Abstract
The HEALthy Brain and Child Development (HBCD) Study, a multi-site prospective longitudinal cohort study, will examine human brain, cognitive, behavioral, social, and emotional development beginning prenatally and planned through early childhood. The acquisition of multimodal magnetic resonance-based brain development data is central to the study’s core protocol. However, application of Magnetic Resonance Imaging (MRI) methods in this population is complicated by technical challenges and difficulties of imaging in early life. Overcoming these challenges requires an innovative and harmonized approach, combining age-appropriate acquisition protocols together with specialized pediatric neuroimaging strategies. The HBCD MRI Working Group aimed to establish a core acquisition protocol for all 27 HBCD Study recruitment sites to measure brain structure, function, microstructure, and metabolites. Acquisition parameters of individual modalities have been matched across MRI scanner platforms for harmonized acquisitions and state-of-the-art technologies are employed to enable faster and motion-robust imaging. Here, we provide an overview of the HBCD MRI protocol, including decisions of individual modalities and preliminary data. The result will be an unparalleled resource for examining early neurodevelopment which enables the larger scientific community to assess normative trajectories from birth through childhood and to examine the genetic, biological, and environmental factors that help shape the developing brain.
Keywords: HBCD, Infant, MRI, MRS, Development, Protocol
1. Introduction
Across the first 10 years of life, the brain undergoes rapid and dynamic change (Alex et al., 2024; Bethlehem et al., 2022), providing the foundation for the brain’s intricate structural, functional, and metabolic architecture that underpins cognitive and behavioral development. Although the brain changes across the lifespan, the first decade of life corresponds to a period of remarkable growth, unparalleled plasticity, and unique sensitivity to one’s own genetic makeup, prenatal conditions, and experiential environments. These factors collectively exert both immediate and long-term influence on neurodevelopmental outcomes.
Magnetic resonance imaging (MRI) has been firmly established as a powerful tool for non-invasively examining the structural and functional organization of the brain and can provide key insights into the trajectory of complex changes that occur across the neurodevelopmental typical-to-atypical continuum and give rise to the variability in outcomes and abilities. Indeed, a growing number of large-scale neuroimaging studies, including the Human Connectome Project (HCP) (Glasser et al., 2016; Van Essen et al., 2013), Lifespan Human Connectome Projects in Development (HCP-D) and Aging (HCP-A) (Somerville et al., 2018; Harms et al., 2018; Bookheimer et al., 2019), Adolescent Brain and Cognitive Development (ABCD) Study (Casey et al., 2018), Developing Human Connectome Project (dHCP) (Makropoulos et al., 2018; Edwards et al., 2022), Baby Connectome Project (BCP) (Howell et al., 2019), and UK biobank (Miller et al., 2016) among others, have led to significant technical advancements in neuroimaging hardware, acquisition protocols, and analysis pipelines. These studies have produced an unprecedented data resource that has transformed our understanding of human brain structure, function, and connectivity. However, few longitudinal brain imaging studies have rigorously examined neurodevelopment using these leading-edge multimodal techniques throughout the window extending from birth through 10 years of age. The relative sparsity of neuroimaging data from a large ethnically and sociodemographically diverse cohort across this age range has limited the scientific community’s ability to discern how early life experiences and exposures, including prenatal exposure to drugs and alcohol, shape individual neurodevelopmental trajectories that ultimately promote or disrupt later childhood outcomes.
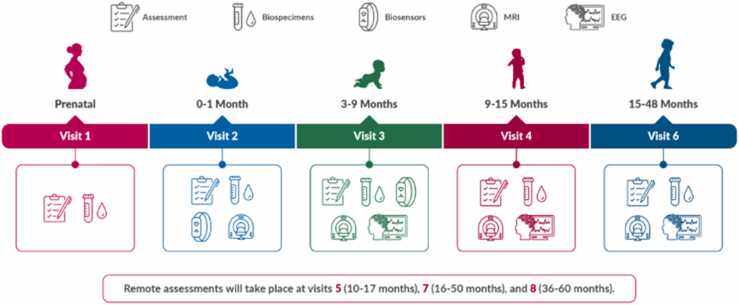
The HEALthy Brain and Child Development (HBCD) Study is a multi-site prospective longitudinal cohort study of human brain, cognitive, behavioral, social, and emotional development beginning prenatally and planned through middle childhood (Volkow et al., 2021). This landmark study will recruit a nationally-representative sample of over 7000 birth parents and children, enriched for prenatal substance exposure, with the primary aim of elucidating how biological and environmental factors influence long-term neurodevelopmental trajectories. Acquisition of longitudinal, state-of-the-art, multi-modal MRI data across infancy and childhood is a central component of the HBCD Study. This includes direct assessment of neurometabolites by magnetic resonance spectroscopy (MRS). The assessment schedule is represented in Fig. 1.
Fig. 1.
Visit schedule for the HBCD protocol. Of note, MRI scans will be collected at each in person assessment visit (visits 2, 3, 4, and 6). Of note, procedures for Visit 4 and beyond are only current as of the time of this manuscript submission and may change during piloting.
Here, we provide an overview of HBCD MRI and MRS procedures and preliminary data, detailing the acquisition protocols, justification for their selection, optimization for the infant brain, and harmonization across the scanner platforms at the 27 HBCD Study recruitment sites. We additionally describe the strategies and procedures deployed across HBCD Study sites for performing MRI in infants and children during natural, non-sedated sleep and outline the proposed HBCD processing and analysis workflows.
2. Materials and methods
The HBCD MRI protocol was directly influenced by recent large-scale neuroimaging initiatives, including HCP, BCP, and ABCD, while also needing to account for the unique challenges when imaging infants and children (Dean et al., 2014, Raschle et al., 2012, Spann et al., 2023). In particular, the NIH Funding Opportunity Announcement for the HBCD Study required the imaging protocol be developmentally sensitive and include the following: 1) multi-vendor (Siemens, Philips, General Electric [GE]) support of MRI scanners with a field strength of at least 3 Tesla; 2) T1-weighted (T1w) and T2-weighted (T2w) structural (sMRI) acquisitions; 3) functional MRI (fMRI) acquisition; 4) diffusion (dMRI) imaging acquisition; 5) MRS acquisition for the measurement of molecules involved in neuronal metabolism, neurotransmission, and oxidative stress; and 6) quantitative relaxometry. These requirements necessitated collaborative innovation. First, collecting data from each of these modalities in a single scanning session required balancing sequence optimization with the time allocated per sequence. Second, rapid neurodevelopmental changes during the first years of life lead to underlying changes in fundamental MR properties (e.g., relaxation times, diffusion and metabolite characteristics) (Lebel et al., 2019, Lebel and Deoni, 2018). These changes lead to differences in image contrast over time, even with the same MRI sequence (Paus et al., 2001). Third, the gold-standard approach for harmonization of sequences across vendors would involve within-infant scanning (across various ages). The logistical challenges of cross-vendor within-infant scanning precluded these tests. Together, the numerous challenges demanded careful consideration of all aspects of the imaging protocol, including scanner hardware, software, and sequence parameters.
To that end, the HBCD MRI Working Group (MRI WG) was formed and tasked with developing an MRI protocol that can be feasibly and reliably obtained in infants and children over the age range of the study and administered across the multiple participating sites. To address the specific requirements of the MRI protocol, five modality-specific sub-groups, including the Structural, Functional, Diffusion, Quantitative, and MRS sub-groups, were formed to devise and implement a strategy for each modality. Importantly, every MRI scan at every timepoint will be collected without the use of sedating medications. Thus, an additional specialized “Scanning Young Populations” sub-group was created to develop guidelines and procedures for non-sedated scanning of infants and children to facilitate consistent acquisition of high-quality, low motion data. A “Processing” workgroup was also formed to determine the best standards and practices to handle HBCD’s “Big Data” and ensure that the study efficiently facilitates reproducible research.
HBCD involves state-of-the-art Siemens, GE, and Philips 3 T scanners, including 19 sites with Siemens 3 T Prisma scanners, two sites with GE MR750 scanners, three sites with 3 T Philips MR7700 scanners, two sites with 3 T Philips Achieva CX dStream DDAS scanners, and one site with a 3 T Philips Ingenia Elition X scanner with a custom built gradient coil. Scanner software platforms across sites are aligned for each vendor, with all software upgrades conducted in a coordinated fashion. MRI data are collected using either 32- or 64-channel head receive coils based upon availability across data collection sites. Framewise Integrated Real-Time MRI Monitoring (FIRMM) software is used for structural (T1w/T2w), functional, and diffusion modalities. FIRMM calculates and displays real-time measures of head motion along with other quality metrics to the MRI technician across modalities during each scan session (Badke D′Andrea et al., 2022, Dosenbach et al., 2017, Fair et al., 2020), allowing technicians to make informed decisions on how to optimize scan sessions for time and required scan repeats. Following collection, all MRI data are transferred from the site to the HBCD Data Coordinating Center following well-established procedures using Flash-memory based Input/Output Network Appliances (FIONA) for quality control and processing. In what follows, we briefly characterize the efforts of the MRI WG for each modality, with the intent of generating more comprehensive descriptions of each modality in subsequent stand-alone publications.
The majority of pulse sequences used for the HBCD MRI protocol are research-specific and some require signing transfer agreements with the academic centers at which the sequences were developed. While our goal was to use product sequences where possible, the decision to use research-specific pulse sequences was motivated by the desire to take advantage of additional non-product features (e.g., motion-tracking navigators) and/or the need for modifications to better harmonize across all three vendors (e.g., ensuring similar RF pulse shapes and timings). The required pulse sequences and protocol specification files are available for scanners with specifications matching those used by the HBCD sites. Instructions for acquiring the necessary sequence and protocols files can be found at https://hbcdsequences.readthedocs.io/en/latest/.
3. HBCD MRI protocol
The HBCD MRI protocol includes sMRI, fMRI, dMRI, MRS, and quantitative relaxometry. Despite the goal to collect all scans from each participant, the MRI WG recognized that not all children would complete the entire protocol given its length and the overall difficulty of scanning infants and toddlers during natural sleep. Therefore, it was necessary to devise a scan hierarchy that balances capturing developmentally sensitive data and aimed to acquire equal numbers of complete datasets across the modalities. To accomplish this, a pseudo-randomized acquisition order was formulated. Following the BCP study (Howell et al., 2019), the protocol begins with a localizer modified to be quieter and minimize waking participants. An age-specific structural MRI sequence is then prioritized as this is needed for radiology review and is an input to many processing pipelines for the other modalities. In particular, the T2w image is prioritized at Visits 2 and 3 as this provides greater contrast compared to T1w images at early ages due to low myelin content and high water content in the infant brain. T1w imaging is prioritized for all subsequent study visits. Functional and diffusion MRI sequences are prioritized following the age-specific structural sequence, with the order of these sequences randomized across study participants. Quantitative relaxometry and MRS follow, again with the scan order randomized across participants. The lower priority structural MRI scan (T1w for Visits 2 and 3; T2w for Visit 4 and beyond) is the final image to get acquired. This pseudo-randomized scan order for each participant is centrally determined and distributed to sites by the HBCD Data Coordinating Center. As detailed below, the minimal criteria for completing a scan session was established by the Scanning Young Populations WG.
3.1. Structural MRI
Recent large-scale developmental neuroimaging studies include high spatial resolution (0.8–1.0 mm isotropic) T1w and T2w structural sequences to support morphometric analyses (Casey et al., 2018, Harms et al., 2018, Hazlett et al., 2012 Howell et al., 2019; Glasser et al., 2016). ABCD provides a well-validated structural 1 mm protocol that has been harmonized across all three major scanner vendors (Casey et al., 2018). However, HCP Lifespan and BCP studies both demonstrate advantages of sub-millimeter resolution for subsequent morphometric analyses (Howell et al., 2019, Glasser et al., 2013). Additionally, recent work has demonstrated the need for infant-specific T2w structural protocols due to incomplete myelination and thus suboptimal grey/white T1w contrast (Howell et al., 2019, Myers et al., 2023).
This prior work shaped the initial HBCD structural protocol proposals, in addition to three key decisions:
-
1.
Approximately seven minutes total was defined as the targeted duration in the MRI protocol for the T1w and T2w structural scans, where previous studies (e.g., ABCD and BCP) took more than 12 minutes.
-
2.
Early pilot scanning determined that scanners available at all sites could achieve 0.8 mm isotropic resolution with acceptable signal-to-noise ratio (SNR) with 32- and 64-channel coils, and that all sites would be able to support navigators for prospective motion detection, real-time monitoring and real-time correction.
-
3.
The Structural WG decided to maintain constant structural protocols over the study duration, as opposed to optimizing protocols by age. In the context of a multi-site and multi-vendor study, we concluded that this approach may better allow longitudinal effects to reflect developmental differences rather than protocol differences.
From these early decisions, the Structural WG established a priority list:
-
1.
Achieve harmonized contrast and acceptable SNR across all three vendors at 0.8 mm resolution;
-
2.
Select acceleration techniques to best achieve the allocated target time for each scan; and
-
3.
Add navigators for prospective motion detection and, when available, real-time correction.
With the adoption of a fixed structural protocol across timepoints, the WG determined that the T1w scan would drive morphometry primarily for children starting at Visit 4 (9–15 months) and beyond, and so adopted contrast parameters from the BCP study. Harmonization of contrast in the T1w scan primarily involved ensuring matched parameters and addressing any remaining technical constraints.
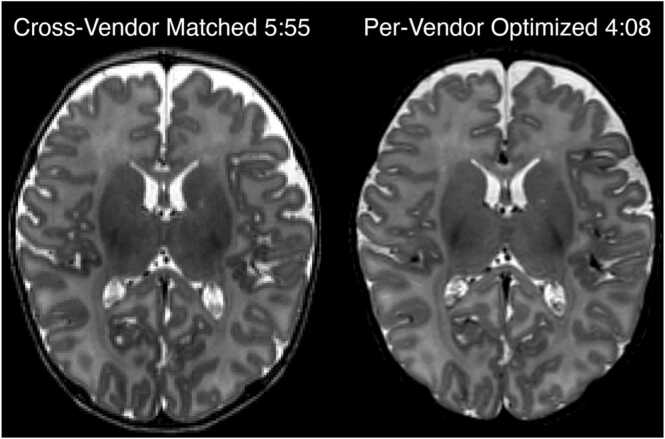
However, the T2w scan provides the morphological contrast for the earliest study timepoints (Visits 2 and 3) and notably is implemented differently across vendors, particularly in the design of the variable flip-angle schemes, making direct harmonization of sequence parameters infeasible. To address this, the WG evaluated infant pilot data with strong cortical contrast (Fig. 2), and then standardized protocols for each vendor giving the highest quality data for morphometry in the youngest subjects.
Fig. 2.
: Pilot data, acquired in a 5 week old participant, showing two alternative T2-weighted protocols evaluated during optimization on the Siemens platform. The vendor-matched protocol harmonized TR, TE, and echo train length (ETL) across all vendors. However, due to variations in how each vendor implements variable-flip-angle turbo spin echo, we could achieve equivalent or superior image contrast in shorter time with vendor-specific choices of TR, TE, and ETL.
