Abstract
Partial deletions within Autographa californica open reading frame 61 (FP25K) alter the expression and accumulation profile of several viral proteins and the transport of occlusion-derived virus (ODV)-E66 to intranuclear membranes during infection (S. C. Braunagel et al., J. Virol. 73:8559–8570, 1999). Here we show the effects of a full deletion and overexpression of FP25K on the transport and expression of two ODV envelope proteins, ODV-E66 (E66) and ODV-E25 (E25). Deletion and overexpression of FP25K substantially altered the levels of expression of E66 during infection. Compared with cells infected with wild-type (wt) virus, the levels of E66 were reduced fivefold in cells infected with a viral mutant lacking FP25K (ΔFP25K) and were slightly increased in cells infected with a viral mutant overexpressing FP25K (FP25Kpolh). In contrast, no significant changes were observed in the levels of E25 among wt-, ΔFP25K-, and FP25Kpolh-infected cells. The changes observed in the levels of E66 among the different viral mutants were not accompanied by changes in either the time of synthesis, membrane association, protein turnover, or steady-state transcript abundance. Deletion of FP25K also substantially altered the transport and localization of E66 during infection. In cells infected with the ΔFP25K mutant virus, E66 accumulated in localized regions at the nuclear periphery and the outer nuclear membrane and did not traffic to intranuclear membranes. In contrast, in cells infected with the FP25Kpolh mutant virus E66 trafficked to intranuclear membranes. For comparison, E25 was normally transported to intranuclear membranes in both ΔFP25K- and FP25Kpolh-infected cells. Altogether these studies suggest that FP25K affects the synthesis of E66 at a posttranscriptional level, probably by altering the translation of E66; additionally, the block in transport of E66 at the nuclear envelope in ΔFP25K-infected cells suggests that the pathway of E66 trafficking to the inner nuclear membrane and intranuclear microvesicles is specifically regulated and must be influenced by factors that do not control the traffic of E25.
Nuclear polyhedrosis viruses produce two types of viral progeny with distinctly different structural and biological properties. The first type of viral progeny made during infection, the budded virus (BV), is produced when nucleocapsids assembled in the nucleus are translocated into the cytoplasm and bud through the plasma membrane, obtaining an envelope in the process. The second form of viral progeny made, the occlusion-derived virus (ODV), is produced when nucleocapsids retained in the nucleus are enveloped using virus-induced intranuclear membranes which appear to be derived from the nuclear membrane (9, 17). The ODV is then occluded within a proteinaceous crystal (polyhedron) composed of the viral protein polyhedrin. Consistent with their different origins, studies on the composition of the viral envelopes of the Autographa californica baculovirus (AcMNPV) show that BV and ODV envelopes exhibit substantial differences in protein and lipid composition and in total protein per lipid content (6). Such differences parallel the functional roles played by ODVs and BVs during the life cycle of the virus: ODVs, responsible for host-to-host dispersal of the virus, are highly infectious for gut cells of the larval host (30) but not for cells maintained in culture; BVs, responsible for cell-to-cell dispersion of the virus within the infected larval host, are highly infectious for cultured cells but not for gut cells. The remarkable differences in protein composition observed between ODV and BV envelopes indicate that the traffic of their protein components must be tightly regulated so that they may efficiently reach their intended destination with the proper stoichiometry for assembly.
Mutations affecting the viral protein FP25K alter the traffic of some ODV envelope proteins during infection. In cells infected with the viral mutant FP-βgal, in which the first 125 N-terminal residues of FP25K were fused to β-galactosidase, the traffic of ODV-E66 (E66) to the nucleus is delayed, while that of ODV-E25 (E25) is normal (4). Furthermore, in cells infected with the viral mutant 480-1, which lacks the first 32 N-terminal residues of FP25K, E66 remains cytoplasmic throughout infection, while E25 trafficking to the nucleus is delayed (4). FP25K was initially identified as a gene located in a region of the baculovirus genome associated with mutations leading to a decrease in the number of occlusions produced within the nucleus of infected cells (the few polyhedra, or FP, phenotype) (1, 11). The FP25K gene is one of the 65 open reading frames (ORFs) present in every one of the seven members of the Baculoviridae family sequenced to date (7) and codes for a structural protein of the viral nucleocapsid highly conserved among members of the Nucleopolyhedrovirus genus (4). FP25K is dispensable in vitro (22) but is essential for natural infections, as it is required for normal ODV envelopment and morphology (10, 15, 21). In addition to producing the FP phenotype, mutations affecting FP25K produce a wide variety of effects, including enhanced production of BVs (10, 15, 23) and a block in the postmortem liquefaction of the larval host (19). Mutations in the FP25K gene also result in significant alterations in the apparent expression and/or accumulation of several viral proteins, including a significant decrease in polyhedrin synthesis (accompanied by a decrease in its steady-state transcript levels) (14) and significant increases in the synthesis of some structural viral proteins of BVs, such as gp67, BV/ODV-E26, and p39 (4). In spite of knowledge on the effects produced by mutations in FP25K, the function of this protein is still unknown.
In this study the effects of deleting and overexpressing FP25K on the synthesis and transport of E25 and E66 were determined. Deletion of FP25K dramatically decreased the amount of E66 protein without affecting E66 steady-state transcript levels, association to cellular membranes, or turnover. Deletion of FP25K also resulted in a block on the traffic of E66 from the endoplasmic reticulum (ER) and outer nuclear membrane to intranuclear membranes (i.e., inner nuclear membrane, virus-induced microvesicles, and viral envelopes). Overexpression of FP25K increased the amount of E66 which accumulated during infection and allowed its normal traffic to intranuclear membranes. In contrast, both deletion and overexpression of FP25K exerted no effect in protein steady-state levels and nuclear trafficking of E25, indicating that the effects observed on E66 were protein specific. These results support a role for FP25K on the posttranscriptional regulation of the expression of E66 during infection and suggest that FP25K itself, or another protein regulated by FP25K, plays a role in the transport of E66 to virus-induced intranuclear membranes.
MATERIALS AND METHODS
Targeted deletion of the FP25K gene.
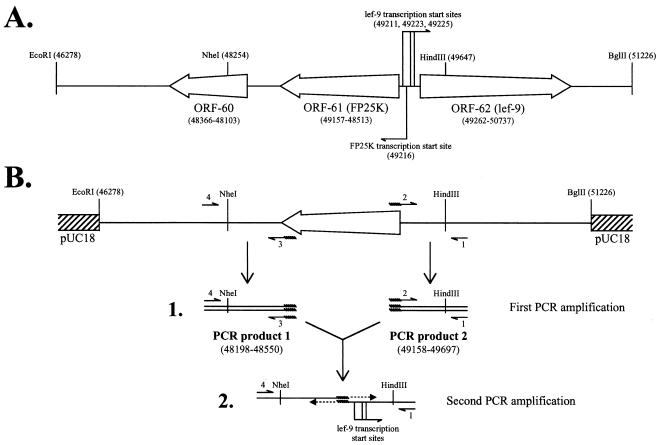
To develop a FP25K deletion mutant, the genomic regions flanking FP25K were joined together using a two-step PCR procedure (Fig. 1) and were cloned into a plasmid, and the resulting plasmid was cotransfected into Sf9 cells with genomic DNA from the baculovirus clone FP-βgal (1). This cloning procedure was designed to keep flanking genes and their promoters intact. Briefly, the AcMNPV (E2 strain) EcoRI-F fragment was digested with EcoRI and BglII and cloned in EcoRI/BamHI-digested pUC18. The resulting clone (pUC18-5kb) contained a region spanning residues 46278 through 51226 of the baculovirus genome (Fig. 1A). Using this clone as template, two rounds of PCR were performed to amplify the regions located immediately upstream and downstream of FP25K (Fig. 1B). In the first round, the FP25K upstream region was amplified using the primers 5′ GAGCGCCCGAAATGTCAACC 3′ (primer 1) and 5′ gctatcgttttATTAACTATATCAACCCGATGCG 3′ (primer 2), and the FP25K downstream region was amplified using the primers 5′ gatatagttaatAAAACGATAGCGAAAAAATGCTTC 3′ (primer 3) and 5′ GCCTTGGCTGCCATTTACCG 3′ (primer 4). Lower-case letters indicate residues added to introduce an overlapping region between the upstream and downstream PCR products, while the complementary region generated is underlined (Fig. 1B-1). In the second round, the purified products from the first round of PCR amplifications were combined and used as templates for the reaction. No additional primers were added for the initial 4 cycles of amplification. Thereafter, primers 1 and 4 were added and the PCR was allowed to continue for 28 additional cycles to give a PCR product in which the upstream and downstream regions of FP25K were directly connected, thus deleting FP25K (residues 48550 to 49158) (Fig. 1B-2). The PCR product obtained was NheI/HindIII digested and cloned into the 4,631-bp fragment produced by the NheI/HindIII digestion of pUC18-5kb. Then a 1,613-bp HindIII fragment excised out of pUC18-5kb during the NheI/HindIII digestion was added back. The resulting clone, named pUC18-FP25Kdel#3.3, was fully sequenced by using internal primers. Cotransfection was performed according to Summers and Smith (28), and the recombinant viruses generated were screened by color selection in plaque assays overlaid with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). White plaques were picked and further purified by three additional rounds of plaque purification. The purified FP25K deletion mutant obtained (referred to as ΔFP25K) was amplified, and the absence of FP25K was confirmed by PCR analyses and Southern blotting of ΔFP25K genomic DNA.
FIG. 1.
