Abstract
Colorectal cancer (CRC) is a significant public health issue owing to its widespread occurrence and substantial morbidity and mortality rates. Recent studies have highlighted serum uric acid (SUA) level as a probable risk factor for CRC; however, the inconsistency in these findings has created doubt. We performed a Mendelian randomization (MR) study utilizing extensive cohort data from the UK BioBank and the NHGRI-EBI Genome-Wide Association Study (GWAS) Catalog to investigate the causal connection between SUA levels and CRC incidence. Our MR study addresses the constraints of earlier studies, including limited sample sizes and inconsistent results. Considering SUA levels as the exposure and CRC as the outcome, the inverse variance-weighted (IVW) approach in MR showed that the odds ratios (ORs) for CRC for each unit increase in SUA were 0.232 (95% confidence interval [CI] of OR 0.094–0.570; P = .001) and 0.551 (95% CI of OR 0.325–0.934; P = .027). Pleiotropic tests and sensitivity analysis confirmed minimal horizontal pleiotropy and the robustness of causality. Our research deepens the understanding of the association between SUA levels and CRC, offering insights into prevention strategies and patient outcomes prediction.
Keywords: colorectal cancer, Mendelian randomization, serum uric acid
1. Introduction
Considering its high prevalence and significant impact on global health, colorectal cancer (CRC) is a pressing public health concern.[1] It ranks among the most prevalent malignancies, and its burden in terms of morbidity and mortality is undeniable.[2] Understanding the risk factors associated with CRC is paramount because this knowledge can potentially guide preventive strategies and improve patient outcomes.[2] Serum uric acid (SUA) levels have emerged as potential contributors to CRC risk.[3]
Recent investigations on the relationship between SUA levels and CRC have revealed a lack of consensus in the scientific literature.[4] In fact, research findings suggest a higher level of SUA is associated with a decreased mortality rate from all types of cancers.[5] Conversely, other studies have indicated an increased risk of CRC in patients with higher SUA levels than in those with lower SUA levels.[6] A review of current studies shows considerable variation in the direction and strength of the link between SUA levels and CRC incidence.[6] The inconclusive nature of these findings has created uncertainty and prompted the need for a comprehensive analysis that can shed light on this intriguing association.[4]
The relationship between SUA levels and the risk of CRC remains a topic of ongoing debate in the scientific community.[6] Even when statistically significant associations have been reported, the direction of these associations has not been consistent across studies.[4] Furthermore, existing research is significantly limited by relatively small sample sizes.[4,7] Recently, extensive cohort data, including genome-wide association study (GWAS) summary data from UK BioBank and NHGRI-EBI GWAS Catalog, have become publicly available.[8,9] However, large-scale cohort studies addressing this debate with sufficient sample size have been conspicuously absent.[4,6] The Mendelian randomization (MR) method, which offers a robust framework for evaluating causal relationships between exposure variables and their associated outcomes, has not been utilized for estimating the causal relationship between SUA levels and CRC.[3,6,10]
To address these challenges, we used large-scale cohort data and the MR approach to determine the causal association between SUA levels and CRC incidence. In this context, SUA levels are treated as the exposure variable and CRC is the designated outcome variable, providing a systematic approach to elucidate the relationships between exposure and outcome.
2. Materials and methods
2.1. Data sources
To assess the causal correlation between SUA levels and CRC, we delineated SUA levels as exposure and CRC as the outcome (Supplementary Table S1, http://links.lww.com/MD/N83). We obtained an SUA-level GWAS summary dataset (GWAS ID: GCST007725) from NHGRI-EBI GWAS Catalog by using “MRInstruments” package (version 0.3.2).[11,12] Also acquired another SUA-level GWAS summary dataset (GWAS ID: ukb-e-30880_CSA) from the MRC Integrative Epidemiology Unit (IEU) OpenGWAS database, and 1 CRC GWAS summary dataset (GWAS ID: ukb-e-208_CSA) from the IEU OpenGWAS database.[12]
The SUA-level GWAS summary data from the NHGRI-EBI GWAS Catalog (GCST007725) encompassed 121,745 individuals of Asian descent, including 5,864,938 single nucleotide polymorphisms (SNPs). The SUA-level GWAS summary data from the IEU OpenGWAS database (GWAS ID: ukb-e-30880_CSA) include 8,411 individuals of Asian descent, comprising 9,811,839 SNPs. The CRC GWAS summary data (GWAS ID: ukb-e-208_CSA) included 8,848 individuals of Asian descent, comprising of 421 cases and 8,427 controls, encompassing 9,809,096 SNPs.
