Abstract
Uterine corpus endometrial carcinoma (UCEC) is one of the most common tumours of the female reproductive system. CCAAT enhancer-binding protein alpha (CEBPA) is a member of the transcription factor family involved in regulating processes such as cell proliferation, differentiation, metabolism, and the immune response. However, the role of CEBPA in UCEC has not been clarified. Here, we performed a comprehensive analysis to explore the expression level, prognostic value, immune infiltration and biological function of CEBPA in UCEC. In this study, we found that CEBPA expression was upregulated and associated with poor prognosis in UCEC patients. KEGG and GO analyses revealed that the genes positively correlated with CEBPA were enriched primarily in immune regulation and oxidative phosphorylation. Immune infiltration analysis revealed that CEBPA is strongly correlated with immune cell infiltration in UCEC. RT-qPCR indicated that CEBPA may regulate the OXPHOS level in Ishikawa cells. CCK-8, cell cycle, Transwell and scratch wound healing assays revealed that CEBPA promoted Ishikawa cell proliferation, invasion and migration. In addition, PPI and survival analyses suggested that CEBPG may be a potential target of CEBPA in UCEC. These results demonstrated that CEBPA may be a potential therapeutic target in UCEC.
Keywords: CEBPA, UCEC, Invasion, Migration, Proliferation, Immune filtration
Subject terms: Cancer microenvironment, Gynaecological cancer, Oncogenes, Tumour immunology, Mechanisms of disease
Introduction
Uterine corpus endometrial carcinoma (UCEC) is a common malignant tumour of the female reproductive system. In Europe and the US, UCEC ranks as the most frequent gynaecological malignant tumour and is associated with high mortality in females. In China, the incidence of UCEC has increased over the past few decades, and the onset of UCEC increases with age1–3. UCEC is classified into oestrogen-dependent (type I) and non-oestrogen-dependent (type II) types according to its clinical and histopathological characteristics. The most common type I UCEC is endometrioid adenocarcinoma, which responds to oestrogen and has a good prognosis. Type II UCEC includes clear cell carcinoma and serous carcinoma, which usually have a poor prognosis4,5. The occurrence and progression of UCEC are complicated, as they involve a variety of genomics and environment factors6,7, and have not yet been fully clarified. Understanding the molecular mechanism of UCEC tumorigenesis is crucial for improving treatment outcomes.
CCAAT enhancer binding protein α (C/EBP-α or CEBPA) is a member of the CCAAT enhancer binding protein (C/EBP) transcription factor family and is a basic region/leucine zipper (bZIP) protein. CEBPA functions in cell proliferation and differentiation and is closely related to the progression of various tumours8. Previous studies have demonstrated that CEBPA is essential for lineage-specific differentiation of blood cells. In acute myeloid leukaemia (AML), disruption of CEBPA blocks the transition of committed myeloid stem cells to granulocyte or monocyte progenitors, which leads to the loss of mature granulocytes9. In chronic myeloid leukaemia (CML), the expression of CEBPA is downregulated and is closely related to the progression of CML10. An in vitro study revealed that overexpression of CEBPA could inhibit proliferation and promote apoptosis of the CML cell line K562 through the Foxo3a-Bin signalling pathway11. In addition, animal experiments have shown that CEBPA negatively regulates the tumour-promoting behaviour and immunosuppressor activity of myeloid-derived suppressor cells (MDSCs)12. These studies suggest that CBEPA may act as a tumour suppressor in haematologic malignancies and that this protein is closely related to immune regulation.
In addition to haematologic tumours, CEBPA is also associated with various solid malignancies. In lung cancer, CEBPA suppresses tumour progression through inactivation of the p38α MAPK signalling pathway13, while in breast cancer, CEBPA can inhibit tumour progression by inhibiting the epithelial‒mesenchymal transition (EMT) process14. In gastric cancer, CEBPA inhibits gastric cancer cell viability15, and mutations in CEBPA are associated with immune-related adverse events in patients with advanced gastric cancer16. In head and neck squamous cell carcinoma (HNSCC), CEBPA is significantly downregulated in the microarray expression profiles of HNSCC samples, and downregulation of CEBPA is associated with poor prognosis17. Recently, a clinical study revealed that the therapeutic upregulation of CEBPA has a strong antitumour effect on hepatocellular carcinoma (HCC) patients and mouse models and that this antitumour effect is mediated by the immune system in mouse models18. Nevertheless, the knock down of CEBPA can inhibit malignant cell growth and viability in glioma and prostate cancer19,20. The overexpression of CEBPA promotes tumour progression by promoting cell invasion and inhibiting apoptosis in oral squamous cell carcinoma (OSCC)21.
Moreover, reports also indicate that CEBPA is involved in the progression of gynaecological cancers. In ovarian cancer, downregulation of CEBPA is associated with poor prognosis. Knockout of CEBPA promotes the proliferation, invasion, migration, and EMT of ovarian cancer cells through the Wnt/β signalling pathway22. In cervical cancer, CEBPA mRNA expression is downregulated, and overexpression of CEBPA has been shown to inhibit the proliferation and migration of cervical cancer cells23. A study of UCEC revealed that CEBPA mRNA was downregulated in 7 samples of endometrial cancer. In response to treatment with a demethylating agent, CEBPA expression then increased in the endometrial cancer cell line Ishikawa, and the overexpression of CEBPA inhibited the colony formation of Ishikawa cells. Given that many tumour suppressor genes are epigenetically silenced in tumours, the authors suggested that CEBPA may be a tumour suppressor gene in endometrial cancer24. However, the prognostic significance, exact role and potential regulatory mechanism of CEBPA in UCEC have not been clarified.
In the present study, we performed a comprehensive analysis to explore the expression level, prognostic value, effects on immune infiltration, and biological function of CEBPA in UCEC. We found that CEBPA expression was upregulated in UCEC and was associated with clinical prognosis. In addition, CEBPA was strongly correlated with immune cell infiltration in UCEC. Moreover, we explored the functions of CEBPA in Ishikawa cells and reported that CEBPA is involved in the regulation of oxidative phosphorylation, proliferation, migration, and invasion in these cells. Our results indicate that CEBPA may be a potential therapeutic target in UCEC.