Deletion of ORF-61 (FP25K) by a two-step PCR procedure. (A) Map of the AcMNPV genomic region cloned into pUC18. The relevant ORFs contained within the cloned region are shown. The transcription start sites for lef-9 (as described by Guarino et al. [13]) and FP25K are indicated. Numbers in parentheses indicate the position (in base pairs) in the AcMNPV-C6 genome. (B) Representation of clone pUC18-5kb, the relative locations of the primers used for PCR amplification, and the products obtained during the first (1) and second (2) rounds of PCR amplification. The regions covered by the PCR products obtained during the first amplification are indicated. The complementary region introduced in primers 2 and 3 is represented by saw-toothed lines. Notice that the strategy followed fully deletes ORF-61 without altering the transcription initiation sites for the adjacent ORFs.
Overexpression of FP25K.
To develop a viral mutant containing a second copy of FP25K in the polyhedrin locus, FP25K was cloned in the pBACgus-1 vector (Novagen, Inc., Madison, Wis.). Sf9 cells were transfected using Bsu361-digested BacPak AcMNPV genomic DNA and the recombinant pBACgus-1 plasmid according to the methods of Summers and Smith (28). Recombinant viruses were selected for glucoronidase activity in plaque assays overlaid with 5-bromo-4-chloro-3-indolyl-β-d-glucoronide cyclohexylammonium salt (X-Gluc). Positive plaques were purified by two additional rounds of plaque purification. The purified recombinant virus obtained (referred to as FP25Kpolh) was amplified, and the presence of a second copy of FP25K in the polyhedrin locus was confirmed by PCR analysis and Southern blotting of FP25Kpolh genomic DNA.
Cell culture, virus infections, and metabolic labeling.
Spodoptera frugiperda IPLB-Sf21 clonal isolate 9 (Sf9) cells were cultured in suspension at 27°C in TNM-FH medium (28) supplemented with 10% fetal bovine serum (complete medium). AcMNPV strain E2 was used as a wild-type (wt) virus control. All infections were performed at a multiplicity of infection of 20, and time zero was set at the time of virus addition. For metabolic labeling, cells were seeded and infected at a density of 3 × 106 cells/flask in 5 ml of complete medium in 25-ml tissue culture flasks. One hour before adding the label the culture medium was collected and replaced with 1 ml of methionine-deficient Grace's medium supplemented with 0.5% fetal bovine serum. To recover dislodged cells and maintain a constant cell number, the collected culture medium was centrifuged (1,000 × g, 5 min, room temperature) and the cell pellet was resuspended in 1 ml of methionine-deficient Grace's medium, added back to the flask, and incubated at 27°C. One hour later, 200 μCi of Tran35S-Label (ICN Pharmaceuticals Inc., Costa Mesa, Calif.) was added and the cells were incubated at 27°C for the desired amount of time. For pulse-chase experiments, after the labeling period the cells were washed twice with complete medium and incubated at 27°C for the desired amount of time. Upon collection the cells were centrifuged (1,000 × g, 5 min, room temperature), washed with 1× phosphate-buffered saline (PBS), centrifuged again, and frozen as wet pellets at −80°C in aliquots of 1.5 × 106 cells per tube. Upon thawing the pellets were processed immediately.
Immunoprecipitation and immunoblotting.
For time course analyses of protein expression, cell extracts were prepared from frozen cell pellets containing 1.5 × 106 metabolically labeled cells collected at different times postinfection. To dissolve virus occlusions, each cell pellet was incubated in 0.04 N NaOH for 10 min at 37°C. The resulting extracts were mixed with 525 μl of 1× radioimmunoprecipitation assay (RIPA) buffer (100 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) and passed through a 25-gauge needle 10 times to shear the DNA. At this stage a 20-μl aliquot was taken, mixed with an equal volume of 4× sample buffer (100 mM Tris-HCl [pH 6.8], 8% SDS, 4% β-mercaptoethanol, 0.04% bromophenol blue, 20% glycerol), incubated for 15 min at 65°C, and used for SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot analyses as described below. In preparation for immunoprecipitation analysis, the residual sample was clarified by centrifugation at 13,000 × g for 10 min at 4°C and the supernatant was preabsorbed with 25 μl of preimmune rabbit serum for 1 h at 4°C, incubated with 40 μl of a 50% slurry of Protein A agarose for 1 h at 4°C, and pelleted at 1,000 × g for 15 min at 4°C. The preabsorbed extract was split into three aliquots of 175 μl (equivalent to 4.4 × 105 cells), each of which was immunoprecipitated with 10 μl of the appropriate rabbit serum overnight at 4°C, followed by incubation with 20 μl of a 50% slurry of Protein A agarose for 1 h at 4°C. The agarose beads were washed three times in 1× RIPA buffer and once in 1× Tris-buffered saline (TBS) (140 mM NaCl, 25 mM Tris [pH 8.0]), mixed with 20 μl of 4× sample buffer, and incubated for 15 min at 65°C and the immunoprecipitated proteins were analyzed by SDS-PAGE.
SDS-PAGE analyses were performed using 4 and 12.5% stacking and resolving gels, respectively, as described by Laemmli (20). Following electrophoresis the proteins were transferred onto Immobilon-P membranes (Millipore, Bedford, Mass.) and the membranes were either processed for immunoblotting or were directly exposed to film or a phosphorscreen for detection of labeled blotted proteins. For immunoblotting the membranes were blocked with 1× TTBS (1× TBS + 0.05% Tween 20) supplemented with 3% nonfat dry milk (1× blocking solution) for 1 h at room temperature. Rabbit serum directed against the appropriate protein (E66, serum 5297; E25, serum 10234; FP25K, serum 2804) was added and incubated with the membrane overnight at 4°C. For time course analyses of protein expression, rabbit antisera were used at a final dilution of 1:10,000 in 50 ml of 1× blocking solution. Upon incubation the membranes were washed three times in 1× TTBS and incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) at a final dilution of 1:12,500 in 50 ml of 1× blocking solution. The membranes were washed three times in 1× TTBS and once in 1× TBS and were developed using the NEN Renaissance Western blot chemiluminescence reagent kit (NEN Life Science Products, Boston, Mass.). To strip the membranes for subsequent rounds of immunoblotting, the membranes were washed three times in 1× TTBS, incubated for 30 min at 55°C in stripping buffer (62.5 mM Tris–HCl [pH 6.8], 2% SDS, 100 mM β-mercaptoethanol), and washed five additional times in 1× TTBS. To test for residual enzymatic activity, the membranes were developed as above. Before reuse the membranes were washed four additional times in 1× TTBS. Subsequent rounds of immunoblotting were started at the blocking step.
To estimate the relative abundance of specific viral proteins, cell extracts were prepared from frozen cell pellets containing 3 × 106 cells collected at 28 h postinfection (hpi). Each cell pellet was resuspended in 500 μl of sterile MilliQ water, mixed with 2 μl of 10 N NaOH, and incubated for 10 min at 37°C. The cell extract was mixed with 500 μl of 4× sample buffer and incubated for 3 min at 100°C. Aliquots (50 μl per lane) of the resulting sample were used for SDS-PAGE analysis. Following electrophoresis the proteins were transferred onto Immobilon-P membranes. Upon transfer the membranes were blocked as above and incubated with the appropriate rabbit antisera at a final dilution of 1:2,000 in 15 ml of 1× blocking solution. Upon incubation the membranes were washed three times in 1× TTBS and incubated for 1 h at room temperature with 2 μCi of 125I-labeled anti-rabbit IgG (ICN Pharmaceuticals, Inc.) in 15 ml of 1× blocking solution. The membranes were washed four times in 1× TTBS, dried out, and exposed to a phosphorscreen at −80°C. All quantitative analyses were performed using ImageQuant v. 0.5.0 software in a Storm FluorImager (Molecular Dynamics, Sunnyvale, Calif.).
Fractionation of integral membrane proteins with Triton X-114.
Detergent fractionations were performed according to the methods of Bordier (3). Briefly, frozen cell pellets containing 3 × 106 cells collected at 24 and 48 hpi were resuspended in 666 μl of ice-cold 1× PBS–1% Triton X-114. The cell extract was incubated for 1 h at 4°C and clarified by centrifugation at 14,000 × g for 10 min at 4°C. The supernatant was loaded onto a 66-μl sucrose cushion (6% sucrose–0.06% Triton X-114 in 1× PBS) and incubated for 3 min at 37°C, and the resulting detergent and aqueous phases were resolved by centrifugation at 500 × g for 5 min at 37°C. The detergent phase was resuspended with 333 μl of ice-cold 1× PBS and incubated for 3 min at 37°C, and the detergent and aqueous phases were resolved again as above. The new aqueous phase was added to the first aqueous phase, and the detergent phase was resuspended in 1 ml of ice-cold 1× PBS and mixed with an equal volume of 4× sample buffer. The pooled aqueous phase was mixed with 33 μl of 11.4% Triton X-114, incubated for 3 min at 37°C, and centrifuged at 500 × g for 5 min at 37°C. The resulting aqueous phase was mixed with an equal volume of 4× sample buffer. The samples were incubated for 15 min at 65°C and analyzed by SDS-PAGE using 35 μl per lane.
Primer extension.
