Abstract
The utilization of X-chromosome short tandem repeats (X-STRs) for human identification particularly in resolving complex kinship cases has been advocated. Since, forensic statistical parameters vary among different populations, and because the X-STRs population data representing the diverse population of Peninsular Malaysia remain unavailable, the specific attempt reported here for the Malays (n = 224) and Chinese (n = 201) populations appears forensically relevant to support the evidential value of the 12 X-STRs markers for human identification in Malaysia. Results derived from the Qiagen Investigator® Argus X-12 kit revealed that DXS10135 as the most polymorphic locus with high genetic diversity, polymorphic information content, heterozygosity as well as power of exclusion. Based on allele frequencies, the combined power of discrimination as well as the mean exclusion chance (MECKrüger, MECKishida, MECDesmarais and MECDesmaraisDuo) values for the Malays and Chinese were individually ≥0.999995532964908. As for the combined power of discrimination as well as the mean exclusion chance (MECKrüger, MECKishida, MECDesmarais and MECDesmaraisDuo) calculated based on haplotype frequencies, the values were ≥0.9999986410567 for the Malays and Chinese populations. In addition, results from the genetic distance, neighbor-joining phylogenetic tree and principal component analysis revealed close biogeographical distributions of the studied populations with other South East Asian populations. Hence, the utilization of the X-STRs data for identifying individuals among the Malays and Chinese populations in Peninsular Malaysia for forensic application appears as highly supported.
Keywords: X-STRs, Forensic statistical parameters, Malay, Chinese, Malaysia
Highlights
-
•
The first population data for X-STRs in Malaysian Chinese.
-
•
12 X-STRs typing using Qiagen Investigator Argus X-12 kit.
-
•
Forensic statistical support for complex kinship investigation.
-
•
High allele and haplotype diversities.
1. Introduction
Malaysia is located in the South East Asia equatorial zone (Latitude: 1° and 7° North; Longitudes: 100° and 119° East) of which it covers an area of 330,534 square kilometres. The federation of Malaysia comprises of Peninsular Malaysia (11 states and 2 Federal Territories) as well as the East Malaysia (2 states and 1 Federal territory), separated by the South China Sea [1]. Being a multi-racial country, the Malaysian population has been estimated at 33.5 million, consisting of Malays and other Bumiputera groups (70.1 %), Chinese (22.6 %), Indians (6.6 %) and others (0.7 %). Out of 70.1 % of Bumiputera, the majority being the Malays (58.0 %) followed by other groups of Bumiputera (12.1 %), residing in the Peninsular and East Malaysia [2]. Previous researchers have described the Malays and Chinese in Peninsular Malaysia as the following. Malay is specifically defined in the Constitution of Malaysia (Article 160(2)) as “a person who professes the religion of Islam, habitually speaks the Malay language, conforms to Malay custom and - (a) was born before Merdeka Day in the Federation or in Singapore or born of parents one of whom was born in the Federation or in Singapore, or is on that day domiciled in the Federation or in Singapore; or (b) is the issue of such a person” [3]. The majority of the Malays and indigenous populations (Negrito, Senoi and Proto-Malay) reside in Peninsular Malaysia and they comprise of various sub-ethnicities namely Banjar, Java, Minangkabau, Bugis and other groups from Austronesian [4], emerged prominently during the Malacca Sultanate (15th century) through the trading activities and seafaring life [5].
Malaysian Chinese on the other hand make up the second-largest overseas Chinese population (after Thailand) and the second-largest ethnic group in Malaysia [6]. Chinese people first came to the Malacca Empire in the early 15th century, when the Sultan Mansur Syah married a Chinese princess named Hang Li Po [6]. This was the beginning of the first wave of migration, when Chinese merchants and young aristocrats from an imperial company started to settle in and around Melaka, assimilated with the local culture and custom through business and marriage [7]. Described as the second wave of migration, the Chinese in PeninsularMalaysia originating from the southern part of China arrived hugely in the 18th to the early 20th centuries due to open migration policy by the British to surplus the labour support for their colonial economic activities [8]. The majority of them are from the southeast coast of Mainland China's Fujian and Guangdong regions [7] and worked in tin mining which later developed into the Chinese civilization in Peninsular Malaysia [9].
