Abstract
We have previously shown that the RNA polymerase 3Dpol of human rhinovirus 2 (HRV2) catalyzes the covalent linkage of UMP to the terminal protein (VPg) using poly(A) as a template (K. Gerber, E. Wimmer, and A. V. Paul, J. Virol. 75:10969–10978, 2001). The products of this in vitro reaction are VPgpU, VPgpUpU, and VPg-poly(U), the 5′ end of minus-strand RNA. In the present study we used an assay system developed for poliovirus 3Dpol (A. V. Paul, E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer, J. Virol. 74: 10359–10370, 2000) to search for a viral sequence or structure in HRV2 RNA that would provide specificity to this reaction. We now show that a small hairpin in HRV2 RNA [cre(2A)], located in the coding sequence of 2Apro, serves as the primary template for HRV2 3Dpol in the uridylylation of HRV2 VPg, yielding VPgpU and VPgpUpU. The in vitro reaction is strongly stimulated by the addition of purified HRV2 3CDpro. Our analyses suggest that HRV2 3Dpol uses a “slide-back” mechanism during synthesis of the VPg-linked precursors. The corresponding cis- replicating RNA elements in the 2CATPase coding region of poliovirus type 1 Mahoney (I. Goodfellow, Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans, J. Virol. 74:4590–4600, 2000) and VP1 of HRV14 (K. L. McKnight and S. M. Lemon, RNA 4:1569–1584, 1998) can be functionally exchanged in the assay with cre(2A) of HRV2. Mutations of either the first or the second A in the conserved A1A2A3CA sequence in the loop of HRV2 cre(2A) abolished both viral growth and the RNA's ability to serve as a template in the in vitro VPg uridylylation reaction.
Human rhinoviruses (HRVs) comprise the largest genus (Rhinovirus) in the Picornaviridae family of plus-strand RNA viruses; the latter contain many different human and animal pathogens (50). Rhinoviruses are divided into a major (about 90 serotypes) and a minor (10 serotypes, including HRV2) group, depending on their binding to ICAM-1 (human intercellular adhesion molecule 1) or to members of the LDLR (low-density lipoprotein receptor) receptors, respec tively (25, 57). These viruses are the major causative agents of the common cold, for which there is no effective treatment (19, 49). Development of vaccines has not been successful due to the large number of serotypes and the poor cross-protection between the serotypes.
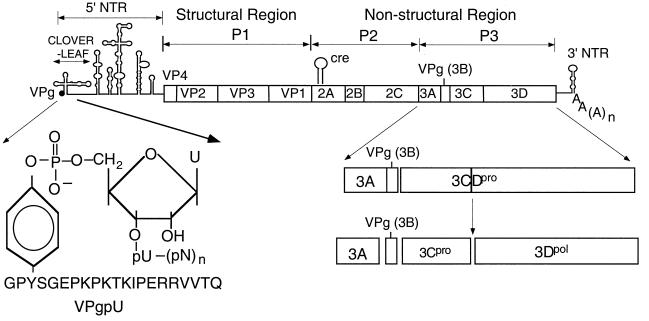
The RNA genome of picornaviruses, including that of HRV2 (7,151 nucleotide [nt]), contains a long 5′ nontranslated region (5′ NTR), a single open reading frame, a short 3′ NTR, and a poly(A) tail (Fig. 1) (28, 55). The 5′-terminal UMP of the RNA is linked in a phosphodiester bond to the O4-hydroxyl group of a tyrosine in the terminal protein VPg (Fig. 1) (2, 31, 48). After entry into the host cell, the viral RNA functions as mRNA directing the synthesis of a large polyprotein that consists of one structural (P1) and two nonstructural (P2 and P3) domains (Fig. 1) (28, 55). The polyprotein is proteolytically cleaved into precursor and mature polypeptides by virus-encoded proteinases 2Apro, 3Cpro, and 3CDpro (reviewed in reference 22). RNA replication takes place in the cytoplasm of the infected host cell on membranous vesicles (8, 29, 66), and the enzyme primarily responsible for RNA synthesis is the viral RNA-dependent RNA polymerase 3Dpol (15, 17, 29, 66). The parental RNA is first transcribed into a minus strand, which in turn is used as a template for the production of the progeny plus strands. Finally, the progeny viral RNA is encapsidated and released from the host cell.
FIG. 1.
Structure of HRV2 genomic RNA. The single-stranded RNA genome of HRV2 is shown with the terminal protein VPg (3B) at the 5′ end of the 5′ NTR. The 3′ NTR is shown with the poly(A) tail. The 5′ NTR consists of a cloverleaf and a large IRES element. The site of attachment of the 5′-terminal UMP of the RNA to the tyrosine of HRV2 VPg is shown enlarged. The polyprotein contains structural (P1) and nonstructural (P2 and P3) domains. Vertical lines within the polyprotein box represent proteinase cleavage sites. Processing of the P3 domain by proteinase 3CDpro is shown enlarged. The location of the cis-replicating element (cre) in the 2Apro coding region is indicated.
Many details of picornaviral minus- and plus-strand RNA replication remain unsolved, particularly those concerning the functions of cis-replicating RNA elements (cre) (reviewed in references 1 and 64). These include the 5′ NTR, the 3′ NTR with the poly(A) tail, and RNA structures located within the coding sequences of the genomes. Most of the information available at this time has been derived from studies of poliovirus, a member of the Enterovirus genus and a prototype of Picornaviridae. The 5′ NTR of entero- and rhinoviruses contains three independent domains: the terminal protein VPg (2, 31), a cloverleaf-like structure containing about 100 nt, and the large internal ribosomal entry site (IRES) (Fig. 1) (10, 27, 43, 46). In the case of poliovirus, the cloverleaf RNA forms a complex with viral proteinase 3CDpro in the presence of either 3AB (23, 65) or the cellular protein PCBP2 (3, 4, 39). These RNPs have been shown to play a role in plus-strand RNA synthesis (3, 4, 23, 39, 65). The 3CDpro/PCBP2 complex has recently been proposed to have an additional function in promoting the switch from translation to replication (16). The primary function of the IRES is to control translation (10, 27, 43), but mutations in this structure also appear to have deleterious effects on RNA replication (11, 53).
The picornaviral 3′ NTR consists of a structured heteropolymeric sequence followed by a poly(A) tail. Numerous biochemical and genetic analyses (1, 34, 35, 44, 64) have demonstrated the importance of the structure of this RNA element in replication. The 3′ NTR therefore was designated as the origin of replication (oriR) for minus-strand RNA synthesis (44). Subsequent studies, however, have been difficult to reconcile with this hypothesis. It was observed that a chimeric poliovirus containing the 3′ NTR of HRV14 or coxsackievirus B4 (CBV4) replicated with nearly normal kinetics (47). In addition, it was found that not even the deletion of the entire poliovirus type 1 Mahoney [PV1(M)] 3′ NTR fully abolished RNA replication (59).
Within the last few years a cis-replicating element was discovered within the coding sequences of several picornaviruses. First it was demonstrated that a small RNA hairpin in the coding sequence of the HRV14 capsid protein VP1 is required for minus-strand RNA synthesis (33). Similar elements were later found in the VP2 coding sequences of mengovirus and Theiler's virus (32) and in the 2CATPase region of poliovirus (18). The internally located cis-replicating elements of these picornaviruses all consist of a simple hairpin structure with widely different nucleotide sequences (18, 32, 33, 42, 45) except for a conserved AAACA motif in the loop (42, 45).
The question of how picornaviral RNA synthesis is initiated has been the subject of numerous studies during the past three decades. The finding of VPg linked to the terminal UMP of both plus and minus strands (37), and the discovery of VPgpUpU in poliovirus-infected cells (12) led to the suggestion that VPg serves as the primer for RNA synthesis (63). This idea was supported by the finding that crude replication complexes isolated from PV1(M) (58, 60) or encephalomyocarditis virus (EMCV)-infected cells (62) could synthesize VPgpU and VPgpUpU. Studies with poliovirus 3Dpol variants implicated this protein as the enzyme responsible for the covalent linkage of UMP to VPg (60). We have recently confirmed this proposal by showing that PV1(M) 3Dpol is able to catalyze in vitro the synthesis of VPgpU, VPgpUpU, and VPg-poly(U) on a poly(A) template (41). At first glance this reaction appears to represent the initiation of minus-strand synthesis, that is, the synthesis of VPg-linked poly(U). However, the homopolymeric template used in this reaction clearly does not contain the specific RNA signals required by the viral RNA polymerase for the recognition of its own RNA. In our search for an RNA sequence or structure in poliovirus RNA that would provide the RNA polymerase with a site of recognition, we have discovered that the first two A's in the A1A2A3CA sequence of PV1(M) cre(2C) RNA, and not the poly(A) tail, serve as the primary template for the synthesis of VPgpU and VPgpUpU (42, 45).
In our accompanying paper we have described a reaction in which purified HRV2 RNA polymerase 3Dpol catalyzes the uridylylation of its cognate VPg on a poly(A) template (17). Using poliovirus 3Dpol as a model system, we have now extended our studies with HRV2 3Dpol to include HRV2 VPg uridylylation on HRV2-specific RNA templates. We have developed an assay with which we were able to identify a cis-replicating element in the coding sequence of protein 2Apro of HRV2. This small RNA hairpin functions as the template for HRV2 3Dpol in the synthesis of VPgpU and VPgpUpU, and the reaction is strongly stimulated by the addition of HRV2 protein 3CDpro. We provide evidence for the importance of the first two A's of the A1A2A3CA motif in the loop of the cre(2A) RNA by correlating the growth properties of mutant genomes in vivo with the ability of the cre element to function as a template in the in vitro VPg uridylylation reaction.
