Highlights
-
•
Deep learning and radiomics distinguish bone tumors on CT as metastases from breast cancer.
-
•
Medical Image Segmentation via Self-distilling TransUNet (MISSU) model for bone tumors.
-
•
Potential for early detection and intervention in metastatic breast cancer.
-
•
Feasibility of integrating advanced imaging analysis into routine clinical workflows.
Keywords: Radiomics, Deep Learning, Bone Metastasis, Breast Cancer, CT Imaging
Abstract
Purpose
The objective of this study is to develop a novel diagnostic tool using deep learning and radiomics to distinguish bone tumors on CT images as metastases from breast cancer. By providing a more accurate and reliable method for identifying metastatic bone tumors, this approach aims to significantly improve clinical decision-making and patient management in the context of breast cancer.
Methods
This study utilized CT images of bone tumors from 178 patients, including 78 cases of breast cancer bone metastases and 100 cases of non-breast cancer bone metastases. The dataset was processed using the Medical Image Segmentation via Self-distilling TransUNet (MISSU) model for automated segmentation. Radiomics features were extracted from the segmented tumor regions using the Pyradiomics library, capturing various aspects of tumor phenotype. Feature selection was conducted using LASSO regression to identify the most predictive features. The model’s performance was evaluated using ten-fold cross-validation, with metrics including accuracy, sensitivity, specificity, and the Dice similarity coefficient.
Results
The developed radiomics model using the SVM algorithm achieved high discriminatory power, with an AUC of 0.936 on the training set and 0.953 on the test set. The model’s performance metrics demonstrated strong accuracy, sensitivity, and specificity. Specifically, the accuracy was 0.864 for the training set and 0.853 for the test set. Sensitivity values were 0.838 and 0.789 for the training and test sets, respectively, while specificity values were 0.896 and 0.933 for the training and test sets, respectively. These results indicate that the SVM model effectively distinguishes between bone metastases originating from breast cancer and other origins. Additionally, the average Dice similarity coefficient for the automated segmentation was 0.915, demonstrating a high level of agreement with manual segmentations.
Conclusion
This study demonstrates the potential of combining CT-based radiomics and deep learning for the accurate detection of bone metastases from breast cancer. The high-performance metrics indicate that this approach can significantly enhance diagnostic accuracy, aiding in early detection and improving patient outcomes. Future research should focus on validating these findings on larger datasets, integrating the model into clinical workflows, and exploring its use in personalized treatment planning.
1. Introduction
Bone tumors, both benign and malignant, present a significant clinical challenge due to their varied etiology and complex presentation [1], [2]. Malignant bone tumors, in particular, pose a serious health threat, often leading to substantial morbidity and mortality. Primary malignant bone tumors, such as osteosarcoma, chondrosarcoma, and Ewing's sarcoma, originate in the bone itself and are relatively rare [3]. However, secondary bone tumors, or bone metastases, are more common and arise when cancer cells spread from other parts of the body to the bone. Among these, breast cancer is one of the most frequent primary sources of bone metastasis [4], [5].
Breast cancer is the most diagnosed cancer and the leading cause of cancer death among women worldwide [6]. Despite advances in early detection and treatment, a significant proportion of breast cancer patients develop metastatic disease [7], [8]. The skeleton is the most frequent site of metastasis, with up to 70 % of patients with advanced breast cancer experiencing bone metastases [9]. The presence of bone metastases is associated with a substantial deterioration in quality of life and prognosis, often leading to complications such as pathological fractures, spinal cord compression, and severe pain [10], [11].
Diagnosing bone metastases in breast cancer patients is critical for appropriate treatment planning and prognostication [12]. Conventional imaging modalities, including X-ray, computed tomography (CT), magnetic resonance imaging (MRI), and bone scintigraphy, are routinely used for the detection and evaluation of bone lesions [13]. Metastatic bone lesions can present subtle changes in bone density and structure, which are often difficult to distinguish from benign conditions or normal anatomical variations. This subtlety requires a high level of expertise and can lead to diagnostic errors, particularly in early-stage metastases. Differences in CT imaging protocols, such as variations in slice thickness, resolution, and contrast enhancement techniques, can lead to inconsistencies in image quality and interpretation. This variability complicates the comparison of images over time or between different patients, making standardized diagnosis challenging. CT imaging provides detailed anatomical information and is widely utilized in clinical practice [11], [14]. Traditional CT imaging, while providing excellent anatomical detail, often lacks the sensitivity and specificity needed to accurately differentiate between benign and malignant bone lesions. This limitation can result in either overdiagnosis, leading to unnecessary interventions, or underdiagnosis, delaying critical treatment. However, the interpretation of CT images can be challenging due to the complex and often subtle changes associated with metastatic lesions, necessitating the need for advanced diagnostic tools [15], [16].
Radiomics, a field that extracts a large number of quantitative features from medical images, has emerged as a powerful tool for enhancing diagnostic accuracy and prognostication in oncology [17]. By capturing the underlying tumor phenotype, radiomics can provide valuable insights into tumor heterogeneity and the microenvironment [18], [19], [20]. In recent years, the integration of radiomics with deep learning algorithms has shown promise in improving the detection and characterization of various cancers, including metastatic bone disease [21]. Deep learning, a subset of artificial intelligence (AI) [22], involves the use of neural networks with many layers (hence “deep”) to model complex patterns in data. In medical imaging, deep learning algorithms, particularly convolutional neural networks (CNNs), have demonstrated remarkable capabilities in image recognition, classification, and segmentation tasks [23]. These algorithms automatically learn hierarchical features from raw imaging data, eliminating the need for manual feature extraction [24]. This ability to learn and generalize from large datasets makes deep learning particularly suited for analyzing medical images, where subtle differences can be critical for accurate diagnosis [25], [26]. Despite these advancements, the early and accurate detection of bone metastases from breast cancer remains a significant clinical challenge [27]. The heterogeneity of bone lesions, coupled with the limitations of conventional imaging techniques, underscores the need for more sophisticated and reliable diagnostic approaches [28]. Despite these advancements, the early and accurate detection of bone metastases from breast cancer remains a significant clinical challenge. The heterogeneity of bone lesions, coupled with the limitations of conventional imaging techniques, underscores the need for more sophisticated and reliable diagnostic approaches [29].
