Abstract
In eubacteria, Holliday junction (HJ) resolvases (HJRs) are crucial for faithful segregation of newly replicated chromosomes, homologous recombination, and repair of stalled/collapsed DNA replication forks. However, compared with the Escherichia coli HJRs, little is known about their orthologs in mycobacterial species. A genome-wide analysis of Mycobacterium smegmatis identified two genes encoding putative HJRs, namely RuvC (MsRuvC) and RuvX (MsRuvX); but whether they play redundant, overlapping, or distinct roles remains unknown. Here, we reveal that MsRuvC exists as a homodimer while MsRuvX as a monomer in solution, and both showed high-binding affinity for branched DNAs compared with unbranched DNA species. Interestingly, the DNA cleavage specificities of MsRuvC and MsRuvX were found to be mutually exclusive: the former efficiently promotes HJ resolution, in a manner analogous to the Escherichia coli RuvC, but does not cleave other branched DNA species; whereas the latter is a versatile DNase capable of cleaving a variety of branched DNA structures, including 3′ and 5′ flap DNA, splayed-arm DNA and dsDNA with 3′ and 5′ overhangs but lacks the HJ resolution activity. Point mutations in the RNase H-like domains of MsRuvC and MsRuvX pinpointed critical residues required for their DNA cleavage activities and also demonstrated uncoupling between DNA-binding and DNA cleavage activities. Unexpectedly, we found robust evidence that MsRuvX possesses a double-strand/single-strand junction-specific endonuclease and ssDNA exonucleolytic activities. Combined, our findings highlight that the RuvC and RuvX DNases play distinct complementary, and not redundant, roles in the processing of branched DNA structures in M. smegmatis.
Keywords: mycobacteria, branched DNAs, RuvC, RuvX/YqgF, junction-specific DNases, complementarity roles
Homologous recombination (HR) is a conserved process that occurs in all domains of life, as well as in viruses (1, 2, 3, 4, 5). In eukaryotic cells, HR is critically important for the repair of DNA double-strand breaks in mitosis and pairing of homologous chromosomes during prophase I of meiosis (6, 7, 8). To successfully complete HR, a structure-specific endonuclease is required to bind and cleave a four-way branched DNA species, also known as Holliday junction (HJ), that occurs as an intermediates in HR. Thus, cells defective in the processing of branched DNA structures, including the HJ, exhibit a plethora of DNA lesions with pathogenic consequences in humans (9, 10, 11). In bacteria and Archaea, branched DNA structures arise during DNA replication, repair, and HR (12, 13, 14). Several lines of evidence suggest that HJ is not only a key intermediate formed during HR but also manifests in certain types of site-specific recombination (15, 16, 17). Structural studies have demonstrated that HJ adopts two global conformations: open and stacked. While the stacked form is induced by the binding of metal ions, open form is a 4-fold symmetric structure with a square, planar configuration and is capable of branch migration between homologous duplexes, a key step in DNA recombination and DNA repair processes (17, 18, 19). They are resolved by structure-specific endonucleases, known as resolvases, which are required for faithful segregation of newly replicated chromosomes, HR, and repair of stalled/collapsed DNA replication forks (20, 21). Not surprisingly, Holliday junction resolvases (henceforth HJRs) have been identified in a wide variety of organisms based on their shared functional characteristics, although they exhibit significant diversity and belong to different classes of nucleases (1, 22, 23).
Phylogenetic analyses using the full-length sequences of bona fide HJRs revealed that they independently originated from four distinct structural folds, namely RNase H-like, RusA, endonuclease, and endonuclease VII colicin E (22, 24, 25). Typically, HJRs can be classified into two groups, canonical and noncanonical HJ resolvases depending upon the nature of cleavage products. While canonical HJRs (e.g., RuvC, Yen1, and GEN1) introduce a pair of coordinated and symmetrical nicks across the branch point, the noncanonical HJRs (e.g., MUS81, EME1, SLX1, and At-HIGLE) generate asymmetric and nonligatable reaction products (19, 21, 26). The most intensively studied HJR is the Escherichia coli RuvC, a member of the RNase H-like subfamily (27, 28, 29, 30, 31). Studies have documented that the prototypic E. coli RuvC dimer (henceforth EcRuvC) resolves the HJs by introducing a pair of symmetric, coordinated nicks in opposite strands across the junction, generating ligatable, nicked duplex DNA products (27, 30, 31, 32, 33). Consistent with EcRuvC, Thermus thermophilus RuvC (TtRuvC) (34), Pseudomonas aeruginosa RuvC (35), Deinococcus radiodurans RuvC (DrRuvC) (36, 37) also function as homodimers, and exhibit HJ-specific endonuclease activities. Besides HJ resolution, EcRuvC cleaves three-stranded junctions, but not splayed-arm DNA, mismatched DNA, heteroduplex loops, and duplex DNA (27, 38). In contrast to EcRuvC, virus-encoded HJRs, such as phage T4 endonuclease VII or T7 endonuclease I display little or no sequence specificity for cleavage (19), whereas eukaryotic HJR GEN1 exhibits a weak specificity (39). Furthermore, structural evidence obtained with apo-EcRuvC, P aeruginosa RuvC, and TtRuvC indicate that they share a common molecular architecture containing two active sites formed by four conserved carboxylates coordinating two metal ions (32, 34, 35, 40). The TtRuvC–HJ complex structure shows that the junction is bound in a tetrahedral conformation with two of its phosphates located near the two active sites of the RuvC dimer, revealing a potential nick-counter nick mechanism of HJ cleavage (39, 41). However, recent studies suggest that, in the HJ–MOC1 complex structure, HJ is lodged in the basic cleft of the dimeric enzyme and that a catalytic tetrad of acidic residues act on the cleavable phosphodiester bond (42).
Several studies have demonstrated that many bacterial genomes harbour genes that encode enzymes belonging to the RNase H/retroviral integrase family of proteins, called YqgF/RuvX proteins (22, 43). Genetic analyses have shown that yqgF/ruvX is essential for the growth of E. coli (44), Mycobacterium tuberculosis (45), Haemophilus influenzae (46), and D. radiodurans (36). Interestingly, the YqgF/RuvX nucleases from different species show important differences in their substrate specificity. For instance, the E. coli and M. tuberculosis YqgF enzymes promote HJ resolution and display nonsequence-specific endonuclease activity on a wide range of substrates such as the replication forks, 5′ and 3′-flap structures, RNA, DNA, and RNA:DNA hybrids (47, 48, 49), whereas Helicobacter pylori DprB (a member of YqgF family) promotes HJ resolution, but does not bind to ssDNA or dsDNA (50). On the other hand, D. radiodurans YqgF lacks the HJ resolution activity, but preferentially degrades poly(A)-containing RNAs via its exonuclease and endonuclease activities (36). These findings collectively underscore considerable mechanistic diversity among YqgF/RuvX orthologs.
Many enzymes and proteins involved in DNA repair and recombination pathways in mycobacteria differ substantially from their E. coli counterparts (51, 52). A few of these proteins have become priority targets in the development of clinically effective antibacterial agents (53). While there is a large body of literature regarding the structure and function of E. coli HJRs, our understanding and knowledge of structural and functional features of mycobacterial HJRs is limited. To address this knowledge gap, we focused on HJRs in Mycobacterium smegmatis, a valuable surrogate for pathogenic Mycobacteria. A bioinformatics analysis revealed that the M. smegmatis genome harbours ruvC (MSMEG_2943) and yqgF (MSMEG_3026) genes that can potentially encode two putative HJRs, namely RuvC and RuvX, respectively. In this work, we have biochemically and biophysically characterized the M. smegmatis putative HJRs in order to understand their roles in processing of diverse branched DNA structures. Affinity binding assays performed using purified proteins revealed that M. smegmatis RuvC (hereafter MsRuvC) binds efficiently to a suite of branched DNA structures, but only cleaves the mobile HJ (mHJ) and does not cleave/degrade other branched DNA species. Conversely, M. smegmatis RuvX (henceforth MsRuvX) does not promote HJ resolution, but cleaves/degrades a variety of branched DNAs, such as 3′ and 5′ flap DNA, splayed-arm DNA, and dsDNA with 5′ and 3′ overhangs, thereby reinforcing the notion that binding alone is insufficient for DNA cleavage activity. Additionally, we show that mutations in the RNase H domain of MsRuvC and MsRuvX resulted in dramatically decreased levels of DNA cleavage activities while their DNA-binding activities were unaffected, indicating that the two activities can be uncoupled. Unexpectedly, we found robust evidence that MsRuvX possesses ssDNA/dsDNA junction–specific endonuclease and exonuclease activities. These results collectively reveal that RuvC and RuvX DNases play complementary roles in the processing of branched DNA structures that has, until now, remained elusive.
Results
Multiple sequence alignments of RuvC and RuvX protein families
The nonpathogenic, fast-growing M. smegmatis shares most DNA metabolic pathways with the medically relevant M. tuberculosis. The multiple sequence alignments of the deduced amino acid sequences of RuvC (Fig. 1A) and RuvX (Fig. 1B) proteins from distant and closely related species indicated different levels of sequence identity between these two pairs of enzymes. Indeed, RuvX is relatively less conserved than RuvC across organisms. However, both RuvC and RuvX share a conserved RNase H-like domain, a hallmark of the RNase H/integrase superfamily of nucleases, and catalytic residues (22, 24).
Figure 1.
Clustal alignment of MsRuvC and MsRuvX/YqgF orthologs, expression, and purification of MsRuvC and MsRuvX.A, multiple sequence alignment of orthologs of Esherichia coli RuvC. Amino acid sequences of RuvC from the indicated bacterial species were retrieved from the UniProt database. The highly conserved acidic residues are enclosed in red rectangles. The amino acid residues are colored according to the ClustalX scheme: the residues are colored based on charge: red, positively charged residues; magenta, negatively charged residues; blue, hydrophobic residues; yellow, prolines; orange, glycines; cyan, aromatic, and green, polar residues. B, multiple sequence alignment of RuvX/YqgF orthologs from different bacterial species. The sequences of RuvC and RuvX/YqgF were obtained from the UniProt SwissProt database. Sequences were compared using the Clustal Omega webserver and visualized in Jalview 2.0. Residues are colored as in Figure 1A. The Cys residue in MtRuvX (serine in MsRuvX) is enclosed in an unfilled black rectangle. C, a 12.5% SDS-polyacrylamide gel stained with Coomassie blue of protein fractions at various stages of purification of WT MsRuvC. Lane 1, molecular mass marker proteins (M); lane 2, whole-cell lysate (US) from uninduced cells (10 μg); lane 3 whole-cell lysate (IS) from induced cells (10 μg); lane 4, 10 μg protein from the SP-Sepharose column; lane 5, 3 μg protein from heparin-agarose affinity column; lane 6, 2 μg from Superdex 75 gel-filtration column; and lane 7, Western blot analysis of purified MsRuvC (2 μg) using polyclonal anti-MtRuvC antibodies. D, 12.5% SDS-polyacrylamide gel stained with Coomassie blue of protein fractions at various stages of purification of WT MsRuvX. Lane 1, molecular mass marker proteins (M); lane 2, whole-cell lysate from uninduced cells (10 μg); lane 3, whole-cell lysate from induced cells (10 μg); lane 4, SP-Sepharose gel-filtration column (5 μg); lane 5, heparin-agarose column (3 μg); and lane 6, Western blot analysis of purified MsRuvX (2 μg) using polyclonal anti-MtRuvX antibodies. MsRuvC, Mycobacterium smegmatis RuvC; MsRuvX, Mycobacterium smegmatis RuvX; MtRuvC, Mycobacterium tuberculosis RuvC; MtRuvX, Mycobacterium tuberculosis RuvX.
A further comparison of the amino acid sequences of M. smegmatis RuvC and RuvX revealed that they share about 77% and 73% sequence identity, respectively, with their M. tuberculosis counterparts (Table S1). Curiously, MtRuvX contains a lone cysteine residue, which is essential for its homodimerization, which, in turn, is required for symmetrical resolution of the HJ (48), whereas MsRuvX is completely devoid of cysteine residues (Fig. 1B). Furthermore, while MtRuvX efficiently cleaved the HJ, M. tuberculosis RuvC (MtRuvC) showed an extremely weak HJ resolution activity (48). These findings provided an impetus to interrogate the biochemical characteristics of M. smegmatis RuvC and RuvX to gain insights into species-specific differences and evolutionary relationships among the HJR.
Purification of MsRuvC and MsRuvX nucleases
To facilitate mechanistic studies on mycobacterial HJRs, the M. smegmatis ruvC (MSMEG_2943) and yqgF (MSMEG_3026) genes were cloned and their gene products were expressed in E. coli and purified to homogeneity (Fig. 1, C and D). The purity of M.smegmatis RuvC (MsRuvC) and MsRuvX proteins was confirmed through SDS-PAGE, wherein both exhibited a single band corresponding to a molecular weight of 20 kDa and 18.1 kDa, respectively. Western blot analysis using polyclonal antibodies against MtRuvC and MtRuvX, respectively, corroborated the identity of MsRuvC and MsRuvX proteins (Fig. 1C, lane 7; Fig. 1D, lane 6).
In solution, MsRuvX exists as a monomer while MsRuvC as a stable homodimer
Previous studies have documented that bacterial HJRs exist in solution as homodimers, whose formation is required for HJ resolution (28, 32, 34, 35). In line with this, we showed that MtRuvX forms homodimers in solution and that residue C38 facilitates its homodimerization (48). Conversely, D. radiodurans and E. coli YqgF proteins exist as monomers (36, 54), likely due to the absence of residues that correspond to C38 of MtRuvX. Curiously, amino acid sequence analysis revealed that MsRuvX is totally devoid of cysteine residues (Fig. 1B). Thus, to investigate the oligomeric state of MsRuvX in solution, together with MtRuvC, we employed two analytical techniques. In the size-exclusion chromatography (SEC) experiments, MsRuvC eluted at a position consistent with a molecular mass of a dimer of about 37.2 kDa (Fig. 2, A and C), whereas MsRuvX eluted at a position corresponding to the mass of a monomer of ∼18.6 kDa (Fig. 2, B and D). In parallel, we used SEC-multiangle light scattering (MALS) to examine the oligomeric state of MsRuvC and MsRuvX in solution. Reassuringly, this analysis yielded a molecular mass of ∼47 kDa across the MsRuvC elution peak, in reasonable agreement with its theoretical mass (Fig. 2E). Similar analysis of MsRuvX showed a single homogeneous peak with a molecular mass of 18.1 kDa (Fig. 2F). These results collectively inform that the former occurs as a stable homodimer in solution, whereas the latter exists as a monomer under similar conditions.
Figure 2.
Determination of native molecular mass of MsRuvC and MsRuvX. Panels (A) and (B) show the SEC elution profiles of MsRuvC and MsRuvX, superimposed upon the elution profiles of protein standards. The blue traces represent the elution profiles of MsRuvC and MsRuvX. Solid blue trace in (B) is merged with the elution profile of protein standards (red trace). Panels (C) and (D) show calibration curves (blue) were obtained with specified proteins as run on the same column (Superose 12 10/300 GL column). The apparent MW of MsRuvC and MsRuvX were estimated by interpolation from the standard curve (red). Panels (E) and (F) show SEC-MALS analysis of MsRuvC and MsRuvX, respectively. The y-axis on the left denotes MW of eluting species, and the y-axis on the right denotes A280 absorbance of eluent. Solid red lines across the peak correspond to the average molecular mass distribution. Data were analyzed and plotted using GraphPad Prism 6.0 software. MALS, multiangle light scattering; MsRuvC, Mycobacterium smegmatis RuvC; MsRuvX, Mycobacterium smegmatis RuvX; MW, molecular weight; SEC, size-exclusion chromatography.
