Abstract
The alphaherpesvirus Us4 gene encodes glycoprotein G (gG), which is conserved in most viruses of the alphaherpesvirus subfamily. In the swine pathogen pseudorabies virus (PRV), mutant viruses with internal deletions and insertions in the gG gene have shown no discernible phenotypes. We report that insertions in the gG locus of the attenuated PRV strain Bartha show reduced virulence in vivo and are defective in their ability to spread from cell to cell in a cell-type-specific manner. Similar insertions in the gG locus of the wild-type PRV strain Becker had no effect on the ability of virus infection to spread between cells. Insertions in the gG locus of the virulent NIA-3 strain gave results similar to those found with the Bartha strain. To examine the role of gG in cell-to-cell spread, a nonsense mutation in the gG signal sequence was constructed and crossed into the Bartha strain. This mutant, PRV157, failed to express gG yet had cell-to-cell spread properties indistinguishable from those of the parental Bartha strain. These data indicated that, while insertions in the gG locus result in decreased cell-to-cell spread, the phenotype was not due to loss of gG expression as first predicted. Analysis of gene expression upstream and downstream of gG revealed that expression of the upstream Us3 protein is reduced by insertion of lacZ or egfp at the gG locus. By contrast, expression of the gene immediately downstream of gG, Us6, which encodes glycoprotein gD, was not affected by insertions in gG. These data indicate that DNA insertions in gG have polar effects and suggest that the serine/threonine kinase encoded by the Us3 gene, and not gG, functions in the spread of viral infection between cells.
The alphaherpesviruses comprise a large number of medically and economically important viruses that include the human pathogens herpes simplex virus (HSV) and varicella-zoster virus, the bovine pathogen bovine herpesvirus 1 (BHV-1), and the swine pathogen pseudorabies virus (PRV). Alphaherpesviruses infect cells by two general mechanisms: free virions can bind to specific receptors on the surface of the cell, resulting in the fusion of the virus envelope with the cell membrane, or virus infection can spread directly from an infected cell to an adjacent, uninfected cell (73). The latter process is referred to as cell-to-cell spread. Cell-to-cell spread can be distinguished from infection by free virions because the former can occur in the presence of neutralizing antibody, whereas the latter cannot. Cell-to-cell spread of virus infection is important for the spread of infection through tissues and is utilized in the transsynaptic passage of virus through the nervous system (reviewed in reference 16). The spread of virus infection from cell to cell allows evasion from the neutralizing antibodies in an immunized host, for example, during reactivation from latency. Furthermore, the spread of infection through the formation of syncytia can occur very quickly. Uninfected cells are recruited into syncytia in much less time than it takes for an infected cell to produce virions. In the face of a competent immune system, it is advantageous for the virus to spread quickly.
Cell-to-cell spread of alphaherpesvirus infection can occur by at least two processes. In the first, the plasma membrane of the infected cell fuses with that of adjacent, uninfected cells, resulting in the formation of giant, multinucleated syncytia (74). Syncytium formation occurs in the lesions caused by PRV, HSV, and varicella-zoster virus in their natural hosts (67). Not all cell types fuse when infected by these viruses, indicating that a specific cellular environment is required. In the second process, infection is thought to spread at cell junctions without the formation of a syncytium (14). This nonsyncytial form of cell-to-cell spread is often seen in cultured cells. The molecular mechanism by which cell-to-cell spread of infection occurs has been shown to be different from the process of infection by free virions in that the specific requirements for viral membrane proteins differ for each process. The spread of infection from cell to cell requires a much larger repertoire of viral proteins than does infection by free virions. Infection by free virions requires glycoprotein B (gB), gD, and the gH/gL complex (73). Similarly, spread from cell to cell also requires gB and gH/gL. The requirements for gD in this process are variable, depending on the alphaherpesvirus (11, 14, 52, 58, 62, 77, 81).
Viral proteins gB, gD, gE/gI, gH/gL, gK, gM, UL20, UL24, and UL45 all participate in the spread of HSV infection between cells (reviewed in references 74 and 77). Precisely how these molecules function in this process is unclear. Deletion of gE and gI has no measurable effect on the entry of virus into cells yet has profound effects on cell-to-cell spread of infection in certain cell types (2, 14, 15, 79, 88). Recent evidence suggests that gE and gI sort virions to epithelial cell junctions, thereby facilitating cell-to-cell spread (24, 47, 86). Mutations in gB, gK, UL20, or UL24 result in viruses that form syncytia on many cultured cell lines (74). Turner and colleagues have used a Cos cell transfection system to study the requirements for HSV type 1 (HSV-1) syncytium formation (81). These studies have shown that expression of gB, gD, gH, and gL is necessary and sufficient to mediate membrane fusion. Similar results were obtained when comparable experiments were performed using the analogous glycoproteins from HSV-2 (51). These studies also highlight the viral fusion machinery as a tightly regulated complex during a viral infection. For example, infection of Cos cells with wild-type virus does not result in extensive syncytium formation, despite high-level expression of gB, gD, gH, and gL in the membranes of infected cells. There is good evidence that gE/gI, gK, gM, and UL45 serve as regulators of the viral fusion machinery and that UL20 is required for efficient processing of gK (2, 11, 12, 17, 20, 22, 28, 31, 61, 88).
We have identified two cell lines that define new requirements for the spread of virus infection between cells. These cell lines are Georgia bovine kidney (GBK) and Madin-Darby bovine kidney (MDBK). Wild-type PRV strains form giant, multinuclear syncytia on GBK or MDBK cell monolayers. When the glycoprotein gE or gI was deleted from PRV, large nonsyncytial plaques formed on these cells, supporting the findings of previous studies (2, 11, 88). The two cell lines also display a cell-type-specific phenotype that involves nonsyncytial cell-to-cell spread. In some viral genetic backgrounds, insertion of the β-galactosidase gene (lacZ) or the gene for enhanced green fluorescent protein (EGFP) (egfp) in the gG gene results in a striking reduction of plaques without affecting syncytium formation. This phenotype is not apparent on monolayers of the swine kidney cell line PK15. Insertions of lacZ or egfp result in a gG-null phenotype but also reduce the expression of the upstream Us3 gene. The plaque size is due not to loss of gG but rather to the reduction of expression of Us3, a unique protein kinase.
MATERIALS AND METHODS
Viruses and cells.
The virus strains used in this study are listed in Table 1. PRV strains expressing β-galactosidase, Becker-Blu, Bartha-Blu, and PRV99-Blu have been described elsewhere (3, 32, 75). The construction of PRV strains expressing EGFP has been described recently (71). Briefly, PRV151, PRV152, and PRV155 were constructed by homologous recombination between a plasmid containing an EGFP expression cassette cloned into the middle of the PRV gG gene and the PRV genome. Briefly, a 2.6-kbp SalI fragment from PRV-Becker containing the 3′ end of the Us3 gene, the entire gG gene, and the 5′end of the gD gene was cloned into the SalI site of pBB3 [a modified pGEM-5zf(+) derivative in which the PstI site and NotI site have been deleted] to generate pBB4. Next, a 2.3-kbp NsiI fragment from pEGFP-N1 (Clontech, Palo Alto, Calif.) containing the cytomegalovirus immediate-early promoter, EGFP sequences, and a simian virus 40 poly(A) signal was cloned into a unique PstI site in pBB4 to generate pII1. This leaves about 840 bp of PRV sequence upstream of the EGFP expression cassette and 1,750 bp downstream of the EGFP expression cassette available for homologous recombination with the viral genome. The plasmid pII1 was digested with SalI and cotransfected with purified PRV-Becker DNA (PRV151), PRV-Bartha DNA (PRV152), or M201 DNA (PRV155) into PK15 cells. Virus produced after cotransfection was plated on PK15 cells, and plaques expressing EGFP were identified with the aid of an inverted epifluorescence microscope. Virus was isolated from EGFP-expressing plaques and subjected to three rounds of purification. Southern blot analysis using the Becker SalI fragment from pBB4 as a probe was performed to verify that the EGFP expression cassette had recombined appropriately into the PRV genome.
TABLE 1.
Genotypes of viruses used in this study and their plaque morphologies on GBK and MDBK cellsa
| PRV strain | Genotype | Comments | Plaque morphology on GBK and MDBK cells
|
||
|---|---|---|---|---|---|
| Syncytial | Large nonsyncytial | Tiny nonsyncytial | |||
| Kaplan | Wild-type | Lab strain | Yes | No | No |
| Becker | Wild-type | Field isolate | Yes | No | No |
| Becker-Blu | lacZ+, gG− | Isogenic with Becker | Yes | No | No |
| PRV151 | EGFP+, gG− | Isogenic with Becker | Yes | No | No |
| PRV91 | gE− | Isogenic with Becker | No | Yes | No |
| PRV98 | gI− | Isogenic with Becker | No | Yes | No |
| PRV99 | gE−, gI− | Isogenic with Becker | No | Yes | No |
| PRV99-Blu | lacZ+, gE−, gI−, gG− | Isogenic with Becker | No | Yes | No |
| Bartha | gC, gM, UL21, gE−, gI−, US9−, US2− | Attenuated vaccine strain | No | Yes | No |
| Bartha-Blu | lacZ+, gC, gM, UL21, gE−, gI−, US9−, US2−, gG− | Isogenic with Bartha | No | No | Yes |
| Bartha-Blu revertant | gC, gM, UL21, gE−, gI−, US9−, US2− | Isogenic with Bartha | No | Yes | No |
| PRV152 | EGFP+, gC, gM, UL21, gE−, gI−, US9−, US2−, gG− | Isogenic with Bartha | No | No | Yes |
| PRV154 | US9-EGFP+, gC, gM, UL21, gE−, gI−, US9−, US2−, gG− | Isogenic with Bartha | No | No | Yes |
| PRV156 | EGFP+, gC, gM, UL21, gE−, gI−, US9−, US2−, gG− | Isogenic with Bartha | No | No | Yes |
| PRV156R | gC, gM, UL21, gE−, gI−, US9−, US2− | PRV156 reverted to Bartha genotype | No | Yes | No |
| PRV157 | gC, gM, UL21, gE−, gI−, US9−, US2−, gG null | Isogenic with Bartha gG nonsense mutation | No | Yes | No |
| NIA-3 | Wild-type | Field isolate | Yes | No | No |
| M201 | gE− | Isogenic with NIA-3 | No | Yes | No |
| PRV155 | EGFP+, gE−, gG− | Isogenic with NIA-3 | No | No | Yes |
The Blu strains express the lacZ gene from the gG locus and are therefore labeled gG−. The EGFP+ strains have an egfp insertion in the gG locus and are also labeled gG−. Point mutations in the genes are italicized, and deletions in the genes are followed by “−”.