The application of X-chromosome short tandem repeats (X-STRs) can be appealing for resolving kinship and deficiency paternity cases, and may prove useful in investigations involving missing persons and mass disasters, in particular when comparison profiles are not available [10]. However, the available of X-STRs population data for Malaysia are limited to Malays (in Kuala Lumpur alone) [11] as well as the minority sub-ethnics [12] with inadequate number of sample size and Kedayan (an ethnicity in the East Malaysia region) [13]. As such the available data may not represent the diverse population of Peninsular Malaysia, limiting their evidential values as evidence in court [10]. Not only that, the complete X-STRs population data for representing the admixture populations (e.g. Malay-Chinese) in Malaysia are also lacking. Considering that the forensic statistical parameters may differ among the different populations [14,15], the statistical support for the use of DNA profiles for one population may not be accurately representing other populations, especially among high diversity ethnic populations and genetic admixture [16,17]. In this context, genetic properties are evaluated based on the relevant populations to determine the rarity of the obtained profile [18]. Therefore, a thorough sampling of populations of interest is paramount for validating and documenting the forensic statistical support for the use of X-STRs analysis for complex kinship investigation as well as proving the evidence as ‘beyond any reasonable doubt’ in the court of law.
Hence, the aim of this research was to provide the empirically justified statistical support for the use of 12 X-STRs population data for the Malays and Chinese in Peninsular Malaysia for human identification in Malaysia, particularly for complex kinship investigation. Importantly, this would be the first attempt to provide such data for the Chinese population in Malaysia.
2. Materials and method
2.1. Ethical requirements and sample collection
The procedures and protocols used in this present research was approved by the Human Research Ethics Committee of Universiti Sains Islam Malaysia (USIM/JKEP/2022-225), dated July 21, 2022, in compliance with the ethical guidelines and requirements for the publication of genetic and genomic data. In this context, it is important to iterate that “Urban and cosmopolitan population studies should involve at least 200 individuals, whereas, regional datasets should contain a minimum of 100 samples. Exceptions are valid for extremely small population sizes.” [19]. The fact that this present research utilized 224 Malay and 201 Chinese participants (amounting to a total of 425 participants), it can be construed that the number of participants examined here is more than sufficient to represent the populations of Malay and Chinese in Peninsular Malaysia. The written informed consents were attained before the finger-pricked blood samples on Flinder's Technology Associates (FTA) cards were collected from the unrelated healthy individuals consisting of (a) 224 Malays (113 females and 111 males) and (b) 201 Chinese (100 females and 101 males) residing in Peninsular Malaysia. For ensuring the representativeness, the stratified purposeful sampling method was used, stratifying the participants according to the different states and territory within the Peninsular Malaysia.
2.2. X-STRs typing
To facilitate international population data exchange, utilization of the X-chromosomal STR kit that has acquired popularity for forensic application must be chosen. The fact that Qiagen Investigator® Argus X-12 QS kit fulfils such a requirement, its utilization to generate suitable population data for the Malays and Chinese in the diverse Malaysian population proves relevant, although there are other kits (e.g. Microreader™ 19X) for X-chromosomal STR that offer more loci for analysis. Since the utilization of Microreader™ 19X Kit appears limited to specific populations and/or geographical location (e.g. Han and Sierra Leone), the ability to enable the international population data exchange may be limited. Furthermore, the lack of population and mutation data prevailed for the other X-STRs kits may impede forensic evaluation on their practical values.
The blood samples on FTA cards were punched (∼1.2 mm each) and subsequently purified. The purified blood samples on FTA cards were then directly amplified using the reduced polymerase chain reaction (PCR) reaction volume of the Qiagen Investigator® Argus X-12 kit (Germany) as described by Alwi et al. [20], co-amplifying the 12 X-STRs loci namely DXS10103, DXS8378, DXS10101, DXS10134, DXS10074, DXS7132, DXS10135, DXS7423, DXS10146, DXS10079, HPRTB and DXS10148 [21]. These markers are grouped into four linkage groups: (LG1:DXS10148, DXS10135, DXS8378; LG2: DXS7132, DXS10079, DXS10074; LG3: DXS10103, HPRTB, DXS10101; LG4: DXS10146, DXS10134, DXS7423).
The PCR amplification was performed on the Applied Biosystems™ ProFlex™ PCR System (Thermo Fisher Scientific). Subsequently, the PCR products were separated via capillary electrophoresis on the Applied Biosystems 3500xL Genetic Analyzer (Thermo Fisher Scientific), and data were analyzed using the GeneMapper™ ID-X v1.5 software (Thermo Fisher Scientific). For confirming the presence of off-ladder alleles (as determined by size), re-injection of the samples into the capillary electrophoresis was performed.
2.3. Quality control
The research was performed in an ISO/IEC 17025:2017 accredited forensic DNA laboratory (Department of Chemistry Malaysia Johor State Laboratory; FT-0084) by the American National Standards Institute (ANSI) National Accreditation Board (ANAB) following the laboratory internal standards and kit controls. The female control DNA 9947A (Thermo Fisher Scientific, USA) was used as the positive control, while deionized water served as the negative ones.