MsRuvC and MsRuvX bind efficiently to various branched DNA structures
A central aspect of RuvC functionality is its high affinity for the HJ, which exceeds its affinity for other branched DNA species, and ssDNA and dsDNA (19, 37, 55, 56). Although previous studies showed that ruvX is required for mycobacterial cell growth and division (45), our mechanistic understanding and knowledge of its function has remained elusive. Using surface plasmon resonance (SPR) spectroscopy, we investigated the substrate specificity and kinetic parameters of MsRuvX using a suite of DNA substrates, including the HJ, DNA replication fork, splayed-arm duplex, 3′ and 5′ flap DNAs, and dsDNA substrate with 5′ and 3′ overhangs, as compared with that of MsRuvC. The common feature of DNA substrates—such as DNA replication fork, splayed-arm duplex, dsDNA with 3′ and 5′ overhangs, and 3′ and 5′ flap DNAs—is that they all contain a 31-bp duplex with 31 nucleotide overhangs either at 3′ or 5′ ends, or at both ends. For each substrate, an oligonucleotide (ODN) with a biotinylated nucleotide in the 5′ terminus was annealed to a complementary ODN template. The biotinylated DNA substrates were tethered individually to the streptavidin-coated microsensor chips. Similar binding modes and protein concentrations (up to 1 μM) with no divalent cations were used for all experiments. The apparent kinetic rate and equilibrium dissociation constants (Kd values) were measured by titrating varying concentrations of MsRuvX (analyte) over the immobilized ligand surface to assess the avidity of its interaction. Inspection of the SPR traces revealed interaction between MsRuvX and various types of DNA substrates (Fig. S1). The sensorgram traces also indicated rapid association followed by slow dissociation phases. The resulting set of curves (both association and dissociation phases) were directly fitted to a 1:1 Langmuir model to analyze the binding kinetics. The data obtained suggest comparable koff, kon and Kd values for the binding of MsRuvX to various DNAs, except 3′ and 5′ flap DNAs, splayed-arm duplex, dsDNA, and ssDNA (Table 1). Our results also revealed that MsRuvX binds to the mobile and immobile HJs with approximately 3-fold stronger affinity than splayed-arm duplex and 3′ and 5′ flap DNAs. On the contrary, MsRuvX bound to ssDNA and dsDNA with about 4- to 5-fold weaker than that of the HJs. The mHJ has a 12 bp homologous core flanked by regions of heterologous sequences, whereas immobile HJ is without a homologous core sequence.
Table 1.
Quantification of the kinetic rate and equilibrium binding constants of the interaction of MsRuvX with various DNA substrates
| DNA substrate | kon (M−1 s−1 × 104) | koff (10−3 s−1) | ka (106 1/M) | Kd (nM) | χ2 |
|---|---|---|---|---|---|
| Mobile HJ | 2.55 ± 0.25 | 5.97 ± 0.60 | 4.97 ± 1.50 | 271 ± 51 | 0.36, 0.28 |
| Immobile HJ | 6.31 ± 2.70 | 5.87 ± 1.7 | 11.4 ± 0.7 | 263 ± 86 | 0.36, 0.23 |
| 3ʹ-flap structure | 0.72 ± 0.36 | 3.07 ± 1.1 | 4.18 ± 0.21 | 346 ± 52 | 1.68, 1.94 |
| Replication fork | 1.33 ± 0.01 | 5.69 ± 0.20 | 3.95 ± 0.18 | 420 ± 53 | 0.39, 0.61 |
| 5′-flap structure | 0.46 ± 0.10 | 2.83 ± 0.01 | 4.07 ± 0.55 | 583 ± 83 | 1.94, 1.06 |
| 3′-ssDNA overhang | 3.47 ± 0.11 | 7.16 ± 1.16 | 3.04 ± 0.44 | 708 ± 61 | 0.89, 0.18 |
| 5′-ssDNA overhang | 2.17 ± 0.31 | 5.37 ± 0.02 | 3.04 ± 0.04 | 711 ± 84 | 1.06, 1.94 |
| Splayed-arm duplex | 0.46 ± 0.10 | 4.26 ± 0.21 | 3.36 ± 0.61 | 898 ± 30 | 0.86, 0.98 |
| dsDNA | 0.54 ± 0.01 | 7.10 ± 0.01 | 1.14 ± 0.07 | 1130 ± 85 | 0.86, 0.75 |
| ssDNA | 0.78 ± 2.1 | 11.9 ± 6.58 | 1.68 ± 0.31 | 1601 ± 130 | 0.59, 0.57 |
MsRuvX, Mycobacterium smegmatis RuvX.
In parallel with MsRuvX, we determined the substrate specificity and kinetic parameters of the binding of MsRuvC to the same panel of DNA substrates using SPR. Consistent with MsRuvX, SPR sensorgrams traces showed rapid association and dissociation kinetics for the interaction of MsRuvC with various branched and unbranched DNA species (Fig. S2). The measured koff, kon and Kd values for the binding of MsRuvC to various types of immobilized DNA substrates are summarized in the Table 2. Curiously, these results indicate that MsRuvC binds to the DNA replication fork and 5′-ssDNA overhang substrates with a ∼1.5- to 2-fold higher affinity than that of the HJ. These results collectively suggest that MsRuvC and MsRuvX exhibit significantly higher affinities for the branched DNAs, than unbranched DNA species.
Table 2.
Quantification of the kinetic rate and equilibrium binding constants of the interaction of MsRuvC with various DNA substrates
| DNA substrate | kon (M−1 s−1 × 104) | koff (10−3 s−1) | ka (106 1/M) | Kd (nM) | χ2 |
|---|---|---|---|---|---|
| 5′-ssDNA overhang | 7.22 ± 1.50 | 7.81 ± 1.70 | 13.5 ± 2.87 | 201 ± 27 | 0.78, 3.2 |
| Replication fork | 7.79 ± 1.81 | 8.33 ± 1.80 | 13.7 ± 2.93 | 203 ± 23 | 0.57, 1.3 |
| 3′-ssDNA overhang | 2.25 ± 0.69 | 4.74 ± 0.69 | 14.9 ± 3.42 | 263 ± 59 | 0.31, 0.17 |
| 3′-flap structure | 2.77 ± 0.48 | 4.82 ± 0.06 | 7.20 ± 3.40 | 296 ± 54 | 0.26, 0.99 |
| Mobile HJ | 4.06 ± 0.29 | 5.92 ± 0.57 | 9.44 ± 0.99 | 312 ± 50 | 0.26, 0.69 |
| Immobile HJ | 3.98 ± 0.37 | 5.42 ± 0.78 | 4.07 ± 0.29 | 382 ± 72 | 0.36, 0.39 |
| 5′-flap structure | 6.15 ± 4.50 | 12.5 ± 2.5 | 11.4 ± 3.74 | 410 ± 57 | 0.17, 0.51 |
| Splayed-arm duplex | 5.10 ± 0.14 | 13.4 ± 2.9 | 7.73 ± 0.31 | 480 ± 81 | 0.88, 0.26 |
| dsDNA | 2.41 ± 0.86 | 24.9 ± 9.9 | 2.62 ± 0.04 | 758 ± 76 | 0.23, 0.17 |
| ssDNA | 2.89 ± 0.09 | 25.3 ± 3.3 | 7.36 ± 0.90 | 985 ± 84 | 0.64, 0.88 |
MsRuvC, Mycobacterium smegmatis RuvC.
MsRuvX, but not MsRuvC, exhibits robust DNA cleavage activity toward various types of DNA structures
It is well established that the HJRs, such as EcRuvC (38, 55, 56), TtRuvC (34, 41) and P. aeruginosa RuvC (35) selectively bind to the HJ and promote its resolution into a pair of nicked DNA duplexes; besides, they also cleave the three-way junctions albeit with reduced efficiency. On the other hand, bacteriophage T4 endonuclease VII, bacteriophage T7 endonuclease I, and RuvC from Lactococcus lactis phage bIL67 exhibit robust cleavage activity toward different types of DNA structures, including the HJ, splayed-arm duplex, mismatches, and heterologous loops (19, 57, 58). To this end, we conducted DNA cleavage experiments using a fixed concentration (0.5 nM) of the indicated 5′ end-labeled DNA substrate and a range of protein concentrations (up to 200 nM of MsRuvC) in the presence of Mg2+. The products of these reactions were analyzed by native 8% PAGE. The results showed that MsRuvC cleaves mHJ efficiently in a manner dependent on its concentration (Fig. 3A), but not immobile HJ resolution (Fig. 3B). Similar results have been reported for EcRuvC (19, 57), suggesting that branch migration is functionally coupled to RuvC-mediated HJ resolution. However, MsRuvC failed to cleave other branched DNAs and unbranched structures such as bubble DNA, ssDNA, and dsDNA (Fig. 3, C–K). Quantification of EMSA data, shown in Figure 3L, indicate that MsRuvC promotes mHJ cleavage in a concentration-dependent manner. We thus used the mHJ as a substrate in experiments described below. Two previous studies have reported the DNA cleavage activity of DrRuvC: One study showed that DrRuvC cleaves mHJ, immobile HJ, and nicked HJ efficiently, but displayed significantly weaker DNase activity toward 3′ overhang, replication fork, and splayed-arm duplex (36); whereas another study observed that DrRuvC cleaves mHJ but not the immobile HJ and other branched DNAs (37). Although, the exact reason for the differences between these two sets of data is not clear, the discordance could be due to differences in the DNA cleavage assay conditions.
Figure 3.
MsRuvC promotes the HJ resolution into nicked duplexes, but does not cleave other DNA structures, including the immobile HJ. The assay was performed in a buffer containing 0.5 nM of 5′-32P-labeled DNA, as specified, in the absence (lane 2) or presence of 10, 20, 40, 60, 80, 100, 120, 140, 160, 180, and 200 nM of MsRuvC (lanes 3–13, respectively). Representative gel images of MsRuvC-mediated cleavage reactions performed using: (A) mobile HJ; (B) immobile HJ; (C) 3′-flap structure; (D) 5′-flap structure; (E) splayed-arm duplex; (F) dsDNA; (G) replication fork; (H) dsDNA with a 3′ ssDNA overhang; (I) bubble containing DNA; (J) ssDNA; and (K) dsDNA with a 5′ ssDNA overhang. The black filled triangle on top of the gel images represents increasing concentrations of MsRuvC. The asterisk on the substrates denotes the position of the 5′ 32P-label. The square on the HJ in panel (A) indicates a 12-bp homologous core. Panels (A–K), lane 1 represents DNA substrate/(Δ) indicates heat-denatured DNA substrate; lane 2 (C), DNA without MsRuvC. L, quantitative analysis of DNA cleavage activity of MsRuvC with the substrates shown in panels A to K. The product bands were quantified by UVI-BandMap v94.04 and expressed as a percentage of DNA cleavage relative to the substrate. The data was plotted using the GraphPad Prism 6.0. The error bars represent the mean and SD from three independent experiments. The curves were fitted to the data points by nonlinear regression. HJ, Holliday junction; MsRuvC, Mycobacterium smegmatis RuvC.
Having defined the DNA cleavage specificity of MsRuvC, experiments were performed to assess the cleavage efficiency of MsRuvX toward the same panel of DNA substrates with varying concentrations of MsRuvX under similar assay conditions. Indeed, we found that MsRuvX failed to cleave mHJ, immobile HJ, dsDNA, and DNA replication fork (Fig. 4, A, B, F, and G), but exhibited robust cleavage activity toward 3′ and 5′ flap DNA, splayed-arm duplex, dsDNA substrate with 3′ or 5′ overhangs, and ssDNA (Fig. 4, C, D, E, H, J, and K). Further, MsRuvX did not cleave the ssDNA region in the bubble DNA substrate (Fig. 4I). The reaction products resulting from MsRuvX-mediated cleavage of 3'/5′ flap DNAs, 3'/5′ ssDNA overhang substrates migrated coincident with that of dsDNA (Fig. 4, C, D, H, and K), implying that cleavage/nicking occurs in the vicinity of dsDNA-ssDNA junction. On the other hand, multiple cleavage products were seen with splayed-arm duplex and ssDNA (Fig. 4, E and J). Interestingly, the quantitative data in Figure 4L shows that the branched DNAs were degraded completely, whereas the HJ, replication fork and dsDNA remained refractory to cleavage by MsRuvX at all concentrations.
Figure 4.
MsRuvX cleaves several branched DNA structures most efficiently, but does not cleave the HJs, dsDNA, replication fork, and bubble DNA. The assay was performed in a buffer containing 0.5 nM of 5′-32P-labeled DNA, as specified, in the absence (lane 2) or presence of 20, 50, 100, 200, 300, 400, 500, 600, 700, 800, and 1000 nM MsRuvX, respectively (lanes 3–12). Representative gel images of MsRuvX-mediated cleavage reaction performed using: (A) mobile HJ; (B) immobile HJ; (C) 3′-flap structure; (D) 5′ flap DNA; (E) splayed-arm duplex; (F) linear duplex; (G) replication fork; (H) dsDNA with a 3′ ssDNA overhang; (I) bubble duplex DNA; (J) ssDNA; and (K) dsDNA with a 5′ ssDNA overhang. Panels (A–K), lane 1 represents DNA substrate and (Δ) indicates heat-denatured DNA substrate; lane 2 (C), DNA without MsRuvX. The filled triangles on top of the gel image denote increasing MsRuvX concentrations. Asterisks represent 32P-label at the 5′ end. The square on the HJ in panel (A) indicates a 12-bp homologous core. L, quantitative analysis of DNA cleavage activity of MsRuvX with the substrates shown in panels A to K. The product bands were quantified by UVI-BandMap v94.04 and expressed as a percentage DNA cleaved relative to the substrate. Data was analyzed and plotted as described in the legend to Figure 3. HJ, Holliday junction; MsRuvX, Mycobacterium smegmatis RuvX.
Interestingly, MsRuvX, unlike MtRuvX (48), did not catalyse the resolution of mHJ (Fig. 4A). This implies that dimer formation is critical for HJ resolution. It is worth noting that E. coli YqgF bound strongly to ssDNA and DNA replication and recombination intermediates, and also cleaved the HJ, replication fork, and 3'/5′- flap DNAs (47). Combined, these findings strongly suggest that MsRuvC is a canonical HJR, whereas the MsRuvX acts on other branched DNA species, raising the possibility that these two DNases might play complementary, and not redundant, roles in vivo.
MsRuvC and MsRuvX DNases require specific divalent cation cofactors for catalysis
While bacterial HJRs usually prefer Mg2+ as a cofactor for DNA cleavage, Mn2+ can replace Mg2+ as a cofactor in catalysis (19, 30, 35, 55, 59). To further understand the DNA binding/cleavage paradox, as noted above, we conducted a systematic study to evaluate the effects of divalent cations on the DNase activities of MsRuvC and MsRuvX using the mHJ and 3′ flap DNA, respectively, as the substrates. The reaction products were visualized after electrophoresis of deproteinized samples on 8% polyacrylamide gels under native and denaturing conditions. We observed that MsRuvC exhibited comparable levels of HJ resolution activity in the presence of either Mg2+ or Mn2+ at a final concentration of 5 mM, but no other divalent cations, including Zn2+, Ca2+, Cu2+, Ni2+, and Fe2+ facilitated DNA cleavage (Fig. 5, A and B). As expected, control reaction showed no evidence of MsRuvC-mediated HJ cleavage when either Mg2+ or Mn2+ ions were omitted (Fig. 5A, lane 3). The MsRuvC-mediated HJ resolution activity was optimal across a broad range of temperature (37–60 °C), and at pH 8.0 (Fig. S3, A–D). It is worth noting that Mn2+ is essential for DrRuvC-mediated HJ cleavage, but not Mg2+ (36, 37). Next, we evaluated the cleavage activity of MsRuvX on 3′ flap DNA, using the same panel of divalent cations at a concentration of 5 mM. We observed that MsRuvX promoted significant cleavage of 3′ flap DNA in the presence of Mg2+, but not in the presence of Mn2+ as well as other divalent cations (Fig. 5, C and D). Curiously, MsRuvX cleaved 3′ flap DNA in the presence of Ca2+ ion, underscoring another point of dissimilarity between MsRuvX and MsRuvC.