MATERIALS AND METHODS
Construction of plasmids.
pT7HRV2 (13), pT7HRV 14 (56), and pT7PV1(M) (61) refer to the full-length plus-strand HRV2, HRV14, and PV1(M) cDNAs, respectively. Mutated nucleotides in the oligonucleotide sequences are shown underlined. Site-directed mutagenesis was carried out using the QuickChange mutagenesis kit from Stratagene. Mutations and constructs were confirmed by sequencing using the ABI Prism DNA sequencing kit.
(i) pProEX HTb/3CDpro (3Cpro C147A).
The C147A mutation was introduced into pT7HRV2 with primers 5′-CCAACTAAATCTGGTTACGCTGGAGGTGTCTTATAC-3′ (plus-strand sequence) and 5′-GTATAAGACACCTCCAGCGTAACCAGATTTAGTTGG-3′ (minus-strand sequence). The coding sequence of HRV2 3CDpro (3Cpro C147A) was amplified from the mutagenized plasmid pT7HRV2(3CproC147A) by PCR using primers 5′-CCGGATCCGGACCAGAGGAGGAATTTG-3′ (plus-strand sequence) and 5′-GGGCTCGAGTTAAAATTTTTCATACCACTC-3′ (minus-strand sequence) and Pfu polymerase. The resulting fragment was cut with BamHI and XhoI and ligated into the same sites of pProEX HTb (Lifetech).
(ii) pProEX HTb/3CDpro(3Cpro C147A/R84S).
Mutation R84S was introduced into pProEX HTb/3CDpro(3CproC147A) by site-directed mutagenesis using primers 5′-GAAATGAAAAATTTAGCGATATCAGGAGATATATACC-3′ (plus-strand sequence) and 5′-GGTATATATCTCCTGATATCGCTAAATTTTTCATTTC-3′ (minus-strand sequence).
(iii) pT7HRV2 cre(2A)(A1C).
An A-to-C mutation (nt 3283) was introduced into pT7HRV2 by site-directed mutagenesis using primers 5′-CATTTACCGA ACCAACACTGTAGGTGATGATTACATTCCC-3′ (plus-strand sequence) and 5′-GGGAATGTAATCATCACCTACAGTGTTGGTTCGGTAAATG-3′ (minus-strand sequence). Plasmid pT7HRV2 (cre2A)(A1C) was cut with ApaI and AgeI, and the resulting fragment was ligated into similarly restricted pT7HRV2.
(iv) pT7HRV2 cre (2A)(A2C).
An A-to-C mutation (nt 3284) was introduced into pT7HRV2 using primers 5′-CATTTACCGAACACACACTGTAGGTGATGATTACATTCCC-3′ (plus-strand sequence) and 5′-GGGAATGTAATCATCACCTACAGTGTGTGTTCGGTAAATG-3′ (minus-strand sequence). Plasmid pT7HRV2 cre(2A)(A2C) was cut with ApaI and AgeI, and the resulting fragment was inserted into the ApaI/AgeI sites of pT7HRV2.
(v) pT7HRV2 VPg(Y3F).
An A-to-T mutation (nt 5076) was introduced into pT7HRV2 by site-directed mutagenesis using primers 5′-GCACATTACAGGGACCATTTTCAGGAGAACCAAAGCCC-3′ (plus-strand sequence) and 5′-GGGCTTTGGTTCTCCTGAAAATGGTCCCTGTAATGTGC-3′ (minus-strand sequence). Plasmid pT7HRV2 VPg(Y3F) was cut with NheI and SphI, and the resulting fragment (1,541 nt) was ligated into similarly cut pT7HRV2.
RNA transcription.
For the production of RNA transcripts in vitro, 1.5 μg of wild-type (wt) or mutant full-length HRV2 cDNAs were linearized with the selected restriction enzyme and transcribed with T7 RNA polymerase. HRV2 RNA, HRV14 RNA, and PV1(M) RNA refer to the RNA generated by in vitro transcription of the corresponding cDNAs linearized at the end of the genome and transcribed with T7 RNA polymerase.
Nested set of pT7HRV2 RNA templates.
The pT7HRV2 cDNA was cut with any one of the following enzymes: EcoRI (nt 743), BssHII (nt 1581), SpeI (nt 2435), NsiI (nt 2536), EcoNI (nt 2655), AgeI (nt 2760), PvuII (nt 3160), ApaI (nt 3454), BsmI (nt 4952), AflII (nt 6638), and KpnI (nt 7153). The resulting fragments of different lengths, containing a T7 promoter at the 5′ end, were used as templates for RNA transcription by T7 RNA polymerase. The DNA template for transcription of the pT7HRV2 sequence nt 3309 to 5187 was obtained through amplification by PCR using primers 5′-GCGAAGCTTAATACGACTCACTATAGGGCCTCTTGTGATTGTACC-3′ (plus-strand sequence with a T7 promoter) and 5′-TGTAATAACACATGAG-3′ (minus-strand sequence). The template for transcription of the pT7HRV2 sequence nt 5118 to 7062 was obtained through amplification by PCR using primers 5′-CCCGGCCTAATACGACTCACTATAGGG-3′ (plus-strand sequence with a T7 promoter) and 5′-CCGAATTCTTAAAATTTTTCATACCACTC-3′ (minus-strand sequence).
Predicted HRV2 RNA stem-loop structures containing an A1A2A3CA motif.
Using the RNA folding program of M. Zucker (http://bioinfo.math.rpi.edu/∼mfold/rna/), stable RNA stem-loop structures containing the A1A2A3CA motif in the loop region were selected from the HRV2 genome and amplified by PCR using the primers shown in Table 1 (first nine rows).
TABLE 1.
Oligonucleotide primers used for the construction of HRV2 RNA stem-loop structures containing an AAACA motif
| No. | Location (nt) | Template | Primer sequencea |
|---|---|---|---|
| 1 | 2915–2955 | pT7 HRV2 | 5′-CGG CCG TAA TAC GAC TCA CTA TAG GGG GAT CAA AAC TAT GGT ACA GC-3′ (+) |
| 5′-GGG GGA CCC CAT GTT ATT TGT G-3′ (*) | |||
| 2 | 2960–2998 | pT7 HRV2 | 5′-CGG CCG TAA TAC GAC TCA CTA TAG GGG GTG CTC TAG GAT AGT AAC AGA G-3′ (+) |
| 5′-GGG GGT ACT TTA TGA ATG TG-3′ (*) | |||
| 3 | 3309–3382 | pT7 HRV2 | 5′-GCG AAG CTT AAT ACG ACT CAC TAT AGG GCC TCT TGT GAT TGT ACC-3′ (+) |
| 5′-GCG CTG CAG GCT TGT AAC TGT AAT TGG G-3′ (*) | |||
| 4 | 3272–3298 | pT7 HRV2 | 5′-CGG CCG TAA TAC GAC TCA CTA TAG GGA TTT ACC GAA CAA ACA C-3′ (+) |
| 5′-GGG ATC ACC TAC AGT GTT TG-3′ (*) | |||
| 5 | 3263–3310 | pT7 HRV2 | 5′-CGG CCG TAA TAC GAC TCA CTA TAC GGG CCG ATT TAA TCA TTT ACC G-3′ (+) |
| 5′-GGG AAT GTA ATC ATC ACC-3′ (*) | |||
| 6 | 3257–3318 | pT7 HRV2 | 5′-CGG CCG TAA TAC GAC TCA CTA TAC GGC CTC ATC AGA TTT AAT CAT TTA C-3′ (+) |
| 5′-CGG TCA CAA GAG GGA ATG TAA TC-3′ (*) | |||
| 7 | 3263–3310 | pT7 HRV2 cre(2A) (A1C) | Same as for no. 5 |
| 8 | 3263–3310 | pT7 HRV2 cre(2A) (A2C) | Same as for no. 5 |
| 9 | 5118–5187 | pT7 HRV2 | 5′-CCC GGC CTA ATA CGA CTC ACT ATA GGG CCT GTA GTA ACA CAG GGA CC-3′ (+) |
| 5′-TGT AAT AAC ACA TGA G-3′ (*) | |||
| 10 | 2318–2414 | pT7 HRV14 | 5′-GGG CCG GCT AAT ACG ACT CAC TAT AGG CAC TCA CTG AAG GCT TAG G-3′ (+) |
| 5′-GGG AAT TCA CGC GTG TGT GTT TTG GAC CAG ATG AG-3′ (*) | |||
| 11 | 4444–4504 | pT7 PV1(M) | 5′-GGG CCG GCT AAT ACG ACT CAC TAT AGG CTC GAG CTA ACT ATT AAC-3′ (+) |
| 5′-CCG AAT TCG CTA GCA AAC ATA CTG GTT C-3′ (*) |
+, plus-strand sequence; *, minus-strand sequence. The T7 promoter site is shown in italics.
Transfection and translation.
RNA transfections were carried out by the DEAE-dextran method as decribed previously (61) with the following exceptions. After transfection of HeLa H1 monolayers in 35-mm plates, cells were incubated at 33.5°C in 1× Dulbecco's modified Eagle medium (Gibco) containing 5% fetal bovine serum, 1× penicillin-streptomycin (Gibco), and 6 mM MgCl2. Three days after transfection, the virus yield in each supernatant was determined by a standard plaque assay (36) containing 6 mM MgCl2 and 5% fetal bovine serum. Plates were incubated at 33.5°C for 3 days. Conditions for in vitro translation have been described previously (36). All mutant RNAs, used for transfections, were first translated to ensure the presence of an intact open reading frame in the genomes (data not shown).