2.4. Forensic statistical analysis
Allele frequencies of male and female samples as well as the haplotype frequencies for each linkage group (LG) of male samples were estimated by simple counting method using Statistics for X-STR (StatsX) v2.0 [22]. The exact test of Hardy-Weinberg equilibrium (HWE) per locus in female samples and the linkage disequilibrium (LD) testing between all pairs of the 12 loci of male and female samples were computed using Arlequin v3.5.2.2 software [23]. The forensic statistical parameters that include the gene diversity (GD), haplotype diversity (HD), polymorphic information content (PIC), power of discrimination in males (PDM) and in females (PDF), mean exclusion chance in (a) deficiency cases (MECKrüger) [24], (b) standard trios involving daughters (MECKishida) [25], (c) standard trios involving daughters (MECDesmarais) and (d) father/daughter or mother/son duos (MECDesmaraisDuo) [26] as well as the combined PDm, PDf, MECKruger, MECKishida, MECDesmarais and MECDesmarais Duo were calculated using StatsX v2.0 [22]. The heterozygosity (Het) and power of exclusion (PE) were calculated using the formula described by the previous researcher [27].
2.5. Population comparisons
The Nei's DA genetic distances [28] among the studied populations (Malays and Chinese) with those of 21 other populations were measured and represented in the form of Neighbor-joining (NJ) tree, constructed using POPTREE2 software [29] with 1000 bootstrap replications. Visualization and interpretation of phylogenetic tree were attempted using Molecular Evolutionary Genetics Analysis version 11 [30]. Subsequently, the principal components analysis (PCA) was performed using the MultiVariate Statistical Package [31].
3. Results and discussion
3.1. Forensic statistical analysis
The allele and haplotype frequencies as well as the forensic parameters were calculated from the Malays and Chinese population in Peninsular Malaysia dataset (Supplementary Tables S1–S4) generated from this present research. Results from the independent sample t-test comparing the allele frequencies between male and female samples indicated no significant differences. However, to provide a clearer understanding of the genetic diversity and structure within and between the populations studied, the allele frequencies and forensic parameters for male and female subjects are separately represented prior to the integration (pooled) (Supplementary Tables S5–S6). A total of 158 and 143 alleles were observed for the Malays and Chinese populations respectively. A total of 12 distinct off-ladder alleles were found at 6 loci in the Malays and Chinese populations. While the majority of the off-ladder alleles were observed at DXS10101 (4 distinct alleles), one allele each at loci DXS10103, DXS10134, DXS10135 and DXS10079 were found in the Malay population. As for the Chinese population, 5 distinct alleles at 4 loci (DXS10101, DXS10134, DXS10079 and DXS10148) were observed. Specific and elaborative discussions pertaining to the aspect of rare and off-ladder alleles in view of its application for forensic intelligence are reported in another research paper. In this present research, the most polymorphic locus was DXS10135 considering the high GD and PIC values, viz. Malays (PIC = 0.9135) and Chinese (PIC = 0.9157). In contrast, locus DXS7423 was found having the lowest PIC values for the Malays (0.4944) and Chinese (0.4322) since only 3–4 alleles were detected at that particular locus. The proportion of heterozygous individuals in the population (Het) as well as the PE values being the highest (DXS10135) as well as the lowest (DXS7423) at the similar loci. These findings were found similar with those reported for other populations [11,32,33] on the same 12 X-STRs markers.
No significant deviations from HWE were observed for the all the female samples (Supplementary Table S7), except in Chinese population (at locus HPRTB, p = 0.0056). However, after applying the Bonferroni's corrections (p < 0.05/12 = 0.0042), no HWE deviation was evident. The results of the LD analysis between all pairs of the 12 loci in both male and female samples are tabulated in Supplementary Tables S8a–b and S9a-b with a p < 0.0008 after an adjustment for multiple testing (0.05/66). Significant LD (p = 0.0000) was observed in both female and male samples of Chinese population within LG3 (between DXS10101-DXS10103) as well as in male samples of Malay population. As for the female samples of Malay population, significant LD was found between HPRTB-DXS10103 (p = 0.0000), comparable with the findings by Samejima et al. [11].
The haplotypes frequencies and forensic statistical parameters corresponding to the four LGs (LG1-DXS10148, DXS10135, DXS8378; LG2- DXS7132, DXS10079, DXS10074; LG3- DXS10103, HPRTB, DXS10101; LG4- DXS10146, DXS10134, DXS7423) are shown in Supplementary Tables S10–S11. The number of unique haplotypes in LG1-LG4 observed were 95, 68, 72 and 81 for the Malays respectively. As for the Chinese population, the same for LG1-LG4 were 90, 68, 75 and 70, correspondingly. For both the Malays (HD = 0.9971) and Chinese (HD = 0.9976), LG1 was found as the most informative LG, while the lowest being LG3 for the Malays (HD = 0.9876) and LG4 for the Chinese (HD = 0.9905).