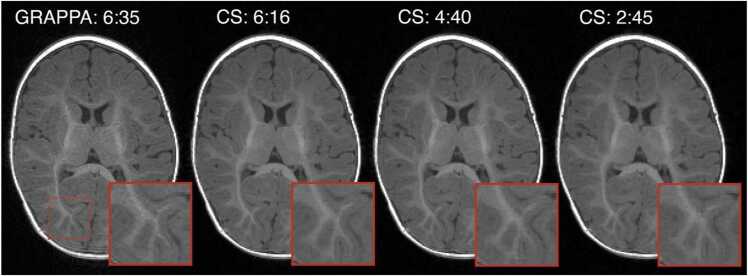
As the WG worked on sequence acceleration, the implementation differences between vendors became further magnified for the T2w scan and new challenges were introduced for the T1w scan. Fortunately, all vendors had product implementations of compressed sensing acceleration available (Lustig et al., 2007), significantly shortening scan times without significantly compromising image quality (Fig. 3). All scanner vendors provided critical assistance in applying this new technology, with support for upgrading the scanner software and modifying sequences in alignment with the study protocol, at every site. However, these acceleration methods were also implemented differently across the vendors precluding direct harmonization of the exact sampling and reconstruction algorithms – the WG concluded that harmonizing on total scan time was ultimately the critical variable, acknowledging that this required different nominal acceleration factors between vendors. Cross-vendor comparisons were made to ensure that different accelerations settings provided comparable levels of image smoothness/noise across the structural protocols. Moreover, the WG also determined that, leveraging the retention of full raw k-space data for all scans, future advanced in reconstruction algorithms could be retrospectively applied to the HBCD Study data.
Fig. 3.
: Pilot data, acquired in a 5 month old participant, showing four acceleration factors that were evaluated for T1-weighted protocols during the optimization on the Siemens platform (either GRAPPA or Compressed SENSE, with total scan duration noted). The region outlined by the red dashed box is zoomed for each volume to highlight the effects of acceleration on fine features. Comparing GRAPPA 6:35 to CS 6:15, we can see that compressed sensing of equivalent scan time preserves fine high-contrast structures with less noise than GRAPPA. However, at CS 2:45 the high degree of acceleration leads to a subtle blurring of fine features. Our final protocol set 4 minutes as the target T1-weighted structural scan duration across all three vendors.
Finally, the Structural WG planned to adopt navigators for prospective motion detection and correction in all protocols as they became available. Using volumetric navigators (vNavs) on Siemens and Philips systems and PROMO on GE systems enables motion detection (White et al., 2010, Tisdall et al., 2016, Andersen et al., 2019). Combined with the decision to save raw k-space data for all scans, it will be possible for researchers to apply retrospective motion-correction methods to the HBCD data in the future. Additionally, real-time motion information from the motion navigators are displayable on the connected FIRMM tablet. Finally, with prospective correction enabled across all sites, the WG anticipates further data quality improvements, especially in older participants in whom have more variability in sleep states.
The final HBCD structural imaging protocol represents a significant advancement on the state-of-the-art; it is both significantly faster (under 9 minutes) and with higher-resolution (0.8 mm isotropic) than any previous multi-vendor and multi-site study. Through pilot protocol exploration together with vendor collaboration, the WG integrated the latest acceleration methods and motion navigators for T1w and T2w protocols, enabling fast and robust structural imaging of the developing brain.
3.2. Functional MRI
Acquiring functional MRI during natural sleep affords the opportunity to characterize spontaneous fluctuations in the BOLD signal, through which we can make inferences about the functional network architecture of the developing brain. The Functional WG aimed to develop a 15-minute protocol appropriate for the HBCD age range that aligned as many acquisition parameters as possible across the three vendors, while optimizing costs. To make preliminary decisions on which fMRI sequences and protocols to test during piloting, the WG considered protocols from prior consortia (e.g., dHCP, BCP, ABCD), extant infant neuroimaging studies (Goksan et al., 2017), and expertise of WG members. Based on these sources, the WG opted against high multi-band (MB) factors (e.g., MB8) given concerns for low subcortical SNR, but also preferred the shorter repetition time (TR) associated with MB factors of 4 or higher (Moeller et al., 2010). To investigate this tradeoff, WG members collected pilot fMRI data in neonates on a Siemens scanner over a narrow range of multi-band and isotropic resolution choices (MB4 2.0 mm, MB4 2.4 mm, MB6 2.0 mm, and MB6 2.4 mm).
The WG ultimately adopted the MB4, 2.0 mm isotropic resolution sequence based on computed SNR, inspection of blood oxygen level-dependent (BOLD) images and functional connectivity seed maps, and preference for higher spatial resolution given the smaller size of the neonatal brain. The decision to not use in-plane acceleration was made based on prior work demonstrating decreased tSNR (Seidel et al., 2020) and increased sensitivity to motion (Ohliger et al., 2003). The shortest TR that could be achieved across vendors with these parameters was 1725 ms. While longer echo times (TE) were considered based on prior work in infants (Goksan et al., 2017), the WG chose to use a constant TE of 37 ms for all the longitudinal timepoints of the study. Echo spacing, bandwidth, and partial Fourier were allowed to differ across vendors to achieve the desired MB factor, resolution, TR, and TE. The WG chose a field of view (FOV) of 224×224 mm2 with 76 slices to ensure whole brain coverage throughout childhood as well as to have an odd number of slice packets with MB4 (Goksan et al., 2017). Further, the WG elected to use a single-phase encoding direction for all the BOLD scans to avoid challenges associated with combining differing amounts of data (following frame censoring) across different phase encoding directions; posterior-anterior (P/A) was selected so that compression artifacts occurred posteriorly rather than anteriorly. Given different T1 values between the infant (∼1700 ms) and adult brain (∼1100 ms) (Gräfe et al., 2021), a flip angle of 62 degrees was chosen as a slightly under-flipped middle point (i.e., 10 % under the average of the Ernst Angle in infants and adults). Fat saturation was enabled/turned on. Finally, although AC/PC alignment was considered for angulation of the FOV (to optimize the efficiency of the brain coverage; (Mennes et al., 2014)), the WG decided instead to acquire strictly axial slices to better align with other HBCD scans and minimize burden on the acquisition technologists across sites.
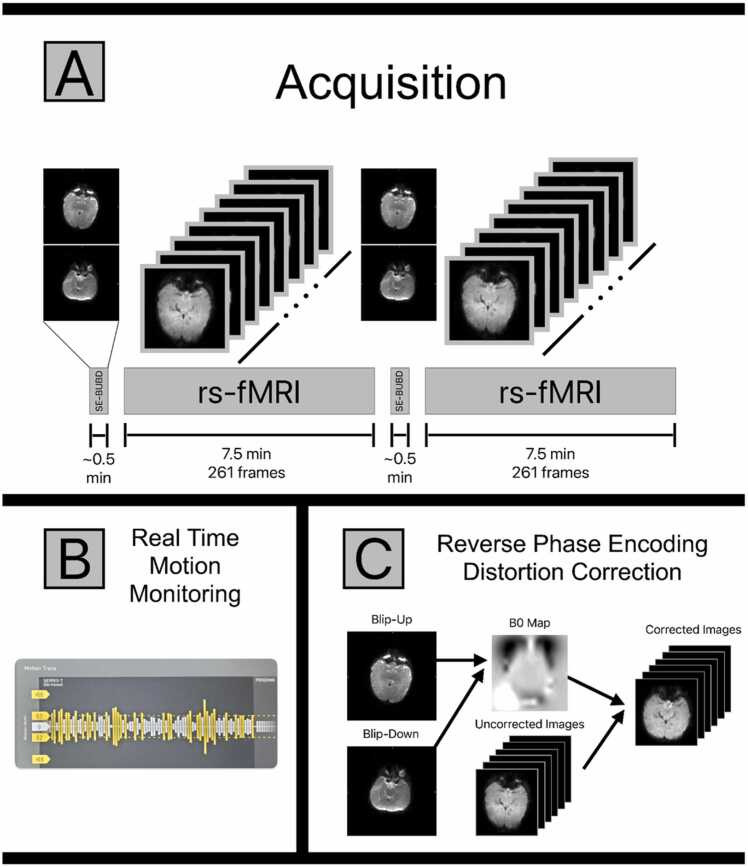
The final fMRI protocol for HBCD consists of two 7.5-minute BOLD runs, each preceded by a blip-up, blip-down phase encoded spin-echo field map pair (Fig. 4). Piloting across all HBCD sites included a third set of field maps after the second BOLD run, but in all cases the first two sets were sufficient, and so the third set was dropped from the main study protocol. The WG considered collecting the BOLD as a single 15-minute run, but not all vendors could achieve this and there were concerns that a single long run in children would be problematic due to potential for more motion at the end of longer runs. FIRMM was enabled to monitor motion in real-time with threshold settings of 0.2 mm and the respiration filter turned off, as this would yield the most accurate information regarding usable data being collected. All sites piloted the two 7.5-minute BOLD runs, adequate image quality was obtained across vendors and sites, and sites consistently obtained low-motion fMRI data. A total of 382 BOLD runs (each 7.5 minutes) were obtained across the consortia during pilot testing, with 73 % having at least 340 seconds of data (5.7 minutes; 75 % of the run length) with framewise displacement (FD) <0.3 mm.
Fig. 4.
(A) A depiction of the HBCD fMRI acquisition protocol. There are two resting state (rs) fMRI acquisitions lasting 7.5 min each. Each rs-fMRI run is preceded by a pair of single shot spin-echo (SE) EPI images with matched bandwidth to the resting state data. One image in the pair of spin-echo images is acquired with reversed polarity phase encoding gradients, allowing for the pair to be used in a blip-up-blip-down (BUBD) distortion correction algorithm. (B) All HBCD fMRI data are monitored in real time at the point of acquisition for motion using FIRMM. If motion is severe enough to prevent further analysis, the pair of SE-BUBD images and a 7.5 min resting state acquisition can be repeated until sufficient data have been acquired. (C) Spin-echo data pairs can be used to estimate the static field as well as the corrections necessary to account for image distortion. Spin echo images are used for this purpose to obtain better estimates in regions of significant inhomogeneity. The calculated corrections are then applied to the gradient echo resting state images, resulting in the distortion corrected rs-fMRI data.
Parameters above were determined based on pilot data from sleeping neonates (<1 month since birth). The current plan is to maintain these imaging parameters at older longitudinal timepoints with minor exceptions (e.g., the respiratory filter may be used in FIRMM at older ages). Additionally, MRI data will be collected in awake participants when children reach a to-be-determined age (i.e., likely at/after 36 months of age), at which point movie stimuli will potentially be curated, standardized, and included to minimize motion artifact. This information will be described in a subsequent publication.
3.3. Diffusion MRI
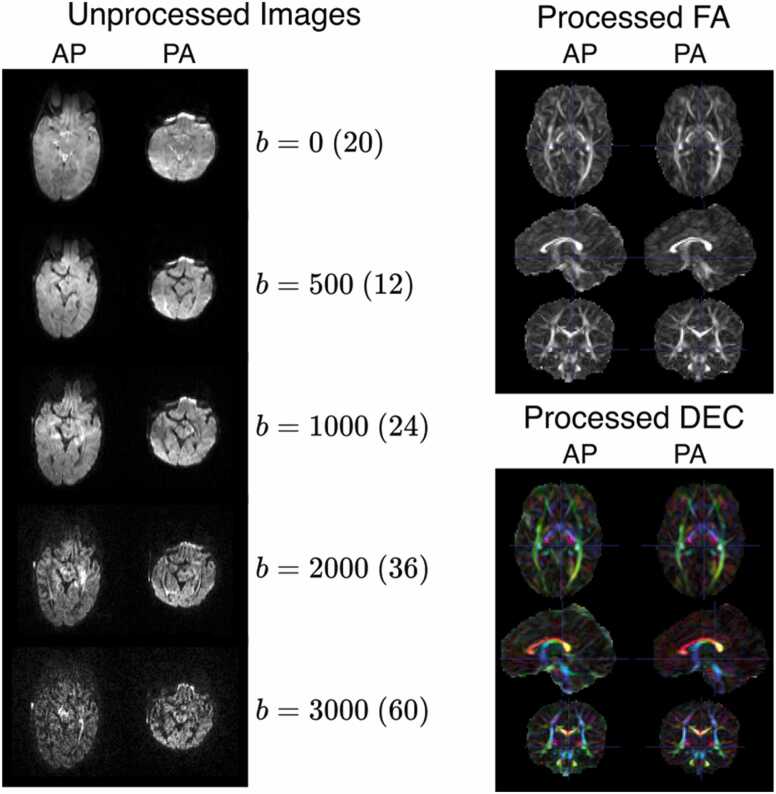
Diffusion MRI (dMRI) is a useful technique for characterizing brain microstructure, mesoscopic organization, and structural brain connectivity (Alexander et al., 2019, Alexander et al., 2011, Basser and Pierpaoli, 1996, Pierpaoli and Basser, 1996, Pierpaoli et al., 1996). dMRI has been specifically effective for delineating these brain properties across perinatal (Huang et al., 2006), infant (Ouyang et al., 2019, Huang, 2022), and childhood (Lebel et al., 2019, Yu et al., 2020) developmental stages. The HBCD dMRI protocol was designed to accommodate a wide range of diffusion models and tractography methods at moderately high spatial resolution in a modest acquisition time. The dMRI protocol was largely patterned after the ABCD protocol, which provided multiple b-value measurements at moderately high spatial resolution (Casey et al., 2018). Identical to ABCD, the HBCD dMRI protocol collects 1.7 mm contiguous axial slices with an FOV of 238 ×238 mm2 with 87 slices and matrix size 140×140, yielding 1.7 mm isotropic spatial sampling. The dMRI acquisition uses MB3 with no additional in-plane acceleration (Feinberg et al., 2010). Other parameters, including TR (4800 ms), TE (88 ms), and partial Fourier sampling in the phase encoding direction (63–75 %) were selected to enable parameter harmonization across the scanner systems. Notably, we chose to use a common TE across vendors, rather than use the minimum TE possible for each vendor (which would be lowest for the Siemens Prisma, given its higher peak gradient strength). Similar to ABCD, multi-shell diffusion encoding was performed at b = 0 (20 gradient orientations), 500 (12 gradient orientations), 1000 (24 gradient orientations), 2000 (36 gradient orientations), and 3000 (58 gradient orientations) s/mm2, though modest changes as compared to the ABCD protocol were made to the number of encodes per shell and the implementation. First, the number of directions was increased for the 500, 1000, and 2000 s/mm2 shells, which should increase the sensitivity to faster diffusing tissue compartments that are more prevalent in the infant brain (Lebel et al., 2019, Alex et al., 2024). Second, the diffusion encoding sets were split between two series with reversed phase encoding (A/P versus P/A). The diffusion gradient orientations were randomly shuffled across the A/P and P/A series, with b0 images interspersed throughout each series (65 unique diffusion encoding measurements per series for a total of 130 directions). This enables both EPI distortion correction using a topup-like approach (Andersson et al., 2003) and better balancing of the local EPI stretching and compression than collecting all of the dMRI encoding with a single phase encoding orientation. There is also evidence that balancing the dMRI encodings across two phase encoding directions will reduce the variance in dMRI measures (Irfanoglu et al., 2021). The total acquisition time for the dMRI protocol is approximately 12.5 minutes. See Fig. 5 for a representation of unprocessed images by b-value, processed fractional anisotropy (FA) images, and processed directionality encoded color (DEC) maps.
Fig. 5.
: Axial slices of unprocessed diffusion-weighted images from an HBCD acquisition. Slices acquired with AP and PA phase encoding directions are shown in the left and right columns, respectively. Gradient strengths in b (s/mm2) are shown per row with the number of images collected at the b value in parenthesis. The AP and PA images shown at b>0 are not the same gradient direction, as the gradient directions are split across the phase encoding directions.
3.4. Quantitative MRI
The MRI tissue contrast arises primarily from differences in the longitudinal (T1) and transverse (T2) relaxation times. Conventional clinical and research neuroimaging techniques have been disproportionately represented by qualitative relaxation time weighted brain images (e.g., T1w, T2w scans). While we include these traditional structural scans in the HBCD protocol, they are a complex function of underlying relaxation properties, pulse sequence parameters, and other participant- and hardware-specific effects, such as participant position. The dependency of the MR signal on these extraneous sources limits interpretation and one’s ability to associate observed contrast changes to biological mechanisms. Consequently, quantitative comparison across participants, longitudinal sessions, and multiple sites becomes difficult. This is especially relevant in pediatric neuroimaging, where the brain is undergoing rapid development that results in changing free water distribution, iron content, and brain myelination. As a result, contrast in conventional MRI changes as a function of age, complicating the study of pediatric populations across varying age ranges. Direct measurement of these relaxation properties can overcome many of these limitations of conventional MRI and allow for improved characterization of brain tissue microstructure (Deoni, 2010, Does, 2018).