Materials and methods
Analysis of CEBPA mRNA expression, prognostic value and clinicopathological features
The clinicopathological data, prognostic information and RNA-seq transcriptome data of 545 UCEC patients and 114 normal samples were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) and the Genotype Tissue Expression (GTEx) database (https://www.gtexportal.org/home/). The expression of CEBPA in normal endometrial tissues and UCEC tumour tissues and in UCEC tumour tissues representing different tumour stages was analysed via the TCGA and GTEX databases. The chi-square test was used to analyse the correlation between CEBPA expression and clinicopathological features in UCEC patients (Table 1). Furthermore, Kaplan–Meier plots were constructed to assess the relationship between the CEBPA expression level and the prognosis of UCEC patients. The survival measures included overall survival (OS), disease-specific survival (DSS), the disease-free interval (DFI), and the progression-free interval (PFI). ROC curves were used to analyse the predictive value of CEBPA for the prognosis of UCEC patients. A P value < 0.05 was considered significant.
Table 1.
Association between mRNA expression of CEBPA and clinical pathological characteristics in UCEC from TCGA.
| CEBPA expression | X2 | P | |||
|---|---|---|---|---|---|
| Low | High | ||||
| Age* | ≤ 65 years | 178 | 128 | 13.596 | < 0.001 |
| > 65 years | 101 | 138 | |||
| Menopause status* | Indeterminate | 8 | 9 | 20.125 | < 0.001 |
| Pre | 30 | 5 | |||
| Peri | 11 | 6 | |||
| Post | 215 | 235 | |||
| Treatment outcome* | CR | 197 | 165 | 11.72 | 0.008 |
| PR | 8 | 3 | |||
| PD | 21 | 42 | |||
| SD | 4 | 5 | |||
| Clinical stage* | Stage I | 189 | 152 | 13.024 | 0.005 |
| Stage II | 29 | 24 | |||
| Stage III | 53 | 72 | |||
| Stage IV | 8 | 21 | |||
| Histologic grade* | G1 | 64 | 34 | 39.983 | < 0.001 |
| G2 | 83 | 37 | |||
| G3 | 127 | 192 | |||
| High grade | 5 | 6 | |||
| Peritoneal wash* | positive | 23 | 34 | 4.203 | 0.04 |
| negative | 194 | 159 | |||
| Vital status | Alive | 240 | 215 | 3.612 | 0.057 |
| Dead | 39 | 54 | |||
| Person cancer status | Tumor free | 208 | 174 | 3.325 | 0.068 |
| With tumor | 37 | 48 | |||
The Asterisk * was considered significant. Significance value bold.
CEBPA-related gene enrichment analysis
Using the Linked Omics portal, CEBPA-related genes were analysed in UCEC. The top 100 genes that were each positively and negatively correlated with CEBPA in UCEC were used for further analysis. Through Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses, we explored the biological functions and pathways enriched in CEBPA-related genes. An FDR of 0.05 or a P value < 0.05 was considered significant.
Immune infiltration analysis
CIBERSORT25 was used to evaluate the relationship between CEBPA mRNA expression and tumour-infiltrating immune cells in UCEC. We compared the abundance of 22 immune cell subtypes between the high CEBPA expression group and the low CEBPA expression group in UCEC. The differences in the infiltration of immune cell subtypes between the high CEBPA expression group and the low CEBPA expression group were further analysed in UCEC. The heatmap was generated to show the correlations among immune cells. A P value < 0.05 was considered significant.
Screening and validation of CEBPA target genes
Since CBEPA is a member of a transcription factor family, the hTFtarget database and the footprintDB database were used to predict CEBPA target genes. The top 100 genes that were positively correlated with CEBPA expression and the top 100 genes that were negatively correlated with CEBPA expression in UCEC intersected with the predicted target genes of CBEPA. We subsequently analysed the protein interactions of the intersecting genes and constructed a protein–protein interaction (PPI) network via the STRING portal. Finally, we used Kaplan–Meier plots and RT-qPCR to analyse the expression of potential target genes obtained from the intersection.
Cell culture and cell transfection
The human UCEC Ishikawa cell line used in this study was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Ishikawa cells were incubated in DMEM (Gibco, USA) supplemented with 10% FBS (Sijiqing, China) and 1% penicillin/streptomycin (Solarbio, China) at 37 °C and 5% CO2. CEBPA overexpression and knockdown lentiviruses (OE-NC, OE-CEBPA, sh-NC, sh-CEBPA-1, and sh-CEBPA-2) were designed and constructed by OBIO (Shanghai, China). Stable cell lines were constructed by the transfection of lentiviruses into Ishikawa cells. After 2 weeks of culture RT-qPCR and Western blotting were performed to verify the knockdown and overexpression efficiency of CEBPA in these cells. The sequences of the sh-CEBPAs used are summarized in Table S1.
RNA extraction, and real-time qPCR
In accordance with the manufacturer’s protocol for Trizol reagent (Solarbio, China), total RNA was isolated from the cells. The purity and concentration of total RNA were tested with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, USA). cDNA was synthesized from total RNA via a PrimeScript II kit (Takara, Japan). Real-time PCR was performed with a SYBR Premix Ex Taq II kit (Takara, Japan). The expression levels of the target genes were determined according to the ΔΔCt method, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the internal control. The sequences of the primers used are summarized in Table S2.
Western blotting
Total protein was extracted with PMSF (Beyotime, China) and RIPA (Beyotime, China) buffer, and the protein concentration was determined with a BCA kit (Beyotime, China). The total protein was mixed with loading buffer and then subjected to SDS-PAGE. The proteins were then separated and transferred onto polyvinylidene difluoride (PVDF) membranes, which were blocked with 5% non-fat milk (Beyotime, China), after which primary and secondary antibody staining was performed before imaging. The primary antibodies used were specific for CEBPA (1:1000, Proteintech, China), β-Tubulin (1:2000, Proteintech, China). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H + L) (1:10,000, Proteintech, China) was used as the secondary antibody. All images were captured using the molecular imager Chemi Doc XRS + (Bio-Rad, CA).
CCK‑8 assay
Transfected cells were seeded in a 96-well plate (1.5 × 103 cells/well) and cultured under standard conditions for 5 days. At specific time points every day, 100 μl of solution containing 10% CCK-8 reagent (Dojindo, Kumamoto, Japan) was added, and the mixture was incubated at 37 °C for 2 h. The OD value (450 nm) was then determined using a microplate reader.