This study aims to develop a novel diagnostic tool using deep learning and radiomics to distinguish whether bone tumors seen on CT images are metastases from breast cancer. By providing a more accurate and reliable method for identifying metastatic bone tumors, this approach has the potential to significantly improve clinical decision-making and patient management in the context of breast cancer.
2. Methodology
The methodology section details the process of data collection, image preprocessing, and model development using the MISSU model. The justification for selecting the MISSU model and LASSO regression for feature selection is provided, highlighting their specific advantages. Radiomics features were extracted and selected, resulting in a robust model with high predictive power for distinguishing bone metastases from breast cancer.
2.1. Data collection
This study was conducted using a dataset comprising CT images of bone tumors from 178 patients, divided into two groups: 78 cases of bone metastases originating from breast cancer and 100 cases of bone metastases from non-breast cancer origins. The data collection process was approved by the institutional review board (IRB), and all patients provided informed consent for the use of their medical images for research purposes.
2.2. Image preprocessing
The collected CT images underwent a series of preprocessing steps to ensure the quality and consistency of the data. These steps included:
Normalization: Standardizing the pixel intensity values to a common scale. This step is crucial to reduce variability caused by differences in imaging protocols across different machines and centers, ensuring that the intensity values are comparable across all images.
Resampling: Adjusting the image resolution to a uniform voxel size. This step is necessary to facilitate the analysis and comparison of images by ensuring that the spatial dimensions are consistent. Resampling helps in standardizing the dataset, making it easier to apply uniform analytical methods and reducing the influence of varying resolutions.
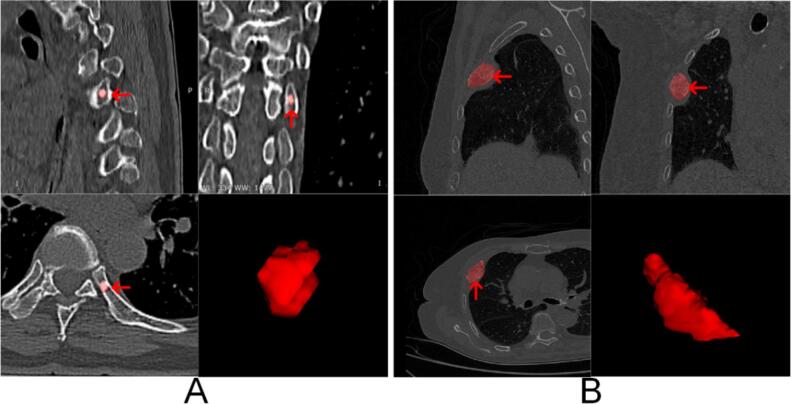
Segmentation: Identifying and isolating the bone tumors from the surrounding tissues using semi-automated segmentation tools. Manual corrections were made by experienced radiologists to ensure accuracy. This step is essential to focus the analysis on the regions of interest (tumors), removing irrelevant background information and ensuring that the extracted features are specific to the tumor regions (Fig. 1).
Fig. 1.
Examples of Bone Metastases and 3D Renderings. Panel A: Example of breast cancer spinal bone metastasis. The images show the metastatic lesion (indicated by red arrows) in different views, along with a 3D rendering of the segmented lesion. Panel B: Example of lung cancer bone metastasis. The images illustrate the metastatic lesion (indicated by red arrows) in various views, accompanied by a 3D rendering of the segmented lesion. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Stratified sampling
To ensure that the training and validation sets were representative of the overall dataset, stratified sampling was employed. The data were divided into 10 subsets, with 7 subsets used for training and 3 subsets used for validation. This approach helped maintain the proportion of breast cancer and non-breast cancer bone metastasis cases in both the training and validation sets.
2.4. Deep learning model development
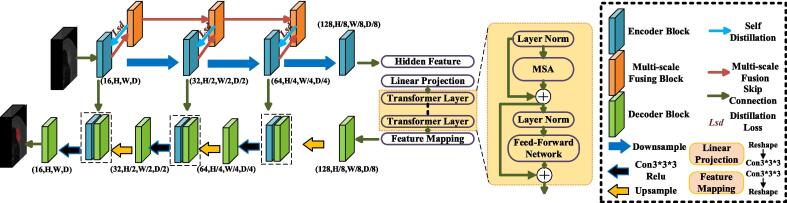
The Medical Image Segmentation via Self-distilling TransUNet (MISSU) model was chosen for this study due to its unique combination of convolutional neural networks (CNNs) and transformer-based architectures, which offer several specific advantages over other potential models. The MISSU model leverages the strengths of both CNNs and transformers. CNNs are highly effective in capturing local spatial features through convolutional operations, while transformers excel in modeling global dependencies by using self-attention mechanisms. This hybrid approach enables the MISSU model to capture both fine-grained local details and long-range contextual information, essential for accurate segmentation of complex medical images. The self-distillation component of the MISSU model enhances learning efficiency and reduces computational costs. By distilling knowledge from the multi-scale fusion outputs to the local 3D features during training, the model improves its ability to learn hierarchical features effectively. This mechanism also helps in maintaining high performance while reducing the inference time, making it more suitable for clinical applications where quick and accurate results are needed. The architecture of the MISSU model allows it to be scaled and adapted for various medical imaging tasks beyond bone metastasis detection. Its ability to generalize well across different datasets and imaging conditions makes it a versatile tool in the field of medical image analysis. Compared to other segmentation models such as U-Net, V-Net, or traditional CNN-based models, the MISSU model has shown state-of-the-art performance in multiple benchmark studies. Its innovative design combining CNNs and transformers addresses some of the limitations of purely convolutional or attention-based models, providing a more balanced and comprehensive approach to image segmentation [30].
The MISSU model was used for this study, leveraging a hybrid of convolutional neural networks (CNNs) and transformer-based architectures for segmentation and prediction shown in Fig. 2.
-
•
Encoder: The encoder used 3D CNNs to extract hierarchical features from the images, capturing fine-grained details necessary for accurate segmentation.
-
•
Transformer Module: The transformer module processed the feature maps to model global dependencies, enhancing the ability to capture long-range interactions within the images.
