Abstract
Emphysema quantification and sub-typing is actively studied on cohorts of full-lung high-resolution CT (HRCT) scans, with promising results. Transfer of quantification and classification tools to cardiac CT scans, which involve 70% of the lungs, is challenging due to lower image resolution and degradation of textural patterns. In this study, we propose an original deep-learning domain-adaptation framework to use a pre-existing dictionary of lung texture patterns (LTP), learned on gold-standard full-lung HRCT scans, to label emphysema regions on cardiac CT scans. The method exploits convolutional neural networks (CNNs) trained for: 1) supervised lung texture classification on synthetic cardiac images, and 2) adversarial learning to discriminate between real and synthetic cardiac images. Combination of the classification and adversarial tasks enables to label real cardiac CT scans, and is evaluated on the MESA cohort (N = 15,357 scans). Our results show that image features derived from the adversarial training preserve the labeling accuracy on synthetic scans. LTP histogram signatures generated on 4,315 longitudinal pairs of cardiac CT scans, show high level of consistency over time and scanner generations. The ability to robustly label emphysema texture patterns on cardiac CT scans will enable large-scale longitudinal studies over 10 years of follow-up, for better understanding of the disease progression.
Keywords: Lung CT, emphysema, texture analysis, CNN, domain adaptation, adversarial learning
1. INTRODUCTION
Pulmonary emphysema can be characterized by specific texture patterns on full-lung CT images. Supervised [1, 2] and unsupervised [3, 4, 5] learning of these texture patterns is an active research field. In our previous study [5] we have established a set of robust emphysema-specific spatially-localized lung texture patterns (sLTPs) on full-lung high-resolution CT (HRCT) scans, using a dedicated parcellation of the lung shape to introduce location information in lung texture learning. So far, largely due to the limited availability of high-quality longitudinal full-lung CT data, emphysema texture patterns have not been studied in longitudinal setting, while this is crucial for understanding disease progression.
The Multi-Ethnic Study of Atherosclerosis (MESA) [6] study consists of 6,814 subjects screened with cardiac CT scans at baseline (Exam 1, 2000-2002) and follow-ups (Exams 2-4, 2002-2008). Among these subjects, 3,965 were enrolled in the MESA Lung study, and underwent cardiac CT and gold-standard full-lung HRCT scanning in Exam 5 (2010-2012). This large dataset provides an invaluable opportunity for population-level longitudinal study of emphysema texture patterns. Emphysema quantification on cardiac CT scans was shown in [7] to have high reproducibility, high correlation with full-lung HRCT-based measures, and significant associations with risk factors of lung disease in population studies [8, 9]. But a straightforward application of either unsupervised learning or labeling of cardiac scans with the predefined sLTPs is hindered by the following factors. 1) MESA is a general population study thus contains insufficient COPD cases to apply the sLTP learning directly; 2) CT image quality in MESA is heterogeneous: cardiac scans in Exam 1-4 are axial CT scans collected using one EBT scanner and six types of MDCT scanners; while scans in Exam 5 were collected with helical CT scanners; 3) Cardiac scanning protocols differ from full-lung HRCT scans; The average slice thickness is 2.82 mm vs. 0.65 mm for cardiac vs. full-lung HRCT scans, which leads to downgraded lung texture details in cardiac scans; 4) The field of view (FOV) in cardiac scans only includes the bottom 70% of the lungs, which prevents generating precise location information of lung textures.
To label sLTPs in cardiac scans, one option could be to register cardiac and full-lung scans, and learn a discriminative model, such as a convolutional neural network (CNN) model, to classify the cardiac regions of interest (ROIs) using the sLTP labels in registered full-lung scans as ground truth. However, such registration is challenging given the differences in FOVs. Moreover, cardiac CT scans in earlier MESA exams do not have paired full-lung HRCT scans, thus generalization to heterogeneous scanner types cannot be guaranteed with such approach. To solve this problem, we propose the following original pipeline: we first synthesize cardiac CT scans from full-lung HRCT scans with ground truth sLTP labels and learn a CNN model for sLTP labeling on these synthetic cardiac scans. The synthetic scans inevitably present domain differences from real cardiac CT scans. For robust sLTP labeling on real cardiac scans, we further propose a CNN-based unsupervised domain adaptation with adversarial learning (UDAA). The UDAA module aims to fool an auxiliary domain discriminator at differentiating synthetic and real cardiac scans, thus enabling to learn domain-invariant feature representations. We apply our proposed UDAA framework to label 4,315 pairs of longitudinal cardiac CT scans, and test its robustness with respect to image domain differences.