We conducted 2 rounds of MR analysis using 2 SUA cohorts and 1 CRC cohort: setting GCST007725 as the exposure and ukb-e-208_CSA as the outcome, and using ukb-e-30880_CSA as the exposure and ukb-e-208_CSA as the outcome.
2.2. Selection of genetic instrumental variables (IVs)
Using the SUA GWAS database, we utilized linkage disequilibrium (LD) clumping to identify independent SNPs serving as IVs in our study. LD clumping was performed with a window size of 10,000 kb and an r2 threshold of < 0.001, using R software (version 4.2.2) and the “TwoSampleMR” package (version 0.5.6). We selected SNPs related to SUA that were independent and significantly associated with LD relationships (P < 5 × 10−8).[12]
The procedure and criteria were applied to both rounds of MR analysis. In the first MR analysis (with GCST007725 as exposure and ukb-e-208_CSA as outcome), LD clumping was conducted on the SUA levels GWAS summary data (GCST007725), followed by harmonization with the CRC GWAS summary data (ukb-e-208_CSA) and exclusion of palindromic SNPs. Fourteen SNPs were selected as IVs for the initial MR analysis.
In the second MR analysis (setting ukb-e-30880_CSA as the exposure and ukb-e-208_CSA as the outcome), the same procedure was used to obtain the 3 SNPs as IVs.
2.3. Measurement of instrument strength
To assess the robustness of the selected SNPs as IVs, we computed F-statistic.[13] Initially, we calculated r2, representing the proportion of phenotypic variance each SNP could account for.[14] The formula used for this calculation is: r2 = 2 × EAF × (1—EAF) × β2, where EAF denotes the effect allele frequency of each SNP, and β represents the effect size.[15,16] Following this, the F-statistic was derived using the formula: F = (N - 2) × r2 ÷ (1 - r2), with N referring to the sample size.[15,16]
An F-statistic exceeding 10 implies that the SNP effectively minimizes bias in the MR analysis. Conversely, if the F-statistic is less than 10, the SNP is identified as a “weak instrument,” indicating potential bias in the analysis.[13]
2.4. Mendelian randomization
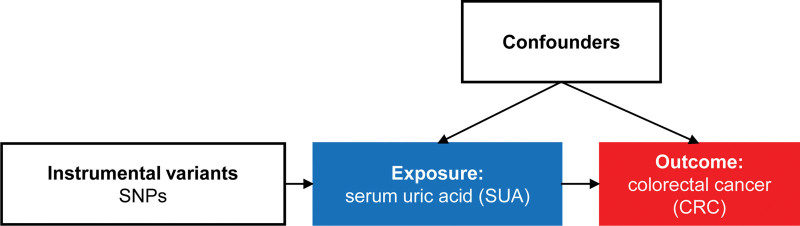
Utilizing the “TwoSampleMR” package (version 0.5.6) in R (version 4.2.2), we conducted MR analysis to assess the causal relationship between SUA levels and CRC.[12] SUA levels served as the exposure variable, while CRC was defined as the outcome (Fig. 1). Significant SNPs associated with SUA levels were used as the IVs. Given that the inverse variance-weighted (IVW) method offers unbiased estimates, the IVW method was employed for the MR analysis.[17] Using the IVW, odds ratio (OR) as the measure of causal effect size, along with its 95% confidence interval (CI).
Figure 1.