Flow cytometry
For cell cycle detection, the transfected cells were fixed in 70% ethanol at -20 °C. These cells were washed with PBS and then stained with propidium iodide for 30 min at room temperature according to the manufacturer’s protocol for the Cell Cycle Detection Kit (MultiSciences, China). Finally, the cells were filtered with a FACSAria II flow cytometer (BD Biosciences, USA), and the data were analysed with FlowJo_V10 software (BD Biosciences, USA).
Invasion assay
Transwell assays were performed to measure the invasion capacity of the transfected cells. Briefly, the upper inserts (8 μm pore size, 6.5 mm diameter; Corning, USA) precoated with 200 μl of Matrigel were incubated at 37 °C for 1 h. Transfected cells (2.5 × 104 cells/well) were seeded in serum-free medium into the upper chamber after the Matrigel was removed. The lower chamber contained 600 µL of DMEM supplemented with 20% FBS. The cells were cultured in 5% CO2 at 37 °C for 48 h. The cells that did not invade through the filters were removed from the upper surfaces with a cotton swab. The inserts were fixed for 30 min in 4% paraformaldehyde and then stained with 0.1% crystal violet at room temperature for 10 min. Staining was captured with an Olympus microscope (BX51 + DP72, Olympus, Japan). The cells that had invaded through the filters to the lower surfaces were counted in 5 randomly selected nonoverlapping fields at a magnification of × 100. Each experiment was performed in triplicate and was repeated at least 3 times.
Scratch wound healing assay
A scratch wound healing assay was performed to measure the migration capacity of the transfected cells. Briefly, the cells were grown to 75–80% confluency and dissociated with trypsin (Solarbio, China) for 5 min at 37 °C. Then, the cells were seeded in a culture-insert (Ibidi, Germany) at a concentration of 5 × 105 cells/well and cultured for 12 h at 37 °C until they reached confluency. The wound was formed after removal of the insert. The cells were subsequently transferred to DMEM containing 10% FBS and incubated in 5% CO2 at 37 °C. The wounds were observed and recorded until they healed. Images were captured with an Olympus microscope (BX51 + DP72, Olympus, Japan).
Statistical analyses
The aforementioned online tools were used to perform statistical analyses automatically. GraphPad Prism 8.0 software was used for statistical analysis. Survival analysis was performed via the Kaplan–Meier method. Student’s t tests were used to determine the differences in data between two comparator groups. Multiple groups were compared via one-way ANOVA or two-way ANOVA. Images were analysed with ImageJ. All the data are presented as the means ± standard errors of the mean (SEMs). P < 0.05 indicated statistical significance.
Results
Higher expression of CEBPA is associated with poor clinical prognosis in UCEC patients
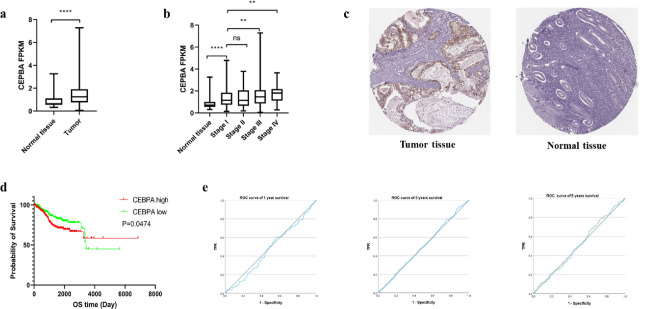
We analysed CEBPA mRNA expression between UCEC tumour and normal tissues from the TCGA and GTEx databases. As shown in Fig. 1a, CEBPA expression was upregulated in UCEC tumour samples compared with normal endometrial tissues. Moreover, the expression of CEBPA was greater in patients with stages III and IV disease than in those with stage I disease (Fig. 1b), which indicates that high expression of CEBPA may be associated with poor prognosis. In addition to mRNA, IHC images from several cases from the HPA website (https://www.proteinatlas.org/) showed that, CEPBA protein was usually positive in UCEC samples, whereas it was almost always negative in the normal endometrium (Fig. 1c, Supplemental Fig. 1a–f). These data indicate that CEPBA is typically highly expressed in UCEC.
Fig. 1.
Expression level and prognostic value of CEBPA in UCEC. (a–b) CEBPA mRNA expression levels in UCEC tumour tissues and normal tissues from TCGA and GTEx. (c) CEBPA protein expression levels in UCEC tumour tissues and normal endometrial tissues from the HPA website (× 200) (immunohistochemistry, hematoxylin-eosin staining). (d) Prognostic significance of CEBPA expression in UCEC according to Kaplan–Meier plotter curves. OS, overall survival. (e) The predictive value of CEBPA for the prognosis of UCEC according to ROC curve. *P < 0.05, **P < 0.01, ***P < 0.001.
The clinical phenotypic data of UCEC patients were also obtained from the TCGA. Patients were divided into two groups according to the CEBPA expression level (with the median of FPKM as the cut-off). The chi-square test was used to analyse the associations of CEBPA expression with clinical and pathological features. As shown in Table 1, patients with higher CEBPA expression were more advanced in terms of age, clinical stage, and histologic tumour grade. The percentage of positive cancer cells in the peritoneal wash fluid was also increased in the high CEBPA expression group, which indicates an increased possibility of tumour metastasis. In addition, patients with high CEBPA expression were more likely to have poor treatment outcomes. Moreover, we generated Kaplan–Meier curves based on the TCGA dataset to evaluate the differences in survival between the high and low CEBPA expression groups. The results revealed that high expression of CEBPA was negatively correlated with overall survival (OS) (p = 0.0474) (Fig. 1d), progression-free interval (PFI) (p = 0.0055) (Supplemental Fig. 1g), and disease-specific survival (DSS) (p = 0.0086) (Supplemental Fig. 1h), although not difference was observed in the disease-free interval (DFI) (Supplemental Fig. 1i). These findings reveal that high CEBPA expression may be linked to poor prognosis in UCEC patients.
To further explore the value of CEBPA in predicting the prognosis of UCEC patients, we generated receiver operating characteristic (ROC) curves for the TCGA-UCEC cluster according to CEBPA expression. However, the results show that CEBPA may not be a good predictor of UCEC prognosis, as the areas under the curve (AUCs) for the 1-, 3-, and 5-year survival rates were all approximately 0.5 (Fig. 1e).