2. METHOD
2.1. Data Cohort and Preprocessing
We exploit ten sLTPs identified by applying the method in [5] to the SPIROMICS [10] full-lung HRCT dataset. SPIROMICS involves 3,200 subjects that are mostly heavy smokers with COPD. In this work, we aim to label the MESA cohort, with N = 15,357 longitudinal cardiac scans as detailed in Table 1. sLTP labeling requires a pre-segmentation of emphysema regions. We use the hidden Markov measure field (HMMF) model [11], which was shown to be able to handle scanner and subject variability in the HRCT and cardiac CT scans [12]. ROIs are extracted from cardiac scans, with a size of 25×25×25 mm3 (36×36×8 voxels) and with percent emphysema larger than the upper limit of normal (ULN) of emphysema. The ULN values were defined by the reference equation derived from healthy never-smokers to account for known demographical differences in percent emphysema [13].
Table 1:
MESA cardiac CT exams along with splits used to train the domain adaptation module.
| Exam (Year) | Ex1 (2000-2002) | Ex2 (2002-2004) | Ex3 (2004-2005) | Ex4 (2005-2008) | Ex5 (2010-2012) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # of Helical scans (train | val | test) | - | - | - | - | - | - | - | - | - | - | - | - | 1,146 | 422 | 414 |
| # of MDCT scans (train | val | test) | 934 | 329 | 2,245 | 408 | 150 | 983 | 423 | 151 | 699 | 152 | 54 | 241 | - | - | - |
| # of EBT scans (train | val | test) | 748 | 260 | 2,167 | 271 | 102 | 967 | 459 | 152 | 772 | 245 | 83 | 380 | - | - | - |
| Total # of scans evaluated | 6,683 | 2,881 | 2,656 | 1,155 | 1,982 | ||||||||||
From the full-lung HRCT scans, we generate synthetic cardiac CT data by down-sampling images along the superior-inferior axis with a factor of 5 (the ratio of average full-lung vs. cardiac scan slice thickness), and keeping the bottom 2/3 of the lung. The superior FOV cutting planes of intra-subject longitudinal cardiac and synthetic cardiac scans are aligned, by segmenting the main bronchi in each scan as the physiological landmark, finding the superior axial slice levels that best match, and cutting all FOVs to the matched levels. Coronal views of a full-lung HRCT scan, a cardiac CT scan, and a synthetic cardiac CT scan are displayed in Fig. 1 (A).
Fig. 1:
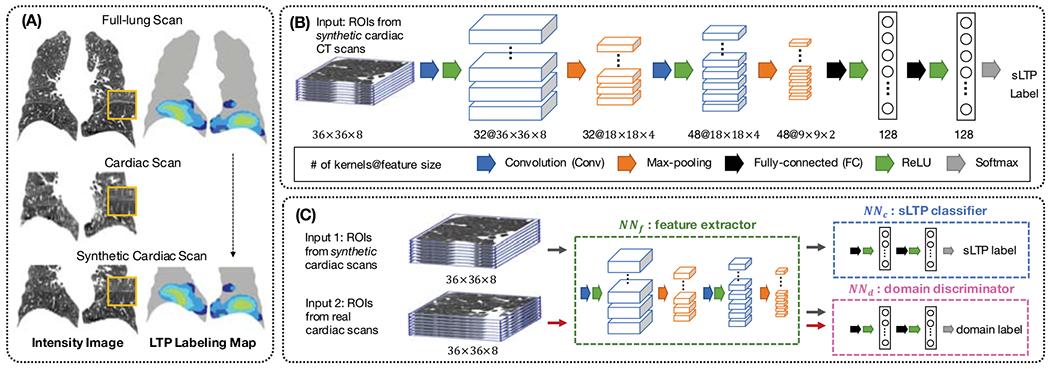
Illustration of the UDAA framework: (A) Generation of synthetic cardiac CT scans; (B) CNN architecture for sLTP labeling on synthetic cardiac CT scans; (C) Domain adaptation component to learn discriminative image features between synthetic and real cardiac scans.