In this present research, the combined PDm, PDf, MECKruger, MECKishida, MECDesmarais and MECDesmarais Duo were calculated based on allele (pooled) and haplotype frequencies. The combined PDm, PDf, MECKruger, MECKishida, MECDesmarais and MECDesmarais Duo calculated based on allele frequencies revealed the values ≥ 0.999995532964908 for the Malays and Chinese population (Supplementary Table S12). When calculated using the haplotype frequencies, the combined PDm, PDf, MECKruger, MECKishida, MECDesmarais and MECDesmarais Duo were ≥0.9999986410567 for the Malays and Chinese population (Supplementary Tables S10–S11).
3.2. Population comparisons
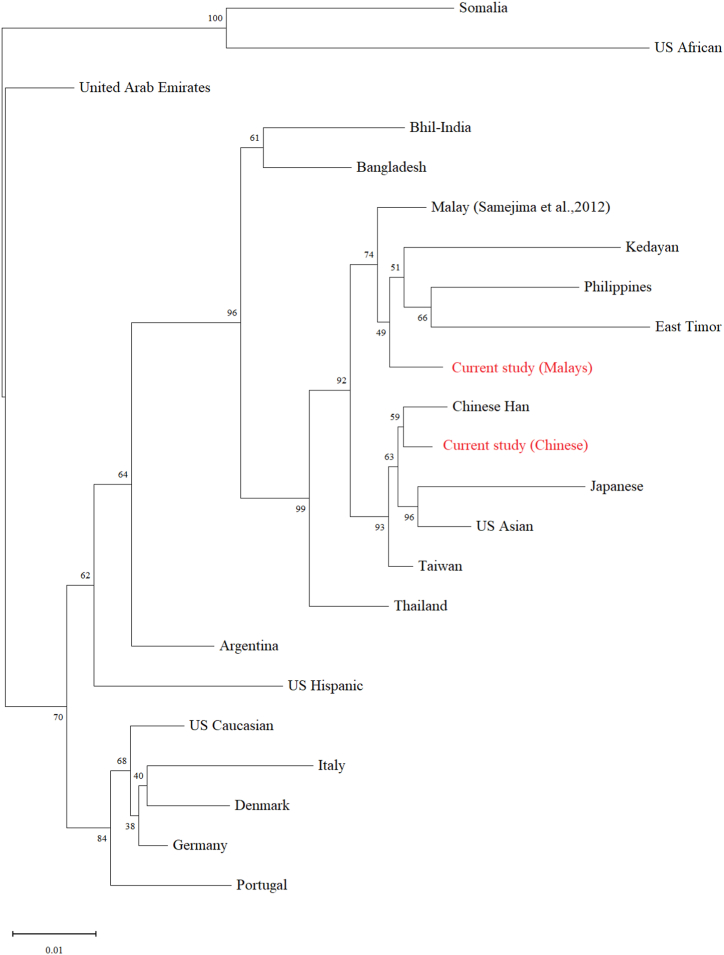
The genetic distance (DA) of all the pairs calculated among the Malaysian Malays and Chinese as well as the 21 other published populations are shown in Supplementary Table S13. The reasons for choosing the 21 other published populations are bipartite in nature. Firstly, all these populations data were derived from the same X-STRs markers and therefore would enable the appropriate discussion to be made with the present data. Secondly, the population data included would represent the different continents, as well as encompassing all the available population data for the South East Asian region, justifying the suitability of the markers for DNA profiling across the different populations. The DA values ranged from 0.0120 to 0.1340 for the studied populations against that of the other populations. A higher DA value was observed when the populations were geographically distant, comparable with the finding reported by previous researchers [34]. Fig. 1 depicts the NJ tree constructed based on the genetic distance (allele frequencies data) among the populations. The NJ tree revealed that the populations of Malays and Chinese studied here are clustered within a separate larger group (bootstrap support: 99 %) than those of the other group that comprised of United Arab Emirates, Somalia, US African, US Caucasian, Italy, Denmark, Germany, Portugal, US Hispanic and Argentina (bootstrap support: 70 %). Furthermore, it can be seen that the Malay and Chinese populations are grouped separately with high bootstrap support (92 %). While the current Malay population is grouped together (bootstrap support: 74 %) with the Malays from the previous study [11], Kedayan [13], Philippines [33] and East Timor [35], the current Chinese population, on the other hand, is grouped together (bootstrap support: 59 %) with the Chinese Han [36], Taiwanese [37], Japanese [38] and US Asian [39].
Fig. 1.
The Neighbor-Joining tree constructed based on the genetic distance (allele frequencies data) among the Malays and Chinese against other populations (as in Supplementary Table S13). Bootstrap values indicate the support for each node. The bar indicates 0.01 substitutions per site.