Figure 5.
Divalent cation requirements for the DNA cleavage activities of MsRuvC and MsRuvX.A, a representative image depicting MsRuvC-mediated HJ resolution as a function of different divalent cations. The assay was performed in a buffer containing 0.5 nM of 5′-32P-labeled mHJ in the absence or presence of 200 nM of MsRuvC. Lane 1, nicked dsDNA; lane 2, mHJ; and lane 3, mHJ and MsRuvC in the absence of divalent cations. Lanes 4 and 5, same as in lane 3, but in the presence of 5 mM Mg2+ or Mn2+, respectively. Lanes 6 to 10, same as in lane 3, but in the presence of different divalent cations (5 mM). B, quantitative analysis of MsRuvC promoted HJ resolution in the absence and presence of different divalent cations. C, a representative image showing the cleavage of 3′-flap DNA by MsRuvX in the absence or presence of various divalent cations. The assay was carried out in a buffer containing 0.5 nM of 5′-32P-labeled 3′-flap DNA in the absence or presence of MsRuvX. Lane 1, DNA alone; lane 2, in the absence of MsRuvX; and lanes 3 to 9, reactions performed in the presence of 5 mM of the indicated divalent cation and MsRuvX. D, quantitative analysis of the 3′-flap DNA cleavage by MsRuvX in the absence or presence of the indicated divalent cations. B and D, data are represented as mean ± SD. The data were quantified using UVI-BandMap v94.04 and plotted using GraphPad Prism v6.0. The error bars represent the mean and the SD of three independent experiments. Nonlinear regression method was used to fit the data. The asterisk denotes the position of the 5′ 32P-label. The square box on the HJ denotes a 12 bp homologous core. HJ, Holliday junction; MsRuvC, Mycobacterium smegmatis RuvC; MsRuvX, Mycobacterium smegmatis RuvX.
Purification and characterization of MsRuvC and MsRuvX variants
The results described above revealed that the MsRuvC and MsRuvX proteins contain amino acid residues that are typically found in the RNase H and the retroviral integrase superfamily of proteins (Fig. 1, A and B) (22, 24). The crystal structures of EcRuvC (32, 60), TtRuvC–HJ complex (40, 41) and chloroplast resolvase MOC1 (42) demonstrated that four conserved carboxylates (D7, E66, D138, and D141) in the active center coordinate with Mg2+ ion to catalyse hydrolysis of the phosphodiester bond. Based on multiple sequence alignment of RuvC proteins, we identified amino acid residues D7 and E68 in MsRuvC, likely to be essential for DNA cleavage activity. To explore the role of these residues in the MsRuvC-mediated HJ resolution, the MsRuvC-D7A and MsRuvC-E68A variants were expressed in the E. coli host strain Rosetta (DE3) pLysS, and purified to >98% homogeneity using the same protocol as that used for WT protein. The purified MsRuvC variants migrated on SDS-PAGE gels with electrophoretic mobility coincident with the WT species (Fig. S4A). Analogously, six MsRuvX variants (MsRuvX-D25A, MsRuvX-D25N, MsRuvX-D115A, MsRuvX-D115N, MsRuvX-E116A, and MsRuvX-D142N) were constructed to examine the role of conserved aspartic acid/glutamic acid residues surrounding the presumed active site of MsRuvX in the cleavage of branched DNA structures. These variants were expressed in and purified from E. coli cell lysates, using the same protocol as the WT MsRuvX. SDS-PAGE analysis of purified variants, followed by staining with Coomassie brilliant blue, indicated the presence of the full-length protein with over 95% purity (Fig. S4B).
Two independent techniques (differential scanning fluorimetry (DSF) and SEC-MALS) were leveraged to probe structural alterations, if any, in the mutant forms of MsRuvC and MsRuvX proteins. DSF experiments were conducted with SYPRO orange as a fluorescent probe, which binds to the hydrophobic patches in the unfolded protein (61), to assess the relative melting temperature of the WT and MsRuvC and MsRuvX variants. It is noteworthy that the WT and mutant proteins of MsRuvC and MsRuvX displayed very similar melting profiles (Fig. S4, C and D). Correspondingly, the Tm values summarized in Fig. S4E indicated no significant differences between the WT and variant forms of MsRuvC/MsRuvX proteins. Furthermore, gel-filtration chromatography and the SEC-MALS elution profiles showed that MsRuvC-D7A, MsRuvC-E68A, and MsRuvX-D25A variants exist as dimers and monomers, respectively (Fig. S5, A–I). These results suggest that mutations of conserved residues in the catalytic center of MsRuvC and MsRuvX had no discernible effect on the thermal stability and oligomeric states of these variants.
Site-specific mutations in the RNase H domain of MsRuvC attenuate its mHJ resolution activity
Since mutation of catalytic residues in MsRuvC had no obvious effect on thermal stability (Fig. S4E), we enquired whether these mutations affect their DNA cleavage activities. To test this idea, a fixed amount of 32P-labeled mHJ was incubated with varying concentrations (10–200 nM) of the WT, MsRuvC-D7A, or MsRuvC-E68A proteins. The results showed that WT MsRuvC could cleave over 90% of the input mHJ substrate, generating nicked duplexes (Fig. 6, A and D); whereas MsRuvC-D7A exhibited some residual cleavage activity (∼10%) that was detectable only at very high concentrations (Fig. 6, B and D), but the MsRuvC-E68A variant was virtually inactive under similar conditions (Fig. 6, C and D). Since the reaction products were analyzed by nondenaturing polyacrylamide gels, it remained possible that products resulting from nicking in only one of the mHJ strands by MsRuvC-D7A or MsRuvC-E68A variants could have escaped detection. To test this premise, reactions were performed under similar conditions but with different mHJs (differ by having 5′-32P label on either strand 1, 2, 3, or 4) in the absence or presence of WT and mutant MsRuvC proteins. The reaction products were analyzed on denaturing (8 M urea) 17% polyacrylamide gels. The results revealed that the WT enzyme resolved the mHJ by symmetrically nicking the strands near the crossover region (Fig. S6A). In contrast, the MsRuvC-D7A variant displayed residual cleavage activity across the junction, whereas the MsRuvC-E68A mutant was completely devoid of such activity (Fig. S6, B and C). Further, both the variants showed cleavage activity at multiple sites. These results are consistent with the data shown in Figure 6, suggesting that residues Asp7 and Glu68 are critical for mHJ resolution by MsRuvC.
Figure 6.
Mutation of active site residues D7 and E68 independently impair MsRuvC-catalyzed HJ resolution. The assay was carried out as described in the legend of Figure 3. A–C, show the representative images of the resolution of mHJ into nicked dsDNA by WT MsRuvC, MsRuvC-D7A, and MsRuvC-E68A nucleases, respectively. D, quantitative analysis of mHJ resolution activity of the nucleases as shown in panels A to C. Data were plotted using the GraphPad Prism v6.0 software. The error bars represent the mean ± SD from three independent experiments. HJ, Holliday junction; mHJ, mobile HJ; MsRuvC, Mycobacterium smegmatis RuvC.
Substitutions in the RNase H domain of MsRuvX abolished its DNA cleavage activity
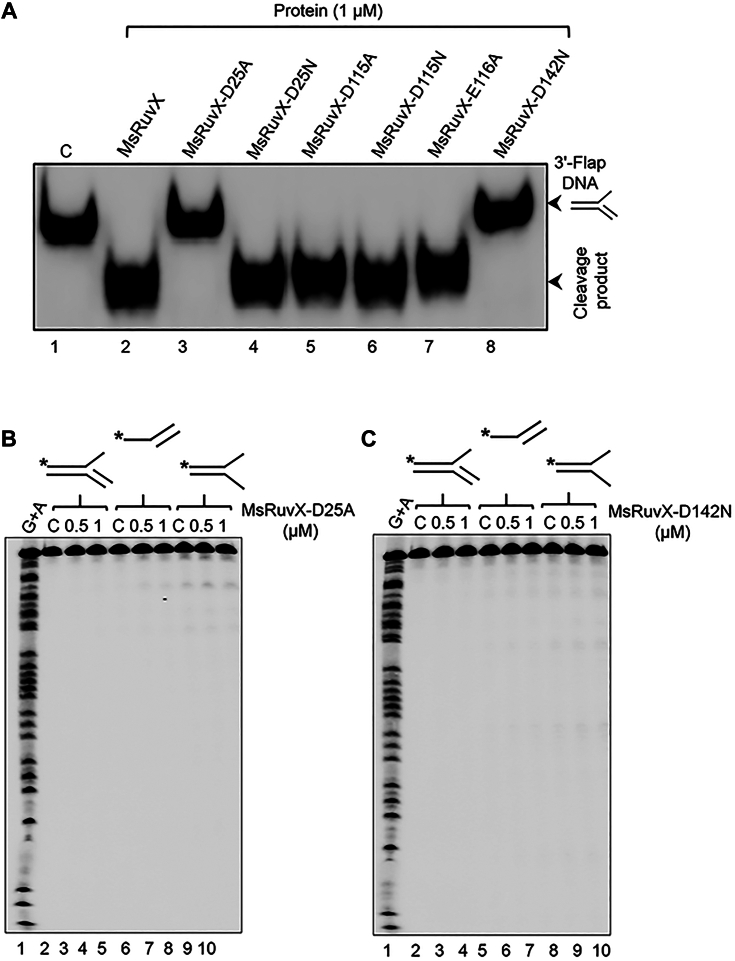
We monitored the catalytic activities of MsRuvX variants (MsRuvX-D25A, MsRuvX-D25N, MsRuvX-D115A, MsRuvX-D115N, MsRuvX-E116A, and MsRuvX-D142N) using 3ʹ flap DNA as the substrate, because it was the most preferred substrate for MsRuvX DNA cleavage activity (Fig. 4C). As anticipated, the results confirmed that WT enzyme displayed robust cleavage activity on 3ʹ flap DNA (Fig. 7A, lane 2). Under similar conditions, the cleavage activities of MsRuvX-D25N, MsRuvX-D115A, MsRuvX-D115N, and MsRuvX-E116A variants were comparable to that of the WT enzyme (Fig. 7A, compare lane 2 with lanes 4–7). However, unlike the MtRuvX-D28N variant (which corresponds to MsRuvX-D25N), which showed approximately 20% of the WT enzyme activity with HJ as the substrate (48), MsRuvX-D25A exhibited no detectable cleavage activity on 3ʹ flap DNA (Fig. 7A, lane 3). Interestingly, substituting Asp25 in MsRuvX with asparagine (D25N) had no impact on cleavage of 3′ flap DNA (Fig. 7A, lane 4). The basis for the differential activity of MsRuvX-D25N (this study) and MtRuvX-D28N (48) enzymes is unclear. To further assess if the MsRuvX mutant variants harbour even very weak nicking endonuclease activity, the cleavage assay was performed by incubating MsRuvX-D25A and MsRuvX-D142N variants with 3′ flap DNA, splayed-arm duplex, or dsDNA with a 5′ ssDNA tail. The reaction products were separated by denaturing urea-PAGE to detect ssDNA fragments. The results showed no DNA bands on the gels, indicating that mutations completely abolished DNA nicking activity of MsRuvX (Fig. 7, B and C). Together, these results support the assertion that cleavage of branched DNA structures is an intrinsic property of MsRuvX.
Figure 7.
Substitution at residues D25 and D142 of MsRuvX abolished its DNA cleavage activity.A, a representative image showing the cleavage of 3′-flap DNA by MsRuvX and its variants. The assay was performed in the cleavage buffer containing 0.5 nM of 5′-32P-labeled 3′-flap DNA in the absence (lane 1) or presence of 1 μM WT MsRuvX or its mutant variants (lanes 2–8). The reaction products were resolve by electrophoresis on an 8% polyacrylamide gel under nondenaturing conditions. B–C, representative images showing the reaction products arising from cleavage of 3ʹ-flap structure, splayed-arm duplex, and dsDNA with overhanging 5′ terminus by MsRuvX-D25A and MsRuvX-D142N variants at the indicated concentrations. Asterisks indicate 32P-label at the 5′ terminus. The reaction products were separated by electrophoresis on 17% polyacrylamide gels under denaturing conditions. MsRuvX, Mycobacterium smegmatis RuvX.
Point mutations in the RNase H domain of MsRuvC and MsRuvX uncouple their DNA-binding and DNA cleavage activities
We next asked whether mutations that attenuate the DNA cleavage activity of MsRuvC and MsRuvX variants also impair their ability to bind to DNA. To address this question, we turned to EMSA, wherein varying concentrations of WT, and MsRuvX variants (D25A and D142N) were incubated with 32P-labeled 3′ flap DNA in the absence of divalent cations. The reaction products were analysed by nondenaturing PAGE and quantified by using a phosphor imager. As anticipated, WT MsRuvX bound robustly to 3′ flap DNA with an apparent binding affinity (Kd) of 36.26 ± 2.6 nM (Fig. 8, A and D). Interestingly, DNA cleavage-deficient MsRuvX variants bound to 3′ flap DNA as robustly as the WT species (Fig. 8, B and C). The apparent Kd values determined from EMSAs indicated that the binding affinity of the MsRuvX-D25A variant (32.70 ± 3.40 nM) was similar to the WT enzyme (36.26 ± 2.6 nM), whereas the MsRuvX-D142N variant displayed an apparent Kd value of 55.10 ± 4.40 nM, which is 1.5-fold weaker than the WT protein (Fig. 8D).
Figure 8.
Mutations at active site residues of MsRuvX and MsRuvC uncouple their DNA binding and DNA cleavage activities.A–C, representative EMSA images showing the binding of increasing concentrations of WT MsRuvX and its variants to a 3′-flap DNA. D, a quantitative analysis of the binding affinities of WT MsRuvX and its variants to a 3′-flap DNA. The EMSA assay was performed as described under Experimental procedures. The reaction mixtures contained 0.5 nM of 32P-labeled 3′-flap DNA in the absence (lane 1) or presence of 0.02, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, and 1 μM of MsRuvX or its variants (lanes 2–12), respectively. The filled triangle on top of the gel image represents increasing concentration of the indicated proteins. The binding efficiency was assessed by quantification of the band intensity in the EMSA assay using UVI-BandMap v94.04 software. Data points were fit with GraphPad Prism v4.0 software, using one-site binding hyperbola model. E–G, represent plots of MST data for the binding of varying concentration of WT MsRuvC and its variants to the 6-FAM-labeled mHJ. Measurements were performed as described under Experimental procedures and background corrected against spectra of buffer alone. Data are presented as mean ± SD of three independent measurements. The data were analyzed using the MO affinity analysis v2.3 (https://shop.nanotempertech.com/en/moaffinity-analysis-software-unlimited-licenses-34), and the datasets were plotted and fitted into the nonlinear regression analysis using one-site binding hyperbola model, and plotted using the GraphPad Prism v6.0. HJ, Holliday junction; mHJ, mobile HJ; MsRuvC, Mycobacterium smegmatis RuvC; MsRuvX, Mycobacterium smegmatis RuvX; MST, microscale thermophoresis.