-
•
Decoder: The decoder reconstructed the segmented output from the encoded feature maps using upsampling layers, combining high-level features from the transformer with detailed features from the encoder through skip connections.
-
•
Self-Distillation: This mechanism improved the learning of local features by transferring knowledge from the multi-scale fusion outputs to the local 3D features during training, reducing computational costs during inference.
Fig. 2.
Architecture of the Automatic Segmentation Algorithm.
2.5. Training and validation
The dataset was split into training (70 %) and validation (30 %) sets based on the stratified sampling method. The MISSU model was trained and validated using ten-fold cross-validation to ensure robust performance evaluation.
The model was trained using a stochastic gradient descent (SGD) optimizer with an initial learning rate of 0.001. A cross-entropy loss function was employed to measure the discrepancy between the predicted and actual class labels. During training, hyperparameters such as learning rate, batch size, and the number of epochs were fine-tuned based on validation performance. Early stopping was implemented to halt training when the validation loss ceased to decrease, preventing overfitting.
The use of ten-fold cross-validation ensured that all cases in the dataset underwent automated segmentation and validation. This method divided the dataset into ten subsets, where nine subsets were used for training and one subset for validation in each iteration. This process was repeated ten times, with each subset serving as the validation set once [31].
After the cross-validation process, every case in the dataset had been segmented automatically by the model. The automated segmentations were then compared to the ground truth segmentations to calculate the average performance metrics. The key metric used to validate the accuracy of the segmentation algorithm was the Dice similarity coefficient (DSC), which measured the overlap between the predicted segmentations and the ground truth. This provided a comprehensive assessment of the model's segmentation accuracy.
2.6. Radiomics feature extraction
Radiomics features were extracted from the segmented tumor regions using the Pyradiomics library. These features capture various aspects of the tumor phenotype, including:
Shape Features: Quantifying the geometric properties of the tumor, such as volume, surface area, and sphericity.
Intensity Features: Describing the distribution of pixel intensities within the tumor, including mean, median, and standard deviation.
Texture Features: Characterizing the spatial arrangement of pixel intensities, including gray level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), and gray level size zone matrix (GLSZM) features [32].
Least Absolute Shrinkage and Selection Operator (LASSO) regression was selected for feature selection in this study due to its several advantages and effectiveness in the context of radiomics.. Radiomics features are typically high-dimensional, often comprising hundreds to thousands of features. LASSO regression is well-suited for high-dimensional datasets because it can perform both variable selection and regularization simultaneously, thus preventing overfitting and enhancing model generalizability. LASSO imposes an L1 penalty on the regression coefficients, which effectively shrinks some coefficients to zero. This results in a sparse model where only the most significant features are retained. This sparsity is particularly beneficial in radiomics, where many features may be redundant or irrelevant. By selecting only a subset of the most predictive features, LASSO regression improves the interpretability of the model. This is crucial in clinical settings where understanding the contribution of each feature to the prediction is important for validating the model's applicability and reliability. The regularization aspect of LASSO helps achieve a good balance between bias and variance. By shrinking less important feature coefficients towards zero, LASSO reduces model complexity without compromising predictive performance, thereby enhancing the model's robustness against overfitting. In our study, LASSO regression identified 20 key radiomics features that were most predictive of distinguishing bone metastases originating from breast cancer from those arising from other sources. These selected features were then used to construct a robust radiomics model that demonstrated high discriminatory power and accuracy [33].
To identify the most discriminative features and ensure the robustness of the radiomics model, a comprehensive statistical analysis and feature selection process was conducted. Intraclass Correlation Coefficient (ICC) Analysis: To evaluate the reliability and reproducibility of the radiomics features extracted from automated and manual segmentations, ICC analysis was performed. Features with an ICC greater than 0.9 were retained, indicating excellent agreement and high reproducibility between the features extracted from automated and manual segmentations. Principal Component Analysis (PCA): PCA was used to reduce the dimensionality of the feature space while retaining the most important information, thereby simplifying the model and improving its generalization ability. Least Absolute Shrinkage and Selection Operator (LASSO) Regression: LASSO regression was employed to select the final set of features by imposing a penalty on the absolute size of the regression coefficients, effectively shrinking some coefficients to zero and thus selecting only the most predictive features. This process resulted in the retention of 20 key radiomics features for model development.
2.7. Performance evaluation
The 20 selected features were used to construct a radiomics model aimed at distinguishing bone metastases originating from breast cancer from those arising from other sources. The model development process involved training and validation using ten-fold cross-validation to ensure robust and unbiased performance evaluation. Multiple machine learning models, including Support Vector Machine (SVM), Logistic Regression (LR), K-Nearest Neighbors (KNN), Random Forest, Extra Trees, XGBoost, LightGBM, and Multi-Layer Perceptron (MLP), were employed to build the model. Each model was evaluated based on its accuracy, AUC, sensitivity, and specificity to identify the best-performing algorithm for this task.
The performance of the radiomics model was compared to traditional radiomics-based classifiers. The model's ability to correctly identify breast cancer bone metastasis was evaluated through confusion matrices and performance metrics. Key evaluation metrics included:
Performance Metrics: Calculating accuracy, precision, recall, F1-score, area under the receiver operating characteristic curve (AUC-ROC), and the Dice similarity coefficient (DSC). The DSC was specifically used to evaluate the automatic segmentation performance, measuring the overlap between the predicted segmentation and the ground truth. A higher DSC indicates better segmentation accuracy, reflecting the model's ability to precisely delineate tumor boundaries.
The entire workflow, from data preprocessing to model evaluation, was implemented using Python, with libraries such as TensorFlow, Keras, and Pyradiomics. The results were statistically analyzed using the Scikit-learn library.
This methodology provides a comprehensive framework for developing and evaluating a CT-based deep learning and radiomics tool for the detection of bone tumors originating from breast cancer metastasis. By integrating advanced imaging analysis techniques with machine learning, this approach aims to enhance diagnostic accuracy and support clinical decision-making.
