Abstract
Dicer is an RNase III enzyme that is responsible for the maturation of small RNAs such as microRNAs. As Dicer’s cleavage products play key roles in promoting cellular homeostasis through the fine-tuning of gene expression, dysregulation of Dicer activity can lead to several human diseases, including cancers. Mutations in Dicer have been found to induce tumorigenesis and lead to the development of a rare pleiotropic tumor predisposition syndrome found in children and young adults called DICER1 syndrome. These patients harbor germline and somatic mutations in Dicer that lead to defective microRNA processing and activity. While most mutations occur within Dicer’s catalytic RNase III domains, alterations within the Platform-PAZ (Piwi-Argonaute-Zwille) domain also cause loss of microRNA production. Using a combination of in vitro biochemical and cellular studies, we characterized the effect of disease-relevant Platform-PAZ-associated mutations on the processing of a well-studied oncogenic microRNA, pre-microRNA-21. We then compared these results to those of a representative from another Dicer substrate class, the small nucleolar RNA, snord37. From this analysis, we provide evidence that mutations within the Platform-PAZ domain result in differential impacts on RNA binding and processing, adding new insights into the complexities of Dicer processing of small RNA substrates.
Graphical Abstract
The RNase III endoribonuclease Dicer, which recognizes and cleaves double-stranded RNA (dsRNA), is an essential enzyme in the RNA interference (RNAi) pathway and in the maturation of microRNAs (miRNAs or miRs).1 MiRNAs are single-stranded, noncoding RNAs that are ~22 nucleotides (nt) in length that function in the regulation of protein-coding genes via post-transcriptional silencing of mRNA translation.2 Expression and biogenesis of miRNA are tightly controlled through a multiprotein pathway with Dicer playing a central role in the final maturation step.1,3,4 Starting in the nucleus, a miRNA is first transcribed by RNA polymerase II to produce a primary miRNA (pri-miRNA), which is subsequently recognized by the Microprocessor complex comprised of the RNase III enzyme Drosha and its cofactor DiGeorge syndrome critical region gene 8 (DGCR8). This complex cleaves the flanking regions of the pri-miRNA to excise a hairpin loop precursor miRNA (pre-miRNA) with a 2-nt overhang on the end of the 3′ (3P) strand and a phosphorylated 5′ (5P) strand. Following export to the cytoplasm by Exportin 5, the pre-miRNA is recognized by Dicer and its cofactor, transactivation response element RNA-binding protein (TRBP), for the final maturation step. Dicer performs this cleavage reaction through the work of its Mg2+-dependent RNase III domains, RNase IIIA and RNase IIIB.1 Interestingly, these RNase III domains act independently with the RNase IIIA domain cleaving the 3P strand and the RNase IIIB domain in control of 5P strand processing.5 The result of these cleavage events leads to removal of the loop region of the pre-miRNA and production of a mature miRNA duplex. A single strand from this duplex is then loaded into Argonaute 2 (Ago2) to form the RNA-induced silencing complex (RISC), which uses the loaded miRNA to target mRNAs on the basis of their complementarity to the miRNA seed sequence.1,2
Because miRNAs regulate a diverse pool of transcripts and, thus, biological processes, dysregulation of miRNA function, sequence integrity, and/or expression level can lead to a variety of diseases.2,6 In particular, alterations of the cleavage activity of Dicer, through either mutation or changes in expression, directly impact miRNA maturation, ultimately leading to tumor development.7,8 Loss-of-function mutations in Dicer have been shown to be recurrent across many cancer types.7-11 Indeed, analyses of human sequencing data have revealed that the prevalence of pathogenic variations in Dicer is more common than would be anticipated: 1:10600 in the general population and 1:4600 in cancer patients from The Cancer Genome Atlas (TCGA).9,12 Mutations are primarily found in metal-binding RNase IIIB domain residues (E1705, D1709, D1810, and E1813), leading to a strand bias in pre-miRNA processing from 5P to 3P.13-15 As most mature miRNAs are active as the 5P strand,2 RNase IIIB domain mutation results in severe impairment of miRNA biogenesis and activity, including a loss of miRNAs that function as tumor suppressors such as let-7 and miR-15/16.10,16 Accordingly, expression of Dicer RNase IIIB mutants in Dicer-null cell lines can induce oncogenic transformation and promote cell proliferation.17,18
While mutation of Dicer has been observed across many cancers, the earliest studies of Dicer mutant cancers emerged from the discovery of a rare pleiotropic tumor predisposition syndrome found in children and young adults called DICER1 syndrome, in which the patients harbor germline and somatic mutations in Dicer that lead to defective miRNA processing and activity.7,8,10,13,19,20 On the basis of the diverse array of miRNA activities throughout human tissues, patients with DICER1 syndrome exhibit multiple pathologic and oncogenic phenotypes. Some of the common cancers found in these patients include pleuropulmonary blastoma (PPB),14,21,22 gynecologic tumors such as ovarian Sertoli-Leydig cell tumors23 and cervical embryonal rhabdomyoscarcoma,24 cystic nephroma,25 etc.7,8,19 While most disease-associated Dicer mutations occur within the catalytic core and affect 5P strand processing via disruption of Mg2+ coordination, Dicer mutations in the Platform-PAZ (Piwi-Argonaute-Zwille) domain have also been shown to alter miRNA maturation and lead to the development of disease phenotypes, although the mechanisms for how they disrupt Dicer activity are not well understood.7
The Platform-PAZ domain plays an important role in substrate recognition as it contains binding pockets for both the 3′ 2-nt overhang and 5′ phosphate of a pre-miRNA substrate.1,26-28 Disruption of the residues associated with these binding pockets directly affects the binding affinity of Dicer for pre-miRNAs and its ability to produce mature miRNAs of an appropriate length.5,27,28 Interestingly, some Platform-PAZ domain disease mutations are not located within the substrate-binding pockets, and unlike disease mutations found in the RNase III domains, preliminary studies have demonstrated that these mutations impact Dicer’s ability to cleave both 5P and 3P strands.7,29 Thus, we became interested in further examining the functional impact of disease-associated mutations within the Platform-PAZ domain, and herein, we describe our efforts to characterize select mutants using a combination of in vitro biochemical analyses and cellular studies.
MATERIALS AND METHODS
Plasmids.
The pCAGGS-Flag-hsDicer plasmid used for expression was a gift from P. Sharp (Addgene plasmid 41584; http://n2t.net/addgene:41584; RRID:Addgene 41584). To obtain the mutant Dicer constructs, site-directed mutagenesis was carried out by GenScript (Piscataway NJ). Mutations were confirmed by sequencing (forward primer, 5′-GGAAT-GGTTTTAACTACACC-3′; reverse primer, 5′-CCCCT-TCTGATTCAAATGTC-3′).
Recombinant Dicer Cell Culture and Transfection.
Expi293 cells were cultured in suspension with Expi293 expression medium in a humidified orbital shaker (120 rpm) at 37 °C with 8% CO2. Expression plasmids pCAGGS-Flag-hs WT and mutant Dicer constructs were transiently transfected into Expi293F cells according to the manufacturer’s instructions. Briefly, Expi293F cells at a density of 3 × 106 viable cells per milliliter were transfected with expression plasmids complexed with ExpiFectamine 293 transfection reagent (Thermo Fisher Scientific). One day after transfection (~18–22 h), the cells were supplemented with a cocktail of transfection enhancers (ExpiFectamine 293 transfection enhancer 1 and enhancer 2). Then, the cultures were incubated for 4 days, cells pelleted gently by centrifugation, and pellets stored at −80 °C for protein purification.
Recombinant Dicer Purification.