2.2. CNN to Label Synthetic Cardiac ROIs
We first train a sLTP classifier on ROIs from synthetic cardiac scans, which are associated with ground-truth labels without requiring registration. Our CNN model used for sLTP labeling is illustrated in Figure 1 (B), which consists of interleaved convolutional (Conv) layers and max-pooling layers, and finally fully-connected (FC) layers. In our implementation, we choose small-sized kernels (3×3×3) in Conv and maxpooling layers to integrate information in small 3D neighborhood, following the size of textons as in [5]. A stride of 2 is used in max-pooling layers. Rectified linear units (ReLU) are used for the non-linear activation functions. We use a relatively shallow architecture, consisting of two 3D Conv layers and two FC layers, which proved sufficient in our experiments to achieve stable and high-accuracy sLTP labeling without introducing too much variance. The number of kernels in each layer is indicated in Figure 1 (B), which were selected via grid search. We carry over this CNN model to the UDAA module in Figure 1 (C), and split it into the fully-convolutional component NNf, as the feature extractor, and the FC component NNc, as the sLTP classifier.
2.3. UDAA Module
2.3.1. Adversarial Domain Discriminator
We propose to use unsupervised and adversarial domain adaptation introduced in [14] for the labeling of ROIs from real cardiac CT scans. Unsupervised domain adaptation is a method that learns from samples and associated labels in a source domain DS (here, ROIs from synthetic cardiac scans):, and applies the learned model to unannotated samples from a different target domain DT (here, ROIs from real cardiac cardiac scans): . The goal of the learning task is to build a classifier with a low target risk .
The proposed adversarial training module is illustrated in Figure 1 (C). The CNN feature extractor NNf is used to generate feature representations h(·) for both synthetic and real cardiac ROIs. Only features from synthetic ROIs are used to train the sLTP classifier NNc. The NNf and NNc form together a standard feed-forward network architecture. Then, a third neural network component, called the domain discriminator (noted NNd), is added into the framework, which takes all h(·) as input and tries to determine whether it comes from DS or DT. Such discrimination serves as an indicator of how much source-specific the representation h(·) is. The domain discriminator NNd is connected to the feature extractor NNf via a gradient reversal layer [14], which multiplies the gradient by a negative constant during the backpropagation to NNf. Gradient reversal ensures that feature distributions over the two domains DS and DT are forced to be similar, thus resulting in domain-invariant features.
NNc is optimized using a categorical cross-entropy loss function Lclass, whereas NNd is optimized using a binary cross-entropy loss function Ldomain. The adversarial training process minimizes the total loss function Ltotal which simultaneously maximizes domain discrimination loss and minimizes sLTP classification loss, as:
| (1) |
where is a positive weight that defines the relative importance of the domain-adaptation task for the sLTP classifier. During training, is initiated at 0 and is gradually increased up to using the following schedule [14]:
| (2) |
where was set to 10, and p is the training progress, linearly increasing from 0 to 1. This strategy allows the NNd to be less sensitive to noisy signal at the early training stages. The weight is determined by maximizing the metric in the validation set, where ACCclass is the sLTP classification accuracy and ACCdomain is the domain classification accuracy.
2.3.2. Domain Discrimination in Longitudinal Setting
While the domain discriminator does not require registered inputs in DS and DT, there is a risk that the learning process is being driven by population-differences rather than domain-differences if the sampling is not regulated. We therefore enforce sampling of ROIs per training batch to come from the same subjects and similar locations, matching the relative distance vectors di between the center voxels of ROIs xi to the lung mask bounding box (hence not using fine registration). In our longitudinal setting, NNd needs to discriminate synthetic cardiac ROIs in MESA Exam 5 from real cardiac ROIs in earlier exams. We further constrain the ROIs sampling for training NNd, such that the percent emphysema difference is less than 5% for two ROIs xi ∈ S and xj ∈ T, if xi and xj come from same subjects longitudinal scans with |di − dj| < 0.1. This excludes pairing ROIs with drastic changes in inflation level or emphysema progression, which may introduce some bias when training NNd.
