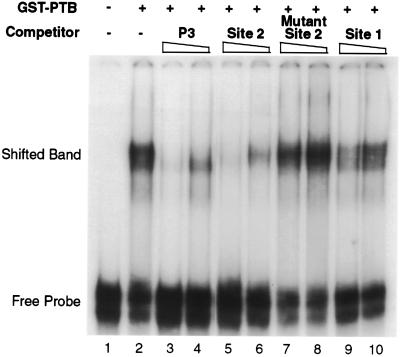
FIG. 5.
EMSA of wild-type PRE fragment III binding to GST-PTB and competition by oligoribonucleotides. 32P-labeled PRE fragment III was incubated with or without (lanes 2 and 1, respectively) 8 ng of GST-PTB and electrophoresed on a native polyacrylamide gel. In addition, unlabeled oligoribonucleotides corresponding to a known PTB site (P3) from intron 2 of α-tropomyosin (lanes 3 and 4) (31), site 2 of fragment III (lanes 5 and 6), mutant site 2 (lanes 7 and 8), and site 1 of fragment III (lanes 9 and 10) were included in the incubation mixture at a 50 or 100-fold molar excess (even- and odd-numbered lanes, respectively) over the amount of labeled probe.
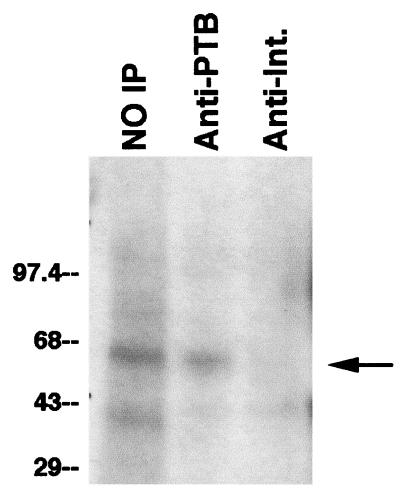
To assess whether cellular PTB can similarly bind fragment III, we cross-linked 32P-labeled fragment III to nuclear extracts and immunoprecipitated the mixture with antibodies to PTB. As shown in Fig. 3, left lane, cross-linking under these conditions revealed two bands, a stronger band corresponding to the 55-kDa protein and a weaker band corresponding to GAPDH (40). Antibodies to PTB precipitated the larger labeled protein but not GAPDH (Fig. 3, middle lane). In contrast, antibodies to an irrelevant protein (integrin α1) precipitated neither protein (Fig. 3, right lane). Together, these data establish that PTB binds to the PRE, both as a purified fusion protein and when present in its native state in a mixture of total nuclear proteins.
FIG. 3.
Immunoprecipitation of nuclear proteins cross-linked to PRE fragment III. Nuclear proteins extracted from HuH-7 cells were cross-linked by UV irradiation to 32P-labeled PRE fragment III. After RNase digestion, the mixture was divided into three portions. One was not further manipulated (NO IP), one was immunoprecipitated with rabbit antibodies to human PTB, and one was immunoprecipitated with rabbit antibodies to human integrin αl (Int.). All three portions were then electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel, and the labeled proteins were detected by autoradiography. The band corresponding to PTB is marked with an arrow. A weak, nonspecific band migrating at ∼43 kDa is present in both immunoprecipitates. Numbers at left are kilodaltons.
Localization of PTB binding sites.
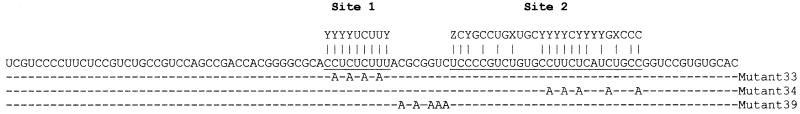
PTB, as its name implies, binds preferentially to pyrimidine-rich RNA sequences. Indeed, examination of the fragment III sequence revealed that it comprises 70% pyrimidine residues (Fig. 4). However, PTB binding also depends on the primary sequence of the RNA. Two groups have used SELEX to obtain putative consensus high-affinity PTB binding sites that differ somewhat from each other (31, 32). Therefore, we used a computer program to search for similar sequences within fragment III. Two such sites were found, with one homologous to each consensus sequence (Fig. 4, sites 1 and 2).
