Abstract
Biallelic pathogenic variants in TARS2 lead to combined oxidative phosphorylation deficiency, subtype 21 (COXPD21, MIM #615918), which is a rare mitochondrial encephalomyopathy (ME) characterized by early-onset severe axial hypotonia, limb hypertonia, psychomotor developmental delay, epilepsy and brain anomalies. To date, approximately 28 individuals with COXPD21 and 28 TARS2 variants have been identified. In this study, we reported additional four individuals from three unrelated Chinese families with mitochondrial encephalomyopathy caused by pathogenic variants in TARS2, and described the novel clinical phenotypes and genotypic information. In addition to two novel variants (c.512G > A, p.Arg171Lys; c.988dup, p.Arg330Lysfs*4), one previously reported variant (c.470 C > G, p.Thr157Arg) recurred in six Chinese individuals with COXPD21 but was not present in populations of other races. Our findings expanded the mutation spectrum of TARS2 and confirmed that c.470 C > G is a Chinese-specific founder mutation. The novel phenotypes, including reduced fetal movement, eye anomalies and sleep irregularities, observed in our patients enriched the clinical characteristics of COXPD21.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-024-03365-w.
Keywords: TARS2, COXPD21, Chinese-specific, Founder mutation, Mitochondrial encephalomyopathy
Introduction
Biallelic pathogenic variants in TARS2 cause combined oxidative phosphorylation deficiency 21 (COXPD21), which is a rare mitochondrial encephalomyopathy characterized by early-onset severe axial hypotonia, limb hypertonia, delayed psychomotor development, epilepsy, brain anomalies and increased serum lactate level, often leading to early death; onset after 6 months results in a milder course and longer survival [1]. TARS2, a mitochondrial aminoacyl-tRNA synthetase (mt-aaRS), generates mitochondrial Thr-tRNAThr by catalyzing the binding of homologous tRNA to specific amino acids and clearing mischarged Ser-tRNAThr during mitochondrial translation, which safeguards normal and accurate mitochondrial protein synthesis. To date, 19 mt-aaRS genes have been identified to be involved in human mitochondrial disorders [2, 3]. Pathogenic variants in mt-aaRS genes lead to a reduction in charged tRNA, which impairs mitochondrial protein synthesis and decreases the amount of mtDNA-encoded subunits of the oxidative phosphorylation system (OXPHOS), ultimately resulting in combined respiratory chain enzyme deficiency [4–7]. TARS2 (MIM 612805) encodes a 718-amino acid mitochondrial threonyl tRNA-synthetase, which contains an N-terminal N1 domain (Leu20-Ser124), an N2 domain (for editing) (Pro125-Asp301), an aminoacylation domain (for amino acid activation and tRNA charging) (His302-Gly605), and an anticodon binding domain (for tRNA binding) (Lys606-Phe718) [8]. To date, only 28 COXPD21 patients and 28 pathogenic variants in TARS2 have been reported; thus, the genotype-phenotype relationships are not well established [1, 9–13]. Here, we present four COXPD21 patients from three unrelated Chinese families. Two novel TARS2 variants were identified, and novel phenotypes were observed; these findings expand the mutation spectrum of TARS2 and enrich the clinical characteristics of this disease. Furthermore, we identified TARS2 c.470 C > G as a Chinese-specific founder mutation.
Materials and methods
Ethical compliance
This study was approved by the Ethics Committee of Maternal and Child Health Hospital of Guangxi Zhuang Autonomous Region, Dongguan Maternal and Child Health Care Hospital, and Shenzhen Luohu People’s Hospital. Written informed consent was obtained from the legal guardians for the publication of any potentially identifiable images or data included in this study.
Whole-exome sequencing (WES)
Whole-exome sequencing was employed for our patients and their parents. Genomic DNA was extracted from peripheral blood samples using QIAamp DNA Blood Mini Kit (Qiagen, Germany). Library preparation was operated using the Agilent SureSelect Human All Exon kit V5 (Agilent, Santa Clara, CA). Bcl2fastq tool (v2.15.0.4) was applied for extracting Fastq files from Illumina bcl sequencing file. BWA (0.7.10-r789), Picard (v1.128) and Genome Analysis Toolkit (GATK v3.5) were performed for genome alignments and variant detection. The Annovar tool was used for variant annotation. Suspected variants were confirmed by Sanger sequencing. The pathogenicity of the sequence variants was assessed according to ACMG/AMP guidelines [14].