To construct PRV154, a 2.6-kbp NsiI fragment from pBB14 containing a Us9-EGFP fusion protein expression cassette was cloned into the PstI site of pBB4, disrupting the gG gene, to generate pAC2. pAC2 was then digested with SalI and cotransfected into PK15 cells with Bartha genomic DNA. Green fluorescent plaques were purified and recombinant viruses were verified by Southern blotting as described above.
To construct PRV156, a 2.6-kbp SalI fragment from the Bartha genome containing the 3′ end of the Us3 gene, the entire gG gene, and the 5′end of the gD gene was cloned into the SalI site of pBB3 to generate pBB33. Next, a 2.3-kbp NsiI fragment from pEGFP-C1 (Clontech) containing an EGFP expression cassette was cloned into a unique PstI site in pBB33 to generate pBB35. This leaves about 840 bp of PRV sequence upstream of the EGFP expression cassette and 1,750 bp downstream of the EGFP expression cassette available for homologous recombination with the viral genome. The plasmid pBB35 was digested with SalI and cotransfected with purified PRV-Bartha DNA into PK15 cells. Green fluorescent plaques were purified and recombinant viruses were verified by Southern blotting as described above.
To construct PRV157, a 2.6-kbp SalI fragment from the Bartha genome containing the 3′ end of the Us3 gene, the entire gG gene, and the 5′end of the gD gene was cloned into the SalI site of pAlter-1 to create pBB31. An amber codon was introduced into the seventh codon of gG in pBB31 by site-directed mutagenesis with the altered-site mutagenesis system (Promega) using the oligonucleotide 5′-TGGGCAACGTAGATCCTCGCC-3′. This mutation also results in the loss of a BamHI site from pBB31; it served as a useful screen for mutant clones. One such clone was named pBB40. To transfer the gG nonsense mutation into the Bartha genome, a 400-bp BssHII/BamHI fragment from pBB40 was cloned into BssHII/BamHI-digested pBB39 (which contains the SalI/NotI fragment from pBB31 cloned into pBSKS+) to create pBB41. pBB41 was sequenced to ensure that the desired mutation in gG was present and that no other mutations had been introduced into the transfer vector. pBB41 was linearized with SalI and cotransfected into PK15 cells with PRV156 DNA. Virus produced after cotransfection was plated on PK15 cells, and plaques that failed to express EGFP were identified with the aid of an inverted epifluorescence microscope. Virus was isolated from non-EGFP-expressing plaques and subjected to three rounds of purification. Southern blot analysis using the SalI fragment from pBB4 as a probe was performed to verify that the EGFP expression cassette had been appropriately excised from the PRV genome. Two types of recombinants were isolated from this cotransfection experiment as determined by PCR and DNA sequencing. Those that had the gG nonsense mutation were designated PRV157 and those that reverted PRV156 to the parental Bartha genotype were designated PRV156R.
All virus strains were propagated and titered on PK15 cells unless otherwise indicated. PK15, GBK, and MDBK cells were maintained at 37°C in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) (Gibco/BRL) in a 5% CO2 environment.
Intraocular injection of chick embryos.
Intraocular injections were performed on E12 White Leghorn chicken embryos exactly as described previously (3). Immediately prior to injection, virus stocks were thawed and sonicated briefly, and cells and cellular debris were removed by brief centrifugation in a microfuge. Animals were injected in the vitreous humor of the right eye with various amounts of virus ranging from 102 to 106 PFU in a total volume of 1 μl. Virulence was measured by determining both the mean time to death after injection of 105 PFU and the 50% lethal dose (LD50). The LD50 was defined as the number of PFU required to kill 50% of the animals within 168 h, a time just prior to hatching. LD50s were calculated using the graphic interpolation method of Reed and Muench (64). Experimental protocols were approved by the Animal Welfare Committee and were consistent with the regulations of the American Association for the Accreditation of Laboratory Animal Care and those in the Animal Welfare Act (Public Law 99-198). All animals were confined to a biosafety-level-2 laboratory.
Primary retinal cultures.
Retinae (∼12) were removed from E8 embryos and were collected in 2 ml of Hanks' buffered salt solution without Ca2+ and Mg2+. Trypsin (Gibco/BRL) was added to the Hanks' buffered salt solution to a final concentration of 1%, and the retinae were incubated at 37°C for 5 min. After trypisinization, 10 ml of DMEM–10% FCS–5% chicken serum (CS) was added to the retinae and the tissue was pelleted by centrifugation at 600 × g for 7 min at room temperature. Cells were resuspended in 2 ml of DMEM–10% FCS–5% CS and were triturated 10 times with a sterile Pasteur pipette. The volume was brought up to 24 ml with DMEM–10% FCS–5% CS, and 2 ml was plated per well onto 6-well culture dishes (Falcon), each well containing a poly-l-lysine-treated 22-mm-diameter glass coverslip. Cells were incubated at 37°C in a 5% CO2 environment for 3 days prior to infection with PRV.
Plaque assays.
Serial 10-fold dilutions of virus were prepared in DMEM–10% FCS. One hundred microliters of each dilution was added to a well of a 6-well cluster dish containing a 85 to 95% confluent monolayer of either PK15, GBK, or MDBK cells, and the dishes were returned to the incubator. The cells and inoculum were rocked every 10 min for 1 h to ensure that the monolayer did not become dry, and then 3 ml of DMEM–10% FCS–1% carboxy-methylcellulose was added to each well and the plates were returned to the incubator for 48 h. Cells were fixed and stained in 70% methanol–0.5% methylene blue. For detection of β-galactosidase activity, infected monolayers were rinsed three times with phosphate-buffered saline and were then fixed in 4% formaldehyde for 10 min at room temperature. Following fixation, cells were rinsed three times with 3 ml of phosphate-buffered saline and 1 ml of substrate buffer added (1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal]/ml, 10 mM potassium ferrocyanide, 10 mM potassium ferricyanide, 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% IGEPAL CA-630 (NP-40) in 0.1 M phosphate buffer, pH 7.4). Cells were incubated in substrate buffer overnight (∼16 h) at 37°C. Substrate buffer was removed and replaced with phosphate-buffered saline until the cells were photographed. For EGFP-expressing viruses, plaques were photographed directly at 48 h postinfection using a Nikon TE200 inverted epifluorescence microscope equipped with a cooled charge-coupled device camera.
Analysis of gG expression.
Expression of gG protein by parental, mutant, and repaired viruses was examined by labeling infected PK15 cells with [35S]methionine. At 3 h postinfection, cells were incubated with [35S]methionine at 50 μCi/ml in methionine-free medium for 30 min. At the end of the labeling period, monolayers were rinsed three times with serum-free medium, serum-free medium was added to the cells, and the plates were returned to the incubator. After 1 h the medium was collected, clarified of cells and cellular debris by centrifugation, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. At the time that the medium was harvested, 4.5 h postinfection, significant amounts of newly synthesized virions were not present in the medium.
RNA analysis.
RNA was isolated from mock-infected or infected (multiplicity of infection = 10) MDBK cells at 3 and 6 h postinfection using the Trizol reagent (Gibco/BRL). Dishes (diameter, 100 mm) containing approximately 5 × 106 cells were used for each sample. Ten micrograms of total RNA from each sample was denatured in 50% formamide, 2.2 M formaldehyde, and 25 mM sodium phosphate buffer pH 7.1, and was separated on a 1% agarose-formaldehyde gel and transferred to a nylon membrane as described elsewhere (45). The blot was hybridized to a biotinylated double-stranded DNA probe corresponding to the Us3b open reading frame, which was prepared as follows: the Us3b open reading frame was amplified by PCR using the forward primer 5′CGGAATTCGTTGTCGCGCGTCCACGCCCAGC3′ and the reverse primer 5′AAGGAAAAAAGCGGCCGCAGGTGTGTGTGTCCTACCGCTCG3′. The PCR product was biotinylated using the NEBlot Phototope Kit (New England Biolabs). Bands hybridizing to the biotinylated Us3b probe were detected using the Phototope-Star detection kit (New England Biolabs) according to the manufacturer's instructions.
Preparation of Us3 antisera.
Antiserum against a Us3 peptide was raised in rabbits and affinity purified by Bethyl Laboratories (Montgomery, Tex.). Briefly, the peptide RRPSADEILNFG corresponding to amino acids 373 to 384 of Us3a and 319 to 330 of Us3b was synthesized, conjugated to keyhole limpet hemocyanin, and used to immunize two New Zealand White rabbits.
Western blot analysis.
MDBK cells growing in 100-mm-diameter culture dishes (∼5 × 106 cells) were infected with PRV at a multiplicity of infection of 10. At 3 and 6 h postinfection, the medium was removed from the dish and the cells were rinsed three times with cold phosphate-buffered saline. Cold lysis buffer (750 μl) (10 mM Tris, 150 mM NaCl, 1% IGEPAL CA-630, 1% sodium deoxycholate, pH 7.4) was added, and the cells were incubated on ice for 10 min. SDS-PAGE sample buffer was added to portions of lysates, and these were subjected to SDS-PAGE on 10% gels as described elsewhere (37). Following electrophoresis, protein in gels was transferred to polyvinylidene difluoride membranes using a Bio-Rad semidry transfer apparatus following the manufacturer's instructions. Membranes were blocked overnight in Tris-buffered saline–Tween (50 mM Tris, 200 mM NaCl, 0.05% Tween 20) containing 3% bovine serum albumin. Proteins were visualized by using rabbit (for Us3 and gD) or goat (for gB) polyclonal primary antibodies and enhanced chemiluminescence detection as recommended by the manufacturer (Renaissance system; NEN).