In the past decade, there has been a significant push towards the development of quantitative magnetic resonance imaging (qMRI) (Ehses et al., 2013, Deoni et al., 2005, Fleysher et al., 2008, Ma et al., 2013, Liao et al., 2017 Kvernby et al., 2014; Kecskemeti et al., 2016, Chen et al., 2018, Piredda et al., 2020) for improved detection, sensitivity, and understanding of pathologies, like epilepsy (Liao et al., 2018, Ma et al., 2019, Salmenpera et al., 2007, Spader et al., 2013) or multiple sclerosis (MacKay et al., 2009, Reitz et al., 2017, Parry et al., 2002). qMRI has enabled consistent vendor-agnostic quantitative images (e.g., T1 and T2 maps) in multi-center studies (Deoni et al., 2008). Such quantitative images are easily compared across sites and offer higher inter-site reproducibility compared to contrast-weighted imaging (Weiskopf et al., 2013). Moreover, quantitative relaxation maps can be used to synthesize the conventional contrast-weighted images (Tanenbaum et al., 2017, Fujita et al., 2019). The deployment of qMRI in clinical and research settings has been limited, potentially because these methods suffer from long scan time and/or lower spatial resolutions. In addition, for multi-site research studies, like HBCD, the chosen method must be available on all scanner platforms. For these reasons, none of the large multi-site MRI studies to date (e.g., ABCD (Casey et al., 2018), HCP (Harms et al., 2018)) have included a qMRI protocol for quantitative T1 and T2 mapping.
Recognizing the unique value of qMRI in measuring brain development, a qMRI module is included in the HBCD MRI protocol, which needed to fulfill the following criteria:
-
1.
Rapid Protocol: Given the overall duration of the HBCD MRI protocol, the duration of the qMRI acquisition had to be less than 5 minutes.
-
2.
Cross-vendor availability and ease of use and processing: The acquisition method had to be harmonized across all MR vendors (and their specific software baselines). It also had to be easy to run on the scanner with the data easily processed.
For HBCD, several qMRI techniques were considered, however, options were limited given the constraints discussed above. Magnetic resonance fingerprinting (MRF) (Ma et al., 2013) enables parametric maps from a single 3D acquisition, but different implementations across vendors are not time-efficient and have not been harmonized. MPnRAGE (Kecskemeti et al., 2016) enables T1 mapping (Kecskemeti and Alexander, 2020), but is also not readily available on all vendors. Other methods, including variable flip angle and DESPOT1–2 (Deoni et al., 2003, Deoni et al., 2005) and Look-Locker (Look and Locker, 1970, Henderson et al., 1999), among others, were considered, but either took more time than allotted for this scan and/or included multiple acquisitions, which could complicate the acquisition should motion occur. For these reasons, we adopted the three-dimensional quantification using an interleaved Look–Locker acquisition sequence with a T2 preparation pulse (3D-QALAS) (Kvernby et al., 2014), which is a time-efficient 3D method capable of simultaneous estimation of T1, T2 and PD maps from a single scan and is available and tested across all three major MRI vendors (Fujita et al., 2019).
Given the protocol time constraints, inherent SNR vs resolution trade-offs, and enhanced compressed sensing acceleration, the WG determined that a 1.3 mm isotropic voxel qMRI protocol that takes approximately 4 minutes to acquire was the best option. Both higher and lower spatial resolutions were tested. Having higher resolution required more aggressive acceleration and resulted in decreased SNR, which significantly hindered the reliability, reproducibility, and the image quality of the estimated relaxometry maps. In contrast, lower resolutions increased the SNR but compromised brain spatial specificity, which is important when monitoring developing brain regions, especially as myelination unfolds.
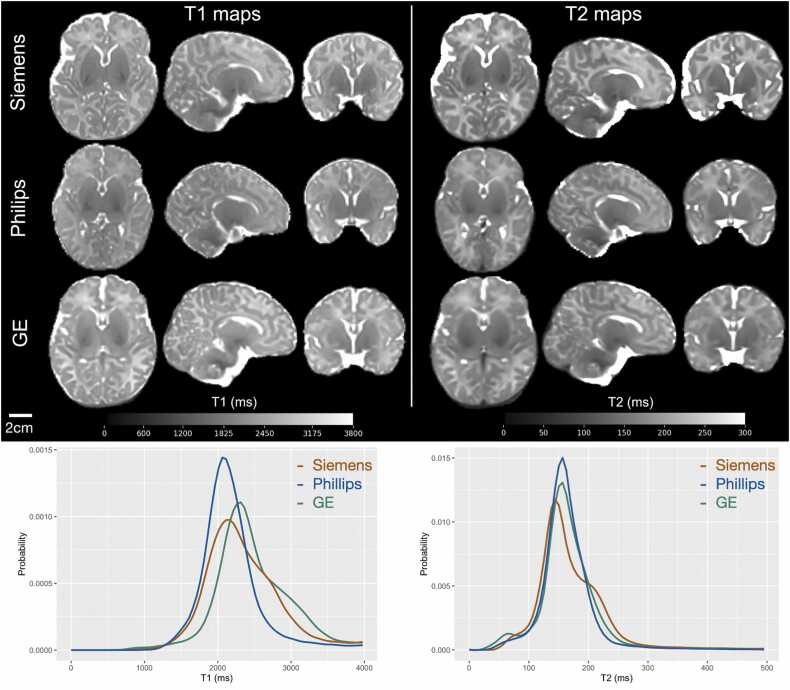
To improve the accuracy of the T1 and T2 maps, a separate transmit inhomogeneity (B1+) field map was included using the Actual Flip Angle (AFI) method (Yarnykh, 2007). Acquiring the B1+ map is important as the participant’s head can end up in different orientation within the coil despite tight padding, and the B1+ field distribution cannot be simply assumed based on the landmarking position and the size of the head. The QALAS and B1+ data is processed using the commercially available SyMRI toolbox. Representative examples of computed T1 and T2 maps on three neonates (scanned at age 3–4 weeks), performed on each of the three vendors are demonstrated in Fig. 6. The images from the five acquired volumes from the 3D-QALAS acquisition used to produce the quantitative T1, T2 and PD maps will be part of the annual HBCD data release.
Fig. 6.
: Representative axial, sagittal, and coronal slices of quantitative T1 and T2 maps acquired in age-matched (3–4 weeks) infants across the three major MRI vendors. Histograms of quantitative T1 and T2 relaxation times of these infants highlight good inter-vendor agreement.
3.5. Magnetic Resonance Spectroscopy
The HBCD funding announcement specified that MRS for measures of molecules involved in neuronal metabolism, neurotransmission, and oxidative stress should be included in the core neuroimaging protocol. The MRS WG identified N-acetylaspartate (NAA), a marker of neuronal mitochondrial metabolism, glutamate and γ-aminobutyric acid (GABA), the principal excitatory and inhibitory neurotransmitters, and glutathione (GSH), the most abundant antioxidant involved in protection against reactive oxygen species in the human brain, as the key metabolites to be measured. There was strong agreement within the WG that generating reliable and reproducible measures of the low-concentration metabolites, GABA and GSH, in particular, would require the use of spectral editing techniques (Choi et al., 2009). Further WG efforts centered on determining which brain region(s) would be interrogated and how, including single vs. multi-voxel localization (i.e., Point-RESolved Spectroscopy PRESS; (Bottomley, 1987)) versus semi-adiabatic localization by adiabatic selective refocusing sLASER (Scheenen et al., 2008, Conolly et al., 1991) and editing (i.e., conventional J-difference techniques versus newer, more efficient editing schemes) techniques to be used. Additionally, the WG considered the constraints specific to HBCD, namely, the need to minimize acquisition time, mitigate the effects of participant motion and frequency drift, harmonization of sequence elements, and timing across MRI vendors, and support for automated data processing.
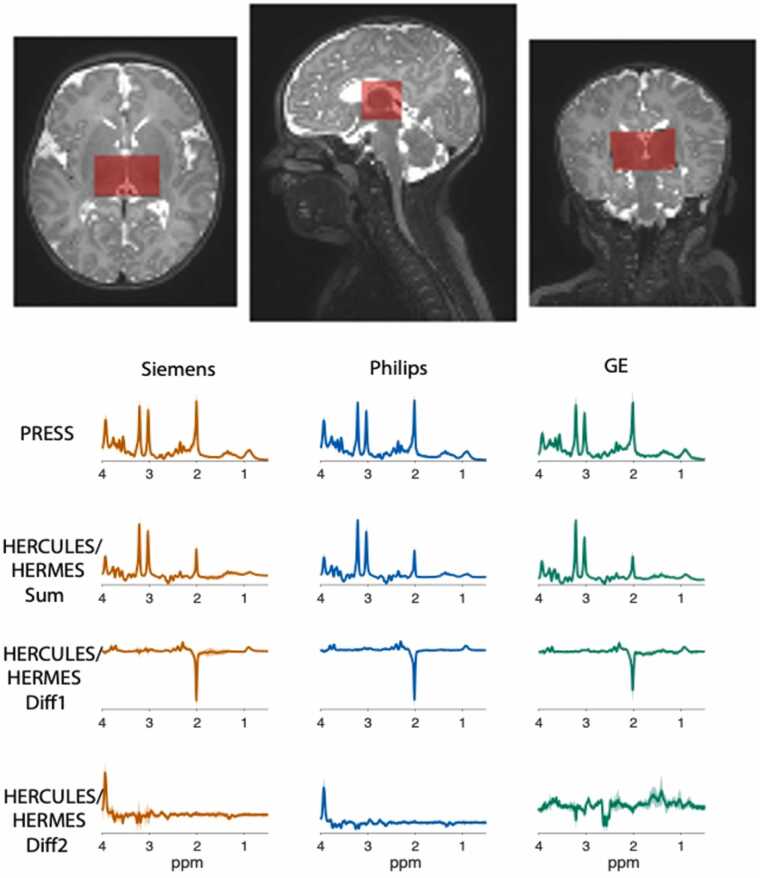
The WG determined that the optimal approach centered on a single-voxel localization (30×23×23 mm3) in the bilateral thalamus (Fig. 7), maximizing signal-to-noise across multiple low-concentration metabolites while maintaining an acquisition time (TA) within the allotted 9 minutes. Additionally, the WG selected an advanced Hadamard-encoded editing scheme developed by Oeltzschner and colleagues to measure glutamate, GABA, and GSH in a single experiment, as well as additional relevant metabolites, including NAA, lactate, ascorbate, creatine, myo-inositol, glutamine, and total choline (Oeltzschner et al., 2019). The WG selected the more conventional PRESS localization over sLASER. Although recent calls to move toward sLASER (Deelchand et al., 2021) were considered, the WG had concern that doing so would not only increase specific absorption rate (SAR), but also necessitate the development and validation of a new sequence on three different vendor platforms in less than 6 months, both impractical and risky in a study like HBCD. Instead, the WG focused its efforts on two other key areas: (1) scanner drift and (2) ensuring robust measurement in pediatric populations. MRS relies on the frequency of the measured signals, and uncorrected frequency drift during data acquisition adversely affects edited MRS quality by changing the contribution of coedited signals and editing efficiency (Harris et al., 2014). This is of particular concern when acquiring MRS after sequences that require high gradient duty-cycle, hence heating (e.g., fMRI, dMRI) (Hui et al., 2021). To mitigate this, it is often suggested that MRS data be acquired first – before fMRI or dMRI (Choi et al., 2021). However, this is not always feasible, and thus, this consideration frequently limits the incorporation of MRS into human connectome studies. The WG addressed this limitation by enabling real-time frequency correction through incorporation of interleaved water referencing (Edden et al., 2016) and laid the foundation for the future incorporation of navigators. Additionally, to mitigate the effects of T2 relaxation on the measured concentrations of NAA and other metabolites (Traber et al., 2004), the WG incorporated a short block of 32 transients acquired at TE = 35 ms without spectral editing, ahead of the main spectral editing block (224 transients, TE = 80 ms). This short-TE block was placed at the beginning of the sequence to ensure that for infants that awoke in the middle of MRS, there would still be sufficient measures of NAA and other high-concentration metabolites available. The final “multi-sequence” is named Integrated Short-TE and Hadamard Multi-Sequence (ISTHMUS) (Hui et al., 2024).
Fig. 7.
: MRS data acquisition is localized to a single voxel (30×23×23 mm3) in the bilateral thalamus as shown in the top panel above. The thalamus is reciprocally interconnected with nearly the entire cerebral cortex and the GABAergic neurons of the thalamus are involved in the generation of normal and abnormal synchronized activity across the various thalamocortical networks. The MRS WG selected the thalamus as the region of interest from which glutamate, GABA and other relevant metabolites would be measured with the goal of determining how differences in the trajectory of thalamic GABA levels related to the trajectory of thalamocortical connectivity and cognitive-behavioral development across the first decade of life. Shown below are averaged spectra (solid lines) and standard deviation (shaded region) across participants acquired during the pilot phase of HBCD on each MRI vendor platform. Note that the pilot GE data were acquired using an older HERMES sequence as the GE ISTHMUS implementation had not yet been finalized at the time.
The inclusion of MRS in the core HBCD neuroimaging protocol was itself innovative not only for the science it will afford – namely, the opportunity to interrogate biochemical mechanisms underlying the observed structural, functional, and behavioral trajectories – but also for the fact that it is the first adult, adolescent, or pediatric study of this magnitude to include such measures. As described, the WG further mitigated the challenges of incorporating edited MRS into a comprehensive pediatric neuroimaging protocol through the development of ISTHMUS (Hui et al., 2024).
3.6. Scanning Young Populations
The Scanning Young Populations (SYP) WG was tasked with a broader charge of supporting the successful acquisition of non-sedated imaging data from infants and young children (i.e., “young populations”). While a growing number of researchers across institutions have been collecting MRI data from infants and toddlers over the past two decades, many investigative teams across the 27 sites had no or limited experience scanning participants in this age range. To train new sites in “the art and science” of scanning infants (Spann et al., 2023, Dean et al., 2014, Raschle et al., 2012, Howell et al., 2019), the SYP WG sought to standardize protocols across sites, maintain flexibility for the heterogeneous imaging center environments across sites, and develop an extensive set of procedures and best practices for obtaining MRI data from infants and toddlers. These procedures and best practices serve as a comprehensive guide for MRI sessions conducted during the non-sedated, natural sleep of young participants with diverse sleep habits (focused initially on visits 2 [ages 0–1 month] and 3 [ages 3–9 months]). These strategies outline the preparation, execution, and follow-up steps necessary for the successful acquisition of high-quality imaging data, including protocols for scheduling and confirming MRI visits with families, preparation of the MRI facility, and techniques for transferring sleeping children to the MRI suite.
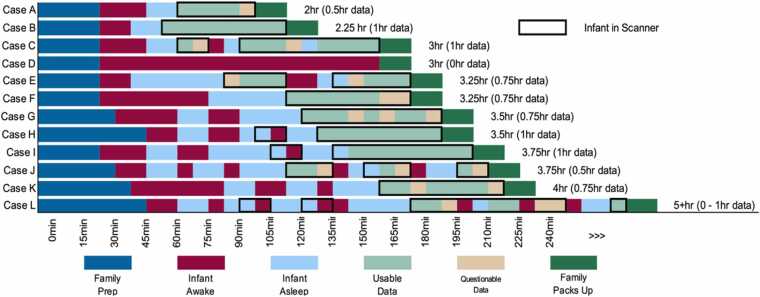
Within the manual, practical advice is provided to ensure an infant’s comfort and to facilitate the onset of sleep, such as minimizing environmental stimuli and allowing the child to fall asleep within the MRI room itself when possible. Recognizing that facilities may vary in their capabilities, recommendations aim to strike a balance between uniformity and site-specific adaptability. The WG also emphasized the importance that sites approach each session with responsive flexibility (Fig. 8), for example, to be ready to adjust to the immediate needs of the participant (and family) within their own unique MRI facility.
