Abstract
Gingiva plays a crucial barrier role at the interface of teeth, tooth‐supporting structures, microbiome, and external agents. To mimic this complex microenvironment, an in vitro microphysiological platform and biofabricated full‐thickness gingival equivalents (gingiva‐on‐chip) within a vertically stacked microfluidic device is developed. This design allowed long‐term and air‐liquid interface culture, and host‐material interactions under flow conditions. Compared to static cultures, dynamic cultures on‐chip enabled the biofabrication of gingival equivalents with stable mucosal matrix, improved epithelial morphogenesis, and barrier features. Additionally, a diseased state with disrupted barrier function representative of gingival/oral mucosal ulcers is modeled. The apical flow feature is utilized to emulate the mechanical action of mouth rinse and integrate the assessment of host‐material interactions and transmucosal permeation of oral‐care formulations in both healthy and diseased states. Although the gingiva‐on‐chip cultures have thicker and more mature epithelium, the flow of oral‐care formulations induced increased tissue disruption and cytotoxic features compared to static conditions. The realistic emulation of mouth rinsing action facilitated a more physiological assessment of mucosal irritation potential. Overall, this microphysiological system enables biofabrication of human gingiva equivalents in intact and ulcerated states, providing a miniaturized and integrated platform for downstream host‐material and host‐microbiome applications in gingival and oral mucosa research.
Keywords: biocompatibility, drug permeation, gingiva, microfluidics, organ‐on‐a‐chip, periodontal disease
Researchers have developed a "gingiva‐on‐chip" system that mimics the human gum tissue. This "gingiva‐on‐chip" model enabled reconstruction of more mature gingival tissues compared to traditional static 3D cultures. It facilitated realistic emulation of mouth rinsing action and physiological assessment of oral‐care products in health and diseased states. This advancement provides a valuable tool for periodontal research and oral‐care product development.
1. Introduction
Gingiva and oral mucosal lining is a distinct microenvironment subject to a myriad of extrinsic factors such as salivary flow, masticatory forces, microbial colonization, habits (such as smoking and drinking), and exposure to dental and oral‐care products such as toothpastes, whitening agents, mouthwashes, anesthetics, and dental restorations.[ 1 , 2 ] In the healthy state, the gingival epithelium along with its keratinized superficial layer and underlying connective tissue termed lamina propria act as barriers against these external factors.[ 3 , 4 ] The barrier function plays a crucial role in the permeation of chemicals, microbes, and their toxins into the underlying proliferating basal epithelial layer and connective tissues.[ 2 , 4 ] Epithelial disruption in diseased states such as periodontal disease, erosion, and ulcer, can lead to direct exposure of the underlying cells and tissues to those external factors. Hence, biomimicry of the complex microenvironment is essential for the spatiotemporal assessment of host‐microbiome and host‐material interactions and responses representative of the health and diseased states.
Current methods for the evaluation of dental biomaterials and oral‐care products are based on standards published by International Organization for Standardization (ISO 10993), American National Standards Institute/American Dental Association (ANSI/ADA 41–2020), American Society for Testing Materials (ASTM F1027‐86(2017)) and Organization for Economic Co‐operation and Development test guidelines (OECD TG431, TG439).[ 1 , 5 ] Existing standards are commonly based on assays using monolayer cultures of a single cell type. In recent years, there has been a significant impetus toward the development and application of in vitro three‐dimensional (3D) organotypic models that emulate the complex microenvironment of the gingiva and oral mucosa such as reconstructed epithelium,[ 6 , 7 , 8 ] connective tissue[ 9 ] and full‐thickness gingival[ 10 , 11 , 12 , 13 , 14 , 15 ] equivalents. These 3D models offer a physiological alternative to understanding the host‐material and host‐microbial interactions. However, 3D cultures under static conditions still have significant limitations in replicating the dynamic microenvironment of the gingiva, such as the effects of fluid flow, shear stress, and long‐term culture.