Peptides and proteins. (i) VPg peptides.
Synthetic VPg peptides were a generous gift from J. H. van Boom (41, 42, 45).
(ii) HRV2 3Dpol.
HRV2 3Dpol was expressed in Escherichia coli strain BL21(DE3)pLysS from plasmid pT5T-HRV2 3D (17), which contains the wt 3D coding sequences preceded by a methionine, and was purified as described previously (40).
(iii) Control proteins.
Control proteins for HRV2 3Dpol were purified like HRV2 3Dpol, with the exception that the expression vector pT5T did not contain the 3D coding sequences (17).
(iv) GST-HRV2 3Dpol.
HRV2 3Dpol with an N-terminal glutathione S-transferase (GST) tag of 26 kDa was expressed in E. coli strain BL21(DE3)pLysS and purified as described previously (17).
(v) His-HRV2 3CDpro(3Cpro C147A).
In these studies we used 3CDpro with an active-site mutation in 3Cpro(C174A) in order to prevent the autoprocessing of the polypeptide. For the sake of simplicity in our discussion, we will call this form of the protein simply 3CDpro. The protein was expressed in E. coli BL21(DE3)pLysS cells from pProEx HTb/HRV2 3CDpro(3CproC147A) in 750 ml of Luria broth with ampicillin at 20°C for 4 h in the presence of 0.02 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were pelleted, resuspended in 40 ml of buffer (50 mM Tris [pH 8.0], 100 mM NaCl, 10 mM dithiothreitol [DTT], 1 mM EDTA, 5% glycerol), and lysed by sonication (with a Branson Sonifier). Cell debris was pelleted, and the proteins from the supernatant were precipitated with 50% ammonium sulfate. The precipitate was resuspended in 3 ml of dialysis buffer (50 mM Tris[pH 8.0], 30 mM NaCl, 1 mM DTT, 1 mM EDTA, 5% glycerol) and was dialyzed against the same buffer at 4°C. The pellet of the sonicated lysate was also resuspended in 3 ml of buffer (50 mM Tris [pH 8.0], 1 mM NaCl, 10 mM DTT, 1 mM EDTA, 5% glycerol, 1% Triton) and incubated with rotation at 4°C for 30 min. The insoluble cell debris was pelleted, and the supernatant was dialyzed in dialysis buffer. Then both of the dialyzed fractions were dialyzed against binding buffer (20 mM Tris [pH 8.5] at 4°C, 100 mM KCl, 10 mM 2-mercaptoethanol, 10% glycerol, 10 mM imidazole, 0.5% Triton) for 7 h at 4°C, filtered, and loaded together onto a Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen). The column was washed with 20 ml of binding buffer, and the His-HRV2 3CDpro(3CproC147A) was eluted with 20 mM Tris (pH 8.5), 100 mM KCl, 10 mM 2-mercaptoethanol, 10% glycerol, and 100 mM imidazole in 500-μl fractions (4°C). The protein concentration was measured (Bio-Rad), and the peak fractions were pooled and dialyzed against 500 ml of buffer (20 mM Tris [pH 7.5], 10 mM NaCl, 2 mM DTT, 5% glycerol) at 4°C. The eluate was run on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE gel) (10% polyacrylamide), and the location of the 70.6-kDa his-HRV2 3CDpro was confirmed by Western blotting with an anti-his antibody (data not shown).
(vi) His-HRV2 3CDpro(3Cpro C147A/R84S).
His-HRV2 3CDpro(3Cpro C147A/R84S) was expressed and purified as described above for His-HRV2 3CDpro(3CproC147A).
RNA-dependent RNA polymerase assays. (i) Assay A: HRV2 VPgpU(pU) synthesis on a poly(A) template by HRV2 3Dpol.
The assay was essentially the same as that described previously (17). The standard reaction mixture (20 μl) for HRV2 VPg uridylylation by HRV2 3Dpol contained 50 mM HEPES (pH 7.6), 0.2 mM manganese (II) acetate (MnAc2; Aldrich), 8% glycerol, 0.5 μg (0.35 μM) of poly(A) RNA template (∼200 nt) (Pharmacia), 2 μg of synthetic HRV2 VPg, 1 μg (1 μM) of purified HRV2 3Dpol, and 1 μCi of [α-32P]UTP (0.017 μM; 3,000 Ci/mmol; Dupont, NEN). Samples were incubated for 1 h at 30°C, reactions were stopped by addition of 5 μl of gel-loading buffer (Bio-Rad), and products were analyzed on Tris–tricine–SDS-PAGE gels (Bio-Rad) with 13.5% polyacrylamide. Gels were dried at 68°C for 2 h without fixing and were autoradiographed for 1 h (Kodak Biomax MS film). Reaction products were quantitated with a PhosphorImager (Molecular Dynamics, Storm 860) by measuring the amount of [α-32P]UMP incorporated into VPgpU(pU) and VPg-poly(U) products. Data were translated into counts per minute with the help of a radioactive marker. VPgpU(pU) refers to the sum of VPgpU and VPgpUpU.
The assay with GST-HRV2 3Dpol was similar to that described above, except that the standard reaction mixture contained 0.05 mM MnAc2.
Assay B: HRV2 VPgpU(pU) synthesis on viral RNA templates by HRV2 3Dpol and GST-HRV2 3Dpol.
The assay was similar to that described for HRV2 VPgpU(pU) synthesis on a poly(A) template (assay A), except that the standard reaction mixture contained 0.5 μg of purified His-HRV2 3CDpro(C147A), 1 mM magnesium acetate (MgAc) instead of MnAc2, and 0.5 μg of HRV2 RNA template [full length or cre(2A)] or other viral RNA templates instead of poly(A).
RESULTS
We have previously shown that both the RNA polymerase 3Dpol of PV1(M) and that of HRV2 catalyze in vitro the uridylylation of their cognate VPgs and synthesis of VPg-poly(U) on a poly(A) template (17, 41, 42): 3Dpol + VPg + poly(A) + Mn2+(Mg2+) + UTP → VPgpU + VPgpUpU → VPg-poly(U). Recently we have developed a new and more relevant assay for the uridylylation of PV1(M) VPg by PV1(M) 3Dpol in which the nonspecific poly(A) template is replaced by transcripts of poliovirus RNA and Mg2+ replaces Mn2+ as a cofactor (42): 3Dpol+ VPg + PV1(M) RNA + 3CDpro + Mg2+ + UTP → VPgpU +VPgpUpU. This reaction is strongly stimulated by addition of purified PV1(M) 3CDpro. To find out whether HRV2 3Dpol also catalyzes the same kind of synthetic reaction, we have tested the enzyme using its cognate RNA and VPg as the template and substrate, respectively, and analyzed the effect of purified HRV2 3CDpro on the reaction.
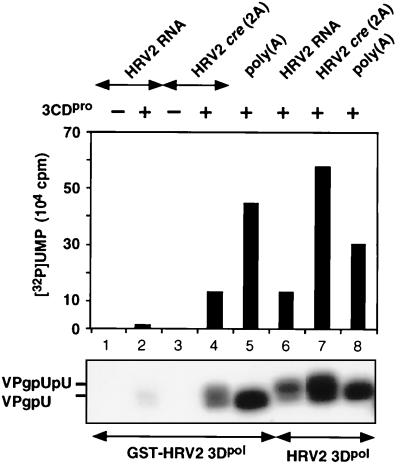
Comparison of VPg uridylylation by HRV2 3Dpol on poly(A) and HRV2 RNA templates.
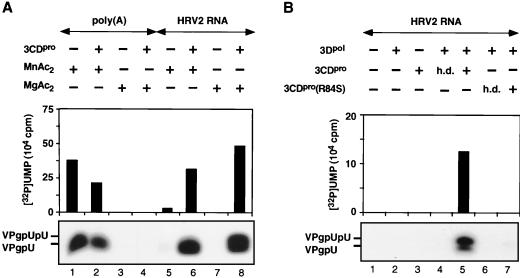
HRV2 3Dpol has only barely detectable HRV2 VPg-uridylylating activity on a poly(A) template with Mg2+ as a cofactor, but it is highly active in the presence of Mn2+ (Fig. 2A; compare lanes 1 and 3). Purified HRV2 3CDpro has no effect on these reactions in the presence of either metal activators (Fig. 2A; compare lane 1 with lane 2 and lane 3 with lane 4). In contrast, when HRV2 RNA is used as a template and Mg2+ is used as a cofactor, there is a very strong stimulation of the reaction by 3CDpro (Fig. 2A; compare lanes 7 and 8), suggesting that under these conditions a viral RNA sequence or structure is being recognized. To a lesser extent the stimulatory effect of 3CDpro can also be seen when the reaction mixture contains Mn2+ (Fig. 2A; compare lanes 5 and 6). Since 3CDpro is the precursor of 3Dpol, we have checked the possibility that this polypeptide also possesses VPg-uridylylating activity. The data clearly indicate that no products are formed when 3Dpol is omitted from the reaction (Fig. 2B; compare lanes 3 and 5), suggesting that 3CDpro only enhances the reaction catalyzed by 3Dpol. We cannot, however, rule out the possibility that 3CDpro possesses activity that is dependent on 3Dpol as a cofactor. A heat-denatured aliquot of 3CDpro (Fig. 2B; compare lanes 4 and 5) has no effect on the reaction, an observation demonstrating that the protein itself is the active component in the enzyme preparation. It should be noted that a catalytically inactive 3CDpro(3Cpro [C147A]) was used in all these experiments to prevent autoprocessing of this precursor to 3Cpro and 3Dpol.