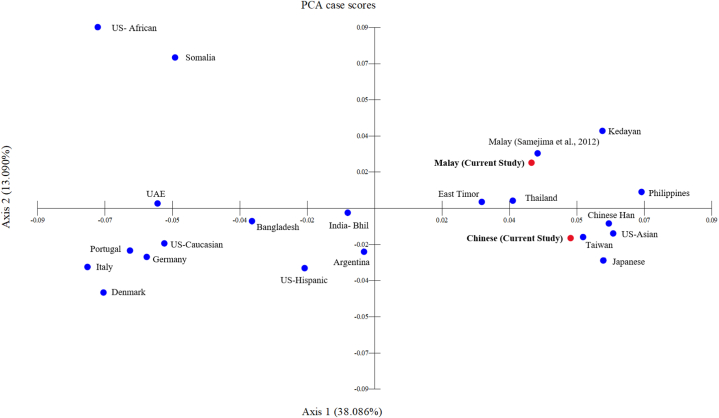
Similar findings on the categorical separation of the populations studied via NJ were also observed in the two-dimensional PCA case scores (Fig. 2). The close proximity observable via NJ and PCA between the Chinese observed in this current research with Chinese Han, Tawian, Japanese and US-Asian populations was within expectation, and it can be explained by several theories. The first theory is “Out of Taiwan” and “Express Train to Polynesian” models, suggesting that Austronesian populations (like the Malays) are originally from the provinces in China namely Taiwan and Yunnan, before arriving in Peninsular Malaysia in about 4500 years ago [40]. The second theory, revolves around the fact that transportation and the globalization especially in Southeast and South Asian populations have resulted in the frequent occurrence of gene flow among the different populations in the surrounding neighbourhoods [41]. The Malays were generally maritime traders, and as such indirectly promoting mixed marriages among the populations of the islands around the peninsular [40]. Deng et al. [42] reported that the population of Malaysia is affected by the genetic admixture from neighbouring East Asian populations. In fact, it was reported that between 4 % and 16 % of Malay genomes are descended from East Asians, with the remainder coming from South Asians, Austronesian, and aboriginal South Asians who interacted with Malays between 175 and 1500 years ago [41]. In point of fact, the assessments of molecular variance and average coefficient of correlation that were carried out by earlier researchers have revealed that the Malay and Chinese populations have the strongest correlation between them, in comparison to those of other populations [43,44].
Fig. 2.
The two-dimensional principal components analysis case scores constructed using the data of allele frequencies across the 12 X-STRs loci among the Malays and Chinese against other populations (as in Supplementary Table S13).
Others studies on genetic markers also demonstrated similar findings attributable to the genetic variations of the Peninsular Malaysia population [5,45]. The fact that Malaysia is a multi-racial country which composed of different genetic lineages, current DNA study should be cautiously interpreted to avoid contradiction with archaeological findings as historical events [45]. It is duly noted that integrating several genetic markers will strengthen the links between ethnic groups in terms of genetic distance, improving the understanding of the genetic makeup of the populations [46].
4. Conclusion
In conclusion, the allele frequencies and forensic statistical parameters of the studied Malaysian Malays and Chinese populations demonstrated that the 12 X-STRs markers were highly informative and discriminating. The population data reported here for the Chinese population in Malaysia appear to be the first of its kind to represent such population in the South East Asian region, substantiating the evidential value of the 12 X-STRs markers for forensic human identification. Therefore, the incorporation of X-STRs analysis for routine DNA casework appears pertinent, especially for complex kinship investigations in multi racial countries like Malaysia. The fact that little emphasis has been given on the admixture populations, suitable effort to provide reliable forensic population data for such populations shall be considered as an interesting future study.
Ethics and consent statement
The ethical approval for this research was awarded by the Human Research Ethics Committee of Universiti Sains Islam Malaysia (USIM/JKEP/2022-225), dated on July 21, 2022. Every participant provided a written informed consent to participate in the research and for their data to be published. The Human Participant Declaration form was provided during submission.
Funding
This research was supported by Ministry of Higher Education Malaysia Fundamental Research Grant Scheme (FRGS/1/2023/STG01/UTM/02/1) awarded to Universiti Teknologi Malaysia, as well as the funding from Faculty of Social Sciences and Humanities, Universiti Kebangsaan Malaysia (PP-FSSK-2024).
Data availability statement
The data are presented in the supplementary materials.
CRediT authorship contribution statement
Aedrianee Reeza Alwi: Writing – original draft, Visualization, Investigation, Formal analysis, Data curation. Naji Arafat Mahat: Writing – review & editing, Supervision, Resources, Project administration, Methodology, Funding acquisition, Conceptualization. Faezah Mohd Salleh: Writing – review & editing, Supervision, Methodology. Seri Mirianti Ishar: Writing – review & editing, Supervision, Methodology. Mohammad Rahim Kamaluddin: Writing – review & editing. Mohd Radzniwan A. Rashid: Writing – review & editing. Sharifah Nany Rahayu Karmilla Syed Hassan: Writing – review & editing. Sasitaran Iyavoo: Writing – review & editing, Formal analysis.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgments
The authors are thankful to the Public Service Department, Malaysia for awarding the Hadiah Latihan Persekutuan (HLP) scholarship to Aedrianee Reeza Alwi, and the Department of Chemistry Malaysia for all the necessary support for this research. The sampling assistance provided by Ahmad Sabri Mohamed, Fadilah Mohd Hassan, Hazwani Hapiz, Nor Wajihan Muda, Nik Ihtisyam Majdah Nik Razi, Salina Mohd Isa and Varohthini K Maharaja are highly appreciated.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e38054.