RESULTS
A PRV vaccine strain, Bartha, with an insertion at the gG locus shows reduced virulence in a chicken embryo eye model.
A chicken embryo eye model was established to study alphaherpesvirus neuronal spread and virulence (3). In this model, virus is injected directly into the right eye of embryonic day-12 embryos through a small window opened in the eggshell. After injection, the window is sealed, eggs are returned to an incubator, and the virulence properties and the capacity of virus to spread from the eye to the brain are measured. The virulence of several PRV strains was tested in two ways: first, by the determination of LD50, and second, by measuring the mean time to death after inoculation with 105 PFU of virus. To facilitate the identification of infected cells and tissue, a number of the strains used express β-galactosidase at the gG locus. The PRV gG locus was chosen as a site to insert the lacZ gene because viruses with deletions or insertions in gG display wild-type virulence in any system studied to date and because it has been used extensively by other groups studying PRV (27, 49, 75, 78; J. P. Card, R. R. Miselis, and L. W. Enquist, unpublished observations).
The results of these virulence studies are summarized in Table 2. As expected, the wild-type derivative Becker-Blu was the most virulent, with the lowest LD50 and the shortest mean time to death. The PRV99-Blu strain, which is isogenic with Becker and is deleted for the glycoproteins gE and gI, displayed reduced virulence compared to Becker-Blu, with a higher LD50 and increased mean time to death. These results were expected because numerous studies have indicated that deletion of gE and gI profoundly affects virulence in numerous animal model systems including the natural host (1, 8, 34–36, 41–43, 48, 50, 53, 54).
TABLE 2.
Virulence properties of PRV strains used in chick embryo eye modela
| Virus strain | Genotype | LD50 (PFU) | Mean time to death in h (105 PFU) |
|---|---|---|---|
| Becker-Blu | gG− | 58 (n = 31) | 55 (n = 45) |
| PRV99-Blu | gE−, gI−, gG− | 200 (n = 31) | 68 (n = 43) |
| Bartha-Blu | gC, gM, UL21, gE−, gI−, US9−, US2−, gG− | 5,600 (n = 56) | 80 (n = 38) |
| Bartha | gC, gM, UL21, gE−, gI−, US9−, US2− | <100 (n = 60) | 80 (n = 26) |
The Blu strains express the lacZ gene from the gG locus and are therefore labeled gG−. The Becker strain is a wild-type, virulent PRV field isolate. PRV99 is isogenic with the Becker strain and has deletions of gE and gI. Point mutations in the genes of the Bartha strain of PRV are italicized. Deletion of genes is followed by “−”.
We also introduced the attenuated vaccine strain Bartha into the chicken embryo eye model. The Bartha strain, which was isolated after repeated passage through chicken embryo fibroblasts and selected for resistance to elevated temperature, has a large deletion that eliminates expression of gI, gE, Us9, and Us2 and point mutations affecting the UL21, gC, and gM genes (Table 1) (4, 13, 30, 40, 66). Because of these changes, it was not surprising that the Bartha-Blu strain had the highest LD50 and mean time to death and proved to be the most attenuated strain tested in the chicken embryo eye model. By contrast, the virulence properties of the parental Bartha strain were surprising. The key observation was that Bartha-Blu was significantly more attenuated than the parental Bartha strain. The Bartha strain had an LD50 that was approximately 60 times lower than that of Bartha-Blu. These data suggested that gG played a role in virulence of the Bartha strain.
A PRV strain with an insertion at the gG locus has defects in cell-to-cell spread.
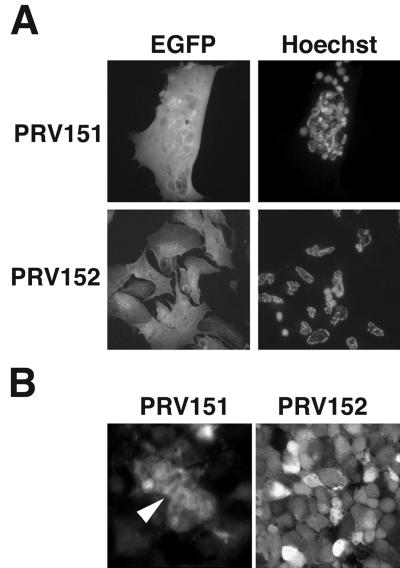
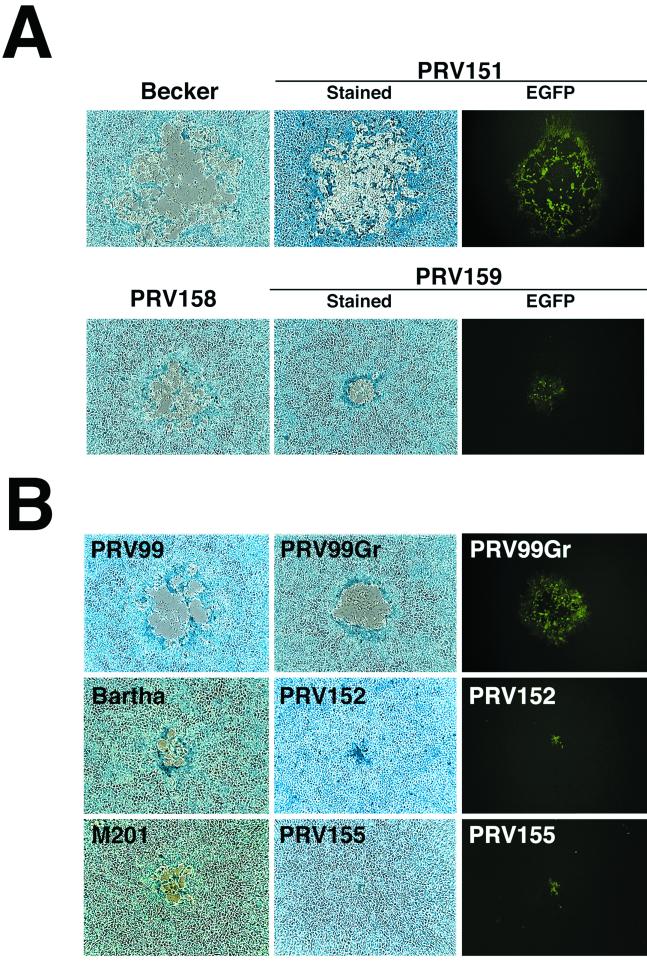
To understand the basis for different virulence properties exhibited by PRV mutants in the chicken embryo eye model, we examined infection of chick primary retinal cultures. The retina is the primary site of viral replication in the chicken embryo eye model. Four days after plating, the cells had established two distinct layers: large flat Müller glial cells grew on the culture dish and neurons spread over this glial cell layer. To facilitate the identification of infected cells, these cultures were infected with PRV strains that express EGFP from the gG locus. Like the virus strains with lacZ insertions, these viruses do not express gG. PRV151 is an EGFP-expressing derivative of the virulent Becker strain, and PRV152 is an EGFP-expressing derivative of the Bartha vaccine strain (71). Four-day-old mixed retinal cultures were infected with PRV151 and PRV152. Twenty-four hours after infection, the cells were fixed and stained with Hoechst 33258 to identify nuclei, and the DNA and EGFP signals were visualized by fluorescence microscopy (Fig. 1A). Infection by PRV151 resulted in the formation of multinuclear syncytia. By contrast, PRV152-infected cells did not form syncytia. This observation provided evidence that the reduced virulence of the Bartha strains in the chicken embryo eye model may reflect defects in cell-to-cell spread.
FIG. 1.
PRV infection of embryonic chicken retinal cells. (A) Infection of primary retinal cultures. Mixed retinal cultures 24 h after infection with PRV151 and PRV152. The EGFP signal is shown on the left, and the same cells stained with Hoechst 33258 are shown on the right. (B) Optical sections midway through PRV-infected chicken embryo retinal whole mounts 48 h after intravitreal infection with PRV151 (left) or PRV152 (right). The EGFP signal was visualized by confocal fluorescence microscopy. The arrowhead indicates fused cells.
To ensure that the observations in the 4-day-old primary cultures were not an artifact of the culture conditions, we tested whether the PRV151 strain caused cell fusion in intact retina in vivo. Forty-eight hours after intraocular infection of embryonic day-12 chick embryos, retinal whole mounts were prepared and infected tissue was visualized by confocal fluorescence microscopy (Fig. 1B). These data demonstrated that the virulent Becker derivative PRV151 fused cells in the retina (arrowhead). The Bartha derivative PRV152 did not fuse cells as evidenced by the clear outline of the boundaries of individual infected cells. These data supported our observations with primary cultures and indicated that syncytial formation by virulent virus strains also occurs in vivo. These observations are consistent with the hypothesis that the fusogenic ability of the wild-type strain influences virulence in the chicken embryo eye model. We next attempted to identify the molecules defective in the Bartha strain that contribute to membrane fusion by the Becker strain.