N-Terminally Flag-tagged hsDicer1 and hsDicer1 mutants were purified from Expi293F cells transiently transfected with pCAGGS-Flag-hs WT or mutant Dicer constructs. The pelleted cell samples were resuspended in 5 pellet volumes of lysis buffer [20 mM Tris (pH 7.4), 150 mM NaCl, 10% glycerol, 1% NP-40, and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) containing an EDTA-free protease inhibitor mixture (Roche)] and lysed by being gently rocked at 4 °C for 45 min. Insoluble material was pelleted by centrifugation at 20000g for 20 min, and the supernatant solution was applied to 250 μL (packed) of anti-FLAG M2 affinity gel (Sigma) in a 15 mL Falcon tube and incubated at 4 °C for 4 h while being gently rocked. After batch binding for 4 h, the resin was pelleted by centrifugation at 500g for 5 min and washed by being resuspended in 5 mL of wash buffer [50 mM Tris (pH 7.4) and 150 mM NaCl]. The resuspended resin was pelleted by centrifugation at 500g for 5 min and washed two additional times with wash buffer. The resin was incubated in 500 μL of elution buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1 mg/mL 3X Flag peptide (Sigma), and 0.5 mM TCEP] for 12 h at 4 °C. The resin was finally pelleted using centrifugation at 500g for 5 min, and the supernatant containing Dicer was concentrated to ~2 mg/mL, flash-frozen using liquid nitrogen, and stored at −80 °C.
Negative Stain Electron Microscopy (EM).
Samples of purified WT or mutant Dicer proteins were prepared for negative stain EM using established methods.30 In brief, 200 mesh copper grids covered with carbon-coated collodion film (EMS, Hatfield, PA) were glow discharged for 30 s at 5 mA in a PELCO (Fresno, CA) easiGlow glow discharge unit. Protein (3.5 μL at ~0.05 mg/mL) was adsorbed onto the grids and incubated for 1 min at room temperature. Samples were washed with two drops of water and then two successive drops of 0.7% (w/v) uranyl formate (EMS) and then blotted until dry. Samples were visualized on a FEI Morgagni instrument operating at an accelerating voltage of 100 keV (Thermo Fisher Scientific, Hillsboro, OR) at a nominal magnification of 22000× (2.1 Å per pixel). Images were collected individually on a Gatan Orius charge-coupled device camera.
Collection and Processing of Negative Stain EM Data.
For further negative stain EM analysis, samples were visualized on a Tecnai Spirit T12 instrument operating at an accelerating voltage of 120 keV (Thermo Fisher Scientific) at a nominal magnification of 42000× (1.45 Å per pixel). Data were collected using Leginon software31,32 on a 4K × 4K Rio complementary metal oxide semiconductor camera (Gatan, Pleasanton, CA) at a −1.5 μm defocus value. Five data collection sessions were performed, and an average of 200 micrographs were collected for WT, S839A, S839F, L881A, and L881P Dicer. CTF estimation was performed on the images using CTFFIND433 in cryoSPARC. Particle picking was carried out in cryoSPARC using a trained Topaz model.34,35 This model was created by first manually picking 500 particles from the WT Dicer data set and then using the extracted particle coordinates as a training set. On average, the trained Topaz model picked ~20000–30000 particles per data set using a box size of 256 pixels (37 nm). Two rounds of two-dimensional (2D) classification with a circular mask diameter of 200 Å were subsequently performed using cryoSPARC. A table of micrograph and particle numbers corresponding to each Dicer construct is presented in Figure S1.
RNA Substrates.
Chemically synthesized RNAs (deprotected, desalted, and HPLC purified) were purchased from Horizon Discovery Biosciences (formerly Dharmacon) and used as received for cleavage and binding assays: pre-microRNA-21, 5′-P-UAGCUUAUCAGACUGAUGUUGAC-UGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCU-GUC-3′; blunt-end pre-microRNA-21, 5′-P-UAGCUU-AUCAGACUGAUGUUGACUGUUGAAUCUCAU-GGCAACACCAGUCGAUGGGCUG-3′; snord37, 5′-AUUCGUGAUGACUGAUCAUUUCUUCACUUU-GACCAGAUGUCUACUGAAGAAAGCCUGCGUCUG-AGG-3′.
Electrophoretic Mobility Shift Assay (EMSA).
pre-miR-21, snord37, or blunt-end pre-miR-21 was first diluted to a working concentration of 2 μM in a 2× master mix solution containing 0.2% NP-40 and 8 mM DTT. After the RNA solution was prepared, 5 μL of the 2× RNA master mix (final concentration of 500 nM) was incubated with WT or mutant Dicer protein (final concentration of 0.1 mg/mL) in a buffer [100 mM Tris (pH 7.6), 100 mM NaCl, 20% glycerol, and 1 mM EDTA] on ice for 0, 10, 30, 60, 90, and 120 min in a total reaction volume of 20 μL. Binding was halted via the addition of 5 μL of 80% glycerol. The RNA–protein interaction was analyzed via gel electrophoresis by running on a 4–15% Tris/glycine gel (Bio-Rad) at 100 V for 1 h, stained using SYBER Green II, and visualized with a Molecular Imager Gel Doc XR+ instrument (Bio-Rad).
Dicer Cleavage Assay.
RNA cleavage was carried out in a total reaction volume of 10 μL. RNA substrates (pre-miR-21, snord37, or blunt-end pre-miR-21; final concentration of 500 nM) were incubated with WT or mutant Dicer protein (final concentration of 0.1 mg/mL) in a buffer [20 mM Tris (pH 7.4), 12 mM NaCl, 2.5 mM MgCl2, and 1 mM DTT] at 37 °C for 0, 10, 30, 60, 90, and 120 min. The reaction was quenched with 2.5 μL of 80% glycerol and analyzed via gel electrophoresis. Samples were run on a 10% 8 M TBE-urea gel (Bio-Rad), and the gel was stained using SYBER Green II (Thermo Fisher Scientific) and visualized with a Molecular Imager Gel Doc XR+ instrument (Bio-Rad).
Dicer1−/− Cell Culture.
Dicer1−/− mouse mesenchymal cells (CRL-3221) were purchased from ATCC. Cells were maintained in α minimum essential medium with ribonucleo-sides and deoxyribonucleosides and 10% fetal bovine serum at 37 °C with 5% CO2 in a humidified incubator.
Plasmid Transfection and Western Blot for Dicer −/− Cell Lines.
First 125000 cells were plated in a six-well tissue culture plate. After 24 h, cells were transfected with 2.5 μg of pCAGGS-Flag-hs wild-type (WT) or mutant Dicer constructs using 7.5 μL of TransIT-LT1 (Mirus Bio, MIR 2304). Two sets of plates were simultaneously transfected. One set was harvested in RIPA buffer [10 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton, 1% sodium deoxycholate, and 0.1% SDS (pH 7.2)]; total protein was quantified using the BCA assay and analyzed by Western blotting. The other set of plates was used for RNA extraction and qRT-PCR (see below). In brief, proteins were resolved on a 4% to 15% Tris/glycine gel and transferred to a PVDF membrane in Towbin’s buffer [25 mM Tris, 192 mM glycine, and 20% (v/v) methanol (pH 8.3)]. The membrane was blocked in 5% milk for 1 h at 25 °C and then incubated with a primary antibody (overnight at 4 °C) and a secondary antibody (1 h at 25 °C). The Dicer antibody was purchased from Cell Signaling Technology (3363), and the actin-HRP antibody was purchased from Santa Cruz Biotechnology (sc-47778). Proteins were visualized using a Bio-Rad ChemiDoc imaging system.
RNA Extraction and qRT-PCR.
