Abstract
The p53 mutation is the most common genetic mutation associated with human neoplasia. TP53 missense mutations, which frequently arise early in breast cancer, are present in over thirty percent of breast tumors. In breast cancer, p53 mutations are linked to a more aggressive course of the disease and worse overall survival rates. TP53 mutations are mostly seen in triple-negative breast cancer, a very diverse kind of the disease. The majority of TP53 mutations originate in the replacement of individual amino acids within the p53 protein's core domain, giving rise to a variety of variations referred to as "mutant p53s." In addition to gaining carcinogenic qualities through gain-of-function pathways, these mutants lose the typical tumor-suppressive features of p53 to variable degrees. The gain-of-function impact of stabilized mutant p53 causes tumor-specific dependency and resistance to therapy. P53 is a prospective target for cancer therapy because of its tumor-suppressive qualities and the numerous alterations that it experiences in tumors. Phenotypic abnormalities in breast cancer, notably poorly differentiated basal-like tumors are frequently linked to high-grade tumors. By comparing data from cell and animal models with clinical outcomes in breast cancer, this study investigates the molecular mechanisms that convert gene alterations into the pathogenic consequences of mutant p53's tumorigenic activity. The study delves into current and novel treatment approaches aimed at targeting p53 mutations, taking into account the similarities and differences in p53 regulatory mechanisms between mutant and wild-type forms, as well.
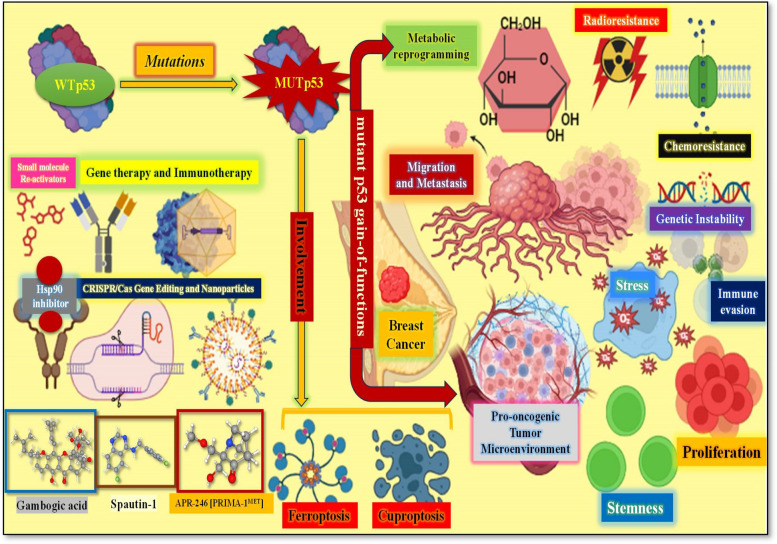
Graphical Abstract
Keywords: Breast cancer, Mutp53, Gain-of-function, HSP90, Ferroptosis, Metabolic reprogramming
Introduction
The breast cancer is characterized by a high global incidence and besides affecting the breast tissue, it has the ability to metastasize to lungs and bones [1, 2]. The genome instability represents one of the cancer hallmarks [3, 4]. The frequently altered gene in cancers in humans is the tumor suppressor TP53, in almost 50% of all cancers. The mutation in TP53 has a strong connection with cancer risk [5]. Located at 17p13.1 locus, the human tumor suppressor gene TP53 encodes a 393 amino acid-long protein, the p53 [6]. P53 is a transcription factor that can be activated by internal and external stressors such as UV radiation and oncogenes. These stressors trigger signals that modify the P53 protein post-translationally, freeing it from its interaction with the MDM2 protein and resulting in P53 activation [7]; Subsequently, p53 may reach the nucleus and stimulate the production of several target genes [8] (Fig. 1). Reported findings suggest that P53 has non-transcriptional activities as well that may contribute to tumor suppression action [9].
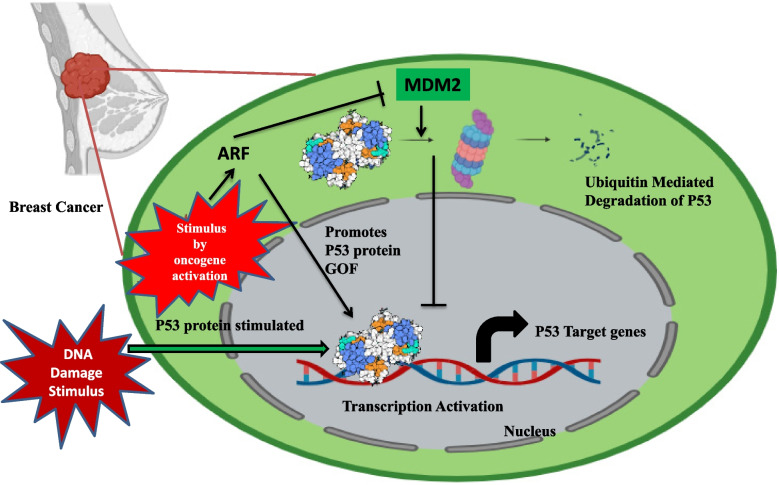
Fig. 1.
The regulation of p53 by MDM2: Highlighting the p53-Mdm2 negative feedback loop, wherein p53 transcriptionally activates Mdm2, which in turn aims p53 for degradation
P53 is an essential tumor suppressor because it represents an important modulator of DNA repair and DNA replication stress [10–12] to achieve the maintenance in genome stability. Be it the DNA damage, oncogenic signaling or oxidative stress, the p53 responds to all these varied cellular stresses [12, 13]. p53’s functional role and expression are hindered by its negative modulator MDM2 protein to be balanced at a low level in case of unstressed, non-transformed cells [14–16]. On the other hand, the interaction of p53-MDM2 is lost and in stressed cells p53 expression upregulation is witnessed [17]. In order to eliminate the damaged cells, upregulated p53 mediates the arrest of cell cycle as well as apoptosis [18]. There is a complex connection of p53 with the survival or death of tumor cells via metabolism modulation [19].
In addition to being a tumor suppressor, the transcription factor p53 is regarded as one of the chief potential targets for curing cancer at molecular level. This is because it modulates a plethora of intracellular metabolic pathways like apoptosis and senescence in addition to DNA damage repair (Fig. 2). Regarded as the “Guardian of the genome”, the p53 protein hampers the cell proliferation carrying genetic anomalies, especially oncogenic mutations. Be it ubiquitination, acetylation, phosphorylation or methylation, the p53 undergoes post-translational modifications in both stressed and unstressed cells. These post-translational modifications help modulate the stability, transcriptional activity as well as cytoplasmic or nuclear localization of p53 protein. And in response to the cellular stress, the Ser or Thr residues undergo phosphorylation and correlates with enhanced p53 activity [20]. The p53’s activation in response to several stresses is significant for normal cells to survive and protect themselves from the process of carcinogenesis. But TP53 is often altered in maximum human cancers ending up in loss of functions (LOFs), important for suppressing tumor and even gain of functions (GOFs) important for growth of tumor [21, 22]. The missense mutation represents the chief type of p53 alteration in the DNA-binding domain (DBD), affecting a solo amino acid in the p53 protein but possessing huge impact on p53’s role in breast cancer [23]. The growing data suggests that additional processes, such as regulating cancer cell metabolism, ferroptosis, and cell stemness, play a significant part in supporting p53's anti-cancer function [19, 24].
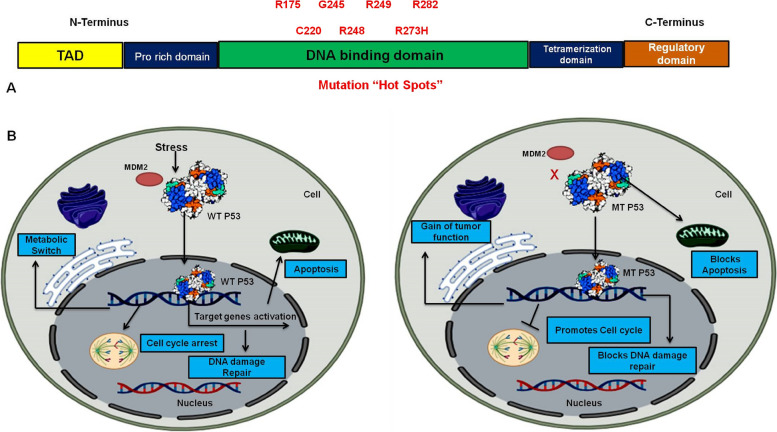
Fig. 2.