Sf9 cells grown in suspension (2 × 106 cells/ml) were infected with either AcMNPV-E2 (wt), ΔFP25K, or FP25Kpolh. At 28 hpi the infected cells were collected and polyadenylated mRNA was purified using the Poly(A) Pure mRNA isolation kit (Ambion, Inc., Austin, Tex.). Primer extensions were performed with 5 μg of mRNA hybridized to specific probes labeled with [γ-32P]ATP. The oligonucleotide sequence of the probes used were the following: E66 probe, 5′ CGGCAAGGGATTGAGATCAATAAAAGC 3′; p39 (capsid) probe, 5′ TTTGTCGCGGCGCCATACCCACGGGCACTAGCGCCATATTG 3′; E25 probe 1, 5′ GCAAAACGATAAGTAACACGATTCCCCAC 3′; E25 probe 2, 5′ GAGATTTAGGTTGTGCAAATGTTTCAAAAGTACAC 3′. To provide internal controls, the primer extension reaction mixtures for E66 and E25 contained a mix of the gene-specific probe and the p39 probe. For E25 primer extensions, only E25 probe 1 was used; E25 probe 2 was used for preliminary primer extension analyses performed to determine transcription start sites for E25. mRNA-primer hybrids were precipitated with 100% ethanol, washed with 70% ethanol, and resuspended in 30 μl of reverse transcription mix (50 mM Tris [pH 8.3], 75 mM KCl, 3 mM MgCl2, 0.666 mM deoxynucleoside triphosphates, 1 mM dithiothreitol, 40 U of RNasin, 50 μg of actinomycin D, and 10 U of SuperScript II RNase H-negative reverse transcriptase [Life Technologies, Inc., Gaithersburg, Md.]). Reverse transcription was performed for 1 h at 42°C. The reaction products were ethanol precipitated and resuspended in 3 μl of 100 mM NaOH and 6 μl of sequencing stop buffer. The samples were boiled for 3 min and analyzed by electrophoresis on a urea–6% polyacrylamide gel together with a sequencing ladder generated with the same oligonucleotides. The gels were dried and the primer extension products were quantified using a Storm FluorImager.
Immunofluorescence confocal microscopy.
Infected Sf9 cells were collected at 36 and 48 hpi, washed, and resuspended in Grace's media, and 2.1 × 105 cells were transferred to a 1-well cytofuge concentrator (StatSpin Technologies, Norwood, Mass.). The cells were allowed to attach for 5 min at room temperature and were fixed with 3.7% paraformaldehyde in PBS (20 mM phosphate, 140 mM NaCl, pH 7.2) for 10 min at room temperature. The cells were washed three times with PBS, permeabilized with methanol (10 min) and 0.5% Triton X-100 in PBS (10 min), and washed twice again with PBS. The cells were blocked for 1 h in blocking solution (1% chicken serum, 3% bovine serum albumin in PBS) and incubated overnight at 4°C with the appropriate antibodies diluted in blocking solution (lamin, monoclonal antibody ADL67 [27], 1:500, provided by P. A. Fisher, Department of Pharmacological Sciences, University of New York at Stony Brook, Stony Brook; E66, serum 5297, 1:1,000; E25, serum [1:2,500] provided by G. Rohrmann, Oregon State University, Corvallis; FP25K, serum 2804, 1:2,500). The cells were rinsed three times with PBS and incubated with Alexa Fluor 488-conjugated anti-rabbit IgG or Alexa Fluor 594-conjugated anti-mouse IgG (both from Molecular Probes, Inc., Eugene, Oreg.) diluted 1:2,000 in blocking solution. After three washes with PBS the cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) at 0.1 μg/ml in PBS for 5 s, washed three additional times with PBS, and viewed with a Zeiss CARV confocal microscope. In each experiment at least 15 large fields of view were observed, each field containing an average of 50 to 60 cells. Then, cells representing the pattern seen in 80% or more of the cells observed were selected to collect confocal Z-stack sections at 0.75-μm intervals. Confocal images of at least five different representative cells were collected per experiment. Each experiment was performed several times. Three-dimensional reconstructions and image deconvolution were performed using the Zeiss KS 400 Imaging System, release 3.0.
Immunoelectron microscopy.
Immunoelectron microscopy was performed as previously reported (5, 16) by using Sf9 cell cultures collected at 48 hpi. Rabbit antisera were used at a final dilution of 1:1,000. Bound rabbit antibodies were detected using anti-rabbit IgG gold-conjugated goat antibodies (25 nm; Electron Microscopy Sciences, Fort Washington, Pa.) at a 1:15 dilution.
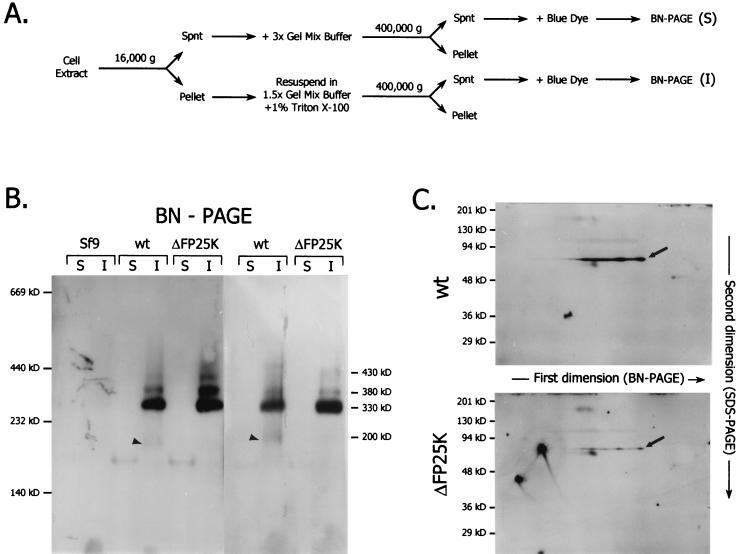
BN-PAGE and two-dimensional BN-PAGE and SDS-PAGE analyses.
Blue native (BN)-PAGE was performed according to the guidelines of Schagger and von Jagow (25) by using linear 6 to 13% acrylamide gradient gels. In preparation for BN-PAGE, frozen pellets of infected (30 hpi) or noninfected Sf9 cells containing 2 × 107 cells were resuspended in sterile MilliQ water supplemented with 200 U of DNase I/ml up to a final volume of 600 μl. The cell suspension was passed through a 27.5-gauge needle several times, sonicated in a water bath sonicator for 45 s, and incubated on ice for 2 h. The cell extract produced was fractionated by differential centrifugation as illustrated in Fig. 10A. Briefly, the extract was centrifuged at 15,000 × g for 10 min at 4°C. The supernatant obtained was mixed with an equal volume of 3× Gel Mix Buffer (1.5 M Aminocaproic acid, 150 mM Bis-Tris, pH 7.0) and centrifuged at 400,000 × g for 30 min at 15°C in a TLA-100 rotor (Beckman Instruments Inc., Palo Alto, Calif.). The resulting supernatant (soluble fraction supernatant) was mixed with blue dye (5% Serva Blue G in 500 mM Aminocaproic acid) at a ratio of 200 μl of supernatant per 10 μl of blue dye. The pellet from the 15,000 × g spin was washed once with MilliQ water, and the resulting pellet was resuspended in 540 μl of a 1:1 dilution of 3× Gel Mix Buffer and MilliQ water and was mixed with 60 μl of a 10% Triton X-100 solution. The solubilized pellet was then centrifuged at 400,000 × g, and the resulting supernatant (insoluble fraction supernatant) was mixed with blue dye, as described above. Sixty microliters of the blue-dye sample mix were loaded per lane. The gels were run overnight at 200 V and 4°C, equilibrated in 1× Transfer Buffer (25 mM Tris, 192 mM glycine, 0.04% SDS, 20% methanol) for 15 min, blotted to Immobilon P membranes, and processed for immunoblotting as described above. For two dimensional analyses, after completion of the first dimension (BN-PAGE) the gel strip corresponding to the lane of interest was cut away, soaked in 50 ml of denaturing solution (1% [wt/vol] SDS and 1% [vol/vol] β-mercaptoethanol) for 2 h, and soaked in 50 ml of 1× SDS-PAGE running buffer for 5 min. Thereafter the gel strip was fixed between two glass plates and excess running buffer was removed, and a discontinuous 10% resolving SDS-PAGE gel was poured so that the immobilized gel strip was surrounded by the stacking gel. The gel was run and blotted into Immobilon P membranes.
FIG. 10.
Immunoblot analysis of protein complexes from noninfected, wt-infected, and ΔFP25K-infected Sf9 cells resolved in one-dimensional (BN-PAGE) or two-dimensional (BN-PAGE and SDS-PAGE) gels. Total cell extracts fractionated into soluble (S) and insoluble (I) fractions (A) were resolved in one dimension (BN-PAGE) (B) or two dimensions (BN-PAGE and SDS-PAGE) (C) and were subsequently analyzed by immunoblot using α-E66. The data shown in panel B corresponds to two independent experiments. Molecular size markers: thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; lactose dehydrogenase, 140 kDa. The location of the molecular size markers and the approximate molecular sizes of the complexes observed by BN-PAGE are indicated. The direction of the first and second dimensions is also indicated. Arrowheads indicate the E66 protein complexes decreased in ΔFP25K-infected cells compared to those in wt-infected cells. Arrows indicate the distribution of E66 upon two-dimensional electrophoresis. Spnt, supernatant.
RESULTS
Deletion of FP25K decreases the levels of ODV-E66 without altering its time of synthesis.
Previous studies indicated that partial deletions of FP25K resulted in altered synthesis and transport of several late and very late baculovirus proteins (4, 14, 15). To further study the role of FP25K, two new virus mutants were developed, one containing a precise deletion of the FP25K gene (ΔFP25K) and another containing an additional copy of FP25K under the polyhedrin promoter (FP25Kpolh). Sf9 cells infected with the ΔFP25K virus exhibited the typical FP phenotype associated with mutations within the FP25K gene (data not shown), as described by Beames and Summers (1). Cells infected with the viral mutant containing an additional copy of FP25K under the polyhedrin promoter (FP25Kpolh) exhibited no significant characteristics by light microscopy other than the occlusion-negative phenotype.