To reinforce the robustness of the data, equilibrium dissociation constants (Kd values) for the binding of WT and the MsRuvC variants toward the mHJ were determined using microscale thermophoresis (MST). The assay was performed using 5′-FAM (6-carboxyfluorescein)-labeled mHJ and varying concentrations of WT or mutant MsRuvC proteins in the presence bovine serum albumin (BSA) (to suppress nonspecific binding to the capillaries). Our data revealed micromolar interactions between the mHJ and WT MsRuvC (Kd = 0.58 ± 0.27 μM) and between the mHJ and MsRuvC-D7A variant (Kd = 0.65 ± 0.17 μM). Curiously, the MsRuvC-E68A variant showed about ∼2-fold stronger affinity (0.30 ± 0.11 μM) than the WT species (Fig. 8, E–G); however, the mechanistic basis for this is unclear. Regardless, these results support our notion that DNA-binding and cleavage activities of MsRuvX and MsRuvC are functionally separable events. A similar observation has previously been reported for EcRuvC (62).
Since MsRuvC and MsRuvX proteins have not yet been characterized at the biochemical or cellular level, in the remainder of this report, we focus on the molecular mechanism by which these enzymes bind and cleave their preferred DNA substrates, as indicated above.
MsRuvC resolves the HJ by making two symmetric incisions in opposite strands
The dimeric HJR, such as EcRuvC and TtRuvC, resolve the HJs by making symmetrically related nicks in strands of like polarity at, or very near, the branch point (19, 40). The prototypic EcRuvC cleaves at a 5′-A/TTT↓G/C-3′ consensus sequence (‘↓’ indicates the position of cleavage site) in opposite strands symmetrically positioned near the core of the HJ (30, 56, 59, 63), through a nick and counter-nick mechanism (56, 62, 64). Intriguingly, our previous work revealed that MtRuvC showed a very weak HJ resolution activity, whereas RuvX from this species exhibited a robust HJ resolution activity, cleaving at the sequence 5′-TT↓GC-3′ in strands 2 and 4 (48). To test the ability of MsRuvC to catalyse HJ resolution, four different mHJs (differ by having 32P-label on strand 1, 2, 3 or 4) were individually incubated with varying concentrations of MsRuvC in the presence of Mg2+. The reaction products were analyzed by denaturing PAGE alongside the Maxam–Gilbert G + A sequencing ladders. In contrast to the weak HJ resolution activity of MtRuvC (48), MsRuvC efficiently cleaved the HJ, and strands 2 and 4 of the mHJ (>95% of substrate was cleaved) more robustly than strands 1 and 3 (Fig. 9, A–D), analogous to the canonical HJRs (19). The cleavage sites were mapped by comparison to reference G + A ladders and depicted schematically in Figure 9E. The results showed that MsRuvC nicked at a specific sequence, 5′-GT↓CC-3′ in stands 2 and 4, and at 5′-TA↓GG-3′ in strands 1 and 3, although the crossover region has the potential to branch migrate within the 12 bp homologous core.
Figure 9.
Mapping of MsRuvC cleavage sites on the Holliday junction.A–D, gels shown are representative examples of the cleavage sites on the mHJ at the nucleotide level by MsRuvC. Four different types of 32P-labeled mHJ (which differ with respect to 32P-label on strand 1, 2, 3, or 4) (1 nM) were incubated in the absence (lane 2) or presence of 100, 200, 400, and 500 nM MsRuvC (lanes 3–6 in panels A–C) or 100, 200 and 400 nM (lanes 3–5 in panel D), respectively. Lane 1, G + A sequencing ladder of strand 1, 2, 3, and 4 in panels A to D, respectively. The filled triangles on top of the gel images indicate increasing concentrations of MsRuvC. E, schematic diagram showing the positions of MsRuvC cleavage sites based on data shown in panels A to D. Blue and the green arrowheads illustrate major and minor cleavage sites, respectively. Asterisks on the mHJ denote the position of the 5′-32P-label. mHJ, mobile HJ; MsRuvC, Mycobacterium smegmatis RuvC.
This result contrast with the EcRuvC, which cleaves the HJs at the sequence 5′-T↓C-3′ or 5′-T↓G-3' (55, 59, 65) or 5′-TTT↓G-3' (66), and the consensus sequence would therefore appear to be (A ∼ T)TT↓(C > G ∼ A) (56). Since a T residue appears to be critical for HJ cleavage, we tested its importance for MsRuvC-mediated cleavage of the HJ. To this end, we constructed a HJ variant lacking A•T base pair in the 12-bp homologous core (Fig. S7A). The cleavage reactions were performed by incubating 32P-labeled HJ (having 32P label either on HJ strand 1, 2, 3, or 4), lacking A•T base pair, in the absence or presence of MsRuvC. The reaction products were analyzed by nondenaturing (Fig. S7B) and denaturing PAGE (Fig. S7C), as described above. Notably, we found that MsRuvC failed to cleave the HJ lacking A•T base pair, as revealed by the absence of expected HJ resolution product (Fig. S7, B and C). From these results, we conclude that MsRuvC cleaves at the 3′-side of thymine moiety in the HJ, and the thymine residue at the core of junction is vital for MsRuvC-mediated HJ resolution function.
MsRuvC-mediated HJ resolution generates nicked duplex products
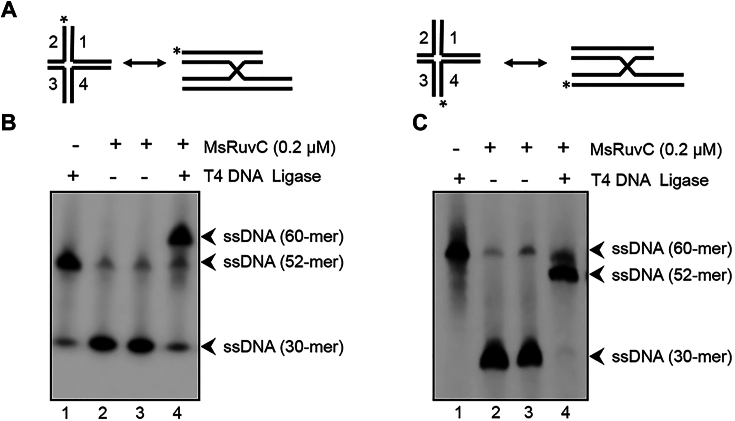
A pair of nicked DNA duplex molecules arising from symmetrical resolution of the HJ by canonical HJRs can be repaired by the action of a DNA ligase (18, 19). To determine if this is the case with MsRuvC, we constructed a mHJ species with a truncated arm, as shown schematically in Figure 10A. We reasoned that nicking of a 52-nt-long 5′-32P-labeled strand (structure shown on the left-hand side), followed by religation, would produce a 60-nt product, whereas nicking of a 60-nt-long 5′-32P-labeled strand (structure shown on the right-hand side), would result in a 52-nt species followed by religation. To test this idea, the HJs were incubated with MsRuvC for 60 min and one reaction mixture was further incubated with T4 DNA ligase for 60 min and were analyzed by denaturing urea PAGE. As shown in Figure 10B, addition of T4 DNA ligase to the products of MsRuvC cleavage reaction resulted in efficient strand religation resulting in the formation of an expected 60-nt species, accompanied by depletion in the levels of 30-nt product (lane 4). Similar results were obtained with 60-nt-long 5′-32P-labeled strand, resulting in the formation of 52-nt product (Fig. 10C, lane 4). These data indicate that the resolution products produced by MsRuvC contain readily ligatable ssDNA termini, revealing that the mechanism of HJ resolution is similar to the canonical HJRs (19).
Figure 10.
The products of MsRuvC-mediated HJ resolution can be ligated by T4 DNA ligase.A, schematic diagrams of the DNA substrates. B and C, representative images showing products of the ligation assay. The assay was performed as described in the Experimental procedures. Plus and minus signs at the top of gel lanes indicate reactions performed in the presence or absence of MsRuvC and/or T4 DNA ligase. The asterisk on mHJ denotes the position of the 5′-32P-label. HJ, Holliday junction; MsRuvC, Mycobacterium smegmatis RuvC.
The two incisions occur independently during MsRuvC-mediated HJ resolution
To further corroborate that MsRuvC is a canonical HJR, we asked whether the pair of incisions inflicted by MsRuvC at the mHJ branch point occur in a sequential or concerted manner. To address this question, we constructed three mHJ variants, in which the cleavage site was modified with a phosphorothioate (PS) linkage (Fig. 11A), which renders DNA resistant to cleavage by endonucleases (67). The three mHJ variants were (1) PS linkage in strand 2 (2), PS linkages in strand 1 and 2, and (3) PS linkages in all four strands. Schematic diagrams above the gel image in Figure 11B shows the positions of PS linkages in mHJ (indicated by yellow box). A similar approach has been previously used to infer the GEN1-mediated HJ cleavage mechanism (68).
Figure 11.
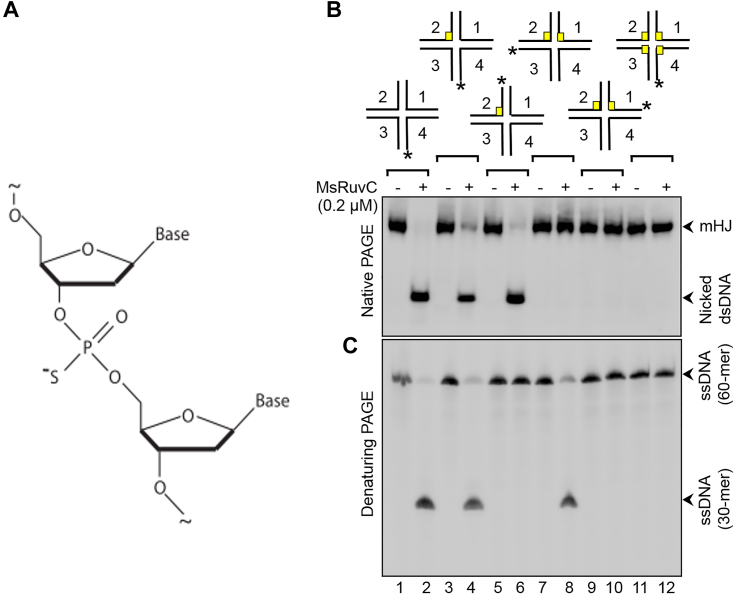
MsRuvC-mediated cleavage of two strands of the HJ can be uncoupled from each other.A, schematic diagram of a dinucleotide containing a PS linkage. The schematic diagrams at the top of the gel image in panel (B and C) illustrate the PS linkages in different strands of the HJ. The assay was performed in reaction mixtures containing 0.5 nM of 32P-labeled, unmodified mHJ (lanes 1 and 2), or 32P-labeled, PS-modified mHJ (lanes 3–12), and the reaction products were analyzed by native PAGE (8% polyacrylamide) (panel B) or denaturing PAGE (12% polyacrylamide, 8 M urea) (panel C), as described in Experimental procedures. Plus or minus signs at the top of gel lanes indicate the absence and presence of 200 nM MsRuvC, respectively. The asterisk on mHJ denotes the position of the 5′-32P-label either on HJ strand 1, 2, 3, or 4. The yellow boxes represent PS inter-nucleotide linkages. HJ, Holliday junction; mHJ, mobile HJ; MsRuvC, Mycobacterium smegmatis RuvC; PS, phosphorothioate.
The mHJs (5′-32P-labeled in the indicated strand) and Mg2+ were incubated in the absence or presence MsRuvC, and the resolution products were analyzed by native and denaturing PAGE. As anticipated, the unmodified mHJ was resolved by MsRuvC, resulting in the generation of a 32P-labeled nicked linear duplex as well as 30-mer ssDNA (Fig. 11, B and C, lane 2). Likewise, MsRuvC cleaved the mHJ containing a PS linkage in strand 2, generating the same products (Fig. 11, B and C, lane 4). Further, cleavage of mHJ containing PS linkage in 32P-labeled strand 2 produced labeled nicked duplex but unlabeled 30-mer ssDNA (Fig. 11, B and C, lane 6). In reaction containing mHJ with PS linkages in strands 1 and 2, but 32P label on strand 3, we observed a labeled 30-mer species as the only reaction product (Figure 11, B and C, lane 8). However, no products were seen in the reaction containing mHJ with PS linkages in strand 1 and 2, with 5′-32P-label in strand 1 (Fig. 11, B and C, lane 10). As anticipated, the mHJ with PS-containing linkages in all four strands was refractory to MsRuvC-mediated resolution (Fig. 11, B and C, lane 12). These results strongly support the notion that the two incisions occur independently during MsRuvC-mediated mHJ resolution, in a manner analogous to the bacterial canonical HJRs and GEN1 (19, 68).
MsRuvX possesses dsDNA/ssDNA junction-specific endonuclease activity
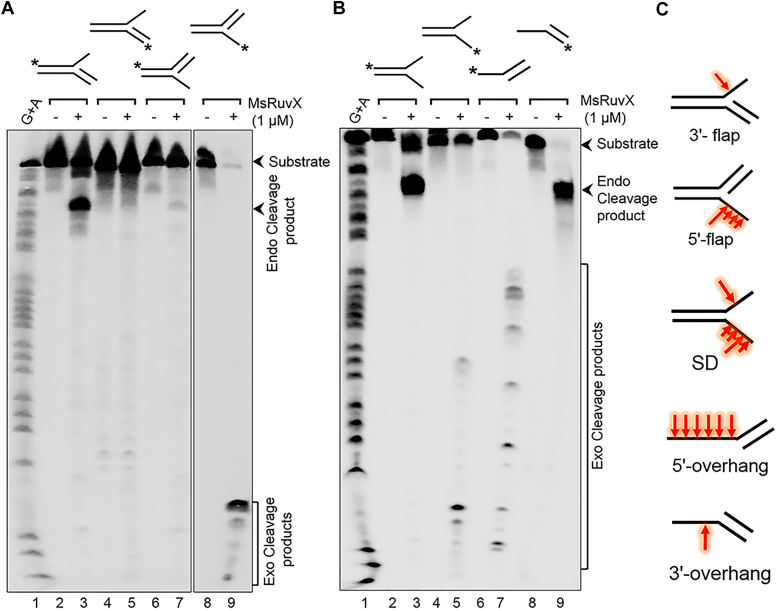
Compared with RuvC, YqgF/RuvX nucleases are less conserved and less studied. As described above, in contrast to MsRuvC, MsRuvX displayed efficient cleavage activity toward certain types of branched DNA structures (Fig. 4, C–E, H, and K). To further describe the mechanism of DNA cleavage, we chose five different DNA substrates, namely 3′ and 5′ flap DNA, splayed-arm DNA, and dsDNA with 3′ and 5′ overhangs. In a series of reactions conducted simultaneously, these substrates (32P-labeled at the 5′ end) were individually incubated in the absence or presence of MsRuvX. The reaction products were run on a denaturing (8 M urea) 17% sequencing gel alongside the Maxam–Gilbert G + A sequencing ladder. The results showed that the 3′ flap DNA was cleaved efficiently by MsRuvX. It makes an initial endonucleolytic incision near the dsDNA/ssDNA junction, releasing a ∼44-mer ssDNA product (Fig. 12A, lane 3). With 5′ flap DNA substrate, the 32P-labeled ssDNA tail liberated by MsRuvX was degraded exonucleolytically to generate a set of short ssDNA fragments (Fig. 12A, lane 9). As expected, this was not apparent with the substrates that contained unlabeled ssDNA tails (Fig. 12A, lanes 5 and 7). Further, MsRuvX did not cleave the duplex region adjacent to the ssDNA/dsDNA junctions (Fig. 12A), even after 60 min of incubation (Fig. S8). In line with this, we found evidence for a similar mechanism, wherein 5′-32P-labeled splayed-arm DNA and 5′-32P-labeled dsDNA with a 3′ ssDNA overhang were cleaved endonucleolytically near the dsDNA/ssDNA junction, yielding a ∼44-mer product (Fig. 12B, lanes 3 and 9). As seen with the 3′ and 5′ flap DNAs, the ssDNA fragments released from the splayed-arm DNA and dsDNA with a 5′ ssDNA overhang with 32P-label on the complementary strand were degraded exonucleolytically, generating several ssDNA fragments of varying length (Fig. 12B, lanes 5 and 7). Schematic diagrams shown in Figure 12C recapitulate the location of incision sites in the 3'/5′ flap DNA, splayed-arm DNA, and dsDNA with 5ʹ and 3ʹ overhangs. Of note, although different in mechanistic detail, MsRuvX functions analogously to those of the eukaryotic Flap endonuclease 1 (69) and Dna2 endonuclease (70).