Results
Clinical phenotypes
Family 1
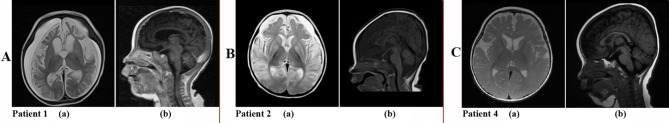
Patient 1 was an 8-month-old boy who was born to a healthy gravida 3, para 2 mother with a negative family history of genetic diseases. He was the first baby of this family, and his delivery at 40 weeks of gestation was uneventful except for obviously reduced fetal movement. He had normal birth measurements: weight 2.8 kg, length 50 cm and head circumference 33 cm. At 3.5 months of age, he exhibited axial hypotonia and could not raise his head. He displayed frequent blinking and nystagmus. Serum lactate level (2.69 mmol/L, normal range 0.5–1.7 mmol/L) was elevated. At 5 months of age, he suffered from intractable epileptic episodes characterized by staring, muscle rigidity, rapid blinking and autonomic symptoms. Electroencephalography (EEG) revealed rhythm weakness on the right side, with diffuse slow waves and epileptic discharges, particularly posteriorly. At 7 months of age, the patient had normal growth development: his height was 69 cm, and his weight was 9 kg. However, he had obvious microcephaly, and his head circumference was 39.5 cm (<-3 SD). Limb hypertonia manifested. Echocardiography revealed an atrial septal defect, diffuse hyperthermic cardiomyopathy and pericardial effusion. MRI showed bilateral underdeveloped insula, frontal, temporal and occipital lobes, microgyria, decreased white-matter volume, dilated bilateral lateral ventricles, bilateral symmetrical T2-weighted imaging (T2WI) multiple hyperintense lesions in the basal ganglia, thalamus and lenticular nucleus and a thin corpus callosum (Fig. 1A). His serum lactate level was increased (8.79 mmol/L). The patient passed away at 8 months of age due to respiratory failure.
Fig. 1.
Brain MRI for our patients. Patient 1 (1 A): MRI showed bilateral underdeveloped insula, frontal, temporal and occipital lobes, microgyria, decreased white-matter volume, dilated bilateral lateral ventricles, bilateral symmetrical T2-weighted imaging (T2WI) multiple hyperintense lesions in the basal ganglia, thalamus and lenticular nucleus (a) and a thin corpus callosum (b); Patient 2 (1B): MRI showed frontal subdural fluid, bilateral hemispheric brain atrophy, multiple hyperintense lesions in the basal ganglia, globus pallidus, thalamus, midbrain and cerebral peduncles (a), and a thin corpus callosum (b); Patient 4 (1 C): Brain MRI revealed hyperintensity of the bilateral basal ganglia on symmetrical T2-weighted imaging (T2WI) (a) and a thin corpus callosum (b)
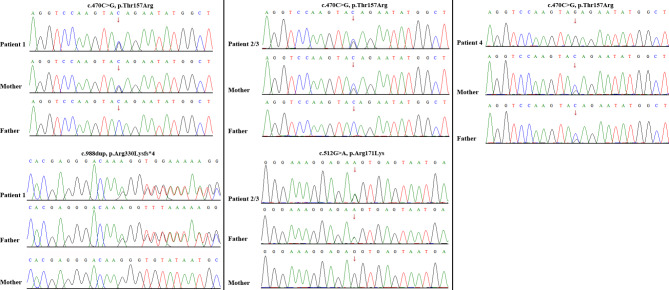
WES identified compound heterozygous variants, c.470 C > G, p.Thr157Arg and c.988dup, p.Arg330Lysfs*4, in TARS2 in the proband. The maternally inherited variant (c.470 C > G, p.Thr157Arg) has been previously reported in the literatures [10, 11]. The paternally inherited variant (c.988dup, p.Arg330Lysfs*4) is novel, and is predicted to truncate the aminoacylation domain of TARS2 (Fig. 2a). Both variants are classified as clinically pathogenic according to the ACMG/AMP guidelines [14].
Fig. 2.