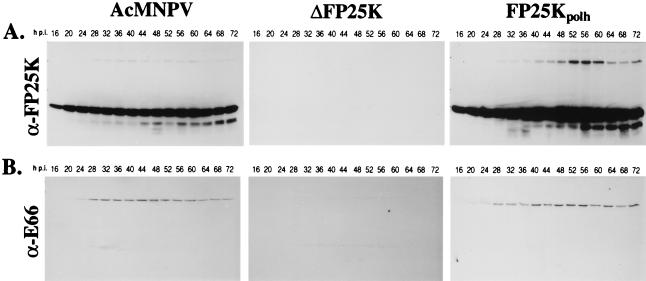
Partial deletions of FP25K decrease the accumulation of E66 but not of E25 (4). To evaluate the effect that the full deletion and the overexpression of FP25K exert on the expression of E66 and E25, immunoblot analyses were performed using metabolically labeled cells infected with wt, ΔFP25K, and FP25Kpolh viruses and antibodies raised against E66 (α-E66), E25 (α-E25), and FP25K (α-FP25K). As expected, there was a noticeable increase in the level of FP25K during FP25Kpolh infection, and FP25K was not detected in ΔFP25K-infected cells (Fig. 2A). E25 accumulated to similar levels in wt-, ΔFP25K-, and FP25Kpolh-infected cells (data not shown). In contrast, significantly lower levels of E66 were detected in ΔFP25K-infected cells compared to those with wt- and FP25Kpolh-infected cells: whereas in wt- and FP25Kpolh-infected cells E66 was easily detected starting at 24 hpi, in ΔFP25K-infected cells E66 was barely detectable throughout infection (Fig. 2B).
FIG. 2.
Accumulation profiles of E66 and FP25K in wt-infected (AcMNPV), ΔFP25K-infected, and FP25Kpolh-infected cells. Cells infected with wt, ΔFP25K, or FP25Kpolh were metabolically labeled for the 4-h interval before collection and were lysed and clarified. Aliquots corresponding to equal cell numbers were run on SDS–12.5% PAGE gels and transferred to Immobilon membranes. The membranes were sequentially probed with α-FP25K, α-E25 (data not shown), and α-E66. (A) Immunoblots performed using α-FP25K. (B) Immunoblots performed using α-E66. Hours postinfection (hpi) indicate the time postinfection at which the samples were labeled.
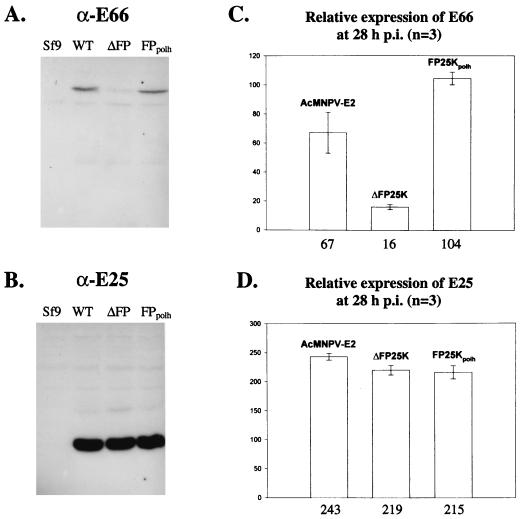
To generate a quantitative estimate of the differences in the accumulation of E66 and E25 synthesized in wt-, ΔFP25K-, and FP25Kpolh-infected Sf9 cells, three independent immunoblotting experiments were performed using total cell extracts from equal numbers of infected cells obtained in three independent infections collected at 28 hpi, treated with NaOH, and solubilized in 4× sample buffer. The relative amounts of E66 and E25 were determined using α-E66 or α-E25, 125I-labeled anti-rabbit IgG, and phosphodensitometry. Large differences were observed in the total accumulation of E66 at 28 hpi among the different viruses. Compared to that in wt-infected cells, E66 accumulation decreased 4.2-fold in ΔFP25K-infected cells and was slightly increased in FP25Kpolh-infected cells (Fig. 3A and C). In contrast, the accumulation of E25 was not significantly altered in cells infected with the different viruses (Fig. 3B and D).
FIG. 3.
Relative amounts of E66 and E25 produced in wt-, ΔFP25K-, and FP25Kpolh-infected cells as determined by quantitative immunoblotting. Equal numbers of infected cells were collected at 28 hpi and processed as indicated in Materials and Methods. The samples were resolved on SDS–12.5% PAGE gels, transferred onto Immobilon membranes, and immunoblotted using α-E66 or α-E25 and 125I-labeled goat anti-rabbit IgG. (A and B) Representative immunoblots performed using α-E66 and α-E25, respectively. Sf9, uninfected Sf9 cells; wt, ΔFP25K, and FP25Kpolh, cells infected with the respective virus. (C and D) Relative amount in arbitrary units of E66 and E25, respectively. Rectangles represent the average values obtained from three independent experiments performed using samples from independent infections. Vertical bars represent the standard error obtained. Numbers under the bars indicate the average value obtained for each set of samples.
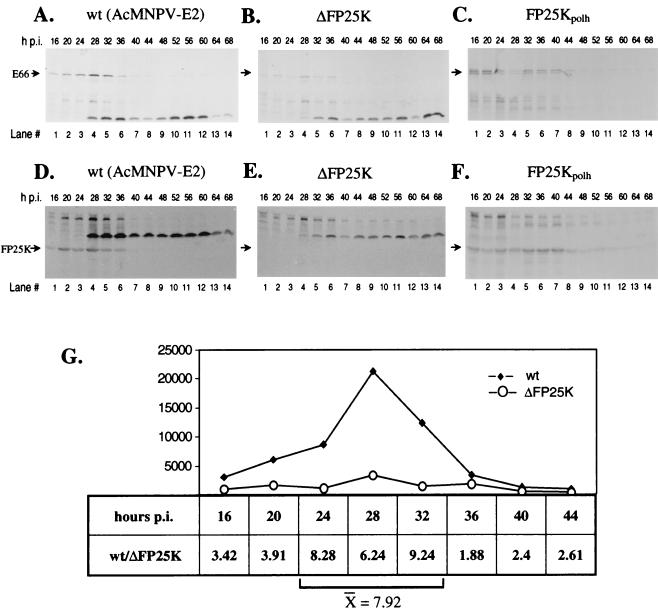
To determine if the decreased accumulation of E66 observed in ΔFP25K-infected cells was due to a delay in the synthesis of E66, the time course of E66 and FP25K synthesis in cells infected with the different viruses was determined by using immunoprecipitation analyses and the same set of labeled total cell extracts used in the first set of immunoblot analyses described above. To make these immunoprecipitations quantitative, control experiments were performed which titrated the amount of antibody required to assure saturation. These experiments showed that as the amount of antibody and Protein A were increased to achieve saturating conditions, the background increased consistently (data not shown); this background is seen throughout Fig. 4. In wt-infected cells, E66 synthesis was detected as early as 16 hpi (Fig. 4A, lane 1), reached maximum levels between 28 and 32 hpi (Fig. 4A, lane 4), and decreased thereafter (Fig. 4A, lanes 5 to 14). The time course of E66 synthesis observed in ΔFP25K-infected cells was identical to that observed in wt-infected cells, with the exception that the amount of E66 detected at each time point was significantly lower (Fig. 4B). To quantify the relative differences in synthesis of E66 between wt- and ΔFP25K-infected cells, the band intensities of the immunoprecipitated E66 were determined by phosphodensitometry. The largest differences in synthesis of E66 between wt- and ΔFP25K-infected cells occurred between 24 and 36 hpi, the interval during which E66 synthesis is maximum. During this period, there was 7.92-fold less E66 produced in ΔFP25K-infected than in wt-infected cells (Fig. 4G). In FP25Kpolh-infected cells, E66 synthesis remained relatively constant between 16 and 44 hpi (Fig. 4C, lanes 1 to 7) and decreased thereafter (Fig. 4C, lanes 8 to 14). The pattern of FP25K synthesis observed in wt- and FP25Kpolh-infected cells was similar to that of E66: in wt-infected cells, FP25K synthesis was detectable at 16 hpi (Fig. 4D, lane 1), reached maximum levels between 28 and 32 hpi (Fig. 4D, lane 4), and decreased thereafter (Fig. 4D, lanes 5 to 14). In FP25Kpolh-infected cells, FP25K synthesis remained relatively constant between 16 and 44 hpi (Fig. 4F, lanes 1 to 7) and decreased thereafter (Fig. 4F, lanes 8 to 14). Synthesis of FP25K was not detected in the ΔFP25K mutant at any time (Fig. 4E).
FIG. 4.
Time course of synthesis of E66 and FP25K in cells infected with wt (AcMNPV-E2), ΔFP25K, or FP25Kpolh. Cell extracts prepared from cells metabolically labeled for the 4-h interval before collection were immunoprecipitated using α-FP25K or α-E66. The position of the respective immunoprecipitated proteins is indicated (arrows). A high background of nonrelated precipitating proteins was obtained due to the large excess of antibodies and Protein A-Sepharose used to make the assay quantitative. (A through C) Immunoprecipitations performed with α-E66. (D through F) Immunoprecipitations performed with α-FP25K. (G) Relative amounts of E66 immunoprecipitated at different times postinfection. The intensity of the E66 signals obtained in the membranes shown in panels A and B were quantified by phosphodensitometry. The values obtained in arbitrary units are represented in the figure, and the wt/ΔFP25K ratios are shown.