Fig. 8.
: Schematic representations of hypothetical infant MRI scan sessions. These are example sequences of events that can occur during infant MRI scan sessions. Note the variation in total session duration (2–5+ hrs, with older infants often taking longer to acquire data), times infants are awake and asleep, and amount of usable data acquired. Scan sessions can be short due to time or staff restrictions (Case A), or because the infant falls asleep quickly and stays asleep for the duration of data acquisition (Case B – ideal situation). Infants may wake shortly after data acquisition begins (Case C), or after some data are acquired (Case E), may stay asleep for all data collection after taking a long time to fall asleep (Case F), or may never go to sleep leading to the family or imaging center policy ending the scan without any data being acquired (Case D). Some children will wiggle throughout a scan session (Case G), or a team may rush getting a sleeping infant into the scanner (Case H), when waiting for them to fall into a deeper sleep in the scanner would have been better (Case I). Others may need to be removed from the scanner and comforted multiple times in order to collect adequate data (Cases J and L). Some extended scan sessions will result in adequate data collection (Case K), while others will seem like anything and everything that can go wrong does (Case L), sometimes with data acquired and other times without. The key is to remain positive and to adapt to each family.
The MRI facility itself presents a set of variables that require attention. Not all sites have access to a dedicated nursery or staging space for the infant to fall asleep in a quiet private room, nor do all scan centers provide opportunities to schedule scans during evening hours when infants are more likely to sleep. Therefore, adaptability in the use of available spaces and equipment has been crucial for preparing sites for considering the unique needs related to imaging this population.
To ensure scan quality and quantity uniformity across diverse environments, the WG considered several factors crucial for the success of the MRI sessions, including the number and relevant expertise of each team member present during scans. At least one team member requires competence in safety and in effectively implementing techniques to induce and maintain sleep amidst the sounds produced during active scanning, as well as the potential physical vibration induced by some sequences (e.g., dMRI) that may rouse infants.
Selecting a recommended list of equipment, suggestions for protocols for inducing sleep and initiating scan acquisition and defining scan success were primary roles for the WG. Success was not merely measured by the completion of a scan sequence or session, but by the quality of data obtained. To this end, the WG established a set of criteria to assess the effectiveness of each visit based on obtaining usable relevant structural scans (e.g., T1w, T2w, depending on child age) and at least two other modalities (i.e., dMRI, fMRI). The WG also recommended that an average success rate across all modalities (as a percentage) be captured and reported to promote collection of all scan modalities beyond those that would allow a specific visit to be determined to be a binary “success.” To inform decision-making for how to support sites for success, the WG recommended sites collect details about each scan visit and interactions with the family, including 1) start and end times for the visit, 2) sequence initiation, scan interruptions and cause, if the child was awoken or remained asleep, and quality metrics for the scan (e.g., from FIRMM for sMRI, dMRI, and fMRI, and 3) visual inspection for the structural and QALAS images and MRS).
In response to the need for standardization across sites, the WG made key decisions on equipment, environmental modifications, and team training protocols for imaging durinal natural, non-sedated sleep. MRI-compatible equipment for infant scanning, such as ear plugs, MiniMuffs®, and DREAMIES® hearing protection followed by MRI-compatible headphones playing white noise throughout the scan, are employed across sites. These measures aim to reduce the overall acoustical sounds perceived by the sleeping child, while the constant white noise helps avoid sudden changes in the acoustical sound from the different sequences used. Additional environmental adjustments, including reducing ambient light levels and playing MRI sounds (e.g., played over an in-room speaker or via tablets in a nursery-space), were suggested to create a conducive atmosphere for maintaining sleep during the MRI acquisition.
Team training included the SYP WG creating videos that demonstrate how to position sleeping infants into the scanner, a “buddy system” in which physically proximal sites could visit and share advice, and bimonthly “office hours” in which questions could be brought to the SYP WG. The WG also developed checklists and reporting tools to be used by HBCD site monitors to ensure consistent data collection and to facilitate communication across sites.
3.7. MRI Processing
Recent large-scale neuroimaging studies reveal that behavioral associations with brain imaging phenotypes may comprise small effects (Dick et al., 2021, Smith and Nichols, 2018, Marek et al., 2022). Furthermore, accurately estimating brain-behavior associations benefits from large samples aggregated across many sites (Marek et al., 2022, Feczko et al., 2021). Big data neuroimaging consortia, such as ABCD study (Casey et al., 2018, Hagler et al., 2019), have been critical for collecting large enough samples to ensure accurate effect size estimation; HBCD will play a pivotal role in the advancement of big data neuroimaging in early life. The Processing WG was formed to outline data management and processing criteria in accordance with the Findable, Accessible, Interoperable, and Reproducible (FAIR) principles (Wilkinson et al., 2016) and NMIND model (Kiar et al., 2023) to ensure best standards and reproducibility.
The WG’s criteria for image processing pipelines were that they be open source, integrative, and modular, utilizing advanced coding standards and validation strategies to maximize reproducibility (Botvinik-Nezer et al., 2020, Goncalves et al., 2021). This includes adherence to BIDS formatting standards (Gorgolewski et al., 2016); version controlled containerized software that minimizes variation in imaging phenotypes due to multiple computing platforms (Kurtzer et al., 2017, Poldrack et al., 2017); and publicly available code with documentation to ensure transparency and invite community evaluation and improvement (Kennedy et al., 2019). All software deployed for HBCD will have gone through the NMIND tool evaluation process, providing the community with information regarding adherence to basic software standards (see www.nmind.org). The image processing pipelines utilized are optimized to work across development and address unique challenges presented by infant data, including differences in head size, reduced SNR, increased motion artifacts, increased partial voluming, and rapid shifts in tissue contrast (Mostapha and Styner, 2019).
For structural pre-processing, BIBSNet (Hendrickson et al., 2023), an automated brain imaging segmentation tool, produces segmentations that are fed into Infant-fMRIprep (Goncalves et al., 2021), an infant-specific extension of fMRIPrep (Esteban et al., 2019a, Esteban et al., 2019b). Functional processing is performed by XCP-D (Ciric et al., 2018, Mehta et al., 2023), a structural and resting state-fMRI (rs-fMRI) post-processing pipeline optimized across the lifespan to extract structural and functional connectivity data, including processed dense and parcellated time-series constructed from several predefined atlases. For diffusion image processing, QSIPrep integrates Nipype’s infant-specific workflow to provide measures related to proportion of restricted, hindered, and free water; neurite density and dispersion; and cellularity (Cieslak et al., 2021). SyMRI is used for quantitative MRI (QALAS) to generate reproducible quantitative measures including T1w and T2w contrasts, proton density, and myelin volume fraction (Fujita et al., 2019, Kvernby et al., 2014).
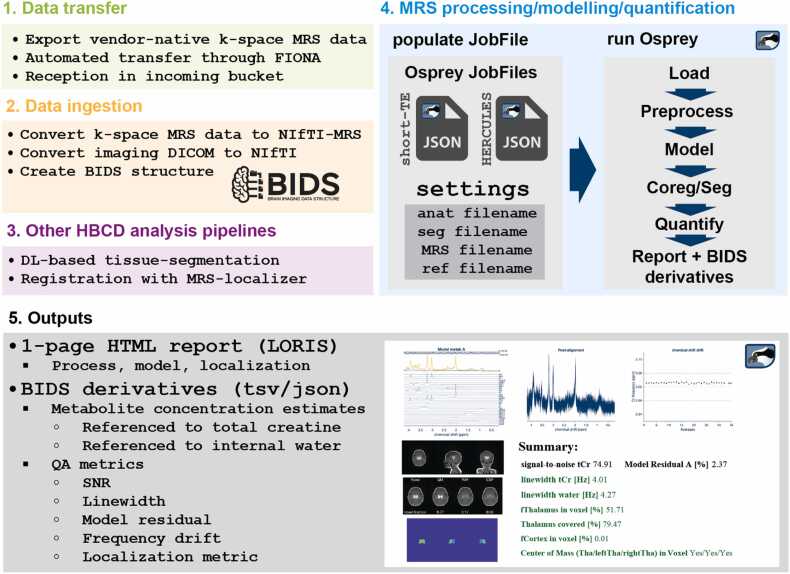
For MR spectroscopy, the WG capitalized on ongoing efforts to develop open-source software fully automated to convert raw data into standardized NIfTI-MRS (Clarke et al., 2022) and BIDS-MRS (Gorgolewski et al., 2016) formats, process, and perform analytics using Python and MATLAB (Fig. 9; (Zollner et al., 2023)). The workflow integrates consensus-recommended preprocessing (Near et al., 2021), quality assurance, linear-combination modeling, voxel-specific tissue segmentation (Hendrickson et al., 2023), quantification (including tissue correction (Gasparovic et al., 2006)) and data visualization. The Osprey tool (Oeltzschner et al., 2020), used for the processing, reconstruction, and estimation of MRS data, quantifies key target metabolites with an expected reliability, ie inter-subject coefficient of variation, of 9 % (NAA), 9 % (Glu), 9 % (GABA+), and 16 % (GSH) (Oeltzschner et al., 2019, Mikkelsen et al., 2017, Mikkelsen et al., 2019).
Fig. 9.
: Summary of the fully automated MRS data processing workflow. The workflow includes automated data transfer and ingestion, integrates derivatives from the HBCD MRI analysis, performs the MRS analysis, and generates quantitative results and summary reports.
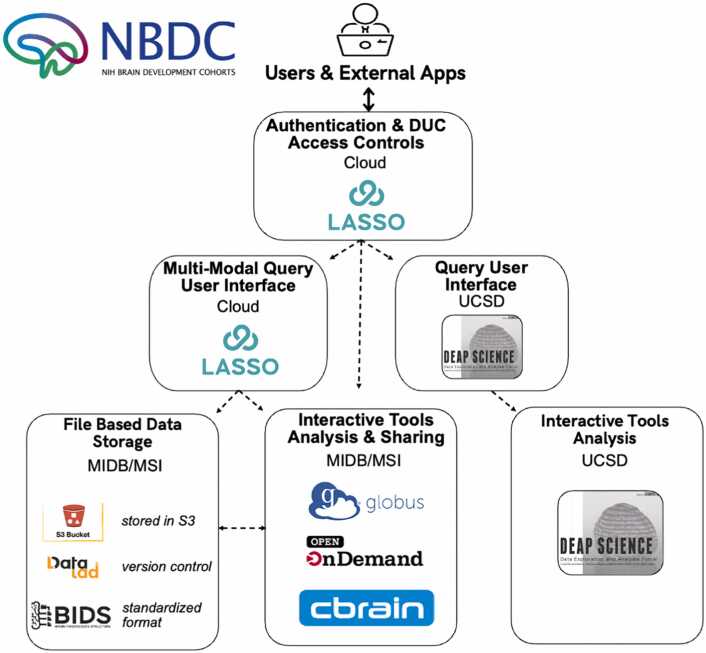
The WG’s criteria for data management include version control and metadata tracking for triage. HBCD data, housed at the Masonic Institute for the Developing Brain (MIDB) and the Minnesota Supercomputing Institute (MSI), are formatted according to BIDS standards (Gorgolewski et al., 2016) and managed with DataLad, an open-source distributed system for data versioning, reproducibility, and collaboration (Halchenko et al., 2021). In addition, computing criteria focused on ensuring reliable, high-throughput processing by leveraging platforms optimized for agile data processing across multiple high performance computing (HPC) clusters (Sherif et al., 2014, Poline et al., 2012). Internally, two key neuroinformatics tools - LORIS (Longitudinal Online Research and Imaging System) (Das et al., 2011) and CBRAIN (Canadian Brain Imaging Research Platform) (Sherif et al., 2014) - were employed for this purpose (Fig. 10). LORIS enables internal investigators to perform filtered queries across neuroimaging data for summary and participant-level information. Over the past 20 years, LORIS has trained thousands of users on its platform to refine its user interface. LORIS implementations have spanned across North America and include multi-site studies such as the Infant Brain Imaging Study (IBIS) Autism Center of Excellence (ACE) network (Hazlett et al., 2017), NIH Pediatric MRI Database (Almli et al., 2007, Evans and Brain Development Cooperative, 2006), and Canadian Consortium on Neurodegeneration in Aging (Chertkow et al., 2019). Coupled with this, CBRAIN (Sherif et al., 2014), internal processes to generate, store, and provide public access to data are well supported. Public release and public access to data will be conducted through the NIH Brain Development Cohort (NBDC) (Fig. 10). This portal is supported by Lasso (the commercial version of LORIS), computing and storage hosted by MIDB and MSI, and tools such as the Data Exploration and Analysis Portal (DEAP). The NBDC will enable seamless search, queries and data selection, data downloads, data sharing, ‘sandbox’ development and computing environments, command line access, and several direct analytic tools.
Fig. 10.
: High-level schematic of the CBRAIN user interface, data management and processing components for HBCD study.
Finally, the WG’s criteria for quality assessment (QA) of processed imaging data include several quantitative measurements for each pipeline, along with crowdsourced visual inspection, the gold standard for neuroimaging QA (Keshavan et al., 2019, Taylor et al., 2023). Crowdsourced qualitative QA is performed via BrainSwipes (https://www.brainswipes.us/), a web application that trains users to evaluate the quality of processed image outputs. Users are presented with brain images to QA and swipe “left” or “right” to “fail” or “pass” brain images, respectively, and can also select the “Help” button to flag images for triage by the HBCD WGs. Leaderboards are used to gamify the process to facilitate usage. High-security standards are maintained to ensure proper authentication and data visualization tools are used to display swipe statistics to ensure QA fidelity. The origin of BrainSwipes comes from SwipesForScience, an open source project that makes it easy to create and customize crowdsourced QA to any dataset (Taylor et al., 2023, Keshavan et al., 2019). Currently, BrainSwipes for HBCD is only available to consortium members; however, public contributions to QA will be made available in thre future as part of the NBDC platform.
The integrated standardized, open source pipelines, indexed and verion controlled data repository (i.e. via DataLad), LORIS/Lasso, CBRAIN, and BrainSwipes system and flexible access options enable an agile data processing lifecycle, where the community identifies data processing issues, relevant HBCD workgroups triage and resolve issues, and the data is reprocessed and updated with transparent and open communication when appropriate. This lifecycle enables the best standards and practices to be continually implemented transparently across all imaging modalities within the HBCD Study.
4. Conclusion
The human brain undergoes rapid and key growth and development throughout the first years of life. Building upon the work of existing large scale neuroimaging efforts and capitalizing on recent technological innovations and expertise across the Consortium, the HBCD MRI WG has established an innovative, vendor-aligned, multimodal MRI and MRS protocol linked with matched leading-edge analysis pipelines that will be employed to comprehensively measure brain structure, function, microstructure, and metabolite levels in infants and young children. The result will be an unparalleled data resource for examining early infant neurodevelopment which enables the larger scientific community to assess and define normative neurodevelopmental trajectories throughout infancy and early childhood and examine the genetic, biological, and environmental factors that help shape the developing brain.