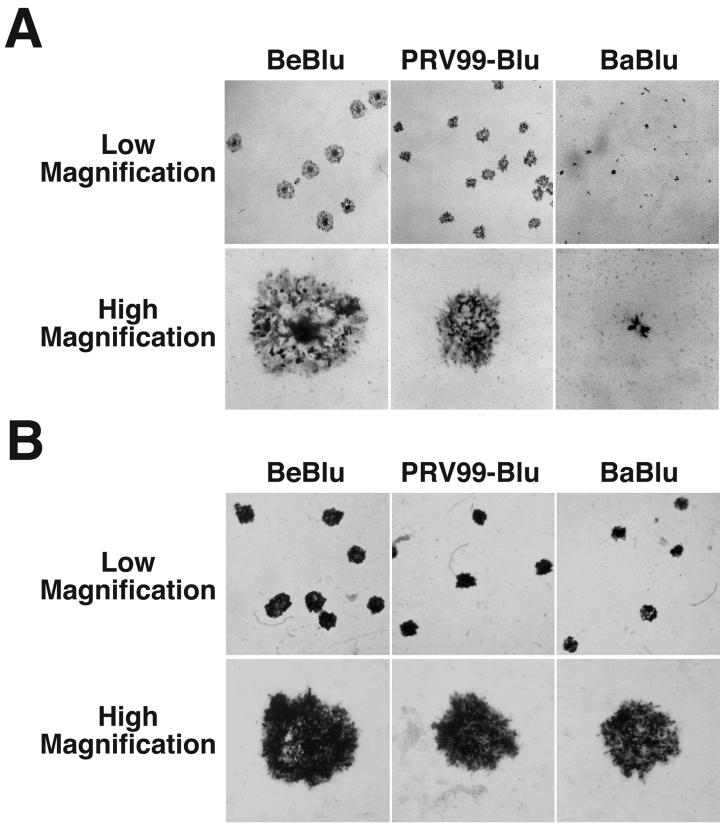
GBK cells and MDBK cells form syncytia upon infection with PRV (79; L. W. Enquist, unpublished observations). When GBK and MDBK cells were infected with Becker-Blu, PRV99-Blu, and Bartha-Blu, remarkable differences in plaque morphologies were observed. Infection with Becker-Blu resulted in the formation of syncytial plaques characterized by large numbers of nuclei in the center of the plaque (Fig. 2A). By contrast, PRV99-Blu formed large nonsyncytial plaques and Bartha-Blu formed tiny nonsyncytial plaques. Interestingly, all three virus strains formed plaques of similar size and morphology on monolayers of the swine kidney cell line PK15; all strains produced large nonsyncytial plaques (Fig. 2B). These data indicated that PRV99-Blu and Bartha-Blu have defects in spread from cell to cell that manifest themselves only when grown on certain cell types.
FIG. 2.
Plaque morphologies of PRV strains with lacZ insertions in gG on GBK and PK15 cells. Low- and high-magnification images of infected GBK cells at 48 h after a low-multiplicity infection. Cells were fixed in 4% formaldehyde for 10 min, rinsed, and incubated in an X-Gal solution to identify infected cells. (A) GBK cells. (B) PK15 cells. BeBlu, Becker-Blu; BaBlu, Bartha-Blu. Results similar to those obtained on GBK cells were observed on monolayers of MDBK cells, another bovine kidney cell line (Fig. 8).
The striking differences in plaque size observed with these viruses on GBK and MDBK cells provided a simple assay to map the viral genes responsible for these different phenotypes. Table 1 summarizes the data from these assays. Deletion of gE and/or gI from the PRV genome abrogates the syncytial phenotype; PRV91, PRV98, PRV99, and PRV99-Blu are isogenic with the Becker strain and have deletions of gE, gI, or both. All four of these strains form large nonsyncytial plaques on GBK and MDBK cells that are similar in size. These data support the observations of others that gE and gI can function to regulate the viral fusion machinery of both PRV and HSV (2, 88). Furthermore, these data suggest that the Bartha strain does not form syncytia on GBK or MDBK cells because it has a deletion of gE and gI.
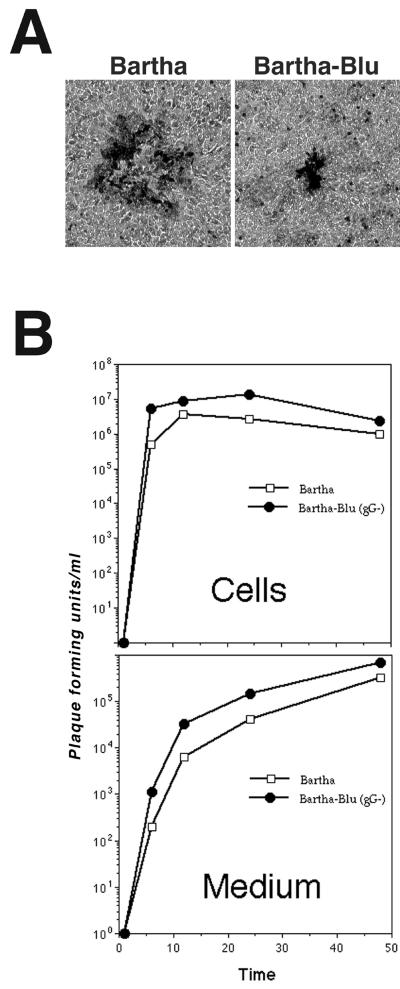
The insertion in gG is responsible for the tiny-nonsyncytial-plaque phenotype demonstrated by the Bartha-Blu strain, because the parental Bartha strain formed large nonsyncytial plaques (Fig. 3A and Table 2). In further support of this conclusion, three other independently isolated Bartha derivatives with insertions in gG, PRV152, PRV154, and PRV156 also formed tiny nonsyncytial plaques similar in morphology and size to those formed by Bartha-Blu. To control for the possibility that mutations outside the gG locus were responsible for the small-plaque phenotype, revertants of the Bartha-Blu strain and the PRV156 strain were constructed. The revertant viruses formed plaques that were indistinguishable from those formed by the parental Bartha strain. Because four independently isolated Bartha strains with insertions in gG form tiny nonsyncytial plaques and because, when two of these viruses were reverted, parental plaque morphologies were restored, we suggest that the tiny-nonsyncytial-plaque phenotype was due to the insertion in the gG gene.
FIG. 3.
Growth of Bartha and Bartha-Blu on GBK cells. (A) Plaque morphologies of Bartha and Bartha-Blu on GBK cell monolayers. Forty-eight hours after infection, cells were fixed and stained with 0.5% methylene blue in 70% methanol. Single plaques are shown. (B) Single-step growth of Bartha and Bartha-Blu on monolayers of GBK cells. Kinetics of infectious virus production in cells is plotted in the top graph, and the rate of release of infectious virus into the medium is plotted on the bottom graph.
Interestingly, Becker strains with insertions in gG did not display the tiny- nonsyncytial-plaque phenotype on GBK or MDBK cells, even if gE and gI had been deleted from the Becker strain, as in the case of PRV99-Blu. This observation indicates that the Becker strain encodes a function that can compensate for insertions in gG and that the Bartha strain lacks this function.
Reduced replication or decreased infection efficiency does not account for tiny-nonsyncytial-plaque phenotype.
Small plaques can arise due to a reduced ability of the virus to replicate, a decrease in the efficiency with which virus infects cells, or a specific defect in the spread of virus from cell to cell. Two of these possibilities were tested directly. To determine if Bartha-Blu had a growth defect relative to Bartha on monolayers of GBK cells, single-step growth analysis was performed. The data indicated that both the rate of infectious virus production and the total amount of infectious virus produced in cells and released into the medium were similar for Bartha and Bartha-Blu (Fig. 3B). In fact, Bartha-Blu consistently produced slightly more infectious virus than Bartha, both in the cells and culture medium. From these data, we concluded that the small-plaque phenotype exhibited by Bartha-Blu was not due to a reduced ability of the virus to replicate in GBK cells.
To eliminate the possibility that Bartha strains with insertions in gG had a reduced ability to infect GBK cells, the specific infectivities of the Becker, Becker-Blu, Bartha, and Bartha-Blu were measured. For every 100 plaques that formed on PK15 cells, approximately 120 plaques formed on monolayers of GBK cells when equivalent inocula were used. This plating efficiency held for all virus strains tested. Decreased efficiency of infection does not account for the reduced ability of plaques to form on GBK cells.
Introduction of a nonsense mutation into the gG gene of the Bartha strain has no effect on cell-to-cell spread.
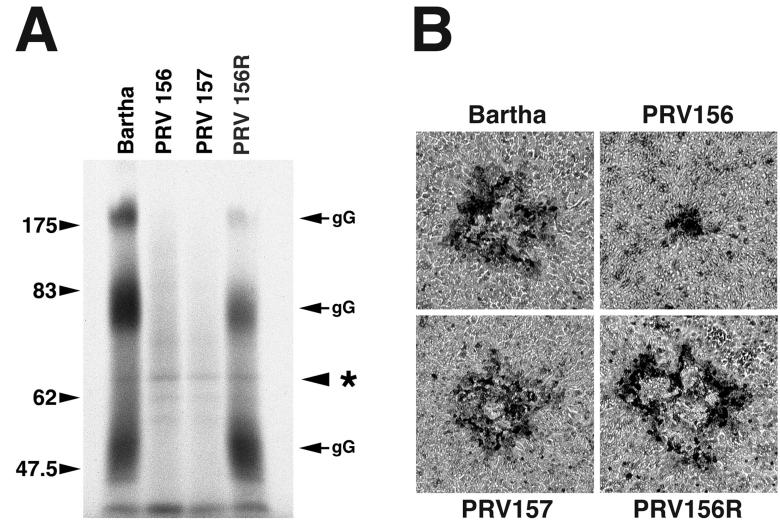
Is the gG protein required for the efficient spread of Bartha viruses from cell to cell? This question was addressed by introducing a stop codon seven codons into the Bartha gG gene by site-directed mutagenesis. PRV156 is a Bartha derivative that expresses EGFP from the gG locus and, like Bartha-Blu, PRV152, and PRV154, produces tiny nonsyncytial plaques on monolayers of GBK cells. The virus with the nonsense mutation in gG is PRV157, and the repaired PRV156 strain is PRV156R.
Because gG is secreted from infected cells in abundance, expression of this glycoprotein can be assayed by analysis of infected-tissue culture supernatants (5, 6). The results from this experiment are shown in Fig. 4A. gG is secreted from cells in three predominant forms of approximately 180, 75, and 50 kDa that arise as a result of proteolytic cleavage and other posttranslational modifications, including sulfation and glycosylation (5, 63, 78). As expected, cells infected with the Bartha strain and the repaired PRV156 strain PRV156R synthesized and secreted gG into the medium. By contrast, cells infected with the EGFP-expressing PRV156 strain or the PRV157 strain, which has a nonsense mutation at codon 7 of gG, did not secrete any detectable gG. Equal loading of samples was confirmed by an internal loading control (Fig. 4A, asterisk). These viruses were next tested for their ability to form plaques on monolayers of GBK cells.
FIG. 4.