KEGG and GO enrichment analyses of CEBPA-related genes in UCEC
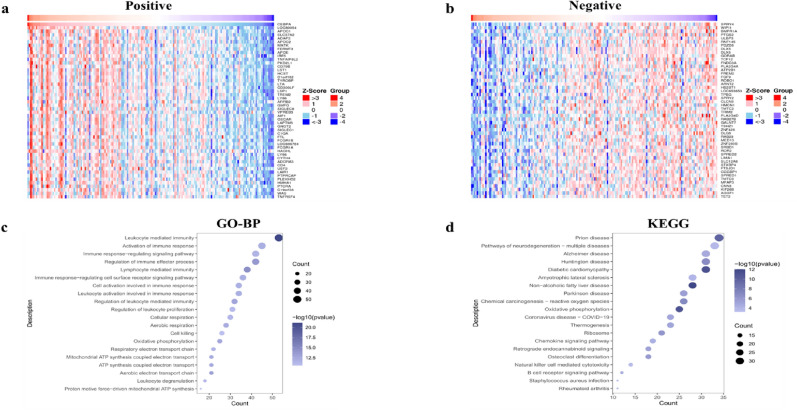
To further investigate the underlying functional mechanism of CEBPA in UCEC, we used the LinkedOmic database (http://www.linkedomics.org/admin.php/) to search for genes whose expression was correlated with CEBPA. The top 100 genes that were each positively and negatively correlated with CEBPA in UCEC are summarized in Table S3. The heatmap (Fig. 2a,b) shows the top 50 genes that were positively or negatively correlated with CBEPA. The protein-encoding gene that was most positively correlated with CEBPA was apolipoprotein C-I (APOC1), which functions as an inhibitor of lipoprotein binding to the low-density lipoprotein (LDL) receptor. The gene whose expression was most negatively correlated was SPRY4, which encodes a protein that suppresses the insulin receptor and the EGFR-transduced MAPK signalling pathway.
Fig. 2.
KEGG and GO enrichment of genes related to the expression of CEBPA in UCEC. (a) Heatmap showing the enrichment patterns of the top 50 genes positively correlated with the CBEPA. (b) Heatmap showing the enrichment patterns of the top 50 genes negatively correlated with the CBEPA. (c) GO-BP enrichment analysis was applied to the top 100 genes that were positively correlated with the CBEPA. (d) KEGG enrichment analysis was applied to the top 100 genes that were positively correlated with the CBEPA.
To further explore the functions of CEBPA-related genes, we performed GO and KEGG enrichment analyses of the positively correlated genes whose correlation indices were > 0.3 and p < 0.05, respectively. The GO enrichment analysis revealed that the genes positively correlated with CEBPA were enriched primarily in terms of immune regulation, such as “leukocyte mediated immunity”, “regulation of immune effector process”, and “regulation of leukocyte mediated immunity” (Fig. 2c), which indicates that CEBPA and its related genes may play important roles in immune cell regulation. Moreover, the GO terms associated with oxidative phosphorylation, such as “aerobic electron transport chain” and “ATP synthesis coupled electron transport”, were also frequently enriched. Moreover, these terms can also be observed in the results of the KEGG enrichment analysis (Fig. 2d). These results suggest that CEBPA and its related genes may exert important functions through immune regulation and oxidative phosphorylation in UCEC.
CEBPA expression is associated with immune cell infiltration in UCEC
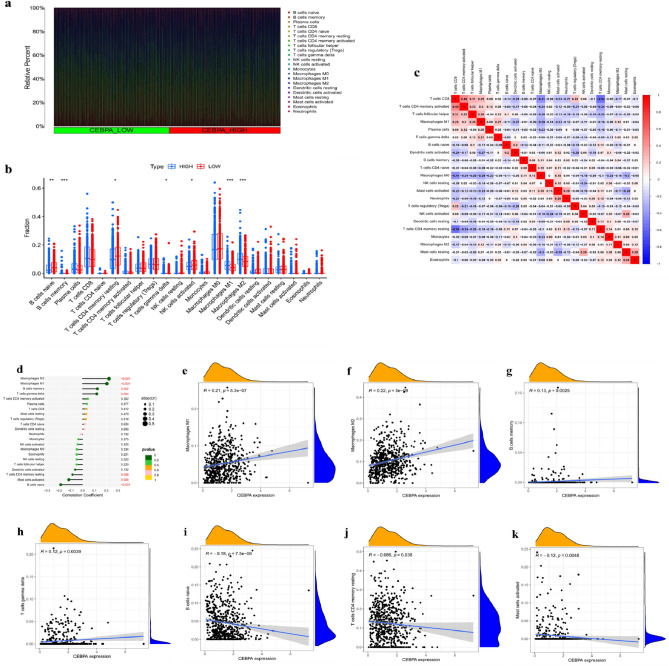
Numerous studies have shown that tumour-infiltrating immune cells play crucial roles in the progression, metastasis, and growth of UCEC26. In this study, GO and KEGG enrichment analyses of CEBPA-related genes indicated that CEBPA was associated with immune regulation. To further explore the potential role of CEBPA in the immune microenvironment, we analysed the relationship between CEBPA expression and immune infiltration via CIBERSORT method in UCEC. Figure 3a shows that the differences in the abundances of 22 immune cell types between the high CEBPA expression group and the low CEBPA expression group in UCEC. The infiltration levels of four cell subtypes, memory B cells, M2 macrophages, M1 macrophages and gamma delta T cells, were greater in the high CEBPA expression group than in the low CEBPA expression group (Fig. 3b). The infiltration levels of three cell subtypes, namely, naive B cells, activated NK cells and memory CD4 T cells, were greater in the low CEBPA expression group than in the high CEBPA expression group (Fig. 3b). A correlation analysis revealed a negative correlation between resting memory CD4 T cells and CD8 T cells (r = − 0.54) (Fig. 3c). In addition, the relationships between CEBPA expression and 22 immune cell types were investigated via Spearman correlation analysis (Fig. 3d). The results indicated that CEBPA was positively correlated with M1 macrophages (p < 0.001), M2 macrophages (p < 0.001), memory B cells (p = 0.002) and gamma delta T cells (p = 0.004) (Fig. 3e–h). In contrast, CEBPA was negatively correlated with naive B cells (p < 0.001), resting memory CD4 T cells (p = 0.038) and activated mast cells (p = 0.005) (Fig. 3i–k). Overall, these findings reveal that CEBPA may play a crucial role in the regulation of tumour immune cell infiltration in UCEC.
Fig. 3.