FIG. 4.
Sequence of PRE fragment III from HBV strain adw2. The two potential PTB sites are underlined. Site 1 shows homology to the consensus sequence UCUU embedded in a pyrimidine-rich region and identified by Perez et al. (31), while site 2 shows homology to a longer consensus sequence identified by Singh et al. (32). The mutated sequences in mutants 33, 34, and 39 are shown below the wild-type sequence. X, Y or G; Y, =C or U; Z, U or G.
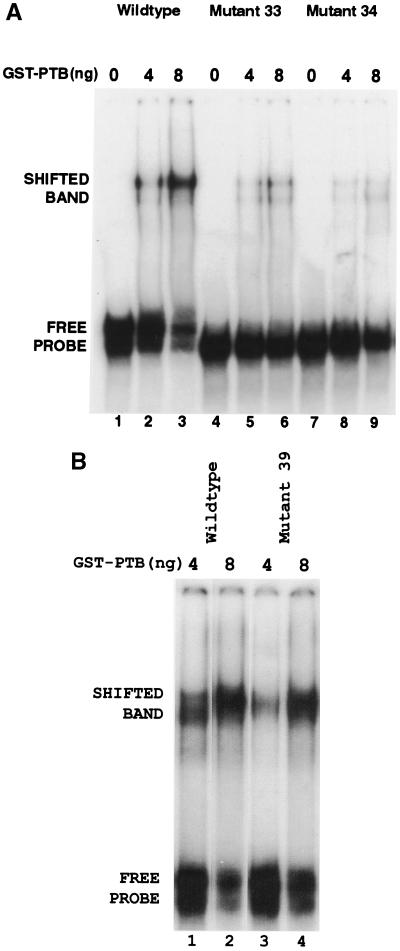
To determine which, if either, of these sites may be a true PTB binding site, we synthesized oligoribonucleotides comprising each site and assessed their ability to compete with fragment III for binding to GST-PTB in an EMSA. As shown in Fig. 5, the oligoribonucleotide corresponding to site 1 weakly competed for GST-PTB binding (lanes 9 and 10), while site 2 competed as efficiently as an authentic high-affinity PTB binding site, namely, P3 from intron 2 of α-tropomyosin (compare lanes 5 and 6 with lanes 3 and 4, respectively) (31). The binding was specific, since a mutant site 2 with five A residues substituted for pyrimidines (mutant 34 in Fig. 4) did not compete (Fig. 5, lanes 7 and 8). Therefore, both sites 1 and 2 bind PTB, with the latter binding more strongly than the former.
Since RNA molecules fold into higher-order structures that may affect binding to proteins, we also tested whether sites 1 and 2 are important for PTB binding in the context of the entire fragment III. To this end, we performed an EMSA on fragment III with either site 1 or site 2 mutated by substitution of A residues for pyrimidines (mutants 33 and 34, respectively, in Fig. 4). As shown in Fig. 6A, both mutant 33 (lanes 4 to 6) and mutant 34 (lanes 7 to 9) showed markedly decreased binding compared with the wild type (lanes 1 to 3). In contrast, substitution of a similar number of A residues for pyrimidines between the two sites (mutant 39 in Fig. 4) did not significantly decrease binding to PTB (Fig. 6B). These results confirm that both sites 1 and 2 in the context of fragment III bind PTB. It is noteworthy that both sites are well conserved, being identical in sequence in 12 HBV genomes (two each from the six genotypes A to F; GenBank accession numbers V00866, X51970, J02203, U95551, D00630, D28880, X97850, X97851, X75657, X75664, X75658, and X75663, respectively).
FIG. 6.