Microfluidic and microfabrication technologies have led to miniaturized cell culture systems termed “organ‐on‐a‐chip” devices.[ 16 ] These organ‐on‐a‐chip devices contain hollow microchannels and microchambers that support fluid control of cell culture media and reagents at the micro‐ and even nanoliter scale. Fluid control systems provide continuous nutrient supply, controlled delivery, and mechanical stimulation through active flow, resulting in a superior representation of the in vivo microenvironment.[ 16 , 17 , 18 ] Further, miniaturization helps to minimize sample size and reagent volumes needed for the assays.[ 19 ] Recent advancements in oral and dental barrier tissue emulation using microfluidic systems are gaining significant traction with platforms such as oral mucosa‐on‐chip,[ 20 , 21 , 22 , 23 ] gingival crevice‐on‐chip,[ 24 ] gingival epithelial‐capillary interface‐on‐chip[ 25 ] and tooth‐on‐chip.[ 15 , 26 , 27 ]
Integration of fluid dynamics and organotypic cultures using perfusion bioreactors,[ 28 , 29 ] oral mucosa‐on‐chip,[ 20 , 21 , 22 , 23 , 30 ] gingival crevice‐on‐chip,[ 24 , 30 ] and gingival epithelial‐capillary interface‐on‐chip[ 25 ] provided unprecedented opportunities to emulate microanatomical features of the gingiva and oral mucosa such as interstitial fluid flow, and controlled exposure to dental materials and bacteria.[ 18 ] These features provided more insights into the host‐material and host‐microbiome interactions under microphysiological flow conditions. However, design features such as horizontally stacked configuration of the microchannels and collagen matrix contraction limited the long‐term culture capabilities,[ 22 , 23 ] epithelial stratification, and the formation of cornified barrier layers,[ 21 , 23 , 24 , 25 , 30 ] which can only be achieved through an air‐liquid interface culture.[ 17 ] Studies have demonstrated that the incorporation of fluid flow enhances the proliferation and metabolic activity of gingival fibroblasts[ 31 ] and keratinocytes.[ 32 ] Besides improving the mass transport of nutrients, active perfusion of culture media also allows the mimicry of interstitial fluid flow, emulating the protective effects of gingival crevicular fluid flow and associated host innate immune responses.[ 24 ] Hence, an in vitro microphysiological platform composed of stratified, keratinized gingival epithelium over a cellular and mechanically stable lamina propria could provide an attractive alternative to current models for a broad variety of oral biology and toxicology studies. Further, microfluidic design features that allow fluid dynamics, an air‐liquid interface culture, and ease of access of the cultured gingival tissues for mechanical and fluidic manipulation would allow to emulate the native tissue‐like features and obtain responses predictive of human tissues.
Hence, the aim of this study was to biofabricate a miniature, full‐thickness gingival equivalents under air‐liquid interface within a microfluidic device (gingiva‐on‐chip). Using this platform, we aimed to understand the impact of flow on epithelial morphogenesis and its downstream application for mucosal irritation and transmucosal permeation studies in healthy and diseased states.
2. Methods
2.1. Device Microfabrication
Gingiva‐on‐chip equivalents were fabricated on a modified version of the microfluidic device described earlier.[ 17 ] The microfluidic device (5 cm × 7.5 cm) is composed of micro‐structured polymethyl methacrylate sheets of different thicknesses that are thermally bonded together (Figure 1A). Each device has four independent culture units for the dynamic culture of the gingival equivalents: each unit includes vertically stacked upper, middle, and lower chambers (Figure 1A). The upper and lower chambers are connected to independent inlet and outlet channels. The upper chamber includes an access opening for seeding the cells and hydrogels into the middle chamber during the culture phase, and for application of test compounds on the cultured gingival equivalents for downstream assays. The middle chamber used for the culture of gingival equivalents, is separated from the lower chamber by a polycarbonate track‐etched porous support membrane. The assembled and sterilized microfluidic organ‐on‐a‐chip device (kindly provided by REVIVO Biosystems, Singapore) was designed to culture four miniature gingival equivalents with a diameter of 7 mm each and an effective diffusion area of 0.4 cm2.