FIG. 2.
(A) Effect of 3CDpro on the uridylylation of VPg by HRV2 3Dpol with either Mn2+ or Mg2+ as a cofactor. The uridylylation of HRV2 VPg was measured by the standard assays for poly(A) or HRV2 RNA templates as described in Materials and Methods (assays A and B, respectively), with either Mn2+ or Mg2+ as a cofactor. Where indicated, HRV2 3CDpro was added to the samples. (B) Effect of 3CDpro or mutant 3CDpro(3Cpro[R84S]) on the uridylylation of VPg by HRV2 3Dpol on a HRV2 RNA template. The uridylylation of HRV2 VPg was measured by the standard assay as described in Materials and Methods (assay B). Where indicated, HRV2 3CDpro or 3CDpro(3Cpro[R84S]) was added to the reaction mixtures in either an active or a heat-denatured (h.d.) form. Both HRV2 3CDpro and HRV2 3CDpro(3Cpro[R84S]) contained a C147A mutation. Sample 3 contained 3CDpro, but 3Dpol was omitted. Both autoradiography of the reaction products and quantitation of the data are shown.
Numerous studies with poliovirus 3CDpro have shown that its 3Cpro domain possesses both the proteinase and RNA binding functions of the polypeptide. Mutations in the RNA binding domain (R84S or I86A) of PV1(M) 3Cpro, which lead to a nonviable phenotype, have no deleterious effect on polyprotein processing (4, 9, 20). We have previously observed that the R84S and I86A mutations in the 3Cpro domain of poliovirus 3CDpro interfered with its ability to stimulate the protein-priming reaction (42). The 3Cpro polypeptides of poliovirus and HRV2 share 35% amino acid identity (55), and they contain a conserved arginine at position 84. Based on the assumption that the RNA binding role of R84 is the same in the 3CDpro of HRV2 as in the 3CDpro of poliovirus, we have tested the stimulatory effect of HRV2 3CDpro(3Cpro[C147A/R84S]) on the uridylylation of HRV2 VPg. A comparison of the effects of wt and mutant HRV2 3CDpro(3Cpro[C147A/R84S]) (Fig. 2B; compare lanes 5 and 7) indicates that the mutation in 3Cpro, predicted to abolish its RNA binding, also eliminates its stimulatory effect. These results are in agreement with our previous conclusion about poliovirus 3CDpro, namely, that the RNA binding activity of this protein is essential for its function in the protein-priming reaction (42).
Identification of HRV2 cre(2A).
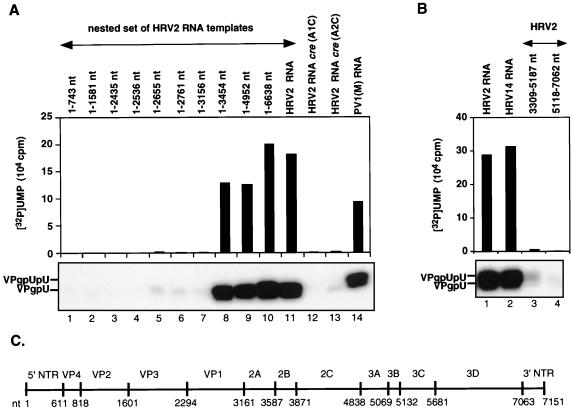
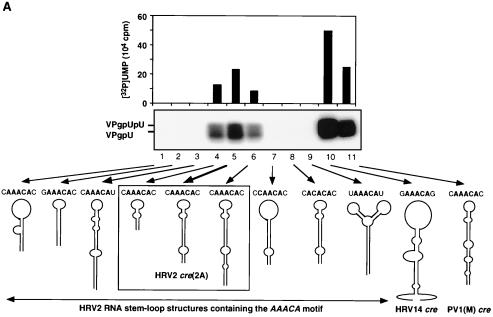
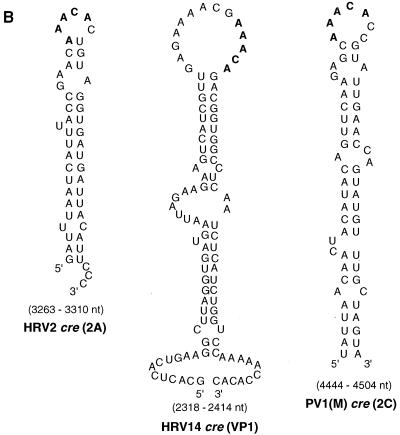
We have recently identified an RNA stem-loop in the coding region of poliovirus 2CATPase [cre(2C)] (18) as the primary template for the synthesis of VPgpU(pU) by PV1(M) 3Dpol in the in vitro reaction (42). This cis-replicating element, along with other recently identified RNA hairpins in the coding regions of picornaviruses (18, 32, 33, 42, 45), contains a conserved sequence of A1A2A3CA in the loop (42, 45). We used two different approaches to locate the corresponding element in the genome of HRV2. First, we made a nested set of in vitro transcripts of HRV2 RNAs, containing various 3′-terminal deletions. These were tested as templates in the HRV2 VPg uridylylation reaction catalyzed by HRV2 3Dpol in the presence of its cognate 3CDpro. As shown in Fig. 3A, transcripts that contained 5′-terminal RNA up to and including HRV2 capsid sequences had no template activities (Fig. 3A; compare lanes 1 to 7 with lanes 8 to 11). Similarly, transcripts that contained sequences downstream from the middle of 2Apro to the end of the 3Dpol coding sequences were inactive as templates in the reaction (Fig. 3B; compare lane 1 with lanes 3 and 4). These results suggested a location for the template nucleotides in 2Apro between nt 3156 and 3309. In the second approach we used an RNA folding program to search for stable stem-loop structures in HRV2 RNA that contained the conserved A1A2A3CA sequence in the loop. Transcripts of these predicted RNA hairpins were tested as templates in the in vitro uridylylation assay using purified HRV2 proteins 3Dpol, 3CDpro, and VPg. Of the five structures selected, two were located in the VP1 domain, two in 2Apro, and one in 3Cpro. None of these transcript RNAs (Fig. 4A, lanes 1 to 3 and 9), except the hairpin in 2Apro, labeled HRV2 cre (2A) (Fig. 4A, lane 5), functioned as a template in the in vitro reaction. Shorter (Fig. 4A, lane 4) or longer (Fig. 4A, lane 6) versions of the RNA element yielded reduced levels of products compared to wt HRV2 cre (2A) (Fig. 4A, lane 5).
FIG. 3.
(A) Nested set of HRV2 RNAs as templates for uridylylation of VPg by HRV2 3Dpol. Uridylylation of HRV2 VPg was measured by the standard assay as described in Materials and Methods (assay B). The lengths of the HRV2 RNA templates used in the reactions were varied as indicated at the top. At the bottom, the autoradiography of the reaction products is shown; above it, the quantitation of the data is displayed. Lanes 1 to 10, nested set of HRV2 RNAs; lane 11, full-length HRV2 RNA; lanes 12 and 13, full-length HRV2 RNA cre(2A)(A1C) and (A2C) with an A-to-C mutation at nt 3283 or 3284 in the conserved A1A2A3CA sequence of the cre, yielding C1A2A3CA (lane 12) and A1C2A3CA (lane 13), respectively; lane 14, full-length PV1(M) RNA. (B) Full-length and shortened viral RNAs as templates in HRV2 VPg uridylylation. Assay B (Materials and Methods) was used to measure HRV2 VPgpU(pU) synthesis except that the RNA templates were varied. Lane 1, full-length HRV2 RNA; lane 2, full-length HRV14 RNA; lanes 3 and 4, HRV2 RNA fragments (nt 3309 to 5187 and nt 5118 to 7062, respectively). (C) Schematic presentation of the HRV2 genome (nt 1 to 7151). Vertical bars indicate the 5′-terminal nucleotide of each segment.
FIG. 4.
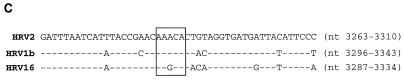
(A) Different HRV2 RNA stem-loop structures, containing the wt or mutant A1A2A3CA sequence motif, as templates for uridylylation of HRV2 VPg by HRV2 3Dpol. With the help of the computer based RNA folding program of Zucker (http://bioinfo.math.rpi.edu/∼mfold/rna/), stable RNA stem-loop structures with the sequence A1A2A3CA in the loop region were selected from the HRV2 RNA genome. Transcripts of these RNAs were made and used as templates for the uridylylation of HRV2 VPg by HRV2 3Dpol in the presence of HRV2 3CDpro, as described in Materials and Methods (assay B). A schematic drawing of the stem-loop structures with the A1A2A3CA sequence and the neighboring nucleotides is shown. The autoradiograph shows results for HRV2 RNA stem-loop structures located in the coding region of VP1 (nt 2915 to 2955 [lane 1] and nt 2960 to 2998 [lane 2]), 2Apro (nt 3309 to 3382 [lane 3]), and 3Cpro (nt 5118 to 5187 [lane 9]); full-length HRV2 cre(2A) (nt 3263 to 3310) (lane 5) and HRV2 cre(2A) with different stem lengths (nt 3272 to 3298 [lane 4] and nt 3257 to 3318 [lane 6]); HRV2 cre(2A) with mutations in the conserved A1A2A3CA sequence, yielding C1A2A3CA (lane 7) and A1C2A3CA (lane 8); HRV14 cre(VP1) (nt 2318 to 2414) (lane 10); and PV1(M) cre(2C) (nt 4444 to 4504) (lane 11). Quantitation of the data is displayed above the autoradiograph. (B) Predicted secondary structure of the proposed HRV2 cre(2A) compared to the HRV14 cre(VP1) and PV1(M) cre(2C) RNAs. Sequences conserved among known picornavirus cis-replicating elements are shown in boldface. Also see Addendum in Proof. (C) Relationship of HRV2, HRV1b, and HRV16 as indicated by a comparison of the cre(2A) domain. The sequence of 48 nt in the cre(2A) domain of HRV2 (55) is compared to similar sequences in the genomes of HRV1b (26) and HRV16 (30). Nucleotide numbers given on the right indicate the locations of the predicted HRV1b and HRV16 cre(2A) structures in the viral genomes. Predicted conserved motifs are boxed.