Figure 12.
MsRuvX cleaves branched DNA structures near the dsDNA/ssDNA junctions.A, a representative image showing the cleavage activity of MsRuvX toward 3′-flap and 5ʹ-flap DNA. Lane 1, G + A ladder. Lanes 2, 4, 6, and 8 are controls containing the indicated 5′-32P-labeled DNA; lanes 3, 5, 7, and 9 represent reactions performed with MsRuvX. Lanes 2 to 5, 3′-flap DNA with 32P-label at the 5′- end of top strand or bottom strand; lanes 6 to 9: 5′-flap DNA with 32P label at the 5′-end of top strand or bottom strand. B, a representative images showing MsRuvX cleavage activity toward splayed-arm DNA and dsDNA with 5ʹ- or 3ʹ ssDNA overhang. Lane 1, G + A ladder. Lanes 2 to 5, splayed-arm DNA with 32P-label at the 5′- end of top or bottom strand; lanes 6 to 9: dsDNA with overhanging 5ʹ or 3ʹ termini with 32P-label at the 5′- end of top strand or bottom strand. Plus or minus signs at the top of gel lanes indicate reactions performed in the presence or absence of MsRuvX. Asterisk denotes the 5′-32P-labeled strand. C, a schematic of each substrate is shown with cleavage sites (red arrows). MsRuvX, Mycobacterium smegmatis RuvX.
MsRuvX degrades ssDNA through initial endonucleolytic cuts, followed by exonucleolytic cleavage at single-nucleotide resolution
One of the hallmarks of processive exonucleases is that the products they produce resolve into a ladder of bands in a polyacrylamide gel after electrophoresis. To further characterize the mechanism of nucleolytic digestion, we speculated that it might cleave ssDNA from either the 5′ or 3′ end, or both ends. To distinguish between these possibilities, a traditional exonuclease assay was carried out, wherein 62-mer ssDNA, 32P-labeled at either the 5′ or 3′ ends were incubated separately with varying concentrations of MsRuvX for 60 min. In parallel, similar reactions were carried out with fixed concentrations of MsRuvX and 32P-labeled 62-mer ssDNA, labeled at either 3′ or 5′ end, for varying time between 2 and 60 min. Reaction mixtures were parsed on a denaturing (8 M urea) 17% sequencing gel along with the Maxam–Gilbert G + A sequencing ladders. The results of protein titration experiments showed that MsRuvX cleaves 5′-32P-labeled 62-mer ssDNA successively at ∼4-nt intervals, resulting in regularly spaced ssDNA fragments, until it reaches at about 8 to 9 nt from the 5′ end (Fig. 13A). The data from kinetic experiments indicated that the amount of input substrate decreased gradually, generating ssDNA fragments of varying sizes (as in Fig. 13A) and their intensities increased as the reaction progressed (Fig. 13B). Several minor products other than fragments with 4-nt intervals were observed, presumably due to exonucleolytic resection.
Figure 13.
MsRuvX exhibits a robust endonucleolytic/exonucleolytic activity on ssDNA. Representative images of denaturing urea-PAGE show the MsRuvX-mediated nucleolytic activity. A, the dose-dependent nuclease activity of MsRuvX. Lanes 3 to 10, reactions were performed with 1 nM 5′-32P-labeled ssDNA and 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, and 1 μM of MsRuvX, respectively. B, the kinetics of ssDNA-specific nuclease activity of MsRuvX (200 nM) on 5′-32P-labeled 62-mer ssDNA. Lane 3 to 10, reactions were stopped at different time intervals as indicated, by adding stop solution as mentioned in the Experimental procedures. C, concentration-dependent nuclease activity of MsRuvX on 3′-32P-labeled ssDNA. Lanes 3 to 10, reactions were performed with 1 nM 3′-32P-labeled ssDNA and 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, and 1 μM of MsRuvX, respectively. D, the kinetics of 3′-32P-labeled ssDNA-specific exonuclease activity of MsRuvX on 3′-32P-labeled 62-mer ssDNA for the indicated time periods. E, the kinetics of ssDNA-specific exonuclease activity of truncated species of Escherichia coli exonuclease VIII on 3′-32P-labeled 62-mer ssDNA for the indicated time periods. In panel (A-E): Lane 1, the Maxam–Gilbert A + G sequencing ladder; lane 2, DNA alone. Asterisks denote the 32P-label. MsRuvC, Mycobacterium smegmatis RuvC; MsRuvX, Mycobacterium smegmatis RuvX.
Next, we assessed the degradation of the same substrate, but with 32P-label at the 3′-end, wherein MsRuvX quantity and incubation time were varied. Indeed, we found that, in contrast to 5′-32P-labeled ssDNA, the reaction products generated by MsRuvX with 32P-label at the 3′-end were different. This notion was supported by the observation that ssDNA was rapidly degraded into progressively shorter DNA fragments as the protein quantities were varied, with the reaction products laddering close to the 3′ terminus (Fig. 13C). This pattern of cleavage implies that MsRuvX makes an incision from the 3′ end of ssDNA, releasing a 10-mer product. It was further shortened by 5′→3′ exonuclease activity to generate a 7-mer product: while its intensity increases with varying concentrations of MsRuvX or incubation time, further resection/cleavage was restrained via an unknown mechanism (Fig. 13, C and D). These findings are in accord with the mechanism of DNA cleavage mediated by the bacteriophage T4 RNase H (71) and P. aeruginosa FAN1 nuclease (72). As shown in Figure 13E, these results are compatible with the 5′→3′ exonucleolytic activity of the truncated form of E. coli exonuclease VIII (NEB# M0545S) (73), which served as a positive control. Collectively, these results support a mechanistic model in which MsRuvX degrades ssDNA through initial endonucleolytic cuts, followed by exonucleolytic resection at single-nucleotide resolution. However, whether MsRuvX exhibits a preference for certain sequences awaits further exploration. Based on these results, we conclude that MsRuvX may play a crucial role in the processing of branched DNA structures that arise during DNA replication, HR, and various DNA repair pathways.
Discussion
The pioneering studies on EcRuvC, the founding member of canonical HJRs, has provided novel insights into the molecular mechanism underlying the processing of intermediates generated during HR and DNA repair (28, 65), and recent studies have expanded the universe of HJRs in eukaryotes (19, 21, 68, 74). These data indicate that HJRs are conserved across all kingdoms of life with important roles in the maintenance of genome stability. Surprisingly, however, RuvC appears to be absent in low-GC content Gram-positive bacterial species (22), suggesting that there may exist distinct strategies for the processing of branched DNA structures that arise during DNA replication, repair, and HR. Consistent with this hypothesis, 90% of bacterial lineages harbour yqgF/ruvX genes that can encode putative HJRs (22); however, whether they are functionally redundant or completely independent of one another remains unclear. Distinguishing between these possibilities is critical for understanding how RuvC and RuvX proteins discriminate between cognate and noncognate substrates in vivo. Intriguingly, as we discuss below, MsRuvC and MsRuvX display strikingly different substrate specificities even though they share structural similarities (22), providing a rich landscape for interrogating structure-function relationships.
Earlier studies from our group revealed that M. tuberculosis RuvC binds robustly to the HJ, but showed very weak HJ resolution activity relative to RuvX from the same species (48). Additionally, we observed a 6- to 8-fold increase in mRNA levels of ruvX and ruvC after treatment of cells with DNA-damaging agents such as methyl methanesulphonate, thereby implicating a role for ruvX in DNA repair and HR (48). In this study, we carried out systematic analyses of the substrate specificity and catalytic properties of purified WT MsRuvC and MsRuvX, and their mutant variants using a broad range of DNA structures that mimic intermediates formed during DNA replication, repair, and HR. We observed that, while the MsRuvC and MsRuvX interact with the same substrates (although with different affinity), but do not share the substrates for cleavage or mechanisms by which they cleave DNA substrates. Although both MsRuvC and MsRuvX bind to the branched DNA structures and unbranched DNAs with affinity constants in the nanomolar and submicromolar range, their DNA cleavage specificities were found to be mutually exclusive. Indeed, we found that, MsRuvC efficiently resolves the HJ, in a manner analogous to the canonical HJRs (19), but does not cleave other branched DNAs, whereas the MsRuvX is a versatile DNase capable of digesting a broad range of branched DNA structures but lacks the HJ resolution activity. Although the significance of high affinity of MsRuvC to the branched DNA species remains unclear, it is possible that such binding may play a key a role in protecting these substrates against a myriad of DNases in vivo.
What might be the reason for the observed differences in DNA substrate-specific cleavage activities of MsRuvC and MsRuvX? One possibility is that MsRuvC may require one or more cofactors to reveal its cleavage activity toward DNA replication fork, splayed-arm DNA, 3'/5′ flap DNAs, and dsDNA with 5′/3′ overhangs. As discussed elsewhere (18, 19), RuvA, a tetrameric DNA-binding protein, unfolds the HJ into a square-planar conformation and facilitates the resolution of the HJ by RuvC. Analogously, MsRuvX might associate with an as yet unidentified auxiliary factor(s) to resolve the HJ into two nicked duplexes. More generally, differences in substrate-specific cleavage activity may arise from steric hindrance caused by bulky substrates. Thus, subtle chemical differences in the active site may not accommodate noncognate substrates with a wide range of different sizes and shapes to facilitate cleavage. To test whether the observed DNA cleavage activities are intrinsic to MsRuvC and MsRuvX, we purified the predicted nuclease-deficient variants and assayed their activity using their preferred DNA substrates. The mutants had no detectable DNA cleavage activity, suggesting that the observed activities are intrinsic to MsRuvC and MsRuvX. These findings indicate that MsRuvC and MsRuvX show broad DNA-binding capabilities, but exhibit distinct cleavage specificities. Curiously, we unexpectedly found that MsRuvX is both a dsDNA/ssDNA junction-specific endonuclease and an exonuclease, reflecting a closer functional resemblance to the eukaryotic Flap endonuclease 1 family of enzymes (69) despite the absence of structural similarity. However, further studies, including single molecule analyses, are required to more accurately determine the mechanism of MsRuvX-mediated cleavage of branched DNA structures.
As noted above, RuvC is structurally related to YqgF and the catalytic residues are fully conserved between the two enzymes (22, 24), but exhibit notable differences in their DNA cleavage activities and cofactor requirements. The notion that reaction conditions with respect to divalent cations can have an impact on the catalytic activity is borne out in our studies on MsRuvX and MsRuvC. Analogous to EcRuvC (59, 60), MsRuvC exhibited robust DNA cleavage activity in the presence of Mg2+ or Mn2+, but not Ca2+ and other divalent cations tested. Using the same assays, we consistently found comparable levels of DNA cleavage activity of MsRuvX in the presence of Mg2+ and Ca2+, but not Mn2+. Broadly speaking, RNase H-like nucleases can utilize Mn2+ as a cofactor in place of Mg2+ for catalysis (75), as the ionic radius of Mn2+ is slightly closer to Mg2+ (76). In most cases, Ca2+ does not support catalysis although it facilitates DNA/RNA-binding activity of nucleases (77). Our finding that Ca2+ serves as a cofactor for the MsRuvX-mediated DNA cleavage activity is not unprecedented. Indeed, previous studies have demonstrated that Ca2+ serves as a cofactor for several enzymes, including restriction endonucleases and sequence nonspecific nucleases (78, 79, 80). Furthermore, the crystal structure of several REases have demonstrated that Ca2+ occupies the same position as that of Mg2+ at the active site (81, 82, 83), thus offering a structural explanation for Ca2+-dependent DNA cleavage activity. Similarly, structural studies are needed to clarify the role played by Ca2+ in MsRuvX-mediated DNA cleavage activity.
The active species of MtRuvX crystalized as a dimer and showed robust HJ resolution activity (48, 49), whereas MsRuvX exists as a monomer in solution, similar to that of DrYqgF (36), the latter two failed to promote HJ resolution. This is in agreement with earlier studies showing that dimer formation is essential for the HJ resolution activity of RuvC (18, 19). Supporting this notion, the dimeric forms of MsRuvC and DrRuvC promoted HJ resolution (this study, ref. (36, 37)). The preferred cleavage site of DrRuvC was identified as 5'-(G/C)TC↓(G/C)-3′, whereas as the consensus cleavage sequence of EcRuvC is 5'-(A/T)TT↓(G/C)-3' (19). We found that MsRuvC, in the absence of RuvAB, analogous to the studies mentioned above, cleaves at 5′-T↓C-3′ consensus sequence located symmetrically around the HJ branch point. Our mutational analysis indicated that two conserved acidic residues D7 and E68 were required for DNA cleavage activity of MsRuvC, but not for its DNA-binding activity. Likewise, the cleavage-deficient MsRuvX variants (D25A and D142N) displayed WT DNA binding activity. Together, these results indicate uncoupling between DNA-binding and DNA cleavage activities of MsRuvC and MsRuvX.
While our studies have focused primarily on understanding of the DNA-binding and DNA cleavage activities of MsRuvC and MsRuvX DNases, the biological implications of our findings merit further discussion. The question is: why some bacteria possess two structurally similar, divalent cation-dependent, structure-specific DNases? The data presented here suggest that the mechanism by which the branched DNA structures are processed by MsRuvC and MsRuvX is distinct. These findings can be rationalized by a mechanistic model in which MsRuvC might be relevant for the resolution of topologically constrained HJs, whereas RuvX acts as housekeeping enzyme to process less complex branched DNA structures, such as DNA flaps, splayed-arm duplex, and dsDNA with 3′ and 5′ overhangs. Mirroring our in vitro data, genetic studies have shown that yqgF and ruvC genes are not interchangeable in Acinetobacter baylyi ADP1 (84). In bacteria, branched DNA structures can arise due to different stresses interfering with DNA replication as a result of DNA damage (85). This raises the possibility that, the MsRuvX-mediated processing of branched DNA structures could in principle a putative mechanism of surveillance to protect cells from the adverse effects of these structures and ensure genomic stability. Supporting this notion, RuvX is indispensable for the growth and survival of E. coli (44), M. tuberculosis (45), H. influenzae (46), and D. radiodurans (36), indicating their critical roles in cellular functions. We envisage that a similar situation could exist in M. smegmatis.
In sum, our work uncovers two mechanisms by which branched DNA structures are processed by RuvC and RuvX nucleases in M. smegmatis. However, several questions remain. Notably, it is obvious that the biological significance of (the activities of) MsRuvC and MsRuvX will have to be determined in knockin and KO mutants of M. smegmatis. In line with this, the interplay between MsRuvC and MsRuvX under normal growth and stress-inducing conditions, including antibiotic stress is worthy of further research. In vivo, branch migration and cleavage of the HJ is mediated by the RuvABC resolvasome complex; therefore, it would be interesting to investigate the mechanism of MsRuvC-mediated HJ cleavage in the context of its cognate RuvAB subcomplex. Nevertheless, the current study opens up avenues for future investigations on the mechanistic and structural studies related to RuvC and RuvX nucleases.