Relationship between CEBPA expression and immune cell infiltration in UCEC. (a) Relative proportions of immune cell infiltration between the high CEBPA expression group and the low CEBPA expression group in UCEC. (b) Diagram of the difference in immune cell infiltration proportions between the high CEBPA expression group and the low CEBPA expression group. (c) Correlation heatmap among immune cell populations; red and blue indicate positive and negative correlations, respectively. (d) Lollipop chart shows the relationships between CEBPA and immune cell infiltration. (e–h) M1 macrophages, M2 macrophages, memory B cells, and gamma delta T cells. (i–k) naive B cells, resting memory CD4 T cells, and activated mast cells.
CEBPA improves the oxidative phosphorylation level in Ishikawa cells
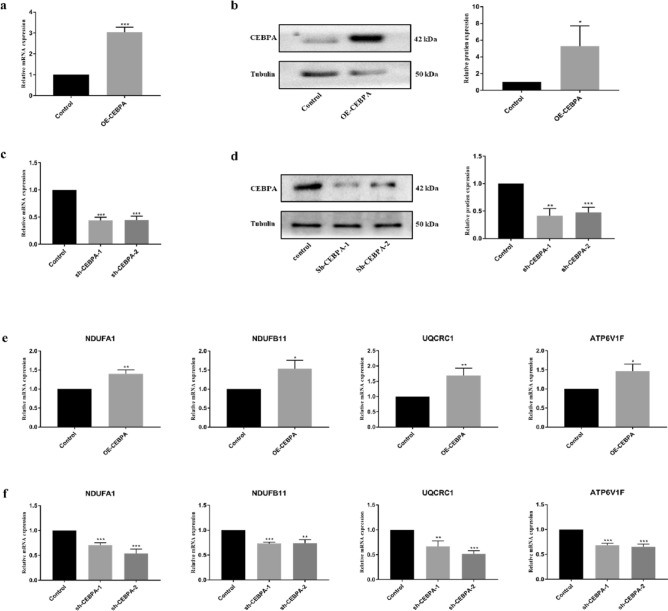
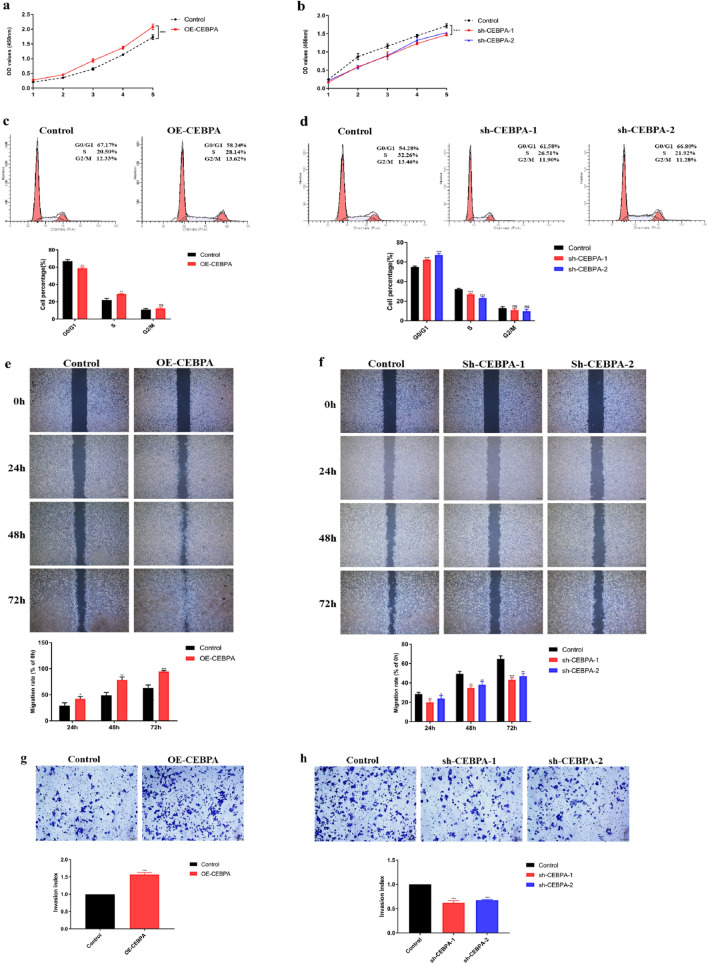
Surprisingly, oxidative phosphorylation (OXPHOS) was also enriched in the genes that were positively correlated with CEBPA. It is well known that cancer cells prefer glycolysis even in the presence of high amounts of oxygen, which is called the Warburg effect. Even so, it is not suggested that OXPHOS is suppressed in all cancers. Previous studies reported that oxidative phosphorylation was increased in several cancers, including UCEC. To confirm whether CEBPA regulates OXPHOS in UCECs, the gene was overexpressed or knocked down in Ishikawa cells (an endometrial cancer cell line) via lentivirus infection. The efficiency of lentivirus infection was confirmed by RT-qPCR and Western blotting. As shown in Fig. 4a–d, CEBPA mRNA and protein levels were significantly increased in CEBPA-overexpressing Ishikawa cells and significantly decreased when two different silencing strategies, sh-CEBPA-1 and sh-CEBPA-2, were used. The results also revealed that the expression levels of the core members of OXPHOS were upregulated and downregulated in CEBPA-overexpressing and sh-CEBPA cells, respectively (Fig. 4e,f), which indicates that CEBPA may regulate the OXPHOS level in UCECs.
Fig. 4.
Effects of CEBPA on the oxidative phosphorylation level in Ishikawa cells. (a–b) The efficiency of CEBPA overexpression in Ishikawa cells was determined via RT-qPCR and western blotting. (c–d) The efficiency of CEBPA knockdown in Ishikawa cells was determined via RT-qPCR and western blotting. (e) Expression of the core members of OXPHOS in OE-CEBPA Ishikawa cells and control cells via RT-qPCR. (f) Expression of the core members of OXPHOS in sh-CEBPA Ishikawa cells and control cells via RT-qPCR. *P < 0.05, **P < 0.01, ***P < 0.001.