The HRV2 cre (2A) RNA forms a stable hairpin structure consisting of about 47 nt (Fig. 4B). It contains the same conserved A1A2A3CA sequence in the loop as the larger cis-replicating elements of either PV1(M) or HRV14 (Fig. 4B) (18, 33). Two HRVs that are closely related to HRV2, HRV1b (26) and HRV16 (30), are predicted to contain a similar element, at essentially the same location in their genomes (Fig. 4C). On the other hand, HRV89 (14), another relative of HRV2, does not appear to possess a similar RNA structure at or near this location (K. Gerber, E. Wimmer, and A. V. Paul, unpublished data).
Surprisingly, the RNA polymerase of HRV2, in the presence of its cognate 3CDpro and VPg, is able to use HRV14 cre (VP1) with high efficiency, and to a lesser extent PV1(M) cre (2C) as well, as a template for VPg uridylylation (Fig. 4A; compare lane 5 with lanes 10 and 11). It is interesting that HRV14 cre(VP1) RNA (33) is also an excellent template for PV1(M) 3Dpol in the uridylylation of PV1(M) VPg (42).
Characterization of mutant HRV2 cre(2A) genomes.
Our previous studies have shown that the first two A's in the conserved A1A2 A3CA sequence of PV1(M) cre(2C) serve as a template for the uridylylation of PV1(M) VPg in vitro (42, 45). The same mutation of A to C at nt 3283 and 3284, respectively, in full-length HRV2 RNA (Fig. 3A; compare lane 11 with lanes 12 and 13) or in HRV2 cre(2A) RNA (Fig. 4A; compare lane 5 with lanes 7 and 8) resulted in a failure of these RNAs to function as templates for HRV2 3Dpol in the in vitro reaction.
Characterization of the VPg uridylylation reaction on an HRV2 cre(2A) template.
The optimal reaction conditions for uridylylation of HRV2 VPg on the HRV2 cre(2A) RNA are similar to what was observed previously for the reaction that used a poly(A) template (17), except for the cation and 3CDpro requirement. As shown in Fig. 5B, the yield of products obtained on a cre(2A) template is about the same with either a Mg2+ or a Mn2+ cofactor (compare lanes 2 and 4), and both reactions are strongly stimulated by 3CDpro (compare lane 1 with lane 2 and lane 3 with lane 4). The optimal concentration of Mg2+ is about 1 mM, under which condition VPg uridylylation on a poly(A) template is negligible (Fig. 5A). The other parameters of the reaction, pH and temperature of incubation, were found to be optimal at 7.0 to 7.6 (Fig. 5C) and 27 to 30°C (Fig. 5D), respectively. HRV2 3CDpro is an essential component of the reaction but by itself has no VPg-uridylylating activity (Fig. 5E; compare lanes 5 and 3). The concentration of 3CDpro was optimal at about 0.5 μM, corresponding to a 3CDpro-to-3Dpol molar ratio of about 0.5:1 (data not shown). A mutation (R84S) in the putative RNA binding domain of HRV2 3CDpro nearly completely eliminated its stimulatory effect (Fig. 5E; compare lanes 5 and 7). Addition of a heat-denatured aliquot of either wt or mutant (R84S) 3CDpro had no stimulatory effect on the VPg uridylylation reaction (Fig. 5E; compare lane 5 with lanes 4 and 6).
FIG. 5.
Optimal reaction conditions for VPg uridylylation by HRV2 3Dpol on an HRV2 cre(2A) template. Uridylylation of HRV2 VPg was measured as described in Materials and Methods (assay B), except as indicated. (A) Optimal MgAc2 concentration. The concentration of MgAc2 in the reaction mixtures was varied as indicated. The template was either poly(A) or HRV2 cre(2A). (B) Effect of 3CDpro on uridylylation of VPg by 3Dpol with either Mn2+ or Mg2+ as a cofactor. Reaction mixtures contained either Mn2+ or Mg2+, and HRV2 3CDpro was added where indicated. (C) Optimal pH. The pH of the HEPES buffers was varied as indicated. (D) Optimal temperature. The incubation temperature was varied as indicated. (E) Effect of 3CDpro on uridylylation of VPg by 3Dpol on the HRV2 cre(2A) template. Lane 5, complete reaction mixture with HRV2 3Dpol and 3CDpro; lane 1, no 3CDpro and no 3Dpol; lane 2, no 3CDpro; lane 3, no 3Dpol; lane 4, heat-denatured (h.d.) 3CDpro; lane 6, heat-denatured 3CDpro(3Cpro[R84S]); lane 7, 3CDpro(3Cpro[R84S]). Both HRV2 3CDpro and HRV2 3CDpro(3Cpro[R84S]) contained a 3Cpro[C147A] mutation. In each panel quantitation of the data is displayed above an autoradiograph of the reaction products.
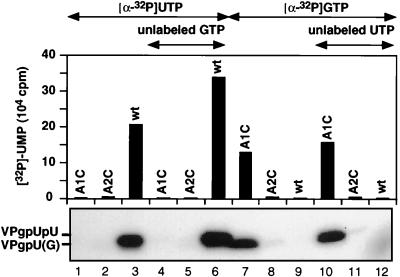
Nucleotide specificity of the VPg uridylylation reaction by HRV2 3Dpol on a HRV2 cre(2A) template.
Since VPg is linked to the 5′-terminal UMP in picornaviral RNAs, one would expect that the RNA polymerase possesses a strict specificity toward UTP in the nucleotidylylation reaction it catalyzes. Based on the polarity of the plus-strand RNA sequence, it would be expected that A2 in the conserved 5′-A1A2A3CA-3′ sequence in the loop of HRV2 cre(2A) is the template for the addition of the first UMP to VPg. Surprisingly, with a mutant C1A2A3CA sequence in the template, HRV2 3Dpol can use only GTP (Fig. 6, lane 7) and not UTP (Fig. 6, lane 1) for the nucleotidylylation of HRV2 VPg. The predicted product, VPgpG, is not elongated into VPgpGpU in the presence of unlabeled UTP (Fig. 6; compare lanes 7 and 10). The mutation of A to C (A1C2A3CA) in the template RNA interferes both with the uridylylation (Fig. 6, lane 2) and the guanylylation (Fig. 6, lane 8) of VPg. These results suggest that A1 is the template nucleotide for both VPgpU and VPgpUpU synthesis and that the addition of the first nucleotide to VPg is not the step that determines the specificity of the overall process (see Discussion).
FIG. 6.
Nucleotidylylation of VPg by HRV2 3Dpol on HRV2 cre(2A) templates in the presence of either [α-32P]UTP (lanes 1 to 6) or [α-32P]GTP (lanes 7 to 12). Standard assay conditions for HRV2 VPg uridylylation (assay B) were used (see Materials and Methods), except that the nucleoside triphosphates and templates were varied as indicated at the top of the figure. Lanes 4 to 6, unlabeled GTP (10 μM) added; lanes 10 to 12, unlabeled UTP (10 μM) added. Three different RNA templates were used: wt HRV2 cre(2A) with the sequence A1A2A3CA (lanes 3, 6, 9, and 12); HRV2 cre(2A)(A1C) with an A-to-C mutation at position 1, resulting in C1A2A3CA (lanes 1, 4, 7, and 10); and HRV2 cre(2A)(A2C) with an A-to-C mutation at position 2, resulting in A1C2A3CA (lanes 2, 5, 8, and 11). Quantitation of the data is displayed above an autoradiograph of the reaction products.
Comparison of VPg uridylylation by HRV2 3Dpol and GST-HRV2 3Dpol.
We have previously analyzed two different preparations of the HRV2 RNA polymerase (17). The form used so far in this work, HRV2 3Dpol, contains an N-terminal methionine or perhaps a polypeptide from which this amino acid has been in part removed. The second form of HRV2 3Dpol, previously analyzed, contains an N-terminal fusion of GST to the 3Dpol polypeptide (GST-HRV2 3Dpol). Of the two enzyme preparations, the GST form is slightly more active than the untagged enzyme in the uridylylation of HRV2 VPg on a poly(A) template (Fig. 7; compare lanes 5 and 8). This is in contrast to the results obtained when either HRV2 full-length or cre (2A) RNAs are used as templates in the reaction. Now the GST-tagged enzyme has much lower activity than HRV2 3Dpol (Fig. 7; compare lane 2 with lane 6 and lane 4 with lane 7). Whether the defect is related directly to the uridylylating activity of the GST-tagged enzyme on cre (2A) or to a lack of 3CDpro stimulation of the reaction catalyzed by GST-HRV2 3Dpol (interference with the formation of a complex) is not yet known. It should be noted that since the concentration of HRV2 RNA is about 100-fold lower than that of HRV2 cre (2A) RNA, the yields of products obtained with these two templates are not directly comparable.
FIG. 7.