CRediT authorship contribution statement
Ralph Noeske: Writing – review & editing, Methodology. Allen T. Newton: Writing – review & editing, Methodology. Thomas Pengo: Writing – review & editing, Methodology. Matthew Cieslak: Writing – review & editing, Investigation. James J. Pekar: Writing – review & editing, Methodology. Ai Wern Chung: Writing – review & editing, Methodology. Todd B. Parrish: Writing – review & editing, Methodology. Yulin Chang: Writing – review & editing, Methodology. Minhui Ouyang: Writing – review & editing, Methodology. Samuel Carpenter: Writing – review & editing, Methodology. Xiawei Ou: Writing – review & editing, Methodology. Bryan Caron: Writing – review & editing, Methodology. Regis Ongaro-Carcy: Writing – review & editing, Methodology. Arvind Caprihan: Writing – review & editing, Methodology. Georg Oeltzschner: Writing – review & editing, Software, Methodology. Elizabeth S. Norton: Writing – review & editing, Methodology. Christopher W. Davies-Jenkins: Writing – review & editing, Investigation. Samir Das: Writing – review & editing, Investigation. Anders Dale: Writing – review & editing, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition. William T. Clarke: Writing – review & editing, Methodology. Helge J. Zöllner: Writing – review & editing, Methodology. Leigh MacIntyre: Writing – review & editing, Methodology. Christopher D. Smyser: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Data curation. Mary Beth Nebel: Writing – review & editing, Methodology. Kimberly B. Weldon: Writing – review & editing, Resources, Methodology. Damien A. Fair: Writing – review & editing, Writing – original draft, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization. Cristian Navarro: Writing – review & editing, Methodology. Chad M. Sylvester: Writing – review & editing, Writing – original draft, Resources, Methodology, Investigation, Conceptualization. Saipavitra Murali-Manohar: Writing – review & editing, Methodology. Tracy Riggins: Writing – review & editing, Writing – original draft, Resources, Methodology, Investigation, Funding acquisition, Conceptualization. Lucille A. Moore: Writing – review & editing, Methodology. Kathryn L. Humphreys: Writing – review & editing, Writing – original draft, Resources, Methodology, Investigation, Conceptualization. Kahini Mehta: Writing – review & editing, Methodology. Hao Huang: Writing – review & editing, Writing – original draft, Methodology, Investigation, Funding acquisition, Conceptualization. Andrew R. Mayer: Writing – review & editing, Methodology. Mary Kate Manhard: Writing – review & editing, Methodology. Cecile Madjar: Writing – review & editing, Methodology. Sergiy Boroday: Writing – review & editing, Methodology. Suchandrima Banerjee: Writing – review & editing, Methodology. Natacha Beck: Writing – review & editing, Methodology. Banu Ahtam: Writing – review & editing, Methodology. Jed T. Elison: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Conceptualization. Sharlene D. Newman: Writing – review & editing, Methodology. Essa Yacoub: Writing – review & editing, Software, Methodology, Investigation. Gizeaddis Simegn: Writing – review & editing, Methodology. Xavier Lecour-Bourcher: Writing – review & editing, Methodology. Eric Feczko: Writing – review & editing, Writing – original draft, Visualization, Supervision, Software, Resources, Methodology, Investigation, Formal analysis, Data curation. Bennett A. Landman: Writing – review & editing, Methodology. Jessica L. Wisnowski: Writing – review & editing, Writing – original draft, Visualization, Validation, Resources, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization. Bidhan Lamichhane: Writing – review & editing, Methodology. M Dylan Tisdall: Writing – review & editing, Writing – original draft, Visualization, Methodology, Formal analysis, Conceptualization. Joshua M. Kuperman: Writing – review & editing, Methodology, Investigation. Douglas C Dean: Writing – review & editing, Writing – original draft, Visualization, Supervision, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization. Tobias Kober: Writing – review & editing, Methodology. Steven Kecskemeti: Writing – review & editing, Methodology. M. Okan Irfanoglu: Writing – review & editing, Methodology. Steve C. N. Hui: Writing – review & editing, Methodology. Yansong Zhao: Writing – review & editing, Methodology. Maarten Versluis: Writing – review & editing, Methodology. Jennifer Vannest: Writing – review & editing, Methodology. Brittany R. Howell: Writing – review & editing, Writing – original draft, Visualization, Resources, Methodology, Investigation, Funding acquisition, Conceptualization. Peter C. M. van Zijl: Writing – review & editing, Methodology. Timothy J. Hendrickson: Writing – review & editing, Methodology, Investigation. Jean Tkach: Writing – review & editing, Methodology. Wei Gao: Writing – review & editing, Writing – original draft, Methodology, Funding acquisition. Barry J. Tikalsky: Writing – review & editing, Methodology. Richard A.E. Edden: Writing – review & editing, Writing – original draft, Visualization, Resources, Methodology, Investigation, Conceptualization. Yulu Song: Writing – review & editing, Methodology, Conceptualization. Xu Li: Writing – review & editing, Methodology. Andrew L. Alexander: Writing – review & editing, Writing – original draft, Methodology, Conceptualization. W. Kyle Simmons: Writing – review & editing, Methodology. Erik G. Lee: Writing – review & editing, Visualization, Methodology. Borjan Gagoski: Writing – review & editing, Writing – original draft, Visualization, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Vidya Rajagopalan: Writing – review & editing, Methodology. Russell A. Poldrack: Writing – review & editing, Methodology. Carlo Pierpaoli: Writing – review & editing, Methodology. Aaron T. Gudmundson: Writing – review & editing, Methodology. Eunkyung Shin: Writing – review & editing, Methodology. Alice M. Graham: Writing – review & editing, Methodology. Lisa S. Scott: Writing – review & editing, Investigation. Guillaume Gilbert: Writing – review & editing, Methodology. Theodore D. Satterthwaite: Writing – review & editing, Methodology. Sandeep K. Ganji: Writing – review & editing, Methodology. Taylor Salo: Writing – review & editing, Methodology. Laetitia Fesselier: Writing – review & editing, Methodology. Jens T. Rosenberg: Writing – review & editing, Investigation. Alan C. Evans: Writing – review & editing, Methodology. Pierre Rioux: Writing – review & editing, Methodology. Alexander J. Dufford: Writing – review & editing, Methodology. Dan W. Rettmann: Writing – review & editing, Methodology. Tom Hilbert: Writing – review & editing, Methodology, Conceptualization. Michael P. Harms: Writing – review & editing, Methodology. Maren Macgregor-Hannah: Writing – review & editing, Methodology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Tobias Kober and Tom Hilbert are employees of Siemens Healthineers International AG, Switzerland. Yulin Chang is an employee of Siemens Medical Solutions USA Inc. Dan Rettmann and Ralph Noeske are employed by GE HealthCare. Guillaume Gilbert, Yansong Zhao, Sandeep Ganji, and Maarten Versluis are employed by Philips Healthcare. Carina Lucena, Lucky Heisler-Roman, and Dhruman Goradia are employed by PrimeNeuro Inc. Under a license agreement between Philips and the Johns Hopkins University, Dr. van Zijl and the University are entitled to fees related to an imaging device used in the study discussed for publication. Dr. van Zijl also is a paid lecturer for Philips and receives research support from Philips. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies. Damien Fair is a patent holder on the Framwise Integrated Real-Time Motion Monitoring (FIRMM) software. He is also a co-founder of Turing Medical Technologies, Inc. The nature of this financial interest and the design of the study have been reviewed by two committees at the University of Minnesota. They have put in place a plan to help ensure that this research is not affected by the financial interest. All other authors report no biomedical financial interests or potential conflicts of interest.
Acknowledgements
Data, plans, and concepts used in the preparation of this article were obtained from the Healthy Brain and Child Development (HBCD) Study (https://hbcdstudy.org/). This is a multisite, longitudinal study designed to recruit over 7000 families and follow them from pregnancy to early childhood. The HBCD Study is supported by the National Institutes of Health and additional federal partners under award numbers U01DA055352, U01DA055353, U01DA055366, U01DA055365, U01DA055362, U01DA055342, U01DA055360, U01DA055350, U01DA055338, U01DA055355, U01DA055363, U01DA055349, U01DA055361, U01DA055316, U01DA055344, U01DA055322, U01DA055369, U01DA055358, U01DA055371, U01DA055359, U01DA055354, U01DA055370, U01DA055347, U01DA055357, U01DA055367, U24DA055325, U24DA055330. A full list of supporters is available at https://hbcdstudy.org/about/federal-partners/. A listing of participating sites and a complete listing of the study investigators can be found at https://hbcdstudy.org/study-sites/. HBCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in the analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or HBCD consortium investigators. MRS sequences were developed as part of NIH grant P41 EB031771, R01 EB016089, R01 EB023963, and R01 EB032788. MRS data processing pipelines were developed with support from NIH grants R00 AG062230, R21 EB033516, and K99 AG080084. Support for title page creation and format was provided by AuthorArranger, a tool developed at the National Cancer Institute.
Contributor Information
Douglas C. Dean, III, Email: deaniii@wisc.edu.
Damien A. Fair, Email: faird@umn.edu.
Christopher D. Smyser, Email: smyserc@wustl.edu.
Jed T. Elison, Email: jtelison@umn.edu.
Data Availability
Data will be made available on request.
References
- Alex A.M., Aguate F., Botteron K., Buss C., Chong Y.S., Dager S.R., Donald K.A., Entringer S., Fair D.A., Fortier M.V., Gaab N., Gilmore J.H., Girault J.B., Graham A.M., Groenewold N.A., Hazlett H., Lin W., Meaney M.J., Piven J., Qiu A., Rasmussen J.M., Roos A., Schultz R.T., Skeide M.A., Stein D.J., Styner M., Thompson P.M., Turesky T.K., Wadhwa P.D., Zar H.J., Zollei L., de Los Campos G., Knickmeyer R.C., group EO A global multicohort study to map subcortical brain development and cognition in infancy and early childhood. Nat. Neurosci. 2024;27(1):176–186. doi: 10.1038/s41593-023-01501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander A.L., Hurley S.A., Samsonov A.A., Adluru N., Hosseinbor A.P., Mossahebi P., Tromp do P.M., Zakszewski E., Field A.S. Characterization of cerebral white matter properties using quantitative magnetic resonance imaging stains. Brain Connect. 2011;1(6):423–446. doi: 10.1089/brain.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D.C., Dyrby T.B., Nilsson M., Zhang H. Imaging brain microstructure with diffusion MRI: practicality and applications. NMR Biomed. 2019;32(4) doi: 10.1002/nbm.3841. [DOI] [PubMed] [Google Scholar]
- Almli C.R., Rivkin M.J., McKinstry R.C., Brain Development Cooperative G The NIH MRI study of normal brain development (Objective-2): newborns, infants, toddlers, and preschoolers. Neuroimage. 2007;35(1):308–325. doi: 10.1016/j.neuroimage.2006.08.058. [DOI] [PubMed] [Google Scholar]
- Andersen M., Bjorkman-Burtscher I.M., Marsman A., Petersen E.T., Boer V.O. Improvement in diagnostic quality of structural and angiographic MRI of the brain using motion correction with interleaved, volumetric navigators. PLoS One. 2019;14(5) doi: 10.1371/journal.pone.0217145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J.L., Skare S., Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20(2):870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- Badke D′Andrea C., Kenley J.K., Montez D.F., Mirro A.E., Miller R.L., Earl E.A., Koller J.M., Sung S., Yacoub E., Elison J.T., Fair D.A., Dosenbach N.U.F., Rogers C.E., Smyser C.D., Greene D.J. Real-time motion monitoring improves functional MRI data quality in infants. Dev. Cogn. Neurosci. 2022;55 doi: 10.1016/j.dcn.2022.101116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser P.J., Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J. Magn. Reson B. 1996;111(3):209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Bethlehem R.A.I., Seidlitz J., White S.R., Vogel J.W., Anderson K.M., Adamson C., Adler S., Alexopoulos G.S., Anagnostou E., Areces-Gonzalez A., Astle D.E., Auyeung B., Ayub M., Bae J., Ball G., Baron-Cohen S., Beare R., Bedford S.A., Benegal V., Vetsa Brain charts for the human lifespan. Nature. 2022;604(7906):525–533. doi: 10.1038/s41586-022-04554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookheimer S.Y., Salat D.H., Terpstra M., Ances B.M., Barch D.M., Buckner R.L., Burgess G.C., Curtiss S.W., Diaz-Santos M., Elam J.S., Fischl B., Greve D.N., Hagy H.A., Harms M.P., Hatch O.M., Hedden T., Hodge C., Japardi K.C., Kuhn T.P., Ly T.K., Smith S.M., Somerville L.H., Ugurbil K., van der Kouwe A., Van Essen D., Woods R.P., Yacoub E. The lifespan human connectome project in aging: an overview. Neuroimage. 2019;185:335–348. doi: 10.1016/j.neuroimage.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley P.A. Spatial localization in NMR spectroscopy in vivo. Ann. N. Y Acad. Sci. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- Botvinik-Nezer R., Holzmeister F., Camerer C.F., Dreber A., Huber J., Johannesson M., Kirchler M., Iwanir R., Mumford J.A., Adcock R.A., Avesani P., Baczkowski B.M., Bajracharya A., Bakst L., Ball S., Barilari M., Bault N., Beaton D., Beitner J., Benoit R.G., Berkers R., Bhanji J.P., Biswal B.B., Bobadilla-Suarez S., Bortolini T., Bottenhorn K.L., Bowring A., Braem S., Brooks H.R., Brudner E.G., Calderon C.B., Camilleri J.A., Castrellon J.J., Cecchetti L., Cieslik E.C., Cole Z.J., Collignon O., Cox R.W., Cunningham W.A., Czoschke S., Dadi K., Davis C.P., Luca A., Delgado M.R., Demetriou L., Dennison J.B., Di X., Dickie E.W., Dobryakova E., Donnat C.L., Dukart J., Duncan N.W., Durnez J., Eed A., Eickhoff S.B., Erhart A., Fontanesi L., Fricke G.M., Fu S., Galvan A., Gau R., Genon S., Glatard T., Glerean E., Goeman J.J., Golowin S.A.E., Gonzalez-Garcia C., Gorgolewski K.J., Grady C.L., Green M.A., Guassi Moreira J.F., Guest O., Hakimi S., Hamilton J.P., Hancock R., Handjaras G., Harry B.B., Hawco C., Herholz P., Herman G., Heunis S., Hoffstaedter F., Hogeveen J., Holmes S., Hu C.P., Huettel S.A., Hughes M.E., Iacovella V., Iordan A.D., Isager P.M., Isik A.I., Jahn A., Johnson M.R., Johnstone T., Joseph M.J.E., Juliano A.C., Kable J.W., Kassinopoulos M., Koba C., Kong X.Z., Koscik T.R., Kucukboyaci N.E., Kuhl B.A., Kupek S., Laird A.R., Lamm C., Langner R., Lauharatanahirun N., Lee H., Lee S., Leemans A., Leo A., Lesage E., Li F., Li M.Y.C., Lim P.C., Lintz E.N., Liphardt S.W., Losecaat Vermeer A.B., Love B.C., Mack M.L., Malpica N., Marins T., Maumet C., McDonald K., McGuire J.T., Melero H., Mendez Leal A.S., Meyer B., Meyer K.N., Mihai G., Mitsis G.D., Moll J., Nielson D.M., Nilsonne G., Notter M.P., Olivetti E., Onicas A.I., Papale P., Patil K.R., Peelle J.E., Perez A., Pischedda D., Poline J.B., Prystauka Y., Ray S., Reuter-Lorenz P.A., Reynolds R.C., Ricciardi E., Rieck J.R., Rodriguez-Thompson A.M., Romyn A., Salo T., Samanez-Larkin G.R., Sanz-Morales E., Schlichting M.L., Schultz D.H., Shen Q., Sheridan M.A., Silvers J.A., Skagerlund K., Smith A., Smith D.V., Sokol-Hessner P., Steinkamp S.R., Tashjian S.M., Thirion B., Thorp J.N., Tinghog G., Tisdall L., Tompson S.H., Toro-Serey C., Torre Tresols J.J., Tozzi L., Truong V., Turella L., van 't Veer A.E., Verguts T., Vettel J.M., Vijayarajah S., Vo K., Wall M.B., Weeda W.D., Weis S., White D.J., Wisniewski D., Xifra-Porxas A., Yearling E.A., Yoon S., Yuan R., Yuen K.S.L., Zhang L., Zhang X., Zosky J.E., Nichols T.E., Poldrack R.A., Schonberg T. Variability in the analysis of a single neuroimaging dataset by many teams. Nature. 2020;582(7810):84–88. doi: 10.1038/s41586-020-2314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey B.J., Cannonier T., Conley M.I., Cohen A.O., Barch D.M., Heitzeg M.M., Soules M.E., Teslovich T., Dellarco D.V., Garavan H., Orr C.A., Wager T.D., Banich M.T., Speer N.K., Sutherland M.T., Riedel M.C., Dick A.S., Bjork J.M., Thomas K.M., Chaarani B., Mejia M.H., Hagler D.J., Jr., Daniela Cornejo M., Sicat C.S., Harms M.P., Dosenbach N.U.F., Rosenberg M., Earl E., Bartsch H., Watts R., Polimeni J.R., Kuperman J.M., Fair D.A., Dale A.M., Workgroup AIA The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 2018;32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu S., Wang Y., Kang Y., Haacke E.M. STrategically Acquired Gradient Echo (STAGE) imaging, part I: creating enhanced T1 contrast and standardized susceptibility weighted imaging and quantitative susceptibility mapping. Magn. Reson Imaging. 2018;46:130–139. doi: 10.1016/j.mri.2017.10.005. [DOI] [PubMed] [Google Scholar]
- Chertkow H., Borrie M., Whitehead V., Black S.E., Feldman H.H., Gauthier S., Hogan D.B., Masellis M., McGilton K., Rockwood K., Tierney M.C., Andrew M., Hsiung G.R., Camicioli R., Smith E.E., Fogarty J., Lindsay J., Best S., Evans A., Das S., Mohaddes Z., Pilon R., Poirier J., Phillips N.A., MacNamara E., Dixon R.A., Duchesne S., MacKenzie I., Rylett R.J. The comprehensive assessment of neurodegeneration and dementia: canadian cohort study. Can. J. Neurol. Sci. 2019;46(5):499–511. doi: 10.1017/cjn.2019.27. [DOI] [PubMed] [Google Scholar]
- Choi C., Dimitrov I., Douglas D., Zhao C., Hawesa H., Ghose S., Tamminga C.A. In vivo detection of serine in the human brain by proton magnetic resonance spectroscopy (1H-MRS) at 7 Tesla. Magn. Reson. Med. 2009;62(4):1042–1046. doi: 10.1002/mrm.22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I.Y., Andronesi O.C., Barker P., Bogner W., Edden R.A.E., Kaiser L.G., Lee P., Marjanska M., Terpstra M., de Graaf R.A. Spectral editing in (1) H magnetic resonance spectroscopy: experts' consensus recommendations. NMR Biomed. 2021;34(5) doi: 10.1002/nbm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslak M., Cook P.A., He X., Yeh F.C., Dhollander T., Adebimpe A., Aguirre G.K., Bassett D.S., Betzel R.F., Bourque J., Cabral L.M., Davatzikos C., Detre J.A., Earl E., Elliott M.A., Fadnavis S., Fair D.A., Foran W., Fotiadis P., Garyfallidis E., Giesbrecht B., Gur R.C., Gur R.E., Kelz M.B., Keshavan A., Larsen B.S., Luna B., Mackey A.P., Milham M.P., Oathes D.J., Perrone A., Pines A.R., Roalf D.R., Richie-Halford A., Rokem A., Sydnor V.J., Tapera T.M., Tooley U.A., Vettel J.M., Yeatman J.D., Grafton S.T., Satterthwaite T.D. QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nat. Methods. 2021;18(7):775–778. doi: 10.1038/s41592-021-01185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric R., Rosen A.F.G., Erus G., Cieslak M., Adebimpe A., Cook P.A., Bassett D.S., Davatzikos C., Wolf D.H., Satterthwaite T.D. Mitigating head motion artifact in functional connectivity MRI. Nat. Protoc. 2018;13(12):2801–2826. doi: 10.1038/s41596-018-0065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke W.T., Bell T.K., Emir U.E., Mikkelsen M., Oeltzschner G., Shamaei A., Soher B.J., Wilson M. NIfTI-MRs: a standard data format for magnetic resonance spectroscopy. Magn. Reson Med. 2022;88(6):2358–2370. doi: 10.1002/mrm.29418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conolly S., Glover G., Nishimura D., Macovski A. A reduced power selective adiabatic spin-echo pulse sequence. Magn. Reson Med. 1991;18(1):28–38. doi: 10.1002/mrm.1910180105. [DOI] [PubMed] [Google Scholar]
- Das S., Zijdenbos A.P., Harlap J., Vins D., Evans A.C. LORIS: a web-based data management system for multi-center studies. Front Neuroinf. 2011;5:37. doi: 10.3389/fninf.2011.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D.C., 3rd, Dirks H., O'Muircheartaigh J., Walker L., Jerskey B.A., Lehman K., Han M., Waskiewicz N., Deoni S.C. Pediatric neuroimaging using magnetic resonance imaging during non-sedated sleep. Pedia Radio. 2014;44(1):64–72. doi: 10.1007/s00247-013-2752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelchand D.K., Berrington A., Noeske R., Joers J.M., Arani A., Gillen J., Schar M., Nielsen J.F., Peltier S., Seraji-Bozorgzad N., Landheer K., Juchem C., Soher B.J., Noll D.C., Kantarci K., Ratai E.M., Mareci T.H., Barker P.B., Oz G. Across-vendor standardization of semi-LASER for single-voxel MRS at 3T. NMR Biomed. 2021;34(5) doi: 10.1002/nbm.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deoni S.C. Quantitative relaxometry of the brain. Top. Magn. Reson Imaging. 2010;21(2):101–113. doi: 10.1097/RMR.0b013e31821e56d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deoni S.C., Peters T.M., Rutt B.K. High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn. Reson Med. 2005;53(1):237–241. doi: 10.1002/mrm.20314. [DOI] [PubMed] [Google Scholar]
- Deoni S.C., Rutt B.K., Peters T.M. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn. Reson Med. 2003;49(3):515–526. doi: 10.1002/mrm.10407. [DOI] [PubMed] [Google Scholar]
- Deoni S.C.L., Williams S.C.R., Jezzard P., Suckling J., Murphy D.G.M., Jones D.K. Standardized structural magnetic resonance imaging in multicentre studies using quantitative T1 and T2 imaging at 1.5 T. Neuroimage. 2008;40(2):662–671. doi: 10.1016/j.neuroimage.2007.11.052. [DOI] [PubMed] [Google Scholar]
- Dick A.S., Lopez D.A., Watts A.L., Heeringa S., Reuter C., Bartsch H., Fan C.C., Kennedy D.N., Palmer C., Marshall A., Haist F., Hawes S., Nichols T.E., Barch D.M., Jernigan T.L., Garavan H., Grant S., Pariyadath V., Hoffman E., Neale M., Stuart E.A., Paulus M.P., Sher K.J., Thompson W.K. Meaningful associations in the adolescent brain cognitive development study. Neuroimage. 2021;239 doi: 10.1016/j.neuroimage.2021.118262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Does M.D. Inferring brain tissue composition and microstructure via MR relaxometry. Neuroimage. 2018;182:136–148. doi: 10.1016/j.neuroimage.2017.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U.F., Koller J.M., Earl E.A., Miranda-Dominguez O., Klein R.L., Van A.N., Snyder A.Z., Nagel B.J., Nigg J.T., Nguyen A.L., Wesevich V., Greene D.J., Fair D.A. Real-time motion analytics during brain MRI improve data quality and reduce costs. NeuroImage. 2017;161:80–93. doi: 10.1016/j.neuroimage.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden R.A., Oeltzschner G., Harris A.D., Puts N.A., Chan K.L., Boer V.O., Schär M., Barker P.B. Prospective frequency correction for macromolecule-suppressed GABA editing at 3T. J. Magn. Reson Imaging. 2016;44(6):1474–1482. doi: 10.1002/jmri.25304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A.D., Rueckert D., Smith S.M., Abo Seada S., Alansary A., Almalbis J., Allsop J., Andersson J., Arichi T., Arulkumaran S., Bastiani M., Batalle D., Baxter L., Bozek J., Braithwaite E., Brandon J., Carney O., Chew A., Christiaens D., Chung R., Colford K., Cordero-Grande L., Counsell S.J., Cullen H., Cupitt J., Curtis C., Davidson A., Deprez M., Dillon L., Dimitrakopoulou K., Dimitrova R., Duff E., Falconer S., Farahibozorg S.R., Fitzgibbon S.P., Gao J., Gaspar A., Harper N., Harrison S.J., Hughes E.J., Hutter J., Jenkinson M., Jbabdi S., Jones E., Karolis V., Kyriakopoulou V., Lenz G., Makropoulos A., Malik S., Mason L., Mortari F., Nosarti C., Nunes R.G., O'Keeffe C., O'Muircheartaigh J., Patel H., Passerat-Palmbach J., Pietsch M., Price A.N., Robinson E.C., Rutherford M.A., Schuh A., Sotiropoulos S., Steinweg J., Teixeira R., Tenev T., Tournier J.D., Tusor N., Uus A., Vecchiato K., Williams L.Z.J., Wright R., Wurie J., Hajnal J.V. The developing human connectome project neonatal data release. Front Neurosci. 2022;16 doi: 10.3389/fnins.2022.886772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses P., Seiberlich N., Ma D., Breuer F.A., Jakob P.M., Griswold MA, Gulani V. IR TrueFISP with a golden-ratio-based radial readout: fast quantification of T1, T2, and proton density. Magn. Reson Med. 2013;69(1):71–81. doi: 10.1002/mrm.24225. [DOI] [PubMed] [Google Scholar]
- Esteban O., Markiewicz C.J., Blair R.W., Moodie C.A., Isik A.I., Erramuzpe A., Kent J.D., Goncalves M., DuPre E., Snyder M., Oya H., Ghosh S.S., Wright J., Durnez J., Poldrack R.A., Gorgolewski K.J. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods. 2019;16(1):111–116. doi: 10.1038/s41592-018-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban O., Wright J., Markiewicz C.J., Thompson W.H., Goncalves M., Ciric R., Blair R.W., Feingold F., Rokem A., Ghosh S. (2019b) NiPreps: enabling the division of labor in neuroimaging beyond fMRIPrep..
- Evans A.C., Brain Development Cooperative G. The NIH MRI study of normal brain development. Neuroimage. 2006;30(1):184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Fair D.A., Miranda-Dominguez O., Snyder A.Z., Perrone A., Earl E.A., Van A.N., Koller J.M., Feczko E., Tisdall M.D., van der Kouwe A., Klein R.L., Mirro A.E., Hampton J.M., Adeyemo B., Laumann T.O., Gratton C., Greene D.J., Schlaggar B.L., Hagler D.J., Jr., Watts R., Garavan H., Barch D.M., Nigg J.T., Petersen S.E., Dale A.M., Feldstein-Ewing S.W., Nagel B.J., Dosenbach N.U.F. Correction of respiratory artifacts in MRI head motion estimates. Neuroimage. 2020;208 doi: 10.1016/j.neuroimage.2019.116400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feczko E., Conan G., Marek S., Tervo-Clemmens B., Cordova M., Doyle O., Earl E., Perrone A., Sturgeon D., Klein R., Harman G., Kilamovich D., Hermosillo R., Miranda-Dominguez O., Adebimpe A., Bertolero M., Cieslak M., Covitz S., Hendrickson T., Juliano A.C., Snider K., Moore L.A., Uriartel J., Graham A.M., Calabro F., Rosenberg M.D., Rapuano K.M., Casey B., Watts R., Hagler D., Thompson W.K., Nichols T.E., Hoffman E., Luna B., Garavan H., Satterthwaite T.D., Ewing S.F., Nagel B., Dosenbach N.U.F., Fair D.A. Adolesc. Brain Cogn. Dev. (ABCD) Community MRI Collect. Uti. 2021 doi: 10.1101/2021.07.09.451638. bioRxiv:2021.2007.2009.451638. [DOI] [Google Scholar]
- Feinberg D.A., Moeller S., Smith S.M., Auerbach E., Ramanna S., Gunther M., Glasser M.F., Miller K.L., Ugurbil K., Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleysher R., Fleysher L., Gonen O. The optimal MR acquisition strategy for exponential decay constants estimation. Magn. Reson Imaging. 2008;26(3):433–435. doi: 10.1016/j.mri.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S., Hagiwara A., Hori M., Warntjes M., Kamagata K., Fukunaga I., Andica C., Maekawa T., Irie R., Takemura M.Y., Kumamaru K.K., Wada A., Suzuki M., Ozaki Y., Abe O., Aoki S. Three-dimensional high-resolution simultaneous quantitative mapping of the whole brain with 3D-QALAS: An accuracy and repeatability study. Magn. Reson Imaging. 2019;63:235–243. doi: 10.1016/j.mri.2019.08.031. [DOI] [PubMed] [Google Scholar]
- Gasparovic C., Song T., Devier D., Bockholt H.J., Caprihan A., Mullins P.G., Posse S., Jung R.E., Morrison L.A. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magn. Reson Med. 2006;55(6):1219–1226. doi: 10.1002/mrm.20901. [DOI] [PubMed] [Google Scholar]
- Glasser M.F., Smith S.M., Marcus D.S., Andersson J.L.R., Auerbach E.J., Behrens T.E.J., Coalson T.S., Harms M.P., Jenkinson M., Moeller S., Robinson E.C., Sotiropoulos S.N., Xu J., Yacoub E., Ugurbil K., Van Essen D.C. The Human Connectome Project's neuroimaging approach. Nat. Neurosci. 2016;19(9):1175–1187. doi: 10.1038/nn.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser M.F., Sotiropoulos S.N., Wilson J.A., Coalson T.S., Fischl B., Andersson J.L., Xu J., Jbabdi S., Webster M., Polimeni J.R., Van Essen D.C., Jenkinson M. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksan S., Hartley C., Hurley S.A., Winkler A.M., Duff E.P., Jenkinson M., Rogers R., Clare S., Slater R. Optimal echo time for functional MRI of the infant brain identified in response to noxious stimulation. Magn. Reson Med. 2017;78(2):625–631. doi: 10.1002/mrm.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves M., Markiewicz C.J., Styner M., Moore L.A., Snider K., Earl E.A., Smyser C.D., Zöllei L., Poldrack R.A., Esteban O. NiBabies: a robust preprocessing workflow tailored for neonate and infant MRI. 27th Annu. Meet. Organ. Hum. Brain Mapp. 2021 [Google Scholar]