Figure 1.
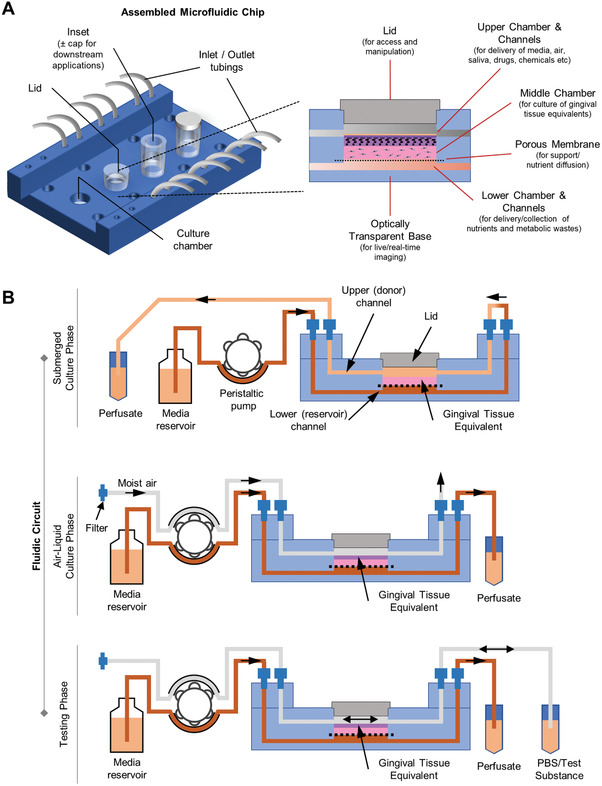
Schematic representation of microfluidic gingiva‐on‐chip device. A) Schematic view of the assembled device that shows the different features such as microchannels, chambers, inlets, and outlets micromilled into polymethyl methacrylate sheets that are vertically stacked and thermally bonded. B) Schematic representation of fluidic circuit used for perfusion of media, air, and test substances at different phases of the organotypic culture.
During the culture and downstream testing phases, continuous perfusion of culture media and receptor solution was maintained in the lower chamber using a peristaltic pump (Ismatec, Cole‐Parmer GmbH) (Figure 1B). Similarly, the upper chamber was perfused with culture media, air, or test substances depending on the culture or testing phase.
2.2. Cell Culture and Biofabrication of Gingival Equivalents under Static and Flow Conditions
Primary human gingival fibroblasts and human oral keratinocytes (OKF6/TERT2) were cultured in fibroblast and keratinocyte medium respectively as described previously.[ 9 , 15 , 33 ] To biofabricate gingiva‐on‐chip equivalents, 2 × 104 gingival fibroblasts were encapsulated within 80 µL of human fibrin‐based mucosal matrix as previously described[ 9 , 15 ] and seeded in the microfluidic device. After 4 days of submerged culture under flow, oral keratinocytes (3 × 105 cells cm−2) were seeded on top of the mucosal matrix and cultured for 2 days under submerged conditions, followed by 14 days of culture at air‐liquid interface (Figure 2A,B). Air‐liquid interface culture was maintained by media perfusion through the microfluidic channels connected to the lower channels (1.5 ± 0.1 µL min−1) and pumping moist air into the upper compartment. Gingival equivalents under static conditions (gingiva‐insert) were fabricated as previously described[ 15 ] (Figure 2A,C).
Figure 2.