3. RESULTS
3.1. Experimental Setting
We use all full-lung HRCT scans and cardiac CT scans in MESA that have ULN values of percent emphysema [13]. This resulted in N = 2,837 subjects in full-lung Exam 5, which we randomly divide into training Strain, validation Sval and test Stest sets with a ratio of 3:1:1. In longitudinal cardiac exams, subjects belonging to Strain and Sval are used to optimize the UDAA framework, while subjects belonging to Stest and subjects not having HRCT full-lung Exam 5 (i.e. unseen during training and validation) are used as longitudinal test sets to evaluate the sLTP labeling performance. The final number of cardiac scans evaluated in this study is reported in Table 1. To measure the influence of the domain-adaptation task in the UDAA module, we also test a source-only CNN training (i.e. training only NNf and NNc using the source synthetic scans, and applying the trained model to real cardiac scans). In validation steps, we measure ACCdomain for the source-only CNN model by adding a domain classifier similar to NNd, but setting its gradient to zero when backpropagating to NNf, so that the domain classifier does not impose any effect on the feature extractor.
3.2. Training and Validation
We trained the UDAA domain adaptation module between synthetic cardiac scans and three target real cardiac scanning conditions (i.e. domains): 1) helical CT scans in Exam 5; 2) MDCT scans in Exam 1-4; and 3) EBT scans in Exam 1-4. We report in Table 2 the overall average values for ACCclass, ACCdomain and the associated for our best model in Sval. From Table 2, we can see that the UDAA module is able to generate CNN features that are less distinguishable by the domain discriminator (lower ACCdomain values), while resulting in only minor sacrifice in the accuracy of sLTP classification task on the source synthetic domain.
Table 2:
Validation accuracy.
| ACCclass | ACCdomain | |||
|---|---|---|---|---|
| Domain | CNN | UDAA | CNN | UDAA () |
| Helical | 0.898 | 0.888 | 0.655 | 0.560 (4.5) |
| MDCT | 0.875 | 0.764 | 0.639 (7.0) | |
| EBT | 0.867 | 0.829 | 0.603 (7.5) | |
ACCclass = sLTP classification accuracy on synthetic ROIs; ACCdomain = domain classification accuracy (real vs. synthetic ROIs).
3.3. sLTP labeling on Longitudinal Cardiac Scans
We labeled all the test cardiac scans in MESA Exam 1-5. Longitudinal consistency of sLTP labeling is evaluated on intrasubject pairs of scans (cardiac vs. synthetic or cardiac vs. cardiac) acquired within a maximum time lapse of 48 months.
3.3.1. Reproducibility of sLTP histograms
We compare sLTP histograms between: 1) Synthetic cardiac scans in Exam 5 (ground truth) vs. real cardiac scans in Exam 5; 2) Synthetic cardiac scans in Exam 5 vs. longitudinal cardiac scans in Exam 4; 3) Longitudinal pairs of real cardiac scans from Exams 1-4 (denoted as ExB for baseline vs. ExF for follow-up), with a time lapse shorter than 48 months. In MESA Exams 1-4, we report separate measures for MDCT and EBT scanners, to evaluate the robustness of our proposed framework versus scanner types. Longitudinal scan pairs with overall non-emphysema proportion changing by more than 20% were excluded from our evaluation. The following metrics are used to compare the sLTP histograms between pairs of scans: 1) χ2 distance, 2) sLTP-level intra-class correlations with respect to the scan in Exam 5, and 3) sLTP-level Pearson correlations for longitudinal scans.
Results are reported in Table 3, with correlation coefficients computed only on the Nk sLTPs that were present in at least twenty pairs of scans (out of Np pairs of scans being tested), for the sake of statistical power. Reported sLTP-level values consist of mean, minimum and maximum among the Nk sLTPs. Statistical differences between the source-only CNN model and the UDAA model were tested using t-test. Significant differences are observed between the two models generally when there is scanner-type change between longitudinal CT exams, and the proposed UDAA model always achieves better reproducibility. If there is no scanner-type change (same domain), reproducibility is similar for both models, which is expected.