A Representing protein sequence of P53 protein where DNA binding domain is mutated with seven hot spot mutations. B Diagrammatic representation of the functional attributes of wild type (WT) P53 and Mutant (MT) P53
The ways through which mutant p53 facilitates cancer progression
There are several ways, through which mutant p53 facilitates cancer progression like inducing genetic instability, high proliferation, metabolism regulation, promoting metastasis, Pro-oncogenic Tumor Microenvironment Facilitation as well as Chemo- and Radio-Resistance Induction. The same are discussed here:
-
Genetic instability induction
Mutp53 has the capacity to cause a genomic abnormality by stopping the replication of DNA. Similar to how some mutp53 proteins trigger Cyclin A to encourage the growth of the intra-S phase checkpoint and the DNA replication origin kinase CHK1 to help duplicate aberrant genomic DNAs by stabilising replication forks [25].
-
Heightened proliferation and metastasis
Numerous studies have demonstrated that the mutp53 increases two of the primary "hallmarks of cancer"—the unbounded ability for replication and the insensitivity to anti-growth signals—during the malignant transformation of a normal cell [4, 26]. To over 90% of cancer-associated deaths are contributed by metastasis, which represents another “hallmark of cancer” [27, 28]. The primary and the most important step in metastasis is represented by EMT (Epithelial-to-mesenchymal transition), and it permits the cells to modify their structure so to further enhance their migration and invasion abilities [29, 30].
Furthermore, it has been observed that mutp53, perhaps in a cell type-dependent manner, stimulates the expression of many important EMT-related transcription factors, including ZEB1, SLUG, and TWIST1, through transcriptional, post-translational, and epigenetic modifications [31–34].
-
Metabolism modulation
The metabolism of glucose, lipids, and nucleotides is the traditional basis for cell viability; during malignant transformation, all of these systems experience dynamic changes. New research indicates that mutp53 proteins are involved in a number of the previously stated processes [35, 36]. The hyperactivation of oncogenic pathways directly affects the metabolic processes that promote tumor development. Interestingly, it has been observed that mutp53 amplifies the Warburg effect, a phenomenon marked by increased absorption of glucose and lactate production even in the presence of oxygen [35, 37]. Furthermore, research indicates that wtp53 promotes oxidative phosphorylation and counteracts the Warburg effect [38, 39]. Furthermore, mutp53-related metabolic alterations and GOF raise ROS levels in cancer cells, which encourage the buildup of ROS [40]. Additionally, p53 functions as a significant metabolic modulator that regulates the glycolysis to oxidative phosphorylation transition, amplifies the biogenesis of Fe-S cluster and coordinates the level of copper chelator GSH (glutathione) [41].
-
Chemo- and radio-resistance induction
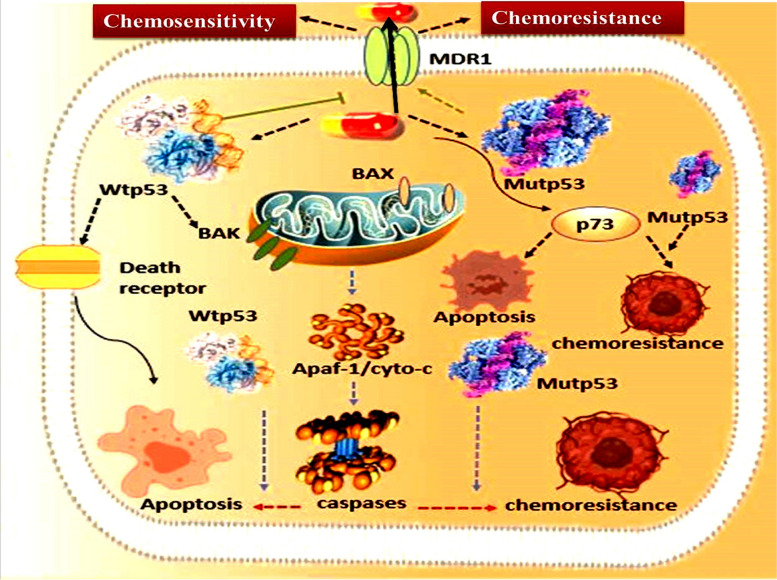
The p53 gene mutation is one of the key attempts made by the tumour cells to establish mechanisms to develop radio- and chemotherapy-resistance in order to survive such treatments [42]. A number of radio- and chemotherapy-resistant genes have their expression altered by the mutp53 proteins. Mutp53 proteins cause MDR1 expression to be greatly elevated. MDR1 encodes an energy-dependent efflux pump that helps cancer cells become resistant to a variety of hydrophobic cytotoxic agents [43, 44] (Fig. 3). Mutp53 has also been shown to be associated with angiogenesis and inflammation. Similar to the GOF mutant, p53 has been shown to promote chronic inflammation and has the capacity to modify the TME [45]. Additionally, the mutp53 encourages angiogenesis within the tumor by altering the pro-angiogenic protein VEGF [46].
Fig. 3.
The involvement of mutp53 towards the development of drug resistance: The mutp53 proteins highly upregulate expression of MDR1, which codes for the efflux pumps that facilitates tumor cell resistance to several hydrophobic cytotoxic agents
-
5.
Pro-oncogenic tumor microenvironment facilitation
The TME, which is comprised chiefly of immune cells, ECM, stromal cells, and blood vessels displays a significant role in the process of carcinogenesis and drug resistance abilities [47, 48]; And mutp53 can regulate the TME by prompting the secretion of pro-inflammatory cytokines as well as angiogenesis [49, 50].
Mutational landscape of p53 in breast cancer
As to the most recent version of the International Agency for Research on Cancer (IARC) TP53 database (http://www-p53.iarc.fr/), which is a part of COSMIC, missense mutations account for over 70% of TP53 changes linked to breast cancer [51]. This ratio matches the p53 mutational pattern seen in other tumors, as does the range of altered codons (the hotspots). There are some significant variations, though. For example, codon 220 ranks seventh in other malignancies (2%), whereas it is the fourth most common missense mutation in breast cancer (3.6%). Corresponding to this, codon 163 is more prevalent in breast cancer (2%) than in other malignancies (1%) [52]. While the reasons for these differences in mutation patterns are not fully explained, geographic or ethnic factors may control the prevalence of specific mutations, potentially due to environmental mutagens [53, 54]. Furthermore, correlations between BRCA1/2 germline mutations and TP53 mutations have been found, most likely as a result of a bias in defective DNA repair pathways [55, 56]. High frequency of TP53 mutations have been substantially associated with two particular polymorphisms in sporadic breast cancers: the presence of the highly active G allele of glutathione-S-transferases (GSTs) [57] and the homozygous Arginine at codon 72 of p53 [58].
The variety and tissue specificity of TP53 mutations in human breast tumors are noteworthy. The DNA binding domain (DBD) has a significant enrichment of mutations, the majority of which are missense (around 80%) [59, 60]. Moreover, variations in specific TP53 mutations are seen across different breast cancer types and grades, and these mutations can impact patient survival, particularly those with certain hotspot mutations. The incidence of missense mutations differs among distinct subclasses of tumours within the same organ, for example. Significant discrepancies have been identified in the occurrence of particular TP53 mutations among different breast cancer kinds and grades. For instance, TP53 point mutations are nearly universally present in luminal breast cancers, but p53 truncations are more commonly observed in basal breast tumours [61]. Additionally, the presence of particular hotspot mutations can influence patient survival outcomes as well.
Molecular Insights into p53 mutations and breast cancer
Tumorigenesis frequently involves the inactivation of the p53 tumor suppressor. Usually, a mutation in the p53 gene produces a persistent mutant protein, which accumulates to be recognized as a characteristic of cancerous cells. In addition to losing their ability to control tumor growth, these mutant p53 proteins frequently take on new carcinogenic properties that aid in cell survival. It is noteworthy that mutations in the p53 gene can arise at different phases of the complex process of malignant transformation, meaning that they influence tumor development, aggressiveness, and metastasis in distinct ways [62].