Contributor Information
Aedrianee Reeza Alwi, Email: aedrianee@kimia.gov.my.
Naji Arafat Mahat, Email: naji.arafat@utm.my.
Faezah Mohd Salleh, Email: faezah@utm.my.
Seri Mirianti Ishar, Email: seri.ishar@ukm.edu.my.
Mohammad Rahim Kamaluddin, Email: rahimk@ukm.edu.my.
Mohd Radzniwan A. Rashid, Email: mradzniwan@usim.edu.my.
Sharifah Nany Rahayu Karmilla Syed Hassan, Email: karmillahassan@unisza.edu.my.
Sasitaran Iyavoo, Email: SIyavoo@lincoln.ac.uk.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Department of Information Malaysia Official year book 2021. 2022. https://dbook.penerangan.gov.my/dbook/?product=official-year-book-2021
- 2.Department of Statistics Malaysia, Media statement for the report of demographic statistics Malaysia, third quarter 2023. 2023. https://www.dosm.gov.my/uploads/release-content/file_20231107114523.pdf
- 3.Ahmad N.M. The definition of Malay according to the federal constitution of Malaysia and its impact on the understanding and islamic practices in Malaysia. Sains Insani. 2020;5(2):9–16. [Google Scholar]
- 4.Yahya P., Sulong S., Harun A., Wan Isa H., Ab Rajab N.S., Wangkumhang P., Wilantho A., Ngamphiw C., Tongsima S., Alwi Z. Analysis of the genetic structure of the Malay population: ancestry-informative marker SNPs in the Malay of Peninsular Malaysia. Forensic Sci Int Genet. 2017;30:152–159. doi: 10.1016/j.fsigen.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Parthipan S., Ishar S.M. Perspectives on ancestral lineages and genetic markers of Malay population in Peninsular Malaysia. Jurnal Sains Kesihatan Malaysia. 2022;20:83–95. doi: 10.17576/jskm-2022-2001-08. [DOI] [Google Scholar]
- 6.Vollmann R., Wooi Soon T. Chinese identities in multilingual Malaysia. 2018. [DOI]
- 7.Zakaria M.F., Ibrahim A. Chinese civilization in Malaysia: history and contribution. Journal of Social Science and Humanities. 2022;5:1–6. doi: 10.26666/rmp.jssh.2022.1.1. [DOI] [Google Scholar]
- 8.Ismail K. Imperialism, colonialism and their contribution to the formation of Malay and Chinese ethnicity: an historical analysis. 2020. http://journals.iium.edu.my/intdiscourse/index.php/islam
- 9.Zakaria M.F., Ibrahim A. Development of Chinese civilisation and education in Malaysia. Journal of Management & Science. 2021;19:6. doi: 10.57002/jms.v19i1.223. [DOI] [Google Scholar]
- 10.Alwi A.R., Mahat N.A., Salleh F.M., Ishar S.M., Kamaluddin M.R., Rashid M.R.A. Applications of X-chromosome short tandem repeats for human identification: a review. J Trop Life Sci. 2023;13:187–212. doi: 10.11594/jtls.13.01.19. [DOI] [Google Scholar]
- 11.Samejima M., Nakamura Y., Nambiar P., Minaguchi K. Genetic study of 12 X-STRs in Malay population living in and around Kuala Lumpur using Investigator Argus X-12 kit. Int. J. Leg. Med. 2012;126:677–683. doi: 10.1007/s00414-012-0705-7. [DOI] [PubMed] [Google Scholar]
- 12.Abdullah N.A. Universiti Sains Malaysia; 2018. Genetic Variations of Six Orang Asli Sub-groups in Peninsular Malaysia Using Autosomal and X-Chromosome Short Tandem Repeats (STRS) Analysis. Master thesis. [Google Scholar]
- 13.Hakim H.M., Khan H.O., Ismail S.A., Lalung J., Kofi A.E., Aziz M.Y., Pati S., Nelson B.R., Chambers G.K., Edinur H.A. Population data and genetic characteristics of 12 X-STR loci using the Investigator® Argus X-12 Quality Sensor kit for the Kedayan population of Borneo in Malaysia. Int. J. Leg. Med. 2021 doi: 10.1007/s00414-021-02577-0. [DOI] [PubMed] [Google Scholar]
- 14.Rashid M.N.A., Mahat N.A., Khan H.O., Wahab R.A., Maarof H., Ismail D., Alwi A.R., SyedHassan S.N.R.K. Population data of 21 autosomal STR loci in Malaysian populations for human identification. Int. J. Leg. Med. 2020;134:1675–1678. doi: 10.1007/s00414-020-02279-z. [DOI] [PubMed] [Google Scholar]