First, 125000 cells were plated in a six-well tissue culture plate. After 24 h, cells were transfected with 2.5 μg of pCAGGS-Flag-hs wild-type (WT) or mutant Dicer constructs using 7.5 μL of TranSIT LT-1 (Mirus Bio, MIR 2304). After 48 h, RNA was extracted using TRIzol reagent (Ambion 15596018) according to the manufacturer’s instructions. Then, 400 ng of RNA was used to prepare cDNA using the TaqMan Advanced miRNA cDNA synthesis kit (Thermo Fisher Scientific A28007). The following TaqMan miRNA assays were used in the study: hsa-miR21-5p 477975_mir (A25576) and U6 snRNA assay (4427975). To quantify mRNA levels, qRT-PCR was performed using the TaqMan Advanced Master Mix (Thermo Fisher Scientific 444557) according to the manufacturer’s instructions on a Quant Studio 5 thermocycler using the fast-qPCR protocol. Standard curves with WT DICER cDNA samples were used to determine the efficiency of the miR-21 and U6 snRNA assays. The relative fold change was calculated using the comparative threshold cycle (CT) method.36 Standard curves with standard cDNA samples were used to determine the efficiency of the miRNA assays. U6 snRNA was used as a control, and all mRNA levels were normalized to U6 snRNA. All experiments were performed in biological quadruplicate.
Quantification and Statistical Analysis.
For quantification of the impact of Dicer PAZ mutations on dicing and binding activity, triplicate experiments were carried out, and the results were analyzed using ImageJ. Each data point represents the average of three independent experiments. Error bars represent the standard deviation.
RESULTS AND DISCUSSION
Effects of Platform-PAZ-Associated Dicer Mutations on Pre-miR-21 Binding and Processing.
Although rarer than RNase III domain mutations, alterations within the Platform-PAZ domain are observed in cancer patients and, in some cases, are associated with disease. Select examples are highlighted in Figure 1A, which provides an overview of Dicer alterations found within the Platform-PAZ domain, their related disease phenotype, and the likelihood of pathogenicity based upon the proposed impact on Dicer structure and, thus, activity.7,10,29,37 While frame shift and deletion mutants would have an obvious impact in altering Platform-PAZ integrity and Dicer activity through loss of amino acids or a shift in the amino acid sequence in sections of the domain (Figure 1B), the effects of the disease-associated point mutations shown are less clear.
Figure 1.
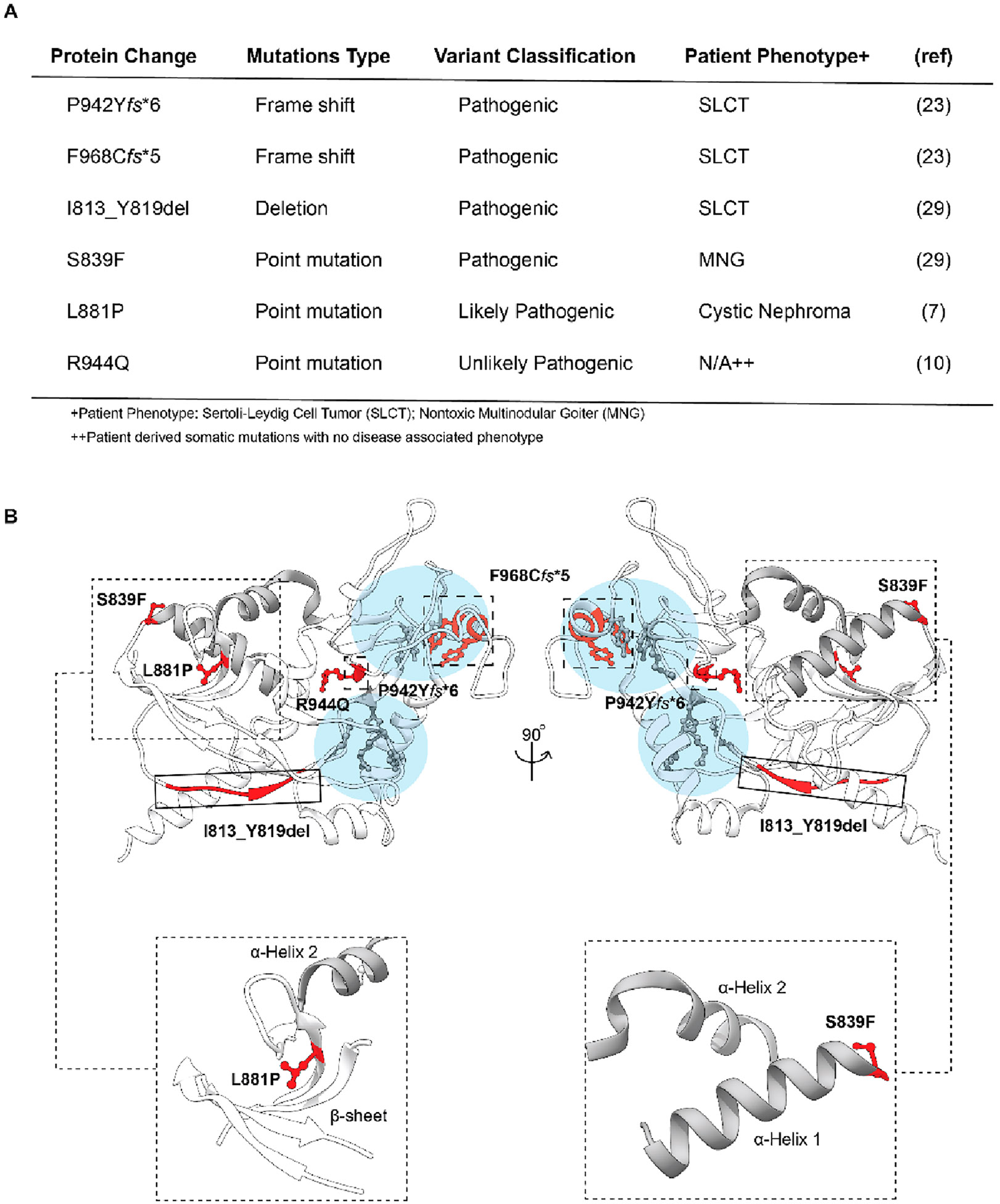
Overview of patient-derived Dicer mutations located within the Platfrom-PAZ domain. (A) Table of patient-derived alterations with corresponding phenotypes. (B) Three-dimensional representation of the Platform-PAZ domain (Protein Data Bank entry 5ZAK) with the location of patient-derived mutations highlighted and close-up views of the secondary structural features surrounding S839 and L881. Residues associated with point mutations are shown as ball and stick models, while frame shift and deletion mutants are grouped by large dashed and solid boxes, respectively. Major RNA-binding pockets are highlighted in light blue with key amino acids colored gray. α-Helices 1 and 2 are colored gray in all models.
We became particularly interested in the germline point mutants, S839F and L881P,7,29 which are located on the opposite side of the Platform-PAZ domain relative to the RNA-binding pockets and are predicted to disrupt secondary structural elements within this domain (Figure 1B). More specifically, the S839F mutation is predicted to disrupt α-helix 1 according to a model generated using Phyre (Protein homology/analogy recognition engine)38 due to the loss of stabilizing hydrogen bonding interactions at the N-terminus of the α-helix from the side chain of serine (Figure 1B).29,39 For the L881P mutation, we proposed that it would alter a nearby β-sheet on the basis of the known propensity of proline to disrupt β-sheet stability (Figure 1B).40 Although S839F and L881P are outside of the 3′ 2-nt overhang- and 5′ phosphate-binding pockets, from analyzing the structure, we hypothesized that they may hinder pre-miRNA binding, particularly because production of both 3P and 5P strands is affected by these alterations.7,29 Thus, we set out to perform biochemical characterizations of these disease-relevant mutant Dicer proteins.
To initiate our investigation, we expressed and purified N-terminally Flag-tagged hsDicer1 and hsDicer1 mutants from Expi293F cells transiently transfected with pCAGGS-Flag-hs WT or mutant Dicer constructs (Figure S1A). In addition to the disease mutants, S839F and L881P, we also prepared alanine mutants to further probe amino acid changes at these sites. Before performing in vitro studies, we visualized WT and Dicer mutants using negative stain electron microscopy to assess the overall protein integrity. For each recombinantly expressed and purified protein, we collected images, picked ~20000–30000 particles, and used 2D class averaging (Figure S1B-D). A representative micrograph and 2D class average for WT and each Dicer mutant are shown in Figure S2. As expected, WT Dicer exhibited an overall L-shaped globular structure as previously established.41 Analysis of the four recombinant Dicer mutants demonstrated the same global fold as that of the WT protein, indicating that these mutations do not destabilize the overall structure of Dicer. Although it is possible that protein structure was altered on a local level because negative stain 2D averages provide only a gross view of protein structure, this analysis afforded confidence that our expression and purification efforts resulted in folded proteins suitable for biochemical characterization.