The TP53 gene in human tumors frequently experiences missense mutations, which are alterations in which one nucleotide is substituted for another. This is in contrast to most tumor suppressor genes, such RB, APC, or BRCA1, which are normally rendered inactive over the course of cancer by deletions or truncating mutations [63].
With only one amino acid changed, a whole protein is generated. The positions of the cancer-associated TP53 mutations within the p53 coding sequence and how they affect the protein's thermodynamic stability differ significantly. Notwithstanding this variation, the great majority of these mutations result in p53 losing its capacity to bind DNA in a way that is particular to a certain sequence and to halt the transcription of classical p53 target genes [64].
The most often changed residues in breast cancer are R248Q and R273H, also known as "contact mutants," which affect how p53 interacts with DNA. On the other hand, alterations such as Y220C and R175H result in "structural mutants" of p53, which under physiological circumstances cause the DNA binding domain (DBD) structure to be deformed. Comprehensive biophysical investigations conducted in vitro have demonstrated a gradient in the level of p53 DBD destabilization resulting from certain hotspot TP53 mutations, suggesting that separate proteins may be functionally associated with different mutants [65]. To assess the activity of mutant p53 proteins, transactivation experiments have been used in yeast or human cultured cells. Numerous mutant p53 variants have been demonstrated to display different promoter activation patterns in research focusing on TP53 mutations frequently seen in breast cancer. The Y220C mutant, for instance, is still able to activate the most sensitive wild-type p53 response element, which is derived from the p21 gene's promoter (WAF1), but it is unable to activate other response elements [66].
A growing body of research using a variety of models, including breast cancer cells, suggests that mutant p53 has specific transactivation and DNA-binding abilities. Hotspot p53 mutants directly control a large number of loci devoid of p53-responsive elements [67, 68].
Furthermore, mutant p53 has the ability to directly stimulate the transcription of certain microRNAs and obstruct microRNA processing, which may change the total amount of microRNAs in cells [69, 70]. Blandino et al., for example, found 40 promoters bound de novo by the R175H p53 mutant utilising studies employing breast cancer cell lines [71], However, ten more genes regulated and bound by the R280K p53 mutant were found by another group [72].
Despite the widespread belief that all p53 mutants are equal, there is growing evidence that certain mutants have unique profiles in terms of how much of their activity is lost compared to wild-type p53, how well they are able to suppress wild-type p53, and how they acquire gain-of-function characteristics [73].
Controversies regarding the gain-of-function of P53 mutation
Since all TP53 mutations are believed to impair the activity of wild-type p53, a great deal of study has been done to determine the function of the ensuing mutant p53 proteins due to the high frequency and recurrence of missense TP53 alleles. Research points to three separate, albeit potentially overlapping, processes by which various p53 mutations affect cancer: They exhibit three main characteristics: (i) they partially or completely lose the capacity to perform the tumor-suppressive functions of wild-type p53; (ii) they operate as dominant negative (DN) inhibitors of these activities; and (iii) they may acquire novel oncogenic roles in addition to p53 inactivation. The gain of function (GOF) theory is the most discussed of these processes, in part because to the variety of biological roles that various mutant p53 proteins are thought to have [74].
It has been discussed for almost thirty years whether mutant p53 proteins may have gain-of-function (GOF) consequences. Levine et al.'s discovery that ectopic expression of certain TP53 mutant alleles could trigger the production of a multidrug resistance gene reporter, but wild-type p53 could not, provided the first clue that p53 mutant proteins could have GOF function. Various TP53 mutants displayed a variety of behaviors, indicating early on that not all TP53 mutations work in the same way [75]. Subsequently, researchers from two different labs found that mice generated with germline missense mutations (R175H and R273H) formed tumor at a rate comparable to that of p53 null mice, but that they had a greater variety of tumor forms and a higher percentage of invasion [76, 77]. These results were seen as undisputed evidence of a gain-of-function (GOF) impact.
Following those first investigations, countless more recent publications have shown further proof that mutant p53 may affect a range of biological processes, including metastasis, stemness, the shift from epithelial to mesenchymal tissue, and more [78–80].
Further evidence supporting the gain-of-function (GOF) acquired by mutant p53 comes from the observation that patients with a TP53 missense mutation (i.e., expressing a mutant p53 protein) in the germline develop cancer substantially earlier than patients with TP53 mutations which culminate in the loss of p53 protein expression [81].
Accordingly, in vivo studies have shown that, in comparison to p53 null or p53 wild-type mice, animals expressing mutant p53 have a more aggressive and metastatic tumor profile [82]. Further research, however, suggests that this impact may be tissue specific. Specifically, when comparable p53 mutations were introduced into the lung, no discernible gain-of-function (GOF) activity was observed in comparison to p53 loss [83]. However, a multitude of in vitro and xenograft models has validated mutant p53's capacity to induce increased invasion and motility. It has been demonstrated that mutant p53 can improve signalling via receptors including MET, TGF-β, and epidermal growth factor receptor [84, 85]. Nevertheless, the community has not yet agreed upon a coherent paradigm to account for these alarming findings. A plethora of effectors have been found, despite the fact that many putative GOF pathways include mutant p53 working with other transcriptional regulators to modify gene expression [86].
Certain TP53 mutations seem to have different GOF capacities, and depending on the situation, even the same mutant may function through different methods [87] indicating that certain discrepancies may be influenced by a person's genetic history. This intricacy is especially noticeable because most research only looks at one or a small number of mutations. With models exhibiting gain-of-function (GOF) activity and displaying R172H or R270H mutations (corresponding to R175H and R273H mutations in humans), some researchers have started to question how well p53 GOF plays a meaningful role in cancer biology. As a result, they are beginning to adopt extensive methods to tackle this question [76, 77]. However, neither the humanized G245S mutant p53 mice models nor the mouse version of the human R249S mutation showed any signs of GOF activity. Nevertheless, R246S might still function as a dominant-negative inhibitor of wild-type p53, enhancing cell survival following radiation exposure [88, 89]. In comparison to p53 null mice, humanized R248Q p53 knock-in mice had an earlier start of tumor growth and a considerably shorter longevity [88], a decrease not observed in other mutant p53 models. Patients with Li-Fraumeni who carry the R248Q mutation typically develop cancer earlier than those who have inherited null mutations or the G245S mutation [88]. These results imply that the function of R248Q p53 differs from that of other p53 mutants under investigation. R248W and R248Q are structural mutations, which is noteworthy, however the humanized R248W p53 knock-in mice does not exhibit a shorter lifetime or an earlier start of illness [90]. Thus, it is a very difficult task to comprehend how each p53 mutation affects the course of the disease and the effectiveness of therapy [91].
Different strategies to target mutant P53 in breast cancer
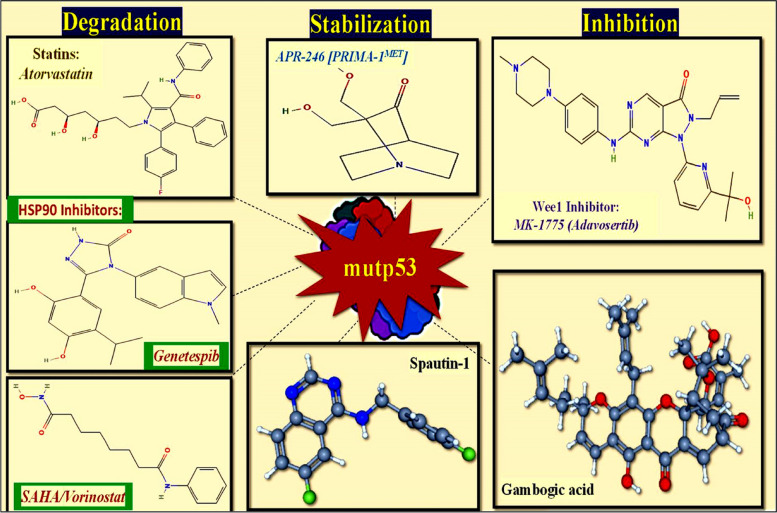
With the aim to eradicate the p53-mutant cancer cells, the general methods are implemented either by reverting the WT onco-suppressor features of p53 or by aiming on elimination of tumor by altering main components of the immune system [92] (Fig. 4). The strategies employed to address same include:
Fig. 4.