- 15.Takić Miladinov D., Vasiljević P., Šorgić D., Podovšovnik Axelsson E., Stefanović A. Allele frequencies and forensic parameters of 22 autosomal STR loci in a population of 983 individuals from Serbia and comparison with 24 other populations. Ann. Hum. Biol. 2020;47:632–641. doi: 10.1080/03014460.2020.1846784. [DOI] [PubMed] [Google Scholar]
- 16.Baeta M., Prieto-Fernández E., Núñez C., Kleinbielen T., Villaescusa P., Palencia-Madrid L., Alvarez-Gila O., Martínez-Jarreta Begoña, De Pancorbo M.M. Study of 17 X-STRs in Native American and Mestizo populations of Central America for forensic and population purposes. Int. J. Leg. Med. 2021;135:1773–1776. doi: 10.1007/s00414-021-02536-9. [DOI] [PubMed] [Google Scholar]
- 17.He G., Li Y., Zou X., Zhang Y., Li H., Wang M., Wu J. X-chromosomal STR-based genetic structure of Sichuan Tibetan minority ethnicity group and its relationships to various groups. Int. J. Leg. Med. 2018;132:409–413. doi: 10.1007/s00414-017-1672-9. [DOI] [PubMed] [Google Scholar]
- 18.Buckleton J.S., Bright J.-A., Curran J.M., Taylor D. In: Forensic DNA Evidence Interpretation. second ed. Buckleton J.S., Bright J.-A., Taylor D., editors. CRC Press; 2016. Validating databases; pp. 133–181. [Google Scholar]
- 19.Parson W., Roewer L. Publication of population data of linearly inherited DNA markers in the International Journal of Legal Medicine. Int. J. Leg. Med. 2010;124:505–509. doi: 10.1007/s00414-010-0492-y. [DOI] [PubMed] [Google Scholar]
- 20.Alwi A.R., Mahat N.A., Mohd Salleh F., Ishar S.M., Kamaluddin M.R., Rashid M.R.A. Internal validation of reduced PCR reaction volume of the Qiagen Investigator® Argus X-12 QS Kit from blood samples on FTA cards. J. Forensic Sci. 2023 doi: 10.1111/1556-4029.15370. [DOI] [PubMed] [Google Scholar]
- 21.Qiagen . Qiagen; Germany: 2022. Investigator ® Argus X-12 QS Handbook. [Google Scholar]
- 22.Lang Y., Guo F., Niu Q. StatsX v2.0: the interactive graphical software for population statistics on X-STR. Int. J. Leg. Med. 2019;133:39–44. doi: 10.1007/s00414-018-1824-6. [DOI] [PubMed] [Google Scholar]
- 23.Excoffier L., Lischer H.E.L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 24.Krüger J., Fuhrmann W., Lichte K., Steffens C. On the utilization of erythrocyte acid phosphatase polymorphism in paternity evaluation. Dtsch. Z. Gesamte Gerichtl. Med. 1968;64:127–146. [PubMed] [Google Scholar]
- 25.Kishida T., Wang W., Fukuda M., Tamaki Y. Duplex PCR of the Y-27H39 and HPRT loci with reference to Japanese population data on the HPRT locus. Nihon Hoigaku Zasshi. 1997;51:67–69. [PubMed] [Google Scholar]
- 26.Desmarais D., Zhong Y., Chakraborty R., Perreault C., Busque L. Development of a highly polymorphic STR marker for identity testing purposes at the human androgen receptor gene (HUMARA) J. Forensic Sci. 1998;43:1046–1049. [PubMed] [Google Scholar]
- 27.Butler J.M. third ed. Academic Press; 2015. Advanced Topics in Forensic DNA Typing: Interpretation. [Google Scholar]
- 28.Nei M., Tajima F., Tateno Y. Accuracy of estimated phylogenetic trees from molecular data II. Gene frequency data. J. Mol. Evol. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- 29.Takezaki N., Nei M., Tamura K. POPTREEW: web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol. Biol. Evol. 2014;31:1622–1624. doi: 10.1093/molbev/msu093. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovach W.L. Kovach Computing Services; Pentraeth, Wales, UK: 1999. MVSP–A Multi-Variate Statistical Package for Windows (2007) Version 3.1, 137(3) [Google Scholar]