- Gorgolewski K.J., Auer T., Calhoun V.D., Craddock R.C., Das S., Duff E.P., Flandin G., Ghosh S.S., Glatard T., Halchenko Y.O., Handwerker D.A., Hanke M., Keator D., Li X., Michael Z., Maumet C., Nichols B.N., Nichols T.E., Pellman J., Poline J.-B., Rokem A., Schaefer G., Sochat V., Triplett W., Turner J.A., Varoquaux G., Poldrack R.A. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data. 2016;3(1) doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräfe D., Frahm J., Merkenschlager A., Voit D., Hirsch F.W. Quantitative T1 mapping of the normal brain from early infancy to adulthood. Pedia Radio. 2021;51(3):450–456. doi: 10.1007/s00247-020-04842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler D.J., Jr., Hatton S., Cornejo M.D., Makowski C., Fair D.A., Dick A.S., Sutherland M.T., Casey B.J., Barch D.M., Harms M.P., Watts R., Bjork J.M., Garavan H.P., Hilmer L., Pung C.J., Sicat C.S., Kuperman J., Bartsch H., Xue F., Heitzeg M.M., Laird A.R., Trinh T.T., Gonzalez R., Tapert S.F., Riedel M.C., Squeglia L.M., Hyde L.W., Rosenberg M.D., Earl E.A., Howlett K.D., Baker F.C., Soules M., Diaz J., de Leon O.R., Thompson W.K., Neale M.C., Herting M., Sowell E.R., Alvarez R.P., Hawes S.W., Sanchez M., Bodurka J., Breslin F.J., Morris A.S., Paulus M.P., Simmons W.K., Polimeni J.R., van der Kouwe A., Nencka A.S., Gray K.M., Pierpaoli C., Matochik J.A., Noronha A., Aklin W.M., Conway K., Glantz M., Hoffman E., Little R., Lopez M., Pariyadath V., Weiss S.R., Wolff-Hughes D.L., DelCarmen-Wiggins R., Feldstein Ewing S.W., Miranda-Dominguez O., Nagel B.J., Perrone A.J., Sturgeon D.T., Goldstone A., Pfefferbaum A., Pohl K.M., Prouty D., Uban K., Bookheimer S.Y., Dapretto M., Galvan A., Bagot K., Giedd J., Infante M.A., Jacobus J., Patrick K., Shilling P.D., Desikan R., Li Y., Sugrue L., Banich M.T., Friedman N., Hewitt J.K., Hopfer C., Sakai J., Tanabe J., Cottler L.B., Nixon S.J., Chang L., Cloak C., Ernst T., Reeves G., Kennedy D.N., Heeringa S., Peltier S., Schulenberg J., Sripada C., Zucker R.A., Iacono W.G., Luciana M., Calabro F.J., Clark D.B., Lewis D.A., Luna B., Schirda C., Brima T., Foxe J.J., Freedman E.G., Mruzek D.W., Mason M.J., Huber R., McGlade E., Prescot A., Renshaw P.F., Yurgelun-Todd D.A., Allgaier N.A., Dumas J.A., Ivanova M., Potter A., Florsheim P., Larson C., Lisdahl K., Charness M.E., Fuemmeler B., Hettema J.M., Maes H.H., Steinberg J., Anokhin A.P., Glaser P., Heath A.C., Madden P.A., Baskin-Sommers A., Constable R.T., Grant S.J., Dowling G.J., Brown S.A., Jernigan T.L., Dale A.M. Image processing and analysis methods for the Adolescent Brain Cognitive Development Study. Neuroimage. 2019;202 doi: 10.1016/j.neuroimage.2019.116091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halchenko Y., Meyer K., Poldrack B., Solanky D., Wagner A., Gors J., MacFarlane D., Pustina D., Sochat V., Ghosh S. DataLad: distributed system for joint management of code, data, and their relationship. J. Open Source Softw. 2021;6(63) [Google Scholar]
- Harms M.P., Somerville L.H., Ances B.M., Andersson J., Barch D.M., Bastiani M., Bookheimer S.Y., Brown T.B., Buckner R.L., Burgess G.C., Coalson T.S., Chappell M.A., Dapretto M., Douaud G., Fischl B., Glasser M.F., Greve D.N., Hodge C., Jamison K.W., Jbabdi S., Kandala S., Li X., Mair R.W., Mangia S., Marcus D., Mascali D., Moeller S., Nichols T.E., Robinson E.C., Salat D.H., Smith S.M., Sotiropoulos S.N., Terpstra M., Thomas K.M., Tisdall M.D., Ugurbil K., van der Kouwe A., Woods R.P., Zollei L., Van Essen D.C., Yacoub E. Extending the Human Connectome Project across ages: imaging protocols for the Lifespan Development and Aging projects. Neuroimage. 2018;183:972–984. doi: 10.1016/j.neuroimage.2018.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A.D., Glaubitz B., Near J., John Evans C., Puts N.A.J., Schmidt-Wilcke T., Tegenthoff M., Barker P.B., Edden R.A.E. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn. Reson. Med. 2014;72(4):941–948. doi: 10.1002/mrm.25009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett H.C., Gu H., McKinstry R.C., Shaw D.W.W., Botteron K.N., Dager S.R., Styner M., Vachet C., Gerig G., Paterson S.J., Schultz R.T., Estes A.M., Evans A.C., Piven J., IBIS Network Brain Volume Findings in 6-Month-Old Infants at High Familial Risk for Autism. Am J Psychiatry. 2012;169(6):601–608. doi: 10.1176/appi.ajp.2012.11091425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett H.C., Gu H., Munsell B.C., Kim S.H., Styner M., Wolff J.J., Elison J.T., Swanson M.R., Zhu H., Botteron K.N., Collins D.L., Constantino J.N., Dager S.R., Estes A.M., Evans A.C., Fonov V.S., Gerig G., Kostopoulos P., McKinstry R.C., Pandey J., Paterson S., Pruett J.R., Schultz R.T., Shaw D.W., Zwaigenbaum L., Piven J., Network I., Clinical S., Data Coordinating C., Image Processing C., Statistical A. Early brain development in infants at high risk for autism spectrum disorder. Nature. 2017;542(7641):348–351. doi: 10.1038/nature21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson E., McKinnon G., Lee T.Y., Rutt B.K. A fast 3D look-locker method for volumetric T1 mapping. Magn. Reson Imaging. 1999;17(8):1163–1171. doi: 10.1016/s0730-725x(99)00025-9. [DOI] [PubMed] [Google Scholar]
- Hendrickson T.J., Reiners P., Moore L.A., Perrone A.J., Alexopoulos D., Lee E.G., Styner M., Kardan O., Chamberlain T.A., Mummaneni A., Caldas H.A., Bower B., Stoyell S., Martin T., Sung S., Fair E., Uriarte-Lopez J., Rueter A.R., Yacoub E., Rosenberg M.D., Smyser C.D., Elison J.T., Graham A., Fair D.A., Feczko E. BIBSNet: a deep learning baby image brain segmentation network for MRI scans. bioRxiv. 2023 doi: 10.1101/2023.03.22.533696. [DOI] [Google Scholar]
- Howell B.R., Styner M.A., Gao W., Yap P.T., Wang L., Baluyot K., Yacoub E., Chen G., Potts T., Salzwedel A., Li G., Gilmore J.H., Piven J., Smith J.K., Shen D., Ugurbil K., Zhu H., Lin W., Elison J.T. The UNC/UMN Baby Connectome Project (BCP): an overview of the study design and protocol development. Neuroimage. 2019;185:891–905. doi: 10.1016/j.neuroimage.2018.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. Oxford University Press; 2022. Imaging the Infant Brain. [DOI] [Google Scholar]
- Huang H., Zhang J., Wakana S., Zhang W., Ren T., Richards L.J., Yarowsky P., Donohue P., Graham E., van Zijl P.C.M., Mori S. White and gray matter development in human fetal, newborn and pediatric brains. NeuroImage. 2006;33(1):27–38. doi: 10.1016/j.neuroimage.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Hui S.C.N., Mikkelsen M., Zollner H.J., Ahluwalia V., Alcauter S., Baltusis L., Barany D.A., Barlow L.R., Becker R., Berman J.I., Berrington A., Bhattacharyya P.K., Blicher J.U., Bogner W., Brown M.S., Calhoun V.D., Castillo R., Cecil K.M., Choi Y.B., Chu W.C.W., Clarke W.T., Craven A.R., Cuypers K., Dacko M., de la Fuente-Sandoval C., Desmond P., Domagalik A., Dumont J., Duncan N.W., Dydak U., Dyke K., Edmondson D.A., Ende G., Ersland L., Evans C.J., Fermin A.S.R., Ferretti A., Fillmer A., Gong T., Greenhouse I., Grist J.T., Gu M., Harris A.D., Hat K., Heba S., Heckova E., Hegarty J.P., 2nd, Heise K.F., Honda S., Jacobson A., Jansen J.F.A., Jenkins C.W., Johnston S.J., Juchem C., Kangarlu A., Kerr A.B., Landheer K., Lange T., Lee P., Levendovszky S.R., Limperopoulos C., Liu F., Lloyd W., Lythgoe D.J., Machizawa M.G., MacMillan E.L., Maddock R.J., Manzhurtsev A.V., Martinez-Gudino M.L., Miller J.J., Mirzakhanian H., Moreno-Ortega M., Mullins P.G., Nakajima S., Near J., Noeske R., Nordhoy W., Oeltzschner G., Osorio-Duran R., Otaduy M.C.G., Pasaye E.H., Peeters R., Peltier S.J., Pilatus U., Polomac N., Porges E.C., Pradhan S., Prisciandaro J.J., Puts N.A., Rae C.D., Reyes-Madrigal F., Roberts T.P.L., Robertson C.E., Rosenberg J.T., Rotaru D.G., O'Gorman Tuura R.L., Saleh M.G., Sandberg K., Sangill R., Schembri K., Schrantee A., Semenova N.A., Singel D., Sitnikov R., Smith J., Song Y., Stark C., Stoffers D., Swinnen S.P., Tain R., Tanase C., Tapper S., Tegenthoff M., Thiel T., Thioux M., Truong P., van Dijk P., Vella N., Vidyasagar R., Vovk A., Wang G., Westlye L.T., Wilbur T.K., Willoughby W.R., Wilson M., Wittsack H.J., Woods A.J., Wu Y.C., Xu J., Lopez M.Y., Yeung D.K.W., Zhao Q., Zhou X., Zupan G., Edden R.A.E. Frequency drift in MR spectroscopy at 3T. Neuroimage. 2021;241 doi: 10.1016/j.neuroimage.2021.118430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S.C.N., Murali-Manohar S., Zöllner H.J., Hupfeld K.E., Davies-Jenkins C.W., Gudmundson A.T., Song Y., Yedavalli V., Wisnowski J.L., Gagoski B., Oeltzschner G., Edden R.A.E. Integrated Short-TE and Hadamard-edited Multi-Sequence (ISTHMUS) for advanced MRS. J Neurosci Methods. 2024;409 doi: 10.1016/j.jneumeth.2024.110206. PMID: 38942238; PMCID: PMC11286357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu M.O., Sadeghi N., Sarlls J., Pierpaoli C. Improved reproducibility of diffusion MRI of the human brain with a four-way blip-up and down phase-encoding acquisition approach. Magn. Reson Med. 2021;85(5):2696–2708. doi: 10.1002/mrm.28624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kecskemeti S., Samsonov A., Hurley S.A., Dean D.C., Field A., Alexander A.L. MPnRAGE: a technique to simultaneously acquire hundreds of differently contrasted MPRAGE images with applications to quantitative T1 mapping. Magn. Reson Med. 2016;75(3):1040–1053. doi: 10.1002/mrm.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kecskemeti S.R., Alexander A.L. Test-retest of automated segmentation with different motion correction strategies: a comparison of prospective versus retrospective methods. Neuroimage. 2020;209 doi: 10.1016/j.neuroimage.2019.116494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D.N., Abraham S.A., Bates J.F., Crowley A., Ghosh S., Gillespie T., Goncalves M., Grethe J.S., Halchenko Y.O., Hanke M., Haselgrove C., Hodge S.M., Jarecka D., Kaczmarzyk J., Keator D.B., Meyer K., Martone M.E., Padhy S., Poline J.B., Preuss N., Sincomb T., Travers M. Everything matters: The ReproNim perspective on reproducible neuroimaging. Front Neuroinf. 2019;13:1. doi: 10.3389/fninf.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavan A., Yeatman J.D., Rokem A. Combining citizen science and deep learning to amplify expertise in neuroimaging. Front Neuroinf. 2019;13:29. doi: 10.3389/fninf.2019.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiar G., Clucas J., Feczko E., Goncalves M., Jarecka D., Markiewicz C.J., Halchenko Y.O., Hermosillo R., Li X., Miranda-Dominguez O., Ghosh S., Poldrack R.A., Satterthwaite T.D., Milham M.P., Fair D. Align with the NMIND consortium for better neuroimaging. Nat. Hum. Behav. 2023;7(7):1027–1028. doi: 10.1038/s41562-023-01647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzer G.M., Sochat V., Bauer M.W. Singularity: Scientific containers for mobility of compute. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvernby S., Warntjes M.J., Haraldsson H., Carlhall C.J., Engvall J., Ebbers T. Simultaneous three-dimensional myocardial T1 and T2 mapping in one breath hold with 3D-QALAS. J. Cardiovasc Magn. Reson. 2014;16(1) doi: 10.1186/s12968-014-0102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C., Deoni S. The development of brain white matter microstructure. Neuroimage. 2018;182:207–218. doi: 10.1016/j.neuroimage.2017.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C., Treit S., Beaulieu C. A review of diffusion MRI of typical white matter development from early childhood to young adulthood. NMR Biomed. 2019;32(4) doi: 10.1002/nbm.3778. [DOI] [PubMed] [Google Scholar]
- Liao C., Bilgic B., Manhard M.K., Zhao B., Cao X., Zhong J., Wald L.L., Setsompop K. 3D MR fingerprinting with accelerated stack-of-spirals and hybrid sliding-window and GRAPPA reconstruction. Neuroimage. 2017;162:13–22. doi: 10.1016/j.neuroimage.2017.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C., Wang K., Cao X., Li Y., Wu D., Ye H., Ding Q., He H., Zhong J. Detection of lesions in mesial temporal lobe epilepsy by using MR fingerprinting. Radiology. 2018;288(3):804–812. doi: 10.1148/radiol.2018172131. [DOI] [PubMed] [Google Scholar]
- Look D.C., Locker D.R. Time saving in measurement of NMR and EPR relaxation times. Rev. Sci. Instrum. 1970;41(2):250–251. doi: 10.1063/1.1684482. [DOI] [Google Scholar]
- Lustig M., Donoho D., Pauly J.M. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson Med. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- Ma D., Gulani V., Seiberlich N., Liu K., Sunshine J.L., Duerk J.L., Griswold MA Magnetic resonance fingerprinting. Nature. 2013;495(7440):187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Jones S.E., Deshmane A., Sakaie K., Pierre E.Y., Larvie M., McGivney D., Blumcke I., Krishnan B., Lowe M., Gulani V., Najm I., Griswold MA, Wang Z.I. Development of high-resolution 3D MR fingerprinting for detection and characterization of epileptic lesions. J. Magn. Reson Imaging. 2019;49(5):1333–1346. doi: 10.1002/jmri.26319. [DOI] [PubMed] [Google Scholar]