With proteins in hand, we first sought to examine binding to a pre-miRNA substrate. As a model, we selected miR-21, a well-studied Dicer-dependent miRNA that is overexpressed in a majority of human tumors and whose maturation was previously shown to be impacted by mutation in Dicer’s PAZ domain.5,6 To characterize the binding of Dicer proteins to pre-miR-21, we performed electrophoretic mobility shift assays (EMSAs). As shown in Figure 2, in line with our hypothesis, negligible binding was observed for the S839F and L881P disease mutants in comparison to that of the WT protein. For the alanine mutants, while S839A demonstrated binding like that of WT as visualized by the gel shift, the L881A mutant showed only weak complex formation. We hypothesize that this is due to the fact that alanine, like proline, can also destabilize β-sheet structures.42
Figure 2.
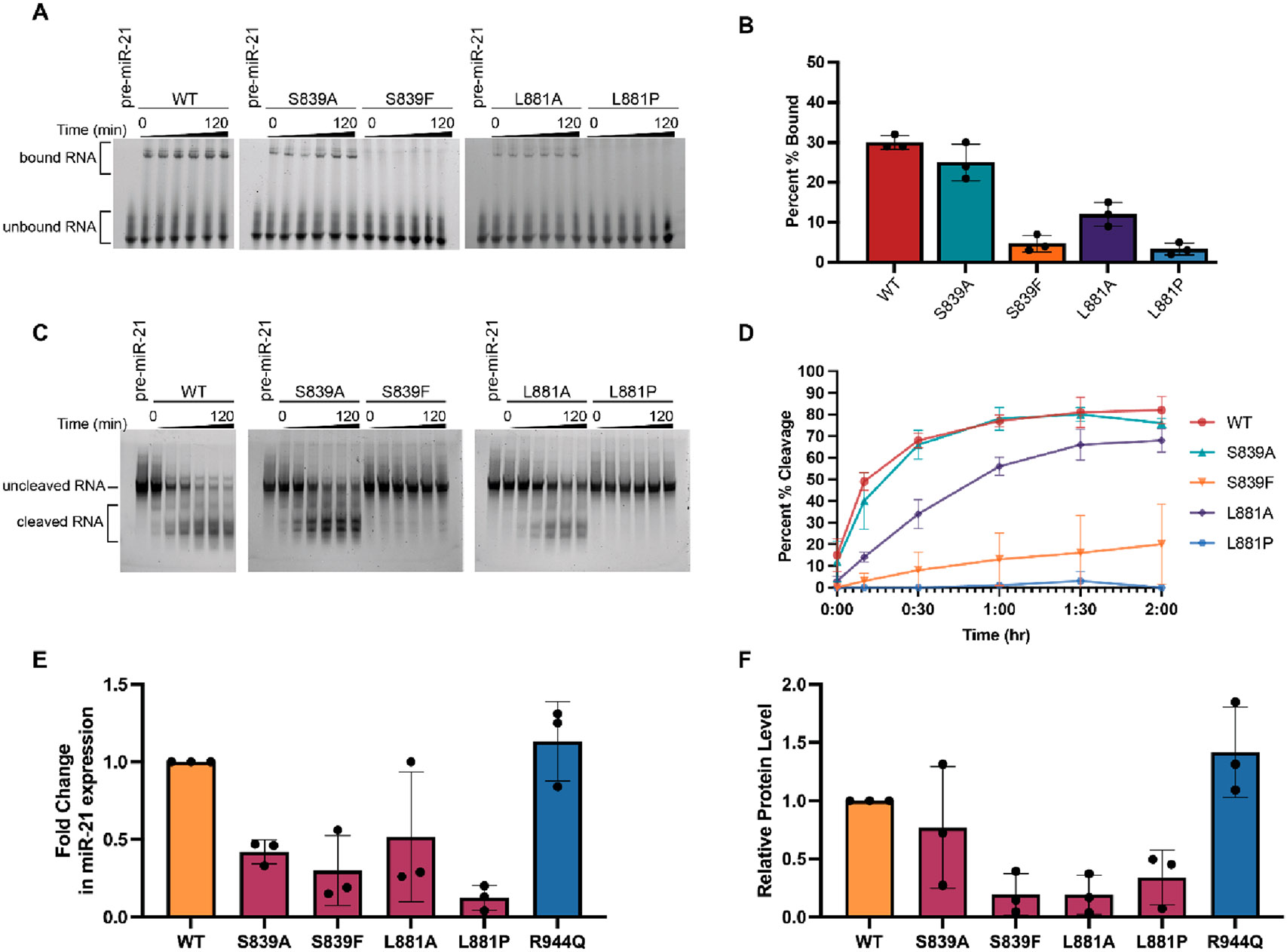
Characterization of the binding and cleavage activity of WT and Dicer mutants for pre-miR-21 in vitro and in cells. (A) Representative EMSA data (Figure S3) shown over the course of 120 min. (B) Quantification of binding at the 120 min time point as determined using the relative difference in the intensity of bound RNA (top band) to unbound RNA (bottom band). (C) Representative time-dependent cleavage visualized via gel electrophoresis (Figure S4). (D) Quantification of Dicer cleavage activity determined on the basis of the relative changes in the band intensity of the uncleaved RNA (top) band and the cleaved RNA (bottom) bands. Bands between these regions, which represent intermediate cleavage products, were not included in our quantification. Gel-based analysis was carried out using ImageJ. Error bars represent the standard deviation from three independent experiments (Tables S1 and S2). (E and F) Impact of Dicer mutations on miR-21 and Dicer protein levels in Dicer1−/− cells. (E) miR-21 expression in WT and Dicer mutant-transfected cells from three biological replicates (Figure S6 and Table S3). RNA levels were normalized to U6 snRNA, and fold changes in miR-21 expression were calculated relative to WT Dicer. (F) Relative protein levels of WT and Platform-PAZ Dicer mutants from three biological replicates (Figure S6 and Table S4). Protein levels were quantified through densitometry analysis from Western blots using ImageJ. Mutant protein levels were normalized to WT Dicer. The variability of the cell-based assays is likely due to deviations in transfection efficiency (Figure S5).
We next analyzed the functional consequence of these mutations by carrying out time-dependent in vitro cleavage assays monitoring pre-miR-21 processing. In agreement with our binding studies, the S839F and L881P Dicer mutants showed a significant decrease in the level of pre-miR-21 processing compared to that of the WT protein. After a 2 h reaction, while WT Dicer was able to cleave 82% of the pre-miR-21 substrate, the S839F and L881P mutants displayed 20% and 0% total cleavage, respectively (Figure 2C,D). Also analogous with our EMSA results, the S839A and L881A mutants exhibited in vitro cleavage activity similar to that of WT Dicer albeit reduced for the L881A protein (76% and 68% cleavage, respectively) (Figure 2C,D).