The various compounds that have been used against mutp53 either by inhibiting; degrading the mutp53 protein or stabilizing the WTp53 protein
Restoring p53 activity in cells with wild-type and mutant p53
The promising small molecules, which possess the ability to revert or restore the function of mutant p53 are the PRIMA-1 (p53 reactivation and induction of massive apoptosis) and its methyl analog APR-246 [PRIMA-1MET] and these two small molecules achieve same by associating with the DNA binding domain, enhancing proper folding/function [93]. As a consequence, there is an increase in the production of pro-apoptotic genes such as Puma, Noxa, and Bax in cells with mutated p53 (Fig. 4). This also leads to the activation of cell-cycle genes and PARP cleavage, regardless of the mutation status of p53. Similar findings were observed in many trials that included multiple forms of cancer such as Breast, Myeloma, and Thyroid [94].
As an example, tiny molecules like PK7088 and PhiKan083, which bind to a particular location in the p53 protein generated by the Y220C mutation, stabilize the structure of this mutant p53. The amount of p53 with a wild-type structure and function is increased by this stabilization [95].
Misfolded p53 may adopt a wild-type shape when metallothioneins, which bind and store intracellular zinc, are lost [96].
Furthermore, it has been demonstrated that incorporating zinc into conformational mutants such as G245C and G245D p53 can partially restore their wild-type shape [97]. As a result, research into the possibility of using zinc to restore wild-type folding has been conducted, and it has been shown that using this strategy can make cells containing endogenous mutant p53 more sensitive to chemotherapy [98]. Furthermore, it has been discovered that the thiosemicarbazone metal ion chelator NSC31926 restores wild-type activity in a variety of cell lines expressing mutant p53, perhaps by making zinc more bioavailable to the mutant p53 [99].
While p53 activity restoration is an intriguing therapeutic strategy, a number of problems must be fixed in order to optimize these treatments' effectiveness for potential clinical use. A hurdle lies in the many biological consequences (such cell cycle arrest, apoptosis, senescence, or differentiation) that arise from p53 activation, which seem to be contingent upon the kind of cell and its surroundings. Surprisingly, research on p53 has mostly concentrated on the advantages of restoring its activity. However, there is evidence to suggest that prolonged p53 activation may be just as damaging as its inactivation. Hyperactivation of p53 has been associated with ischaemic damage from strokes or cardiac arrest [100], as well as cell death in degenerative illnesses such arthritis, multiple sclerosis [101], and neuropathies [102].
Under these circumstances, it was discovered that p53 pathway overexpression caused apoptotic death that resulted in the loss of important cell types, whereas p53 function suppression provided protection. It is still unknown if newly developed substances meant to increase p53 activity have these harmful consequences in entire animals. But when p53-restoring medications enter clinical trials, it would be wise to investigate any possible side effects these medications could have in non-target tissues.
Inhibiting and degrading mutant forms of p53
To efficiently remove the mutant p53 proteins, increasing its turnover is an alternate strategy for targeting it. The ubiquitin ligase MDM2 has the ability to target both wild-type and mutant p53 for proteasomal destruction in otherwise normal cells. The overexpression of mutant p53 seen in cancer cells is thought to be caused by inhibition of MDM2, which is also responsible for the activation of wild-type p53 in response to stress. In fact, it seems that the stabilization of mutant p53 brought on by stress is required for its gain-of-function (GOF) consequences [103]. Apart from MDM2, it has been demonstrated that CHIP, an additional chaperone-associated E3 ubiquitin ligase, plays a crucial role in the breakdown of mutant p53 [104, 105]. Mutant p53 interacts with the Hsp70 and Hsp90 chaperone complex for stabilization; however, normal functioning of this complex requires interaction with HDAC6 [106]. Heat shock proteins (HSP) separate from mutant p53 when HDAC6 interaction is disrupted, enabling MDM2 and CHIP to degrade p53 [106]. HDAC inhibitors that stop HDAC6 from interacting with Hsp90, like SAHA, have the potential to destabilize mutant p53 [107].
Nevertheless, it has recently been demonstrated that SAHA and the pan-HDAC inhibitor NaB control both the transcription and stability of mutant p53 through the p53 activator HoxA5 [108]. When employing HDAC inhibitors as therapeutic drugs, caution should be used to ascertain the p53 status of tumors since this action was not limited to reducing the expression of mutant p53 but also decreased the expression of wild-type p53 [108], It has also been demonstrated that small molecule SIRT1 activators cause p53 to become deacetylated and lower the total amount of mutant p53 [109]. Furthermore, in previous investigations, Stathmin—a transcriptional target of both mutant and wild-type p53—boosted the activity of mutant p53 in ovarian tumors by controlling its phosphorylation and stability (by the control of miR-223) [110].
Autophagy has a multifaceted function in cancer, as it may either stimulate or impede the growth of tumors based on the targets of the autophagic process and the stage at which the tumor is evolving [111]. Mutant p53 was specifically encouraged to be degraded by macro-autophagy triggered by glucose restriction, but wild-type p53 was stabilized in comparable circumstances [112]. Protease inhibition also aided in the degradation of mutant p53, which also required the presence of functioning autophagy machinery [113].
In addition, a specialized kind of autophagy called chaperone-mediated autophagy may facilitate the degradation of mutant p53 when glucose shortage and confluent growth conditions are present [114]. Interestingly, the degradation of mutant p53 via this specific autophagy route was improved by the suppression of macro-autophagy, indicating that glucose-deprived mutant p53 degradation is conditional, in contrast to the findings of Rodriguez et al. (2012) [112, 114] Moreover, when located in the cytoplasm, both mutant and wild-type p53 may suppress autophagy [115], suggesting a complicated interaction between autophagy and mutant p53.
Consequently, even while it is possible to target mutant p53 for degradation, there are still questions regarding the efficacy of merely eliminating mutant p53 without substituting it with wild-type p53 that is resistant to degradation in order to trigger a therapeutic response. Nevertheless, a number of investigations have demonstrated that increasing apoptosis follows reductions in mutant p53 levels (either by siRNA or spautin treatment), suggesting that these cells may have grown reliant on mutant p53 for survival [116–118]. More research is needed to determine whether lowering mutant p53 levels is adequate for long-term and in vivo treatment.
P53-oriented cancer immunotherapy to inhibit GOF activity
Recently, cancer immunotherapy has attracted a lot of interest because of its exceptional efficacy in treating a variety of malignancies. There is a revived interest in p53-based therapies as well, especially those that improve the immune system's capacity to identify and destroy cancer cells that have p53 deregulation. These tactics are justified by the fact that cancer cells harbouring TP53 missense mutations frequently overexpress p53, which may result in an increased amount of p53-derived peptides being presented on their surface via major histocompatibility complexes (MHCs). But a major problem is that, rather than just increased p53 mRNA expression and translation, the excess of mutant p53 (mutp53) proteins in cancer cells is mostly caused by their poor degradation by the ubiquitin–proteasome system. Since the immunoproteasome uses proteolytic degradation to create MHC-displayed peptides, ineffective mutp53 degradation may actually lessen the presentation of p53-derived peptides. Despite this, studies conducted over the previous 20 years give promise that p53-based immunotherapy may eventually find use in clinical settings [119–121].
It is generally anticipated that the overexpression of missense mutp53 proteins in cancer cells will highlight different peptides from the whole p53 protein. Normal cells produce relatively low quantities of p53, which underlies the immune system's preference for cancer cells, even if some of these peptides may overlap with those from wild-type p53 (wtp53) [122]. The fact that natural self-tolerance does not restrict the T cell response to p53 adds credence to the viability of this strategy. But it's dangerous to assume that healthy cells won't be impacted. Rapidly growing normal progenitor cells may have more TP53 mRNA expression than differentiated cells, which may have little p53 mRNA and thus less p53 protein production [123].
