Abstract
The identification and epitope mapping of broadly neutralizing anti-human immunodeficiency virus type 1 (HIV-1) antibodies (Abs) is important for vaccine design, but, despite much effort, very few such Abs have been forthcoming. Only one broadly neutralizing anti-gp41 monoclonal Ab (MAb), 2F5, has been described. Here we report on two MAbs that recognize a region immediately C-terminal of the 2F5 epitope. Both MAbs were generated from HIV-1-seropositive donors, one (Z13) from an antibody phage display library, and one (4E10) as a hybridoma. Both MAbs recognize a predominantly linear and relatively conserved epitope, compete with each other for binding to synthetic peptide derived from gp41, and bind to HIV-1MN virions. By flow cytometry, these MAbs appear to bind relatively weakly to infected cells and this binding is not perturbed by pretreatment of the infected cells with soluble CD4. Despite the apparent linear nature of the epitopes of Z13 and 4E10, denaturation of recombinant envelope protein reduces the binding of these MAbs, suggesting some conformational requirements for full epitope expression. Most significantly, Z13 and 4E10 are able to neutralize selected primary isolates from diverse subtypes of HIV-1 (e.g., subtypes B, C, and E). The results suggest that a rather extensive region of gp41 close to the transmembrane domain is accessible to neutralizing Abs and could form a useful target for vaccine design.
Eliciting broadly neutralizing antibodies (Abs) to human immunodeficiency virus (HIV-1) is a major goal of vaccine research but one that has proved extraordinarily elusive (8, 11, 77). This probably reflects the low antigenicity and immunogenicity of the HIV-1 envelope spike and most especially of relatively conserved regions of the spike. It is clear that much of the protein surface of the gp120 and gp41 protein molecules in the heterotrimeric envelope spike (gp1203-gp413) is directly or indirectly occluded from Ab binding by protein-protein interaction. Thus, for example, extensive surfaces on gp41 appear to be involved in interaction with other gp41 molecules and with gp120 (62, 63). Reciprocally, a portion of the surface of gp120 is occluded by the interaction with gp41 and by trimer formation (28, 32, 75, 76). The relatively low immunogenicity of HIV-1 envelope trimers is also inferred from the low titers of neutralizing Abs, particularly cross-isolate neutralizing Abs, elicited during natural infection (31, 39, 40). This follows since a good correlation has been established between Ab neutralization and binding to envelope spikes, at least for T-cell-line-adapted viruses (50, 57, 61), suggesting that the deficit in neutralization originates from a deficit in spike binding. Low immunogenicity presumably arises, at least in part, from the weakly stimulating properties of the exposed regions of the envelope trimer. These include extensive regions of carbohydrate. A caveat here is that one cannot generally be sure of the eliciting antigen: Abs reactive with the trimer may have been elicited by other forms of envelope such as monomeric gp120 or gp160.
The most highly conserved functional sites on gp120 within the envelope spikes appear to evade Ab recognition and do so most probably via multiple mechanisms. The CD4 binding site is a narrow, recessed cavity to which antibody access on the trimer is severely limited (52). Only one monoclonal Ab (MAb), b12, against an epitope overlapping the CD4 binding site has been described that potently neutralizes a range of isolates. The coreceptor binding site on gp120 is revealed only following CD4 binding and allows interaction with Abs such as 17b and 48d (56, 68). However, these MAbs do not neutralize primary HIV-1 in the absence of soluble CD4 (sCD4), suggesting that the revealed epitopes have restricted accessibility to Ab during envelope activation and fusion. A large part of the surface of gp120 is covered by carbohydrate (particularly on the so-called silent face) and is unlikely to stimulate an Ab response. Nevertheless, a broadly neutralizing MAb (2G12) has been described (70) that recognizes an epitope, consisting at least in part of carbohydrate chains, at the junction of the silent and neutralizing faces.
gp41 appears to be almost completely occluded from Ab recognition in the HIV-1 envelope spike (60, 62, 63). Only one neutralizing MAb (2F5) binding an epitope at the C-terminal part of the extracellular domain of gp41 has been described (45). This MAb neutralizes a broad array of primary HIV-1 (17, 20, 69). Binding involves the linear sequence ELDKWA, as defined by numerous studies (7, 45, 54). However, attempts to elicit Abs having the properties of 2F5 by immunization with this peptide sequence expressed in a number of contexts have failed (15, 23, 35, 44). This has led to the hypothesis that the recognition of ELDKWA by 2F5 is critically dependent on the environment in which it is present on gp41 in the trimer or that ELDKWA is not the complete epitope. The binding of 2F5 to envelope spikes on infected cells as measured by flow cytometry is relatively weak compared to, for example, that of b12 and 2G12. This has invited speculation that the epitope for 2F5 may be best presented on a fusion intermediate form of gp41 (27), although this remains controversial.
The potent and broadly neutralizing MAbs b12, 2G12, and 2F5 are all unique Abs, whose specificity has not been replicated despite the huge number of MAbs that have been generated against the HIV envelope. This has raised concerns about the feasibility of eliciting these specificities by vaccination. Here we describe two human MAbs that recognize a region immediately adjacent and C-terminal to the 2F5 epitope on gp41. The MAbs show considerable cross-isolate neutralizing ability and emphasize the membrane-proximal external region of gp41 as a promising target for vaccine design.
MATERIALS AND METHODS
Materials.
The following materials were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (NARRRP): peptides from gp160MN including numbers 2015, 2016, 2017, 2030, 2031, 2032, and 2047 (>80% pure by high-pressure liquid chromatography [HPLC]; contributed by Anaspec Inc. [Table 1]); HIV-1 isolates including 92RW009, 92RW026, DJ259, JR-FL, SF2, DH123, HC4, 2044, 2076 cl.3, NL4-3, 89.6, MN, HxBc2, 94ZW109, 94ZW106, DJ258, ZAM20, DU151, 92BR025, 92UG035, UG270, 92UG024, CM235, 92TH001, BZ162, R1, RU570, and G3 (contributed in some cases by the World Health Organization Network for HIV-1 Isolation and Characterization); sCD4 (amino acids [aa] 1 to 370; contributed by N. Schuelke); and U87R5 cells (4035; contributed by HongKui Deng and Dan Littman). Peptides 2031c, ND, and KGND were synthesized by Multiple Peptide Systems (San Diego, Calif.) and were >90% pure by reverse phase-HPLC); their amino acid sequences are shown in Table 1. The human MAb 50–69 (26) was a gift from Susan Zolla-Pazner (New York University, New York, N.Y.), human anti-HLA (DR) MAb L-243 was obtained from Luc Teyton (The Scripps Research Institute), and murine MAb T3 was provided by Pat Earl (National Institute of Allergy and Infectious Diseases, Bethesda, Md.). Recombinant gp120JR-FL was kindly donated by Bill Olson and Paul Maddon (Progenics, Tarrytown, N.Y.); gp160 (PS)IIIB was purchased from Protein Sciences Corp. (Meriden, Conn.); gp41(INT)IIIB (99.5% pure) was purchased from Intracel (Issaquah, Wash.) and is produced in Escherichia coli; and gp41(VT)IIIB (aa 546 to 682; >95% pure) was purchased from Viral Therapeutics Inc. (Ithaca, N.Y.) and is produced in Pichia pastoris. HIV immune globulin HIVIG was provided by John Mascola (VRC, Bethesda, Md.). Highly concentrated (1,000-fold in TNE) and purified HIV-1MN particles, MN-1000×, were generously provided by Larry Arthur (clone 4–12, lot P3423; NCI-Frederick Cancer Research Facility, Frederick, Md.). HIV-1-neutralizing serum from patient FDA-2 (51) was prepared from blood drawn on 29 September 1999.
TABLE 1.
Amino acid sequences of peptides used in this study
| Peptidea | Amino acid sequence |
|---|---|
| MN peptides | |
| 2015 | TKAKRRVVQREKRAAIGALF |
| 2016 | LGFLGAAGST |
| 2017 | GAAGSTMGAAS |
| 2030 | QTQQEKNEQELLELDKWASL |
| 2031 | LLELDKWASLWNWFDITNWLW |
| 2031c | LLELDKWASLWNWFDITNWLWGC |
| 2032 | DITNWLWYIKI |
| 2047 | AVAEGTDRVIEVLQRAGRAI |
| Other | |
| ND | Ac-NWFDITK-NH2b |
| KGND | KGWNWFDITNWGK-NH2 |
| SIVmac239 (671-690) | KLNSWDVFGNWFDLASWIKY |
See Materials and Methods for sources of peptides.
Ac-, N terminus is acetylated; -NH2, C terminus is amidated.
Isolation of Z13 Fab from a phage display library.
The Fab libraries from the patient, FDA-2, were prepared essentially as described previously (2, 3, 9), except that an immunoglobulin G3 (IgG3)-specific primer (1963: 5′-TGTGTGACTAGTGTCACCAAGTGGGGTTTT-3′) was used in addition to the IgG1-specific primer (3). The RNA was prepared from 5 ml of bone marrow (drawn in August 1996) from an HIV-1-seropositive individual with exceptionally broad HIV-1 primary-isolate neutralizing Ab titers (patient FDA-2) (24, 40, 51, 72) and used to prepare libraries of >107 clones using the phagemid pComb3H as a vector (2, 3, 9). Two separate Fab phage libraries were prepared, one with kappa light chains and one with lambda light chains. In addition, we prepared single-chain Fv libraries essentially as described previously (3) except that a redesigned primer set based on that of Sblattero and Bradbury (64) was used (D. R. Burton and P. W. H. I. Parren, unpublished data). Libraries of either kappa or lambda light-chain variable-region fragments were combined separately with the variable-region fragments from heavy chains primed by a generic IgG cDNA primer. The libraries were subjected to six rounds of affinity selection against the MN peptide, 2031 (LLELDKWASLWNWFDITNWLW). The enriched Fab phage pools were engineered to secrete soluble Fab, and positive clones were identified by enzyme-linked immunosorbent assay (ELISA) by screening bacterial supernatants against the 2031 peptide.