To extend our analysis, we performed cellular assays to determine the activity of these mutants in Dicer1−/− mouse mesenchymal cells, an established model for studying Dicer activity.5,26 As an additional mutant for testing, we chose the point mutant R944Q, which was found only in hypermutated tumors and not anticipated to be a driver of disease despite being located in the proximity of the RNA-binding pockets of the Platform-PAZ domain.10 Because this mutant was previously shown to have a negligible impact on Dicer function in cells,10 we selected it as an additional control for our assays. Dicer1−/− cells were transfected with WT and mutant Dicer constructs using the TransIT-LT1 transfection reagent, which was found to yield suitable transfection efficiency and to cause minimal toxicity to the cells (Figure S5). Mature miR-21 levels were then quantified from extracted RNA via qRT-PCR. As shown in Figure 2E, miR-21 levels were reduced for most of the mutants, and in line with our in vitro cleavage assays, Dicer mutants S839F and L881P showed the greatest decrease in comparison to WT Dicer. The R944Q mutant exhibited activity similar to that of WT as expected. To determine if changes in miR-21 were due to enzymatic activity, we measured protein levels of each mutant via Western blot analysis. Interestingly, quantification of protein levels via densitometry analysis and normalization to WT Dicer revealed significant changes in protein levels for the S839F, L881P, and L881A mutants (Figure 2F). All other Dicer mutants were present at protein levels with no significant difference from WT Dicer in the transfected cells (Figure 2F). Thus, in cells, additional regulatory mechanisms may be utilized to reduce the level of production of these loss-of-function Dicer mutants and/or the mutant proteins could be degraded in cells. Thus, the measured decreases in activity are likely due to a combination of the weakened ability to bind substrate and enzymatic activity as revealed by our biochemical analyses, and protein levels.
Effects of Platform-PAZ-Associated Dicer Mutations on snord37 Binding and Processing.
While the primary emphasis of Dicer characterization studies has been on its function in pre-miRNA processing, Dicer is capable of binding and processing a variety of RNA substrates, including small nucleolar RNAs (snoRNAs).43-45 The canonical role of snoRNAs is to guide post-transcriptional modification of rRNA and small nuclear (snRNA) targets.46 However, snoRNAs can also be cleaved into sno-derived RNAs (snordRNA), which perform functions similar to those of miRNAs acting as Ago2-bound guides in the RNA interference (RNAi) pathway.43,47 Further investigations have revealed that Dicer plays a role in snordRNA production and is capable of precisely cleaving certain snoRNAs to a predetermined length.44,45 As this type of cleavage pattern mirrors that observed for pre-miRNAs, we were curious if the Platform-PAZ mutants also altered snoRNA processing.
As a model snoRNA, we chose snord37, which was previously shown to be processed by Dicer and cleaved in a pattern similar to that of pre-miRNAs despite containing distinct predicted secondary structure motifs.44 As shown in Figure 3A, while pre-miR-21 exists as a hairpin loop with minimal perturbation within the stem region,48 snord37 is predicted to contain larger internal loops within its stem–loop structure. Additionally, and of relevance to Platform-PAZ binding, snord37 lacks the canonical 5′ phosphate and 3′ 2-nt overhang found in pre-miRNA substrates. To determine how these distinct RNA structures affect binding to WT and Platform-PAZ Dicer mutants, we utilized EMSA-based binding assays (Figure 3B,C). As expected, because both are known Dicer substrates, the WT enzyme showed no significant differences in binding across the two RNA substrates (30% and 22% for pre-miR-21 and snord37, respectively) (Figure 3D). For the disease mutants, S839F and L881P, although we observed significant impairment of binding to pre-miR-21, this was not observed with snord37 (Figure 3D). Indeed, snord37 exhibited a level of binding similar to that of WT across all mutants examined (Figure 3B,C).
Figure 3.
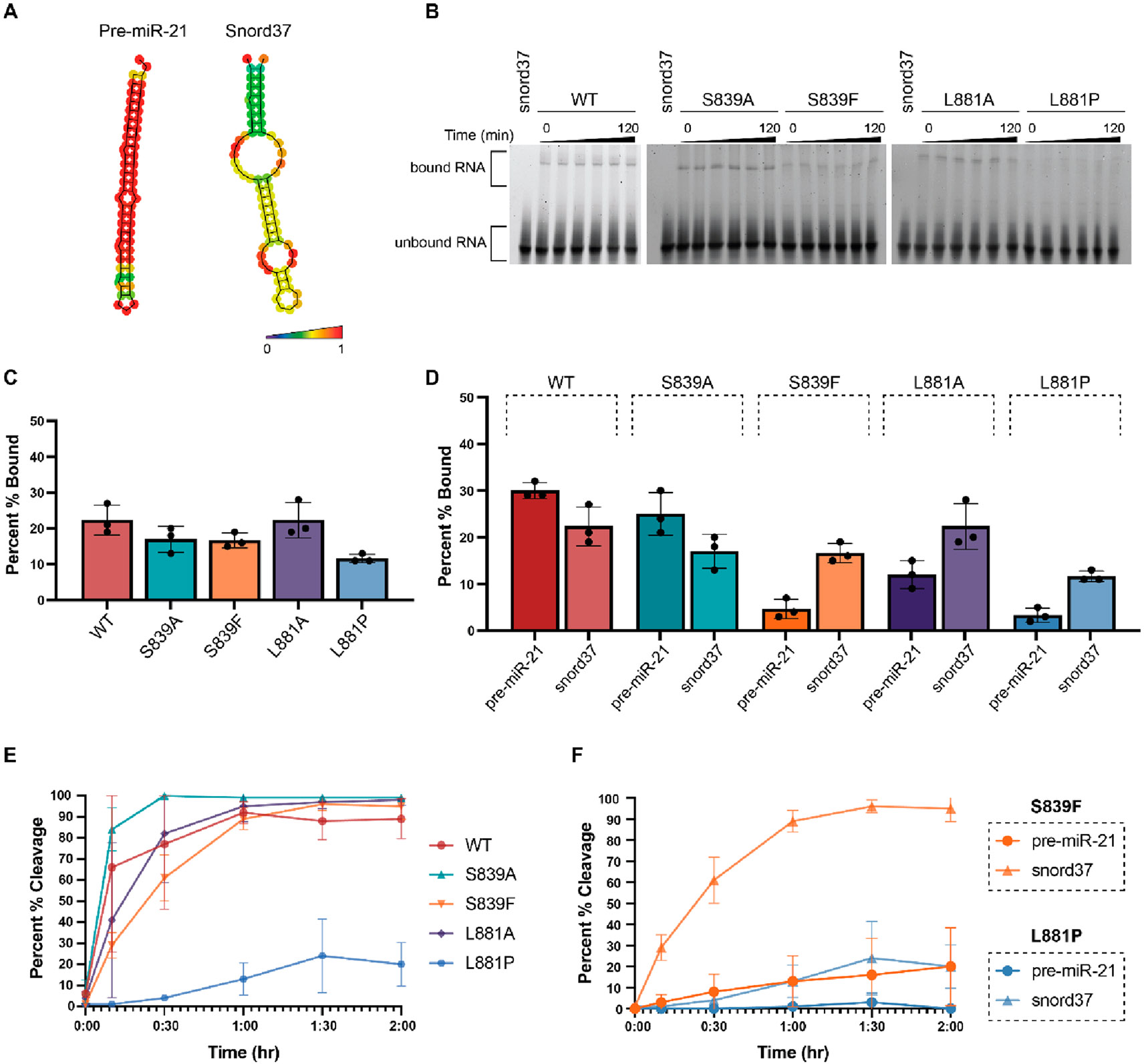
Characterization of the binding and cleavage activity of WT and Dicer mutants for snord37. (A) Predicted secondary structures of pre-miR-21 and snord37 as determined via RNAfold 2.4.18. The color scale indicates base-pair probabilities. (B) Representative EMSA data from three independent experiments (Figure S3) shown over the course of 120 min. (C) Quantification of binding at the 120 min time point as determined using the relative difference in intensity of bound RNA (top band) to unbound RNA (bottom band). (D) Comparison of binding to pre-miR-21 and snord37 at 120 min. (E) Quantification of time-dependent Dicer cleavage activity (Figure S4). Gel-based analysis was carried out using ImageJ. Error bars represent the standard deviation from three independent experiments (Tables S1 and S2). (F) Comparison of cleavage of pre-miR-21 and snord37 at 120 min by Dicer disease mutants S839F and L881P.