- MacKay A.L., Vavasour I.M., Rauscher A., Kolind S.H., Madler B., Moore G.R., Traboulsee A.L., Li D.K., Laule C. MR relaxation in multiple sclerosis. Neuroimaging Clin. N. Am. 2009;19(1):1–26. doi: 10.1016/j.nic.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Makropoulos A., Robinson E.C., Schuh A., Wright R., Fitzgibbon S., Bozek J., Counsell S.J., Steinweg J., Vecchiato K., Passerat-Palmbach J., Lenz G., Mortari F., Tenev T., Duff E.P., Bastiani M., Cordero-Grande L., Hughes E., Tusor N., Tournier J.D., Hutter J., Price A.N., Teixeira R., Murgasova M., Victor S., Kelly C., Rutherford M.A., Smith S.M., Edwards A.D., Hajnal J.V., Jenkinson M., Rueckert D. The developing human connectome project: A minimal processing pipeline for neonatal cortical surface reconstruction. Neuroimage. 2018;173:88–112. doi: 10.1016/j.neuroimage.2018.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Tervo-Clemmens B., Calabro F.J., Montez D.F., Kay B.P., Hatoum A.S., Donohue M.R., Foran W., Miller R.L., Hendrickson T.J., Malone S.M., Kandala S., Feczko E., Miranda-Dominguez O., Graham A.M., Earl E.A., Perrone A.J., Cordova M., Doyle O., Moore L.A., Conan G.M., Uriarte J., Snider K., Lynch B.J., Wilgenbusch J.C., Pengo T., Tam A., Chen J., Newbold D.J., Zheng A., Seider N.A., Van A.N., Metoki A., Chauvin R.J., Laumann T.O., Greene D.J., Petersen S.E., Garavan H., Thompson W.K., Nichols T.E., Yeo B.T.T., Barch D.M., Luna B., Fair D.A., Dosenbach N.U.F. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603(7902):654–660. doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta K., Salo T., Madison T., Adebimpe A., Bassett D.S., Bertolero M., Cieslak M., Covitz S., Houghton A., Keller A.S., Luo A., Miranda-Dominguez O., Nelson S.M., Shafiei G., Shanmugan S., Shinohara R.T., Sydnor V.J., Feczko E., Fair D.A., Satterthwaite T.D. XCP-D: a Robust Pipeline for the post-processing of fMRI data. bioRxiv. 2023 doi: 10.1101/2023.11.20.567926. [DOI] [Google Scholar]
- Mennes M., Jenkinson M., Valabregue R., Buitelaar J.K., Beckmann C., Smith S. Optimizing full-brain coverage in human brain MRI through population distributions of brain size. Neuroimage. 2014;98:513–520. doi: 10.1016/j.neuroimage.2014.04.030. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M., Barker P.B., Bhattacharyya P.K., Brix M.K., Buur P.F., Cecil K.M., Chan K.L., Chen D.Y., Craven A.R., Cuypers K., Dacko M., Duncan N.W., Dydak U., Edmondson D.A., Ende G., Ersland L., Gao F., Greenhouse I., Harris A.D., He N., Heba S., Hoggard N., Hsu T.W., Jansen J.F.A., Kangarlu A., Lange T., Lebel R.M., Li Y., Lin C.E., Liou J.K., Lirng J.F., Liu F., Ma R., Maes C., Moreno-Ortega M., Murray S.O., Noah S., Noeske R., Noseworthy M.D., Oeltzschner G., Prisciandaro J.J., Puts N.A.J., Roberts T.P.L., Sack M., Sailasuta N., Saleh M.G., Schallmo M.P., Simard N., Swinnen S.P., Tegenthoff M., Truong P., Wang G., Wilkinson I.D., Wittsack H.J., Xu H., Yan F., Zhang C., Zipunnikov V., Zöllner H.J., Edden R.A.E. Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage. 2017;159:32–45. doi: 10.1016/j.neuroimage.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M., Rimbault D.L., Barker P.B., Bhattacharyya P.K., Brix M.K., Buur P.F., Cecil K.M., Chan K.L., Chen D.Y., Craven A.R., Cuypers K., Dacko M., Duncan N.W., Dydak U., Edmondson D.A., Ende G., Ersland L., Forbes MA, Gao F., Greenhouse I., Harris A.D., He N., Heba S., Hoggard N., Hsu T.W., Jansen J.F.A., Kangarlu A., Lange T., Lebel R.M., Li Y., Lin C.E., Liou J.K., Lirng J.F., Liu F., Long J.R., Ma R., Maes C., Moreno-Ortega M., Murray S.O., Noah S., Noeske R., Noseworthy M.D., Oeltzschner G., Porges E.C., Prisciandaro J.J., Puts N.A.J., Roberts T.P.L., Sack M., Sailasuta N., Saleh M.G., Schallmo M.P., Simard N., Stoffers D., Swinnen S.P., Tegenthoff M., Truong P., Wang G., Wilkinson I.D., Wittsack H.J., Woods A.J., Xu H., Yan F., Zhang C., Zipunnikov V., Zöllner H.J., Edden R.A.E. Big GABA II: Water-referenced edited MR spectroscopy at 25 research sites. Neuroimage. 2019;191:537–548. doi: 10.1016/j.neuroimage.2019.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.L., Alfaro-Almagro F., Bangerter N.K., Thomas D.L., Yacoub E., Xu J., Bartsch A.J., Jbabdi S., Sotiropoulos S.N., Andersson J.L., Griffanti L., Douaud G., Okell T.W., Weale P., Dragonu I., Garratt S., Hudson S., Collins R., Jenkinson M., Matthews P.M., Smith S.M. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 2016;19(11):1523–1536. doi: 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S., Yacoub E., Olman C.A., Auerbach E., Strupp J., Harel N., Uğurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson Med. 2010;63(5):1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostapha M., Styner M. Role of deep learning in infant brain MRI analysis. Magn. Reson Imaging. 2019;64:171–189. doi: 10.1016/j.mri.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M.J., Labonte A.K., Gordon E.M., Laumann T.O., Tu J.C., Wheelock M.D., Nielsen A.N., Schwarzlose R., Camacho M.C., Warner B.B., Raghuraman N., Luby J.L., Barch D.M., Fair D.A., Petersen S.E., Rogers C.E., Smyser C.D., Sylvester C.M. (2023) Functional parcellation of the neonatal brain. bioRxiv. doi:〈10.1101/2023.11.10.566629〉.
- Near J., Harris A.D., Juchem C., Kreis R., Marjańska M., Öz G., Slotboom J., Wilson M., Gasparovic C. Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: experts' consensus recommendations. NMR Biomed. 2021;34(5) doi: 10.1002/nbm.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltzschner G., Saleh M.G., Rimbault D., Mikkelsen M., Chan K.L., Puts N.A.J., Edden R.A.E. Advanced Hadamard-encoded editing of seven low-concentration brain metabolites: principles of HERCULES. Neuroimage. 2019;185:181–190. doi: 10.1016/j.neuroimage.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltzschner G., Zollner H.J., Hui S.C.N., Mikkelsen M., Saleh M.G., Tapper S., Edden R.A.E. Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data. J. Neurosci. Methods. 2020;343 doi: 10.1016/j.jneumeth.2020.108827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohliger M.A., Grant A.K., Sodickson D.K. Ultimate intrinsic signal-to-noise ratio for parallel MRI: electromagnetic field considerations. Magn. Reson. Med. 2003;50(5):1018–1030. doi: 10.1002/mrm.10597. [DOI] [PubMed] [Google Scholar]
- Ouyang M., Dubois J., Yu Q., Mukherjee P., Huang H. Delineation of early brain development from fetuses to infants with diffusion MRI and beyond. Neuroimage. 2019;185:836–850. doi: 10.1016/j.neuroimage.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry A., Clare S., Jenkinson M., Smith S., Palace J., Matthews P.M. White matter and lesion T1 relaxation times increase in parallel and correlate with disability in multiple sclerosis. J. Neurol. 2002;249(9):1279–1286. doi: 10.1007/s00415-002-0837-7. [DOI] [PubMed] [Google Scholar]
- Paus T., Collins D.L., Evans A.C., Leonard G., Pike B., Zijdenbos A. Maturation of white matter in the human brain: a review of magnetic resonance studies. Brain Res Bull. 2001;54(3):255–266. doi: 10.1016/s0361-9230(00)00434-2. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C., Basser P.J. Toward a quantitative assessment of diffusion anisotropy. Magn. Reson Med. 1996;36(6):893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- Pierpaoli C., Jezzard P., Basser P.J., Barnett A., Chiro G.D. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201(3):637–648. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- Piredda G.F., Hilbert T., Granziera C., Bonnier G., Meuli R., Molinari F., Thiran J.-P., Kober T. Quantitative brain relaxation atlases for personalized detection and characterization of brain pathology. Magn. Reson. Med. 2020;83(1):337–351. doi: 10.1002/mrm.27927. [DOI] [PubMed] [Google Scholar]
- Poldrack R.A., Baker C.I., Durnez J., Gorgolewski K.J., Matthews P.M., Munafo M.R., Nichols T.E., Poline J.B., Vul E., Yarkoni T. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci. 2017;18(2):115–126. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poline J.B., Breeze J.L., Ghosh S., Gorgolewski K., Halchenko Y.O., Hanke M., Haselgrove C., Helmer K.G., Keator D.B., Marcus D.S., Poldrack R.A., Schwartz Y., Ashburner J., Kennedy D.N. Data sharing in neuroimaging research. Front Neuroinf. 2012;6:9. doi: 10.3389/fninf.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle N., Zuk J., Ortiz-Mantilla S., Sliva D.D., Franceschi A., Grant P.E., Benasich A.A., Gaab N. Pediatric neuroimaging in early childhood and infancy: challenges and practical guidelines. Ann. N. Y Acad. Sci. 2012;1252(1):43–50. doi: 10.1111/j.1749-6632.2012.06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz S.C., Hof S.M., Fleischer V., Brodski A., Groger A., Gracien R.M., Droby A., Steinmetz H., Ziemann U., Zipp F., Deichmann R., Klein J.C. Multi-parametric quantitative MRI of normal appearing white matter in multiple sclerosis, and the effect of disease activity on T2. Brain Imaging Behav. 2017;11(3):744–753. doi: 10.1007/s11682-016-9550-5. [DOI] [PubMed] [Google Scholar]
- Salmenpera T.M., Symms M.R., Rugg-Gunn F.J., Boulby P.A., Free S.L., Barker G.J., Yousry T.A., Duncan J.S. Evaluation of quantitative magnetic resonance imaging contrasts in MRI-negative refractory focal epilepsy. Epilepsia. 2007;48(2):229–237. doi: 10.1111/j.1528-1167.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- Scheenen T.W., Heerschap A., Klomp D.W. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. MAGMA. 2008;21(1-2):95–101. doi: 10.1007/s10334-007-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel P., Levine S.M., Tahedl M., Schwarzbach Jens V. Temporal Signal-to-Noise Changes in Combined Multislice- and In-Plane-Accelerated Echo-Planar Imaging with a 20- and 64-Channel Coil. Sci. Rep. 2020;10(1):5536. doi: 10.1038/s41598-020-62590-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherif T., Rioux P., Rousseau M.E., Kassis N., Beck N., Adalat R., Das S., Glatard T., Evans A.C. CBRAIN: a web-based, distributed computing platform for collaborative neuroimaging research. Front Neuroinform. 2014;8:54. doi: 10.3389/fninf.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Nichols T.E. Statistical Challenges in "Big Data" Human Neuroimaging. Neuron. 2018;97(2):263–268. doi: 10.1016/j.neuron.2017.12.018. [DOI] [PubMed] [Google Scholar]
- Somerville L.H., Bookheimer S.Y., Buckner R.L., Burgess G.C., Curtiss S.W., Dapretto M., Elam J.S., Gaffrey M.S., Harms M.P., Hodge C., Kandala S., Kastman E.K., Nichols T.E., Schlaggar B.L., Smith S.M., Thomas K.M., Yacoub E., Van Essen D.C., Barch D.M. The Lifespan Human Connectome Project in Development: A large-scale study of brain connectivity development in 5-21 year olds. Neuroimage. 2018;183:456–468. doi: 10.1016/j.neuroimage.2018.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spader H.S., Ellermeier A., O'Muircheartaigh J., Dean D.C., 3rd, Dirks H., Boxerman J.L., Cosgrove G.R., Deoni S.C. Advances in myelin imaging with potential clinical application to pediatric imaging. Neurosurg. Focus. 2013;34(4) doi: 10.3171/2013.1.FOCUS12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann M.N., Wisnowski J.L., Group HPISYPW, Smyser CD, Fetal I, Toddler Neuroimaging G, Howell B, Dean DC, 3rd The Art, Science, and Secrets of Scanning Young Children. Biol. Psychiatry. 2023;93(10):858–860. doi: 10.1016/j.biopsych.2022.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum L.N., Tsiouris A.J., Johnson A.N., Naidich T.P., DeLano M.C., Melhem E.R., Quarterman P., Parameswaran S.X., Shankaranarayanan A., Goyen M., Field A.S. Synthetic MRI for Clinical Neuroimaging: Results of the Magnetic Resonance Image Compilation (MAGiC) Prospective, Multicenter, Multireader Trial. AJNR Am. J. Neuroradiol. 2017;38(6):1103–1110. doi: 10.3174/ajnr.A5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.A., Glen D.R., Reynolds R.C., Basavaraj A., Moraczewski D., Etzel J.A. Editorial: Demonstrating quality control (QC) procedures in fMRI. Front Neurosci. 2023;17 doi: 10.3389/fnins.2023.1205928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdall M.D., Reuter M., Qureshi A., Buckner R.L., Fischl B., van der Kouwe A.J.W. Prospective motion correction with volumetric navigators (vNavs) reduces the bias and variance in brain morphometry induced by subject motion. Neuroimage. 2016;127:11–22. doi: 10.1016/j.neuroimage.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traber F., Block W., Lamerichs R., Gieseke J., Schild H.H. 1H metabolite relaxation times at 3.0 tesla: Measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J. Magn. Reson Imaging. 2004;19(5):537–545. doi: 10.1002/jmri.20053. [DOI] [PubMed] [Google Scholar]
- Van Essen D.C., Smith S.M., Barch D.M., Behrens T.E., Yacoub E., Ugurbil K., Consortium WU-MH The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N.D., Gordon J.A., Freund M.P. The Healthy Brain and Child Development Study-Shedding Light on Opioid Exposure, COVID-19, and Health Disparities. JAMA Psychiatry. 2021;78(5):471–472. doi: 10.1001/jamapsychiatry.2020.3803. [DOI] [PubMed] [Google Scholar]
- Weiskopf N., Suckling J., Williams G., Correia M.M., Inkster B., Tait R., Ooi C., Bullmore E.T., Lutti A. Quantitative multi-parameter mapping of R1, PD(*), MT, and R2(*) at 3T: a multi-center validation. Front Neurosci. 2013;7:95. doi: 10.3389/fnins.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N., Roddey C., Shankaranarayanan A., Han E., Rettmann D., Santos J., Kuperman J., Dale A. PROMO: Real-time prospective motion correction in MRI using image-based tracking. Magn. Reson Med. 2010;63(1):91–105. doi: 10.1002/mrm.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E., Bouwman J., Brookes A.J., Clark T., Crosas M., Dillo I., Dumon O., Edmunds S., Evelo C.T., Finkers R., Gonzalez-Beltran A., Gray A.J., Groth P., Goble C., Grethe J.S., Heringa J., t Hoen P.A., Hooft R., Kuhn T., Kok R., Kok J., Lusher S.J., Martone M.E., Mons A., Packer A.L., Persson B., Rocca-Serra P., Roos M., van Schaik R., Sansone S.A., Schultes E., Sengstag T., Slater T., Strawn G., Swertz M.A., Thompson M., van der Lei J., van Mulligen E., Velterop J., Waagmeester A., Wittenburg P., Wolstencroft K., Zhao J., Mons B. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3 doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnykh V.L. Actual flip-angle imaging in the pulsed steady state: a method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn. Reson Med. 2007;57(1):192–200. doi: 10.1002/mrm.21120. [DOI] [PubMed] [Google Scholar]
- Yu Q., Peng Y., Kang H., Peng Q., Ouyang M., Slinger M., Hu D., Shou H., Fang F., Huang H. Differential White Matter Maturation from Birth to 8 Years of Age. Cereb. Cortex. 2020;30(4):2673–2689. doi: 10.1093/cercor/bhz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner H.J., Davies-Jenkins C.W., Lee E.G., Hendrickson T.J., Clarke W.T., Edden R.A.E., Wisnowski J.L., Gudmundson A.T., Oeltzschner G. Continuous Automated Analysis Workflow for MRS Studies. J. Med Syst. 2023;47(1):69. doi: 10.1007/s10916-023-01969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.