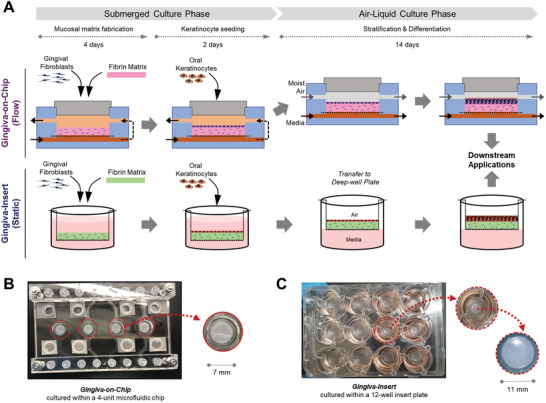
Biofabrication of organotypic full‐thickness gingival equivalents underflow and static conditions. A) Schematic representation of the organotypic culture under flow within the microfluidic device (gingiva‐on‐chip) and under static conditions using porous culture inserts (gingiva‐insert). The culture under both conditions includes mucosal matrix fabrication, keratinocyte seeding, and air‐liquid interface culture followed by their downstream applications. Macroscopic views of the gingival tissues fabricated within the B) microfluidic device and C) in static insert culture systems. The gingiva‐on‐chip equivalents within the microfluidic device B) are visible and easily accessible through the lid opening in the upper chamber (inset).
2.3. Oral Mucosal Irritation Test
Commercially available alcohol‐based (LISTERINE Cool Mint) and alcohol‐free (LISTERINE Gum Care Zero) mouthwashes were used as representative oral‐care products. One percent sodium lauryl sulfate (SLS, MatTek Corporation) and phosphate‐buffered solution (PBS, Vivantis Technologies) were used as positive and negative controls, respectively.
For on‐chip cultures, the test substances were actively pumped through the upper channel at 50 µL min−1 using a peristaltic pump‐driven forward and backward flow to mimic the rinsing action. The exposure was repeated after a 10‐h interval. After each exposure, the test substances were flushed out using PBS and incubated for 24 h after the second exposure.
For gingiva‐insert cultures, the epithelial surface was exposed to 130 µL of liquid test substance in triplicate for 30 s twice at 10‐h intervals. After each exposure, the test substances were washed three times with PBS, transferred to a 12‐well plate with fresh culture media and incubated at 37 °C for 24 h after the second exposure.
After incubation, media from on‐chip and insert cultures were collected for lactate dehydrogenase (LDH) release assay and tissues were harvested for histology or viability assays.
2.4. Dental Anesthetic Permeation Studies
Dental anesthetics, lidocaine hydrochloride (HCl) (Sigma‐Aldrich), and articaine HCl solution (Ubistesin Forte, 3M) were reconstituted to working concentrations of 1.67 mg mL−1 and 3.33 mg mL−1 in PBS respectively. To investigate the permeation kinetics of the dental anesthetics through tissue equivalents on‐chip, the tissues were equilibrated overnight with PBS, and lidocaine HCl (1.25 mg cm−2) or articaine HCl (2.5 mg cm−2) was applied to the tissue surface through the access openings. Perfusate flowing out of the lower compartments was collected directly into a 96‐well plate at 30‐min sampling intervals a 4.5‐h perfusion period as described previously.[ 17 , 19 ] The amount of lidocaine HCl and articaine HCl in the perfusates collected from the receptor compartments was quantified using spectroscopic measurement of peak absorbance at 221 nm and 272 nm, respectively.[ 19 , 34 ]
2.5. Statistical Analysis
Comparative evaluation of the parameters between two groups was performed using unpaired two‐tailed t‐tests, and those between more than two groups using one‐way ANOVA followed by Bonferroni post hoc analysis.
3. Results
3.1. Microfluidic Device Enables the Reconstruction of Gingival Equivalents under Flow Conditions
The fibrin‐based gingival tissue equivalents biofabricated within the microfluidic device was stable and maintained its initial proportions, without macroscopic contraction or degradation over the 3‐week culture period (Figures 1, 2). Macroscopically, the surface of the tissues looked flat, and dry reminiscent of keratinized epithelial surface (Figure 2B). The flat surface without wrinkles further demonstrated the stable non‐contractile nature of the matrix.