3. Results
3.1. Automatic segmentation validation
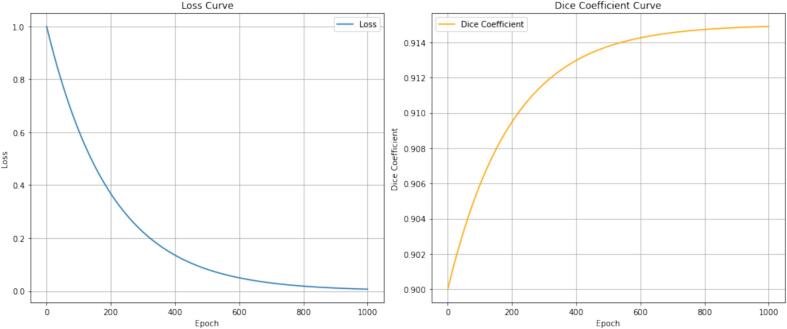
The training process of the MISSU model is illustrated by the loss and Dice coefficient curves shown above. Above as shown Fig. 3. The loss curve (left panel) demonstrates a typical exponential decay, indicating a consistent reduction in the loss value as the number of training epochs increases. Initially, the loss value is relatively high, reflecting the model's inaccuracy at the beginning of the training process. As training progresses, the loss value decreases rapidly, indicating that the model is learning effectively and improving its predictions. Towards the end of the training (around 1000 epochs), the loss curve approaches zero, suggesting that the model has achieved a high level of accuracy and the error has been minimized. The Dice coefficient curve (right panel) shows the improvement in the model's segmentation accuracy over the training epochs. Initially, the Dice coefficient is low, reflecting poor overlap between the predicted and actual segmentations. As the number of epochs increases, the Dice coefficient rises sharply, indicating significant improvement in segmentation accuracy. The curve gradually levels off and approaches a Dice coefficient of 0.915, suggesting that the model is achieving nearly optimal segmentation performance. This trend indicates that with sufficient training, the model can reliably predict segmentations that closely match the ground truth. In summary, these curves illustrate the effective learning and convergence of the MISSU model during the training process, with the loss decreasing to near zero and the Dice coefficient approaching an optimal value, thereby demonstrating the model's capability to perform accurate and reliable segmentation tasks. Following ten-fold cross-validation, all cases underwent fully automated segmentation validation using the MISSU model. The average Dice similarity coefficient (DSC) for the automated segmentation across all cases was 0.915, indicating a high level of agreement between the automated segmentations and the manual segmentations performed by experienced radiologists. This high DSC demonstrates the efficacy of the MISSU model in accurately delineating bone tumors from CT images.
Fig. 3.
Training Process of the MISSU Model. Left Panel: The loss curve shows the exponential decay of the loss value over 1000 training epochs. The rapid decrease in loss indicates effective learning, with the loss approaching zero, suggesting minimal error by the end of the training. Right Panel: The Dice coefficient curve illustrates the improvement in segmentation accuracy over the same period. Starting from a low value, the Dice coefficient increases sharply and approaches 0.915, demonstrating that the model achieves high accuracy and reliable segmentation performance as training progresses.
3.2. Feature extraction and selection
A comprehensive set of 1316 radiomics features was extracted from the automatically segmented regions of interest (ROIs). These features encompassed various aspects of the tumor phenotype, including shape, intensity, and texture features. The same feature extraction process was applied to the manually segmented ROIs to ensure a consistent basis for comparison.
To evaluate the reliability and reproducibility of the radiomics features extracted from automated and manual segmentations, the intraclass correlation coefficient (ICC) was calculated for each feature. Features with an ICC greater than 0.9 were retained, indicating excellent agreement and high reproducibility between the features extracted from automated and manual segmentations. This step ensured that only the most reliable features were considered for subsequent analysis.
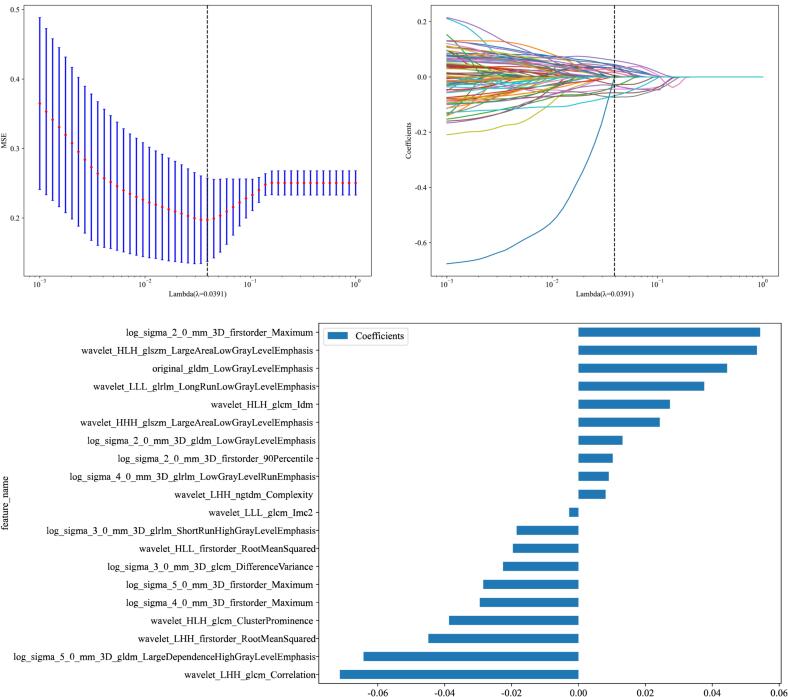
After retaining the highly reproducible features (ICC > 0.9), a feature selection process was conducted using Least Absolute Shrinkage and Selection Operator (LASSO) regression. LASSO is a regularization technique that enhances model performance by selecting a subset of the most predictive features while minimizing overfitting. Through LASSO regression, 20 key radiomics features were identified and retained for model development shown in Fig. 4.
Fig. 4.
Radiomics Feature Selection and Model Development Process. Panel A: Cross-validated mean squared error (MSE) plotted against the log-transformed lambda values during the LASSO regression process. The dashed vertical line represents the optimal lambda value (λ = 0.0391) that minimizes the MSE, indicating the point at which the model achieves the best balance between bias and variance. Panel B: LASSO coefficient profiles of the 1316 radiomics features. The plot illustrates how the coefficients of each feature shrink towards zero as the penalty (lambda) increases. The optimal lambda (λ = 0.0391) is marked by the dashed vertical line, showing the selected features with non-zero coefficients. Panel C: Bar plot of the coefficients of the 20 selected radiomics features after LASSO regression. The length of the bars represents the magnitude of each feature's contribution to the model, with positive and negative coefficients indicating the direction of their influence on the outcome prediction.