EMSA of wild-type and mutant PRE fragment III binding to GST-PTB. (A) 32P-labeled wild-type fragment III, mutant 33, or mutant 34 was incubated with the indicated amount of GST-PTB and then electrophoresed on a native polyacrylamide gel. Mutants 33 and 34 contain clustered point mutations in sites 1 and 2, respectively (Fig. 4). (B) 32P-labeled wild-type fragment III or mutant 39 was incubated with the indicated amount of GST-PTB and then electrophoresed on a native polyacrylamide gel. Mutant 39 contains clustered point mutations between sites 1 and 2 (Fig. 4).
Role of PTB binding in PRE function.
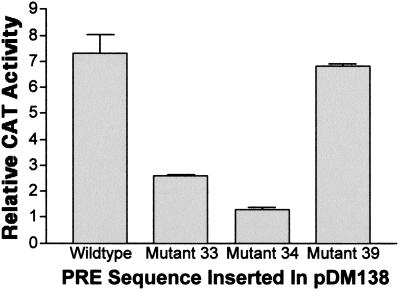
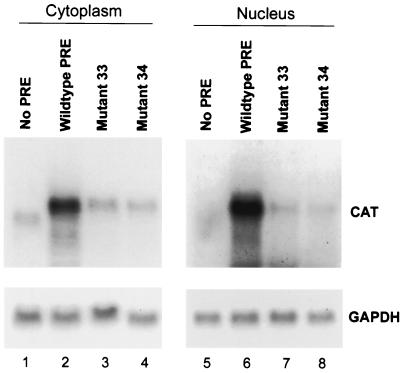
The binding of PTB to fragment III does not necessarily reflect a role for PTB in PRE function. To assess the functional significance of PTB binding, we performed two sets of experiments. First, we determined the effect of diminishing PTB binding on PRE function by individually mutating sites 1 and 2 in the context of the entire PRE and inserting these mutations into pDM138. This plasmid contains the SV40 enhancer-early promoter driving the bacterial CAT gene, which is embedded within a poorly utilized intron from HIV-1 (15). Consequently, the CAT message is largely degraded in the nucleus rather than exported to the cytoplasm, unless an export element is inserted downstream of the CAT gene. The activity of this export element in transfected cells can be easily quantitated by enzymatic assays of CAT activity. In HuH-7 human hepatoma cells, the PRE activates CAT expression from this plasmid by approximately sevenfold (Fig. 7) (20). Mutating either PTB site of PRE fragment III by substituting A residues for pyrimidines significantly reduced PRE function, with a greater effect of mutating site 2 than of mutating site 1 (mutants 34 and 33, respectively, in Fig. 7; sequences are shown in Fig. 4). In contrast, similar substitutions introduced into a region between sites 1 and 2 (mutant 39 in Fig. 4) had no significant effect on PRE function (Fig. 7). Northern blotting confirmed a significant decrease in cytoplasmic unspliced CAT mRNA amounts in mutant 33- and mutant 34-transfected cells as well as a decrease in nuclear CAT RNA (Fig. 8), indicating decreases in both nuclear stability and export of unspliced RNA, as expected from previous data (30, 41). Note that the PRE has no effect on transcription from the SV40 promoter in this plasmid (20). Thus, these results show that both PTB sites in fragment III are important for PRE to function posttranscriptionally.
FIG. 7.
Effect of mutations in PTB sites on the function of the PRE in plasmid pDM138, as assessed by CAT activity. HuH-7 cells were transfected with parental pDM138 or pDM138 containing either wild-type full-length PRE or PRE with mutations in PTB site 1 (mutant 33), PTB site 2 (mutant 34), or an irrelevant site (mutant 39). After 2 days, the cell lysates were assayed for CAT activity with thin-layer chromatography. The data are the mean and standard deviation for three transfections and represent the relative CAT activity normalized to that for cells transfected with pDM138.
FIG. 8.
Effect of mutations in PTB sites on the function of the PRE in plasmid pDM138, as assessed by CAT RNA levels in the nucleus and cytoplasm. HuH-7 cells were transfected with parental pDM138 or pDM138 containing either wild-type full-length PRE or PRE with mutations in PTB site 1 (mutant 33) or PTB site 2 (mutant 34). After 2 days, total RNA was extracted from the nuclei and cytoplasm of these cells and used for Northern blotting with a 32P-labeled CAT probe. As an internal control, the blot was washed and reprobed for GAPDH RNA. Similar results were obtained from a duplicate experiment.