Experimental procedures
Biochemicals, plasmids, bacterial strains, enzymes, and oligonucleotides
Fine chemicals (analytical grade) were purchased from GE Healthcare Life Sciences and Sigma-Aldrich. Restriction endonucleases, T4 DNA ligase, T4 polynucleotide kinase, terminal deoxynucleotidyl transferase and Phusion High-Fidelity DNA polymerase, Taq DNA polymerase, 1 kb DNA ladder, gel extraction kit, and plasmid isolation kits were procured from New England Biolabs or Thermo Fischer Scientific. DNA oligonucleotides and SYPRO orange dye were obtained from Sigma-Aldrich and SynBio technologies. [γ-32P] ATP and [α-32P] dCTP were procured from the Board of Radiation and Isotope Technology or PerkinElmer Life Sciences. E. coli strains DH5α, Rosetta (DE3) pLysS, the plasmids pET21a (+), and pUC19 were purchased from Novagen and New England Biolabs. SP-Sepharose, Heparin-agarose resin, prepacked Superdex 75 10/300, and Superose 12 10/300 GL columns were obtained from GE Healthcare Life Sciences. Gel filtration standards were procured from Bio-Rad.
Bioinformatics analysis
The sequences of RuvC and RuvX/YqgF orthologs from different species were retrieved from the UniProt database. The RuvC sequences of M. smegmatis mc2 155 (spA0QWH4), M. tuberculosis (spP9WGV9), Mycobacterium bovis (spC1AF63), Mycobacterium leprae (spP40834), E. coli (spP0A814), D. radiodurans (spQ9RX75), Streptomyces griseus (spB1W3G2), T. thermophiles (spQ72JP3), H. pylori (spO25544), Agrobacterium fabrum (spQ8U9K4), Neisseria gonorrhoeae (spQ5FA76), and Thermotoga maritime (spQ9WZ45) were retrieved from UniProt server (https://www.uniprot.org/uniprotkb?query=ruvC). Similarly, the sequences of RuvX/YqgF orthologs from M. smegmatis (spA0QWQ5), M. tuberculosis (spP9WGV7), M. bovis (spC1AF23), M. leprae (spB8ZUJ7), E. coli (spP0A8I1), Bacillus subtilis (spO34634), D. radiodurans (spQ9RRI2), S. griseus (trA0A2X2M8E1), T. thermophiles (spQ5SHA1), H. pylori (spB2USG5), A. fabrum (spQ8UFT1), N. gonorrhoeae (spQ5F936), and T. maritime (spQ9X1N1) were retrieved from the UniProt database (https://www.uniprot.org/uniprotkb?query=YqgF). Multiple sequence alignment was performed using the Clustal Omega (version 1.2.4) (https://www.ebi.ac.uk/jdispatcher/msa/clustalo) and visualized using Jalview v2.0 (https://www.jalview.org/download/).
Construction of recombinant plasmids carrying M. smegmatis mc2155 ruvC and ruvX gene
The M. smegmatis ruvC and ruvX ORFs (accession number, MSMEG_2943 and MSMEG_3026, respectively) were identified from Mycobrowser database (https://mycobrowser.epfl.ch/). The MSMEG_2943- and MSMEG_3026-encoding genes in M. smegmatis mc2155 were amplified via PCR from the genomic DNA using the forward and reverse primers with NheI and HindIII restriction site, as shown in Table S2. Reaction mixture (20 μl) contained 1× Phusion high-fidelity buffer, forward and reverse primers (0.5 μM each), 50 ng of M. smegmatis mc2155 genomic DNA, 0.3 mM dNTPs, Phusion High-Fidelity DNA polymerase (0.4 U/μl), and 10% dimethyl sulphoxide. The ruvC and ruvX genes were amplified using a standard PCR protocol. Briefly, PCR was carried out at 98 °C for 5 min, followed by 30 cycles of amplification: each cycling step involved denaturation (95 °C for 30 s), annealing (70 °C for 40 s), and extension (72 °C for 45 s), followed by a final 10 min extension at 70 °C. The amplified 558 bp (ruvC) and 513 bp (ruvX) products were gel purified using the GeneJET gel extraction kit (Thermo Fisher Scientific) and digested with NheI and HindIII (0.2 U/μl each). The resulting products were cloned into the pET21a (+) expression vector (Novagen) by mixing 100 ng of linearized vector with 300 ng of PCR product and T4 DNA ligase (10 CEU/μl; Thermo Fisher Scientific). The ligation mix was transformed into E. coli DH5α cells. The transformants were selected on LB agar plates supplemented with 100 mg/ml of ampicillin. Plasmid DNA was isolated from the transformants using the GeneJET plasmid midiprep kit (Thermo Fisher Scientific). The insertions in the recombinant plasmids were verified by colony PCR using the above primers, restriction digestion, and Sanger sequencing. The resulting plasmids were designated pMsRuvC and pMsRuvX.
Site-directed mutagenesis of putative active site residues in ruvC and ruvX of M. smegmatis
A PCR-based site-directed mutagenesis method was used to introduce mutations as previously described (86). Briefly, mutations were created by the Quick-change method using Phusion High-Fidelity DNA polymerase (NEB), DpnI (Thermo Fisher Scientifi), plasmid pMsRuvC, Phusion High-Fidelity DNA polymerase (NEB), and DpnI (Thermo Scientific MA, USA). Plasmid pMsRuvX was used as template to generate its variants using the same protocol. In the case of MsRuvC, Asp7, and Glu68 were substituted with alanine. Likewise, in MsRuvX, residues D25, D115, and E116 were substituted with alanine. Similarly, MsRuvX polypeptide sequence, aspartic acid residues were substituted with asparagine at position 25, 115, and 142. The primers used for alanine and asparagine substitutions are shown in Table S3 and S4. PCR reactions were carried out in a final volume of 50 μl using 1× Phusion High-Fidelity buffer, forward and reverse primers (0.5 μM each), 100 ng of pMsRuvX or pMsRuvC, 0.6 mM dNTPs, 1 u of Phusion High-Fidelity DNA polymerase, and 10% dimethyl sulphoxide. The PCR procedure was as follows: initial denaturation at 98 °C for 5 min, 35 cycles (denaturation at 94 °C for 30 s, annealing at 70 °C for 30 s and extension (72 °C for 5 min), and final extension at 72 °C for 10 min. PCR products were isolated from gels using GeneJET PCR clean-up kit (Thermo Fisher Scientific) and were incubated with DpnI (1 U) for 6 h at 37 °C. The ligation mixtures were transformed into competent E. coli DH5α cells and the transformants were selected on LB agar plates containing 100 μg/ml of ampicillin. Plasmid DNA was isolated from the transformants using the GeneJET plasmid Midiprepep kit (Thermo Fisher Scientific). The mutations in the variants (D7A, E68A, D25A, D25N, D115A, D115N, E116A, and D142N) were verified by DNA sequencing. The resulting plasmids were designated pMsRuvC-D7A pMsRuvC-E68A, pMsRuvX-D25A, pMsRuvX-D25N, pMsRuvX-D115A, pMsRuvX-D115N, pMsRuvX-E116A, and pMsRuvX-D142N.
Expression and purification of the WT and variant MsRuvC and MsRuvX proteins
The WT MsRuvC, MsRuvX, and their variants were expressed and purified using similar protocols. Briefly, the transformed E. coli host Rosetta (DE3) pLysS cells carrying plasmids encoding either WT MsRuvC, MsRuvX, or the DNA cleavage-deficient variants were grown in LB medium (4 L) supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37 °C with constant shaking at 180 rpm until the A600 reached 0.5. Expression of genes encoding those proteins was induced by adding IPTG to a final concentration of 0.5 mM, and the cultures were further incubated with constant shaking (250 rpm) at 37 °C for 8 h. Cells were collected by centrifugation at 6000 rpm for 20 min, at 4 °C, using a Beckman JA-10 rotor. The cell pellet was resuspended in buffer A [20 mM Tris–HCl (pH 8), 50 mM NaCl, 10% glycerol, and 5 mM 2-mercaptoethanol] and stored at −80 °C. The frozen cell pellets were thawed at 4 °C and resuspended in 50 ml of buffer A. Cells were lysed by sonication at 4 °C (GEX750 ultrasonic processor, 5 cycles at 55% amplitude, length of pulses = 1 s). Cell debris was removed by centrifugation in a Beckman Ti-45 rotor at 30,000 rpm at 4 °C for 1 h. The resulting supernatant fraction was loaded on a 10 ml column of SP-Sepharose, which had been equilibrated with buffer A. Proteins from SP-Sepharose resins were eluted by a linear NaCl gradient from 100 mM→500 mM in buffer A. The fractions containing MsRuvC or MsRuvX or their variants (as determined from its SDS/PAGE mobility) were pooled and dialyzed at 4 °C for 6 h against buffer B [20 mM Tris–HCl (pH 8), 10% glycerol] containing 50 mM NaCl. The dialysate containing MsRuvC, MsRuvX, or their variants were loaded separately on a 5 ml column of heparin-agarose that had been equilibrated with buffer B containing 50 mM NaCl. The bound proteins were eluted with 100 mM→500 mM linear gradient of NaCl in buffer B. At this stage, the fractions containing MsRuvX or its mutant variants were pooled, proteins were concentrated to 5 ml, using Amicon Ultra-15 centrifugal filter unit with 10-kDa molecular mass cut-off (Millipore). MsRuvX and variants were dialyzed against the 1 L of 20 mM Tris–HCl (pH 8), 200 mM NaCl, 20% glycerol, and 1 mM DTT] for 4 h at 4 °C. Aliquots were stored in −80 °C.
In case of MsRuvC and its variants, the concentrated samples were subjected to gel-filtration chromatography (Superdex 75 increase 10/300 GL, GE Healthcare) with a buffer containing 20 mM Tris–HCl (pH 8) containing 200 mM NaCl, 20% glycerol, and 1 mM DTT. The fractions containing MsRuvC or its variants were pooled and dialyzed against the 1 L of 20 mM Tris–HCl (pH 8), 200 mM NaCl, 20% glycerol, and 1 mM DTT] for 4 h at 4 °C. Aliquots of MsRuvC or its variants were stored in −80 °C. Samples of purified proteins were analyzed by SDS-PAGE (12.5% gel) and found to be ∼ 95% pure. The concentrations of purified proteins were determined by the dye binding method using BSA as a standard. The identity of MsRuvC, MsRuvX, and their variants were confirmed by Western blotting using the anti-MtRuvC and anti-MtRuvX antibody, respectively (48).
Analytical gel-filtration chromatography
Analytical SEC on Superose 12 10/300 GL column (GE Healthcare) was performed using fast protein liquid chromatography system (Bio-Rad) at a flow rate of 0.3 ml/min at 4 °C, which had been equilibrated with a buffer A [50 mM Tris/HCl (pH 8), 100 mM NaCl and 5% glycerol]. The column was calibrated with molecular weight markers, comprising of vitamin B12 (1.3 kDa), myoglobin (17 kDa), ovalbumin (44 kDa), bovine γ-globulin (158 kDa), and thyroglobulin (670 kDa), and the resulting chromatograms were used to construct the standard curve. Samples (200 μl from 1 mg/ml stock) of either WT MsRuvC and MsRuvX (Fig. 2) or their variants (MsRuvC-D7A, MsRuvC-E68A, and MsRuvX-D25A (Fig. S5), dialyzed against the buffer A for 6 h at 4 °C, were applied onto a Superose 12 10/300 column. The fractions were monitored at 280 nm for protein. Apparent molecular masses of MsRuvC and MsRuvX and their variants were calculated by interpolation using the calibration curve obtained with standards, as previously described (86).
SEC coupled with MALS
The SEC-MALS experiments were performed using a Superose 12 10/300 GL SEC column (GE Healthcare) attached to a Shimadzu HPLC system with a UV detector (SPD-10A VP Shimadzu, UV-Visible detector), a miniDAWN TREOS MALS detector and a Waters 2414 RI detector (Wyatt Technology Corp). Experiments were carried out at 18 °C in tris-NaCl-glycine buffer (50 mM Tris–HCl (pH 8), 100 mM NaCl, and 5% glycerol). Samples (200 μl, 1 mg/ml) of MsRuvC, MsRuvX, MsRuvC-D7A, MsRuvC-E68A, or MsRuvX-D25A were applied to a Superose 12 10/300 Gl column, which had been equilibrated with the same buffer at a flow rate of 0.3 ml/min. Molecular mass calculations were performed using the ASTRA 6.1 software (Wyatt Technology; https://www.wyatt.com/products/software/astra.html).
Preparation of radiolabeled DNA substrates
The sequences of ODNs used in this study are listed in Table S5. The ODNs were labeled at the 5′ or 3′ end by using either [γ-32P] ATP and T4 polynucleotide kinase or [α-32P] dCTP and terminal deoxynucleotidyl transferase. The unincorporated radioisotopes were removed as previously described (86). DNA substrates (Table S6) were constructed by annealing equimolar amounts of appropriate ODNs in a 100 μl buffer containing 0.3 M sodium citrate (pH 7) and 3 M NaCl as previously described (49, 86). Samples were incubated at 95 °C for 5 min and then slowly cooled to 4 °C over a period of 2 h. The annealed products were resolved by electrophoresis on 8% polyacrylamide gels using 89 mM Tris-borate buffer (pH 8) containing 2 mM EDTA at a constant voltage of 100 V for 8 to 10 h. The bands corresponding to annealed substrates were excised from the gels and DNA was eluted by soaking gel slices in tris-EDTA buffer [10 mM Tris–HCl, pH 7.5, 1 mM EDTA] for 12 h.
SPR measurements
The binding kinetics and affinities of MsRuvC and MsRuvX for various DNA substrates were determined by SPR using a Biacore 2000 optical biosensor instrument (GE Healthcare) at 25 °C. The streptavidin functionalized SA (GE Healthcare) chips were washed with 1 M NaCl. The DNA substrates were labeled at the 5ʹ-end with biotin (Table S5). These were constructed by mixing 5ʹ-biotinylated ODNs with appropriate combination of complementary ODNs (Table S6) in a buffer containing 0.3 M sodium citrate (pH 7) and 3 M NaCl. Samples were incubated at 95 °C for 5 min and then gradually cooled to 4 °C. Aliquots containing 100 nM of 5ʹ-biotinylated DNA substrate in buffer A (25 mM Hepes, pH 8.0, 200 mM NaCl, 3 mM EDTA, and 0.005% P-20 surfactant) were passed through the surface of four streptavidin-derivatized sensor chips at a flow rate of 50 μl/min until the target of response units (RUs) on the surface was achieved using standard procedures. Under these conditions, approximately 400 RU, 700 RU, 600 RU, and 500 RU were immobilized on chip 1, chip 2, chip 3, and chip 4, respectively. After washing off the unbound DNA substrate from the sensor chips using buffer A, increasing concentrations of purified MsRuvC or MsRuvX were passed over the chips at a flow rate of 30 μl/min, allowing 100 s contact and 200 s of dissociation time. The kinetic rates of the binding of MsRuvC and MsRuvX were derived from global Langmuir fitting for 1∶1 binding kinetics to sensorgrams collected at different ligand concentrations using BIAcore evaluation v3.0 software (licensed to IISc Biochemistry with serial no. BIA-0043: http://www.molecular-interactions.si/data/equipment/BIAeval3-AC.pdf). The affinities of MsRuvC and MsRuvX (Kd values) were determined by averaging the Kd values obtained at various concentration of MsRuvC and MsRuvX. The Kd values were calculated from the Koff and Kon values using the equation Kd = Koff/Kon. Residual plots and Chi2 were used as a measure of the curve fitting efficiency.