A Bartha derivative with a nonsense mutation in gG is not affected in cell-to-cell spread. (A) Autoradiogram of tissue culture supernatants from infected cells pulse-labeled with [35S]methionine for 30 min at 2 h postinfection and chased in the presence of nonlabeled methionine for 1 h. Samples were analyzed by SDS-PAGE on an 8% gel. Asterisk denotes internal loading control. gG is secreted from cells in three predominant forms of approximately 180, 75, and 50 kDa. (B) Plaque morphologies of Bartha, PRV156, PRV157, and PRV156R on GBK cell monolayers. Forty-eight hours after infection, cells were fixed and stained with 0.5% methylene blue in 70% methanol.
Figure 4B shows the plaque morphologies of Bartha, PRV156, PRV157, and PRV156R. Bartha, PRV157, and PRV156R formed large nonsyncytial plaques, whereas PRV156 formed tiny nonsyncytial plaques. The important point is that elimination of gG expression through the introduction of a nonsense mutation did not result in the formation of tiny plaques on GBK cell monolayers. The effect of insertion into the gG locus of the Bartha strain was not due to elimination of gG expression alone.
Us3 mRNA levels are not affected by insertions in gG locus.
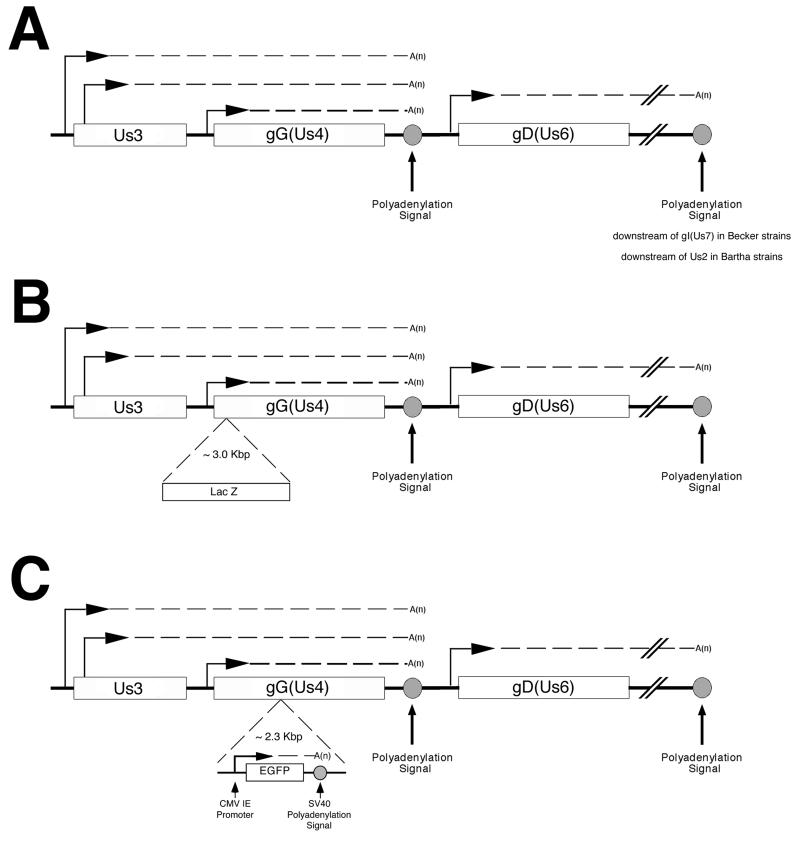
A cartoon of the gG locus and surrounding genes is shown in Fig. 5. The transcription of the Us3 and Us4 genes has been characterized in detail for both PRV and HSV, and the patterns of transcription from this region are strikingly similar between viruses (65, 82, 87).
FIG. 5.
Expected transcripts from Us3, gG (Us4), and gD (Us6) in Becker or Bartha (A), viruses with lacZ insertions (B), and viruses with egfp insertions (C). Horizontal arrows denote the position of a promoter. The Us3/Us4 transcripts are a 3′nested family of three mRNAs. The Us3 gene encodes two transcripts, designated Us3a and Us3b, that both terminate at a polyadenylation site located downstream of the gG (Us4) open reading frame. The slightly larger (by ∼150 bases) Us3a transcript represents about 5% of the total Us3 transcription, and the smaller Us3b transcript represents about 95% (83). In PRV, these mRNAs encode two different proteins using the same reading frame. It follows that both proteins have identical carboxy termini. The larger Us3a transcript encodes a protein with an additional 54 N-terminal amino acids. Both of these proteins are expressed in PRV-infected cells, as shown by Western blot analysis (83). It is unclear if the two products of the Us3 gene have different functions. The mRNA that directs the translation of gG is initiated just upstream of the gG (Us4) open reading frame and terminates at a polyadenylation site located immediately downstream of the gG (Us4) open reading frame. Located immediately downstream of gG (Us4) is the essential gD (Us6) gene. gD (Us6) belongs to a family of transcripts different from that of Us3 and gG. SV40, simian virus 40; CMV IE, cytomegalovirus immediate-early promoter.
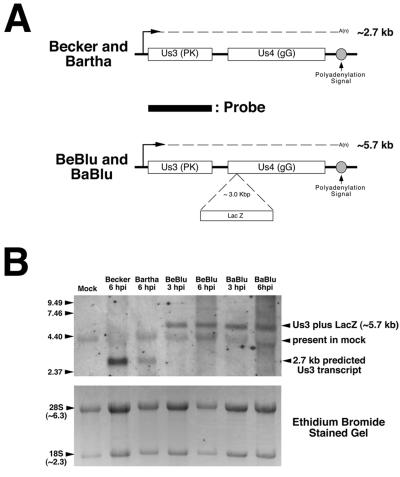
Because the tiny-nonsyncytial-plaque phenotype of Bartha strains with insertions in gG is not due to disruption of gG function, we next studied the effect of these insertions on Us3 transcripts. To do this, we examined the steady-state levels of Us3 mRNA 3 and 6 h after infection (Fig. 6). The probe used was the Us3b open reading frame. The location of the probe relative to the predicted Us3 transcripts from parental and lacZ insertion viruses is shown in Fig. 6A. At 6 h postinfection the Us3 probe hybridized to a 2.7-kb RNA isolated from cells infected with Becker and Bartha (Fig. 6B). When RNA isolated from Becker-Blu- and Bartha-Blu-infected cells at 3 and 6 h postinfection was analyzed by Northern blotting, using the Us3b probe described above, an expected 5.7-kb RNA was detected. The relative levels of Us3 RNA detected in Becker- and Bartha-infected cells versus those in Becker-Blu- and Bartha-Blu-infected cells were not significantly different. From these data we conclude that insertion of lacZ into the gG locus of Blu strains does not markedly affect Us3 transcript levels. We next examined the levels of Us3 protein in infected cells.
FIG. 6.
Analysis of Us3 transcription in PRV strains containing lacZ insertions in gG. (A) Cartoon illustrating the predicted size of Us3 transcripts from parental and lacZ insertion viruses and the location of the double-stranded DNA probe used to detect Us3 RNA by Northern blotting. Only the Us3 transcript is shown. (B) Northern blot of total RNA isolated from MDBK cells infected with Becker, Bartha, Becker-Blu (BeBlu), or Bartha-Blu (BaBlu) at 3 and 6 h postinfection (top panel). The blot was hybridized to a biotinylated double-stranded DNA probe corresponding to the Us3b open reading frame and hybridizing RNAs detected, as described in Materials and Methods. The positions of RNA size markers (in kilobases) are shown on the left of the gel. The bottom panel shows the ethidium bromide-stained gel prior to transfer of RNA to the blot and is meant to serve as a loading control. hpi, hours postinfection.
Us3 protein expression is substantially reduced in PRV strains with insertions in gG.
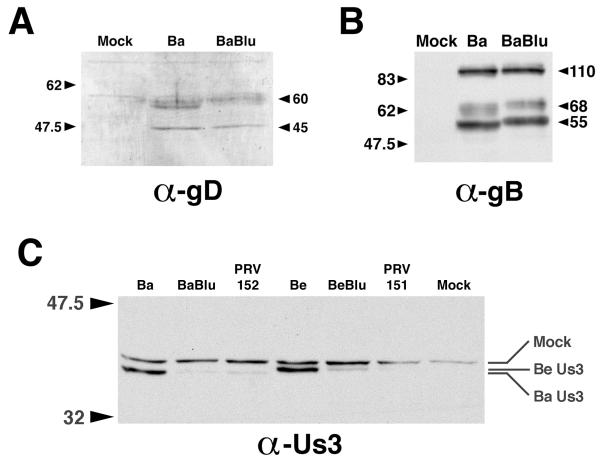
Protein extracts were prepared from PRV-infected MDBK cells at 6 h postinfection and were run on an SDS–10% PAGE gel. Protein in gels was transferred to polyvinylidene difluoride membranes and was probed using antiserum raised against gD, gB, or Us3 (Fig. 7). Insertions in gG had no effect on the steady-state levels of either gD or gB, as evidenced by the fact that similar amounts of protein were detected in Bartha- and Bartha-Blu-infected cell extracts (Fig. 7A and B). By contrast, steady-state levels of Us3 were significantly reduced in PRV strains that had either lacZ or egfp insertions in gG, regardless of whether the strain was derived from Becker or Bartha (Fig. 7C). Curiously, the relative mobility of Us3 identified in Becker- versus Bartha-derived strains differed in that Bartha's Us3 migrated more rapidly in the SDS-PAGE gel. The significance of this finding is under investigation. Results obtained with infected PK15 cell extracts were indistinguishable from those obtained with MDBK cells.
FIG. 7.
Reduced Us3 expression in cells infected with PRV strains that have insertions in gG. (A) Western blot of proteins extracted from MDBK cells (Mock) or MDBK cells infected with Bartha (Ba) or Bartha-Blu (BaBlu) probed with rabbit polyclonal antisera to PRV gD. (B) Western blot of proteins extracted from MDBK cells (Mock) or MDBK cells infected with Bartha (Ba) or Bartha-Blu (BaBlu) probed with goat polyclonal antisera to PRV gB. (C) Western blot of proteins extracted from MDBK cells (Mock) or MDBK cells infected with Bartha (Ba), Bartha-Blu (BaBlu), PRV152, Becker (Be), Becker-Blu (BeBlu), or PRV151, probed with an affinity-purified rabbit polyclonal antiserum raised against a synthetic peptide corresponding to the carboxy terminus of Us3. All infected cell extracts were prepared at 6 h postinfection. The positions of protein size markers (in kilodaltons) are shown to the left of the gels.