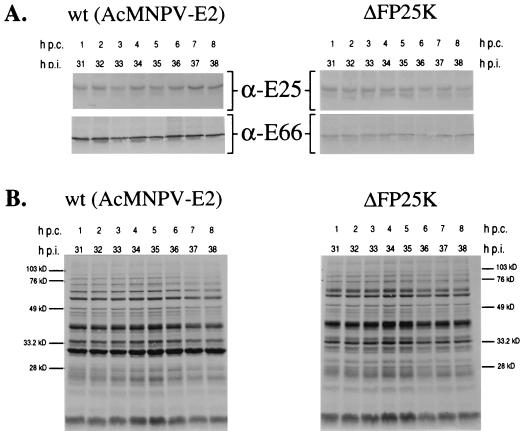
E66 and E25 exhibit minimal turnover in wt- and ΔFP25K-infected cells.
To determine if the decreased levels of E66 in ΔFP25K-infected cells were due to an increased turnover rate of E66, immunoprecipitation analyses were performed using pulse-labeled wt- and ΔFP25K-infected cell extracts collected at different times postchase. In both wt- and ΔFP25K-infected cells no significant differences were observed in the amount of E66 and E25 precipitated throughout the 8-h period analyzed (Fig. 5A). Aliquots from the samples used for immunoprecipitation were also run on SDS-PAGE gels to determine the overall profile of protein synthesis and degradation. Indeed, the overall protein profile of the pulse-labeled cell extracts indicates very little turnover for those proteins synthesized between 28 and 30 hpi in both wt- and ΔFP25K-infected cells during the 8-h period analyzed (Fig. 5B). The relative amounts of E66 and E25 precipitated from each virus and the overall protein profiles obtained resembled those observed when the samples were collected immediately after the labeling period, suggesting that minimal protein turnover occurred between 0 and 1 h postchase.
FIG. 5.
Analysis of E25 and E66 turnover in cells infected with wt (AcMNPV-E2) or ΔFP25K. ΔFP25K- and wt-infected cells were pulse-labeled between 28 and 30 hpi, chased with cold medium, and collected at 1-h intervals from 1 to 8 h postchase. Total cell extracts were prepared and immunoprecipitated with α-E66 or α-E25. (A) Immunoprecipitations performed using α-E25 and α-E66. (B) Total cell extracts of samples used for immunoprecipitations. The positions of the molecular size protein markers are indicated. h p.c., hours postchase.
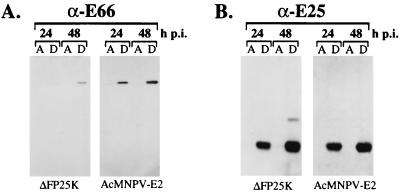
E66 and E25 expressed in ΔFP25K-infected cells are integral membrane proteins.
E66 and E25 are predicted to be integral membrane proteins of the ODV envelope, and in vitro translation assays performed in the presence of microsomal membranes support such a prediction for E66 (17). To evaluate if the absence of FP25K affected the association of E66 and E25 with membranes, wt- and ΔFP25K-infected cells collected at 24 and 48 hpi were fractionated into aqueous and detergent fractions by using Triton X-114 and were examined by immunoblot analysis. In both wt- and ΔFP25K-infected cells E66 and E25 partitioned with the detergent phase (Fig. 6A and B). In comparison, p39 (capsid), a major structural protein of the virus capsid that lacks transmembrane domains, partitioned exclusively with the aqueous phase in wt- and ΔFP25K-infected cells (data not shown).
FIG. 6.
Triton X-114 partitioning of E66 and E25 in cells infected with wt (AcMNPV-E2) or ΔFP25K. Infected cells collected at 24 and 48 hpi were partitioned with Triton X-114, and the resulting phases were resolved by SDS-PAGE and probed by immunoblotting with α-E66 (A) or α-E25 (B). A, aqueous phase; D, detergent phase.
Steady-state levels of E66 and E25 transcripts are not significantly altered by deleting or overexpressing FP25K.
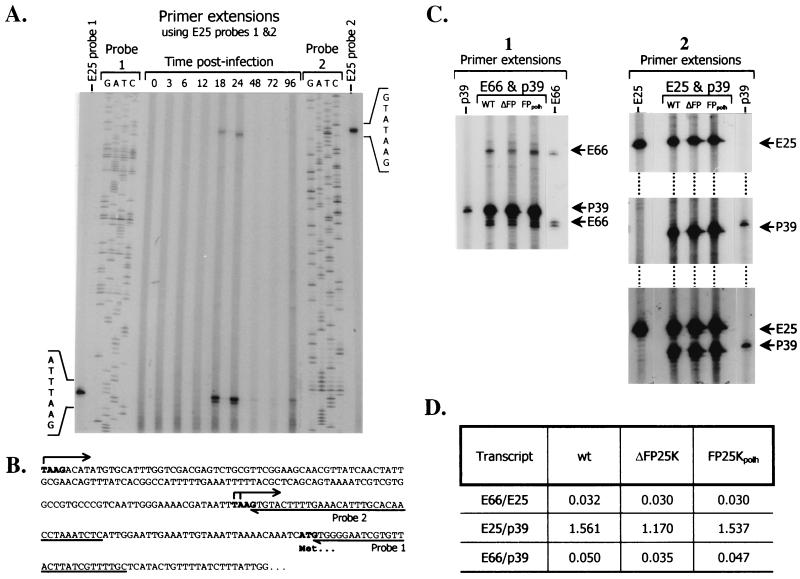
To determine if deletion of FP25K alters the steady-state levels of E66 and/or E25 transcripts, primer extension analyses were performed. Since no data were available on the transcription start sites used for E25, a temporal series of primer extension analyses of E25 were performed using two different probes and mRNA from wt-infected cells collected at different times postinfection. These analyses found that E25 transcripts initiated at two TAAG motifs at positions −70 and −219 and were made throughout the late and very late phases of infection, being first detected at 18 hpi (Fig. 7A and B). Two additional transcription initiation sites located at least 350 bp further upstream from the −219 start site were detected (data not shown). Transcripts initiated at those positions clearly exhibited the same temporal pattern but could not be resolved on the gels and therefore were not used in the quantitative analyses. For quantitative comparisons of steady-state levels of E66 and E25 transcripts, mRNA was isolated from wt-, ΔFP25K-, and FP25Kpolh-infected cells collected at 28 hpi, and primer extension analyses were performed using equal amounts of purified mRNA. p39 primer extensions were performed as internal controls. The relative intensity of the signals produced by the different E66, E25, and p39 transcripts were determined by phosphodensitometry, and the values obtained were averaged for each gene. No significant differences were observed in the usage of specific transcriptional start sites for E66, E25, or p39 among the different viruses (Fig. 7C). Two ratios were calculated and used to measure differences in E66 steady-state transcript levels, E66/E25 and E66/p39. E25 was chosen because its protein levels did not exhibit significant variations among the different viral mutants (Fig. 3B and D), while p39 was chosen because it has been used as a control for other similar studies (4). The E66/E25 transcript ratios obtained indicated no differences in relative E66 transcript abundance among wt-, ΔFP25K-, and FP25Kpolh-infected cells (Fig. 7D). Similarly, the E66/p39 ratios indicated only minor differences in the steady-state levels of E66 transcripts among the different viruses. A direct comparison of the values obtained for the E66 and E25 primer extension products in wt-, ΔFP25K-, and FP25Kpolh-infected cells also revealed very small differences in the steady state of such transcripts among the different virus mutants.
FIG. 7.
Determination of steady-state levels of E66, E25, and p39 transcripts in wt-, ΔFP25K-, and FP25Kpolh-infected cells. GATC indicate sequencing ladders used to determine the location of the corresponding transcripts. Control primer extension reactions performed using wt mRNA and primers corresponding exclusively to either E66, E25, or p39 are labeled in vertical orientation. The probes used in each experiment are indicated above the lanes. WT, ΔFP, and FPpolh indicate the source of the mRNA used. (A) Primer extension analysis of E25. (B) E25 upstream genomic region showing sites of transcription initiation for E25 as determined by primer extension analysis. (C) Primer extension analyses of E66 (panel 1), E25 (panel 2), and p39, showing the transcripts used for quantitative analysis. Only relevant regions of the gels are shown. (D) Transcript ratios calculated for the different viruses.
Intranuclear transport of E66 in ΔFP25K-infected cells is blocked at the level of the nuclear envelope.