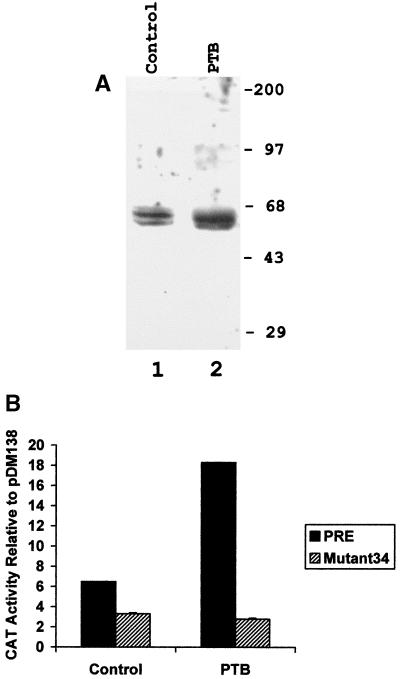
We next performed the converse experiment by determining if the overexpression of PTB would increase PRE-mediated gene expression. We used a murine cell line which overexpresses human PTB from a stably integrated plasmid (Fig. 9A). Another cell line stably transfected with the empty vector plasmid was used as a control. Both of these cell lines were transfected with either pDM138-PRE or parental pDM138, and the amounts of CAT expression in the cells were quantitated. As shown in Fig. 9B, CAT expression from pDM138-PRE was approximately sixfold higher than that from pDM138, similar to the sevenfold activation seen with HuH-7 cells (Fig. 7). However, in PTB-overexpressing cells, this ratio increased to approximately 18 (Fig. 9B). Thus, the expression of exogenous PTB caused a threefold increase in the function of the PRE. In contrast, the CAT activity expressed from mutant 34 was not increased by the expression of PTB (Fig. 9B). These results show that PTB is a positive trans-acting factor for the PRE.
FIG. 9.
PRE function in murine cells overexpressing PTB. (A) Western blot of DBT cells stably transfected with a plasmid expressing human PTB (lane 2) or empty vector plasmid (lane 1). The former cells express approximately twice as much of the long form of PTB as the latter cells. The short form is not expressed from the cDNA expression plasmid, since it is generated by alternative splicing. Numbers at right are kilodaltons. (B) Cells were transfected with pDM138, pDM138-PRE, or mutant 34. After 2 days, the cells were harvested and assayed for CAT activity. The data are the mean and standard deviation for three transfections and represent the relative amount of CAT activity in cells transfected with either pDM138-PRE or mutant 34, normalized to the amount of CAT activity in cells transfected with pDM138.
PTB as a potential RNA export factor.
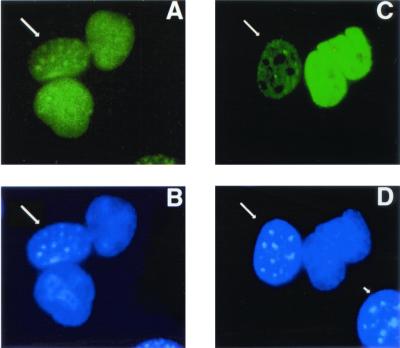
While PTB is a nuclear protein at steady state, it has been previously shown to shuttle rapidly between the nucleus and the cytoplasm and thus potentially can play a direct role in the export of PRE-containing RNA (26). If such is the case, then its export from the nucleus should show the same pattern of sensitivity to inhibitors as that seen with PRE. It was previously shown that PRE-mediated export is not blocked by leptomycin B, an antibiotic that blocks the function of a leucine-rich NES, such as the one present in Rev (38). Therefore, we determined the effect of leptomycin B on PTB export. This was accomplished with a heterokaryon fusion assay (25), in which we fused human HuH-7 cells with murine NIH 3T3 cells in the presence of cycloheximide and stained the heterokaryons with a monoclonal antibody that recognizes human but not murine PTB (17). The murine nuclei can be identified by the presence of bright granules with nuclear staining by DAPI (25) (compare the nuclei marked by arrows with the other nuclei in Fig. 10B and D). Even in the presence of leptomycin B, at 15 min after fusion, there was already a substantial amount of human PTB in the murine nucleus (Fig. 10A), similar to the situation seen in the absence of leptomycin B (Fig. 10C). A positive control experiment showed that leptomycin B prevented Rev nuclear export under the conditions used (data not shown). Therefore, PTB export is not blocked by leptomycin B, just like PRE function.