Bispecific antibodies represent a highly promising avenue in cancer immunotherapy [124]. A newly created synthetic single-chain bispecific antibody targets the TCR–CD3 complex and a neo-antigen produced from the p53(R175H) hotspot mutant [125]. Neo-antigens on cancer cell surfaces provide a problem because of their low density, which usually impedes efficient immune clearance. This bispecific antibody, however, gets around this challenge by binding with great affinity to the TCR-CD3 complex on T cells as well as the p53(R175H) peptide–HLA complex on cancer cells. Both in vitro and in vivo, this dual binding efficiently reroutes T lymphocytes to identify and assault cancer cells presenting the mutant peptide, producing remarkably specific cytotoxicity against p53(R175H)-expressing cancer cells [125]. These encouraging results suggest that mutp53-selective immunotherapy strategies—like the use of this bispecific antibody—will become more and more common in the years to come.
Issues and considerations of p53 treatment strategies
Despite ongoing advancements in p53-based cancer therapies, numerous challenges persist, and the quest for effective and selective drugs that can eventually be used in clinical settings continues. One major issue with p53-based treatments, as with all anticancer therapies, is the risk of developing resistance. Although knowledge in this area is still limited, some potential resistance mechanisms are already predictable. For instance, mutations in the TP53 gene could lead to resistance to MDM2 inhibitors, as demonstrated in experiments with prolonged nutlin treatment [126]. Other mechanisms could include activation of anti-apoptotic genes, inherent resistance to apoptosis, and increased levels of MDM4 [127]. The impact of these factors on the effectiveness of p53-based therapies remains uncertain.
Furthermore, p53-based medications are not expected to work well as stand-alone therapies. The goal of research has been to find efficient medication combinations. For example, nutlin 3a plus a BET inhibitor increased p53 activation in a wild-type p53 acute myeloid leukaemia (AML) mouse model [128]. In a similar vein in primary AML cells, combining the BCL-2 inhibitor venetoclax with idasanutlin produced encouraging outcomes, surpassing resistance to either medication alone by reducing the apoptotic threshold [129]. Clinical trials for young adults and children with solid tumors and relapsed or refractory leukaemia have been initiated using this combination (NCT04029688). Furthermore, p53 activation with immunotherapy is a compelling combination that may minimize required medication dosages and, in certain situations, overcome resistance [130]. The fact that mice models are used largely for in vivo testing of p53-based medications presents another difficulty. Although mice are commonly used in drug development, there are notable distinctions between mice and humans, such as variances in the p53, MDM2, and MDM4 protein sequences and variations in the p53 signalling pathway [8]. Cutting-edge methods like organ-on-a-chip and other ex vivo models might help resolve these interspecies variations and speed up the development of p53-targeted medications for clinical use [131, 132].
Our limited comprehension of human biology and the intricate cellular mechanisms induced by medication administration is a significant obstacle. While screening techniques are becoming more sophisticated and producing a greater number of compounds targeted to the p53, it is still difficult to anticipate off-target effects and possible harmful on-target effects. Unacceptable toxicity is frequently only identified in patient trials. More precise testing models and a greater comprehension of these processes are desperately needed.
Discussion
There are very significant roles associated with WT-p53 within breast cancer cells. And the resulting mutations in p53 from varied causes could lead to several types of malignancies. Thus, particular therapeutic ways to aim at p53 anomalies and pathways linked with this protein have drawn significant focus in recent times. Here we have reviewed the recent advances in the understanding of p53 roles and associated anomalies as well as the role it primarily plays as a tumor-suppressor in the body, with special focus on breast cancer. Over half of all human tumors and seventy-five percent of tumors with changed transitions have p53 missense mutations, which are pro-tumorigenic factors. The aberrant stabilization and accumulation of p53 mutations in tumor cells is one of the primary factors contributing to the malignant growth of tumors. Our lab has recently suggested through an in silico approach the two PhiKan-083 analogues, which could possibly address the p53 misfolding after performing proper in vitro and in vivo experiments and validate the same [133]. We think that these methods have promising clinical application prospects and that significant developments based on them are about to come. New advances in the area of mutp53 cancer treatment will become physically apparent if they pass the pre-clinical and clinical tests, which will be difficult but very rewarding. The resurgence of tumor immunotherapy is also igniting interest in p53-based tactics, primarily those that try to strengthen the immune system's capacity to identify and eliminate cancer cells that have p53 that is dysregulated. Overall, TP53 represents an important therapeutic target for cancer and since more treatments aiming at p53 are being explored, it will be suitable to develop personalized treatment plans as per the patients’ p53 mutation status. Future studies will shed more light on the full mechanisms of the complex modulatory interplay between the cancer TP53 status and the pathways that get engaged in the overall pathophysiology of cancers especially Breast Cancer.
Tumor-specific dependency and chemotherapy resistance are induced by the GOF effect, which is brought on by mutant p53 stabilization. When malignant tumors have heterozygous mutant/wild-type p53 alleles, the oncogenic effects of the GOF impact of mutant p53 are effectively mitigated by p53 degradation. Consequently, there is considerable promise for enhancing prognosis, extending cancer patient survival, and advancing cancer therapy through controlled degradation of mutant p53 protein. Due to its inherent tumor suppressor role and the high prevalence of p53 mutations in tumors, p53 is a potentially attractive target for cancer therapy. Cancer-related stress is caused by a relatively small number of factors, including nutritional imbalance, hypoxia, reactive oxygen or nitrogen compounds, DNA damage, and somatic mutations. However, the situation is extremely complex due to the multiple pathways involved and the changing conditions a tumor encounters during its clinical evolution. It will need further research and careful assessment of experimental conditions to determine the best individualized treatment plans for cancer patients. To sum up, p53 is an extremely topical therapeutic target for BC and needs more research at clinical level, particularly for patients with high p53 mutational burden who also have TNBC and HER2-positive subtypes. An important future focus will be the optimization of p53-associated treatment techniques, including logical combinations.
Acknowledgements
The author Burhan Ul Haq would like to thank the Department of Science and Technology, Government of India for supporting this work under INSPIRE Fellowship Program [Research Student- IF220332]. The authors would also like to thank BioRender for Figures and PubChem for structures of the compounds used in this Review.
Abbreviations
- TNBC
Triple-negative breast cancer
- CRISPR/Cas9
Clustered regularly interspaced short palindromic repeats/ Caspase 9
- ROS
Reactive oxygen species
- HDAC
Histone deacetylase
- Bid
BH3 interacting-domain death agonist
- Bax
Bcl-2 Associated X-protein
- Bim
Bcl-2 Interacting Mediator of cell death
- CHK1
Checkpoint kinase 1
- ZEB1
Zinc finger E-box binding homeobox 1
- SNAI2/SLUG
Snail Family Transcriptional Repressor 2
- TWIST1
Twist-related protein 1
- MDR1
Multidrug resistance mutation 1
- TME
Tumour microenvironment
- ECM
Extracellular matrix
- PARP
Poly (ADP-ribose) polymerases
- STAT
Signal Transducer and Activator of Transcription
- PRIMA-1
P53 reactivation and induction of massive apoptosis-1
- DSF
Disulfiram
- MEFs
Mouse Embryonic Fibroblasts
- CSCs
Cancer Stem Cells
- VEGF
Vascular endothelial growth factor
- ABC Transporters
ATP binding cassette
- SAHA
Suberoylanilide Hydroxamic Acid
- DM2
Type II diabetes mellitus
- CHIP
Carboxyl terminus of Hsp70-interacting Protein
Authors’ contributions
M.A.M designed and supervised the work; B.U.H, H.Q and S.S contributed equally to the work. M.A.M and B.U.H prepared Graphical Abstract and Figures 1 and 4; H.Q, S.S and N.J prepared Figures 2 and 3. All the authors edited and reviewed the manuscript.
Funding
This work has been sponsored by the Science Engineering and Research Board (SERB-DST) Govt of India vide grant No. SERB/CRG/2023/008460 sanctioned to Dr Manzoor Ahmad Mir.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All contributing authors agree to the publication of this article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hina Qayoom, Burhan Ul Haq and Shazia Sofi contributed equally to this work.