- 32.Xing J., Adnan A., Rakha A., Kasim K., Noor A., Xuan J., Zhang X., Yao J., McNevin D., Wang B. Genetic analysis of 12 X-STRs for forensic purposes in Liaoning Manchu population from China. Gene. 2019;683:153–158. doi: 10.1016/j.gene.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 33.Salvador J.M., Apaga D.L.T., Delfin F.C., Calacal G.C., Dennis S.E., De Ungria M.C.A. Filipino DNA variation at 12 X-chromosome short tandem repeat markers. Forensic Sci Int Genet. 2018;36:e8–e12. doi: 10.1016/j.fsigen.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Veselinović I., Petrić G., Vapa D., Djan M., Veličković N., Veljović T. Genetic analysis of 12 X-STR loci in the Serbian population from Vojvodina province. Int. J. Leg. Med. 2018;132:405–408. doi: 10.1007/s00414-017-1677-4. [DOI] [PubMed] [Google Scholar]
- 35.Moreira H., Costa H., Tavares F., Souto L. Genetic variation of 12 X-chromosomal STR loci in an East Timor sample. Int. J. Leg. Med. 2015;129:257–258. doi: 10.1007/s00414-014-1126-6. [DOI] [PubMed] [Google Scholar]
- 36.Zeng X.P., Ren Z., Chen J.D., Lv D.J., Tong D.Y., Chen H., Sun H.Y. Genetic polymorphisms of twelve X-chromosomal STR loci in Chinese Han population from Guangdong Province. Forensic Sci Int Genet. 2011;5 doi: 10.1016/j.fsigen.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Chen M.Y., Ho C.W., Pu C.E., Wu F.C. Genetic polymorphisms of 12 X-chromosomal STR loci in Taiwanese individuals and likelihood ratio calculations applied to case studies of blood relationships. Electrophoresis. 2014;35:1912–1920. doi: 10.1002/elps.201300645. [DOI] [PubMed] [Google Scholar]
- 38.Uchigasaki S., Tie J., Takahashi D. Genetic analysis of twelve X-chromosomal STRs in Japanese and Chinese populations. Mol. Biol. Rep. 2013;40:3193–3196. doi: 10.1007/s11033-012-2394-1. [DOI] [PubMed] [Google Scholar]
- 39.Diegoli T.M., Linacre A., Vallone P.M., Butler J.M., Coble M.D. Allele frequency distribution of twelve X-chromosomal short tandem repeat markers in four U.S. population groups. Forensic Sci Int Genet Suppl Ser. 2011;3 doi: 10.1016/j.fsigss.2011.09.102. [DOI] [Google Scholar]
- 40.Hanis Zainal Abidin N.W., Mohd Nor N., Sundararajulu P., Zafarina Z. Understanding the genetic history of Malay populations in Peninsular Malaysia via KIR genes diversity. Am. J. Hum. Biol. 2021;33 doi: 10.1002/ajhb.23545. [DOI] [PubMed] [Google Scholar]
- 41.Deng L., Hoh B.P., Lu D., Saw W.Y., Twee-Hee Ong R., Kasturiratne A., Janaka De Silva H., Zilfalil B.A., Kato N., Wickremasinghe A.R., Teo Y.Y., Xu S. Dissecting the genetic structure and admixture of four geographical Malay populations. Sci. Rep. 2015;5 doi: 10.1038/srep14375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng L., Hoh B.P., Lu D., Fu R., Phipps M.E., Li S., Nur-Shafawati A.R., Hatin W.I., Ismail E., Mokhtar S.S., Jin L., Zilfalil B.A., Marshall C.R., Scherer S.W., Al-Mulla F., Xu S. The population genomic landscape of human genetic structure, admixture history and local adaptation in Peninsular Malaysia. Hum. Genet. 2014;133:1169–1185. doi: 10.1007/s00439-014-1459-8. [DOI] [PubMed] [Google Scholar]
- 43.Chang Y.M., Perumal R., Keat P.Y., Kuehn D.L.C. Haplotype diversity of 16 Y-chromosomal STRs in three main ethnic populations (Malays, Chinese and Indians) in Malaysia. Forensic Sci. Int. 2007;167:70–76. doi: 10.1016/j.forsciint.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura Y., Samejima M., Minaguchi K., Nambiar P. Population genetics of identifiler System in Malaysia. Bull. Tokyo Dent. Coll. 2016;57:233–239. doi: 10.2209/tdcpublication.2016-1400. [DOI] [PubMed] [Google Scholar]
- 45.Norhalifah H.K., Syaza F.H., Chambers G.K., Edinur H.A. The genetic history of Peninsular Malaysia. Gene. 2016;586:129–135. doi: 10.1016/j.gene.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z., Song M., Lyu Q., Ying J., Wu Q., Song F., Jiang L., Wei X., Wang S., Wang F., Zhou Y., Song X., Luo H. A forensic population database of autosomal STR and X-STR markers in the Qiang ethnic minority of China. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e21823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are presented in the supplementary materials.