FIG. 10.
Effect of leptomycin B on PTB nuclear export in a heterokaryon fusion assay. Human HuH-7 and murine NIH 3T3 cells were cocultured in the presence of cycloheximide and fused for 15 min. (A and B) Cells were pretreated with leptomycin B to block leucine-rich NES-mediated export. Human PTB was localized with a monoclonal antibody that recognizes human but not murine PTB and fluorescein isothiocyanate-labeled secondary antibody (A and C), while nuclei were stained with DAPI (B and D). Murine nuclei (arrows) can be identified by the presence of bright DAPI-positive granules that are absent in human nuclei. In the fused cells, even in the presence of leptomycin B, murine nuclei stained for human PTB (long arrows), while the murine nucleus in an unfused cell (short arrow) did not stain.
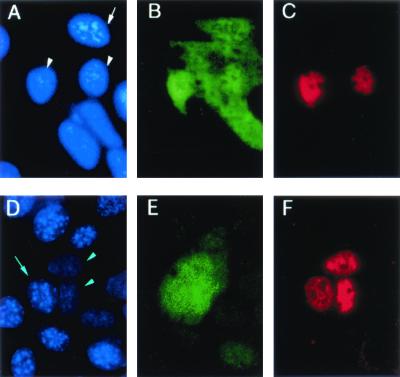
On the other hand, PRE-mediated gene expression is blocked by a mutant form of RBP1 that lacks its NES (41). To determine if mutant RBP1 also blocks PTB export, we again used the heterokaryon fusion assay. However, because mutant RBP1 must be expressed from a transfected plasmid, we looked for export of myc-tagged PTB expressed from a cotransfected plasmid rather than export of native human PTB. A murine nucleus (Fig. 11A) that had imported mutant RBP1 from the cytoplasm (Fig. 11B) failed to import PTB (Fig. 11C) from the transfected human nuclei (Fig. 11A) in the heterokaryon. Thus, mutant RBP1 must have blocked the export of PTB from the nucleus to the cytoplasm. This effect was not due to the GFP moiety fused to mutant RBP1, since GFP by itself (Fig. 11E) did not prevent the export of PTB from human nuclei (Fig. 11D) into a mouse nucleus (Fig. 11F). Taken together, these data are consistent with PTB being an export factor for PRE-containing RNA.
FIG. 11.
Effect of mutant RBP1 on PTB nuclear export in a heterokaryon fusion assay. Human HuH-7 cells were cotransfected with plasmids that express mutant RBP1 fused to GFP and human PTB tagged with a myc epitope. After 2 days, the cells were cocultured with murine NIH 3T3 cells in the presence of cycloheximide and fused for 15 min. RBP1 was localized by the green fluorescence of GFP (B), PTB was localized to the myc epitope by a monoclonal antibody and Cy3-labeled secondary antibody (C), while nuclei were stained with DAPI (A). Two transfected human nuclei (A, arrowheads) can be identified by the presence of red staining for PTB. As expected, mutant RBP1 was present in both the cytoplasm and these two human nuclei. In addition, however, mutant RBP1 was imported into a murine nucleus (A, arrow), indicating that a human-mouse heterokaryon had formed. This murine nucleus remained negative for myc-tagged PTB, whose export from the human nuclei thus must have been blocked by mutant RBP1. A negative control experiment using unfused GFP (E) confirmed that GFP itself has no effect on the export of PTB (F) from human nuclei (D, arrowheads) into murine nuclei (D, arrow).