Variant identification by Sanger sequencing. Compound heterozygous TARS2 variants, c.470 C > G (p.Thr157Arg) (maternally inherited) and c.988dup (p.Arg330Lysfs*4) (paternally inherited), in patient 1 (a); c.470 C > G (p.Thr157Arg) (maternally inherited) and c.512G > A (p.Arg171Lys) (paternally inherited), in patient 2 and his sibling (patient 3) (b); a homozygous variant, c.470 C > G (p.Thr157Arg) in TARS2, in patient 4. Both parents were asymptomatic heterozygous carriers (c)
Family 2
Patient 2 was the second child of a nonconsanguineous Chinese couple. The pregnancy was normal except for significantly reduced fetal movement. He was born at full term with normal birth measurements: his length was 49 cm, his weight was 3.64 kg and his head circumference was 34 cm. His Apgar scores were 8, 9, and 9. After birth, he was referred to the clinic due to pneumonia and jaundice. At 1 month of age, he displayed axial hypotonia and feeding difficulties. At 2.5 months of age, he was diagnosed with focal epilepsy, with each seizure lasting for approximately one minute and characterized by staring and rigidity of the upper extremities. EEG showed epileptiform discharges with frequent polyspikes and slow waves. The symptoms could not be effectively alleviated by administering antiepileptic drugs. Serum lactate (7.03 mmol/L) and blood NH3 (73 µmol/L, normal range 9–47 µmol/L) levels were elevated. At 4 months of age, he developed limb hypertonia. MRI showed frontal subdural fluid, bilateral hemispheric brain atrophy, multiple hyperintense lesions in the basal ganglia, globus pallidus, thalamus, midbrain and cerebral peduncles, and a thin corpus callosum (Fig. 1B). He had a growth delay at 6 months of age: his height was 63 cm (<-2 SD), his weight was 6.2 kg (<-2 SD), and his head circumference was 40 cm (-3 SD). He died due to severe respiratory failure at 1 year and 10 months of age.
His elder brother (Patient 3) was also born after an uneventful pregnancy, during with markedly reduced fetal movement was observed. His birth measurement parameters were normal: his weight was 2.6 kg, length was 50 cm, and head circumference was 34 cm. He also exhibited clinical features of mitochondrial encephalopathy that were strikingly similar to those of his younger brother. He had never achieved the ability to raise his head alone and exhibited no eye contact. Increased serum lactate level was regularly monitored. He exhibited delayed growth: his height was 73 cm (<-2 SD), his weight was 9 kg (<-2 SD), and his head circumference was 42.5 cm (<-3 SD) at 1 year and 3 months of age. He died due to respiratory problems, lactic acidosis and severe developmental delay at 2 years and 4 months of age.
WES revealed the recurrent missense variant, c.470 C > G, p.Thr157Arg (maternally inherited), and a novel variant, c.512G > A, p.Arg171Lys (paternally inherited), in TARS2 in the two affected children. The variant c.512G > A, p.Arg171Lys was located in the N2 domain and was highly conserved among different species, and was absent in in the Genome Aggregation Database (Fig. 2b). The variant was confirmed in trans with the previously identified pathogenic variant c.470 C > G, p.Thr157Arg. Thus, the variant was classified as likely pathogenic according to the ACMG/AMP guidelines [14].
Family 3
Patient 4 was a 2-year and 2-month-old female born to unrelated parents. She was born at full term after an uneventful pregnancy. She had normal birth measurements: her length was 50 cm, her weight was 3.3 kg, and her head circumference was 33 cm. She was able to raise her head at 5 months of age. However, the patient presented with progressive developmental regression, including unstable head control and axial hypotonia at 7 months of age, and she gradually displayed limb spasticity after eight months of age. She had obvious feeding difficulties. An increased serum lactate level was detected (4.9 mmol/L). Furthermore, plasma amino acid analysis revealed increased free carnitine (70.28 µmol/L, normal range 9-53.7 µmol/L) and hyperammonemia (98 µmol/L, normal range 18–72 µmol/L). At 5 months of age, she displayed infantile spasms, characterized by brief generalized muscle spasm with straight arms extended and bent torso and legs. The events lasted for 15 s–1 min each time, and eight episodes usually occurred every day. EEG revealed hypsarrhythmia, including a large number of irregular high-voltage, sharp spikes/polyspikes/slow waves. The patient’s epilepsy was not controlled even when sodium valproate or a ketogenic diet was applied. The serum lactate concentration was maintained at a constant level according to real-time monitoring. Brain MRI revealed hyperintensity of the bilateral basal ganglia on symmetrical T2-weighted imaging (T2WI) and a thin corpus callosum at 7 months of age (Fig. 1C). On recent physical examination at 2 years and 2 months of age, she exhibited growth delay: her height was 82 cm (-2 SD), her weight was 10 kg (-2 SD), and her head circumference was 43.5 cm (<-3 SD). She was not able to raise her head and had no language or cognitive development.