Haematoxylin‐esoin (H‐E) stained sections showed the formation of a multi‐layered, orthokeratinized, squamous epithelium over a cellular lamina propria (Figure 3A), characterized by distinct basal, spinous, granular, and corneal layers (Figure 3A and Figure S1, Supporting Information). The pattern of columnar to cuboidal basal keratinocytes, and Ki‐67 positive proliferating cells restricted to the basal keratinocytes, indicated the viability and epithelial homeostasis of the tissue constructs.
Figure 3.
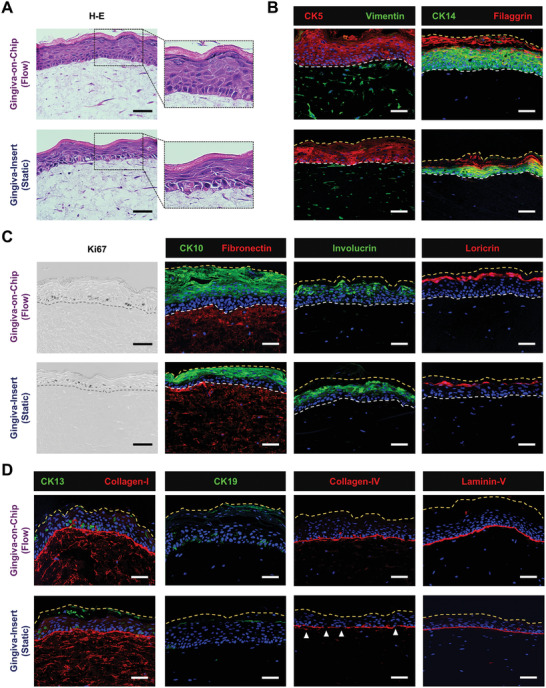
Characterization of the full‐thickness gingiva equivalents reconstructed underflow and static conditions. A) Hematoxylin‐eosin H–E) stained sections demonstrate the multi‐layered, orthokeratinized gingival epithelium over fibroblast‐populated lamina propria‐like matrix. B–D) Immunostained sections show the expression of cytokeratins (CK5, CK14, CK10, CK13, and CK19), stratified epithelial differentiation markers (involucrin, filaggrin, loricrin), proliferation marker (Ki67), vimentin, matrix proteins (collagen‐I and fibronectin) and basement membrane markers (collagen‐IV, laminin‐V) (Scale bar: 50 µm).
The epithelium showed a strong expression of stratified epithelial cytokeratins (CK5 and CK14) across the basal and suprabasal layers, and early epithelial differentiation markers (CK10 and involucrin) in the suprabasal cells (Figure 3B–D). The epithelium was devoid of CK13 and CK19 expression, similar to the native keratinized gingival or palatal epithelium.[ 4 ] The formation of an epithelial barrier was indicated by the strong expression of late differentiation markers (filaggrin and loricrin) at the junction of granular and corneal layers.
The lamina propria‐like mucosal matrix expressed fibronectin and collagen‐I, and contained vimentin‐positive gingival fibroblasts with dendritic morphology (Figure 3A–D). The fibrous pattern fibronectin and collagen‐I expression demonstrated the authentic de novo production of a lamina propria‐like matrix. The formation of a well‐defined basement membrane that anchors the epithelium to the lamina propria was indicated by the strong expression of collagen‐IV and laminin‐V at the junction between the epithelium and the underlying lamina propria‐like matrix (Figure 3D).
These results suggest that the gingival tissue equivalents reconstructed under flow conditions within the microfluidic device closely resemble native human gingival tissues.
3.2. Influence of Microfluidic Perfusion on Epithelial Morphogenesis and Basement Membrane Maturation
To understand the impact of active perfusion of culture media, the development of gingival equivalents fabricated on‐chip was compared to those cultured on culture inserts under static conditions. Both conditions produced a well‐differentiated stratified squamous epithelium with an orthokeratinized corneal layer, but distinct differences were evident in the gingiva‐on‐chip equivalents (Figure 3A).