Scheme of MR study. Research layout for an MR linking SUA levels and CRC. SNPs serve as genetic IVs, to investigate the causal link between exposure and outcome. CRC = colorectal cancer, IV = instrumental variable, MR = Mendelian randomization, SNP = single nucleotide polymorphism, SUA = serum uric acid.
2.5. Leave-one-out sensitivity analysis and bias of horizontal pleiotropy
To assess potential bias in the causal association of each SNP on the findings, leave-one-out sensitivity analyses were conducted for every SNP using “TwoSampleMR” package (version 0.5.6).[12]
Additionally, to evaluate whether IVs related to SUA levels might impact CRC through pathways other than SUA levels, a horizontal pleiotropy test was performed using the MR-Egger intercept test, also with the “TwoSampleMR” package (version 0.5.6).[12]
3. Results
3.1. Assessing the robustness of chosen SNPs as IVs
F-statistics were used to assess the robustness of selected SNPs as IVs, the F-statistics were used. F-statistics were computed for the 14 SNPs in the first MR analysis (rs11202346, rs11952102, rs1260326, rs13230625, rs1886603, rs2220970, rs2281293, rs244423, rs6774054, rs73575095, rs7679724, rs811372, rs963837, and rs9895661). Similarly, for the second MR analysis, F-statistics were calculated for the 3 selected SNPs (rs2231142, rs2360872, and rs9994216). Because the F-statistics for individual SNPs in both MR analyses exceeded 10, biased results were unlikely to be obtained in the MR analyses.
3.2. MR analysis
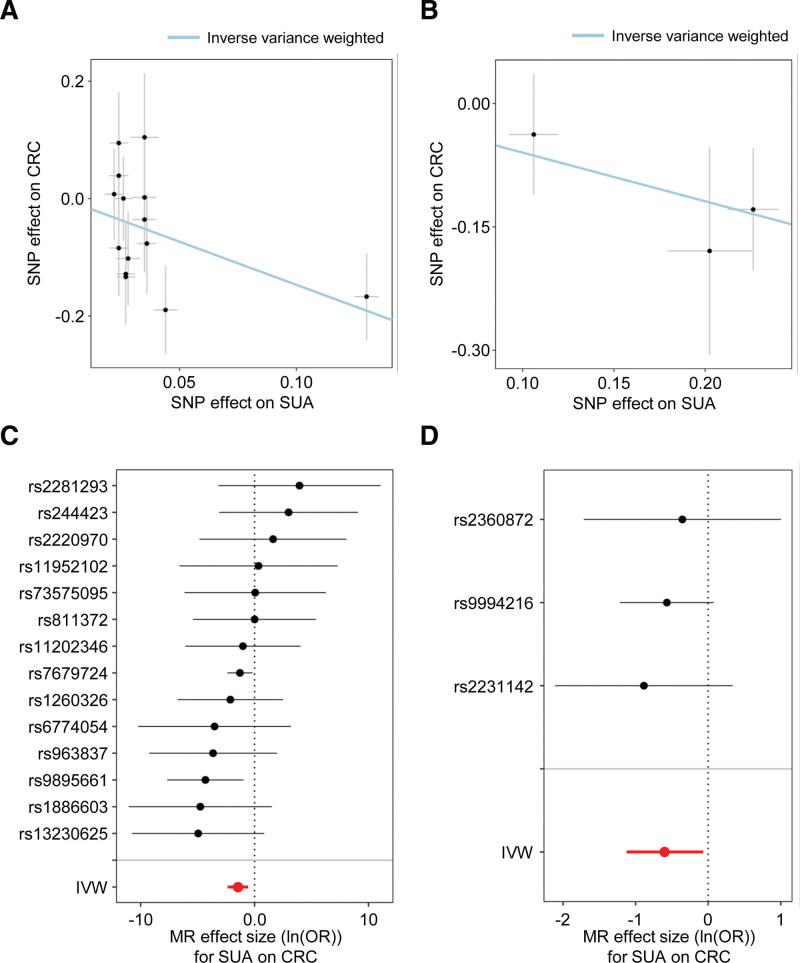
We conducted MR analyses to demonstrate a causal relationship between SUA levels and CRC. Both analyses yielded similar results in the same direction. Figure 2A and B show scatter plots of the estimated causal relationship between SUA levels and CRC using the IVW method.