WES revealed a homozygous variant, c.470 C > G, p.Thr157Arg in TARS2, in the patient. Both parents were asymptomatic heterozygous carriers (Fig. 2c).
Discussion
Biallelic pathogenic variants in TARS2 are the genetic cause of combined oxidative phosphorylation deficiency 21 (COXPD21). To date, only 28 variants in TARS2 have been reported in 28 COXPD21 patients from 24 unrelated families [1, 9–13]. Here, we described additional four patients from three unrelated Chinese families who displayed psychomotor delay, axial hypotonia, limb hypertonia, intractable epilepsy, increased serum lactate levels and abnormal brain MRI. Patients were diagnosed with mitochondrial encephalomyopathy by experienced clinical specialists. WES revealed a recurrent missense variant (c.470 C > G, p.Thr157Arg) and two novel variants (c.512G > A, p.Arg171Lys and c.988dup, p.Arg330Lysfs*4). These variants were classified as clinically likely pathogenic/ pathogenic and were related to our patients’ manifestations. These newly identified variants further expand the TARS2 mutation spectrum and improve the molecular diagnosis of COXPD21.
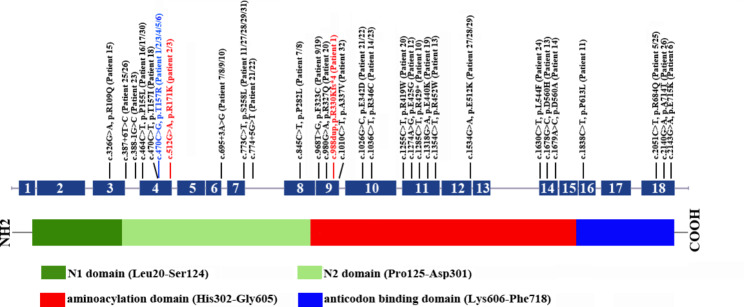
To date, a total of 30 TARS2 variants, including the two novel variants reported in this study, have been identified [1, 9–13]. TARS2 null variants (nonsense, frameshift and splicing) and missense variants accounted for 20% and 80%, respectively. These variants are distributed in the N1 domain (3.3%), N2 domain (33.3%), aminoacylation domain (50%) and anticodon binding domain (13.3%) (Fig. 3). Patients carrying TARS2 null variants (Patients 1, 7, 8, 9, 10), especially biallelic null variants (Patient 10), are more likely to suffer from early-onset severe clinical phenotypes with early death (usually less than one year old). Individuals carrying TARS2 biallelic missense variants presented with later-onset manifestations with longer survival, except for Patients 2, 3, 15, 18 and 30, who may have been affected by complications that were not promptly corrected [1, 9–13]. Null variants may lead to more severe clinical outcomes than missense variants owing to complete loss of protein function. According to previous reports in Chinese patients, the c.470 C > G, p.Thr157Arg variant recurred in six Chinese patients [10, 11]. According to the Genome Aggregation Database, this variant was detected only in East Asians, with an allele frequency of 0.00001193 (3/251430), but it recurred in affected patients who were either heterozygous or homozygous. This result suggests that c.470 C > G is a Chinese-specific founder mutation with a frequency of 1.8/104 in the Chinese population based on the cumulative allele frequency in one commercial Chinese genetic testing database (n = 152,423).
Fig. 3.