For affinity selections against virion particles of HIV-1 (MN-1000×), microwells were coated overnight at 4°C with 50 μl of phosphate-buffered saline (PBS) containing 2 μg of the anti-HLA MAb L-243 per ml, washed twice with PBS containing 0.05% Tween 20, and blocked with 4% nonfat dry milk (NFDM) for 1 h at 37°C (49). The wells were washed twice with PBS alone, MN-1000× (diluted 1:4 in 50 μl of PBS containing 1% bovine serum albumin [BSA]) was added, and the plate was incubated for 2 h at 37°C. The wells were washed three times with PBS, and the phage library (∼1011 phage diluted in 50 μl of PBS containing 1% BSA) was added. After the plate was incubated for a further 2 h at 37°C, the wells were washed (twice, twice, 5 times, 5 times, 10 times, and 10 times for rounds 1 through 6, respectively) and phage were eluted as described previously (2, 49). A total of six rounds of panning on MN virus were performed before clones were screened as above.
Preparation of purified Fab fragments.
A single colony of each Fab-expressing clone was used to inoculate 8 liters of superbroth [for 1 liter, 10 g of 3-(N-morpholino)propanesulfonic acid (MOPS), 30 g of tryptone, and 20 g of yeast extract (pH 7.0)] containing 20 mM MgCl2, tetracycline (10 μg/ml), and carbenicillin (50 μg/ml). The flasks were incubated at 37°C with shaking at 250 rpm until an optical density at 600 nm of 0.8 was reached, after which time 1 mM isopropyl-β-d-thiogalactopyranoside was added to each culture. These cultures were then incubated for a further 16 h at 30°C. The cultures were pelleted by centrifugation at 8,000 rpm for 15 min in a Sorvall SLA-3000 rotor at 4°C, and the pellets were resuspended in 30 ml of PBS containing phenylmethylsulfonyl fluoride (0.3 μM). The bacteria were lysed by sonication (Misonix sonicator, 1/2-in. horn) four times at 50% cycle time (power 7 for 20 s). The bacterial debris was centrifuged at 17,000 rpm in a Sorvall SS-34 rotor for 35 min at 4°C and stopped without braking. The supernatant was filtered (pore size, 0.8 μm and then 0.2 μm) and then loaded onto a 5-ml rabbit anti-human Fab column, preequilibrated with PBS. The Fab was eluted with 0.2 M glycine-HCl (pH 2.2), neutralized with 2 M Tris base (pH 9.0), concentrated with a Centriprep YM-30 concentrator (Millipore Corp., Bedford, Mass.), and dialyzed against PBS (pH 7.0).
ELISAs.
Microplate wells (no. 3690; Corning, Corning, N.Y.) were coated overnight at 4°C with 50 μl of PBS containing synthetic peptide (4 μg/ml), gp41(VT)IIIB (2 μg/ml), SOS-gp140JRFL (6 μg/ml) (6), or gp160(PS)IIIB (4 μg/ml); all other antigens were coated at 4 μg/ml. In cases requiring denaturation, proteins were diluted in 1% sodium dodecyl sulfate–50 mM dithiothreitol (Sigma, St. Louis, Mo.), boiled for 5 min, and then diluted 1:10 in PBS before being used to coat microplate wells (5). The wells were washed twice with PBS containing 0.05% Tween 20 and blocked with 3% BSA for 1 h at 37°C. After a single wash, MAbs were added to the wells in PBS containing 1% BSA and 0.02% Tween and allowed to incubate at 37°C for 2 h. The wells were washed four or five times, goat anti-human IgG F(ab′)2 alkaline phosphatase (no. 31312; Pierce, Rockford, Ill.) diluted 1:500 in PBS containing 1% BSA was added, and the plate was incubated for 40 min at room temperature. The wells were washed five times and developed by adding 50 μl of alkaline phosphatase substrate, prepared by adding one tablet of disodium p-nitrophenyl phosphate (no. 104–105; Sigma) to 5 ml of alkaline phosphatase staining buffer (pH 9.8), as specified by the manufacturer. After 30 min, the optical density at 405 nm was read on a microplate reader (Molecular Devices).
For competition ELISA, 45-μl volumes of the competing MAbs were added to the blocked and washed wells, followed by 5 μl of biotinylated (BIO) MAb previously determined to result in an ELISA signal that was 50 to 75% of maximum without competitor. The wells containing BIO MAbs were blocked and washed as above, probed with a streptavidin-alkaline phosphate conjugate (Pierce) diluted 1:3,000 in 1% BSA containing 0.025% Tween 20, and detected as above.
For virus ELISA, microwells were coated overnight at 4°C with 50 μl of PBS containing 2 μg of the anti-major histocompatibility complex MAb L-243 per ml, washed twice with PBS containing 0.05% Tween 20, and blocked with 4% NFDM for 1 h at 37°C. The wells were washed twice with PBS alone, MN-1000× (diluted at various concentrations in 50 μl PBS containing 1% BSA) was added, and the plate was incubated for 2 h at 37°C. The wells were then washed three times with PBS, the MAbs were added, and the plate was incubated for 2 h at 37°C. The wells were washed four more times with PBS and probed with goat anti-human lgG F(ab′)2 alkaline phosphatase (Pierce), with incubation for 1 h at 37°C. After being washed four times, the wells were developed with substrate as above.
Screening of gp160 phage-displayed fragment library.
A fragment library of the gene encoding gp160 (HxBc2) was prepared, essentially as described previously (74). Briefly, a KpnI-BamHI fragment of the gp160 gene encoding most of gp120 (except the first 12 amino acids) and the gp41 ecto- and transmembrane domains was cut from the vector pSVIIIenv HXBc2 (provided by Joe Sodroski [67]) and cloned into the pUC19 vector (80). The plasmid clone containing both pUC19 vector sequences and gp160 DNA was randomly digested with DNase I. The resulting fragments were blunt-end ligated in three separate reactions to a flanking sequence containing a SfiI endonuclease restriction site and encoding either a GG, GC, or PP linker sequence, respectively. The three ligation products were separated using Tris-borate-EDTA–polyacrylamide gel electrophoresis (10% polyacrylamide) (Bio-Rad, Hercules, Calif.), and fragments in the range of 50 to 250 bp were electroeluted from gel slices. The fragments were cut with SfiI and cloned into the vector pFRAG, which is a modified version of pComb3 (74). The SfiI cloning site in pFRAG is located immediately upstream of gene 3, enabling expression of a library of gp160 fragments linked to pIII on the surface of bacteriophage M13. The three libraries each contained 6 × 107 to 7 × 107 independent clones. For the initial round of affinity selection, approximately 1011 clones from each linker library were separately panned and then pooled for subsequent rounds. The panning was performed as described for the Fab-phage libraries using stringent washing throughout (10 washes over the course of an hour with PBS containing 0.1% Tween 20) and continued for a total of four rounds.
Flow cytometry.
All experiments with live HIV-1 were performed in a biosafety level 3 facility. Experiments were performed essentially as described by Sattentau et al. (63). Briefly, H9 cells were infected with MN or HxBc2 virus and grown for ∼10 days at 37°C in RPMI 1640 medium (Gibco-BRL, Rockville, Md.) containing 10% heat-inactivated fetal bovine serum. The infected cells were centrifuged at 800 × g for 5 min and resuspended in fluorescence-activated cell sorter (FACS) buffer (PBS containing 1% fetal bovine serum and 0.05% sodium azide) at 7 × 106 cells per ml. The cell suspension was divided into two equal portions. To one portion, sCD4 (NARRRP) was added to a final concentration of 25 μg/ml; the other portion was left untreated. The cells were incubated for 1 h at 37°C while slowly rocking. They were washed once in FACS buffer, distributed into microwells in 30-μl aliquots, treated with Abs at 100 μg/ml (except MAb 50–69, which was used at 20 μg/ml), and incubated for 2 h as above. The cells were washed twice, fixed with 1% formaldehyde in PBS, and washed another two times. They were incubated for 1 h at 4°C in the presence of the F(ab′)2 fragment of goat anti-human lgG F(ab′)2-fluorescein isothiocyanate conjugate (Jackson ImmunoResearch Laboratories Inc., West Grove, Pa.) diluted 1:100 in FACS buffer. They were washed twice and analyzed with a FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.) with CellQuest software (Version 3.1f). A total of 10,000 events were measured per sample. Values are reported in median fluorescent-intensity units.
Neutralization assays. (i) Protocol A. 4E10 screen against a panel of isolates of HIV-1.
Human PBMCs were stimulated in three separate pools treated either with phytohemagglutinin (PHA) at 5 μg/ml, PHA at 0.5 μg/ml or anti-CD3. After 3-days stimulation, the pools were mixed immediately prior to their use in the assay. Aliquots (100 μl) of peripheral blood mononuclear cells (PBMCs) (2 × 106 cells per ml) were mixed in flat-bottom plates with MAbs, which had been preincubated with 10 50% tissue culture infective doses of virus for 1 h at 37°C, and incubated for 7 days. The presence of p24 was then determined by ELISA using the Ab EH12E-1, which was diluted 1:800 in Tris-buffered saline containing 20% sheep serum and 0.05% Tween 20, and color development was achieved using TROPIX substrate (CSPD). All experiments were performed at least twice using duplicate samples.
(ii) Protocol B. Side-by-side comparison of Z13 with the other anti-HIV-1 MAbs.