Figure 2.
MR using the IVW method demonstrates negative associations between SUA levels and CRC. The correlations of genetic instruments for SUA (depicted as the effect size on the X axis) with CRC (represented on the Y axis) were charted based on the GWAS summary data in two MR analyses: (A) the causal relationship of the first MR analysis (setting GCST007725 as exposure and ukb-e-208_CSA as the outcome), and (B) the causal relationship of the second MR analysis (setting ukb-e-30880_CSA as exposure and ukb-e-208_CSA as the outcome). The gradient of the blue line indicates the MR estimation of the causal effect, using the IVW method. Each black dot indicates the causal effect size of each SNP, acting as genetic IVs, determined through the Wald ratio test in the first MR analysis (C) and second MR analysis (D). The red dot represents the effect size for the causality of the amalgamated SNPs, utilizing the IVW method. Error bars illustrate 95% CIs. CI = confidence interval, CRC = colorectal cancer, GWAS = genome-wide association study, IV = instrumental variable, IVW = inverse variance-weighted, MR = Mendelian randomization, SNP = single nucleotide polymorphism, SUA = serum uric acid.
In the first MR analysis (setting GCST007725 as the exposure and ukb-e-208_CSA as the outcome; Fig. 2C), the IVW method estimated that a higher SUA-level significantly reduced the risk of CRC (OR = 0.232; 95% CI of OR = 0.094–0.570; P = .001; Table 1).
Table 1.
Estimates from MR utilizing the IVW method to determine the causal relationship of SUA levels (as exposure) on CRC (as outcome).
| Exposure cohort | Outcome cohort | # of SNPs | SE | P value | OR (95% CI) | |
|---|---|---|---|---|---|---|
| GCST007725 | ukb-e-208_CSA | 14 | −1.46 | 0.46 | .001 | 0.232 (0.094–0.570) |
| ukb-e-30880_CSA | ukb-e-208_CSA | 3 | −0.60 | 0.27 | .027 | 0.551 (0.325–0.934) |
CI = confidence interval, CRC = colorectal cancer, IVW = inverse variance-weighted, MR = Mendelian randomization, OR = odds ratio, SE = standard error of , SNPs = single nucleotide polymorphisms, SUA = serum uric acid.
In the second MR analysis (using ukb-e-30880_CSA as the exposure and ukb-e-208_CSA as the outcome), these 3 SNPs were confirmed as IVs. In the second MR analysis, since both exposure and outcome originate from the same cohort (i.e., UK Biobank), this constitutes a 1-sample MR design. A 1-sample MR within a large cohort can effectively estimate the causal effect between exposure and outcome using the IVW method typically employed in 2-sample analyses.[18] In the second MR analysis, employing the IVW results indicated that a higher SUA-level significantly reduced the risk of CRC (OR = 0.551; 95% CI of OR 0.325–0.934; P = .027; Fig. 2D; Table 1).
Overall, the MR analysis revealed a significant causal relationship between SUA levels and CRC.