The basal cells in the gingiva‐on‐chip exhibited a polarized, columnar morphology with their long axes aligned perpendicular to the basement membrane, compared to the cuboidal basal cells in the gingiva‐insert. The gingiva‐on‐chip exhibited significantly thicker viable epithelium supported by more layers of CK5, CK14, and CK10 positive keratinocytes (Figure 3A–C). Additionally, a regular arrangement of columnar basal keratinocytes, a thicker viable epithelium, and a higher Ki67 proliferation index in the gingiva‐on‐chip indicated an increased stability of the epithelial architecture (Figures 3A,C and 4A,B). Increased viable epithelial thickness in the gingiva‐on‐chip was further supported by larger area and higher intensity of stratification marker CK14 (Figure 4C). The gingiva‐on‐chip also exhibited a stronger and increased area of expression of early differentiation marker CK10 (Figure 4D). The impact of flow conditions on epithelial maturation was further evidenced by pronounced keratohyalin granules at the junction of viable epithelium and corneal layer (Figure 3A, inset) and higher expression of late differentiation markers filaggrin and loricrin (Figures 3B,C and 4E). Furthermore, the gingiva‐on‐chip also exhibited a continuous distribution of filaggrin and loricrin at the junction of upper spinous and granular layers of the epithelium (Figures 3A–D and 4E). These results suggest an enhanced terminal differentiation of the keratinocytes under flow conditions.
Figure 4.
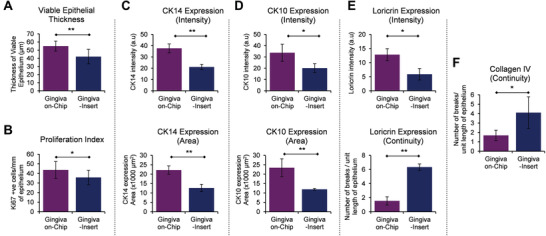
Morphometric analysis of the full‐thickness gingiva equivalents reconstructed underflow and static conditions. The bar graphs show the quantification and comparison of A) thickness of viable epithelium, B) Ki67 proliferation index, C–E) intensity and area of CK14, CK10, and loricrin expression, and E,F) number of breaks in the expression of loricrin and collagen IV among the gingival equivalents cultured under flow and static conditions. Area of CK14 expression corresponds to the area of viable epithelium (without the cornified layers), while the area of CK10 expression corresponds to suprabasal layers including the cornified layers. The number of breaks in loricrin and collagen IV are normalized to unit length (400 µm) of the epithelium. (n≥3; *p<0.05, **p<0.01, Student t‐test).
The junction between epithelium and lamina propria‐like matrix in the gingiva‐on‐chip was marked by a stronger expression of basement membrane proteins collagen‐IV and laminin‐V (Figure 3D). Further, a continuous and a thicker collagen‐IV expression in gingiva‐on‐chip equivalents, compared to a patchy expression in gingiva‐insert cultures, suggest an improved maturation of the basement membrane and a better anchorage of the epithelium to the underlying lamina propria‐like tissue under flow conditions (Figure 4F).
Overall, the results suggest that the gingiva‐on‐chip provides an in vitro system with improved epithelial morphogenesis and maturation, which is ideally suited for downstream pharmaco‐toxicological testing.
3.3. Gingiva‐on‐Chip Enables Mucosal Irritation Tests under Near‐Physiological Conditions
With growing oral hygiene awareness, new oral‐care formulations target specific benefits like cavity protection, tartar reduction, gum health improvement, and teeth whitening. However, these formulations may contain active ingredients and excipients that may potentially cause mucosal irritation. Traditional assessment of mucosal irritation involves human subjects,[ 35 ] but the advent of reconstructed oral mucosal epithelial models has provided an alternative in vitro model.[ 1 , 14 , 36 ] Current methods for assessment of oral mucosal irritation on reconstructed oral mucosal epithelial models are based on skin irritation tests (OECD TG439), wherein the impact of the mechanical action of the mouth rinse is poorly understood and not represented in the current test methods.