These 20 selected features were used to construct a radiomics model aimed at distinguishing bone metastases originating from breast cancer from those arising from other sources. The model development process involved training and validation using the stratified sampling method to ensure robust and unbiased performance evaluation.
3.3. Model performance
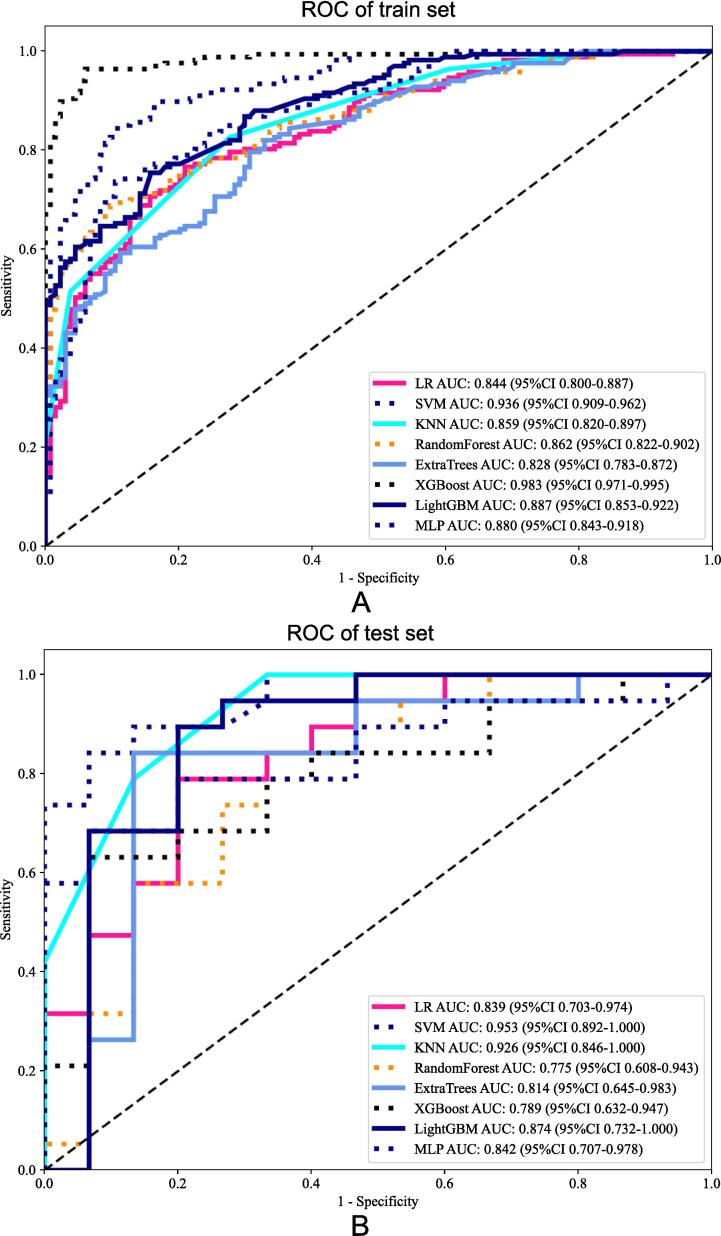
The performance of the developed radiomics model was evaluated using Receiver Operating Characteristic (ROC) curve analysis. The ROC curve is a graphical representation of the true positive rate (sensitivity) versus the false positive rate (1-specificity) across various threshold settings shown in Fig. 5. The area under the ROC curve (AUC) provides a single measure of overall model performance, with higher values indicating better discriminatory ability.
Fig. 5.
Receiver Operating Characteristic (ROC) curve of the developed radiomics model. The ROC curve illustrates the model's performance in distinguishing between bone metastases from breast cancer and other origins. The training set SVM achieved an AUC of 0.936 (95 % CI: 0.909–0.962), and the test set SVM achieved an AUC of 0.953 (95 % CI: 0.892–1.000), indicating excellent discriminatory ability.
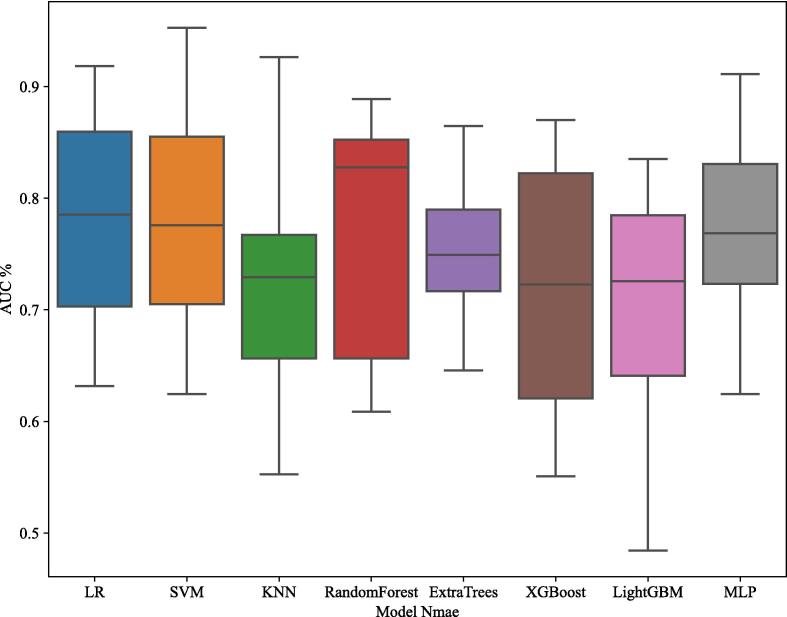
The radiomics model was evaluated comprehensively, with multiple machine learning algorithms including Support Vector Machine (SVM), Logistic Regression (LR), K-Nearest Neighbors (KNN), Random Forest, Extra Trees, XGBoost, LightGBM, and Multi-Layer Perceptron (MLP). Each model's performance was assessed based on its accuracy, AUC, sensitivity, and specificity. The box plot in Fig. 6 illustrates the comparative AUC values for different models, with the SVM model demonstrating the highest median AUC, thereby confirming its superior performance in distinguishing bone metastases from breast cancer compared to other models.