References
- 1.Urooj T, Wasim B, Mushtaq S, Shah SNN, Shah M. Cancer cell-derived secretory factors in breast cancer-associated lung metastasis: their mechanism and future prospects. Curr Cancer Drug Targets. 2020;20(3):168–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qayoom H, Alshehri B, Haq BU, Almilaibary A, Alkhanani M, Mir MA. Decoding the molecular mechanism of stypoldione against breast cancer through network pharmacology and experimental validation. Saudi J Biol Sci. 2023;30(12):103848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–8. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 5.Willenbrink TJ, Ruiz ES, Cornejo CM, Schmults CD, Arron ST, Jambusaria-Pahlajani A. Field cancerization: Definition, epidemiology, risk factors, and outcomes. J Am Acad Dermatol. 2020;83(3):709–17. [DOI] [PubMed] [Google Scholar]
- 6.Lane DP, Crawford LV. T antigen is bound to a host protein in SY40-transformed cells. Nature. 1979;278(5701):261–3. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Tavana O, Gu W. p53 modifications: exquisite decorations of the powerful guardian. J Mol Cell Biol. 2019;11(7):564–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36(28):3943–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho T, Tan BX, Lane D. How the other half lives: What p53 does when it is not being a transcription factor. Int J Mol Sci. 2019;21(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15(5):276–89. [DOI] [PubMed] [Google Scholar]
- 11.Adriaens C, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22(8):861–8. [DOI] [PubMed] [Google Scholar]
- 12.Lindström MS, Bartek J, Maya-Mendoza A. p53 at the crossroad of DNA replication and ribosome biogenesis stress pathways. Cell Death Differ. 2022;29(5):972–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutelle AM, Attardi LD. p53 and tumor suppression: it takes a network. Trends Cell Biol. 2021;31(4):298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–9. [DOI] [PubMed] [Google Scholar]
- 15.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. [DOI] [PubMed] [Google Scholar]
- 16.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91(3):325–34. [DOI] [PubMed] [Google Scholar]
- 17.Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25(1):104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018;25(1):114–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Gu W. The complexity of p53-mediated metabolic regulation in tumor suppression. Semin Cancer Biol. 2022;85:4–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse J-P, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabapathy K, Lane DP. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat Rev Clin Oncol. 2018;15(1):13–30. [DOI] [PubMed] [Google Scholar]
- 22.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotler E, et al. A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol Cell. 2018;71(1):178–90. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Gu W. p53 in ferroptosis regulation: the new weapon for the old guardian. Cell Death Differ. 2022;29(5):895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S, Vaughan CA, Frum RA, Grossman SR, Deb S, Deb SP. Mutant p53 establishes targetable tumor dependency by promoting unscheduled replication. J Clin Investig. 2017;127(5):1839–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- 27.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–95. [DOI] [PubMed] [Google Scholar]
- 28.Dong P, et al. Mutant p53 gain-of-function induces epithelial–mesenchymal transition through modulation of the miR-130b–ZEB1 axis. Oncogene. 2013;32(27):3286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qayoom H, Wani NA, Alshehri B, Mir MA. “An insight into the cancer stem cell survival pathways involved in chemoresistance in triple-negative breast cancer,” (in eng). Future Oncol. 2021;17(31):4185–206. 10.2217/fon-2021-0172. [DOI] [PubMed] [Google Scholar]
- 30.Sofi S, Jan N, Qayoom H, Alkhanani M, Almilaibary A, Ahmad Mir M. "Elucidation of interleukin-19 as a therapeutic target for breast cancer by computational analysis and experimental validation," (in eng). Saudi J Biol Sci. 2023;30(9):103774. 10.1016/j.sjbs.2023.103774. [DOI] [PMC free article] [PubMed]
- 31.Wang S-P, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11(6):694–704. [DOI] [PubMed] [Google Scholar]
- 32.Kogan-Sakin I, et al. Mutant p53R175H upregulates Twist1 expression and promotes epithelial–mesenchymal transition in immortalized prostate cells. Cell Death Differ. 2011;18(2):271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C, El-Deiry WS. Targeting p53 for enhanced radio-and chemo-sensitivity. Apoptosis. 2009;14:597–606. [DOI] [PubMed] [Google Scholar]
- 34.Sofi S, et al. "Cyclin-dependent kinases in breast cancer: expression pattern and therapeutic implications," (in eng), Med Oncol. 2022;39(6);106. 10.1007/s12032-022-01731-x. [DOI] [PubMed]
- 35.Eriksson M, et al. Effect of mutant p53 proteins on glycolysis and mitochondrial metabolism. Mol Cell Biol. 2017;37(24):e00328-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Zhang C, Hu W, Feng Z. Tumor suppressor p53 and metabolism. J Mol Cell Biol. 2019;11(4):284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340–4. [DOI] [PubMed] [Google Scholar]
- 38.Zhou G, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54(6):960–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernández-Reséndiz I, et al. Dual regulation of energy metabolism by p53 in human cervix and breast cancer cells. Biochim Biophys Acta. 2015;1853(12):3266–78. [DOI] [PubMed] [Google Scholar]
- 40.Liu DS, et al. Inhibiting the system xC−/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat Commun. 2017;8(1):14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong C, Ling H, Hao Q, Zhou X. Cuproptosis: p53-regulated metabolic cell death? Cell Death Differ. 2023;30(4):876–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takara K, Sakaeda T, Okumura K. An update on overcoming MDR1-mediated multidrug resistance in cancer chemotherapy. Curr Pharm Des. 2006;12(3):273–86. [DOI] [PubMed] [Google Scholar]
- 43.Chin K-V, Ueda K, Pastan I, Gottesman MM. Modulation of activity of the promoter of the human MDR 1 gene by Ras and p53. Science. 1992;255(5043):459–62. [DOI] [PubMed] [Google Scholar]
- 44.Stein Y, Aloni-Grinstein R, Rotter V. Mutant p53—a potential player in shaping the tumor–stroma crosstalk. J Mol Cell Biol. 2019;11(7):600–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooks T, et al. Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell. 2013;23(5):634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khromova NV, Kopnin PB, Stepanova EV, Agapova LS, Kopnin BP. p53 hot-spot mutants increase tumor vascularization via ROS-mediated activation of the HIF1/VEGF-A pathway. Cancer Lett. 2009;276(2):143–51. [DOI] [PubMed] [Google Scholar]
- 47.Cordani M, Pacchiana R, Butera G, D’Orazi G, Scarpa A, Donadelli M. Mutant p53 proteins alter cancer cell secretome and tumour microenvironment: Involvement in cancer invasion and metastasis. Cancer Lett. 2016;376(2):303–9. [DOI] [PubMed] [Google Scholar]
- 48.Mir MA, Aisha S, Sofi S, Rasheid S. Chapter 2 - The tumor microenvironment. In: Mir MA, editor. Role of Tumor Microenvironment in Breast Cancer and Targeted Therapies. Academic Press; 2022. p. 31–58. 10.1016/B978-0-443-18696-7.00007-5.