Neutralization of primary isolates was assessed using PHA-activated PBMC as indicator cells and determination of p24 antigen production as the end point. PBMC from three CCR5 wild-type donors were isolated, pooled, and stimulated with PHA (5 μg/ml) for 48 h followed by PHA plus interleukin-2 (40 U/ml) for 72 h in RPMI 1640 containing 10% heat-inactivated FBS, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM l-glutamine. The Abs were diluted, and 50 μl per well was pipetted into round-bottom microtiter plates, after which an equal volume containing 100 50% tissue culture infective doses of primary-isolate stock was added. The Ab-virus mixture was incubated for 1 h at 37°C. Next, 100 μl of PHA-activated PBMC (5 × 105/ml) was added to each well. The calculated neutralization titers refer to the Ab concentration present during this incubation step. After an overnight incubation, the cells were washed twice with tissue culture medium. On day 4,100 μl of the medium was replaced with fresh tissue culture medium. On day 7, the cultures were collected and treated with 1% (vol/vol) Empigen (Calbiochem). Triplicate samples were then tested for p24 content using an in-house ELISA, as originally described by Moore et al. (41). In brief, sheep anti-p24 Ab D7320 (Aalto Bioreagents) was coated overnight onto 96-well polystyrene EIA plates (Costar) in 100 mM NaHCO3 (pH 8.5). The plates were washed in PBS, and p24 was captured from serial dilutions of the HIV-1-containing samples in PBS–0.1% Empigen. After a 3-h incubation, unbound p24 was washed away and bound p24 was detected with alkaline phosphatase-labeled Ab BC1071 (International Enzymes) diluted 1:3,000 in PBS containing 20% sheep serum and 2% NFDM. After a 1-h incubation, the plates were washed and developed with an AMPAK kit (Dako Diagnostics) as recommended by the manufacturer. p24 production antigen in the Ab-containing cultures was compared to p24 production in cultures without Ab run in the same assay, and the Ab concentrations resulting in a 50 and 90% reduction in p24 content were determined.
Nucleotide sequence accession numbers.
The DNA sequences of Z13 have been deposited in GenBank (VH, AY035845; and VL, AY035846).
RESULTS
Identification of Fab Z13.
In previous studies it was found that serum drawn from the donor FDA-2 had exceptionally broad neutralizing activity against HIV-1 (24, 40, 51, 72). For this reason, Fab phage libraries were prepared from the bone marrow of this patient by using primers amplifying the heavy chains of the IgG1 and IgG3 isotypes and lambda and kappa light chains. Two single-chain (sc) Fv phage libraries (FDA-2) were also prepared using a generic first-strand cDNA IgG primer for the heavy chains; one library contained lambda light chain fragments, and the other contained kappa light chain fragments. The FDA-2 libraries, as well as four other Fab phage libraries prepared from bone marrow of HIV-1-infected individuals (RW, L, P, and M) (47, 51), were selected against the MN peptide 2031. The 2031 peptide (Table 1) contains the amino acid motif ELDKWA that corresponds to the core epitope of the potent and broadly HIV-1-neutralizing MAb 2F5 (54). Following six rounds of affinity selection, 2031-specific clones were identified from both FDA-2 Fab -libraries and the lambda scFv-library; none of the other libraries yielded 2031-specific clones. As shown in Fig. 1, a number of clones, isolated from independent libraries, have a similar heavy chain with a number of point mutations found throughout the variable region. Figure 1 also reveals that a single clone, designated Z13 (κ), was enriched from the kappa library whereas three different clones having unique light chains were selected from the lambda libraries (two from the Fab library and one from the scFv library). This indicates that there is considerable heavy-chain promiscuity for this MAb, which is not uncommon for Fabs isolated from phage display libraries (4).
FIG. 1.
Deduced amino acid sequences of the variable regions of Fabs selected against MN peptide 2031 from FDA-2 kappa and lambda phage display libraries.
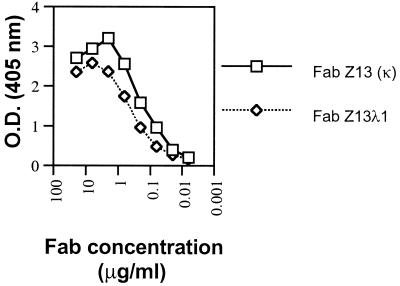
In a separate panning experiment, the FDA-2 Fab phage libraries were selected against whole HIV-1MN virus (MN-1000×) in an attempt to isolate Fabs able to bind to surface-exposed epitopes on HIV-1 particles. After six rounds of affinity selection, 2 of 30 clones selected for sequencing were identical to Z13 (κ). This indicates that the epitope to which Z13 (κ) binds is available in some form on captured MN virions. This was later verified using soluble Fab Z13 (κ) in a whole-virus ELISA (see below). A side-by-side titration of purified Fab Z13 (κ) and one Fab from the lambda library (Z13λ1) revealed that the Z13 (κ) clone bound the 2031 peptide with slightly greater affinity (half-maximal binding at ∼0.2 μg/ml; [Fig. 2]). Due to the slightly stronger binding of Z13 (κ) for 2031 and the fact that Z13 (κ) phage was also selected in the HIV-1MN panning, Z13 (κ) was chosen for further study; Z13 (κ) is referred to as Z13 below. Sequence analysis in the CH1 region of Z13 indicates that it is derived from the IgG1 isotype. The HCDR3 loop of Z13 contains mostly hydrophobic residues and is 17 aa in length. The VH gene for Z13 is from the VH4 family, the J segment belongs to the JH6 family, and no alignment for the D segment was found using available databases (IMGT, http://imgt.cnusc.fr:8104/dnaplot/; V-base, http://www.mrc-cpe.cam.ac.uk/imt-doc/public/INTRO.html). The kappa light chain of Z13 derives from the VK3 family with a J segment that most closely aligns to the JK1 family.
FIG. 2.
ELISA binding curve of Fab Z13 (κ) and Fab Z13λ1 (λ) against immobilized 2031 peptide. O.D., optical density.
Specificity of Fab Z13 and comparison with IgG 4E10.
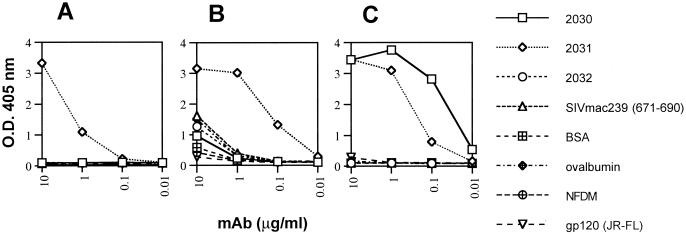
We noticed in early experiments that another anti-gp41 MAb 4E10, generated by combined polyethylene glycol-electrofusion of PBMC from a seropositive donor and heteromyeloma cells (7), reacted strongly with peptide 2031, even though it had been tentatively mapped to another epitope on gp41 (7). To explore the antigenicity of this peptide in more detail, a series of overlapping peptides and other protein antigens were tested for binding to Fab Z13, IgG 4E10, and IgG 2F5 by ELISA. As shown in Fig. 3, all three MAbs bind strongly to MN peptide 2031c whereas none of the MAbs bind significantly to an SIV peptide analogous to 2031 (SIVmac239, aa 671 to 690). In contrast to the other two MAbs, 2F5 also binds tightly to the MN peptide 2030 (Fig. 3C), consistent with previous studies in which the core of the epitope for this MAb was identified as the linear sequence, ELDKWA (7, 45, 54). IgG 4E10 exhibits some nonspecific binding to a range of antigens at higher concentrations (e.g., 10 μg/ml) (Fig. 3B), which may be responsible for difficulties in prior attempts to map the 4E10 epitope (7). We observed a similar weak binding of 4E10, for example to MN peptide 2047 containing the motif AEGTDRV, to which 4E10 had been previously mapped (reference 7 and data not shown). Significantly, neither Z13 nor 4E10 bound to peptide 2032 (Fig. 3A and B), which, together with their lack of reactivity with 2030, suggests that the core of the epitopes of these mAbs is C-terminal to the 2F5 epitope but upstream of the C terminus of the 2031 peptide. No significant binding of any of the three MAbs was observed against the gp41MN peptides 2015, 2016, or 2017 (peptide sequences shown in Table 1 and data not shown), and no binding of an irrelevant MAb (b12) was seen against peptide 2031 (data not shown).
FIG. 3.
Specificities of Fab Z13, IgG 4E10, and IgG 2F5 for a range of immobilized proteins and peptides by ELISA. Fab Z13 (A), IgG 4E10 (B), and IgG 2F5 (C) were titrated against MN peptides 2030, 2031c, and 2032; the SIV mac239 peptide (aa 671 to 690); BSA; ovalbumin; NFDM; or recombinant gp120JR-FL. O.D., optical density.
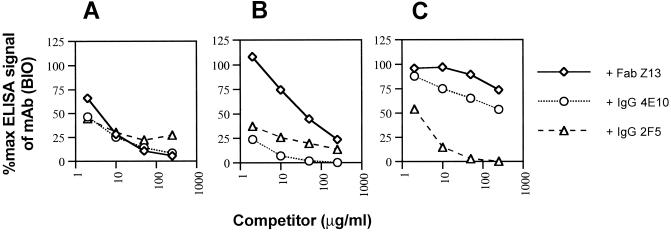
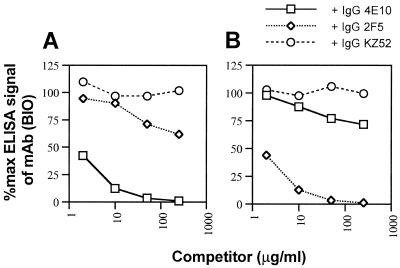
To further explore the overlapping nature of the Z13, 4E10, and 2F5 epitopes, these MAbs were biotinylated and used in competition ELISAs against immobilized 2031 peptide. The results clearly demonstrate that these MAbs compete with each other, albeit to differing degrees, for binding to peptide 2031 (Fig. 4). It is noteworthy that while 4E10 inhibited >90% of the signal generated by Fab Z13 (BIO), 2F5 was able to inhibit only a maximum of 70% of the signal produced by Fab Z13 (BIO) on 2031. Although Z13 did not bind to IIIB gp41 by ELISA (see below), partial competition between 2F5 and 4E10 for immobilized, recombinant gp41 (VT)IIIB was verified (Figs. 5A and B) whereas an irrelevant MAb (KZ52) did not inhibit binding even at 250 μg/ml. The degree to which 2F5 and 4E10 competed for each other against gp41, however, was less than the level of competition observed for these MAbs against immobilized 2031 (Fig. 4B and 4C). This result suggests that the 4E10 epitope is presented somewhat differently on the peptide and gp41. Taken together, the results above suggest that Z13 and 4E10 have closely overlapping epitopes whereas the epitope for 2F5 only partially overlaps those of Z13 and 4E10.
FIG. 4.