Comparison of the VPg-uridylylating activities of GST-HRV2 3Dpol (lanes 1 to 5) and HRV2 3Dpol (lanes 6 to 8) on different RNA templates. Standard assays for HRV2 VPg uridylylation (Assays A or B) were used as described in Materials and Methods, but the RNA templates were varied as follows: HRV2 RNA (lanes 1, 2, and 6); HRV2 cre(2A) (lanes 3, 4, and 7); and poly(A) (lanes 5 and 8). HRV2 3CDpro was added to samples 2 and 4 to 8. Quantitation of the data is displayed above an autoradiograph of the reaction products.
Comparison of wt and mutant VPg peptides as substrates for uridylylation by HRV2 3Dpol.
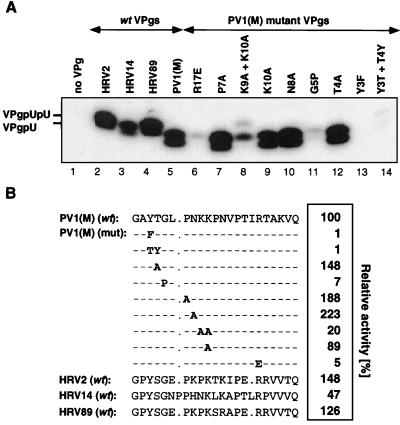
Using mutant synthetic PV1(M) VPg peptides as substrates for HRV2 3Dpol, we have previously shown that several amino acids are essential for the peptide's ability to function in a reaction with a poly(A) template (17). In the present study we characterized the VPg specificity of the uridylylation reaction catalyzed by HRV 2 3Dpol on the cre(2A) template (Fig. 4B) in the presence of HRV2 3CDpro. As expected, no uridylylated products are formed in the absence of the VPg substrate (Fig. 8A; compare lanes 1 and 2). The enzyme is able to utilize as substrates, in addition to its own VPg, the terminal peptides of other picornaviruses such as HRV89, PV1(M), and, to a lesser extent, HRV14 (Fig. 8A; compare lane 2 with lanes 4, 5, and 3, respectively). It is not surprising that HRV89 VPg is an excellent substrate for the HRV2 enzyme, since the two VPgs have the same length (21 amino acids) and have nearly identical amino acid sequences (Fig. 8B) (14). However, PV1(M) VPg was not expected to be such a good substrate for uridylylation by HRV2 3Dpol, since this peptide is 22 residues long, and of these it shares only 8 amino acids (G1, Y3, G5, P7, K10, P14, R17, and Q22) with HRV2 VPg (Fig. 8B) (28, 55). The VPg of HRV14 (Fig. 8B) (56), the least active of the viral VPgs tested, is 24 amino acids long, and 13 of these are identical with those of HRV2 (55).
FIG. 8.
(A) Comparison of various wt and poliovirus mutant VPg's as substrates for uridylylation by HRV2 3Dpol. The standard assay was used for VPg uridylylation (assay B) (see Materials and Methods) except that the synthetic VPg peptide was varied as indicated at the top of the panel. An autoradiograph of the reaction products is shown. (B) The incorporation of [α-32P]UMP into products was quantitated and is shown as relative activity. The amino acid sequences of the wt and mutant VPgs are given. The mutant VPg's are derived from wt PV1(M) VPg (mutations shown in boldface).
The fact that PV1(M) VPg works very well as a substrate for HRV2 3Dpol has enabled us to use mutant PV1(M) peptides to determine those amino acids that are essential for the function of VPg function in vitro. Among the mutant VPgs (Fig. 8A), those containing the Y3F (lane 13), Y3T T4Y (lane 14), G5P (lane 11), and R17E (lane 6) changes were found to have less than 10% of the template activity of the wt peptide (Fig. 8A, lane 2, and 8B). It is interesting that Y3, G5, and R17 are fully conserved among the different picornavirus VPgs (see above). Surprisingly, the substrate activity of some of the mutant PV1(M) VPg peptides for HRV2 3Dpol was higher than that of HRV2 VPg. This group includes the P7A and N8A mutant peptides (Fig. 8A; compare lane 2 with lanes 7 and 10, respectively). Essentially all of the results described here with the PV1(M) VPg peptides are in good agreement with those observed by using poly(A) as a template with either HRV2 3Dpol (17) or PV1(M) 3Dpol (A. V. Paul, J. Peters, and E. Wimmer, unpublished data).
It has previously been shown that the migration of the uridylylated form of PV1(M) or EMCV VPg (2 kDa) in SDS-polyacrylamide gels is highly aberrant and resembles that of a 7- to 12-kDa peptide (31, 48, 58, 62). The same appears to be true for the migration of the other uridylylated viral VPg's or mutant PV(M) VPg peptides. As shown in Fig. 8A, the shorter HRV2 and HRV89 peptides (21 amino acids) migrate even more slowly than PV1(M) VPg (22 amino acids) or HRV14 VPg (24 amino acids) (compare lanes 2 and 4 with lanes 3 and 5). A change of 1 or 2 amino acids (R17E, K9A K10A, G5P) in PV1(M) VPg is sufficient to cause a visible upward shift in the location of the mutant VPgpU bands (Fig. 8A; compare lane 5 with lanes 6, 8, and 11). The reason for the aberrant migration patterns of these peptides is not known.
Effects of mutations in HRV2 VPg and in cre(2A) RNA on the viability of HRV2.
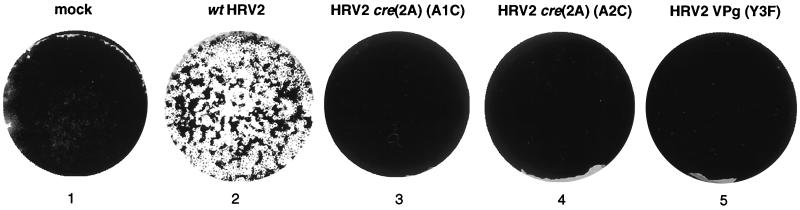
Our experiments have shown that the uridylylation of HRV2 VPg by HRV2 3Dpol in vitro is totally dependent on the presence of a Y3 in VPg and on the integrity of the first two A's of the A1A2A3CA sequence in the loop of the cre(2A) RNA. To correlate our in vitro data with virus viability, we have examined the effect of the Y3F change in VPg, and of the A1C and A2C mutations in cre(2A) RNA, on the ability of HRV2 to replicate (Fig. 9). Transfection of HeLa H1 cell monolayers with RNA transcripts derived from wt HRV2 cDNA gave complete cytopathic effect (CPE) within 3 days (Fig. 9, well 2). In contrast, there was no CPE following transfections with RNA transcripts derived from the mutant plasmids (Fig. 9, wells 3 to 5), just as with mock-transfected cells (Fig. 9, well 1). Supernatants recovered after 3 days were used to reinfect fresh HeLa cell monolayers in five consecutive passages. Cells infected with the wt gave CPE within 3 days, but no CPE was observed with supernatants derived from the mutant viruses. These results strongly suggest that the lethal phenotypes of HRV2 (VPgY3F), HRV2 cre(2A)(A1C), and HRV2 cre(2A)(A2C) relate to a defect in the synthesis of HRV2 VPgpU(pU). The inability of the HRV2 cre(2A)(A1C) RNA to yield virus cannot be due to a defect in 2Apro function, since this mutation does not lead to an amino acid change in the polyprotein. In this respect no firm conclusions can be drawn about the HRV2 cre(2A)(A2C) mutant RNA, in which 2Apro contains an asparagine-to-histidine change.
FIG. 9.
Plaque assays of wt and mutant HRV2 RNAs. HeLa H1 monolayers were transfected with transcripts derived from wt and mutant pT7HRV2 DNA, and the virus yield of each lysate (500 μl) was measured by a plaque assay as described in Materials and Methods. Well 1, mock-transfected; well 2, wt HRV2 RNA; well 3, HRV2 cre(2A)(A1C) RNA; well 4, HRV2 cre(2A)(A2C) RNA; well 5, HRV2 VPg(Y3F) RNA.
DISCUSSION
The primary aim of the experiments reported in this paper was to determine whether a conserved mechanism exists for the initiation of minus-strand RNA synthesis among members of different genera within the Picornaviridae virus family. Since most of our knowledge about this important event in genome replication has been derived from poliovirus, a member of the genus Enterovirus, we have selected HRV2, a member of the genus Rhinovirus, as the subject of this study. In our accompanying paper we describe a simple in vitro assay to measure the uridylylation of HRV2 VPg and VPg-poly(U) synthesis on a poly(A) template by HRV2 3Dpol (17). Using PV1(M) as a model system (42), we have now developed an improved assay for HRV2 VPg uridylylation by HRV2 3Dpol. This assay has allowed us to search for a cis-replicating element in HRV2 RNA required for the protein-priming reaction. The new assay provides specificity to the reaction by use of HRV2 3CDpro and full-length HRV2 RNA or cre(2A) RNA, instead of poly(A), as a template for 3Dpol. In addition, Mg2+ is now used as a cofactor for the polymerase instead of Mn2+. This is an important change because use of Mn2+ has the disadvantage of relaxing the specificity of polymerases for their templates and substrates (5) (Paul, Peters, and Wimmer, unpublished). The exact role of 3CDpro in the reaction is not yet known, but it is presumed to enhance the binding of VPg/3Dpol to the template. This might occur by 3CDpro binding to and stabilizing the structure of the cre RNA or by 3CDpro forming a complex with the other proteins and assisting in their binding to the element.