- 49.Bykov VJN, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8(3):282–8. [DOI] [PubMed] [Google Scholar]
- 50.Bykov VJN, et al. PRIMA-1MET synergizes with cisplatin to induce tumor cell apoptosis. Oncogene. 2005;24(21):3484–91. [DOI] [PubMed] [Google Scholar]
- 51.Petitjean A, et al. “Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database,” (in eng). Hum Mutat. 2007;28(6):622–9. 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 52.Feki A, Irminger-Finger I. “Mutational spectrum of p53 mutations in primary breast and ovarian tumors,” (in eng). Crit Rev Oncol Hematol. 2004;52(2):103–16. 10.1016/j.critrevonc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Olivier M, Hainaut P. “TP53 mutation patterns in breast cancers: searching for clues of environmental carcinogenesis,” (in eng). Semin Cancer Biol. 2001;11(5):353–60. 10.1006/scbi.2001.0390. [DOI] [PubMed] [Google Scholar]
- 54.Hill KA, Sommer SS. “p53 as a mutagen test in breast cancer,” (in eng). Environ Mol Mutagen. 2002;39(2–3):216–27. 10.1002/em.10065. [DOI] [PubMed] [Google Scholar]
- 55.Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. “TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution,” (in eng). Cancer Res. 2001;61(10):4092–7. [PubMed] [Google Scholar]
- 56.Manié E, et al. “High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors,” (in eng). Cancer Res. 2009;69(2):663–71. 10.1158/0008-5472.Can-08-1560. [DOI] [PubMed] [Google Scholar]
- 57.Nedelcheva Kristensen V, et al., "Single tube multiplex polymerase chain reaction genotype analysis of GSTM1, GSTT1 and GSTP1: relation of genotypes to TP53 tumor status and clinicopathological variables in breast cancer patients," (in eng). Pharmacogenetics. 1998;8(5):441–7. 10.1097/00008571-199810000-00009. [DOI] [PubMed]
- 58.Langerød A, et al. “The TP53 codon 72 polymorphism may affect the function of TP53 mutations in breast carcinomas but not in colorectal carcinomas,” (in eng). Cancer Epidemiol Biomarkers Prev. 2002;11(12):1684–8. [PubMed] [Google Scholar]
- 59.Hainaut P, Pfeifer GP. "Somatic TP53 Mutations in the Era of Genome Sequencing," (in eng). Cold Spring Harb Perspect Med. 2016;6(11), 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed]
- 60.Olivier M, Hollstein M, Hainaut P. “TP53 mutations in human cancers: origins, consequences, and clinical use,” (in eng). Cold Spring Harb Perspect Biol. 2010;2(1): a001008. 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dumay A, et al. “Distinct tumor protein p53 mutants in breast cancer subgroups,” (in eng). Int J Cancer. 2013;132(5):1227–31. 10.1002/ijc.27767. [DOI] [PubMed] [Google Scholar]
- 62.Rivlin N, Brosh R, Oren M, Rotter V. “Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis,” (in eng). Genes Cancer. 2011;2(4):466–74. 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hainaut P, Hollstein M. “p53 and human cancer: the first ten thousand mutations,” (in eng). Adv Cancer Res. 2000;77:81–137. 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 64.Bullock AN, Fersht AR. “Rescuing the function of mutant p53,” (in eng). Nat Rev Cancer. 2001;1(1):68–76. 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 65.Bullock AN, Henckel J, Fersht AR. “Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy,” (in eng). Oncogene. 2000;19(10):1245–56. 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 66.Jordan JJ, et al. “Altered-function p53 missense mutations identified in breast cancers can have subtle effects on transactivation,” (in eng). Mol Cancer Res. 2010;8(5):701–16. 10.1158/1541-7786.Mcr-09-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fontemaggi G, et al. “The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis,” (in eng). Nat Struct Mol Biol. 2009;16(10):1086–93. 10.1038/nsmb.1669. [DOI] [PubMed] [Google Scholar]
- 68.Weisz L, et al. “Transactivation of the EGR1 gene contributes to mutant p53 gain of function,” (in eng). Cancer Res. 2004;64(22):8318–27. 10.1158/0008-5472.Can-04-1145. [DOI] [PubMed] [Google Scholar]
- 69.Donzelli S, et al. “MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function,” (in eng). Cell Death Differ. 2012;19(6):1038–48. 10.1038/cdd.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. “Modulation of microRNA processing by p53,” (in eng). Nature. 2009;460(7254):529–33. 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 71.Dell’Orso S, et al. “ChIP-on-chip analysis of in vivo mutant p53 binding to selected gene promoters,” (in eng). OMICS. 2011;15(5):305–12. 10.1089/omi.2010.0084. [DOI] [PubMed] [Google Scholar]
- 72.Girardini JE, et al. “A Pin1/mutant p53 axis promotes aggressiveness in breast cancer,” (in eng). Cancer Cell. 2011;20(1):79–91. 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Halevy O, Michalovitz D, Oren M. “Different tumor-derived p53 mutants exhibit distinct biological activities,” (in eng). Science. 1990;250(4977):113–6. 10.1126/science.2218501. [DOI] [PubMed] [Google Scholar]
- 74.Kennedy MC, Lowe SW. “Mutant p53: it’s not all one and the same,” (in eng). Cell Death Differ. 2022;29(5):983–7. 10.1038/s41418-022-00989-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dittmer D, et al. “Gain of function mutations in p53,” (in eng). Nat Genet. 1993;4(1):42–6. 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 76.Lang GA, et al. “Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome,” (in eng). Cell. 2004;119(6):861–72. 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 77.Olive KP, et al. “Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome,” (in eng). Cell. 2004;119(6):847–60. 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 78.Aschauer L, Muller PA. “Novel targets and interaction partners of mutant p53 Gain-Of-Function,” (in eng). Biochem Soc Trans. 2016;44(2):460–6. 10.1042/bst20150261. [DOI] [PubMed] [Google Scholar]
- 79.Bargonetti J, Prives C. “Gain-of-function mutant p53: history and speculation,” (in eng). J Mol Cell Biol. 2019;11(7):605–9. 10.1093/jmcb/mjz067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muller PA, Vousden KH. “Mutant p53 in cancer: new functions and therapeutic opportunities,” (in eng). Cancer Cell. 2014;25(3):304–17. 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bougeard G, et al. “Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families,” (in eng). J Med Genet. 2008;45(8):535–8. 10.1136/jmg.2008.057570. [DOI] [PubMed] [Google Scholar]
- 82.Doyle B, Morton JP, Delaney DW, Ridgway RA, Wilkins JA, Sansom OJ. “p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma,” (in eng). J Pathol. 2010;222(2):129–37. 10.1002/path.2748. [DOI] [PubMed] [Google Scholar]
- 83.Jackson EL, et al. “The differential effects of mutant p53 alleles on advanced murine lung cancer,” (in eng). Cancer Res. 2005;65(22):10280–8. 10.1158/0008-5472.Can-05-2193. [DOI] [PubMed] [Google Scholar]
- 84.Adorno M, et al. “A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis,” (in eng). Cell. 2009;137(1):87–98. 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Chen JX, Liu JP, You C, Liu YH, Mao Q. “Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide,” (in eng). Ann Surg Oncol. 2014;21(4):1337–44. 10.1245/s10434-013-3380-0. [DOI] [PubMed] [Google Scholar]
- 86.Pfister NT, Prives C. "Transcriptional Regulation by Wild-Type and Cancer-Related Mutant Forms of p53," (in eng), Cold Spring Harb Perspect Med. 2017;7(2), 10.1101/cshperspect.a026054. [DOI] [PMC free article] [PubMed]
- 87.Freed-Pastor WA, et al. “Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway,” (in eng). Cell. 2012;148(1–2):244–58. 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. “Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis,” (in eng). Cell Death Differ. 2013;20(7):898–909. 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee MK, Teoh WW, Phang BH, Tong WM, Wang ZQ, Sabapathy K. “Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo,” (in eng). Cancer Cell. 2012;22(6):751–64. 10.1016/j.ccr.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 90.Song H, Hollstein M, Xu Y. “p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM,” (in eng). Nat Cell Biol. 2007;9(5):573–80. 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 91.Muller PA, et al. “Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion,” (in eng). Oncogene. 2013;32(10):1252–65. 10.1038/onc.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chasov V, et al. Key players in the mutant p53 team: Small molecules, gene editing, immunotherapy. Front Oncol. 2020;10:1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bykov VJN, Wiman KG. Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett. 2014;588(16):2622–7. [DOI] [PubMed] [Google Scholar]