Table 3:
Evaluation of reproducibility of sLTP histograms on longitudinal scan pairs acquired within a time lapse <= 48 months.
| Np | Nk | χ2 Distance | Correlation: Mean [Min, Max] | |||
|---|---|---|---|---|---|---|
| CNN | UDAA | CNN | UDAA | |||
| Ex5-Syn. Ex5-Helical |
369 | 8 | 2.81 | 2.41 | 0.73 [0.46, 0.90] | 0.79 [0.52, 0.90] |
| Ex5-Syn. Ex4-MDCT |
51 | 6 | 4.61 | 3.33 | 0.50 [0.19, 0.79] | 0.60 [0.28, 0.81] |
| Ex5-Syn. Ex4-EBT |
73 | 6 | 5.10 | 4.60 | 0.57 [0.14, 0.81] | 0.59 [0.17, 0.82] |
| ExB-MDCT ExF-MDCT |
1,812 | 7 | 1.81 | 1.76 | 0.80 [0.67, 0.86] | 0.82 [0.68, 0.90] |
| ExB-EBT ExF-EBT |
1,839 | 10 | 2.15 | 2.10 | 0.76 [0.55, 0.90] | 0.77 [0.59, 0.92] |
| ExB-EBT ExF-MDCT |
171 | 8 | 4.69 | 3.12 | 0.53 [0.15, 0.84] | 0.67 [0.42, 0.85] |
Bold = significantly better performance (p < 0.05).
3.3.2. Spatial consistency of sLTP labels
we divide each left and right lung bounding boxes into 8 lung zones (superior/inferior, anterior/posterior, medial/lateral), and construct a histogram of 8 bins for each sLTP per lung. Then we compute the χ2 distances between the longitudinal pairs of cardiac scans in Exam 1-4 studied in Table 3. We again compare separately ExB-MDCT vs. ExF-MDCT, ExB-EBT vs. ExF-EBT and ExB-EBT vs. ExF-MDCT scans. We found that χ2 distances are significantly smaller (i.e. better) with the proposed UDAA model for respectively 3, 4, 3 sLTPs, while the other sLTPs do not show significant difference between UDAA and the source-only CNN model.
4. DISCUSSION
In this work, we proposed an unsupervised domain adaptation with adversarial learning (UDAA) framework to label cardiac CT scans with emphysema-specific lung texture patterns (sLTPs) previously learned on full-lung HRCT scans. Translation to cardiac scans relies on synthetic cardiac scans generated from MESA HRCT scans previously labeled with sLTPs. These labels are used as our ground-truth for the supervised training component. Domain adaptation is exploited in an unsupervised context to transition from synthetic to real cardiac scans. Comparison of sLTP labeling on intra-subject pairs of (cardiac, full-lung HRCT) scans from the same scanning session, and on longitudinal cardiac scans acquired within a maximum time lapse of 48 months, show significantly better consistency with the UDAA framework than naively training a CNN model with synthetic-only data. To our knowledge, this is the first study on longitudinal subtyping of emphysema patterns on cardiac CT scans. Such tool can enable large-scale multi-sites longitudinal studies of emphysema subtypes and could potentially advance the understanding of emphysema progression and COPD. Future work will focus on the association of sLTP longitudinal progression patterns with clinical measures and genomic data.
Acknowledgments
Funding provided by NIH/NHLBI R01-HL121270, R01-HL077612, RC1-HL100543, R01-HL093081 and N01-HC095159 through N01-HC-95169, UL1-RR-024156 and UL1-RR-025005.