DISCUSSION
The PRE is important for high-level HBV gene expression by acting at the posttranscriptional level to increase the amount of cytoplasmic mRNA (16, 19, 39). It can also increase the amount of unspliced HIV-derived mRNA in the cytoplasm (19), similar to the RRE in the presence of Rev, which are known cis- and trans-acting effectors of RNA export, respectively. Therefore, the PRE has the attributes of an RNA export element. To begin to understand the molecular mechanism of PRE function, we have looked for cellular proteins that bind to PRE. Two cellular proteins, approximately 35 and 55 kDa in mass, were previously shown to bind to the PRE (21). The smaller protein was identified as GAPDH (40), whose role in PRE function is as yet undefined. The data presented here show that the other PRE binding protein is PTB, a well-known RNA binding nuclear protein. PTB binds to two pyrimidine-rich regions of the critical central region of PRE. These two sites are entirely conserved among 12 HBV isolates of all six genotypes. When either PTB binding site is mutated, the function of PRE is markedly compromised. In contrast, mutation of a region between the two sites has no effect. Furthermore, mutation of the higher-affinity site (site 2) has a greater detrimental effect on PRE function than mutation of the other site. These results are all consistent with PTB playing an important role in PRE function. Nevertheless, it remained possible that another cellular factor that is the true effector of PRE function also binds to these sites and/or that mutation of these sites changes the secondary or tertiary structure of the PRE and thus prevents the binding of another factor at other sites within the PRE. However, we have also been able to demonstrate a strong positive effect of the expression of exogenous PTB on PRE function. This result confirms that PTB binding to PRE is functionally important. It is pertinent that a putative export element in the herpesvirus thymidine kinase mRNA has been shown to bind to hnRNP L (24), which is the hnRNP showing the greatest sequence homology to PTB.
PTB is known to play a role in pre-mRNA metabolism. Specifically, it has been implicated in regulating the selection of alternative exons by repressing the usage of splice sites in the vicinity of cognate binding sites (37). Therefore, PTB can potentially increase the export of mRNA by an indirect mechanism, e.g., by blocking an alternative fate of the pre-mRNA (interaction with the splicing machinery). On the other hand, PTB also shuttles rapidly between the nucleus and the cytoplasm by virtue of possessing a nuclear localization signal and an NES (26); hence, it has the capacity of being more directly involved in mRNA export. Indeed, Xenopus PTB may play a role in the localization of Vg1 mRNA in the oocyte cytoplasm (5). Furthermore, we have shown here that the export of PTB is blocked by mutant RBP1 but not by leptomycin B, just like PRE-mediated mRNA export (41). Taken together, these data suggest that PTB directly plays a role in PRE-mediated export, although they do not rule out the possibility that PTB also has other effects on HBV pre-mRNA metabolism.
Our data show that PTB is important for PRE function. Based on this observation, we have attempted to examine whether the introduction of consensus PTB sites into plasmid pDM138 would increase CAT gene expression. However, no significant increase in CAT activity was observed (data not shown). One interpretation of this result is that for PTB to function as an export factor, its cognate sites must be in a specific spatial array in the RNA. On the other hand, this result raises the possibility that other cellular proteins (such as GAPDH) are required to cooperate with PTB for PRE function. These roles may include helping PTB to bind PRE, preventing an alternative fate of the pre-mRNA in the nucleus, functioning as a cofactor in nuclear export, and/or stripping PTB from mRNA that has reached the cytoplasm.
In summary, we have presented data showing that PTB plays an important role in PRE function by binding to cis sequences in PRE. Further experiments are needed to understand the detailed mechanism of action of PTB and the possible role of other cellular factors in PRE function.
ACKNOWLEDGMENTS
W.-Q.Z. and B.L. contributed equally to this work.
We thank B. Felber, A. Gil, T. Hope, S. Huang, T. Parslow, P. Sharp, H. Shida, D. Spector, and M. Yoshida for plasmids, reagents, and/or antibodies.
This work was supported by a VA merit review grant and BioSTAR grant S97-11 from the University of California and Immune Response Corporation.