3.3. Leave-one-out sensitivity analysis
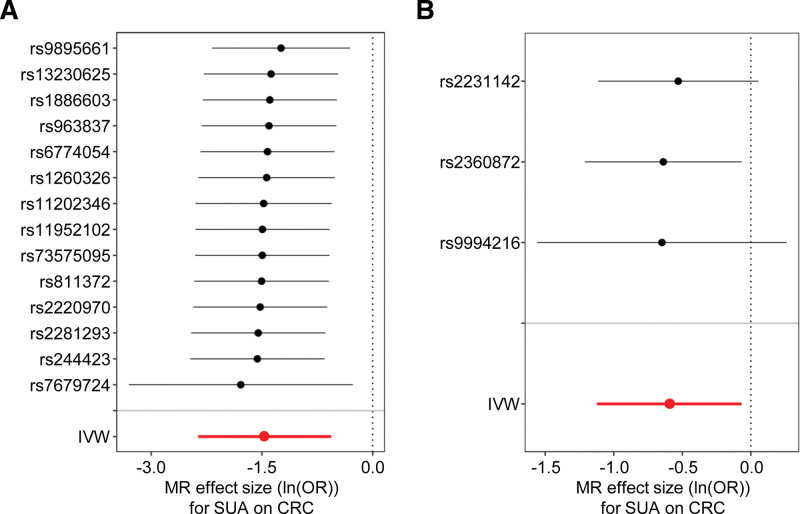
To evaluate the potential bias in the inferred causal relationships from both MR analyses, we performed a leave-one-out sensitivity analysis, indicating the robustness of our MR results (Fig. 3A and B).
Figure 3.
The sensitivity analyses indicate no bias. The leave-one-out sensitivity analyses are portrayed in the forest plots. (A) Causal estimates in the first MR analysis (setting GCST007725 as exposure and ukb-e-208_CSA as the outcome). (B) Causal estimates in the second MR analysis (setting ukb-e-30880_CSA as exposure and ukb-e-208_CSA as the outcome). Each black dot indicates the calculated causal effect size employing all SNPs, except for the SNP indicated on the y axis. The red dot symbolizes IVW estimates incorporating all SNPs. Error bars represent the 95% CI in each MR analysis. CI = confidence interval, IVW = inverse variance-weighted, MR = Mendelian randomization, SNP = single nucleotide polymorphism.
3.4. Horizontal pleiotropy test
We conducted further horizontal pleiotropy tests to investigate whether SNPs serving as IVs demonstrated horizontal pleiotropic effects. In the first MR analysis, we found no indications of horizontal pleiotropy (P = .93). Similarly, in the second MR analysis, we observed no evidence of horizontal pleiotropy (P = .80). The results of these horizontal pleiotropy tests indicated that the SNPs chosen as IVs strictly affected CRC via SUA levels.
4. Discussion
This study aimed to identify a causal association between SUA levels and CRC using MR based on GWAS summary data. The results of studies on the association between SUA levels and the risk of CRC are still disputable; although the association was significant, the direction of the association was not consistent across studies. In this context, our results suggest a significant causal relationship between SUA levels and CRC incidence through MR analysis using large cohorts, indicating a negative relationship. Furthermore, our study shows that the results were validated for robustness through sensitivity analysis.
SUA acts as an antioxidant in the extracellular environment but promote oxidation intracellularly.[19] Thus, SUA has long been suggested to suppress cancer onset due to its antioxidant properties.[20] Previous research indicates that individuals with lower SUA levels have a higher risk of all-cancer mortality compared to those with higher levels.[19] This aligns with our study findings of a negative correlation between SUA levels and CRC.[19] However, research also suggests that SUA, as an oxidation promoter, aid in the proliferation, migration, and survival of normal cells into tumor cells, and that CRC patients with higher SUA levels have lower survival rates compared to those with lower levels.[21] Thus, the role of SUA levels in CRC remains unclear and controversial. Our study supports the hypothesis that, despite the controversial role of SUA on CRC, SUA levels reduce CRC through their antioxidant properties.