To determine if this greater binding capacity was correlated with enhanced substrate processing for snord37, we performed time-dependent in vitro cleavage assays (Figure 3E). In line with our binding data, WT Dicer showed little difference in cleavage activity between pre-miR-21 (82% cleavage) and snord37 (89% cleavage) after 2 h. Moreover, each of the mutants, including disease-associated mutants S839F and L881P, could process snord37 better than pre-miR-21. In particular, S839F showed a major increase in activity, cleaving 95% of snord37 compared to 20% for pre-miR-21 (Figure 3F). L881P, which exhibited negligible activity with pre-miR-21, could cleave 20% of the snord37 substrate (Figure 3F). This diminished cleavage activity in comparison to those of the other mutants is likely due its reduction in the level of RNA binding (Figure 3B,C). Thus, these results, overall, indicate a direct correlation between binding and enzymatic activity for the Dicer substrates tested.
Effects of Platform-PAZ-Associated Dicer Mutations on the Integrity of Small RNA Processing.
As described, the Platform-PAZ domain plays an important role in pre-miRNA substrate recognition via binding to the 3′ 2-nt overhang and 5′ phosphate, and docking of these motifs within the domain enables Dicer to produce mature miRNAs of a precise length.27-28 To determine how the disease-relevant Platform-PAZ mutants affect the integrity of small RNA processing, we were eager to probe the ability of the Dicer mutants to produce the expected 5P and 3P cleavage products.
As an additional substrate for this analysis, we included a 5′-phosphorylated pre-miR-21 lacking a 3′ 2-nt overhang (blunt pre-miR-21). Previous characterization studies of human Dicer have demonstrated the significance that the 3′ 2-nt overhang, specifically, plays in Dicer’s ability to cleave pre-miRNA substrates.49,50 To determine if the absence of this motif also impacts activity with the S839 and L881 mutants, we first examined binding of blunt pre-miR-21 to WT and Platform-PAZ mutants in comparison to that of pre-miR-21 and snord37. As shown in Figure 4A, binding to WT Dicer was similar across the substrates (30%, 22%, and 26% for pre-miR-21, snord37, and blunt pre-miR-21, respectively) and diminished for most of the mutant proteins. For the disease-associated S839F and L881P mutants, blunt pre-miR-21 exhibited binding that was intermediate between those measured for pre-miR-21 and snord37. Most notable was an improvement in binding of blunt pre-miR-21 to L881P in comparison to that of pre-miR-21 (9% and 3%, respectively), indicating that the lack of a 3′ 2-nt overhang may aid in binding to this mutant. No significant difference was observed for S839F in binding to blunt pre-miR-21 and pre-miR-21.
Figure 4.
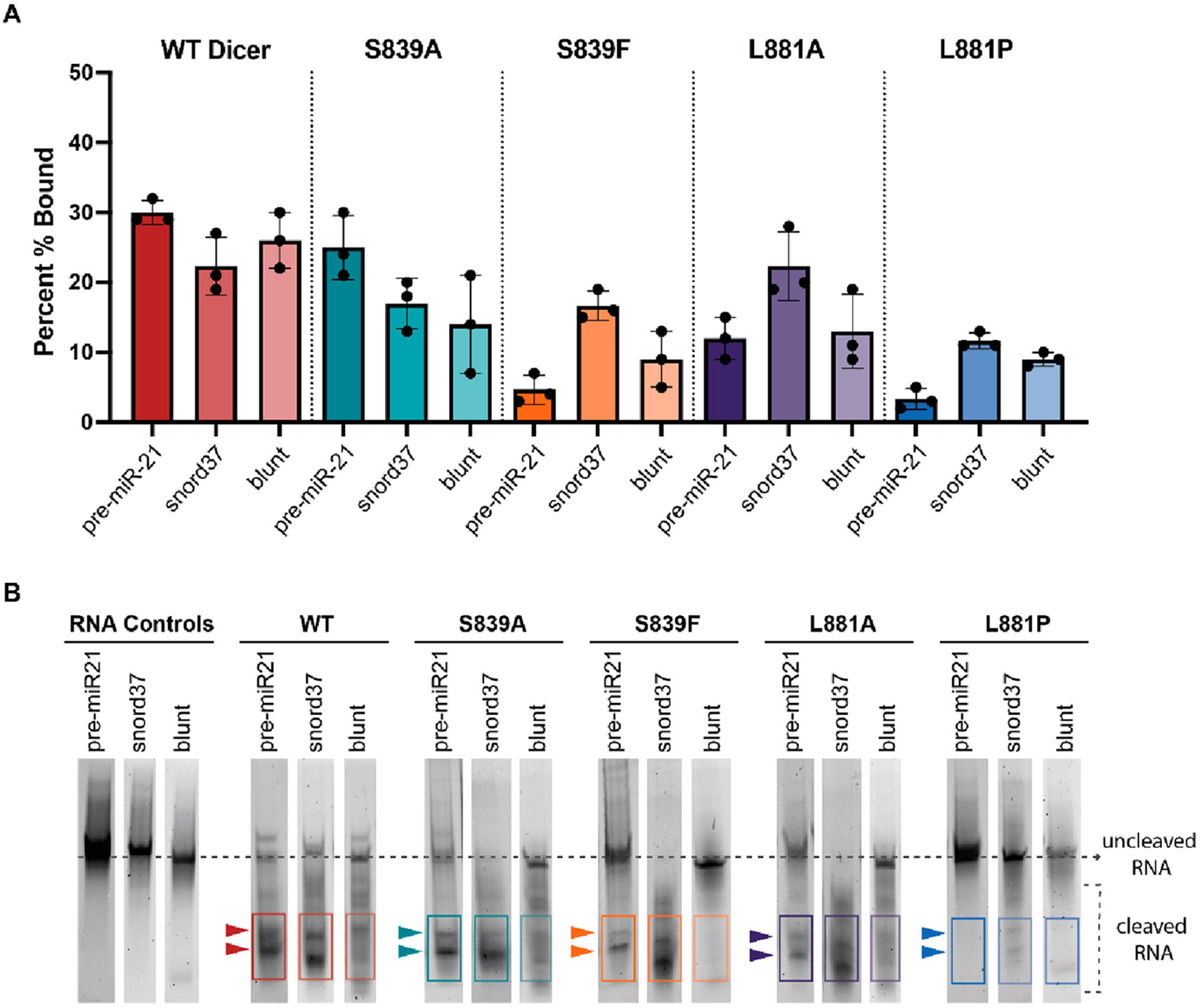
Comparison of the binding, cleavage activity, and cleavage product formation of WT and Dicer mutants for pre-miR-21, snord37, and blunt pre-miR-21. (A) Comparison of binding across substrates at 120 min. Quantified data from three independent experiments (Figure S3 and Table S1). (B) Characterization of Dicer cleavage product formation; representative data from three independent experiments (Figure S4). Cleavage products were observed for pre-miR-21 and snord37 after a 120 min reaction. Blunt pre-miR-21 showed an increase in the levels of multiple cleavage products of varying sizes based on positions in the gel.
We next compared cleavage activity and cleavage product formation across the three Dicer substrates. As shown in Figure 4B, across WT Dicer and disease-associated Platform-PAZ mutants, we observed distinct RNA bands for the cleavage products of both pre-miR-21 and snord37. However, despite sharing a nearly identical secondary structure with pre-miR-21 and similar binding and cleavage patterns across the Dicer enzymes, blunt pre-miR-21 showed a drastic change in product formation. As observed with the WT enzyme, cleavage of blunt pre-miR-21 resulted in an increase in the levels of RNA products of varying lengths and a decrease in the level of formation of the canonical 5P and 3P cleavage products observed with pre-miR-21. These results suggest that proper cleavage of pre-miR-21 is influenced by the presence of its 3′ 2-nt overhang. However, as evidenced by the inactivity of the S839F and L881P mutants with blunt pre-miR-21, the integrity of the Platform-PAZ domain is also critical for processing of this substrate lacking an overhang, potentially indicating disrupted 5′ phosphate interactions with these mutants. This effect is specific to the disease mutants, as both alanine mutants were found to process blunt pre-miR-21 to a level similar to that of WT Dicer.