Competition ELISA using Fab Z13, IgG 4E10, and IgG 2F5 against their BIO counterparts for recognition of immobilized MN 2031c peptide. Fab Z13 (BIO) (A), IgG 4E10 (BIO) (B), and IgG 2F5 (BIO) (C) were coincubated with various concentrations of competing MAb and added to wells containing immobilized 2031c. Each data point represents the mean of duplicate samples.
FIG. 5.
Competition ELISA of IgG 4E10 and IgG 2F5 against their BIO counterparts for recognition of immobilized gp41(VT)IIIB. IgG 4E10 (BIO) (A) and IgG 2F5 (BIO) (B) were coincubated with various concentrations of competing MAb and added to wells containing immobilized gp41(VT)IIIB. No competition was observed for either of the BIO MAbs using an irrelevant MAb, IgG KZ52. Each data point represents the mean of duplicate samples.
As mentioned above, in contrast to IgGs 4E10 and 2F5, Fab Z13 did not recognize gp41 of the IIIB strain. Table 2 summarizes the reactivity of the three MAbs with different HIV-1 envelope proteins and peptides. The lack of binding of Z13 to gp41IIIB is probably due to the Asn 674 residue: Asn 674 is an Asp residue in the MN, JR-FL, and 89.6 envelope proteins, which are all positively recognized by Z13. We also found that Fab Z13 binds specifically to gp41JR-FL by Western blot analysis (data not shown). Also summarized in Table 2 are the results of an ELISA in which 4E10, 2F5, and Z13 were used to probe two shorter peptides, ND and KGND. Indeed, Z13 reacted very well with the KGND peptide (roughly equal to the reactivity with 2031c) and also with the ND peptide, although the binding was much weaker to the latter, shorter peptide derived from the sequence of the isolate JR-FL. MAb 4E10 also recognized peptides KGND and ND, but the reactivity decreased in strength from 2031c to KGND and then again to ND. In combination with the results of the overlapping-peptide analysis (see above), the weaker binding of both Z13 and 4E10 to the shorter peptides strongly suggests that although the core epitopes of these MAbs are contained within the central region of peptide 2031, additional binding energy is provided either directly by flanking amino acid residues in 2031 or indirectly by the same residues providing a particular favored conformation of the central core epitope.
TABLE 2.
Mapping the epitopes of anti-gp41 MAbs using synthetic peptides and recombinant proteins
| Antigen | Amino acid sequence | Reactivity by ELISA
|
||
|---|---|---|---|---|
| Z13 | 4E10 | 2F5 | ||
| Peptides | ||||
| 2030 (MN) | QTQQEKNEQELLELDKWASL | − | − | + |
| 2031 (MN) | LLELDKWASLWNWFDITNWLW | + | + | + |
| 2032 (MN) | DITNWLWYIKI | − | − | − |
| KGND | KGWNWFDITNWGK-NH2 | + | + | − |
| ND (JR-FL) | Ac-NWFDITK-NH2 | ±b | ± | − |
| SIVmac239 (671–690) | KLNSWDVFGNWFDLASWIKY | − | − | − |
| Recombinant proteins | ||||
| gp41(INT)IIIB | ...LLELDKWASLWNWFNITNWLV... | − | + | + |
| gp41(VT)IIIB | ...LLELDKWASLWNWFNITNWLW... | − | + | + |
| gp160(PS)IIIB | ...LLELDKWASLWNWFNITNWLW... | − | + | + |
| gp140JR-FL, SOS-gp140JR-FL | ...LLELDKWASLWNWFDITNWLW...a | + | + | + |
| gp14089.6 | ...LLELDKWASLWNWFDITNWLW... | + | + | + |
Sequence sometimes appears with a K at the underlined position; the K variant was not tested.
±, weak reactivity by ELISA.
Screening of an HxBc2 gp160 fragment expression library to define MAb epitopes.
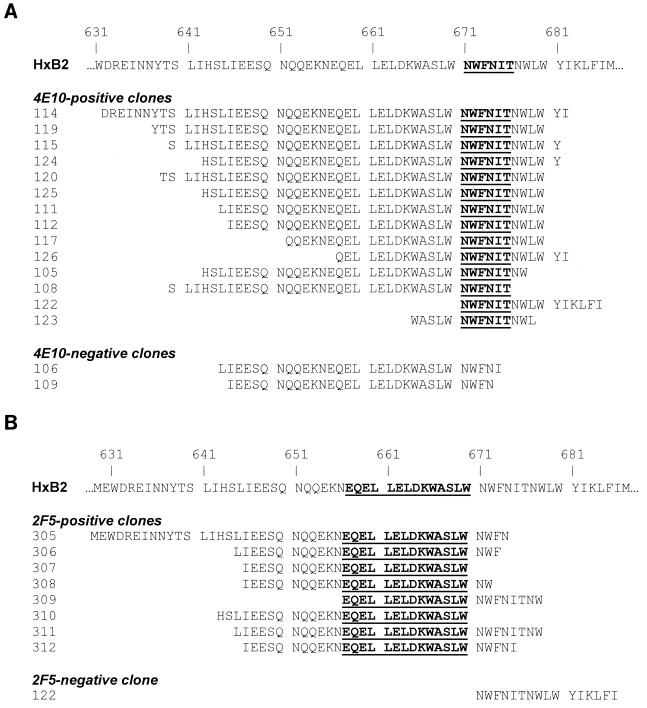
We decided to further characterize the epitopes of the anti-gp41 MAbs by screening a gene fragment expression library. The gene fragment library contains random fragments of the HxBc2 (IIIB) env gene fused to gene 3 of the filamentous phage, such that the corresponding peptides are displayed on the surface of each phage clone in the library (library size, ∼107). The approach was possible for 4E10 and 2F5, which both recognize IIIB gp41, but not for Z13, which does not recognize this strain of HIV-1. In addition, we reasoned that since the core of the 2F5 epitope has been well defined, this would provide a measure of control on the approach. After four rounds of affinity selection with immobilized 4E10 or 2F5, random phage clones were tested individually by ELISA for reactivity with each of the MAbs, and the DNA sequences of positively binding clones were determined. The results of the selection are shown in Fig. 6. In Fig. 6A, a minimal epitope of 6 aa (NWFNIT) for 4E10 was identified by overlapping peptides displayed by positive clones. Also shown in Fig. 6A are two clones (selected with 2F5) that were not recognized by 4E10 and whose peptides do not have all six of the residues within the minimal epitope identified above. The results of this analysis are in good agreement with the ELISA results in Fig. 3B and Table 2, which place the epitope of 4E10 proximal to the C terminus of MN peptide 2031. In Fig. 6B, a minimal epitope of 14 residues (EQELLELDKWASLW) for 2F5 was identified from the screening. This motif is longer than the minimal epitope identified from previous studies in which it was determined that the residues ELDKWA were sufficient for 2F5 binding (45) and that LDKW was a highly conserved core of the epitope (53). The presence of additional flanking residues in the overlapping peptides selected using 2F5 (under the highly stringent conditions used in this panning experiment) may suggest that these residues are directly or indirectly involved in binding to 2F5.
FIG. 6.
Amino acid sequences of peptides affinity selected from an HxB2 gp160 fragment library using IgG 4E10 or IgG 2F5. The HxBc2 gp160 protein fragment phage display library was subjected to four rounds of panning using either IgG 4E10 (A) or IgG 2F5 (B) as selecting agents. Random clones were selected from the enriched pools of phage and tested for reactivity against IgG 4E10 and/or IgG 2F5 by ELISA, and the positive clones were sequenced in the insert region. The residues that are in bold text and underlined represent the minimal motif that is shared by overlapping peptides.
Anti-gp41 MAb binding to HIV-1MN virions by ELISA.
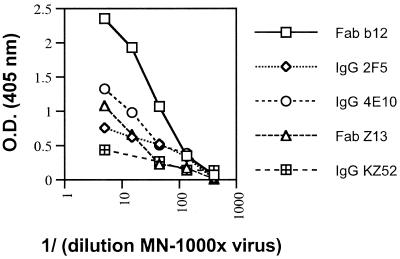
The three MAbs Z13, 4E10, and 2F5 were next compared for their ability to bind to HIV-1MN virions. An ELISA was performed in which concentrated MN virus was first captured with an anti-major histocompatibility complex (class II) MAb and then probed with either Fab Z13, IgG 4E10, IgG 2F5, Fab b12 (2), or a negative control, IgG KZ52 (36). Figure 7 shows the results of this assay. Clearly, the best binding to captured virus was seen with Fab b12, although the anti-gp41 MAbs were also positive. This result is consistent with previous studies that show that 2F5 binds only weakly, compared to anti-CD4bs Abs, to envelope on virions and infected cells (63). This experiment shows that binding of Fab Z13 and IgG 4E10 to HIV-1MN is significant and qualitatively similar to that of IgG 2F5.
FIG. 7.
Binding by ELISA of Fab b12, IgG 2F5, IgG 4E10, Fab Z13, and IgG KZ52 to various concentrations of HIV-1MN (MN-1000×) viral particles captured by human anti-HLA MAb L-243. All MAbs were used at 10 μg/ml. O.D., optical density.
Anti-gp41 MAb binding to infected cells by flow cytometry.
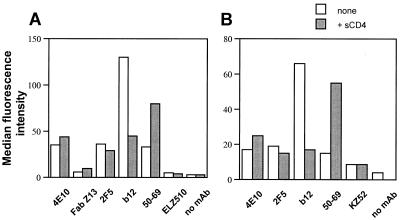
Next, we wanted to use flow cytometry to compare Fab Z13, IgG 4E10, and IgG 2F5 for their ability to bind to HIV-1-infected cells. Figure 8 summarizes the results of an analysis of both HIV-1MN-infected (Fig. 8A) and HIV-1HxBc2-infected (Fig. 8B) H9 cells probed with several anti-HIV-1 MAbs following a 1-h preincubation in the presence or absence of sCD4 (25 μg/ml) at 37°C. In this analysis, 4E10 generated a similar signal to that of 2F5 when used to probe both MN- and HxBc2-infected cells in the absence of preincubation with sCD4. Fab Z13 bound weakly to MN-infected cells (IIIB-infected cells were not tested with Fab Z13), generating a signal approximately twice that produced by an irrelevant control Fab, ELZ510. The pretreatment with sCD4 caused only a small perturbation in the binding of Z13 and 4E10, something which is also characteristic of 2F5 (63) and is in dramatic contrast to the large increase in binding that was seen with the anti-gp41 cluster I MAb, 50–69.