CEBPA promotes the proliferation, cell cycle, invasion, and migration of Ishikawa cells
Cancer cells with high OXPHOS levels usually present a more malignant phenotype. Therefore, we performed a series of experiments to investigate the role of CEBPA in UCEC progression. CCK-8 assays revealed that CEBPA overexpression significantly promoted the proliferative capacity of Ishikawa cells, whereas CEBPA knockdown significantly inhibited the proliferative capacity of these cells (Fig. 5a,b). Since CEBPA could affect Ishikawa cell proliferation, we used flow cytometry to detect the cell cycle phase of Ishikawa cells after interference with CEBPA expression. The results revealed that CEBPA knockdown resulted in G0/G1 arrest, whereas CEBPA overexpression alleviated the arrest in G0/G1 phase (Fig. 5c and d). Hence, CEBPA may promote Ishikawa cell proliferation by regulating cell cycle progression in these cells.
Fig. 5.
Effects of CEBPA on the proliferation, invasion and migration ability of Ishikawa cells. (a–d) CCK-8 and cell cycle assays were performed to evaluate the proliferation of sh-CEBPA Ishikawa cells, OE-CEBPA Ishikawa cells, and control cells. (e–f) Scratch wound healing assay was applied to evaluate the migration of sh-CEBPA Ishikawa cells, OE-CEBPA Ishikawa cells, and control cells. (g–h) Transwell assay was performed to determine the invasiveness of sh-CEBPA Ishikawa cells, OE-CEBPA Ishikawa cells, and control cells. *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate the function of CEBPA in cancer cell behaviour, we performed scratch wound healing and Transwell assays to evaluate the invasive and migratory capabilities of Ishikawa cells after CEBPA interference. A scratch wound healing assay revealed that overexpression of CEBPA promoted migration, whereas knockdown of CEBPA inhibited migration (Fig. 5e,f). Transwell assays also revealed that overexpression of CEBPA increased the number of invading cells, whereas knockdown of CEBPA decreased the number of invading cells (Fig. 5g,h). Hence, CEBPA promotes the invasion and migration of Ishikawa cells. These data indicate that CEBPA may act as a tumour promoter in UCEC and may partly explain the poor prognosis of TCGA-UCEC patients with high CEBPA expression.
CEBPG may be a functional target of CEBPA in UCEC
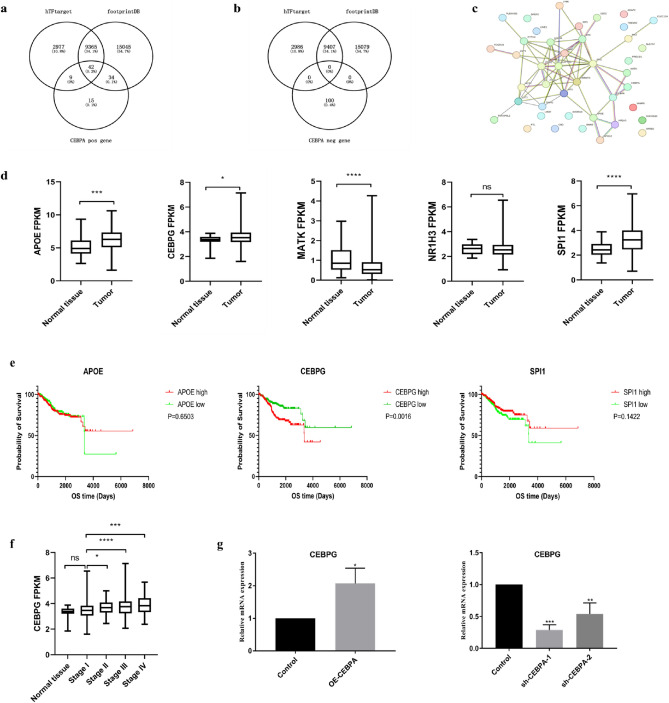
CEBPA is a transcription factor that can regulate the transcription of other genes. Therefore, we performed further analysis to identify CEBPA target genes that may be functional in UCEC. We used the hTFtarget and footprintDB transcription factor databases to predict the target genes of CEBPA, after which the predicted target genes were intersected with genes that were positively or negatively correlated with CEBPA. As shown in Fig. 6a,b, 42 of the predicted target genes overlapped with genes that were positively correlated with CEBPA, whereas 0 predicted target gene overlapped with genes that were negatively correlated with CEBPA, which indicates that CEBPA may function predominantly as a transcriptional activator in UCEC. We then constructed a PPI network map of the 42 genes using the STRING database (https://string-db.org/cgi/input.pl) (Fig. 6c). The map revealed that five genes, namely, CEBPG, APOE, NR1H3, MATK, and SPI1, were directly functionally associated with CEBPA. We compared the expression of these five genes between normal tissues and tumours in the TCGA-UCEC cluster and found that CEBPG, APOE, and SPI1 (Fig. 6d) were highly expressed in tumours. However, only CEBPG showed significant prognostic value in the survival analysis (Fig. 6e). Moreover, the expression of CEBPG was upregulated with increasing tumour stage (Fig. 6f), similar to the pattern of CEBPA. In addition, we verified the relationship between CEBPA and CEBPG expression in Ishikawa cells after CEBPA interference. The results revealed that CEBPG expression was upregulated in CEBPA-overexpressing cells, and downregulated in sh-CEBPA cells (Fig. 6g). These results indicated that CEBPG may be a functional target of CEBPA in UCEC.
Fig. 6.
Screening and validation of CEBPA target genes in UCEC. (a–b) The intersection between genes that were positively or negatively related to CEBPA, hTF target genes and footprintDB target genes. (c) The PPI network map of 42 intersecting genes in the STRING portal. (d) Comparison of CEBPA-related genes expression between normal tissue and tumour tissue in the TCGA-UCEC cluster. (e) The impact of APOE, CEBPG and SPI1 on the overall survival (OS) of UCEC patients was determined via Kaplan–Meier plotter curves. (f) CEBPG mRNA expression levels in UCEC tumour tissues and normal tissues from TCGA and GTEx. (g) mRNA expression of CEBPG in Ishikawa cells after silencing or overexpressing CEBPA via RT-qPCR. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In this study, we performed a comprehensive analysis of CEBPA on the prognosis of patients with UCEC and its function in UCEC. According to the TCGA dataset, the expression level of CEBPA was upregulated in UCEC, and high expression of this protein was significantly associated with poor prognosis. Furthermore, we found that CEBPA is closely related to immune cell infiltration in UCEC. In addition, CEBPA improved oxidative phosphorylation levels and promoted the proliferation, invasion and migration of Ishikawa cells. Therefore, our results indicate that CEBPA may be a potential therapeutic target in UCEC.