We identified 17 SNPs used in the 2 MR analyses and 16 adjacent genes (Supplementary Table S2, http://links.lww.com/MD/N84 and Supplementary Table S3, http://links.lww.com/MD/N85). We investigated the biological association of these genes with SUA levels and cancer. Among the 17 SNPs, rs11202346 was adjacent to Shieldin Complex Subunit 2 (SHLD2), a gene reported in GWAS for SUA levels.[22] SHLD2 regulates the DNA repair pathway, closely related to tumorigenesis.[23] For rs11952102 adjacent to MAX Dimerization Protein 3 (MXD3), which is involved in transcriptional regulation as one of the MAX binding partners. A genome-wide meta-analysis revealed an association between increased SUA levels and gout in a Japanese population.[11] MXD3 is, reportedly, an immune-oncogenic molecule and a potential prognostic marker for multiple cancer types.[24] The rs1260326 SNP is near the glucokinase regulator (GCKR). GCKR polymorphism was shown to be significantly correlated with hyperuricemia in a recent GWAS.[25] Homozygous carriers of GCKR variants showed a decreased risk of developing CRC.[26] Also, rs244423 is adjacent to the nuclear factor of activated T cell 5 (NFAT5). Previous studies have shown that SUA triggers oxidative stress, which activates the NFAT5 gene, a process linked to the stimulation of aldose reductase.[27] Additionally, it has been demonstrated in the research that enterotoxigenic Bacteroides fragilis (ETBF) enhances the inherent CRC cell characteristics. NFAT5 boosts the presence of the JmjC-domain-containing histone demethylase 2 B via the Toll-like receptor 4 pathway, activated by ETBF, potentially increasing the incidence of CRC.[28]
These multifaceted pathophysiological connections elucidate the causal association between SUA levels and CRC onset and progression, suggesting that uric acid management may play a crucial role in cancer prevention and treatment.
This study had some limitations. The evaluation utilized data from Asian populations. Therefore, to confirm such causal relationships in other ethnicities, MR should be performed using GWAS data from non-Asian populations for exposure and outcomes. Additionally, the results should be interpreted with caution as unknown confounding factors may reduce the reliability of the results.
5. Conclusion
In conclusion, the MR analysis revealed a significant association between SUA levels and CRC incidence.
Author contributions
Conceptualization: Seungyoon Nam.
Data curation: Miseon Lee.
Formal analysis: Miseon Lee.
Investigation: Miseon Lee.
Methodology: Miseon Lee.
Project administration: Seungyoon Nam.
Resources: Miseon Lee.
Software: Miseon Lee.
Supervision: Seungyoon Nam.
Validation: Miseon Lee, Seungyoon Nam.
Visualization: Miseon Lee.
Writing – original draft: Miseon Lee, Seungyoon Nam.
Writing – review & editing: Seungyoon Nam.
Supplementary Material
Abbreviations:
- CI
- confidential interval
- CRC
- colorectal cancer
- ETBF
- enterotoxigenic Bacteroides fragilis
- GCKR
- glucokinase regulator
- GWAS
- genome-wide association study
- IEU
- MRC integrative epidemiology unit
- IV
- instrumental variable
- IVW
- inverse variance-weighted
- LD
- linkage disequilibrium
- MR
- Mendelian randomization
- MXD3
- MAX dimerization protein 3
- NFAT5
- nuclear factor of activated T cells 5
- OR
- odds ratio
- SHLD2
- Shieldin complex subunit 2
- SNP
- single nucleotide polymorphism
- SUA
- serum uric acid
This work was supported by the Gachon University research fund of 2023 (GCU-202303710001 to SN) and the Gachon University Gil Medical Center (Grant number FRD2023-13 to SN).
Ethics approval and consent to participate: Not applicable because of the lack of patient recruitment.
The authors have no conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental Digital Content is available for this article.
How to cite this article: Lee M, Nam S. The causal relationship of serum uric acid on colorectal cancer: A Mendelian randomization study. Medicine 2024;103:26(e38722).