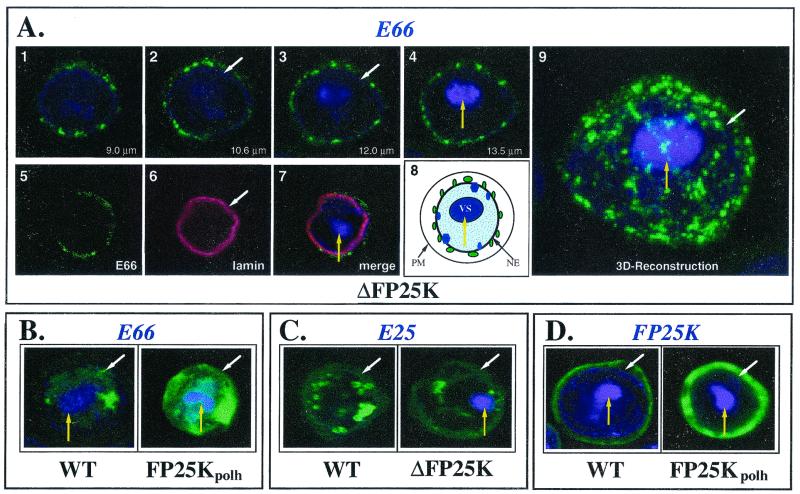
Mutations affecting FP25K block the transport of E66 to intranuclear vesicles but only delay the transport of E25 to such structures (4). To assess the effects of deleting and overexpressing FP25K on the intracellular transport of E66 and E25, immunofluorescence confocal microscopy analyses of wt-, ΔFP25K-, and FP25Kpolh-infected cells collected at 36 and 48 hpi were performed. In these experiments hundreds of cells were viewed, multiple confocal images were obtained, and representative Z-section images obtained with each viral mutant are shown. As expected, less E66 was observed in ΔFP25K- than in wt-infected cells (compare Fig. 8A, images 1 to 4, with Fig. 8B, wt). However, the most noticeable effect of deleting FP25K was that E66 was detected only at punctate regions around the nucleus, thus indicating that E66 was not transported to virus-induced intranuclear membranes (Fig. 8A, numbers 1 to 8). This perinuclear punctate pattern of E66 was very evident in three-dimensional reconstructions (Fig. 8A, image 9). In FP25Kpolh-infected cells E66 was efficiently transported to the nucleus and seemed somewhat more abundant and diffuse than in wt- and ΔFP25K-infected cells (Fig. 8B, FP25Kpolh). The transport of E25 was not affected by the deletion or the overexpression of FP25K. In ΔFP25K-infected (Fig. 8C, ΔFP25K) and FP25Kpolh-infected (data not shown) cells, E25 trafficked to intranuclear vesicles as efficiently as in wt-infected cells (Fig. 8C, wt). FP25K was detected predominantly in the cytoplasm in both wt- and FP25Kpolh-infected cells, even though a significant signal increase was observed in FP25Kpolh-infected cells (Fig. 8D).
FIG. 8.
Representative Z-section images showing the intracellular distribution of E66, E25, and FP25K in wt-, ΔFP25K-, and FP25Kpolh-infected cells. The different panels exemplify the intracellular distribution of E66, E25, and FP25K in the different mutant viruses, as determined by immunofluorescence confocal microscopy. The nucleus was visualized by using DAPI; however, under the exposure conditions used for capturing the confocal images presented, the DAPI signal appears concentrated at the virogenic stroma. Thus, most of the nucleoplasm appears as a void area around the virogenic stroma. (A, images 1 to 4) Z-section series showing the distribution of E66 in a representative ΔFP25K-infected cell 48 hpi. (A, images 5 to 7) Single Z-section showing the distribution of E66 in relation to that of lamin in a ΔFP25K-infected cell 48 hpi. Images 1 to 4 and 5 to 7 of panel A were obtained in separate experiments. (A, image 8) Schematic showing the approximate location of the plasma membrane (PM), nuclear envelope (NE), and virogenic stroma (VS) in the cell presented in the Z-section series (A, images 1 to 4). (A, image 9) Three-dimensional reconstruction of the cell presented in the Z-section series (images 1 to 4). (B) Representative cells showing the location of E66 in wt- and FP25Kpolh-infected cells at 48 hpi. (C) Representative cells showing the location of E25 in wt- and ΔFP25K-infected cells at 48 hpi. (D) Representative cells showing the location of FP25K in wt- and FP25Kpolh-infected cells at 36 hpi. The white arrows indicate the nuclear envelope. Yellow arrows indicate the virogenic stroma. Notice that the E66 signal shown in panel A precisely delineates the nuclear envelope. Also notice that the distribution of FP25K shown in panel D provides a good indication of the distribution of the cytoplasm in infected cells.
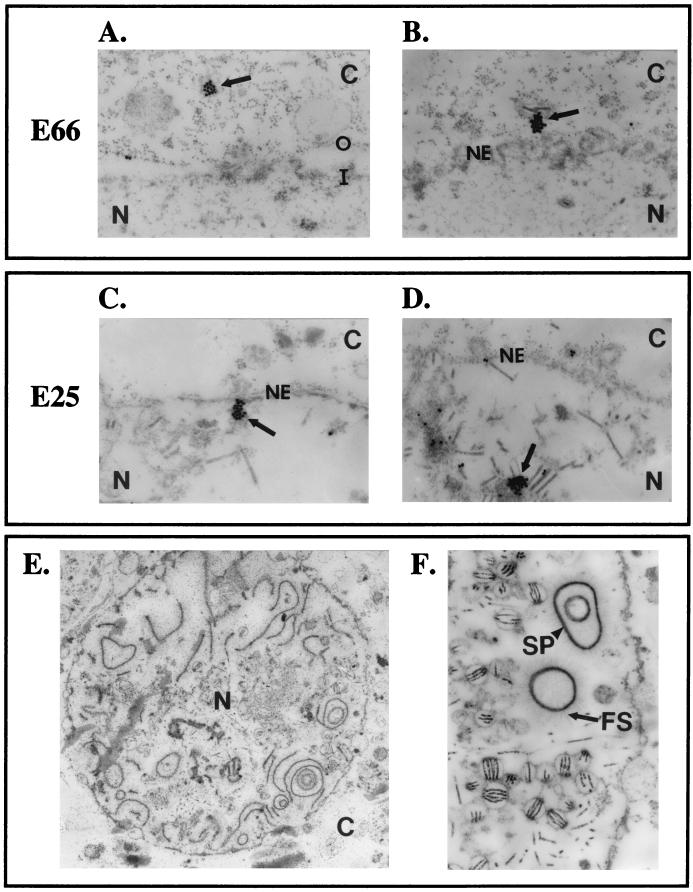
To more precisely determine the localization of E66 and E25 in the ΔFP25K mutant, immunoelectron microscopy (IEM) studies were performed using wt-infected (data not shown) and ΔFP25K-infected cells collected 48 hpi (Fig. 9). Consistent with previous observations of other FP25K virus mutants, the membranes of the nuclear envelope are more irregular and vesiculated in ΔFP25K- than in wt-infected cells. The irregularity of the membranes of the nuclear envelope and the decreased amount of E66 in ΔFP25K-infected cells makes conclusive results difficult. However, considerable attention was given to the region adjacent to the nuclear envelope during IEM analysis, and a clear trend was observed: in ΔFP25K-infected cells, E66 was prominently detected in association with cytoplasmic membranes located in close proximity to the nuclear envelope. Convincing E66 label was not seen inside the nucleus, either at the region of the nuclear envelope or in virus-induced intranuclear microvesicles (Fig. 9A and B). This pattern was not true for E25: in ΔFP25K-infected cells, E25 label was easily detected at both the cytoplasmic and the nuclear sides of the nuclear envelope and within virus-induced intranuclear microvesicles (Fig. 9C and D). Both E25 and E66 were visualized as clusters of label in ΔFP25K-infected cells (Fig. 9 A through D), a pattern that is not routinely observed in wt-infected cells.
FIG. 9.
Representative IEM images showing the distribution of E66 and E25 around the nuclear envelope in ΔFP25K-infected cells and the presence of abundant electron-dense and fibrous structures in the nucleus of FP25Kpolh-infected cells. (A to D) ΔFP25K-infected cells. (A and B) E66. (C and D) E25. N, nucleus; C, cytoplasm; O, outer nuclear membrane; I, inner nuclear membrane; NE, nuclear envelope. (E and F) FP25Kpolh-infected cells. FS, fibrous structures; SP, electron-dense spacers.
IEM studies performed using FP25Kpolh-infected cells revealed normal viral envelopment (Fig. 9F) in spite of the formation of large amounts of fibrillar and electron-dense structures within the nucleus (Fig. 9E and F). The electron-dense structures appeared associated to the fibrous structures and frequently formed large circles that sometimes contained smaller electron-dense circular structures inside. These electron-dense and fibrous structures closely resemble the electron-dense spacers (SP) and the fibrous structures rich in p10 described by van der Wilk et al. (29). Such structures are normally seen in wt- and polyhedrin deletion virus-infected cells, but their quantity is clearly enriched during FP25Kpolh infection.
Deletion of FP25K produces small changes in the profile of protein complexes containing E66.
To determine the size and diversity of protein complexes containing E66, total cell extracts from uninfected, wt-infected, and ΔFP25K-infected Sf9 cells collected at 30 hpi were fractionated by differential centrifugation (Fig. 10A) and BN-PAGE and were analyzed by immunoblotting using α-E66. In both wt- and ΔFP25K-infected cells, most of E66 was found in a protein complex of approximately 330 kDa, and lesser amounts were observed in complexes of approximately 380 and 430 kDa, all of which were found in the insoluble fraction (Fig. 10B). While the complexes referred to above appeared equally or slightly more abundant in ΔFP25K- than in wt-infected cells, a minor complex of approximately 200 kDa was visually more abundant in wt- than in ΔFP25K-infected cells (Fig. 10B, arrowheads). When the proteins separated on a BN-PAGE gel were further resolved on a second-dimension SDS-PAGE gel and analyzed by immunoblotting, E66 was detected spanning the approximate molecular sizes referred to above for the first dimension and was more abundant in wt- than in ΔFP25K-infected cells (Fig. 10C, arrows).