REFERENCES
- 1.Bachi A, Braun I C, Rodrigues J P, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bear J, Tan W, Zolotukhin A S, Tabernero C, Hudson E A, Felber B K. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchman A R, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cote C A, Gautreau D, Denegre J M, Kress T L, Terry N A, Mowry K L. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 6.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 7.Donello J E, Beeche A A, Smith III G J, Lucero G R, Hope T J. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J Virol. 1996;70:4345–4351. doi: 10.1128/jvi.70.7.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals [see comments] Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 10.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2703–2737. [Google Scholar]
- 11.Ge L, Rudolph P. Simultaneous introduction of multiple mutations using overlap extension PCR. BioTechniques. 1997;22:28–30. doi: 10.2144/97221bm03. [DOI] [PubMed] [Google Scholar]
- 12.Gil A, Sharp P A, Jamison S F, Garcia-Blanco M A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 13.Hirano N, Fujiwara K, Hino S, Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 14.Hope T J. Viral RNA export. Chem Biol. 1997;4:335–344. doi: 10.1016/s1074-5521(97)90124-1. [DOI] [PubMed] [Google Scholar]
- 15.Hope T J, McDonald D, Huang X J, Low J, Parslow T G. Mutational analysis of the human immunodeficiency virus type 1 Rev transactivator: essential residues near the amino terminus. J Virol. 1990;64:5360–5366. doi: 10.1128/jvi.64.11.5360-5366.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang S, Deerinck T J, Ellisman M H, Spector D L. The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Wimler K M, Carmichael G G. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Z M, Yen T S. Hepatitis B virus RNA element that facilitates accumulation of surface gene transcripts in the cytoplasm. J Virol. 1994;68:3193–3199. doi: 10.1128/jvi.68.5.3193-3199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z M, Yen T S. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Z M, Zang W Q, Yen T S. Cellular proteins that bind to the hepatitis B virus posttranscriptional regulatory element. Virology. 1996;217:573–581. doi: 10.1006/viro.1996.0152. [DOI] [PubMed] [Google Scholar]
- 22.Izaurralde E, Mattaj I W. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 23.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Mertz J E. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 25.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 26.Michael W M, Siomi H, Choi M, Pinol-Roma S, Nakielny S, Liu Q, Dreyfuss G. Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harbor Symp Quant Biol. 1995;60:663–668. doi: 10.1101/sqb.1995.060.01.071. [DOI] [PubMed] [Google Scholar]
- 27.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 28.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 29.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 30.Otero G C, Harris M E, Donello J E, Hope T J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez I, McAfee J G, Patton J G. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- 32.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 33.Soros V, Cochrane A. Alterations in HIV-1 Rev transport in response to cell stress. Virology. 2001;280:199–210. doi: 10.1006/viro.2000.0752. [DOI] [PubMed] [Google Scholar]
- 34.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 35.Stauber R, Gaitanaris G A, Pavlakis G N. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology. 1995;213:439–449. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 36.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 37.Valcarcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 38.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 39.Yen T S B. Posttranscriptional regulation of gene expression in hepadnaviruses. Semin Virol. 1998;8:319–326. [Google Scholar]
- 40.Zang W Q, Fieno A M, Grant R A, Yen T S. Identification of glyceraldehyde-3-phosphate dehydrogenase as a cellular protein that binds to the hepatitis B virus posttranscriptional regulatory element. Virology. 1998;248:46–52. doi: 10.1006/viro.1998.9255. [DOI] [PubMed] [Google Scholar]
- 41.Zang W Q, Yen T S B. Distinct export pathway utilized by the hepatitis B virus posttranscriptional regulatory element. Virology. 1999;259:299–304. doi: 10.1006/viro.1999.9777. [DOI] [PubMed] [Google Scholar]
- 42.Zolotukhin A S, Felber B K. Mutations in the nuclear export signal of human ran-binding protein RanBP1 block the Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Biol Chem. 1997;272:11356–11360. doi: 10.1074/jbc.272.17.11356. [DOI] [PubMed] [Google Scholar]
- 43.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]