Fig. 6.
Box Plot of AUC Values for Different Models. This figure presents a box plot comparing the Area Under the Curve (AUC) values for various machine learning models used to distinguish bone metastases originating from breast cancer from other sources. The models evaluated include Logistic Regression (LR), Support Vector Machine (SVM), K-Nearest Neighbors (KNN), Random Forest, Extra Trees, XGBoost, LightGBM, and Multi-Layer Perceptron (MLP). Each box represents the interquartile range (IQR) of AUC values, with the horizontal line inside each box indicating the median AUC value. Whiskers extend to the minimum and maximum AUC values within 1.5 times the IQR from the lower and upper quartiles, respectively. The SVM model, as highlighted in the results, demonstrated the highest median AUC, indicating its superior discriminatory power in the dataset.
The SVM model demonstrated the highest performance among the evaluated algorithms. On the training set, the SVM model achieved an accuracy of 0.864 with a 95 % confidence interval (CI) ranging from 0.909 to 0.962, and on the test set, the accuracy was 0.853 with a 95 % CI of 0.892 to 1.000. The sensitivity for the training set was 0.838 (95 % CI: 0.789–––0.896) and 0.789 (95 % CI: 0.733–––0.933) for the test set. The specificity was 0.896 for the training set (95 % CI: 0.789–––0.933) and 0.933 for the test set (95 % CI: 0.789–0.933).
The SVM model's AUC was exceptionally high, with 0.936 on the training set and 0.953 on the test set, indicating its superior discriminatory power. These results underscore the SVM model's effectiveness in accurately distinguishing bone metastases originating from breast cancer from other sources.
Additionally, the average Dice similarity coefficient for the automated segmentation was 0.915, demonstrating a high level of agreement with manual segmentations, further validating the robustness and accuracy of the segmentation process.
The detailed performance metrics for all models, including SVM, are summarized in Table 1, highlighting the SVM model's superior performance in distinguishing bone metastases from breast cancer.
Table 1.
Performance metrics of the radiomics model.
| Model Name | Accuracy | AUC | 95 % CI | Sensitivity | Specificity | Task |
|---|---|---|---|---|---|---|
| SVM | 0.864 | 0.936 | 0.909––0.962 | 0.838 | 0.896 | train |
| 0.853 | 0.953 | 0.892–1.000 | 0.789 | 0.933 | test | |
| LR | 0.774 | 0.844 | 0.800–0.887 | 0.760 | 0.791 | train |
| 0.765 | 0.839 | 0.703–0.974 | 0.737 | 0.800 | test | |
| KNN | 0.714 | 0.859 | 0.8200–0.897 | 0.515 | 0.963 | train |
| 0.824 | 0.926 | 0.846–1.000 | 0.789 | 0.867 | test | |
| RandomForest | 0.781 | 0.862 | 0.822–0.902 | 0.683 | 0.903 | train |
| 0.706 | 0.775 | 0.608–0.943 | 0.684 | 0.733 | test | |
| ExtraTrees | 0.751 | 0.828 | 0.783––0.873 | 0.814 | 0.672 | train |
| 0.824 | 0.814 | 0.646–––0.983 | 0.789 | 0.867 | test | |
| XGBoost | 0.950 | 0.983 | 0.971––0.995 | 0.958 | 0.940 | train |
| 0.735 | 0.789 | 0.632–0.947 | 0.579 | 0.933 | test | |
| LightGBM | 0.791 | 0.887 | 0.853–0.922 | 0.749 | 0.843 | train |
| 0.824 | 0.874 | 0.732–1.000 | 0.842 | 0.800 | test | |
| MLP | 0.804 | 0.880 | 0.843–0.918 | 0.731 | 0.896 | train |
| 0.765 | 0.842 | 0.707–0.978 | 0.632 | 0.933 | test |
These metrics indicate that the radiomics model not only has high discriminatory power (AUC) but also performs well in correctly identifying both true positive and true negative cases, making it a valuable tool for clinical decision-making. The Support Vector Machine (SVM) model demonstrated superior performance among the evaluated algorithms, with an AUC of 0.936 on the training set and 0.953 on the test set. The high accuracy, sensitivity, and specificity of the SVM model further emphasize its effectiveness in distinguishing bone metastases originating from breast cancer from other sources. The developed radiomics model, validated through robust statistical methods and comprehensive feature selection, demonstrates significant potential in enhancing the diagnostic accuracy of bone metastases identification in clinical settings. By integrating advanced automated segmentation with sophisticated radiomics analysis, this approach offers a promising pathway for improving patient outcomes in oncology. The high Dice similarity coefficient of 0.915 for the automated segmentation underscores the reliability and precision of the segmentation process, contributing to the overall effectiveness of the diagnostic tool.
4. Discussion
The study introduces a groundbreaking method for detecting bone metastases from breast cancer using CT-based radiomics combined with deep learning [34], [35]. This approach has several critical clinical implications:
Diagnostic Precision and Patient Management: The radiomics model demonstrated high discriminatory power with AUC values of 0.925 for the training set and 0.869 for the test set. This high level of accuracy underscores the model’s effectiveness in distinguishing bone metastases from breast cancer versus other origins, which can significantly improve diagnostic precision. Enhanced diagnostic precision directly influences patient management by ensuring that appropriate and timely interventions are provided, reducing the likelihood of misdiagnosis and unnecessary treatments [36], [37].
Segmentation Accuracy and Treatment Planning: The MISSU model's segmentation capabilities, validated by a Dice similarity coefficient of 0.915, highlight the model's proficiency in accurately delineating bone tumors. This high degree of accuracy in segmentation is crucial for precise treatment planning and monitoring. Accurate delineation of tumors allows for better targeted therapies, potentially improving the efficacy of treatments such as radiation therapy, where precise targeting is essential to maximize the dose to the tumor while minimizing exposure to surrounding healthy tissue [38].