Relationship between gE/gI and insertions in gG in spread of infection between cells.
The data so far suggest that Us3 functions in the spread of viral infection between MDBK or GBK cells. gE and gI also participate in the cell-to-cell spread of infection in MDBK and GBK cells. To investigate the relationship between the role of gE/gI versus that of Us3 in cell-to-cell spread, we first examined the effect of insertions in gG in viruses that encode gE and gI. PRV158 is a derivative of the Bartha strain in which the deletion of gI, gE, Us9, and Us2 has been repaired with Becker sequences. PRV159 is a derivative of PRV158 that has an egfp insertion in the gG locus. Plaque morphologies of Becker, PRV151, PRV158, and PRV159 were examined on monolayers of MDBK cells (Fig. 8A). Restoration of the gI, gE, Us9, and Us2 genes in the Bartha strain does not fully rescue plaque size on MDBK cells, as evidenced by the observation that PRV158 plaques are considerably smaller than plaques formed by Becker on MDBK cells. This data suggest that other mutations in the Bartha strain contribute to its small plaque size on MDBK cells. Insertion of egfp into these strains causes a modest reduction in plaque size (compare Becker versus PRV151 and PRV158 versus PRV159, Fig. 8A). These data indicate that expression of gE/gI, Us9, and Us2 in the Bartha strain can overcome the effects of an insertion in gG (compare PRV159, Fig. 8A, to PRV152, Fig. 8B).
FIG. 8.
Plaque morphologies of PRV strains with insertions in gG in the presence or absence of gE and gI. Forty-eight hours after infection with the indicated virus strain, MDBK cells were either fixed and stained with 0.5% methylene blue in 70% methanol or examined by fluorescence microscopy for EGFP expression. (A) egfp insertions in gG (Us4) in the presence of gE and gI. (B) egfp insertions in gG (Us4) in the absence of gE and/or gI.
Next, we examined the effects of egfp insertions in strains that lack gE and/or gI (Fig. 8B). PRV99 is a Becker derivative with deletions of gE and gI, and PV99Gr is a PRV99 derivative with an egfp insertion. Insertion of egfp into gG had a modest effect on plaque size on MDBK cells. By contrast, an egfp insertion in the Bartha strain (PRV152) had a striking affect on plaque size. The M201 strain, a derivative of the virulent wild-type strain NIA-3 that has a deletion of gE, makes plaques on MDBK cells similar in size to those formed by Bartha. An egfp insertion was introduced into M201 to construct PRV155. PRV155 forms tiny plaques on MDBK cells similar to those formed by PRV152. These data indicate that, in the absence of gE or gI, the Becker strain can compensate for insertions in the gG locus and spread from cell to cell efficiently, whereas the Bartha and NIA-3 strains cannot.
DISCUSSION
We report that insertions in the gG locus of PRV can affect the spread of virus infection from cell to cell in a cell-type-specific manner. Interestingly, this phenotype was not due to loss of gG expression, because a mutant with a nonsense mutation in the gG signal sequence had no spread defect. We then noted that Us3 protein expression was dramatically reduced in PRV strains with either lacZ or egfp insertions in gG. By contrast, the gD gene, located immediately downstream of gG and belonging to a family of transcripts different from that of Us3 and gG, was not affected by insertions in the gG gene. Analysis of mRNA synthesized in cells infected with gG insertion mutants indicated that, although Us3 transcript size increased as expected, the steady-state amount of Us3 transcript was not markedly reduced. Apparently, insertions in the gG locus of PRV affect export of the altered Us3 message from the nucleus to the cytoplasm or result in inefficient translation of the altered Us3 transcripts. Further experiments will be required to determine the mechanism by which Us3 expression is attenuated in viruses with insertions in gG.
Early studies on HSV and PRV gG indicated that it was not required for efficient growth in culture; mutant viruses had no discernible growth defects (2, 44, 78, 84). Modest phenotypes were observed in vivo for HSV gG mutants, however. For example, an HSV-1 strain with a transposon insertion in gG was attenuated after intracranial inoculation of mice and an HSV-1 strain with a lacZ insertion in gG had reduced ability to invade the peripheral nervous system after inoculation of the mouse ear pinna (2, 84). By contrast, PRV strains with mutations in gG have had no discernible phenotypes in most model systems or in the natural host (27, 49, 75, 78; Card et al., unpublished). Recently, Kim and colleagues described differences in the virulence properties between the Bartha and the Bartha-Blu strain in the rat eye model. Bartha-Blu, which has a lacZ insertion in gG, took longer to kill animals, and infected animals had increased time until the appearance of symptoms (25). In the present study, we noted a significant difference in the virulence properties between the Bartha and Bartha-Blu strains in the chicken embryo eye model. Bartha-Blu had an LD50 that was 60-fold greater than that of the parental Bartha strain. Whether the in vivo phenotypes described here or previously are a result of a loss of gG function or reduced expression of Us3 remains to be clarified.
In other alphaherpesviruses, such as HSV and BHV-1, the Us3 message polyadenylation signal is located downstream of gG (39, 65). Accordingly, one might predict that insertions or deletions in the gG genes of these viruses might also affect the expression of Us3. Recently, two groups have described functions for gG using mutant viruses with insertions or deletions in the gG open reading frame (55, 56, 80). Tran and coworkers have described a gG mutant of HSV-1 that has a defect in the entry of virus into polarized MDCK cells from the apical surface (80). MDCK cells are extraordinarily refractory to infection compared to Vero or HEp-2 cells. Indeed, roughly 20,000-fold more virus is required to infect MDCK cells from the apical surface and greater than 100,000-fold more virus is required to infect these cells from the basolateral surface than are required for Vero cells, for example. Perhaps alternative or “suboptimal” virus entry pathways are functioning in the infection of MDCK cells by HSV. The gG mutant RAS104 used in the study by Tran et al. (80) is a gG deletion mutant. In light of the present study, it is possible that the deletion affects mRNA structure and stability. It may be that the upstream Us3 mRNA is affected by the gG deletion. Examination of Us3 expression in the RAS104 strain or the analysis of a HSV-1 strain with a nonsense mutation in gG would clarify this point. Nakamichi and colleagues have ascribed two functions to BHV-1 gG (55, 56). A mutant BHV-1 strain, BHV-1/TF9-5, has a small deletion in gG into which the PRV thymidine kinase gene was inserted. This virus has a defect in cell-to-cell spread on MDBK cells, which is similar to what we observed in this study (56). In addition, the BHV-1/TF9-5 strain fails to protect RK13 rabbit kidney cells from BHV-1-induced apoptosis (56). It may be that the effects on BHV-1 biology observed by these scientists are due at least in part to effects on Us3 expression. In the aforementioned studies, the gG insertion and deletion viruses were repaired, which resulted in complete reversion to the parental phenotype. However, if the gG insertion-deletion phenotype were repaired, any effect of these mutations on Us3 expression would also be repaired.
Smith and Enquist noted that insertion of either an EGFP expression cassette or bacterial artificial chromosome sequences into an intergenic region immediately downstream of the PRV Us9 gene markedly attenuated Us9 expression (72). Removal of these foreign sequences restored Us9 expression. These observations further emphasize the risk of unanticipated alterations in gene expression when inserting new sequences into the viral genome.
The Us3 gene encodes a serine/threonine kinase (18, 46). Us3 has been shown to play a role in the inhibition of HSV-induced apoptosis (19, 21, 23, 38). The specific functions of this molecule are not known; however, it is not essential for growth of PRV or HSV in many cultured cell lines (26, 44, 57). Interestingly, a PRV strain with a mutation in Us3 grows well in swine kidney cell lines but displays a strong reduction in growth in an immortalized swine B-cell line (26). This cell-type-specific defect is intriguing because the cell-to-cell spread defects of gG insertion mutants that we observe are evident only in some cell types. Moreover, the Us3 protein kinase is important for viral egress from the nucleus in specialized cell types (59, 83). For example, a PRV strain with an inactivated Us3 gene has a defect in the maturation of virions in porcine nasal mucosa explant cultures (59). Why do we see cell-type-specific effects of insertions in gG? It may be that cell types in which gG insertion mutants spread normally synthesize a kinase that can replace Us3. Alternatively, cells that make small plaques may produce an inhibitor of Us3 function that is absent in cells that show normal virus spread.
A major substrate of the HSV Us3 protein kinase is the membrane-associated UL34 protein (60). UL34 is an essential protein in HSV and PRV required for the primary envelopment of nucleocapsids at the inner nuclear membrane (29, 68). There is no evidence to support the idea that UL34 functions in cell-to-cell spread, however. Other viral molecules that are substrates for the HSV Us3 protein kinase are the UL12 alkaline nuclease and the protein encoded by the Us9 gene (9, 10). As observed with UL34 mutants, mutations in UL12 affect capsid envelopment at the nuclear membrane (70). Us9 has been shown to be required for the spread of PRV in certain neuronal circuits (7). However, previous studies have shown that deletion of Us9 has no influence on the ability of PRV to spread from cell to cell in GBK or MDBK cells. Our hypothesis is that one or more viral proteins required for cell-to-cell spread are modified by the Us3 protein kinase. It is likely that as-yet-unidentified viral and cellular substrates exist for Us3 and that one or more of these contribute to cell-to-cell spread.