DISCUSSION
Previous studies of the AcMNPV 480-1 and FP-βgal mutants show that partial deletions of FP25K have significant effects on the accumulation of several proteins during infection, decreasing the levels of polyhedrin (18) and E66 (4), and increasing the levels of gp67, BV/ODV-E26, and p39 (4). Those studies also demonstrated that partial deletions of FP25K result in altered transport of soluble and membrane-associated viral proteins into the nucleus. In 480-1-infected cells, the transport of polyhedrin and E25 to the nucleus is delayed (4, 18), while that of E66 is blocked (4); in FP-βgal-infected cells, the transport of E66 to intranuclear membranes is delayed, while that of E25 is not altered (4). The possible presence of other mutations in the 480-1 mutant (a natural virus isolate) and the presence of most of FP25K as a β-galactosidase fusion protein in the FP-βgal mutant hindered a more direct assessment of the role of FP25K relative to the effects described with these mutants. Thus, to further characterize the role of FP25K and its effects on the synthesis and transport of E66 and E25, we developed a viral mutant containing a precise deletion of FP25K. Additionally, since previous studies showed that mutants containing a second copy of the gene in question under the control of the polyhedrin promoter provide valuable insights on the traffic (17) and potential function (2) of the encoded protein, we also developed a viral mutant containing an additional FP25K gene under the control of the polyhedrin promoter.
Immunoblotting and immunoprecipitation analyses showed that deletion of FP25K decreased the synthesis and accumulation of E66 approximately fivefold but did not affect the synthesis and accumulation of E25 (Fig. 3 and 4). Overexpression of FP25K increased the synthesis and accumulation of E66 but had no effect on the synthesis and accumulation of E25 (Fig. 3 and 4). These results indicated that FP25K exerts gene-specific effects on virus protein expression. The effects of deleting FP25K on the synthesis and accumulation of E66 could be due to (i) delayed synthesis of E66; (ii) decreased E66 steady-state transcripts levels; (iii) increased rate of E66 protein turnover; (iv) decreased rate of E66 translation; or (v) combinatorial effects of any of the above. Immunoprecipitation analyses of metabolically labeled cells collected at different times postinfection showed that E66 synthesis is maximal at 28 to 32 hpi in both wt- and ΔFP25K-infected cells, thus the temporal synthesis of E66 is not affected in the absence of FP25K (Fig. 4). Primer extension analyses did not detect significant differences in E66 steady-state transcript levels during the time of maximal synthesis of E66 among wt and mutant FP25K viruses (Fig. 7). These results were similar to those reported with the FP-βgal mutant (4) and indicate that E66 transcription is not affected by FP25K. Immunoprecipitation analyses using cells pulse-labeled during the time of maximum synthesis of E66 showed no detectable turnover of E66 in ΔFP25K- and wt-infected cells (Fig. 5); thus, the turnover rate of E66 is also not altered in the absence of FP25K. Furthermore, Triton X-114 extraction analyses showed that E66 is associated with intracellular membranes in ΔFP25K- and wt-infected cells (Fig. 6). Hence, the lack of FP25K does not affect membrane insertion of E66. Altogether these data showed that deletion of FP25K decreases E66 protein synthesis without producing significant changes on the temporal pattern of synthesis, transcription, transcript stability, protein turnover, and membrane insertion of E66. Therefore, we postulate that FP25K probably affects E66 synthesis at the translational level.
Studies by Harrison et al. indicated that mutations affecting FP25K alter polyhedrin expression at the transcriptional level, specifically by decreasing its transcriptional rate (14). If FP25K does indeed affect translational rates of specific mRNAs within infected cells, as suggested for E66, FP25K would represent a protein with transcriptional and translational regulatory abilities. However, computer-assisted analyses of the primary structure of FP25K do not suggest the presence of RNA binding motifs or any significant homology between FP25K and known transcriptional or translational regulatory proteins. Thus, it is possible that FP25K achieves these functions by interacting with transcriptional and/or translational regulatory proteins, which in turn affect regulatory cascades at hierarchical points. The apparent ability of FP25K to regulate gene expression at both transcriptional and translational levels may be responsible for the multiple phenotypes observed in FP25K mutant viruses, as they may be related to changes in the synthesis, accumulation, and traffic of specific proteins during infection. For instance, the altered envelopment of nucleocapsids in the nucleus and consequent decreased production of ODV may be associated with decreased synthesis and transport of ODV-E66 and perhaps other ODV proteins; the decreased degradation of Bombyx mori larvae infected by subcutaneous injection of BmNPV FP25K mutants (19), a phenomenon that we also observed in Trichoplusia ni larvae infected by subcutaneous injection with the ΔFP25K mutant (data not shown), may be explained by a decrease in the expression of the chitinase and cysteine protease genes, as suggested by Katsuma et al. (19); and the substantial morphological alterations observed in cells infected with the FP25Kpolh virus, such as the increased production of electron-dense spacers and fibrous structures in the nucleus (Fig. 9E), may be related to altered expression of p10 and other virus genes. Remarkably, the expression of FP25K under the control of the polyhedrin promoter did not result in expression of FP25K at polyhedrin levels but in a two- to threefold increase in FP25K (Fig. 1); however, such an increase was enough to produce the morphological effects referred to above. While little is known about translational regulation during baculovirus infection, such mechanisms might help maintain appropriate levels of specific virus and/or host factors required for essential processes during infection.
Deletion of FP25K results in the accumulation of E66 in distinct, concentrated regions of the nuclear envelope (Fig. 8). In cells infected with the ΔFP25K mutant, IEM analyses detected E66 associated with cytoplasmic membranes in close proximity to the nuclear envelope but failed to detect E66 associated with membranes inside the nucleus (i.e., inner nuclear membrane, virus-induced microvesicles, and viral envelopes) (Fig. 9A and B). In contrast, trafficking of E25 in ΔFP25K-infected cells appeared normal, as E25 was detected in membranes associated with the nuclear envelope as well as in intranuclear microvesicles (Fig. 9C and D). Our previous study showed that E25 localization to intranuclear microvesicles was delayed in 480-1- but not in FP-βgal-infected cells. Since both mutant viruses contain significant portions of the FP25K gene yet only 480-1 infection resulted in a delay of E25 localization to intranuclear microvesicles, this result may be revealing functions that still exist within the truncated FP25K protein expressed during 480-1 infection. It is also possible that since 480-1 is a naturally occurring viral isolate, other spontaneous, as-yet uncharacterized mutations exist within the 480-1 genome. The use of the precise deletion of the FP25K gene shows that E66 localization is specifically affected, and this effect cannot be generalized to other ODV envelope proteins.
Due to the differences observed in E66 trafficking between ΔFP25K- and wt-infected cells, we considered it important to compare the size of E66 protein complexes in cells infected with these viruses. To this end we developed a fractionation procedure that removes nucleocapsids and virions and solubilizes membrane-associated protein complexes by using nonionic detergents, thus selecting for complexes representing those interactions involved in protein traffic prior to assembly (Fig. 10A). The solubilized complexes were analyzed by BN-PAGE and immunoblotting. Such analyses showed that in both wt- and ΔFP25K-infected cells E66 is present in at least four major protein complexes, ranging in size from 200 to 430 kDa. Except for the 200-kDa complex, all the E66 protein complexes observed seemed equally or slightly more abundant in ΔFP25K- than in wt-infected cells (Fig. 10B). Such a result was unexpected, as E66 is produced in lesser quantities in ΔFP25K- than in wt-infected cells (Fig. 3 and 4). When proteins separated by BN-PAGE were further resolved by an SDS-PAGE second dimension, significantly stronger E66 bands were detected in wt- than in ΔFP25K-infected cells for all the complexes observed. The similarity in the profiles obtained in the two-dimensional analyses confirmed that E66 forms protein complexes of equal size in wt- and ΔFP25K-infected cells (Fig. 10C). The differences observed between the one- and two-dimensional analyses may relate to changes in the predominant epitopes exposed by E66 under native and denatured conditions and the type of protein complexes formed by E66 in wt- and ΔFP25K-infected cells. The 200-kDa complex appears to be an E66-protein complex down-regulated in the absence of FP25K and therefore may constitute an important target for future studies.
Based on the above data, we considered two possible mechanisms of how FP25K may function to regulate the traffic of E66. First, it is possible that, in the absence of FP25K, decreased quantity or improper folding of E66 results in altered transport. It is possible that the decreased amount of E66 produced in cells infected with the ΔFP25K mutant might affect its trafficking. However, in our previous study (4) we showed that, during infection with the FP-βgal mutant, the quantity of E66 decreased to levels equivalent to those observed in ΔFP25K-infected cells, but still some E66 trafficked normally to intranuclear microvesicles. Thus, the block in the intranuclear trafficking of E66 should not be entirely due to the decreased amount of protein. Alternatively, in the absence of FP25K, E66 may not be folded properly, thus affecting its trafficking. Indeed, IEM analyses showed that, in the absence of FP25K, E66 is detected in clusters (Fig. 9A and B) seldom found in wt-infected cells, which might represent misfolded or aggregated E66. However, like E66, E25 was also detected in similar clusters, yet its traffic was unaffected (Fig. 9C and D). Additionally, E66 was found in protein complexes of similar size in ΔFP25K- and wt-infected cells (Fig. 10). Furthermore, while molecular size may limit the traffic of large proteins from the outer to the inner nuclear membrane (26), a large integral membrane fusion protein (125-β-galactosidase, molecular size of >110 kDa) traffics to the nuclear envelope and to virus-induced intranuclear microvesicles as efficiently as ODV envelope proteins during baculovirus infection (17). These observations suggest that although size limitations and folding requirements cannot be discounted, the differences seen in the traffic of E66 and E25 in ΔFP25K-infected cells do not represent an intrinsic limitation of transport due to molecular size. While protein misfolding is usually accompanied by an increased turnover, the turnover rate of E66 appeared to be identical in wt- and ΔFP25K-infected cells.