- 94.Perdrix A, et al. PRIMA-1 and PRIMA-1Met (APR-246): From mutant/wild type p53 reactivation to unexpected mechanisms underlying their potent anti-tumor effect in combinatorial therapies. Cancers. 2017;9(12):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. “Targeted rescue of a destabilized mutant of p53 by an in silico screened drug,” (in eng). Proc Natl Acad Sci U S A. 2008;105(30):10360–5. 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Puca R, et al. “Restoring wtp53 activity in HIPK2 depleted MCF7 cells by modulating metallothionein and zinc,” (in eng). Exp Cell Res. 2009;315(1):67–75. 10.1016/j.yexcr.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 97.Pintus SS, et al. “The substitutions G245C and G245D in the Zn(2+)-binding pocket of the p53 protein result in differences of conformational flexibility of the DNA-binding domain,” (in eng). J Biomol Struct Dyn. 2013;31(1):78–86. 10.1080/07391102.2012.691364. [DOI] [PubMed] [Google Scholar]
- 98.Puca R, et al. “Restoring p53 active conformation by zinc increases the response of mutant p53 tumor cells to anticancer drugs,” (in eng). Cell Cycle. 2011;10(10):1679–89. 10.4161/cc.10.10.15642. [DOI] [PubMed] [Google Scholar]
- 99.Yu X, Vazquez A, Levine AJ, Carpizo DR. “Allele-specific p53 mutant reactivation,” (in eng). Cancer Cell. 2012;21(5):614–25. 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Komarova EA, Gudkov AV. “Chemoprotection from p53-dependent apoptosis: potential clinical applications of the p53 inhibitors,” (in eng). Biochem Pharmacol. 2001;62(6):657–67. 10.1016/s0006-2952(01)00733-x. [DOI] [PubMed] [Google Scholar]
- 101.Wosik K, Antel J, Kuhlmann T, Brück W, Massie B, Nalbantoglu J. “Oligodendrocyte injury in multiple sclerosis: a role for p53,” (in eng). J Neurochem. 2003;85(3):635–44. 10.1046/j.1471-4159.2003.01674.x. [DOI] [PubMed] [Google Scholar]
- 102.Mattson MP, Duan W, Pedersen WA, Culmsee C. "Neurodegenerative disorders and ischemic brain diseases," (in eng). Apoptosis 2001;6(1–2):69–81. 10.1023/a:1009676112184. [DOI] [PubMed]
- 103.Suh YA, et al. “Multiple stress signals activate mutant p53 in vivo,” (in eng). Cancer Res. 2011;71(23):7168–75. 10.1158/0008-5472.Can-11-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Esser C, Scheffner M, Höhfeld J. “The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation,” (in eng). J Biol Chem. 2005;280(29):27443–8. 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 105.Lukashchuk N, Vousden KH. “Ubiquitination and degradation of mutant p53,” (in eng). Mol Cell Biol. 2007;27(23):8284–95. 10.1128/mcb.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li D, et al. “Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells,” (in eng). Mol Cancer Res. 2011;9(5):577–88. 10.1158/1541-7786.Mcr-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li D, Marchenko ND, Moll UM. “SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis,” (in eng). Cell Death Differ. 2011;18(12):1904–13. 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yan W, et al. “Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8,” (in eng). Oncogene. 2013;32(5):599–609. 10.1038/onc.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yi YW, Kang HJ, Kim HJ, Kong Y, Brown ML, Bae I. “Targeting mutant p53 by a SIRT1 activator YK-3-237 inhibits the proliferation of triple-negative breast cancer cells,” (in eng). Oncotarget. 2013;4(7):984–94. 10.18632/oncotarget.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sonego M, et al. “Stathmin regulates mutant p53 stability and transcriptional activity in ovarian cancer,” (in eng). EMBO Mol Med. 2013;5(5):707–22. 10.1002/emmm.201201504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu EY, Ryan KM. “Autophagy and cancer–issues we need to digest,” (in eng). J Cell Sci. 2012;125(Pt 10):2349–58. 10.1242/jcs.093708. [DOI] [PubMed] [Google Scholar]
- 112.Rodriguez OC, et al. “Dietary downregulation of mutant p53 levels via glucose restriction: mechanisms and implications for tumor therapy,” (in eng). Cell Cycle. 2012;11(23):4436–46. 10.4161/cc.22778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Choudhury S, Kolukula VK, Preet A, Albanese C, Avantaggiati ML. “Dissecting the pathways that destabilize mutant p53: the proteasome or autophagy?,” (in eng). Cell Cycle. 2013;12(7):1022–9. 10.4161/cc.24128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vakifahmetoglu-Norberg H, et al. “Chaperone-mediated autophagy degrades mutant p53,” (in eng). Genes Dev. 2013;27(15):1718–30. 10.1101/gad.220897.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morselli E, et al. “Mutant p53 protein localized in the cytoplasm inhibits autophagy,” (in eng). Cell Cycle. 2008;7(19):3056–61. 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 116.Ali A, et al. “Differential regulation of the REGγ-proteasome pathway by p53/TGF-β signalling and mutant p53 in cancer cells,” (in eng). Nat Commun. 2013;4:2667. 10.1038/ncomms3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu H, Mao Q, Lin Y, Yang K, Xie L. “RNA interference targeting mutant p53 inhibits growth and induces apoptosis in DU145 human prostate cancer cells,” (in eng). Med Oncol. 2011;28(Suppl 1):S381–7. 10.1007/s12032-010-9679-9. [DOI] [PubMed] [Google Scholar]
- 118.Zhu HB, Yang K, Xie YQ, Lin YW, Mao QQ, Xie LP. "Silencing of mutant p53 by siRNA induces cell cycle arrest and apoptosis in human bladder cancer cells," (in eng). World J Surg Oncol. 2013;11:22. 10.1186/1477-7819-11-22. [DOI] [PMC free article] [PubMed]
- 119.Hassin O, Oren M. Drugging p53 in cancer: one protein, many targets. Nat Rev Drug Discovery. 2023;22(2):127–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mir MA, Qayoom H, Mehraj U, Nisar S, Bhat B, Wani NA. “Targeting Different Pathways Using Novel Combination Therapy in Triple Negative Breast Cancer,” (in eng). Curr Cancer Drug Targets. 2020;20(8):586–602. 10.2174/1570163817666200518081955. [DOI] [PubMed] [Google Scholar]
- 121.Qayoom H, Sofi S, Mir MA. “Targeting tumor microenvironment using tumor-infiltrating lymphocytes as therapeutics against tumorigenesis,” (in eng). Immunol Res. 2023;71(4):588–99. 10.1007/s12026-023-09376-2. [DOI] [PubMed] [Google Scholar]
- 122.Lauwen MM, et al. Self-tolerance does not restrict the CD4+ T-helper response against the p53 tumor antigen. Can Res. 2008;68(3):893–900. [DOI] [PubMed] [Google Scholar]
- 123.Xue Y, et al. Bortezomib stabilizes and activates p53 in proliferative compartments of both normal and tumor tissues in vivo. Can Res. 2019;79(14):3595–607. [DOI] [PubMed] [Google Scholar]
- 124.You G, et al. Bispecific antibodies: a smart arsenal for cancer immunotherapies. Vaccines. 2021;9(7):724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hsiue EHC, et al. Targeting a neoantigen derived from a common TP53 mutation. Science. 2021;371(6533):eabc8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Michaelis M, et al. Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis. 2011;2(12):e243–e243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chapeau EA, et al. Resistance mechanisms to TP53-MDM2 inhibition identified by in vivo piggyBac transposon mutagenesis screen in an Arf−/− mouse model. Proc Natl Acad Sci. 2017;114(12):3151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Latif A-L, et al. BRD4-mediated repression of p53 is a target for combination therapy in AML. Nat Commun. 2021;12(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pan R, et al. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell. 2017;32(6):748–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Daver NG, et al. Safety, efficacy, pharmacokinetic (PK) and biomarker analyses of BCL2 inhibitor venetoclax (Ven) plus MDM2 inhibitor idasanutlin (idasa) in patients (pts) with relapsed or refractory (R/R) AML: a phase Ib, non-randomized, open-label study. Blood. 2018;132:767.30115637 [Google Scholar]
- 131.Horejs C. Organ chips, organoids and the animal testing conundrum. Nat Rev Mater. 2021;6(5):372–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gavert N, et al. Ex vivo organotypic cultures for synergistic therapy prioritization identify patient-specific responses to combined MEK and Src inhibition in colorectal cancer. Nature cancer. 2022;3(2):219–31. [DOI] [PubMed] [Google Scholar]
- 133.Haq BU, Qayoom H, Ahmad I, Ahmad F, Mir MA. Targeting p53 misfolding conundrum by stabilizing agents and their analogs in breast cancer therapy: a comprehensive computational analysis. Front Pharmacol. 2024;14:1333447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.