FIG. 8.
MAb binding to H9 cells infected with HIV-1MN and HIV-1HxB2 as measured by flow cytometry. (A) H9 cells infected with HIV-1MN were probed with IgG 4E10 (100 μg/ml), Fab Z13 (200 μg/ml), IgG 2F5 (100 μg/ml), IgG b12 (50 μg/ml), IgG 50–69 (10 μg/ml), and Fab ELZ510 (200 μg/ml). (B) H9 cells infected with HIV-1HxB2 were probed with IgG 4E10, IgG 2F5, IgG b12, and IgG KZ52 (100 μg/ml each) or IgG 50–69 (20 μg/ml). Infected cells were incubated in the presence or absence of 25 μg of sCD4 per ml for 1 h at 37°C prior to the addition of MAbs. Uninfected H9 cells were also probed by each MAb at the same concentration and always yielded background signals that were lower than that observed for the negative control MAbs on infected cells.
Effect of denaturation of gp41 and SOS-gp140 on the binding of Fab Z13, IgG 4E10, and IgG 2F5 by ELISA.
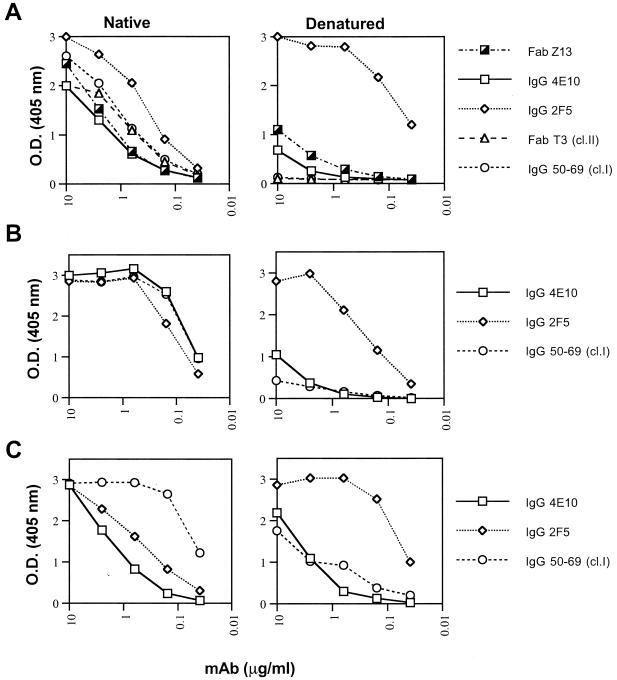
The results of the peptide-mapping studies of Z13 and 4E10, as well as the fragment library screening of 4E10, suggest that these MAbs bind to predominantly linear epitopes on gp41, just as has been proposed for 2F5 (45). In contrast to 2F5, cluster I, II, and III MAbs against gp41 are sensitive to denaturation of this protein by ELISA (5). We compared the effects of denaturation of SOS-gp140JRFL (6), gp41 (VT)IIIB, or gp160(PS)IIIB on the binding of MAbs Z13 and 4E10 with the effects on the binding of other anti-gp41 MAbs. As shown in Fig. 9A, the signal of 2F5 is enhanced by denaturation of SOS-gp140JRFL. The cluster I MAb 50–69, as well as the cluster II Fab T3 (5), lost all ability to bind SOS-gp140JRFL when this protein was denatured, whereas Z13 and 4E10 showed intermediate behavior. Note that although cluster I and II MAbs bind to SOS-gp140 by ELISA, these epitopes were poorly expressed on SOS-gp140 when assayed by radioimmunoprecipitation (6). The results with gp41(VT)IIIB and gp160(PS)IIIB were slightly different but the trend was the same: 2F5 binding was slightly reduced (or enhanced) whereas 50–69 binding was greatly diminished on denaturation (Fig. 9B and C); the binding of 4E10 was intermediate in that it was reduced relative to that observed with 2F5 but the reduction was less than observed with 50–69. These experiments demonstrate that Z13 and 4E10 are intermediate between 2F5 and cluster I (50–69) and II (Fab T3) MAbs with respect to sensitivity to the denaturation of envelope protein.
FIG. 9.
Binding of anti-gp41 MAbs to SOS-gp140JRFL(A), gp41(VT)IIIB (B), and gp160(PS)IIIB (C) coated directly on polystyrene plates either untreated (Native) or following treatment with SDS, dithiothreitol, and boiling (Denatured [for more details, see Materials and Methods]), as measured by ELISA. IgG 50–69 and Fab T3 are cluster I (cl.I) and cluster II (cl.II) MAbs, respectively.
Analysis of amino acid diversity within the anti-gp41 MAb binding epitopes.
The region of gp41 to which Z13 and 4E10 bind is relatively conserved among diverse isolates of HIV-1. The amino acid sequence NWFDIT lies within the core of the Z13 and 4E10 epitopes, as defined by our peptide-mapping studies, and is the MN homolog of the motif identified from the 4E10 screening of the HxBc2 fragment library. Table 3 shows the observed frequency of each residue for each position in the NWFDIT motif among 250 sequences in the Los Alamos database (http://hiv-web.lanl.gov/) and compares it with the ELDKWA motif to which the broadly neutralizing MAb 2F5 had previously been mapped (45, 53). Both are 6-mer motifs, and both are entirely composed of residues that are most prevalent at each position. Furthermore, the probability of an exact match to each motif—equal to the product of the observed frequencies of each residue for each position—is the same for each motif: 76 expected matches out of 250 sequences. The number of clones in the Los Alamos database having an exact match to both motifs is 38. The number of observed matches for the separate motifs, however, is higher for ELDKWA (89 of 250) than for NWFDIT (68 of 250). Nevertheless, these numbers are both high. By comparison, there are 55 of 250 exact matches to the V3-loop motif GPGRAF, but only 15% of these are from non-B subtypes, whereas exact sequence matches to ELDKWA and NWFDIT are found in non-B subtypes 30 and 45% of the time, respectively. These observations suggest that the sequence NWFDIT may be an attractive target for vaccine design.
TABLE 3.
Observed frequencies of amino acid residues at each position in the gp41 6-mer motifs NWFDIT and ELDKWA out of 250 sequences in the Los Alamos database
| Motif | Amino acid | Frequency of amino acid residue in motif:
|
|||||
|---|---|---|---|---|---|---|---|
| N | W | F | D | I | T | ||
| NWFDIT (gp160, aa 671–676) | |||||||
| A | 0 | 0 | 0 | 1 | 0 | 1 | |
| D | 2 | 0 | 0 | 153 | 0 | 0 | |
| E | 0 | 0 | 0 | 3 | 0 | 0 | |
| F | 0 | 0 | 241 | 0 | 0 | 0 | |
| G | 0 | 0 | 0 | 10 | 0 | 0 | |
| I | 0 | 0 | 0 | 0 | 249 | 0 | |
| L | 0 | 0 | 6 | 0 | 0 | 0 | |
| M | 0 | 0 | 0 | 0 | 1 | 0 | |
| N | 187 | 0 | 0 | 27 | 0 | 0 | |
| P | 0 | 0 | 1 | 0 | 0 | 0 | |
| S | 49 | 0 | 1 | 52 | 0 | 75 | |
| T | 12 | 0 | 0 | 3 | 0 | 174 | |
| W | 0 | 249 | 0 | 0 | 0 | 0 | |
| Y | 0 | 0 | 1 | 1 | 0 | 0 | |
| Stop | 0 | 1 | 0 | 0 | 0 | 0 | |
| ELDKWA (gp160, aa 662–667) | |||||||
| E | L | D | K | W | A | ||
| A | 107 | 0 | 0 | 0 | 0 | 198 | |
| D | 0 | 0 | 245 | 0 | 0 | 7 | |
| E | 122 | 0 | 1 | 9 | 0 | 6 | |
| F | 0 | 1 | 0 | 0 | 0 | 0 | |
| G | 2 | 0 | 0 | 0 | 0 | 3 | |
| H | 0 | 0 | 0 | 1 | 0 | 0 | |
| K | 6 | 0 | 0 | 202 | 0 | 7 | |
| L | 0 | 249 | 0 | 0 | 0 | 0 | |
| N | 0 | 0 | 4 | 6 | 0 | 4 | |
| P | 0 | 0 | 0 | 0 | 0 | 1 | |
| Q | 11 | 0 | 0 | 10 | 0 | 14 | |
| R | 0 | 0 | 0 | 1 | 0 | 0 | |
| S | 1 | 0 | 0 | 18 | 0 | 3 | |
| T | 12 | 0 | 0 | 3 | 0 | 7 | |
| V | 1 | 0 | 0 | 0 | 0 | 0 | |
| W | 0 | 0 | 0 | 0 | 248 | 0 | |
| Stop | 0 | 0 | 0 | 0 | 2 | 0 | |
Neutralization assays with the anti-gp41 MAbs.
A panel of isolates of HIV-1, including T-cell-line-adapted strains and primary isolates, were tested in two sets of assays for their susceptibility to neutralization by anti-gp41 MAbs. In the first set, 4E10 alone was tested against a range of isolates from different subtypes of HIV-1. Remarkably, as shown in Table 4, 4E10 was able to neutralize (90% inhibition) across subtypes of HIV-1 at concentrations lower than 50 μg/ml, including isolates from subtypes A (DJ259, 92RW026 and 92RW009), B (JR-FL and HxBc2), C (94ZW106, DJ258, and DU151), E (CM235 and 92TH001), F (BZ162 and R1), and G (G3 and RU570). Some isolates from subtypes B (SF2, DH123, HC4, 2044, 2076, NL4-3, 89.6, and MN) and D (UG270 and 92UG024) were not neutralized at the highest concentrations tested. Nevertheless, the degree of cross-neutralizing activity of IgG 4E10 against HIV-1 is substantial.
TABLE 4.