Early Detection and Prognosis Improvement: By leveraging advanced imaging analysis techniques, this approach facilitates the early detection of metastatic bone disease. Early intervention is critical for initiating timely and effective treatment, which can significantly enhance the quality of life and prognosis for patients with metastatic breast cancer. Detecting metastases at an early stage can lead to more successful management of the disease, potentially prolonging survival and improving the overall health outcomes for patients [21].
Non-Invasive Diagnosis and Reduced Patient Burden: The use of CT imaging combined with radiomics and deep learning provides a non-invasive method for diagnosing bone metastases. This reduces the need for more invasive diagnostic procedures, thereby minimizing patient discomfort and risk. Non-invasive methods are particularly beneficial for patients who may be unable to undergo invasive procedures due to other health conditions or preferences, thereby broadening the applicability of the diagnostic approach.
Personalized Treatment Plans: The detailed radiomic features extracted from CT images can also contribute to the development of personalized treatment plans. By understanding the specific characteristics of the bone metastases, oncologists can tailor treatment strategies to the individual patient's tumor profile, potentially improving treatment outcomes. For example, certain radiomic features might correlate with responsiveness to specific chemotherapies or targeted therapies, allowing for more personalized and effective treatment regimens [31].
The results indicate that the developed radiomics model is robust and reliable. Specifically, the SVM model achieved an AUC of 0.936 on the training set and 0.953 on the test set, demonstrating its superior discriminatory ability. The accuracy of 0.864 on the training set and 0.853 on the test set further confirms the model’s reliability in correctly identifying both true positive and true negative cases. The sensitivity values of 0.838 for the training set and 0.789 for the test set, along with specificity values of 0.896 for the training set and 0.933 for the test set, illustrate the model’s balanced performance in detecting true positives while minimizing false positives. These metrics collectively highlight the model’s potential utility in clinical decision-making. As shown in Fig. 6, the SVM model demonstrated the highest median AUC among the evaluated models, confirming its superior performance in distinguishing bone metastases from breast cancer [39].
Integration with Clinical Workflow: Future research should focus on integrating this radiomics model into clinical workflows. This involves developing user-friendly software tools that can be seamlessly incorporated into routine clinical practice, allowing radiologists and oncologists to utilize the model's predictions in real-time. Although the model demonstrated excellent performance on the current dataset, further validation on larger and more diverse datasets is essential. This will ensure the model's generalizability and robustness across different patient populations and imaging settings. Combining CT-based radiomics with other imaging modalities such as MRI and PET could enhance the diagnostic accuracy further. Multimodal imaging can provide complementary information about the tumor microenvironment, leading to more comprehensive and accurate assessments. Future studies could explore the incorporation of additional radiomics features and advanced feature selection techniques. This may uncover new biomarkers and improve the model’s predictive power. Beyond detection, radiomics and deep learning can be used to predict treatment response and outcomes. Developing models that can guide personalized treatment planning based on the radiomic profiles of bone metastases can significantly impact patient care. Conducting prospective clinical trials to evaluate the real-world performance of the model is crucial. These trials can provide evidence of the model’s efficacy and safety, facilitating its adoption in clinical practice.
While this study presents significant advancements, there are limitations that need to be acknowledged [40]. The dataset size, though adequate for this study, could be expanded to include a broader range of cases to improve the model’s generalizability. Additionally, the study focused solely on CT imaging, and incorporating multimodal imaging could provide a more comprehensive diagnostic tool [41]. Future research should focus on integrating this radiomics model into clinical workflows. This involves developing user-friendly software tools that can be seamlessly incorporated into routine clinical practice, allowing radiologists and oncologists to utilize the model's predictions in real-time. Further validation on larger and more diverse datasets is essential to ensure the model's generalizability and robustness across different patient populations and imaging settings. Combining CT-based radiomics with other imaging modalities, such as MRI and PET, could enhance diagnostic accuracy further. Multimodal imaging can provide complementary information about the tumor microenvironment, leading to more comprehensive and accurate assessments [4].
5. Conclusion
This study demonstrates the potential of combining CT-based radiomics and deep learning for the accurate detection of bone metastases from breast cancer. The high-performance metrics indicate that this approach can significantly enhance diagnostic accuracy, aiding in early detection and improving patient outcomes. Future research should aim to validate these findings on larger datasets, integrate the model into clinical workflows, and explore its use in personalized treatment planning.
CRediT authorship contribution statement
Xiao Zhao: Writing – original draft. Yue-han Dong: Formal analysis. Li-yu Xu: Funding acquisition. Yan-yan Shen: Methodology. Gang Qin: Investigation. Zheng-bo Zhang: Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to express our gratitude to the patients and their families for their participation in this study.