The occurrence and progression of UCEC is a complicated and dynamic process that involves a variety of gene and signalling pathway abnormalities6,7. With the development of molecular biology, many genes related to UCEC have been discovered. Exploring the functions of these genes is crucial for the prevention and treatment of UCEC. Previous studies have shown that CEBPA is abnormally expressed and plays a different regulatory role in a variety of tumours. Several studies have shown that low expression of CEBPA is associated with poor prognosis and that CEBPA usually functions as a tumour suppressor gene, such as in ovarian cancer22, cervical cancer23, lung cancer27, and HNSCC17. However, some studies have suggested that CEBPA mRNA expression is upregulated in hepatoblastoma and HCC and that high expression is correlated with poor prognosis. Furthermore, the overexpression of CEBPA promotes the growth of cells, which suggests that CEBPA functions as a carcinogenic factor in HCC28,29. Here, we showed that CEBPA mRNA was increased and that the mRNA expression level of CEBPA was closely related to tumour stage in UCEC. High CEBPA expression predicted poor prognosis in patients with UCEC. The IHC sample images from the HPA database also indicated that CEBPA is usually strongly positive in UCEC (Figs. 1c and S1). More importantly, we found that the overexpression of CEPBA promoted the proliferation, migration and invasion of Ishikawa cells, whereas the knockdown of CEBPA suppressed the proliferation, migration and invasion of Ishikawa cells. These results indicate that CEBPA is more likely to function as a tumour promoter in UCEC. This contradiction with a previous study is probably due to its small sample size24.
Recently, several studies have reported that CEBPA is closely related to the regulation of immune cell function in various tumours. CEBPA participates in the occurrence and development of liver diseases through the regulation of CD4 + T cells or CD8 + T cells18,30. One study revealed that many immune cells are present in UCEC tissues24. However, the relationship between CEBPA and the immune microenvironment has not been studied sufficiently in UCEC. To further explore the function of CEBPA in UCEC, we analysed the genes whose expression was correlated with that of CEBPA. Surprisingly, the genes that were positively correlated with CEBPA were enriched in many terms associated with immune cell regulation, including “leukocyte-mediated immunity”, “regulation of immune effector process”, and “leukocyte cell–cell adhesion”. Furthermore, we found that CEBPA overexpression was significantly correlated with the infiltration of several types of immune cells, including M1 and M2 macrophages, in UCEC. Previous studies have shown that M1 macrophages play an antitumour role in the early stages of tumour development, whereas M2 macrophages play a protumour role during tumour progression31. Moreover, the number of M2 macrophages is positively correlated with clinical stage and lymphatic metastasis in UCEC32. Our results revealed that the infiltration level of M2 macrophages was greater than that of M1 macrophages in UCEC and that M2 macrophages were positively correlated with CEBPA. In addition, we found that CEBPA expression was positively correlated with the clinical stage of UCEC. Hence, these findings revealed that CEBPA may play a crucial role in the regulation of tumour immune cell infiltration in UCEC, and we speculated that CEBPA may function as an immune regulator in UCEC progression.
Historically, glycolysis was believed to be predominant in cancer, a phenomenon known as the “Warburg effect”. However, recent studies have revealed that many cancers, including UCEC, have increased OXPHOS33. OXPHOS leads to many advanced capabilities in cancer cells, including the ability to produce enough energy, participate in many facets of cancer progression, and even the involvement in immune escape34. We found that the genes that are positively correlated with CEBPA were also enriched in terms related to OXPHOS, which suggests that CEBPA may be involved in the regulation of OXPHOS in UCEC. Our results revealed that several genes encoding OXPHOS complexes, such as NDUFA1, NDUFB11, UQCRC1, and ATP6V1F, were regulated by CEBPA interference, which suggests that increased CEBPA expression may be associated with increased OXPHOS in UCEC. Targeting OXPHOS is a novel treatment strategy for cancer; therefore, CEBPA may be a target for cancer treatment, which relies on an anti-OXPHOS strategy.
By overlapping the CEBPA-correlated genes with target genes predicted in two transcription factor databases, we identified dozens of genes that may be target of CEBPA. Further analysis suggested that CEBPG, another number of the CCAAT/enhancer binding protein family, may be a functional target of CEBPA in UCEC. The function of CEBPG in the progression of UCEC has not been reported. In oesophageal squamous cell carcinoma, CEBPG promotes cancer progression by enhancing PI3K-AKT signalling35, which suggests a role for tumour promoter. In addition, CEBPG was reported to be negatively regulated by CEBPA in acute myeloid leukaemia36, which indicates a relationship between these two proteins, although our analysis suggested their positive regulation. Given that no gene overlap was observed between the genes that were negatively correlated with CEBPA and the predicted target genes, this suggests a transcriptional activation role of CEBPA in UCEC; it is therefore not surprising that CEBPA positively regulates CEBPG in UCEC. However, the way in which CEBPA regulates CEBPG and the function of CEBPG in UCEC require further investigation.
Conclusion
In summary, CEBPA expression was significantly associated with poor prognosis and immune cell infiltration in UCEC patients. Furthermore, CEBPA increased the oxidative phosphorylation level and promoted the proliferation, invasion and migration of Ishikawa cells. Therefore, CEBPA may be a potential therapeutic target in UCEC. Our study provides new insights into the underlying mechanisms and potential therapeutic targets for UCEC.
Supplementary Information
Acknowledgements
We acknowledge the support from the National Natural Science Foundation of China. We thank Prof. Huizhao Su and Prof. Yuanhua Huang for useful advice.
Author contributions
Ping Long, Jiaxing Wang and Lihong Pang provided financial support, designed the research and edited the manuscript. Jiaxing Wang, Weiyu Huang and Shiwei Chai conducted the experiments and analyzed the data. Jiaxing Wang wrote the manuscript. Jiayi Gan and Yulu Zeng helped cell culture and western blot experiments. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 81960281and No. 82260306), the Guizhou Basic Research Project (ZK [2023] General 456), the Guizhou Provincial Health Commission Science and Technology Fund (gzwkj2022-067), and the Open Project of NHC Key Laboratory of Thalassemia Medicine (NO. GJWJWDP202201).
Data availability
The datasets used and/or analysed during the present study are openly available in The Cancer Genome Atlas (TCGA) at https://portal.gdc.cancer.gov/, Genotype Tissue Expression (GTEx) at https://www.gtexportal.org/home/, and the HPA website at https://www.proteinatlas.org/. The original data or findings presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jiaxing Wang, Weiyu Huang and Shiwei Chai.