5. REFERENCES
- [1].Depeursinge Adrien, Foncubierta-Rodriguez Antonio, Ville Dimitri Van De, and Müller Henning, “Three-dimensional solid texture analysis in biomedical imaging: Review and opportunities,” Medical Image Analysis, vol. 18, no. 1, pp. 176–196, 2014. [DOI] [PubMed] [Google Scholar]
- [2].Yang Jie, Feng Xinyang, Angelini Elsa D, and Laine Andrew F, “Texton and sparse representation based texture classification of lung parenchyma in CT images,” in IEEE 38th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC), 2016, pp. 1276–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Binder Polina, Batmanghelich Nematollah K, Estepar Raul San Jose, and Golland Polina, “Unsupervised discovery of emphysema subtypes in a large clinical cohort,” in International MICCAI Workshop on Machine Learning in Medical Imaging. Springer, 2016, pp. 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Song Jingkuan, Yang Jie, Smith Benjamin, Balte Pallavi, Hoffman Eric A, Barr R Graham, Laine Andrew F, and Angelini Elsa D, “Generative method to discover emphysema subtypes with unsupervised learning using lung macroscopic patterns (LMPS): The MESA COPD study,” in IEEE 14th International Symposium on Biomedical Imaging (ISBI), 2017, pp. 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang Jie, Angelini Elsa D, Balte Pallavi P, Hoffman Eric A, Austin John HM, Smith Benjamin M, Song Jingkuan, Barr R Graham, and Laine Andrew F, “Unsupervised discovery of spatially-informed lung texture patterns for pulmonary emphysema: The MESA COPD study,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2017, pp. 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bild Diane E, Bluemke David A, Burke Gregory L, Detrano Robert, Roux Ana V Diez, Folsom Aaron R, Greenland Philip, Ja-cobs David R Jr, Kronmal Richard, and Liu Kiang, “Multi-ethnic study of atherosclerosis: objectives and design,” American Journal of Epidemiology, vol. 156, no. 9, pp. 871–881, 2002. [DOI] [PubMed] [Google Scholar]
- [7].Hoffman Eric A, Jiang Rui, Baumhauer Heather, Brooks Michael A, Carr J Jeffrey, Detrano Robert, Reinhardt Joseph, Rodriguez Josanna, Stukovsky Karen, Wong Nathan D, et al. , “Reproducibility and validity of lung density measures from cardiac ct scans - the multi-ethnic study of atherosclerosis (MESA) lung study,” Academic Radiology, vol. 16, no. 6, pp. 689–699, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Oelsner Elizabeth C, Hoffman Eric A, Folsom Aaron R, Carr J Jeffrey, Enright Paul L, Kawut Steven M, Kronmal Richard, Lederer David, Lima Joao AC, Lovasi Gina S, et al. , “Association between emphysema-like lung on cardiac computed tomography and mortality in persons without airflow obstruction a cohort study emphysema-like lung on ct and all-cause mortality,” Annals of Internal Medicine, vol. 161, no. 12, pp. 863–873, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aaron Carrie P, Schwartz Joseph E, Hoffman Eric A, Angelini Elsa, Austin John HM, Cushman Mary, Jacobs David R Jr, Kaufman Joel D, Laine Andrew, Smith Lewis J, et al. , “A longitudinal cohort study of aspirin use and progression of emphysema-like lung characteristics on ct imaging: the MESA lung study,” Chest, vol. 154, no. 1, pp. 41–50, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Couper David, LaVange Lisa M, Han MeiLan, Barr R Graham, Bleecker Eugene, Hoffman Eric A, Kanner Richard, Kleerup Eric, Martinez Fernando J, Woodruff Prescott G, et al. , “Design of the subpopulations and intermediate outcomes in COPD study (SPIROMICS),” Thorax, pp. thoraxjnl-2013, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hame Yrjo, Angelini Elsa D, Hoffman Eric A, Barr R Graham, and Laine Andrew F, “Adaptive quantification and longitudinal analysis of pulmonary emphysema with a hidden Markov measure field model,” IEEE Transactions on Medical Imaging, vol. 33, no. 7, pp. 1527–1540, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yang Jie, Angelini Elsa D, Balte Pallavi P, Hoffman Eric A, Wu Colin O, Venkatesh Bharath A, Barr R Graham, and Laine Andrew F, “Emphysema quantification on cardiac CT scans using hidden markov measure field model: The MESA Lung study,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2016, pp. 624–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hoffman Eric A, Ahmed Firas S, Baumhauer Heather, Budoff Mathew, Carr J Jeffrey, Kronmal Richard, Reddy S, and Barr R Graham, “Variation in the percent of emphysema-like lung in a healthy, nonsmoking multiethnic sample. the MESA lung study,” Annals of the American Thoracic Society, vol. 11, no. 6, pp. 898–907, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ganin Yaroslav, Ustinova Evgeniya, Ajakan Hana, Germain Pascal, Larochelle Hugo, Laviolette François, Marchand Mario, and Lempitsky Victor, “Domain-adversarial training of neural networks,” Journal of Machine Learning Research, vol. 17, no. 59, pp. 1–35, 2016. [Google Scholar]