Second, the transport of E66 may be mediated by specific factors whose synthesis or activity is regulated by FP25K. It is possible that after E66 is inserted into membranes (a feature that is not disrupted in cells infected with the ΔFP25K mutant [Fig. 6]), a combination of factors may help it to progress toward the nuclear envelope. Transport models for resident proteins of the inner nuclear membrane suggest that these proteins diffuse laterally along the continuous membranes of the ER, outer nuclear membrane, and inner nuclear membrane, where they are retained via interactions with chromatin, lamins, or other resident proteins (8, 12, 24, 26). Although none of the ODV envelope proteins contain inner nuclear membrane retention signals, the continued movement of ODV envelope proteins to membranes inside the nucleus during infection may serve to remove proteins from the diffusional pool. However, altered retention does not explain why E66 would locate to discrete membranes at the exterior of the nucleus in the absence of FP25K. It is possible that a lipid gradient from the ER to the nuclear envelope and intranuclear vesicles could provide directionality to the lateral diffusion of proteins along the membrane. However, if such a gradient were the only factor regulating protein movement along the continuous membranes of the ER and nuclear envelope, then mutations affecting the trafficking of one envelope protein (E66) potentially could alter the trafficking of others (E25). The specific effects observed on the trafficking of E66 suggest that, in the absence of FP25K, proteins or protein activities that facilitate movement of E66 to intranuclear membranes are altered. This could include factors that generate a facilitated diffusion pathway or a more complex, active transport pathway.
Our data clearly argue against an unregulated lateral diffusion model for the transport of E66 to intranuclear microvesicles. It suggests that during AcMNPV infection the transport of E66 to intranuclear microvesicles is specifically regulated and that the factors controlling this trafficking include FP25K or a protein(s) regulated by FP25K. Further characterization of the components of complexes containing E66 may identify transport factors and help discern the mechanism involved in the trafficking of E66 from its site of insertion at the ER to the nuclear envelope and to virus-induced intranuclear microvesicles. Considering that viruses are very efficient manipulators of cellular pathways but rarely invent entirely new pathways not used by their host, we expect that studies on the integral membrane proteins of the baculovirus ODV envelope may reveal insights into the mechanism governing the transport of membrane proteins to the nuclear envelope in uninfected cells.
ACKNOWLEDGMENTS
We thank Paul A. Fisher (Department of Pharmacological Sciences, University of New York at Stony Brook, Stony Brook) for providing the monoclonal antibody ADL67 and George Rohrmann (Oregon State University, Corvallis) for providing the α-E25 rabbit serum. We thank Jared Burks for his expert assistance with confocal microscopy. Electron microscopy was performed using the facilities of the Electron Microscopy Center at Texas A&M University.
This work was supported in part by National Institutes of Health Grant 2RO1GM47552 (M.D.S., S.C.B.) and the Texas Agricultural Experimental Station Project TEXO8078 (M.D.S.).
REFERENCES
- 1.Beames B, Summers M D. Location and nucleotide sequence of the 25K protein missing from baculovirus few polyhedra (FP) mutants. Virology. 1989;168:344–353. doi: 10.1016/0042-6822(89)90275-4. [DOI] [PubMed] [Google Scholar]
- 2.Beniya H, Braunagel S C, Summers M D. Autographa californica nuclear polyhedrosis virus: subcellular localization and protein trafficking of BV/ODV-E26 to intranuclear membranes and viral envelopes. Virology. 1998;240:64–75. doi: 10.1006/viro.1997.8903. [DOI] [PubMed] [Google Scholar]
- 3.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 4.Braunagel S C, Burks J K, Rosas-Acosta G, Harrison R L, Ma H, Summers M D. Mutations within the Autographa californica nucleopolyhedrovirus FP25K gene decrease the accumulation of ODV-E66 and alter its intranuclear transport. J Virol. 1999;73:8559–8570. doi: 10.1128/jvi.73.10.8559-8570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunagel S C, Parr R, Belyavskyi M, Summers M D. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology. 1998;244:195–211. doi: 10.1006/viro.1998.9097. [DOI] [PubMed] [Google Scholar]
- 6.Braunagel S C, Summers M D. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology. 1994;202:315–328. doi: 10.1006/viro.1994.1348. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Ijkel W F, Tarchini R, Sun X, Sandbrink H, Wang H, Peters S, Zuidema D, Lankhorst R K, Vlak J M, Hu Z. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J Gen Virol. 2001;82:241–257. doi: 10.1099/0022-1317-82-1-241. [DOI] [PubMed] [Google Scholar]
- 8.Ellenberg J, Siggia E D, Moreira J E, Smith C L, Presley J F, Worman H J, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser M J. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J Ultrastruct Mol Struct Res. 1986;95:189–195. [Google Scholar]
- 10.Fraser M J, Hink W F. Comparative sensitivity of several plaque assay techniques employing TN-368 and IPLB-SF 21AE insect cell lines for plaque variants of Galleria mellonella nuclear polyhedrosis virus. J Invertebr Pathol. 1982;40:89–97. [Google Scholar]
- 11.Fraser M J, Smith G E, Summers M D. Acquisition of host cell DNA sequences by baculoviruses: relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa K, Fritze C E, Gerace L. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J Biol Chem. 1998;273:4213–4219. doi: 10.1074/jbc.273.7.4213. [DOI] [PubMed] [Google Scholar]
- 13.Guarino L A, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol. 1998;72:7985–7991. doi: 10.1128/jvi.72.10.7985-7991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison R L, Jarvis D L, Summers M D. The role of the AcMNPV 25K gene, “FP25” in baculovirus polh and p10 expression. Virology. 1996;226:34–46. doi: 10.1006/viro.1996.0625. [DOI] [PubMed] [Google Scholar]
- 15.Harrison R L, Summers M D. Mutations in the Autographa californica multinucleocapsid nuclear polyhedrosis virus 25 kDa protein gene result in reduced virion occlusion, altered intranuclear envelopment and enhanced virus production. J Gen Virol. 1995;76:1451–1459. doi: 10.1099/0022-1317-76-6-1451. [DOI] [PubMed] [Google Scholar]
- 16.Hong T, Braunagel S C, Summers M D. Transcription, translation, and cellular localization of PDV-E66: a structural protein of the PDV envelope of Autographa californica nuclear polyhedrosis virus. Virology. 1994;204:210–222. doi: 10.1006/viro.1994.1525. [DOI] [PubMed] [Google Scholar]
- 17.Hong T, Summers M D, Braunagel S C. N-terminal sequences from Autographa californica nuclear polyhedrosis virus envelope proteins ODV-E66 and ODV-E25 are sufficient to direct reporter proteins to the nuclear envelope, intranuclear microvesicles and the envelope of occlusion derived virus. Proc Natl Acad Sci USA. 1997;94:4050–4055. doi: 10.1073/pnas.94.8.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis D L, Bohlmeyer D A, Garcia A., Jr Enhancement of polyhedrin nuclear localization during baculovirus infection. J Virol. 1992;66:6903–6911. doi: 10.1128/jvi.66.12.6903-6911.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsuma S, Noguchi Y, Zhou C L, Kobayashi M, Maeda S. Characterization of the 25K FP gene of the baculovirus Bombyx mori nucleopolyhedrovirus: implications for postmortem host degradation. J Gen Virol. 1999;80:783–791. doi: 10.1099/0022-1317-80-3-783. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.MacKinnon E A, Henderson J F, Stoltz D B, Faulkner P. Morphogenesis of nuclear polyhedrosis virus under conditions of prolonged passage in vitro. J Ultrastruct Res. 1974;49:419–435. doi: 10.1016/s0022-5320(74)90055-0. [DOI] [PubMed] [Google Scholar]
- 22.Potter J N, Faulkner P, MacKinnon E A. Strain selection during serial passage of Trichoplusia ni nuclear polyhedrosis virus. J Virol. 1976;18:1040–1050. doi: 10.1128/jvi.18.3.1040-1050.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potter K N, Jaques R P, Faulkner P. Modification of Trichoplusia ni nuclear polyhedrosis virus passaged in vivo. Intervirology. 1978;9:76–85. doi: 10.1159/000148925. [DOI] [PubMed] [Google Scholar]
- 24.Rolls M M, Stein P A, Taylor S S, Ha E, McKeon F, Rapoport T A. A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J Cell Biol. 1999;146:29–44. doi: 10.1083/jcb.146.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 26.Soullam B, Worman H J. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuurman N, Maus N, Fisher P A. Interphase phosphorylation of the Drosophila nuclear lamin: site-mapping using a monoclonal antibody. J Cell Sci. 1995;108:3137–3144. doi: 10.1242/jcs.108.9.3137. [DOI] [PubMed] [Google Scholar]
- 28.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agric Exp Station Bull. 1987;1555:10–48. [Google Scholar]
- 29.van F. der Wilk J W, van Lent M, Vlak J M. Immunogold detection of polyhedrin, p10, and virion antigens in Autographa californica nuclear polyhedrosis virus-infected Spodoptera frugiperda cells. J Gen Virol. 1987;68:2615–2623. [Google Scholar]
- 30.Volkman L E, Summers M D. Autographa californica nuclear polyhedrosis virus: comparative infectivity of the occluded, alkali-liberated, and nonoccluded forms. J Invertebr Pathol. 1977;30:102–103. doi: 10.1016/0022-2011(77)90045-3. [DOI] [PubMed] [Google Scholar]