Contributor Information
Ping Long, Email: 1071545806@qq.com.
Lihong Pang, Email: panglihong@sr.gxmu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-74242-6.
References
- 1.Makker, V. et al. Endometrial cancer. Nat. Rev. Dis. Primers7(1), 88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eskander, R. N. & Powell, M. A. Immunotherapy as a treatment strategy in advanced stage and recurrent endometrial cancer: Review of current phase III immunotherapy clinical trials. Ther. Adv. Med. Oncol.13, 17588359211001200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer. J. Clin.71(3), 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Bokhman, J. V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol.15(1), 10–17 (1983). [DOI] [PubMed] [Google Scholar]
- 5.McCluggage, W. G., Naveena Singh, C. & Gilks, B. Key changes to the World Health Organization (WHO) classification of female genital tumours introduced in the 5th edition (2020). Histopathology80(5), 762–778 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Wild, P. J. et al. p53 suppresses type II endometrial carcinomas in mice and governs endometrial tumour aggressiveness in humans. EMBO Mol. Med.4(8), 808–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, L. W. et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov.1(2), 170–185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koschmieder, S., Halmos, B., Levantini, E. & Tenen, D. G. Dysregulation of the C/EBP alpha differentiation pathway in human cancer. J. Clin. Oncol.27(4), 619–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radomska, H. S. et al. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell Biol.18(7), 4301–4314 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagita, S., Uppalapati, S., Gundeti, S. & Digumarti, R. Correlation of C/EBPα expression with response and resistance to imatinib in chronic myeloid leukaemia. Jpn. J. Clin. Oncol.45(8), 749–754 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Zhang, G. et al. Overexpression of CCAAT enhancer-binding protein α inhibits the growth of K562 cells via the Foxo3a-bim pathway. Acta Haematol.136(2), 65–70 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Mackert, J. R. et al. Dual negative roles of C/EBPα in the expansion and pro-tumor functions of MDSCs. Sci Rep.7(1), 14048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato, A., Yamada, N., Ogawa, Y. & Ikegami, M. CCAAT/enhancer-binding protein-α suppresses lung tumor development in mice through the p38α MAP kinase pathway. PloS one.8(2), e57013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lourenço, A. R. et al. C/EBPɑ is crucial determinant of epithelial maintenance by preventing epithelial-to-mesenchymal transition. Nat. Commun.11(1), 785 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomizawa, M. et al. CCAAT/enhancer-binding protein α decreases the viability of gastric cancer cells. Oncol. Lett.13(6), 4322–4326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin, Y. et al. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol Cancer.19(1), 154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roepman, P. et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet.37(2), 182–186 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto, A. et al. Upregulation of C/EBPα inhibits suppressive activity of myeloid cells and potentiates antitumor response in mice and patients with cancer. Clin. Cancer Res.27(21), 5961–5978 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, X. et al. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theragnostic8(4), 1084–1105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue, M. et al. Upregulation of long non-coding RNA urothelial carcinoma associated 1 by CCAAT/enhancer binding protein α contributes to bladder cancer cell growth and reduced apoptosis. Oncol. Rep.31(5), 1993–2000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, Y. et al. Long non-coding RNA CEBPA-AS1 correlates with poor prognosis and promotes tumorigenesis via CEBPA/Bcl2 in oral squamous cell carcinoma. Cancer Biol. Ther.19(3), 205–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, L. et al. CCAAT enhancer binding protein α suppresses proliferation, metastasis, and epithelial-mesenchymal transition of ovarian cancer cells via suppressing the Wnt/β-catenin signaling. Neoplasma68(3), 602–612 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Pan, Z. et al. Down-regulation of the expression of CCAAT/enhancer binding protein α gene in cervical squamous cell carcinoma. BMC Cancer14, 417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takai, N. et al. Discovery of epigenetically masked tumor suppressor genes in endometrial cancer. Mol. Cancer Res.3(5), 261–269 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods12(5), 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giatromanolaki, A., Kouroupi, M., Kontomanolis, E. N. & Koukourakis, M. I. Regulatory tumor-infiltrating lymphocytes prevail in endometrial tumors with low vascular survival ability. Immunobiology226(3), 152078 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Yong, K. J. et al. Targeted BMI1 inhibition impairs tumor growth in lung adenocarcinomas with low CEBPα expression. Sci Transl Med.8(350), 350ra104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomizawa, M. et al. Reciprocal expression of CCAAT/enhancer binding proteins alpha and beta in hepatoblastomas and its prognostic significance. Oncol. Rep.17(2), 341–344 (2007). [PubMed] [Google Scholar]
- 29.Lu, G. D. et al. CCAAT/enhancer binding protein α predicts poorer prognosis and prevents energy starvation-induced cell death in hepatocellular carcinoma. Hepatology61(3), 965–978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, J. et al. C/EBPα/miR-7 controls CD4+ T-cell activation and function and orchestrates experimental autoimmune hepatitis in mice. Hepatology74(1), 379–396 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Gottlieb, C. E., Mills, A. M., Cross, J. V. & Ring, K. L. Tumor-associated macrophage expression of PD-L1 in implants of high grade serous ovarian carcinoma: A comparison of matched primary and metastatic tumors. Gynecol. Oncol.144(3), 607–612 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Cassetta, L. et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer cell.35(4), 588-602.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashton, T. M., McKenna, W. G., Kunz-Schughart, L. A. & Higgins, G. S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res.24(11), 2482–2490 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Qiu, X., Li, Y. & Zhang, Z. Crosstalk between oxidative phosphorylation and immune escape in cancer: a new concept of therapeutic targets selection. Cell Oncol. (Dordr).46(4), 847–865 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Huang, Y. et al. CEBPG promotes esophageal squamous cell carcinoma progression by enhancing PI3K-AKT signaling. Am. J. Cancer Res.10(10), 3328–3344 (2020). [PMC free article] [PubMed] [Google Scholar]
- 36.Alberich-Jordà, M. et al. C/EBPγ deregulation results in differentiation arrest in acute myeloid leukemia. J. Clin. Invest.122(12), 4490–4504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the present study are openly available in The Cancer Genome Atlas (TCGA) at https://portal.gdc.cancer.gov/, Genotype Tissue Expression (GTEx) at https://www.gtexportal.org/home/, and the HPA website at https://www.proteinatlas.org/. The original data or findings presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.