On the contrary, snord37, which lacks a 5′ phosphate and 3′ 2-nt overhang, was not as strongly affected by the presence of disease-associated mutations S839F and L881P. Indeed, this substrate was the only RNA tested that showed binding to both Dicer disease mutants. These results imply that other structural features of snord37 likely play a role in determining why mutations affecting the structural integrity of the Platform-PAZ domain have a minimized effect on binding and processing of this substrate. One potential hypothesis is that snord37 uses an alternative counting rule. Typically, counting to determine the precise site at which to cleave mature miRNA products begins at the 3′ or 5′ binding pockets within the Platform-PAZ domain, and cleavage is dependent upon the physical distance between these binding pockets and the RNase III domains.1,26-28,44,51 However, more recent studies have revealed a third counting rule, termed the loop counting rule, which uses the single-stranded regions of the RNA substrate, either an internal bulge or a terminal loop, to anchor the cleavage site 2 nt upstream of this structural motif.44,52 Thus, it is possible that loop interactions drive the binding and processing of snord37, and future studies should be directed at investigating this question as well as global analysis of small RNA processing affected by these Dicer mutants in cellular disease models.
CONCLUSION
While it is known that Dicer plays an important role in miRNA biogenesis, we are just beginning to understand the mechanisms behind Dicer processing and its interactions with RNA substrates beyond pre-miRNAs. This extends to Platform-PAZ Dicer mutations and disease-associated Dicer mutations, in general, which are usually characterized only via examination of their effects on miRNA processing. The focus on studying one type of substrate has limited our understanding of how Dicer engages and processes RNA substrates outside of the canonical miRNA pathway. Using clinically relevant and alanine mutations at residues S839 and L881, we tested the significance of the Platform-PAZ domain in Dicer activity between a canonical pre-miRNA, pre-miR-21, and a non-miRNA substrate, snord37. From these analyses, we observed that mutations in the Platform-PAZ domain do not affect Dicer binding and activity uniformly between these substrates. As our study focused on only a representative pre-miRNA and snoRNA, future efforts should focus on expanding this analysis across Dicer’s small RNA substrates to determine the generality of our findings. Nonetheless, our work contributes new knowledge to how Dicer activity can be altered by changes in one of its noncatalytic domains and adds to recent findings characterizing the significance of cancer-associated mutations in Dicer’s Platform-PAZ domain.28
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R01 GM135252 to A.L.G. and F31 GM139291 to R.M.T.).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.2c00687.
Recombinant WT and mutant hsDicer purification with corresponding negative stain TEM analysis; replicate experiments for binding, cleavage, and qRT-PCR studies and transfection efficiency of Dicer1−/− cells; and quantification and analysis of binding, cleavage, qRT-PCR results, and relative protein levels of WT and mutant Dicer in transfected Dicer1−/− cells (PDF)
Accession Codes
Dicer, Q9UPY3.
Contributor Information
Rachel M. Torrez, Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States; Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, United States
Shruti Nagaraja, Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States.
Arya Menon, Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States.
Louise Chang, Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, United States.
Melanie D. Ohi, Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, United States; Department of Cell and Developmental Biology, University of Michigan Medical School, Ann Arbor, Michigan 48109, United States.
Amanda L. Garner, Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States.
REFERENCES
- (1).Torrez RM; Ohi MD; Garner AL Structural insights into the advances and mechanistic understanding of human Dicer. Biochemistry 2023, 62, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bartel DP Metazoan microRNAs. Cell 2018, 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ha M; Kim VN Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol 2014, 15, 509–524. [DOI] [PubMed] [Google Scholar]
- (4).Treiber T; Treiber N; Meister G Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol 2019, 20, 5–20. [DOI] [PubMed] [Google Scholar]
- (5).Gurtan AM; Lu V; Bhutkar A; Sharp PA In vivo structure-function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA 2012, 18, 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Rupaimoole R; Slack FJ MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Disc 2017, 16, 203–221. [DOI] [PubMed] [Google Scholar]
- (7).de Kock L; Wu MK; Foulkes WD Ten years of DICER1 mutations: provenance, distribution, and associated phenotypes. Hum. Mutat 2019, 40, 1939–1953. [DOI] [PubMed] [Google Scholar]
- (8).Foulkes WD; Priest JR; Duchaine TF DICER1: mutations, microRNAs and mechanisms. Nat. Rev. Cancer 2014, 14, 662–672. [DOI] [PubMed] [Google Scholar]
- (9).Kim J; Schultz KAP; Hill DA; Stewart DR The prevalence of germline DICER1 pathogenic variation in cancer populations. Mol. Genet. Genomic Med 2019, 7, No. e555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Vedanayagam J; Chatila WK; Aksoy BA; Majumdar S; Skanderup AJ; Demir E; Schultz N; Sander C; Lai EC Cancer-associated mutations in DICER1 RNase IIIa and IIIb domain exert similar effects on miRNA biogenesis. Nat. Commun 2019, 10, 3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kamburov A; Lawrence MS; Polak P; Leshchiner I; Lage K; Golub TR; Lander ES; Getz G Comprehensive assessment of cancer missence mutation clustering in protein structures. Proc. Natl. Acad. Sci. U. S. A 2015, 112, E5486–E5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kim J; Field A; Schultz KAP; Hill DA; Stewart DR The prevalence of DICER1 pathogenic variation in population databases. Int. J. Cancer 2017, 141, 2030–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Anglesio MS; Wang Y; Yang W; Senz J; Wan A; Heravi-Moussavi A; Salamanca C; Maines-Bandiera S; Huntsman DG; Morin GB Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J. Pathol 2013, 229, 400–409. [DOI] [PubMed] [Google Scholar]
- (14).Pugh TJ; Yu W; Yang J; Field AL; Ambrogio L; Carter SL; Cibulskis K; Giannikopoulos P; Kiezun A; Kim J; McKenna A; Nickerson E; Getz G; Hoffher S; Messinger YH; Dehner LP; Roberts CWM; Rodriguez-Galindo C; Williams GM; Rossi CT; Meyerson M; Hill DA Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene 2014, 33, 5295–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wu MK; de Kock L; Conwell LS; Stewart CJR; King BR; Choong CS; Hussain K; Sabbaghian N; MacRae IJ; Fabian MR; Foulkes WD Functional characterization of multiple DICER1 mutations in an adolescent. Enodcr. Rel. Cancer 2016, 23, L1–L5. [DOI] [PubMed] [Google Scholar]
- (16).JnBaptiste CK; Gurtan AM; Thai KK; Lu V; Bhutkar A; Su M-J; Rotem A; Jacks T; Sharp PA Dicer loss and recovery induce an oncogenic switch driven by transcriptional activation of the oncofetal Imp1–3 family. Genes Dev. 2017, 31, 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chen J; Wang Y; McMonechy MK; Anglesio MS; Yang W; Senz J; Maines-Bandiera S; Rosner J; Trigo-Gonzalez G; Cheng SWG; Kim J; Matzuk MM; Morin GB; Huntsman DG Recurrent DICER1 hotspot mutations in endometrial tumours and their impact on microRNA biogenesis. J. Pathol 2015, 237, 215–225. [DOI] [PubMed] [Google Scholar]