We have previously described a comparison of two different preparations of HRV2 3Dpol, with either a methionine or GST fused to the N terminus of the polypeptide, both in the oligonucleotide elongation reaction and in VPg-poly(U) synthesis on a poly(A) template (17). Surprisingly, the two versions of the enzyme had similar activities in these synthetic reactions, although the bulky N-terminal tag was expected to interfere with polymerase functions. Based on the amino acid sequence similarity of HRV2 3Dpol and PV1(M) 3Dpol (54), the two polypeptides would be expected to have similar structures. X-ray crystallographic analyses of PV1(M) 3Dpol by Hansen et al. (21) and recent studies by Hobson et al. (24) suggest that the N-terminal segment of one 3Dpol molecule directly interacts with the thumb subdomain of another polymerase molecule. This prediction seemed supported by the finding that mutations in the N-terminal domain of PV1(M) 3Dpol or the deletion of the first 6 N-terminal amino acids of the polypeptide abolished the enzyme's elongation activity (21, 24). The observation that the N-terminal GST tag has a deleterious effect on the VPg-uridylylating activity of HRV2 3Dpol on its cognate cis-replicating element could be considered to be consistent with such a model. It is not clear, though, why the GST tag on HRV2 3Dpol does not significantly reduce VPg uridylylation on a poly(A) template and does not interfere at all with elongation. One possible explanation of these results is that the GST tag does not interfere with the covalent linkage of UMP to VPg per se but rather with an essential 3Dpol-3CDpro interaction required only for the reaction on the cre(2A) RNA. In any event, it would be fascinating to solve the crystal structure of the GST-tagged 3Dpol and to learn about the position of the enzyme's N-terminal sequence.
cis-replicating elements located at different sites in the open reading frames of picornaviral genomes have been described for viruses of the genera Rhinovirus, Enterovirus, and Cardiovirus. All of the elements characterized thus far [HRV14 cre(VP1), poliovirus cre(2C), Theiler's murine encephalomyelitis virus cre(VP2), and Mengovirus cre(VP2)] consist of an RNA hairpin with widely varying RNA sequences (18, 32, 33, 42, 45) except for a conserved motif (A1A2A3CA) in the loop (42, 45). Mutational analyses of the structures previously identified led to the conclusion that sequences in the loop and top of the stem are required for their function (18, 32, 33, 45). For HRV14 and PV3, these elements have been directly shown to be involved in minus-strand RNA synthesis (18, 33). The first step in minus-strand synthesis is the uridylylation of VPg, yielding VPgpU (41). Using genetic and biochemical analyses, we have recently identified the first two A's in the A1A2A3CA sequence of PV1(M) cre(2C) as the primary template for the in vitro synthesis of VPgpU and VPgpUpU (42, 45).
The new assay system we developed for HRV2 VPg uridylylation by HRV2 3Dpol offered a simple way to search for a cis-replicating element in the nucleotide sequence of HRV2 RNA that possesses the same function as the PV1(M) cre(2C) element (18, 42, 45). Using a nested set of 3′-terminal deletions of HRV2 RNA as templates for HRV2 3Dpol, we have shown that an essential RNA element is located upstream of the middle of 2Apro coding sequences. This was confirmed by the finding that sequences downstream from the center of 2Apro are not required for VPg uridylylation. Finally, with the aid of computer-based prediction of RNA folding, we have located several stable RNA hairpins with an A1A2A3CA sequence in the loop in the coding sequences of HRV2 RNA. Of these only one was found to function as a template for 3Dpol in the in vitro VPg uridylylation reaction. This small hairpin, containing only 47 nt, is located in the 5′-terminal half of the 2Apro sequence. Interestingly, two other closely related rhinoviruses, HRV1b (26) and HRV16 (30), but not HRV89 (14), are predicted to contain similar stable stem-loops in 2Apro at essentially the same positions as in HRV2 (Fig. 4C). The predicted sequence (A1A2G3CA) in the loop of HRV16 differs by 1 nucleotide from the A1A2A3CA motif of the HRV2 and HRV1b hairpins. The G at position 3 of the A1A2G3CA motif resembles that predicted for an enterovirus, BEV1 (A1A2G3AA) (18).
We selected the first two A residues of the A1A2A3CA motif in HRVcre(2A) for mutation. The effect on VPg uridylylation of an A-to-C change in either of these two nucleotides correlated well with the replication phenotypes of the mutant viral genomes. These mutations totally abolished both viral growth and the template activity of the cre(2A) RNAs in the in vitro VPg uridylylation reaction. The lethal phenotype of the A1C mutation can be directly linked to a defect in VPg uridylylation rather than to an altered function of 2Apro, since the mutation did not introduce an amino acid change in the polypeptide. We cannot be equally certain that the effect of the A2C change on the growth phenotype is purely due to a defect in the protein-priming reaction. This mutation introduced an asparagine-to-histidine change, and its effect on 2Apro function(s) is not yet known.
Our previous studies suggested the possibility that the synthesis of VPgpU(pU) on the PV1(M) cre(2C) template by PV1(M) 3Dpol might occur by a “slide-back” mechanism (45). Such a mechanism has been described for the nucleotidylylation reactions catalyzed by the DNA polymerases of phages φ29 and PRD1 and adenovirus (reviewed in reference 51). Because of the polarity of the conserved 5′-A1A2A3CA-3′ sequence, A2 would be expected to serve as a template for the synthesis of VPgpU, followed by the addition of the second UMP on the A1 template nucleotide. According to the slide-back mechanism, the addition of the first UMP to VPg would occur on the A1 template nucleotide, followed by a translocation of VPgpU to A2. The second UMP would then be again transcribed from A1. Our experiments that were designed to test the nucleotide specificity of HRV2 3Dpol in VPg uridylylation strongly favor such a slide-back mechanism. Using mutated cre(2A) RNAs (A1C and A2C) as templates for HRV2 3Dpol, we show that A1, and not A2, of the conserved motif, directs the linkage of the first UMP to VPg. Surprisingly, HRV2 3Dpol does not posses stringent specificity toward the NMP it links to VPg in vitro. This is shown by the fact that it is able to covalently link GMP to VPg when provided with GTP and a C1A2A3CA mutant template. However, only the C1A2A3CA and not an A1 C2A3CA sequence serves as a template for 3Dpol for the synthesis of VPgpG. This precursor cannot be elongated to VPgpGpU in the presence of UTP, presumably because the mismatched nucleotides prevent slide-back. The finding that the C1A2A3CA sequence does not support the synthesis of VPgpU with UTP further confirms that the attachment of the first UMP to VPg is templated by the A1 nucleotide. Although A2 is not used as a template nucleotide for the synthesis of either VPgpU or VPgpUpU, it appears to be the nucleotide that provides specificity to the overall process of VPgpUpU synthesis. This might be achieved in two ways. First, A2 appears be essential either for the specific recognition of the RNA element by the viral proteins or for the formation of the initial ribonucleoprotein complex. These possibilities are suggested by the finding that only negligible amounts of VPgpU are formed in the presence of UTP on an A1C2A3CA mutant template. The second function of A2 is predicted to be the specific binding of VPgpU by hydrogen bonding with A2 during the slide-back step.
The amino acid sequences of PV1(M) and HRV2 3Dpol share 56% identity (54). This evolutionary relationship is reflected in their nearly identical in vitro synthetic activities. These include elongation of an oligonucleotide primer on an RNA template, the uridylylation of VPg on a poly(A) template, followed by VPg-poly(U) synthesis, and finally the synthesis of VPgpU and VPgpUpU on their cognate cis-replicating elements in the presence of 3CDpro. Both of these RNA polymerases lack the ability in vitro to use VPgpUpU, made on their cognate cre RNAs, as primers for minus-strand synthesis (data not shown). These precursors are expected to be translocated to the 3′ end of the poly(A) tail prior to elongation. At this time we do not yet know whether it is the translocation step or the elongation reaction itself that is inefficient under our in vitro assay conditions. The translocation step, for example, might be blocked because the mutant 3CDpro(C147A) used in our assay is unable to undergo autoprocessing, possibly a prerequisite for the release of VPgpUpU from the cre RNA. Since translation and replication are coupled (38), our failure to achieve elongation of VPgpUpU might be due to the fact that the RNA element has to be located on the same genome from which 3Dpol, VPg, and/or 3CDpro were translated.
Our model for PV1(M) or HRV2 RNA synthesis, discussed above, is based primarily on that used by the reverse transcriptase of hepatitis B virus (HBV RT) (52). Both the picornaviral RNA polymerases and HBV RT catalyze the covalent linkage of a nucleotide to a protein using an internal RNA hairpin as their template in the reaction. The substrate for nucleotidylylation by PV1(M) and HRV2 3Dpol is the terminal protein VPg; that for nucleotidylylation by HBV RT is its own polypeptide. It has been directly shown that HBV RT, covalently linked to a short oligonucleotide, is transferred to near the 3′ end of the RNA, where it serves as a primer for cDNA synthesis (52). For optimal cDNA synthesis the HBV polymerase recruits a chaperone complex with several proteins (52). The possibility that picornaviral minus-strand RNA synthesis also requires additional viral or cellular factors has been suggested by Barton and Flanegan (6, 7) on the basis of their studies of the in vitro translation replication system that synthesizes viable poliovirus (36). Their results suggest that the guanidine-sensitive ATPase activity of protein 2CATPase and a soluble HeLa cell factor are required for minus-strand RNA synthesis (6, 7). Our in vitro assay of VPg uridylylation offers a useful system to search for additional viral or cellular factors that might be required for the initiation of minus-strand RNA synthesis of HRV2 and of other picornaviruses.
ACKNOWLEDGMENTS
We thank K. Kirkegaard for the generous gift of plasmid pT5T3D and T. Skern for the gift of plasmid pT7HRV2. We are indebted to J. H. Van Boom for the synthetic VPg peptides. We also thank E. Rieder and T. Pfister for helpful suggestions and J. Mugavero and F. Maggiore for excellent technical assistance.