HIV-1 neutralization assays using IgG 4E10 against a range of isolates from different subtypes
| Isolate | Subtype | IgG 4E10 concna:
|
|
|---|---|---|---|
| 50% | 90% | ||
| 92RW009 | A | 0.9 | 4.7 |
| 92RW026 | A | 0.7 | 12 |
| DJ259 | A | 3.5 | 16 |
| JR-FL | B | 9.2, 25 | 15, >25 |
| SF2 | B | 38 | >50 |
| DH123 | B | >50 | >50 |
| HC4 | B | >50 | >50 |
| 2044 | B | >50 | >50 |
| 2076 cl.3∗ | B | 2 | >50 |
| NL4-3 | B | >50 | >50 |
| 89.6∗ | B | >25 | >25 |
| MN | B | 5 | >50 |
| HxBc2 | B | 0.15 | 10 |
| 94ZW109 | C | 2.0, >50 | 6.7, >50 |
| 94ZW106 | C | 7.2 | 42 |
| DJ258 | C | 2.0 | 6.6 |
| ZAM20 | C | 7.9 | >50 |
| DU151 | C | 3.9 | 34 |
| 92BR025∗ | C | 10 | —b |
| 92UG035∗ | D | 20 | — |
| UG270 | D | >50 | >50 |
| 92UG024∗ | D | >50 | >50 |
| CM235 | E | 5.2, 7.5 | 11.4, 25 |
| 92TH001 | E | 2.7 | 8.2 |
| BZ162 | F | 4.4 | 2.1 |
| R1 | F | 0.3 | 5.1 |
| RU570 | G | 0.35 | 7.6 |
| G3 | G | 0.2 | 1.3 |
50% and 90% refer to the concentration of MAb (micrograms per milliliter) required for 50 or 90% inhibition of maximum p24 levels. Underlined values were determined in experiments using protocol B (see Materials and Methods); all other values were determined using protocol A. Asterisks indicate that the assay was performed only once for that isolate.
—, not done.
Encouraged by the results from this first set of assays, we selected several 4E10-susceptible isolates for a second set of neutralization assays using Fab Z13 as well as IgG 4E10, IgG 2F5, and IgG b12 controls. Because monovalent Fab is usually between about 1 and 100 times less effective in neutralization assays than its bivalent IgG counterpart (47), higher concentrations of Fab Z13 were used in these assays. Table 5 summarizes the results of this second set of assays. Indeed, Fab Z13 was able to neutralize (90% inhibition) several isolates from different subtypes of HIV-1 including subtypes B (JR-FL at 125 μg/ml), E (CM235 at 250 μg/ml), and C (94ZW109 at 250 μg/ml). Fab Z13 failed to neutralize the laboratory-adapted strain HxBc2 at 40 μg/ml, as expected, since it did not bind to the sequence-related gp41IIIB (Table 3). On the other hand, HxBc2 was found to be sensitive to neutralization by 4E10 (90% neutralized at 10 μg/ml). Fab Z13 was not able to neutralize MN at 40 μg/ml, and because even 4E10 exhibited only partial neutralization of MN at 50 μg/ml (<90% inhibition), Fab Z13 was not further tested with this T-cell-line-adapted subtype B isolate. 89.6 was not neutralized by either Z13 at 100 μg/ml or 4E10 at 25 μg/ml. Significantly, we observed no neutralization of the subtype C virus 94ZW109 by either b12 or 2F5 (50 μg/ml) and no neutralization of the subtype E isolate CM235 by b12 (50 μg/ml); however, these isolates were susceptible to neutralization by 4E10 and Z13. Taken together, these results demonstrate that primary isolates from a range of subtypes of HIV-1 are susceptible to neutralization by both Z13 and 4E10 and that these MAbs display antiviral activity in some cases for which the broadly neutralizing MAbs b12 and 2F5 do not.
TABLE 5.
HIV-1 neutralization assays including IgG b12, IgG 2F5, Fab Z13, and IgG 4E10
| Isolate | Subtype | Concn of MAb (μg/ml) required for 50 or 90% inhibition of maximum p24 levels
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| IgG b12
|
IgG 2F5
|
Fab Z13
|
IgG 4E10
|
||||||
| 50% | 90% | 50% | 90% | 50% | 90% | 50% | 90% | ||
| MN | B | 0.0015 | 0.015 | 0.13 | 1 | >40 | >40 | 5 | >50 |
| HxBc2 | B | 0.025 | 0.5 | 0.75 | 5 | >40 | >40 | 0.15 | 10 |
| JR-FL | B | 0.4 | 0.8 | 2 | 13 | 95 | 130 | 25 | >25 |
| 89.6b | B | —a | 0.8 | — | — | >100 | >100 | >25 | >25 |
| 94ZW109 | C | >50 | >50 | >50 | >50 | 190 | 250 | >50 | >50 |
| CM235 | E | >50 | >50 | 19 | 25 | 170 | 250 | 7.5 | 25 |
—, experiment was not done.
The results for 89.6 are from a single experiment only.
DISCUSSION
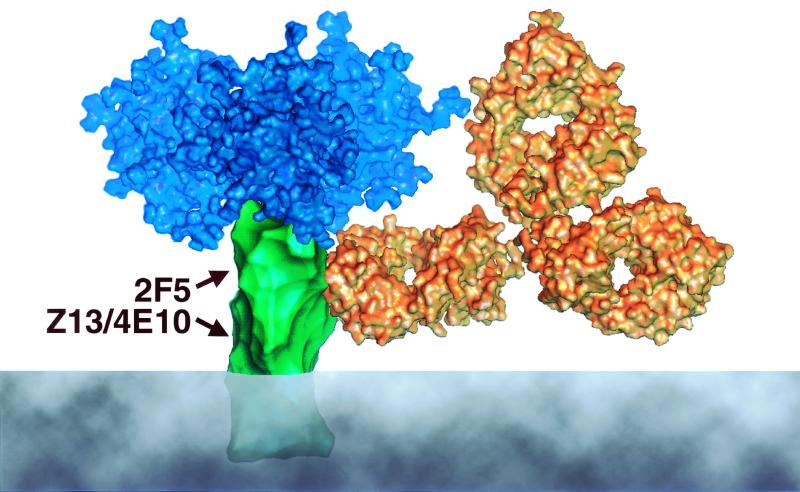
MAbs against HIV-1 have proven to be extremely powerful tools for a variety of applications including probing the structure and function of envelope glycoproteins (5, 18, 42, 78), exploring the antigenicity of novel HIV-1 vaccine candidates (8, 10, 77), and establishing the conditions for protection against HIV-1 infection in vivo (1, 16, 25, 37, 38, 48). We have identified a novel human MAb, Z13, which binds to the C-terminal extracellular portion of gp41 that now expands the current repertoire of anti-HIV-1 MAbs and, along with MAb 4E10, will allow further characterization of a region that is nearly unexplored by immunological techniques. Figure 10 shows a model of this region of gp41 and indicates the proximity of the epitopes of Z13 and 4E10 to both the 2F5 epitope and the viral (or host cell) membrane.
FIG. 10.
Cartoon model of the HIV-1 putative trimeric envelope spike. Most of the surface of gp41 is believed to be occluded by gp120 and other molecules of gp41. A region of gp41 close to the membrane defined by MAbs 2F5, Z13, and 4E10 is suggested to be exposed to antibody binding. An IgG molecule is shown to scale in proximity to the proposed binding region. The trimer is based on the structure proposed by Kwong et al. (32). The IgG molecule dimensions are taken from those of a human IgG1 molecule (46). Surfaces were calculated using the msms program (59).
Our results from screening overlapping synthetic peptides map the epitopes of Z13 and 4E10 within the peptide 2031 (aa 660 to 680), and the selection of the phage fragment library using 4E10 maps the core epitope of this MAb to between aa 671 and 676 (NWFNIT). We were unable to screen the HxBc2 fragment library with Z13 since Z13 does not bind gp41 from HxBc2. The only difference in amino acid sequence between HxBc2 and MN in this region is at position 674, which also strongly suggests that the core of the Z13 epitope closely overlaps that of 4E10. Moreover, using the synthetic peptides KGND and ND, the epitopes of Z13 and 4E10 were confirmed to center around the NWFDIT motif (Table 2), although additional flanking residues are clearly important for full reactivity of these MAbs with peptide 2031. Although sequence analysis identifies a potential N-glycosylation site at Asn 674, this residue is not glycosylated in HxBc2 gp160 produced in COS-7 cells (34).
A murine MAb, T3 (22), not to be confused with Fab T3 (Fig. 9), was previously proposed to bind to the same region where we now map the epitopes of Z13 and 4E10 (21). There are key differences between the specificity of Z13, 4E10, and MAb T3. For example, we found that MAb T3 does not bind peptide 2031 (data not shown), nor does it bind gp140JR-FL (21). In addition, MAb T3 was unable to inhibit IgG 4E10 (BIO) binding to gp41 (VT)IIIB, even at 250 μg/ml, by ELISA (data not shown). Fab Z13, however, did show partial inhibition of MAb T3 binding to gp14089.6 (24% reduction in ELISA signal at 20 μg of Z13 per ml [data not shown]). Thus, MAb T3 does appear to map to a region proximal to the Z13 epitope, but, as described above, the two epitopes are distinct and the precise binding determinant of MAb T3 remains to be defined.
Despite the apparent linear nature of the epitopes of Z13 and 4E10, denaturation of recombinant envelope protein reduced the binding of these MAbs, and not of 2F5. Whether these effects are due to direct changes in the conformation of these epitopes or to epitope -masking via the immobilization to polystyrene remains to be investigated. Moreover, further mutagenesis studies are required to determine the binding contributions of individual amino acid residues to Z13 and 4E10. Along these lines, the fact that 4E10 neutralizes HIV-1 isolates with sequences that differ from the above NWFDIT motif at positions 1, 4, and 6 (e.g., DJ258 contains the motif, SWFGIT; 92BR025 contains TWFGIT; DJ259 contain NWFSIT), as well as the fact that Z13 and 4E10 do not neutralize some isolates that contain the above motif (e.g., 89.6, NWFDIT; NL4-3, NWFNIT), suggests that conformation and other determinants on the native envelope spike influence the exposure of the 4E10 and Z13 epitopes. These issues would be far better addressed if structural data were available for this region. Indeed, solving the atomic structure of the membrane-proximal region, alone and/or complexed with Z13 or 4E10, would be of great interest, considering that, to our knowledge, the only currently available structures of gp41 are derived from studies done on truncated peptides, predominantly of the N and C helices (14, 73) of gp41, most probably in the postfusion conformation.