- (18).Wang Y; Chen J; Yang W; Mo F; Senz J; Yap D; Anglesio MS; Gilks B; Morin GB; Huntsman DG The oncogenic roles of DICER1 RNase IIIb domain mutations in ovarian sertoli-leydig cell tumors. Neoplasia 2015, 17, 650–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Stewart DR; Best AF; Williams GM; Harney LA; Carr AG; Harris AK; Kratz CP; Dehner LP; Messinger YH; Rosenberg PS; Hill DA; Schultz KAP Neoplasm risk among individuals with a pathogenic germline variant in DICER1. J. Clin. Oncol 2019, 37, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Brenneman M; Field A; Yang J; Williams G; Doros L; Rossi CT; Schultz KAP; Rosenberg A; Ivanovich J; Turner J; Gordish-Dressman H; Stewart DR; Yu W; Harris AK; Schoettler P; Goodfellow PJ; Dehner LP; Messinger Y; Hill DA Temporal order of RNAase IIIb and loss-of-function mutations during development determines phenotype in pleuropulmonary blastoma/DICER1 syndrome: a unique variant of the two-hit tumor suppression model. F1000Research 2015, 4, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hill DA; Ivanovich J; Priest JR; Gurnett CA; Dehner LP; Desruisseau D; Jarzembowski JA; Wikenheiser-Brokamp KA; Suarez BK; Whelan AJ; Williams G; Bracamontes D; Messinger Y; Goodfellow PJ DICER1 mutations in familial pleuropulmonary blastoma. Science 2009, 325, 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Seki M; Yoshida K; Shiraishi Y; Shimamura T; Sato Y; Nishimura R; Okuno Y; Chiba K; Tanaka H; Kato K; Kato M; Hanada R; Nomura Y; Park M-J; Ishida T; Oka A; Igarashi T; Miyano S; Hayashi Y; Ogawa S; Takita J Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res. 2014, 74, 2742–2749. [DOI] [PubMed] [Google Scholar]
- (23).Heravi-Moussavi A; Anglesio MS; Cheng SWG; Senz J; Yang W; Prentice L; Fejes AP; Chow C; Tone A; Kalloger SE; Hamel N; Roth A; Ha G; Wan ANC; Maines-Bandiera S; Salamanca C; Pasini B; Clarke BA; Lee AF; Lee C-H; Zhao C; Young RH; Aparicio SA; Sorensen PH; Woo MMM; Boyd N; Jones SJM; Hirst M; Marra MA; Gilks B; Shah SP; Foulkes WD; Morin GB; Huntsman DG Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N. Engl. J. Med 2012, 366, 234–242. [DOI] [PubMed] [Google Scholar]
- (24).Doros L; Yang J; Dehner L; Rossi CT; Skiver K; Jarzembowski JA; Messinger Y; Schultz KA; Williams G; André N; Hill DA DICER1 mutation in embryonal rhabdomyo-sarcomas from children with and without familial PPB-tumor predisposition syndrome. Pediatr. Blood Cancer 2012, 59, 558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Doros LA; Rossi CT; Yang J; Field A; Williams GM; Messinger Y; Cajaiba MM; Perlman EJ; Schultz KAP; Cathro HP; Legallo RD; LaFortune KA; Chikwava KR; Faria P; Geller JI; Dome JS; Mullen EA; Gratias EJ; Dehner LP; Hill DA DICER1 mutations in childhood cystic nephroma and its relationship to DICER1-renal sarcoma. Mod. Pathol 2014, 27, 1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Park J-E; Heo I; Tian Y; Simanshu DK; Chang H; Jee D; Patel DJ; Kim VN Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tian Y; Simanshu DK; Ma J-B; Park J-E; Heo I; Kim VN; Patel DJ A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol. Cell 2014, 53, 606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lee Y-Y; Lee H; Kim H; Kim VN; Roh S-H Structure of the human DICER-pre-miRNA complex in a dicing state. Nature 2023, 615, 331–338. [DOI] [PubMed] [Google Scholar]
- (29).Rio Frio T; Bahubeshi A; Kanellopoulou C; Hamel N; Niedziela M; Sabbaghian N; Pouchet C; Gilbert L; O’Brien PK; Serfas K; Broderick P; Houlston RS; Lesueur F; Bonora E; Muljo S; Schimke RN; Bouron-Dal Soglio D; Arseneau J; Schultz KA; Priest JR; Nguyen V-H; Harach HR; Livingston DM; Foulkes WD; Tischkowitz M DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA 2011, 305, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ohi M; Li Y; Cheng Y; Walz T Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online 2004, 6, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Potter CS; Chu H; Frey B; Green C; Kisseberth N; Madden TJ; Miller KL; Nahrstedt K; Pulokas J; Reilein A; Tcheng D; Weber D; Carragher B Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultra-microscopy 1999, 77, 153–161. [DOI] [PubMed] [Google Scholar]
- (32).Carragher B; Kisseberth N; Kriegman D; Milligan RA; Potter CS; Pulokas J; Reilein A Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol 2000, 132, 33–45. [DOI] [PubMed] [Google Scholar]
- (33).Rohou A; Grigorieff N CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 2015, 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bepler T; Kelley K; Noble AJ; Berger B Topaz-Denoise: general deep denoising models for cryoEM and cryoET. Nat. Commun 2020, 11, 5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Punjani A; Rubinstein JL; Fleet DJ; Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [DOI] [PubMed] [Google Scholar]
- (36).Schmittgen TD; Livak KJ Analyzing real-time PCR data by the comparative CT method. Nat. Protoc 2008, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- (37).De Paolis E; Paragliola RM; Concolino P Spectrum of DICER1 germline pathogenic variants in ovarian Sertoli-Leydig cell tumor. J. Clin. Med 2021, 10, 1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kelley LA; Mezulis S; Yates CM; Wass MN; Sternberg MJE The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 2015, 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Andrews MJI; Tabor AB Forming stable helical peptides using natural and artificial amino acids. Tetrahedron 1999, 55, 11711–11743. [Google Scholar]
- (40).Bhattacharjee N; Biswas P Position-specific propensities of amino acids in the beta-strand. BMC Struct. Biol 2010, 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lau P-W; Potter CS; Carragher B; MacRae IJ Structure of the human Dicer-TRBP complex by electron microscopy. Structure 2009, 17, 1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Smith CK; Withka JM; Regan L A thermodynamic scale for the beta-sheet forming tendencies of the amino acids. Biochemistry 1994, 33, 5510–5517. [DOI] [PubMed] [Google Scholar]
- (43).Song M-S; Rossi JJ Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem. J 2017, 474, 1603–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Luo Q-J; Zhang J; Li P; Wang Q; Zhang Y; Roy-Chaudhuri B; Xu J; Kay MA; Zhang QC RNA structure probing reveals the structural basis of Dicer binding and cleavage. Nat. Commun 2021, 12, 3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Rybak-Wolf A; Jens M; Murakawa Y; Herzog M; Landthaler M; Rajewsky N A variety of Dicer substrates in human and C. elegans. Cell 2014, 159, 1153–1167. [DOI] [PubMed] [Google Scholar]
- (46).Wajahat M; Bracken CP; Orang A Emerging functions for snoRNAs and snoRNA-derived fragments. Int. J. Mol. Sci 2021, 22, 10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ender C; Krek A; Friedlander MR; Beitzinger M; Weinmann L; Chen W; Pfeffer S; Rajewsky N; Meister G A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [DOI] [PubMed] [Google Scholar]
- (48).Shortridge MD; Walker MJ; Pavelitz T; Chen Y; Yang W; Varani G A macrocyclic peptide ligand binds the oncogenic microRNA-21 precursor and suppresses Dicer processing. ACS Chem. Biol 2017, 12, 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Zhang H; Kolb FA; Jaskiewicz L; Westhof E; Filipowicz W Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [DOI] [PubMed] [Google Scholar]
- (50).Feng Y; Zhang X; Graves P; Zeng Y A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA 2012, 18, 2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).MacRae IJ; Zhou K; Li F; Repic A; Brooks AN; Cande WZ; Adams PD; Doudna JA Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [DOI] [PubMed] [Google Scholar]
- (52).Gu S; Jin L; Zhang Y; Huang Y; Zhang F; Valdmanis PN; Kay MA The loop position of shRNAs and pre-miRNAs is critical for the accuracy of Dicer processing in vivo. Cell 2012, 151, 900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