K. Gerber was an exchange student between the graduate programs of the University of Konstanz, Konstanz, Germany, and SUNY at Stony Brook. This work was supported in part by an NIH grant (AI15122).
ADDENDUM IN PROOF
Genetic experiments carried out by S. Lemon and colleagues have indicated that the essential motif in the loop of HRV14 cre(VP1) is AAACG. These results were confirmed by biochemical studies (Y. Yang, A. V. Paul, E. Wimmer, and S. Lemon, unpublished results).
REFERENCES
- 1.Agol V I, Paul A V, Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. doi: 10.1016/s0168-1702(99)00037-4. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V, Baltimore D. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J Biol Chem. 1978;253:5263–5266. [PubMed] [Google Scholar]
- 3.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 4.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold J J, Ghosh S K B, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol). Divalent cation modulation of primer, template, and nucleotide selection. J Biol Chem. 1999;274:37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- 6.Barton D J, Black E P, Flanegan J B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton D J, Flanegan J B. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–8489. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienz K, Egger D, Pfister T, Troxler M. Structural and functional characterization of the poliovirus replication complex. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blair W S, Parsley T B, Bogerd H P, Towner J S, Semler B L, Cullen B R. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA. 1998;4:215–225. [PMC free article] [PubMed] [Google Scholar]
- 10.Borman A, Jackson R J. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology. 1992;188:685–696. doi: 10.1016/0042-6822(92)90523-r. [DOI] [PubMed] [Google Scholar]
- 11.Borman A M, Deliat F G, Kean K M. Sequences within the poliovirus internal ribosomal entry segment control viral RNA synthesis. EMBO J. 1994;13:3149–3157. doi: 10.1002/j.1460-2075.1994.tb06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford N M, Baltimore D. Genome-linked protein VPg of poliovirus is present as free VPg and VPgpUpU in poliovirus-infected cells. Proc Natl Acad Sci USA. 1983;80:7452–7455. doi: 10.1073/pnas.80.24.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duechler M, Skern T, Blaas D, Berger B, Sommergruber W, Kuechler E. Human rhinovirus serotype 2: in vitro synthesis of an infectious RNA. Virology. 1989;168:159–161. doi: 10.1016/0042-6822(89)90414-5. [DOI] [PubMed] [Google Scholar]
- 14.Duechler M, Skern T, Sommergruber W, Neubauer C, Gruendler P, Fogy I, Blaas D, Kuechler E. Evolutionary relationships within the human rhinovirus genus: comparison of serotypes 89, 2 and 14. Proc Natl Acad Sci USA. 1987;84:2605–2609. doi: 10.1073/pnas.84.9.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flanegan J B, Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A) Proc Natl Acad Sci USA. 1977;74:3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber K, Wimmer E, Paul A V. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: purification and enzymatic analysis of the RNA-dependent RNA polymerase 3Dpol. J Virol. 2001;75:10969–10978. doi: 10.1128/JVI.75.22.10969-10978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond J W, Barclay W, Evans D J. Identification of a cis-acting replication element within the poliovirus coding region. J Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gwaltney J M., Jr Rhinovirus infection of the normal human airway. Am J Respir Crit Care Med. 1995;152:S36–S39. doi: 10.1164/ajrccm/152.4_Pt_2.S36. [DOI] [PubMed] [Google Scholar]
- 20.Hammerle T, Molla A, Wimmer E. Mutational analysis of the proposed FG loop of poliovirus proteinase 3C identifies amino acids that are necessary for 3CD cleavage and might be determinants of a function distinct from proteolytic activity. J Virol. 1992;66:6028–6034. doi: 10.1128/jvi.66.10.6028-6034.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 22.Harris K S, Hellen C U T, Wimmer E. Proteolytic processing in the replication of picornaviruses. Semin Virol. 1990;1:323–333. [Google Scholar]
- 23.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of the poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome: identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 24.Hobson S D, Rosenblum E S, Richards O C, Richmond K, Kirkegaard K, Schultz S. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 2001;20:1153–1163. doi: 10.1093/emboj/20.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blaas D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91:1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes P J, North C, Jellis C H, Minor P D, Stanway G. The nucleotide sequence of human rhinovirus 1B: molecular relationships within the rhinovirus genus. J Gen Virol. 1988;69:49–58. doi: 10.1099/0022-1317-69-1-49. [DOI] [PubMed] [Google Scholar]
- 27.Jang S K, Krausslich H G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitamura N, Semler B L, Rothberg P G, Larsen G R, Adler C J, Dorner A J, Emini E A, Hanecak R, Lee J J, van der Werf S, Anderson C W, Wimmer E. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 29.Koliais S I, Dimmock N J. Rhinovirus RNA polymerase: products and kinetics of appearance in human diploid cells. J Virol. 1974;14:1035–1039. doi: 10.1128/jvi.14.5.1035-1039.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee W M, Wang W, Rueckert R R. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes. 1995;9:177–181. doi: 10.1007/BF01702661. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y F, Nomoto A, Detjen B M, Wimmer E. A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci USA. 1977;74:59–63. doi: 10.1073/pnas.74.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lobert P E, Escriou N, Ruelle J, Michiels T. A coding RNA sequence acts as a replication signal in cardioviruses. Proc Natl Acad Sci USA. 1999;96:11560–11565. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKnight K L, Lemon S M. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melchers W J G, Hoenderop J G, Bruins Slot H J, Pleij C W A, Pilipenko E V, Agol V I, Galama J M D. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirmomeni M H, Hughes P J, Stanway G. An RNA structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 37.Nomoto A, Detjen B, Pozzatti R, Wimmer E. The location of the polio genome protein in viral RNAs and its implication for RNA synthesis. Nature. 1977;268:208–213. doi: 10.1038/268208a0. [DOI] [PubMed] [Google Scholar]
- 38.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 39.Parsely T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly(rC) binding protein 2 forms a ternary complex with the 5′ terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 40.Pata J D, Schultz S C, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 41.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 42.Paul A V, Rieder E, Kim D W, van Boom J H, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelletier J, Sonnenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 44.Pilipenko E V, Poperechny K V, Maslova S V, Melchers W J, Slot H J B, Agol V I. cis-element oriR, involved in the initiation of (−)strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (kissing) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 45.Rieder E, Paul A V, Kim D W, van Boom J H, Wimmer E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J Virol. 2000;74:10371–10380. doi: 10.1128/jvi.74.22.10371-10380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rivera V M, Welsh J D, Maizel J V. Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology. 1988;165:42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- 47.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rothberg P G, Harris T J R, Nomoto A, Wimmer E. O4-(5′-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc Natl Acad Sci USA. 1978;75:4868–4872. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowlands D J. Rhinoviruses and cells: molecular aspects. Am J Respir Crit Care Med. 1995;152:531–535. doi: 10.1164/ajrccm/152.4_Pt_2.S31. [DOI] [PubMed] [Google Scholar]
- 50.Rueckert R R. Picornaviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 609–654. [Google Scholar]
- 51.Salas M, Miller J T, Leis J, DePamphilis M L. Mechanisms for priming DNA synthesis. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 131–176. [Google Scholar]
- 52.Seeger C, Mason W S. Replication of the hepatitis virus genome. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 815–831. [Google Scholar]
- 53.Shiroki K, Ishii T, Aoki T, Kobashi M, Ohka S, Nomoto A. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J Virol. 1995;69:6825–6832. doi: 10.1128/jvi.69.11.6825-6832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skern T, Sommergruber W, Blaas D, Pieler C H, Kuechler E. Relationship of human rhinovirus strain 2 and poliovirus as indicated by comparison of the polymerase gene regions. Virology. 1984;136:125–132. doi: 10.1016/0042-6822(84)90253-8. [DOI] [PubMed] [Google Scholar]
- 55.Skern T, Sommergruber W, Blaas D, Gruendler P, Fraundorfer F, Pieler C H, Fogy I, Kuechler E. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985;13:2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanway G, Hughes P J, Mountford R C, Minor P D, Almond J W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984;12:7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer J A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 58.Takegami T, Kuhn R J, Anderson C W, Wimmer E. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc Natl Acad Sci USA. 1983;80:7447–7451. doi: 10.1073/pnas.80.24.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. Virology. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toyoda H, Yang C F, Takeda N, Nomoto A, Wimmer E. Analysis of RNA synthesis of type 1 poliovirus by using an in vitro molecular genetic approach. J Virol. 1987;61:2816–2822. doi: 10.1128/jvi.61.9.2816-2822.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van der Werf S, Bradley J, Wimmer E, Studier F W, Dunn J J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vartapetian A B, Koonin E V, Agol V I, Bogdanov A A. Encephalomyocarditis virus RNA synthesis in vitro is protein-primed. EMBO J. 1984;3:2593–2598. doi: 10.1002/j.1460-2075.1984.tb02179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wimmer E. Genome-linked proteins of viruses. Cell. 1982;28:199–201. doi: 10.1016/0092-8674(82)90335-x. [DOI] [PubMed] [Google Scholar]
- 64.Xiang W, Paul A V, Wimmer E. RNA signals in entero- and rhinovirus genome replication. Semin Virol. 1997;8:256–273. [Google Scholar]
- 65.Xiang W, Harris K S, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin F H, Knight E., Jr In vivo and in vitro synthesis of human rhinovirus type 2 ribonucleic acid. J Virol. 1972;10:93–98. doi: 10.1128/jvi.10.1.93-98.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]