Schematic representation of TARS2 variants identified to date. The structure of TARS2 contained 18 exons (dark blue rectangles), and introns (grey horizontal line); lower side: the TARS2 protein domains: N1 domain; N2 domain; Aminoacylation domain; Anticodon binding domain; motif. The localization of variants and substitutions identified is signed with vertical line. Black: Variants identified in the literature; Red: Novel variants detected in this study. Blue: the founder mutation in Chinese patients with COXPD21
Next, we analyzed the clinical phenotypes of all COXPD21 patients (Supplementary Table 1) [1, 9–13]. The main clinical features included psychomotor development delay (30/31), axial hypotonia (27/29), limb hypertonia (21/27), developmental regression (12/32), absent speech (21/30), epilepsy (17/28), growth delay (14/26), microcephaly (10/24), eye anomalies (10/25), increased serum lactate (21/23), brain MRI anomalies (25/28) and EEG anomalies (11/13). The other phenotypes included hearing loss (4/25), feeding difficulties (9/9), no/poor eye-eye contact (8/8), respiratory problems (6/8), reduced fetal movement (4/4), hyperhidrosis (2/7), sleep irregularities (1/2) and echocardiography anomalies (6/24). Neuropsychiatric anomalies and increased serum lactate levels occur in almost all reported COXPD21 patients. Epilepsy is a common phenotype of COXPD21. All of patients with epilepsy except for Patient 24 suffered from intractable epilepsy with no response to antiepileptic drugs [1, 10, 11, 13]. Developmental regression, absent speech, growth delay, microcephaly, feeding difficulties, no/poor eye-eye contact and respiratory problems were frequently described in COXPD21 patients, which should be added in OMIM phenotypic spectrum of COXPD21 [1, 9–13]. Notably, eye anomalies, including nystagmus and strabismus, have been observed in previously reported patients [1, 13]. Patient 5 underwent further follow-up by Yuan (Haiming Yuan is the corresponding author of the literature) [11]. Now, Patient 5 was 4.5 years old. Brief episodes of tonic upward eye deviation and visual inattention were observed. Three patients (Patients 10, 13 and 27) also exhibited the similar features [1, 13]. It added to the known eye anomaly features of COXPD21 [1]. Reduced fetal movement was previously reported in Patient 5, which was also observed in three patients (Patients 1, 2 and 3) in this study, which further confirmed that reduced fetal movement is one of the characteristics of COXPD21. Furthermore, Patient 5 displayed obvious sleep irregularities beginning at 2 years of age. This phenotype was not described in other patients either because some patients were too young to exhibit the trait or because the phenotype was ignored during clinical evaluation, which deserves further investigation. In summary, the novel phenotypes greatly enriched the clinical features of COXPD21.
Conclusions
We identified two novel TARS2 variants in our COXPD21 patients, which expands the genetic variant spectrum of TARS2. Our study suggested that TARS2 c.470 C > G is a Chinese-specific founder mutation and should be considered for inclusion in a Chinese carrier screening panel. Our patients exhibited novel phenotypes, including eye anomalies, reduced fetal movement and sleep irregularities. These findings will deepen our understanding of the clinical characteristics of COXPD21 patients and guide their clinical management and genetic counseling.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors appreciate the patients and her parents for their cooperation.
Abbreviations
- COXPD21
Combined oxidative phosphorylation deficiency 21
- ME
Mitochondrial encephalomyopathy
- TARS2
Threonyl tRNA-synthetase 2
- WES
Whole-exome sequencing
- ACMG/AMP
American College of Medical Genetics and Genomics/Association for Molecular Pathology
Authors’ contributions
QMW. Conception and design: SJZ, HSQ, HMY and BLL. Clinical evaluation and data collection: SJZ, HMY, BLL, HSQ, QMW, YHL, ZLQ, JSL, QY and JH. Data analysis: SJZ, HMY, ZLQ, QMW and QY. Sanger sequencing: QLC. Biochemical and mass spectrometry detection: GXG, XJ and LJK. MRI diagnosis: YFW. Sample collection and DNA extraction: GXG and SKW. Manuscript writing: SJZ and HMY. Picture processing: HMY, HSQ and SJZ. Manuscript editing: HMY and YPS. Supervision throughout the study: SJZ, HMY and BLL. Final approval: ALL.
Funding
This study was supported by the Guangxi Natural Science Foundation under Grant No.2024GXNSFBA010072; the Young Scientists Fund of the National Natural Science Foundation of China (No. 82201312); the National Natural Science Foundation of China (No. 82071276); the Health Department of Guangxi Zhuang Autonomous Region (No. Z-A20230362 and No. Z-A20240323).
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The data are not publicly available due to privacy or ethical restrictions.
Declarations
Ethics approval and consent to participate
This clinical and genetic study was approved by each of the participating institutions’ review boards. Informed consent was obtained from each study subject (or their guardian) prior to enrollment.
Consent for publication
Consent was obtained from all patients for publication of any potentially identifiable images or data involved in this study.
Competing interests
All the authors declare no conflicts of interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shujie Zhang, Haisong Qin and Qingming Wang contributed equally to this work.
Contributor Information
Haiming Yuan, Email: haimingyuan@sina.cn.
Baoling Lai, Email: angell_hood95@sina.com.