References
- [1].de Martel C, Georges D, Bray F, et al. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8:e180–90. [DOI] [PubMed] [Google Scholar]
- [2].Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mi N, Huang J, Huang C, et al. High serum uric acid may associate with the increased risk of colorectal cancer in females: a prospective cohort study. Int J Cancer. 2022;150:263–72. [DOI] [PubMed] [Google Scholar]
- [4].Oh YJ, Lee YJ, Lee E, et al. Cancer risk in Korean patients with gout. Korean J Intern Med. 2022;37:460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Taghizadeh N, Vonk JM, Boezen HM. Serum uric acid levels and cancer mortality risk among males in a large general population-based cohort study. Cancer Causes Control. 2014;25:1075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li W, Liu T, Siyin ST, et al. The relationship between serum uric acid and colorectal cancer: a prospective cohort study. Sci Rep. 2022;12:16677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chang W, Zhong Q, Liang S, et al. A high spatial resolution dataset for anthropogenic atmospheric mercury emissions in China during 1998-2014. Sci Data. 2022;9:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liao W, Wang Y, Zhang W. Serum uric acid and the risk of colorectal cancer: a meta-analysis. Eur J Cancer Prev. 2024;33:19–28. [DOI] [PubMed] [Google Scholar]
- [11].Nakatochi M, Kanai M, Nakayama A, et al. Genome-wide meta-analysis identifies multiple novel loci associated with serum uric acid levels in Japanese individuals. Commun Biol. 2019;2:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Burgess S, Thompson SG, Collaboration CCG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40:755–64. [DOI] [PubMed] [Google Scholar]
- [14].Pu B, Gu P, Luo L, et al. Causal effects of tea intake on multiple types of fractures: a two-sample Mendelian randomization study. Medicine (Baltim). 2023;102:e33542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang X, Theodoratou E, Li X, et al. Genetically predicted physical activity levels are associated with lower colorectal cancer risk: a Mendelian randomisation study. Br J Cancer. 2021;124:1330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wu E, Ni J, Tao L, et al. A bidirectional Mendelian randomization study supports the causal effects of a high basal metabolic rate on colorectal cancer risk. PLoS One. 2022;17:e0273452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Minelli C, Del Greco MF, van der Plaat DA, et al. The use of two-sample methods for Mendelian randomization analyses on single large datasets. Int J Epidemiol. 2021;50:1651–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yiu A, Van Hemelrijck M, Garmo H, et al. Circulating uric acid levels and subsequent development of cancer in 493,281 individuals: findings from the AMORIS Study. Oncotarget. 2017;8:42332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kuhn T, Sookthai D, Graf ME, et al. Albumin, bilirubin, uric acid and cancer risk: results from a prospective population-based study. Br J Cancer. 2017;117:1572–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mao L, Guo C, Zheng S. Elevated urinary 8-oxo-7,8-dihydro-2’-deoxyguanosine and serum uric acid are associated with progression and are prognostic factors of colorectal cancer. Onco Targets Ther. 2018;11:5895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ko YL. Genetics of hyperuricemia and gout: Insights from recent genome-wide association studies and Mendelian randomization studies. Tzu Chi Med J. 2022;34:261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Findlay S, Heath J, Luo VM, et al. SHLD2/FAM35A co-operates with REV7 to coordinate DNA double-strand break repair pathway choice. EMBO J. 2018;37:e100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wu SY, Lin KC, Lawal B, et al. MXD3 as an onco-immunological biomarker encompassing the tumor microenvironment, disease staging, prognoses, and therapeutic responses in multiple cancer types. Comput Struct Biotechnol J. 2021;19:4970–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ho LJ, Lu CH, Su RY, et al. Association between glucokinase regulator gene polymorphisms and serum uric acid levels in Taiwanese adolescents. Sci Rep. 2022;12:5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ollberding NJ, Cheng I, Wilkens LR, et al. Genetic variants, prediagnostic circulating levels of insulin-like growth factors, insulin, and glucose and the risk of colorectal cancer: the Multiethnic Cohort study. Cancer Epidemiol Biomarkers Prev. 2012;21:810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sanchez-Lozada LG, Andres-Hernando A, Garcia-Arroyo FE, et al. Uric acid activates aldose reductase and the polyol pathway for endogenous fructose and fat production causing development of fatty liver in rats. J Biol Chem. 2019;294:4272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu QQ, Li CM, Fu LN, et al. Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut Microbes. 2020;12:1788900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.