Z13 and 4E10 appear to bind with similar affinity to HIV-1MN virions by capture ELISA (Fig. 7). While these data clearly show that these MAbs are capable of binding virion-associated envelope protein, it should be noted that virions probably contain gp41 moieties from which gp120 has been shed. Fab Z13 yielded poorer signals than did IgG 4E10 against HIV-1MN-infected cells by flow cytometry. This correlates with the inability of Fab Z13 to neutralize HIV-1MN and could be due to a difference in avidity of the Fab fragment compared to the whole antibody or to particular contraints imposed by the native envelope spike on the access of Fab Z13 to its epitope.
The conserved, C-terminal extracellular region of gp41 that contains the epitopes of Z13 and 4E10 is important for many processes in the HIV-1 infection cycle. In one study, Helseth et al. (29) showed that a K683I mutation adjacent to the membrane reduces syncytium formation by 95% and the resulting envelope is no longer capable of supporting viral transmission. In contrast, Cao et al. (13) showed that certain mutations within a region spanning residues 669 to 775 of HxB2 gp160 enhanced syncytium formation relative to that in the wild type. In more recent studies, it has been demonstrated that the membrane-proximal region is important for env-mediated fusion and virus infectivity (43, 58): mutations to Ala of three of five conserved Trp residues in this region are sufficient to abrogate syncytium formation (58). Biophysical studies have also shown this region has distinctive properties. For example, Lawless et al. (33) constructed a mutant version of the anti-HIV-1 peptide, T20, in which the motif WNWF (aa 670 to 673) was changed to ANAA. It was found by circular dichroism spectroscopy that the mutant peptide lost the ability to interact with its N-helix partner, T21 (33). Suarez et al. (65, 66) showed that a peptide spanning residues 664 to 683 of IIIB gp160 associates with lipid membranes and causes membrane leakage. The ability of this peptide to bind and disrupt membranes is suggestive of a similar role for the homologous region of gp41 during the env-mediated membrane fusion event. Whether any of these findings have implications for the mechanism of neutralization of Z13 and 4E10 remains to be explored.
In one study, it was proposed that the region to which Z13 and 4E10 bind is an immunodominant site on gp41 (12). These authors screened a panel of HIV-1 patient sera using overlapping peptides covering aa 647 to 684 of gp41 and identified an immunodominant site in which the residues WNWFDI were most critical for Ab recognition. The “immunodominance” of this region must, however, be put into context since previous serum panel screens with overlapping peptides failed to identify the membrane-proximal region as an immunodominant site (30, 71). Furthermore, we were unable to observe any inhibition of Z13 or 4E10 binding to peptide 2031c using either HIVIG (1 mg/ml) or autologous patient (FDA-2) serum (1:10 serum dilution) (data not shown). In the study by Calarota et al., different residues within the WNWFDI motif were found to be critical for recognition by different patient serum samples (12). It would be interesting to determine whether other Abs against this region are neutralizing and whether particular critical residue “patterns” are important. Defining such residue patterns for Z13 and 4E10 would help determine which gp41 residues are exposed on the native envelope spike.
The observation that Z13 binding to peptide 2031c is not inhibited by autologous FDA-2 serum, furthermore, indicates that Z13 is a minor specificity in the serum antibody repertoire of the subject FDA2 and cannot by itself account for the broad neutralizing activity of the serum of patient FDA2. We are continuing to characterize the antibody response of FDA-2 to try to explain its neutralizing activity.
Z13 and 4E10 were able to neutralize primary isolates from diverse subtypes of HIV-1 (e.g., subtypes B, C, and E). Recent results indicate that Z13 can neutralize the autologous FDA-2 virus, R2 (>90% neutralization at 200 μg/ml) (G. Quinnan, personal communication). The R2 virus bears the sequence NWFDIS (55), which is very similar to the MN motif NWFDIT. There is an apparent difference in neutralization potency between IgG 4E10 and Fab Z13. It should be noted, however, that Z13 has so far been tested only as a monovalent Fab fragment. The neutralization potency of the corresponding whole antibody may be considerably greater (up to 100-fold [47]); this is currently being investigated. In a previous study by D'Souza et al. (19), it was suggested that 4E10 was able to neutralize certain T-cell-line-adapted subtype B isolates of HIV-1. More recently, Xu et al. (79) showed that 4E10 was able to potently neutralize the subtype C isolate BW11. Moreover, Stiegler et al. (G. Stiegler, R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger, submitted for publication) demonstrated broad neutralizing activity of 4E10 against primary isolates of HIV-1 from subtypes A, B, C, D, and E. Taken together with the fact that 2F5 is a potent and broadly neutralizing MAb, the collected results on 4E10 and Z13 provide encouragement to search for yet more, possibly cryptic epitopes on gp41 and suggest that a sizeable portion of the gp41 protein surface close to the transmembrane domain is accessible to neutralizing Abs. Since this region is quite highly conserved (Table 3), it is a potential target for drug and vaccine design. However, some caution is necessary, given unsuccessful attempts to elicit HIV-1-neutralizing Abs in immunization studies using the putative 2F5-target, ELDKWA (15, 23, 35, 44). Our results from affinity selecting the HxBc2 fragment library with 2F5 suggest that the 2F5 epitope involves residues flanking the ELDKWA sequence. Thus, it is likely that reproducing the specific presentation and/or conformation of the NWFDIT target, as it exists on gp41, will be critical if there is to be any chance of eliciting HIV-1-neutralizing Abs using an epitope-targeted immunogen. Experiments to determine whether this is the case are under way.
ACKNOWLEDGMENTS
We thank Darren Michaels and Andrew Phillips for Fab purification, Maxime Moulard for help with flow cytometry experiments, and Erik Karrer for Fab library preparation. We also thank Harvey Alter, Jorge Tavel, and Bob Walker for assistance with obtaining the serum and bone marrow samples. We are grateful to patient FDA-2 for donating serum and bone marrow; without his cooperation, this study would not have been possible.
M.B.Z. was supported by a fellowship from the Natural Science and Engineering Research Council of Canada (NSERC). This work was supported by grants from the NIH to D.R.B. (AI33292), to J.P.M. (AI36082, AI39420, and HL59735), and to P.W.H.I.P. (AI40377 and AI44293). J.P.M. is an Elizabeth Glazer Scientist of the Pediatric AIDS Foundation and a Stavros S. Niarchos Scholar. The Department of Microbiology and Immunology at the Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation. E.O.S. thanks Ian A. Wilson for his continued support and acknowledges funding from NIH grant GM46192 (I.A.W.). We acknowledge the assistance of the GCRC of The Scripps Research Institute (M01 RR00833).
REFERENCES
- 1.Baba T W, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini L A, Posner M R, Katinger H, Stiegler G, Bernacky B J, Rizvi T A, Schmidt R, Hill L R, Keeling M E, Lu Y, Wright J E, Chou T C, Ruprecht R M. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6:200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 2.Barbas C F, III, Bjorling E, Chiodi F, Dunlop N, Cababa D, Jones T M, Zebedee S L, Persson M A, Nara P L, Norrby E, Burton D R. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc Natl Acad Sci USA. 1992;89:9339–9343. doi: 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbas C F, III, Burton D R, Scott J K, Silverman G J. Phage display: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 4.Barbas C F, III, Collet T A, Amberg W, Roben P, Binley J M, Hoekstra D, Cababa D, Jones T M, Williamson R A, Pilkington G R, Haigwood N L, Cabezas E, Satterthwait A C, Sanz I, Burton D R. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 5.Binley J M, Ditzel H J, Barbas III C F, Sullivan N, Sodroski J, Parren P W H I, Burton D R. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retrovir. 1996;12:911–924. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- 6.Binley J M, Sanders R W, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma D J, Maddon P J, Olson W C, Moore J P. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauder A, Katinger H. Generation of human monoclonal antibodies against HIV-1 proteins: electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retrovir. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 8.Burton D R. A vaccine for HIV type 1: the antibody perspective. Proc Natl Acad Sci USA. 1997;94:10018–10023. doi: 10.1073/pnas.94.19.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton D R, Barbas III C F, Persson M A A, Koenig S, Chanock R M, Lerner R A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton D R, Montefiori D C. The antibody response in HIV-1 infection. AIDS. 1997;11(Suppl. A):S87–S98. [PubMed] [Google Scholar]
- 11.Burton D R, Moore J P. Why do we not have an HIV vaccine and how can we make one? Nat Med. 1998;4:495–498. doi: 10.1038/nm0598supp-495. [DOI] [PubMed] [Google Scholar]
- 12.Calarota S, Jansson M, Levi M, Broliden K, Libonatti O, Wigzell H, Wahren B. Immunodiminant glycoprotein 41 epitope identified by seroreactivity in HIV type 1-infected individuals. AIDS Res Hum Retrovir. 1996;12:705–713. doi: 10.1089/aid.1996.12.705. [DOI] [PubMed] [Google Scholar]
- 13.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 15.Coëffier E, Clément J M, Cussac V, Khodaei-Boorane N, Jehanno M, Rojas M, Dridi A, Latour M, El Habib R, Barré-Sinoussi F, Hofnung M, Leclerc C. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine. 2000;19:684–693. doi: 10.1016/s0264-410x(00)00267-x. [DOI] [PubMed] [Google Scholar]
- 16.Conley A J, Kesslerll J A, Boots L J, McKenna P M, Schleif W A, Emini E A, Mark G E I, Katinger H, Cobb E K, Lunceford S M, Rouse S R, Murthy K K. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J Virol. 1996;70:6751–6758. doi: 10.1128/jvi.70.10.6751-6758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conley A J, Kessler J A I, Boots L J, Tung J-S, Arnold B A, Keller P M, Shaw A R, Emini E A. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc Natl Acad Sci USA. 1994;91:3348–3352. doi: 10.1073/pnas.91.8.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ditzel H J, Parren P W H I, Binley J M, Sodroski J, Moore J P, Barbas C F, Burton D R. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J Mol Biol. 1997;267:684–695. doi: 10.1006/jmbi.1997.0912. [DOI] [PubMed] [Google Scholar]