References
- 1.Zheng WQ, Pedersen SV, Thompson K, Bellacchio E, French CE, Munro B, Pearson TS, Vogt J, Diodato D, Diemer T, Ernst A, Horvath R, Chitre M, Ek J, Wibrand F, Grange DK, Raymond L, Zhou XL, Taylor RW, Ostergaard E. Elucidating the molecular mechanisms associated with TARS2-related mitochondrial disease. Hum Mol Genet. 2022;31(4):523–34. [DOI] [PubMed] [Google Scholar]
- 2.Ni M, Black LF, Pan C, Vu H, Pei J, Ko B, Cai L, Solmonson A, Yang C, Nugent KM, Grishin NV, Xing C, Roeder E, DeBerardinis RJ. Metabolic impact of pathogenic variants in the mitochondrial glutamyl-tRNA synthetase EARS2. J Inherit Metab Dis. 2021;44(4):949–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei N, Zhang Q, Yang XL. Neurodegenerative Charcot-Marie-tooth disease as a case study to decipher novel functions of aminoacyl-tRNA synthetases. J Biol Chem. 2019;294(14):5321–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boczonadi V, Ricci G, Horvath R. Mitochondrial DNA transcription and translation: clinical syndromes. Essays Biochem. 2018;62(3):321–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diodato D, Ghezzi D, Tiranti V. The mitochondrial aminoacyl tRNA synthetases: genes and syndromes. Int J Cell Biol. 2014;2014:787956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sissler M, González-Serrano LE, Westhof E. Recent advances in mitochondrial Aminoacyl-tRNA synthetases and Disease. Trends Mol Med. 2017;23(8):693–708. [DOI] [PubMed] [Google Scholar]
- 7.Musante L, Püttmann L, Kahrizi K, Garshasbi M, Hu H, Stehr H, Lipkowitz B, Otto S, Jensen LR, Tzschach A, Jamali P, Wienker T, Najmabadi H, Ropers HH, Kuss AW. Mutations of the aminoacyl-tRNA-synthetases SARS and WARS2 are implicated in the etiology of autosomal recessive intellectual disability. Hum Mutat. 2017;38(6):621–36. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Zhou XL, Ruan ZR, Liu RJ, Eriani G, Wang ED. A human disease-causing point mutation in mitochondrial Threonyl-tRNA synthetase induces both structural and functional defects. J Biol Chem. 2016;291(12):6507–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diodato D, Melchionda L, Haack TB, Dallabona C, Baruffini E, Donnini C, Granata T, Ragona F, Balestri P, Margollicci M, Lamantea E, Nasca A, Powell CA, Minczuk M, Strom TM, Meitinger T, Prokisch H, Lamperti C, Zeviani M, Ghezzi D. VARS2 and TARS2 mutations in patients with mitochondrial encephalomyopathies. Hum Mutat. 2014;35(8):983–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Peng B, Hou C, Li J, Zeng Y, Wu W, Liao Y, Tian Y, Chen WX. Novel compound heterozygous TARS2 variants in a Chinese family with mitochondrial encephalomyopathy: a case report. BMC Med Genet. 2020;21(1):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He P, Wang Q, Hong X, Yuan H. Novel TARS2 variant identified in a Chinese patient with mitochondrial encephalomyopathy and a systematic review. Am J Med Genet A. 2023;191(1):70–6. [DOI] [PubMed] [Google Scholar]
- 12.Gao X, Xin G, Tu Y, Liang X, Yang H, Meng H, Wang Y. TARS2 variants cause combination oxidative phosphorylation Deficiency-21: a Case Report and Literature Review. Neuropediatrics. 2024;55(3):178–182. [DOI] [PubMed]
- 13.Accogli A, Lin SJ, Severino M, Kim SH, Huang K, Rocca C, Landsverk M, Zaki MS, Al-Maawali A, Srinivasan VM, Al-Thihli K, Schaefer GB, Davis M, Tonduti D, Doneda C, Marten LM, Mühlhausen C, Gomez M, Lamantea E, Mena R, Nizon M, Procaccio V, Begtrup A, Telegrafi A, Cui H, Schulz HL, Mohr J, Biskup S, Loos MA, Aráoz HV, Salpietro V, Keppen LD, Chitre M, Petree C, Raymond L, Vogt J, Sawyer LB, Basinger AA, Pedersen SV, Pearson TS, Grange DK, Lingappa L, McDunnah P, Horvath R, Cognè B, Isidor B, Hahn A, Gripp KW, Jafarnejad SM, Østergaard E, Prada CE, Ghezzi D, Gowda VK, Taylor RW, Sonenberg N, Houlden H, Sissler M, Varshney GK, Maroofian R. Clinical, neuroradiological, and molecular characterization of mitochondrial threonyl-tRNA-synthetase (TARS2)-related disorder. Genet Med. 2023;25(11):100938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The data are not publicly available due to privacy or ethical restrictions.