Microscale thermophoresis
The binding affinities (Kd values) of MsRuvC and its mHJ cleavage-deficient mutants was investigated using MST. Experiments were performed on a Monolith NT.115 instrument (Nano Temper Technologies) as previously described (87). Briefly, 5′-FAM (6-carboxyfluorescein)-labeled mHJ was diluted in the MST buffer, which contained 10 mM Hepes (pH 7.5), 150 mM NaCl, and 0.1 mg/ml BSA, to a final concentration of 50 nM. In parallel, 16 dilutions of MsRuvC or its variants (where specified) were prepared in the same buffer as above to a final concentration ranging from 1.5 nM to 12.5 μM. For each experiment, 10 μl of 50 nM FAM-labeled DNA in MST buffer was mixed with 10 μl (25 μM) MsRuvC (or its variants) of each dilution. After incubation at 37 °C for 30 min, 10 μl from the reaction mixture was loaded into capillaries (NanoTemper, Inc.), and MST measurements were performed using 60% blue light-emitting diode power and 80% MST power. The results were analyzed, and the values were normalized and plotted as a function of MsRuvC, MsRuvC-D7A, and MsRuvC-E68A. The data were analyzed using MO. Control v2.3 (https://shop.nanotempertech.com/en/mocontrol-2-software-1-license-38) and MO. Affinity Analysis v2.3 software (https://shop.nanotempertech.com/en/moaffinity-analysis-software-unlimited-licenses-34).
Differential scanning fluorimetry
The DSF was performed as previously described (88). The thermal denaturation profiles of proteins (MsRuvC, MsRuvX, MsRuvC-D7A, MsRuvC-E68A, MsRuvX-D25A, MsRuvX-D25N, MsRuvX-D115A, MsRuvX-D115N, MsRuvX-E116A, and MsRuvX-D142N) were determined using a fluorescence microplate reader (CFX96 Touch Real-Time PCR System, Bio-Rad) as a function of temperature. The measurements were carried out as follows: reaction mixtures (25 μl) containing 15 μg of the WT MsRuvC, MsRuvX or their variants were incubated in a buffer containing 10 mM Hepes (pH 8) and 4× of SYPRO-Orange (Sigma-Aldrich) in a 96-well PCR plate (Multiplate 96-Well PCR Plate, Bio-Rad) for 5 min at 4 °C. Sealed PCR plates (Microseal PCR plate sealing film, Bio-Rad) were centrifuged at 4000 rpm for 5 min to remove air bubbles. Subsequently, samples were subjected to the melt runs, ranging from 10 °C to 95 °C, in increments of 0.5 °C for 10 s, and the fluorescence intensities (excitation at 470 nm and emission at 570 nm) were measured. The melting curves were tracked with the CFX Maestro software (Bio-Rad) (https://www.bio-rad.com/en-in/product/cfx-maestro-software-for-cfx-real-time-pcr-instruments?ID=OKZP7E15) and plotted as relative fluorescence unit versus temperature (°C). Thereafter, the derivative of the plots (-d(RFU)/dT) were used to determine the Tm.
Western blot analysis
Protein samples (2 μg) were resolved by SDS-PAGE was transferred to polyvinylidene difluoride membranes (Bio-Rad), as previously described (48). The polyclonal antibodies against M. tuberculosis RuvC and RuvX were raised in rabbits and their specificity was characterized as previously described (48). Prior to immunostaining, polyvinylidene difluoride membranes were blocked with 5% skimmed milk in tris-buffered saline with Tween 20 (TBST) buffer [50 mM Tris–HCl (pH 8.0), 150 mM NaCl, and 0.2% Tween 20] at 25 °C for 2 h, and washed with TBST buffer for 1 min and incubated with polyclonal anti-MtRuvC or anti-MtRuvX antibodies at 4 °C for 12 h. After washing the blots with TBST buffer for 3 times for 15 min each, they were incubated with anti-rabbit IgG antibody at 4 °C for 6 h. The antibodies were used at the following dilutions: anti-MtRuvC (1:4000), anti-MtRuvX (1:4000), and anti-rabbit IgG antibody (1:40,000). After washing the membranes with TBST buffer 3 times for 15 min each, they were incubated with horseradish peroxidase–conjugated anti-rabbit secondary antibody (Sigma-Aldrich) at 4 °C for 3 h. The blots were developed using chemiluminescent substrates (Clarity Western ECL substrate, Bio-Rad Laboratories) and imaged using the ChemiDoc ImageQuant system (GE LAS 4000) (https://timothyspringer.org/files/tas/files/imagequantlas4000.pdf).
Electrophoretic mobility shift assay
The assay was performed as previously described (89). The reaction mixtures (20 μl) contained 20 mM Tris–HCl (pH 8), 100 mg/ml BSA, 0.5 nM of 5′-32P-labeled 3′-flap DNA, and increasing concentrations of MsRuvX (or its variants). After incubation at 37 °C for 30 min, the reaction was stopped by adding 2 μl loading dye [0.1% (w/v) of bromophenol blue and xylene cyanol in 20% glycerol]. Samples were analyzed by electrophoresis on a 6% polyacrylamide gel using 0.25× tris-borate-EDTA (TBE) buffer (22.5 mM Tris-borate, and 0.5 mM EDTA) at 70 V at 4 °C for 12 h. The dried gels were exposed to phosphorimaging screens and bands were visualized using a Fuji FLA-9000 phosphorimager. The band intensities were quantified in an UVItech gel documentation imaging system using the UVI-Band Map software (v97.04; Copyright 1997 UVI Tech, software no. 8610) and plotted using GraphPad Prism software (v6.0; https://www.graphpad.com/rf/7982103771/). Nonlinear regression analysis was used to fit the data in one-site binding hyperbola model.
DNA cleavage assay
The reaction mixture (20 μl) contained 20 mM Tris–HCl (pH 8), 5 mM MgCl2, 100 μg/ml BSA, 0.5 nM of the indicated 5′-32P- labeled DNA substrate, and different concentration of either MsRuvX (or its variants) or MsRuvC (or its variants) as specified in the figure legends. In the case of WT MsRuvC and its variants, the following reaction condition was used: 20 mM Tris–HCl (pH 8), 3 mM MgCl2, 100 μg/ml BSA, 2 mM DTT, along with the 5′-32P-labeled DNA (0.5 nM). After incubation at 37 °C for 1 h, reaction was stopped by adding EDTA, SDS, and proteinase K to a final concentration of 10 mM, 1% and 10 μg/ml, respectively, and incubation was continued at 37 °C for an additional 20 min. Samples were analyzed by electrophoresis on native 8% polyacrylamide gels using TBE buffer (89 mM Tris-borate buffer (pH 8) and 2 mM EDTA) at 100 V for 4 to 8 h at 4 °C (the time required varied depending on the complexity of the DNA substrates). In the case of denaturing PAGE, samples were further incubated at 95 °C for 5 min along with the formamide loading dye [(0.3% (w/v) bromophenol blue, 0.3% (w/v) xylene cyanol, 10 mM EDTA (pH 7.5), and 80% (w/v) formamide] and were separated by electrophoresis on 8% polyacrylamide gels containing 8 M urea using TBE buffer (89 mM Tris-borate buffer (pH 8) and 2 mM EDTA) at 150 V for 2 to 3 h at 24 °C. The dried gels were exposed to phosphorimaging screens, and the bands were visualized using the Typhoon FLA-9000 phosphorimager. The band intensities were quantified in UVI-tech gel documentation station using UVI-BandMap v97.04 software. Data were plotted using GraphPad Prism v6.0 software. Nonlinear regression analysis was used to fit the data.
To perform the dual incision reaction, the ODNs (Table S5, Sl. No: 15–18) containing a PS linkage in which one of the nonbridging oxygens of the phosphodiester bond between 31 to 32 nt in strand 1, 29 to 30 nt in strand 2, 32 to 33 nt in strand 3, and 30 to 31 nt in strand 4 was replaced by sulphur. These ODNs were obtained from Synbio Technologies. Nuclease assays were carried out as described above and the deproteinized samples were analyzed by nondenaturing PAGE (8% polyacrylamide) and denaturing PAGE (17% polyacrylamide and 8 M urea) using TBE buffer. The reaction products were visualized in gels as described above.
Mapping of MsRuvC cleavage site on the mHJ
The assay was performed as previously described (49). The reaction mixtures (20 μl) contained 20 mM Tris–HCl (pH 8), 3 mM MgCl2, 100 μg/ml BSA, 2 mM DTT, 1 nM of 5′-32P-labeled HJ and indicated concentration of WT MsRuvC or its variants. After incubation at 37 °C for 1 h, reaction was stopped by adding 5 μl of stop solution (10 mg/ml proteinase K, 200 mM EDTA and 2.5% w/v SDS), followed by incubation for an additional 20 min at 37 °C. Samples were dried under vacuum and the dried pellets were resuspended into 5 μl of buffer [0.3% (w/v) bromophenol blue, 0.3% (w/v) xylene cyanol, 10 mM EDTA (pH 7.5), and 80% (w/v) formamide]. The resuspended sample denatured by boiling at 95 °C for 5 min. The Maxam–Gilbert G + A sequencing ladder was obtained for each 5′-32P-labeled HJ strand as previously described (90). Samples were then loaded onto a denaturing (8 M urea) and 17% polyacrylamide gel and electrophoresed using TBE buffer (89 mM Tris-borate buffer (pH 8) and 2 mM EDTA) at 1600 V for 3 h. The dried gels were exposed to phosphorimaging cassettes and visualized using a Typhoon FLA-9000 phosphor imager. The specific position of cleavage sites was mapped with reference to the G + A Maxam–Gilbert sequencing ladder.
Mapping of MsRuvX cleavage sites on various DNA substrates
For cleavage site mapping, reactions (20 μl) were performed in a buffer containing 20 mM Tris–HCl (pH 8), 5 mM MgCl2, 100 μg/ml BSA, and 1 nM of 5′-32P-labeled DNA substrate in the absence or presence of MsRuvX (or its variants) as indicated in the figure legend. After incubation at 37 °C for 1 h, reaction was stopped by adding 5 μl of stop solution (10 mg/ml proteinase K, 200 mM EDTA and 2.5% w/v SDS), and incubation was extended for an additional 20 min at 37 °C. Five microliters of gel loading solution [0.3% (w/v) bromophenol blue, 0.3% (w/v) xylene cyanol, 10 mM EDTA (pH 7.5), and 80% (w/v) formamide] was added to the reaction mixtures and heated at 95 °C for 5 min. The reaction products together with G + A ladders were analyzed by denaturing (8 M urea) and 17% polyacrylamide gel electrophoresis at 1600 V and 32 W using TBE buffer (89 mM Tris-borate buffer (pH 8) and 2 mM EDTA) for 3 h. Dried gels were exposed to phosphorimaging screens, and the bands were visualized using the Typhoon FLA-9000 phosphor imager. Cleavage sites were mapped with reference to the Maxam–Gilbert G + A sequencing ladder.
Religation of mHJ resolution products
The assay was performed using 5′-32P-labeled HJs in which one arm is shortened by 8 bp (Table S6), with 32P label on a different DNA strand. Each reaction mixture (20 μl) contained 20 mM Tris–HCl (pH 8), 3 mM MgCl2, 100 μg/ml BSA, 2 mM DTT, 0.5 nM of the indicated 5′-32P-labeled HJ, and 200 nM MsRuvC. After incubation at 37 °C for 1 h, 1× T4 DNA ligase buffer was added to the indicated reaction mixture along with 100 CEU of T4 DNA ligase (Thermo Fisher Scientific) and further incubated at 37 °C for 1 h. Reactions were stopped by adding 5 μl of gel loading solution [0.3% (w/v) bromophenol blue, 0.3% (w/v) xylene cyanol, 10 mM EDTA (pH 7.5), and 80% (w/v) formamide] and heated at 95 °C for 5 min. Samples were electrophoresed on a 8% polyacrylamide gel containing 8 M urea using TBE buffer (89 mM Tris-borate buffer (pH 8) and 2 mM EDTA) at a constant voltage of 150 V for 2 h at room temperature. The gels were dried and phosphor-imaged, as described in the previous sections.
ssDNA exonuclease assay
The assay were performed in a buffer (20 μl) containing 20 mM Tris–HCl (pH 7.5), 2 mM DTT, 5 mM MgCl2, 100 μg/ml BSA, 1 nM of 5′- or 3′-32P-labeled 62-mer ssDNA and a fixed amount (200 nM) or varying concentrations of MsRuvX. After incubation for 1 h, or for different time intervals, at 37 °C, reaction was stopped by adding 5 μl of stop solution (10 mg/ml proteinase K, 2.5% SDS, and 100 mM EDTA) to each sample, and incubation was continued for an additional 20 min. Subsequently, 5 μl of gel loading solution [0.3% (w/v) bromophenol blue, 0.3% (w/v) xylene cyanol] was added to each sample and were incubated at 95 °C for 10 min. Samples were then analyzed on to the 17% polyacrylamide gels in the presence of 8 M urea using TBE buffer (89 mM Tris-borate buffer (pH 8) and 2 mM EDTA) at 1600 V for 3 h. The gels were then dried and were exposed to phosphorimaging screens, and the bands were visualized using the Typhoon FLA-9000 phosphor imager.
Data availability
All data generated or analyzed during the course of this work are included in this article and will be made available upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
A. A. and K. M. writing–original draft; A. A. and K. M. visualization; A. A. validation; A. A. investigation; A. A. methodology; A. A. formal analysis; A. A. data curation; K. M. writing–review and editing; K. M. supervision; K. M. funding acquisition; K. M. conceptualization; K. M. project administration.
Funding and additional information
This work was funded by the Bhatnagar Fellowship research grant (CSIR/KM/425) to K. M. from the Council of Scientific & Industrial Research, New Delhi.