It is well established that gE and gI participate in cell-to-cell spread of infection in polarized epithelial cells and in the nervous system (14, 15, 16, 24, 33, 47, 79, 85, 86). Recent results from Johnson and colleagues suggest that the carboxy terminus of gE is required for targeting virus to adherens junctions in polarized epithelial cells (24). Takahashi and coworkers have reported that the alphaherpesvirus receptor nectin-1α/HveC localizes to adherens junctions (76). Moreover, these workers have determined that interaction of the cytoplasmic tail of nectin-1α/HveC with afadin, which serves to link nectin-1α/HveC to the actin cytoskeleton, is required for efficient cell-to-cell spread of HSV-1 but not for virus entry (69). Taken together, these data strongly suggest that cell-to-cell spread of infection occurs at adherens junctions. We report here that expression of gE and gI is sufficient to overcome the effects of insertions in gG in the Bartha strain, whereas in the absence of gE and gI, both Bartha and NIA-3 are sensitive to insertions in gG (Fig. 8). One interpretation of these data is that an alternative, gE/gI-independent cell-to-cell-spread pathway exists in GBK and MDBK cells. It follows that insertions in gG disrupt this gE/gI-independent spread pathway and that Us3 activity is required for this mode of cell-to-cell spread. Alternatively, Us3 may also function at cell-to-cell spread at adherens junctions, but expression of gE and gI is sufficient to overcome any requirement for Us3 in GBK and MDBK cells. Examination of the effects of Us3 expression on the structure and function of adherens junctions may provide clues to the role of Us3 in cell-to-cell spread.
Because both the Bartha and NIA-3 strains are affected by insertions in gG, it may be that the Becker strain encodes a function that can compensate for this insertion. Alternatively, Bartha and NIA-3 may encode a function that perturbs cell-to-cell spread in the absence of Us3. Suppressor mutants of Bartha-Blu, PRV152, and PRV156 that tolerate insertions in gG arise at high frequency (about 1/2,000 plaques). These suppressors retain the gG insertion, do not have genomic reorganization at the Us3 or gG loci, synthesize reduced amounts of Us3 relative to the Bartha strain, and do not have enhanced replication kinetics (G. L. Demmin and B. W. Banfield, unpublished observations). Taken together, these observations suggest that the suppressor mutations reside in a second site and affect cell-to-cell spread, specifically. It may be that the Becker strain is genotypically similar to the suppressor mutants at this second site. Marker transfer experiments are under way to identify genes from the Becker strain and the suppressor mutants that compensate for insertions in gG. Our prediction is that genes with suppressor mutations will participate in a Us3-dependent cell-to-cell spread pathway.
ACKNOWLEDGMENTS
We thank Kevin Durand for expert technical assistance and Ilya Iofin and Alexander Costa for providing plasmids pII1 and pAC2, respectively. We thank L. Jacobs for kindly providing PRV strain M201. We acknowledge Robert Ho and Jean Schwarzbauer for generous use of microscopy equipment and Joe Goodhouse for help with confocal microscopy. We are grateful to Tony Minson for suggesting the possibility that insertions in the PRV gG gene might affect Us3 expression. Many thanks go to Renée Finnen and David Wentworth for critical reviews of the manuscript.
While at Princeton University, B.W.B. was supported by a postdoctoral fellowship from the Medical Research Council of Canada. This work was supported by NINDS grant 1R0133506 to L.W.E. and NIAID grant 1RO1AI48626 to B.W.B.
REFERENCES
- 1.Babic N, Klupp B, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 2.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 3.Banfield B W, Yap G S, Knapp A C, Enquist L W. A chicken embryo eye model for the analysis of alphaherpesvirus neuronal spread and virulence. J Virol. 1998;72:4580–4588. doi: 10.1128/jvi.72.6.4580-4588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartha A, Belák S, Benyeda J. Trypsin and heat resistance of some strains of the virus group. Acta Vet Hung. 1969;19:97–99. [PubMed] [Google Scholar]
- 5.Bennett L M, Timmins J G, Thomsen D R, Post L E. The processing of pseudorabies virus glycoprotein gX in infected cells and in an uninfected cell line. Virology. 1986;155:707–715. doi: 10.1016/0042-6822(86)90230-8. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Porat T, Kaplan A S. Synthesis of proteins in cells infected with herpesvirus. V. Viral glycoproteins. Virology. 1970;41:265–273. doi: 10.1016/0042-6822(70)90078-4. [DOI] [PubMed] [Google Scholar]
- 7.Brideau A D, Card J P, Enquist L W. Role of pseudorabies virus us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J Virol. 2000;74:834–845. doi: 10.1128/jvi.74.2.834-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daikoku T, Kurachi R, Tsurumi T, Nishiyama Y. Identification of a target protein of US3 protein kinase of herpes simplex virus type 2. J Gen Virol. 1994;75:2065–2068. doi: 10.1099/0022-1317-75-8-2065. [DOI] [PubMed] [Google Scholar]
- 10.Daikoku T, Yamashita Y, Tsurumi T, Nishiyama Y. The US3 protein kinase of herpes simplex virus type 2 is associated with phosphorylation of the UL12 alkaline nuclease in vitro. Arch Virol. 1995;140:1637–1644. doi: 10.1007/BF01322537. [DOI] [PubMed] [Google Scholar]
- 11.Davis-Poynter N, Bell S, Minson T, Browne H. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J Virol. 1994;68:7586–7590. doi: 10.1128/jvi.68.11.7586-7590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietz P, Klupp B G, Fuchs W, Kollner B, Weiland E, Mettenleiter T C. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J Virol. 2000;74:5083–5090. doi: 10.1128/jvi.74.11.5083-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkstra J M, Mettenleiter T C, Klupp B G. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology. 1997;237:113–122. doi: 10.1006/viro.1997.8766. [DOI] [PubMed] [Google Scholar]
- 14.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwell K S, Doering L C, Johnson D C. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol. 1995;69:7087–7098. doi: 10.1128/jvi.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1999;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 17.Foster T P, Kousoulas K G. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J Virol. 1999;73:8457–8468. doi: 10.1128/jvi.73.10.8457-8468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frame M C, Purves F C, McGeoch D J, Marsden H S, Leader D P. Identification of the herpes simplex virus protein kinase as the product of viral gene US3. J Gen Virol. 1987;68:2699–2704. doi: 10.1099/0022-1317-68-10-2699. [DOI] [PubMed] [Google Scholar]
- 19.Galvan V, Roizman B. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc Natl Acad Sci USA. 1998;95:3931–3936. doi: 10.1073/pnas.95.7.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haanes E J, Nelson C M, Soule C L, Goodman J L. The UL45 gene product is required for herpes simplex virus type 1 glycoprotein B-induced fusion. J Virol. 1994;68:5825–5834. doi: 10.1128/jvi.68.9.5825-5834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hata S, Koyama A H, Shiota H, Adachi A, Goshima F, Nishiyama Y. Antiapoptotic activity of herpes simplex virus type 2: the role of US3 protein kinase gene. Microbes Infect. 1999;1:601–607. doi: 10.1016/s1286-4579(99)80059-8. [DOI] [PubMed] [Google Scholar]
- 22.Hutchinson L, Roop-Beauchamp C, Johnson D C. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J Virol. 1995;69:4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jerome K R, Fox R, Chen Z, Sears A E, Lee H-Y, Corey L. Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3. J Virol. 1999;73:8950–8957. doi: 10.1128/jvi.73.11.8950-8957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson D C, Webb M, Wisner T W, Brunetti C. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J Virol. 2001;75:821–833. doi: 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J S, Enquist L W, Card J P. Circuit-specific coinfection of neurons in the rat central nervous system with two pseudorabies virus recombinants. J Virol. 1999;73:9521–9531. doi: 10.1128/jvi.73.11.9521-9531.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimman T G, De Wind N, De Bruin T, de Visser Y, Voermans J. Inactivation of glycoprotein gE and thymidine kinase or the US3-encoded protein kinase synergistically decreases in vivo replication of pseudorabies virus and the induction of protective immunity. Virology. 1994;205:511–518. doi: 10.1006/viro.1994.1672. [DOI] [PubMed] [Google Scholar]
- 27.Kimman T G, de Wind N, Oei-Lie N, Pol J M A, Berns A J M, Gielkens A L J. Contributions of single genes within the unique short region of Aujeszky's disease virus (suid herpes virus 1) to virulence, pathogenesis and immunogenicity. J Gen Virol. 1992;73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- 28.Klupp B G, Baumeister J, Dietz P, Granzow H, Mettenleiter T C. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J Virol. 1998;72:1949–1958. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klupp B G, Granzow H, Mettenleiter T C. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol. 2000;74:10063–10073. doi: 10.1128/jvi.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klupp B G, Lomniczi B, Visser N, Fuchs W, Mettenleiter T C. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology. 1995;212:466–473. doi: 10.1006/viro.1995.1504. [DOI] [PubMed] [Google Scholar]
- 31.Klupp B G, Nixdorf R, Mettenleiter T C. Pseudorabies virus glycoprotein M inhibits membrane fusion. J Virol. 2000;74:6760–6768. doi: 10.1128/jvi.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knapp A C, Enquist L W. Pseudorabies virus recombinants expressing functional virulence determinants gE and gI from bovine herpesvirus 1.1. J Virol. 1997;71:2731–2739. doi: 10.1128/jvi.71.4.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knapp A C, Husak P J, Enquist L W. The gE and gI homologs from two alphaherpesviruses have conserved and divergent neuroinvasive properties. J Virol. 1997;71:5820–5827. doi: 10.1128/jvi.71.8.5820-5827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kritas S K, Nauwynck H J, Pensaert M B. Dissemination of wild-type gC-, gE-, and gI-deleted mutants of Aujeszky's disease virus in the maxillary nerve and trigeminal ganglion of pigs after intranasal inoculation. J Gen Virol. 1995;76:2063–2066. doi: 10.1099/0022-1317-76-8-2063. [DOI] [PubMed] [Google Scholar]