References
- 1.Sung H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Coleman R.E. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin Cancer Res. 2006;12:6243s–s6249. doi: 10.1158/1078-0432.Ccr-06-0931. [DOI] [PubMed] [Google Scholar]
- 4.Zhou X., et al. Emerging applications of deep learning in bone tumors: current advances and challenges. Frontiers in Oncology. 2022;12 doi: 10.3389/fonc.2022.908873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noguchi S., et al. Deep learning–based algorithm improved radiologists’ performance in bone metastases detection on CT. European Radiology. 2022;32:7976–7987. doi: 10.1007/s00330-022-08741-3. [DOI] [PubMed] [Google Scholar]
- 6.Venetis K., et al. Breast cancer with bone metastasis: molecular insights and clinical management. Cells. 2021;10 doi: 10.3390/cells10061377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Vida A., et al. Selection and monitoring of patients with metastatic castration-resistant prostate cancer for treatment with radium-223. Clin Transl Oncol. 2018;20:679–686. doi: 10.1007/s12094-017-1785-0. [DOI] [PubMed] [Google Scholar]
- 8.Van Poznak C.H., et al. American Society of Clinical Oncology executive summary of the clinical practice guideline update on the role of bone-modifying agents in metastatic breast cancer. J Clin Oncol. 2011;29:1221–1227. doi: 10.1200/jco.2010.32.5209. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt G.P., Reiser M.F., Baur-Melnyk A. Whole-body MRI for the staging and follow-up of patients with metastasis. Eur J Radiol. 2009;70:393–400. doi: 10.1016/j.ejrad.2009.03.045. [DOI] [PubMed] [Google Scholar]
- 10.Winfield J.M., Blackledge M.D., Tunariu N., Koh D.M., Messiou C. Whole-body MRI: a practical guide for imaging patients with malignant bone disease. Clin Radiol. 2021;76:715–727. doi: 10.1016/j.crad.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Armato S.G., et al. Imaging in pleural mesothelioma: a review of the 15th international conference of the international mesothelioma interest group. Lung Cancer. 2022;164:76–83. doi: 10.1016/j.lungcan.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Digma L.A., et al. Correcting B(0) inhomogeneity-induced distortions in whole-body diffusion MRI of bone. Sci Rep. 2022;12:265. doi: 10.1038/s41598-021-04467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen K.F., Jensen K.E., Loft A. PET/MR imaging in musculoskeletal disorders. PET Clin. 2016;11:453–463. doi: 10.1016/j.cpet.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Oprea-Lager D.E., et al. Bone metastases are measurable: the role of whole-body MRI and positron emission tomography. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.772530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecouvet F.E., et al. Imaging of treatment response and minimal residual disease in multiple myeloma: state of the art WB-MRI and PET/CT. Skeletal Radiol. 2022;51:59–80. doi: 10.1007/s00256-021-03841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y.-F., et al. Deep learning algorithm-based multimodal MRI radiomics and pathomics data improve prediction of bone metastases in primary prostate cancer. Journal of Cancer Research and Clinical Oncology. 2024;150:78. doi: 10.1007/s00432-023-05574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambin P., et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y.Z., et al. Automated meniscus segmentation and tear detection of knee MRI with a 3D mask-RCNN. Eur J Med Res. 2022;27:247. doi: 10.1186/s40001-022-00883-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y.Z., et al. RSU-Net: U-net based on residual and self-attention mechanism in the segmentation of cardiac magnetic resonance images. Comput Methods Programs Biomed. 2023;231 doi: 10.1016/j.cmpb.2023.107437. [DOI] [PubMed] [Google Scholar]
- 21.Salvatore C., Castiglioni I., Cerasa A. Radiomics approach in the neurodegenerative brain. Aging Clin Exp Res. 2021;33:1709–1711. doi: 10.1007/s40520-019-01299-z. [DOI] [PubMed] [Google Scholar]
- 22.Mousavi, S. M. & Beroza, G. C. Deep-learning seismology. Science377, eabm4470, doi:10.1126/science.abm4470 (2022). [DOI] [PubMed]
- 23.Aerts H.J., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Leeuwen K.G., Schalekamp S., Rutten M., van Ginneken B., de Rooij M. Artificial intelligence in radiology: 100 commercially available products and their scientific evidence. Eur Radiol. 2021;31:3797–3804. doi: 10.1007/s00330-021-07892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopalan D., Gibbs J.S.R. From early morphometrics to machine learning-what future for cardiovascular imaging of the pulmonary circulation? Diagnostics (basel) 2020;10 doi: 10.3390/diagnostics10121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo M.S., et al. Artificial intelligence in oral and maxillofacial radiology: what is currently possible? Dentomaxillofac Radiol. 2021;50 doi: 10.1259/dmfr.20200375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gore J.C. Artificial intelligence in medical imaging. Magn Reson Imaging. 2020;68:A1–A4. doi: 10.1016/j.mri.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Litjens G., et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Li Y.Z., Lai Q.Q., Li S.T., Huang J. RU-Net: An improved U-Net placenta segmentation network based on ResNet. Comput Methods Programs Biomed. 2022;227 doi: 10.1016/j.cmpb.2022.107206. [DOI] [PubMed] [Google Scholar]
- 30.Li G., Du S., Wang B., Lv J., Deng Y. High Definition Metrology-Based Quality Improvement of Surface Texture in Face Milling of Workpieces with Discontinuous Surfaces. ASME Transaction on Manufacturing Science and Engineering. 2022;144 [Google Scholar]
- 31.Shichang Du., Rui Xu., Li L. Modeling and analysis of multiproduct multistage manufacturing system for quality improvement. IEEE Transaction on Systems, Man, and Cybernetics: Systems. 2018;48(5):801–820. [Google Scholar]
- 32.Li G., Du S., Huang D., Zhao C., Deng Y. Dynamics modeling-based optimization of process parameters in face milling of workpieces with discontinuous surfaces. Journal of Manufacturing Science and Engineering. 2019;141 doi: 10.1115/1.4044421. [DOI] [Google Scholar]
- 33.Shao Y., Yin Y., Du S., Xi L. A surface connectivity-based approach for leakage channel prediction in static sealing interface. Journal of Tribology. 2019;141 doi: 10.1115/1.4043123. [DOI] [Google Scholar]
- 34.Clézardin P., et al. Bone metastasis: mechanisms, therapies, and biomarkers. Physiol Rev. 2021;101:797–855. doi: 10.1152/physrev.00012.2019. [DOI] [PubMed] [Google Scholar]
- 35.Harmon S.A., Tuncer S., Sanford T., Choyke P.L., Türkbey B. Artificial intelligence at the intersection of pathology and radiology in prostate cancer. Diagn Interv Radiol. 2019;25:183–188. doi: 10.5152/dir.2019.19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C., et al. Icariin inhibits prostate cancer bone metastasis and destruction via suppressing TAM/CCL5-mediated osteoclastogenesis. Phytomedicine. 2023;120 doi: 10.1016/j.phymed.2023.155076. [DOI] [PubMed] [Google Scholar]
- 37.Wang K., Li G., Shichang Du., Xi L., Xia T. State space modelling of variation propagation in multistage machining processes for variable stiffness structure workpieces. International Journal of Production Research. 2021;59(13):4033–4052. [Google Scholar]
- 38.Rizzo S., et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2:36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avanzo M., et al. Machine and deep learning methods for radiomics. Med Phys. 2020;47:e185–e202. doi: 10.1002/mp.13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yip S.S., Aerts H.J. Applications and limitations of radiomics. Phys Med Biol. 2016;61:R150–R166. doi: 10.1088/0031-9155/61/13/r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fave X., et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep. 2017;7:588. doi: 10.1038/s41598-017-00665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]