Reviewed by members of the JBC Editorial Board. Edited by Craig Cameron
Supporting information
References
- 1.Wright W.D., Heyer W.D. Rad54 Functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D Loop formation. Mol. Cell. 2014;53:420–432. doi: 10.1016/j.molcel.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrical S.W. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect. Biol. 2015;7:1–20. doi: 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray S., Cohen P.E. Control of meiotic crossovers: from double-strand break formation to designation. Annu. Rev. Genet. 2016;50:175–210. doi: 10.1146/annurev-genet-120215-035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weller S.K., Sawitzke J.A. Recombination promoted by DNA viruses: phage λ to Herpes simplex virus. Annu. Rev. 2014;68:237–258. doi: 10.1146/annurev-micro-091313-103424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Šimičić P., Židovec-Lepej S. A glimpse on the evolution of RNA viruses: implications and lessons from SARS-CoV-2. Viruses. 2022;15:19–21. doi: 10.3390/v15010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinks-Robertson S., Petes T.D. Mitotic recombination in yeast: what we know and what we don’t know. Curr. Opin. Genet. Dev. 2021;71:78–85. doi: 10.1016/j.gde.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Massy B. Spp1 links sites of meiotic DNA double-strand breaks to chromosome axes. Mol. Cell. 2013;49:3–5. doi: 10.1016/j.molcel.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Mehta A., Haber J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect. Biol. 2014;6:1–17. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyatt H.D., Sarbajna S., Matos J., West S.C. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 10.Dehe P.M., Gaillard P.H.L. Control of structure-specific endonucleases to maintain genome stability. Nat. Rev. Mol. Cell Biol. 2017;18:315–330. doi: 10.1038/nrm.2016.177. [DOI] [PubMed] [Google Scholar]
- 11.Giaccherini C., Gaillard P.H. Control of structure-specific endonucleases during homologous recombination in eukaryotes. Curr. Opin. Genet. Dev. 2021;71:195–205. doi: 10.1016/j.gde.2021.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Azeroglu B., Leach D.R.F. RecG controls DNA amplification at double-strand breaks and arrested replication forks. FEBS Lett. 2017;591:101–1113. doi: 10.1002/1873-3468.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubnau D., Blokesch M. Mechanisms of DNA uptake by naturally competent bacteria. Annu. Rev. Genet. 2019;53:217–237. doi: 10.1146/annurev-genet-112618-043641. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L., Jiang D., Wu M., Yang Z., Oger P.M. New insights into DNA repair revealed by NucS endonucleases from hyperthermophilic Archaea. Front. Microbiol. 2020;11:1263. doi: 10.3389/fmicb.2020.01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grindley N.D., Whiteson K.L., Rice P.A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 16.Tian X., Zhou B. Strategies for site-specific recombination with high efficiency and precise spatiotemporal resolution. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West S.C. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 18.Lilley D.M.J. Holliday junction-resolving enzymes-structures and mechanisms. FEBS Lett. 2017;591:1073–1082. doi: 10.1002/1873-3468.12529. [DOI] [PubMed] [Google Scholar]
- 19.Wyatt H.D.M., West S.C. Holliday junction resolvases. Cold Spring Harb Perspect. Biol. 2014;6:1–29. doi: 10.1101/cshperspect.a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarbajna S., West S.C. Holliday junction processing enzymes as guardians of genome stability. Trends Biochem. Sci. 2014;39:409–419. doi: 10.1016/j.tibs.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 21.West S.C., Blanco M.G., Chan Y.W., Matos J., Sarbajna S., Wyatt H.D.M. Resolution of recombination intermediates: mechanisms and regulation. Cold Spring Harb. Symp. Quant. Biol. 2015;80:103–109. doi: 10.1101/sqb.2015.80.027649. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L., Makarova K.S., Koonin E.V. Survey and Summary: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West S.C. The search for a human Holliday junction resolvase. Biochem. Soc. Trans. 2009;37:519–526. doi: 10.1042/BST0370519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majorek K.A., Dunin-Horkawicz S., Steczkiewicz K., Muszewska A., Nowotny M., Ginalski K., et al. The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014;42:4160–4179. doi: 10.1093/nar/gkt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verma P., Kumari P., Negi S., Yadav G., Gaur V. Holliday junction resolution by At-HIGLE: an SLX1 lineage endonuclease from Arabidopsis thaliana with a novel in-built regulatory mechanism. Nucleic Acids Res. 2022;50:4630–4646. doi: 10.1093/nar/gkac239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahagi M., Iwasaki H., Nakata A., Shinagawa H. Molecular analysis of the Escherichia coli ruvC gene, which encodes a Holliday junction-specific endonuclease. J. Bacteriol. 1991;173:5747–5753. doi: 10.1128/jb.173.18.5747-5753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandal T.N., Mahdi A.A., Sharples G.J., Lloyd R.G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J. Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett R.J., Dunderdale H.J., West S.C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- 31.Dunderdale H.J., Sharples G.J., Lloyd R.G., West S.C. Cloning, overexpression, purification, and characterization of the Escherichia coli RuvC Holliday junction resolvase. J. Biol. Chem. 1994;269:5187–5194. [PubMed] [Google Scholar]
- 32.Ariyoshi M., Vassylyev D.G., Iwasaki H., Nakamura H., Shinagawa H., Morikawa K. Atomic structure of the RuvC resolvase : a Holliday junction-specific endonuclease from E . coli. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 33.Shah R., Cosstick R., West S.C. The RuvC protein dimer resolves Holliday junctions by a dual incision mechanism that involves base-specific contacts. EMBO J. 1997;16:1464–1472. doi: 10.1093/emboj/16.6.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L., Shi K., Yin Z., Aihara H. Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution. Nucleic Acids Res. 2013;41:648–656. doi: 10.1093/nar/gks1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y., He Y., Lin Z. Biochemical and structural characterization of the Holliday junction resolvase RuvC from Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 2020;525:265–271. doi: 10.1016/j.bbrc.2020.02.062. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y., Yang J., Xu G., Cheng K. Biochemical and structural study of RuvC and YqgF from Deinococcus radiodurans. MBio. 2022;13 doi: 10.1128/mbio.01834-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin C., Han W., Xu Y., Zhao Y., Xu H., Tian B., et al. Structural and functional characterization of the Holliday junction resolvase RuvC from Deinococcus radiodurans. Microorganisms. 2022;10:1160. doi: 10.3390/microorganisms10061160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benson F.E., West S.C. Substrate specificity of the Escherichia coli RuvC protein. Biochemistry. 1994;269:5195–5201. [PubMed] [Google Scholar]
- 39.Punatar R.S., Martin M.J., Wyatt H.D.M., Chan Y.W., West S.C. Resolution of single and double Holliday junction recombination intermediates by GEN 1. Proc. Natl. Acad. Sci. U. S. A. 2017;114:443–450. doi: 10.1073/pnas.1619790114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorecka K.M., Komorowska W., Nowotny M. Crystal structure of RuvC resolvase in complex with Holliday junction substrate. Nucleic Acids Res. 2013;41:9945–9955. doi: 10.1093/nar/gkt769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Górecka K.M., Krepl M., Szlachcic A., Poznański J., Šponer J., Nowotny M. RuvC uses dynamic probing of the Holliday junction to achieve sequence specificity and efficient resolution. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-019-11900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan J., Hong S., Guan Z., He W., Zhang D., Yin P. Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-15242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponting C.P. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freiberg C., Wieland B., Spaltmann F., Ehlert K., Brötz H., Labischinski H. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J. Mol. Microbiol. Biotechnol. 2001;3:483–489. [PubMed] [Google Scholar]
- 45.Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 46.Zalacain M., Biswas S., Ingraham K.A., Ambrad J., Bryant A., Chalker A.F., et al. A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J. Mol. Microbiol. Biotechnol. 2003;6:109–126. doi: 10.1159/000076741. [DOI] [PubMed] [Google Scholar]
- 47.Kumar A., Beloglazova N., Bundalovic-Torma C., Phanse S., Deineko V., Gagarinova A., et al. Conditional epistatic interaction maps reveal global functional rewiring of genome integrity pathways in Escherichia coli. Cell Rep. 2016;14:648–661. doi: 10.1016/j.celrep.2015.12.060. [DOI] [PubMed] [Google Scholar]
- 48.Nautiyal A., Rani P.S., Sharples G.J., Muniyappa K. Mycobacterium tuberculosis RuvX is a Holliday junction resolvase formed by dimerisation of the monomeric YqgF nuclease domain. Mol. Microbiol. 2016;100:656–674. doi: 10.1111/mmi.13338. [DOI] [PubMed] [Google Scholar]
- 49.Thakur M., Mohan D., Singh A.K., Agarwal A., Gopal B., Muniyappa K. Novel insights into ATP-stimulated cleavage of branched DNA and RNA substrates through structure-guided studies of the Holliday Junction resolvase RuvX. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167014. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X.S., Blaser M.J. DprB facilitates inter- and intragenomic recombination in Helicobacter pylori. J. Bacteriol. 2012;194:3891–3903. doi: 10.1128/JB.00346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muniyappa K., Vaze M.B., Ganesh N., Sreedhar Reddy M., Guhan N., V.R Comparative genomics of Mycobacterium tuberculosis and Escherichia coli for recombination (rec) genes. Microbiology. 2000;146:2093–2095. doi: 10.1099/00221287-146-9-2093. [DOI] [PubMed] [Google Scholar]
- 52.Mizrahi V., Andersen S.J. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol. Microbiol. 1998;29:1331–1339. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 53.Miggiano R., Rizzi M., Ferraris D.M. Mycobacterium tuberculosis pathogenesis, infection prevention and treatment. Pathogens. 2020;9:10–13. doi: 10.3390/pathogens9050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu D., Wang Y.-S., Wyss D.F. Solution structure of the hypothetical protein YqgF from Escherichia coli reveals an RNAse H fold. J. Biomol. NMR. 2003;27:389–392. doi: 10.1023/a:1025840121177. [DOI] [PubMed] [Google Scholar]
- 55.Takahagi M., Iwasaki H., Shinagawa H. Structural requirements of substrate DNA for binding to and cleavage by RuvC, a Holliday junction resolvase. J. Biol. Chem. 1994;269:15132–15139. [PubMed] [Google Scholar]
- 56.Fogg J.M., Schofield M.J., White M.F., Lilley D.M.J. Sequence and functional-group specificity for cleavage of DNA junctions by RuvC of Escherichia coli. Biochemistry. 1999;38:11349–11358. doi: 10.1021/bi990926n. [DOI] [PubMed] [Google Scholar]
- 57.Sharples G.J. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 2001;39:823–834. doi: 10.1046/j.1365-2958.2001.02284.x. [DOI] [PubMed] [Google Scholar]
- 58.Curtis F.A., Reed P., Sharples G.J. Evolution of a phage RuvC endonuclease for resolution of both Holliday and branched DNA junctions. Mol. Microbiol. 2005;55:1332–1345. doi: 10.1111/j.1365-2958.2004.04476.x. [DOI] [PubMed] [Google Scholar]
- 59.Shah R., Bennett R.J., West S.C. Activation of RuvC Holliday junction resolvase in vitro. Nucleic Acids Res. 1994;22:2490–2497. doi: 10.1093/nar/22.13.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito A., Iwasaki H., Ariyoshi M., Morikawa K., Shinagawa H. Identification of four acidic amino acids that constitute the catalytic center of the RuvC Holliday junction resolvase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huynh K., Partch C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015;79:28.9.1–28.9.14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fogg J.M., Lilley D.M.J. Ensuring productive resolution by the junction-resolving enzyme RuvC: large enhancement of the second-strand cleavage rate. Biochemistry. 2000;39:16125–16134. doi: 10.1021/bi001886m. [DOI] [PubMed] [Google Scholar]
- 63.Shah R., Bennett R.J., West S.C. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994;79:853–864. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 64.Osman F., Gaskell L., Whitby M.C. Efficient second strand cleavage during Holliday junction resolution by RuvC requires both increased junction flexibility and an exposed 5' phosphate. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunderdale H.J., Benson F.E., Parsons C.A., Sharpies G.J., Lloyd R.G., West S.C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- 66.Shida T., Iwasaki H., Saito A., Kyogoku Y., Shinagawa H. Analysis of substrate specificity of the RuvC Holliday junction resolvase with synthetic Holliday junctions. J. Biol. Chem. 1996;271:26105–26109. doi: 10.1074/jbc.271.42.26105. [DOI] [PubMed] [Google Scholar]
- 67.Iannitti T., Morales-Medina J.C., Palmieri B. Phosphorothioate oligonucleotides: effectiveness and toxicity. Curr. Drug Targets. 2014;15:663–673. doi: 10.2174/1389450115666140321100304. [DOI] [PubMed] [Google Scholar]
- 68.Chan Y.W., West S. GEN1 promotes Holliday junction resolution by a coordinated nick and counter-nick mechanism. Nucleic Acids Res. 2015;43:10882–10892. doi: 10.1093/nar/gkv1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gloor J.W., Balakrishnan L., Bambara R.A. Flap Endonuclease 1 mechanism analysis indicates flap base binding prior to threading. J. Biol. Chem. 2010;285:34922–34931. doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng L., Meng Y., Campbell J.L., Shen B. Multiple roles of DNA2 nuclease/helicase in DNA metabolism, genome stability and human diseases. Nucleic Acids Res. 2020;48:16–35. doi: 10.1093/nar/gkz1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhagwat M., Hobbs L.J., Nossal N.G. The 5'-exonuclease activity of bacteriophage T4 RNase H is stimulated by the T4 gene 32 single-stranded DNA-binding protein, but its flap endonuclease is inhibited. J. Biol. Chem. 1997;272:28523–28530. doi: 10.1074/jbc.272.45.28523. [DOI] [PubMed] [Google Scholar]
- 72.Jin H., Roy U., Lee G., Schärer O.D., Cho Y. Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease. J. Biol. Chem. 2018;293:6482–6496. doi: 10.1074/jbc.RA118.002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joseph J.W., Kolodner R. Exonuclease VIII of Escherichia coli. mechanism of action. J. Biol. Chem. 1983;258:10418–10424. [PubMed] [Google Scholar]
- 74.Ip S.C., Rass U., Blanco M.G., Flynn H.R., Skehel J.M., West S.C. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 75.Pang J., Guo Q., Lu Z. The catalytic mechanism, metal dependence, substrate specificity, and biodiversity of ribonuclease H. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1034811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mills J.V., Barnhart H.A., DePaolo D.J., Lammers L.N. New insights into Mn2+ and Mg2+ inhibition of calcite growth. Geochim. Cosmochim. Acta. 2022;334:338–367. [Google Scholar]
- 77.Yang W., Lee J.Y., Nowotny M. Making and breaking nucleic acids: two- Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 78.Hewish D.R., Burgoyne L.A. The calcium dependent endonuclease activity of isolated nuclear preparations. Relationships between its occurrence and the occurrence of other classes of enzymes found in nuclear preparations. Biochem. Biophys. Res. Commun. 1973;52:475–481. doi: 10.1016/0006-291x(73)90736-5. [DOI] [PubMed] [Google Scholar]
- 79.Chandrashekaran S., Saravanan M., Radha D.R., Nagaraja V. Ca2+-mediated site-specific DNA cleavage and suppression of promiscuous activity of KpnI restriction endonuclease. J. Biol. Chem. 2004;279:49736–49740. doi: 10.1074/jbc.M409483200. [DOI] [PubMed] [Google Scholar]
- 80.Reddi K.K. Micrococcal nuclease. Meth. Enzymol. 1967;12-A:257–262. [Google Scholar]
- 81.Viadiu H., Aggarwal A.K. The role of metals in catalysis by the restriction endonuclease BamHI. Nat. Struct. Biol. 1998;5:910–916. doi: 10.1038/2352. [DOI] [PubMed] [Google Scholar]
- 82.Horton J.R., Cheng X. PvuII endonuclease contains two calcium ions in active sites. J. Mol. Biol. 2000;300:1049–1056. doi: 10.1006/jmbi.2000.3938. [DOI] [PubMed] [Google Scholar]
- 83.Martin A.M., Sam M.D., Reich N.O., Perona J.J. Structural and energetic origins of indirect readout in site-specific DNA cleavage by a restriction endonuclease. Nat. Struct. Biol. 1999;6:269–277. doi: 10.1038/6707. [DOI] [PubMed] [Google Scholar]
- 84.McClung D.J., Calixto A., Mosera M.N., Kumar R., Neidle E.L., Elliott K.T. Novel heterologous bacterial system reveals enhanced susceptibility to DNA damage mediated by yqgF, a nearly ubiquitous and often essential gene. Microbiology. 2016;162:1808–1821. doi: 10.1099/mic.0.000355. [DOI] [PubMed] [Google Scholar]
- 85.Kuzminov A. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sambrook J., Russell D.W. 3rdedn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 87.Jerabek-willemsen M., André T., Wanner R., Marie H., Duhr S., Baaske P., et al. MicroScale thermophoresis : interaction analysis and beyond. J. Mol. Struct. 2014;1077:101–113. [Google Scholar]
- 88.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 89.Thakur M., Agarwal A., Muniyappa K. The intrinsic ATPase activity of Mycobacterium tuberculosis UvrC is crucial for its damage-specific DNA incision function. FEBS J. 2021;288:1179–1200. doi: 10.1111/febs.15465. [DOI] [PubMed] [Google Scholar]
- 90.Maxam A.M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the course of this work are included in this article and will be made available upon request.