- 35.Kritas S K, Pensaert M B, Mettenleiter T C. Invasion and spread of single glycoprotein deleted mutants of Aujeszky's disease virus (ADV) in the trigeminal nervous pathway of pigs after intranasal inoculation. Vet Microbiol. 1994;40:323–334. doi: 10.1016/0378-1135(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 36.Kritas S K, Pensaert M B, Mettenleiter T C. Role of envelope glycoproteins gI, gp63 and gIII in the invasion and spread of Aujeszky's disease virus in the olfactory nervous pathway of the pig. J Gen Virol. 1994;75:2319–2327. doi: 10.1099/0022-1317-75-9-2319. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Leopardi R, Van Sant C, Roizman B. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc Natl Acad Sci USA. 1997;94:7891–7896. doi: 10.1073/pnas.94.15.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung-Tack P, Audonnet J C, Riviere M. The complete DNA sequence and the genetic organization of the short unique region (US) of the bovine herpesvirus type 1 (ST strain) Virology. 1994;199:409–421. doi: 10.1006/viro.1994.1139. [DOI] [PubMed] [Google Scholar]
- 40.Lomniczi B, Blankenship M L, Ben-Porat T. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the genomes. J Virol. 1984;49:970–979. doi: 10.1128/jvi.49.3.970-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomniczi B, Kaplan A S, Ben-Porat T. Multiple defects in the genome of pseudorabies virus can affect virulence without detectably affecting replication in cell culture. Virology. 1987;161:181–189. doi: 10.1016/0042-6822(87)90184-x. [DOI] [PubMed] [Google Scholar]
- 42.Lomniczi B, Watanabe S, Ben-Porat T, Kaplan A S. Genetic basis of the neurovirulence of pseudorabies virus. J Virol. 1984;52:198–205. doi: 10.1128/jvi.52.1.198-205.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lomniczi B, Watanabe S, Ben-Porat T, Kaplan A S. Genome location and identification of functions defective in the Bartha vaccine strain of pseudorabies virus. J Virol. 1987;61:796–801. doi: 10.1128/jvi.61.3.796-801.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longnecker R, Roizman B. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science. 1987;236:573–576. doi: 10.1126/science.3033823. [DOI] [PubMed] [Google Scholar]
- 45.Maniatis T C, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 46.McGeoch D J, Davison A J. Alphaherpesviruses possess a gene homologous to the protein kinase gene family of eukaryotes and retroviruses. Nucleic Acids Res. 1986;14:1765–1777. doi: 10.1093/nar/14.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMillan T N, Johnson D C. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J Virol. 2001;75:1928–1940. doi: 10.1128/JVI.75.4.1928-1940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mettenleiter T C, Lukacs N, Rziha H J. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J Virol. 1985;56:307–311. doi: 10.1128/jvi.56.1.307-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mettenleiter T C, Rauh I. A glycoprotein gX-β-galactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J Virol Methods. 1990;30:55–66. doi: 10.1016/0166-0934(90)90043-f. [DOI] [PubMed] [Google Scholar]
- 50.Mettenleiter T C, Zsak L, Kaplan A S, Ben-Porat T, Lomniczi B. Role of a structural glycoprotein of pseudorabies in virus virulence. J Virol. 1987;61:4030–4032. doi: 10.1128/jvi.61.12.4030-4032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muggeridge M I. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J Gen Virol. 2000;81:2017–2027. doi: 10.1099/0022-1317-81-8-2017. [DOI] [PubMed] [Google Scholar]
- 52.Mulder W, Pol J, Kimman T, Kok G, Priem J, Peeters B. Glycoprotein D-negative pseudorabies virus can spread transneuronally via direct neuron-to-neuron transmission in its natural host, the pig, but not after additional inactivation of gE or gI. J Virol. 1996;70:2191–2200. doi: 10.1128/jvi.70.4.2191-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mulder W A, Jacobs L, Priem J, Kok G L, Wagenaar F, Kimman T G, Pol J M. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J Gen Virol. 1994;75:3095–3106. doi: 10.1099/0022-1317-75-11-3095. [DOI] [PubMed] [Google Scholar]
- 54.Mulder W A, Pol J M, Gruys E, Jacobs L, De Jong M C, Peeters B P, Kimman T G. Pseudorabies virus infections in pigs. Role of viral proteins in virulence, pathogenesis and transmission. Vet Res. 1997;28:1–17. [PubMed] [Google Scholar]
- 55.Nakamichi K, Kuroki D, Matsumoto Y, Otsuka H. Bovine herpesvirus 1 glycoprotein G is required for prevention of apoptosis and efficient viral growth in rabbit kidney cells. Virology. 2001;279:488–498. doi: 10.1006/viro.2000.0740. [DOI] [PubMed] [Google Scholar]
- 56.Nakamichi K, Ohara K, Kuroki D, Otsuka H. Bovine herpesvirus 1 glycoprotein G is required for viral growth by cell-to-cell infection. Virus Res. 2000;68:175–181. doi: 10.1016/s0168-1702(00)00171-4. [DOI] [PubMed] [Google Scholar]
- 57.Nishiyama Y, Yamada Y, Kurachi R, Daikoku T. Construction of a US3 lacZ insertion mutant of herpes simplex virus type 2 and characterization of its phenotype in vitro and in vivo. Virology. 1992;190:256–268. doi: 10.1016/0042-6822(92)91212-d. [DOI] [PubMed] [Google Scholar]
- 58.Peeters B, Pol J, Gielkens A, Moormann R. Envelope glycoprotein gp50 of pseudorabies virus is essential for virus entry but is not required for viral spread in mice. J Virol. 1993;67:170–177. doi: 10.1128/jvi.67.1.170-177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pol J M A, Wagenaar F, Gielkens A. Morphogenesis of three pseudorabies virus strains in porcine nasal mucosa. Intervirology. 1991;32:327–337. doi: 10.1159/000150217. [DOI] [PubMed] [Google Scholar]
- 60.Purves F C, Spector D, Roizman B. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J Virol. 1991;65:5757–5764. doi: 10.1128/jvi.65.11.5757-5764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajcani J, Kudelova M. Glycoprotein K of herpes simplex virus: a transmembrane protein encoded by the UL53 gene which regulates membrane fusion. Virus Genes. 1999;18:81–90. doi: 10.1023/a:1008025520655. [DOI] [PubMed] [Google Scholar]
- 62.Rauch D A, Rodriguez N, Roller R J. Mutations in herpes simplex virus glycoprotein D distinguish entry of free virus from cell-cell spread. J Virol. 2000;74:11437–11446. doi: 10.1128/jvi.74.24.11437-11446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rea T J, Timmins J G, Long G W, Post L E. Mapping and sequence of the gene for the pseudorabies virus glycoprotein which accumulates in the medium of infected cells. J Virol. 1985;54:21–29. doi: 10.1128/jvi.54.1.21-29.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 65.Rixon F J, McGeoch D J. Detailed analysis of the mRNAs mapping in the short unique region of herpes simplex virus type 1. Nucleic Acids Res. 1985;13:953–973. doi: 10.1093/nar/13.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robbins A K, Ryan J P, Whealy M E, Enquist L W. The gene encoding the gIII envelope protein of pseudorabies virus vaccine strain Bartha contains a mutation affecting protein localization. J Virol. 1989;63:250–258. doi: 10.1128/jvi.63.1.250-258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roizman B. Polykaryocytosis. Cold Spring Harbor Symp Quant Biol. 1962;27:327–342. doi: 10.1101/sqb.1962.027.001.031. [DOI] [PubMed] [Google Scholar]
- 68.Roller R J, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J Virol. 2000;74:117–129. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakisaka T, Taniguchi T, Nakanishi H, Takahashi K, Miyahara M, Ikeda W, Yokoyama S, Peng Y F, Yamanishi K, Takai Y. Requirement of interaction of nectin-1α/HveC with afadin for efficient cell-cell spread of herpes simplex virus type 1. J Virol. 2001;75:4734–4743. doi: 10.1128/JVI.75.10.4734-4743.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shao L, Rapp L M, Weller S K. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology. 1993;196:146–162. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- 71.Smith B N, Banfield B W, Smeraski C A, Wilcox C L, Dudek F E, Enquist L W, Pickard G E. Pseudorabies virus expressing enhanced green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc Natl Acad Sci USA. 2000;97:9264–9269. doi: 10.1073/pnas.97.16.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith G A, Enquist L W. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc Natl Acad Sci USA. 2000;97:4873–4878. doi: 10.1073/pnas.080502497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spear P G. Entry of alphaherpesvirus into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 74.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 75.Standish A, Enquist L W, Miselis R R, Schwaber J S. Dendritic morphology of cardiac related medullary neurons defined by circuit-specific infection by a recombinant pseudorabies virus expressing β-galactosidase. J Neurovirol. 1995;1:359–368. doi: 10.3109/13550289509111025. [DOI] [PubMed] [Google Scholar]
- 76.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Terry-Allison T, Montgomery R I, Whitbeck J C, Xu R, Cohen G H, Eisenberg R J, Spear P G. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J Virol. 1998;72:5802–5810. doi: 10.1128/jvi.72.7.5802-5810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomsen D R, Marchioli C C, Yancey R J, Jr, Post L E. Replication and virulence of pseudorabies virus mutants lacking glycoprotein gX. J Virol. 1987;61:229–232. doi: 10.1128/jvi.61.1.229-232.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tran L C, Kissner J M, Westerman L E, Sears A E. A herpes simplex virus 1 recombinant lacking the glycoprotein G coding sequences is defective in entry through apical surfaces of polarized epithelial cells in culture and in vivo. Proc Natl Acad Sci USA. 2000;97:1818–1822. doi: 10.1073/pnas.020510297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Zijl M, van der Gulden H, de Wind N, Gielkens A, Berns A. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. J Gen Virol. 1990;71:1747–1755. doi: 10.1099/0022-1317-71-8-1747. [DOI] [PubMed] [Google Scholar]
- 83.Wagenaar F, Pol J M, Peeters B, Gielkens A L, de Wind N, Kimman T G. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J Gen Virol. 1995;76:1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 84.Weber P C, Levine M, Glorioso J C. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science. 1987;236:576–579. doi: 10.1126/science.3033824. [DOI] [PubMed] [Google Scholar]
- 85.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wisner T, Brunetti C, Dingwell K, Johnson D C. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J Virol. 2000;74:2278–2287. doi: 10.1128/jvi.74.5.2278-2287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang G, Stevens R, Leader D P. The protein kinase encoded in the short unique region of pseudorabies virus: description of the gene and identification of its product in virions and in infected cells. J Gen Virol. 1990;71:1757–1765. doi: 10.1099/0022-1317-71-8-1757. [DOI] [PubMed] [Google Scholar]
- 88.Zsak L, Zuckermann F, Sugg N, Ben-Porat T. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J Virol. 1992;66:2316–2325. doi: 10.1128/jvi.66.4.2316-2325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]