- 19.D'Souza M P, Geyer S J, Hanson C V, Hendry R M, Milman G. Evaluation of monoclonal antibodies to HIV-1 envelope by neutralization and binding assays: an international collaboration. AIDS. 1994;8:169–181. [PubMed] [Google Scholar]
- 20.D'Souza M P, Livnat D, Bradac J A, Bridges S The AIDS Clinical Trials Group Antibody Selection Working Group; Collaborating Investigators. Evaluation of monoclonal antibodies to HIV-1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J Infect Dis. 1997;175:1056–1062. doi: 10.1086/516443. [DOI] [PubMed] [Google Scholar]
- 21.Earl P L, Broder C C, Doms R W, Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earl P L, Broder C C, Long D, Lee S A, Peterson J, Chakrabarti S, Doms R W, Moss B. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68:3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckhart L, Raffelsberger W, Ferko B, Purtscher K A, Katinger H, Ruker F. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J Gen Virol. 1996;77:2001–2008. doi: 10.1099/0022-1317-77-9-2001. [DOI] [PubMed] [Google Scholar]
- 24.Fenyö E M, Albert J, McKeating J. The role of the humoral immune response in HIV infection. AIDS. 1996;10(Suppl. A):S97–S106. doi: 10.1097/00002030-199601001-00014. [DOI] [PubMed] [Google Scholar]
- 25.Gauduin M C, Allaway G P, Maddon P J, Barbas III C F, Burton D R, Koup R A. Evaluation of the protective role of two recombinant immunoglobulin molecules in passive protection against primary isolates of HIV-1. Keystone Symp. 1996;E6:39. [Google Scholar]
- 26.Gorny M K, Gianakakos V, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorny M K, Zolla-Pazner S. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J Virol. 2000;74:6186–6192. doi: 10.1128/jvi.74.13.6186-6192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horal P, Svennerholm B, Jeansson S, Rymo L, Hall W W, Vahlne A. Continuous epitpes of the human immunodeficiency virus type 1 (HIV-1) tranmembrane glycoprotein and reactivity of human sera to synthetic peptides representing various HIV-1 isolates. J Virol. 1991;65:2718–2723. doi: 10.1128/jvi.65.5.2718-2723.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostrikis L G, Cao Y, Ngai H, Moore J P, Ho D D. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J Virol. 1996;70:445–458. doi: 10.1128/jvi.70.1.445-458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong P D, Wyatt R, Sattentau Q J, Sodroski J, Hendrickson W A. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawless M K, Barney S, Guthrie K I, Bucy T B, Petteway S R, Jr, Merutka G. HIV-1 membrane fusion mechanism: Structural studies of the interactions between biologically-active peptides from gp41. Biochemistry. 1996;35:13697–13708. doi: 10.1021/bi9606962. [DOI] [PubMed] [Google Scholar]
- 34.Lee W R, Yu X F, Syu W J, Essex M, Lee T H. Mutational analysis of conserved N-linked glycosylation sites of human immunodeficiency virus type 1 gp41. J Virol. 1992;66:1799–1803. doi: 10.1128/jvi.66.3.1799-1803.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang X, Munshi S, Shendure J, MarkIII G, Davies M E, Freed D C, Montefiori D C, Shiver J W. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope proteins. Vaccine. 1999;17:2862–2872. doi: 10.1016/s0264-410x(99)00125-5. [DOI] [PubMed] [Google Scholar]
- 36.Maruyama T, Rodriguez L L, Jahrling P B, Sanchez A, Khan A S, Nichol S T, Peters C J, Parren P W H I, Burton D R. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J Virol. 1999;73:6024–6030. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mascola J R, Lewis M G, Stiegler G, Harris D, VanCott T C, Hayes D, Louder M K, Brown C, Sapan C V, Frankel S S, Lu Y, Robb M L, Katinger H, Birx D L. Protection of macaques against pathogenic SHIV-89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mascola J R, Stiegler G, VanCott T C, Katinger H, Carpenter C B, Hanson C E, Beary H, Hayes D, Frankel S S, Birx D L, Lewis M G. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 39.Moog C, Fleury H J A, Pellegrin I, Kirn A, Aubertin A M. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J Virol. 1997;71:3734–3741. doi: 10.1128/jvi.71.5.3734-3741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore J P, Cao Y, Leu J, Qin L, Korber B, Ho D D. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J Virol. 1996;70:427–444. doi: 10.1128/jvi.70.1.427-444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 42.Moore J P, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munoz-Barroso I, Salzwedel K, Hunter E, Blumenthal R. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J Virol. 1999;73:6089–6092. doi: 10.1128/jvi.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muster T, Guinea R, Trkola A, Purtscher M, Klima A, Steindl F, Palese P. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Rüker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ollmann Saphire E, Parren P W H I, Pantophlet R, Zwick M B, Morris G M, Rudd P M, Dwek R A, Stanfield R L, Burton D R, Wilson I A. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 47.Parren P W H I, Burton D R. Antibodies against HIV-1 from phage display libraries: mapping of an immune response and progress towards antiviral immunotherapy. Chem Immunol. 1997;65:18–56. doi: 10.1159/000319346. [DOI] [PubMed] [Google Scholar]
- 48.Parren P W H I, Ditzel H J, Gulizia R J, Binley J M, Barbas III C F, Burton D R, Mosier D E. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS. 1995;9:F1–F6. doi: 10.1097/00002030-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 49.Parren P W H I, Fisicaro P, Labrijn A F, Binley J M, Yang W P, Ditzel H J, Barbas III C F, Burton D R. In vitro antigen challenge of human antibody libraries for vaccine evaluation: the human immunodeficiency virus type 1 envelope. J Virol. 1996;70, 12:9046–9050. doi: 10.1128/jvi.70.12.9046-9050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parren P W H I, Mondor I, Naniche D, Ditzel H J, Klasse P J, Burton D R, Sattentau Q J. Neutralization of HIV-1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parren P W H I, Wang M, Trkola A, Binley J M, Purtscher M, Katinger H, Moore J P, Burton D R. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type-1. J Virol. 1998;72:10270–10274. doi: 10.1128/jvi.72.12.10270-10274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poignard P, Ollmann Saphire E, Parren P W H I, Burton D R. GP120: biologic aspects of structural features. Annu Rev Immunol. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- 53.Purtscher M, Trkola A, Grassauer A, Schulz P M, Klima A, Dropper S, Gruber G, Buchacher A, Muster T, Katinger H. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS. 1996;10:587–593. doi: 10.1097/00002030-199606000-00003. [DOI] [PubMed] [Google Scholar]
- 54.Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, Tauer C, Berger R, Barrett N, Jungbauer A, Katinger H. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retrovir. 1994;10:1651–1658. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- 55.Quinnan G V, Jr, Zhang P F, Fu D W, Dong M, Alter H J, International Collaborators Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res Hum Retrovir. 1999;15:561–570. doi: 10.1089/088922299311088. [DOI] [PubMed] [Google Scholar]
- 56.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 57.Roben P, Moore J P, Thali M, Sodroski J, Barbas III C F, Burton D R. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salzwedel K, West J T, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanner M F, Olson A J, Spehner J C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 60.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sattentau Q J, Moore J P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sattentau Q J, Zolla-Pazner S, Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology. 1995;206:713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 64.Sblattero D, Bradbury A. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology. 1998;3:271–278. doi: 10.1016/s1380-2933(97)10004-5. [DOI] [PubMed] [Google Scholar]
- 65.Suarez T, Gallaher W R, Agirre A, Goni F M, Nieva J L. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J Virol. 2000;74:8038–8047. doi: 10.1128/jvi.74.17.8038-8047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suarez T, Nir S, Goni F M, Saez-Cirion A, Nieva J L. The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: a novel fusogenic sequence. FEBS Lett. 2000;477:145–149. doi: 10.1016/s0014-5793(00)01785-3. [DOI] [PubMed] [Google Scholar]
- 67.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trkola A, Pomales A P, Yuan H, Korber B, Maddon P J, Allaway G, Katinger H, Barbas III C F, Burton D R, Ho D D, Moore J P. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type I. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ugen K E, Goedert J J, Boyer J, Refaeli Y, Frank I, Williams W V, Wiloughby A, Landesman S, Mendez H, Rubinstein A, et al. Vertical transmission of human immunodeficiency virus (HIV) infection. Reactivity of maternal sera with glycoprotein 120 and 41 peptides from HIV type 1. J Clin Investig. 1992;89:1923–1930. doi: 10.1172/JCI115798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vujcic L K, Quinnan G V., Jr Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res Hum Retrovir. 1995;11:783–787. doi: 10.1089/aid.1995.11.783. [DOI] [PubMed] [Google Scholar]
- 73.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 74.Williamson R A, Peretz D, Pinilla C, Ball H, Bastidas R B, Rozenshteyn R, Houghten R A, Prusiner S B, Burton D R. Mapping the prion protein using recombinant antibodies. J Virol. 1998;72:9413–9418. doi: 10.1128/jvi.72.11.9413-9418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wyatt R, Desjardin E, Olshevsky U, Nixon C, Binley J, Olshevsky V, Sodroski J. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J Virol. 1997;71:9722–9731. doi: 10.1128/jvi.71.12.9722-9731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 77.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 78.Xu J Y, Gorny M K, Palker T, Karwowska S, Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol. 1991;65:4832–4838. doi: 10.1128/jvi.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu W, Smith-Franklin B A, Li P L, Wood C, He J, Du Q, Bhat G J, Kankasa C, Katinger H, Cavacini L A, Posner M R, Burton D R, Chou T C, Ruprecht R M. Potent neutralization of primary HIV clade C isolates with a synergistic combination of human monoclonal antibodies raised against clade B. J Hum Virol. 2001;4:55. [PubMed] [Google Scholar]
- 80.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectos. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]