Hence, we next aimed to evaluate the application of the gingiva‐on‐chip system for in vitro assessment of oral‐care products. The gingiva‐on‐chip and gingiva‐insert cultures were exposed to PBS, 1% SLS, and commercially available alcohol‐based and alcohol‐free mouthwashes. To mimic the actual use case scenario, the cultures were exposed to the test substances twice a day at 10‐h interval, washed with PBS after each exposure, and cultured for 24 h prior to downstream analysis (Figure 5A,B). H‐E and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining of cultures exposed to negative control (PBS) showed no disruption in epithelial integrity. However, exposure to 1% SLS (positive control) caused a dramatic disruption of the epithelium, resulting in TUNEL‐positive cells in the epithelium and in the lamina propria. Interestingly, the gingiva‐on‐chip cultures showed complete disruption of the epithelial cohesion leading to separation of the epithelium from the underlying matrix (Figure 5C). Low tissue viability levels (<20%) and high LDH release from these tissues confirmed the strong mucosal irritant potential of 1% SLS (Figure 5D,E).
Figure 5.
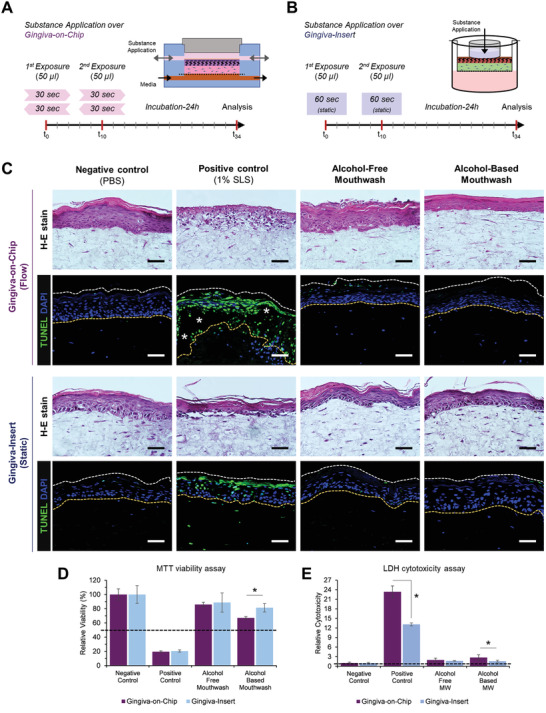
Application and comparison of gingival equivalents for in vitro assessment of oral‐care products underflow and static conditions. Schematic representation of exposure of the surface of gingival equivalents to oral‐care formulations under A) flow and B) static conditions on the gingival‐on‐chip and gingiva‐insert respectively. C) Hematoxylin‐eosin H–E) and TUNEL‐stained sections of the gingival equivalents after exposure to oral‐care formulations and controls (Scale bar: 50 µm; * represent epithelial disruption). D) Relative cell viability and E) relative LDH release following exposure to oral‐care formulations. Dotted lines represent the respective thresholds. (n≥3; *p<0.05).
The gingiva‐on‐chip and gingiva‐insert cultures exposed to alcohol‐free and alcohol‐based mouthwashes showed minimal disruption of the corneal layers of the epithelium, and absence of TUNEL‐positive cells (Figure 5C). High tissue viability levels that are above the 50% threshold and low amounts of LDH release compared to positive control confirmed their classification as non‐irritants (Figure 5D,E). However, the gingiva‐on‐chip cultures exposed to alcohol‐based mouthwash exhibited a thinner epithelium with flattened keratinocytes and slightly higher amounts (≈3‐fold) of LDH release.
Overall, the flow‐based exposure using gingiva‐on‐chip system provided a near‐physiological environment for in vitro testing of oral‐care products.
3.4. Application of Gingiva‐on‐Chip for Disease Modeling and Drug Permeation Studies
Toward modeling diseased states, we simulated oral mucosal ulcers within the microfluidic device (ulcer‐on‐chip), by culturing gingival fibroblast‐populated mucosal matrix under submerged conditions (without the addition of keratinocytes) under constant flow conditions (Figure 6A).
Figure 6.
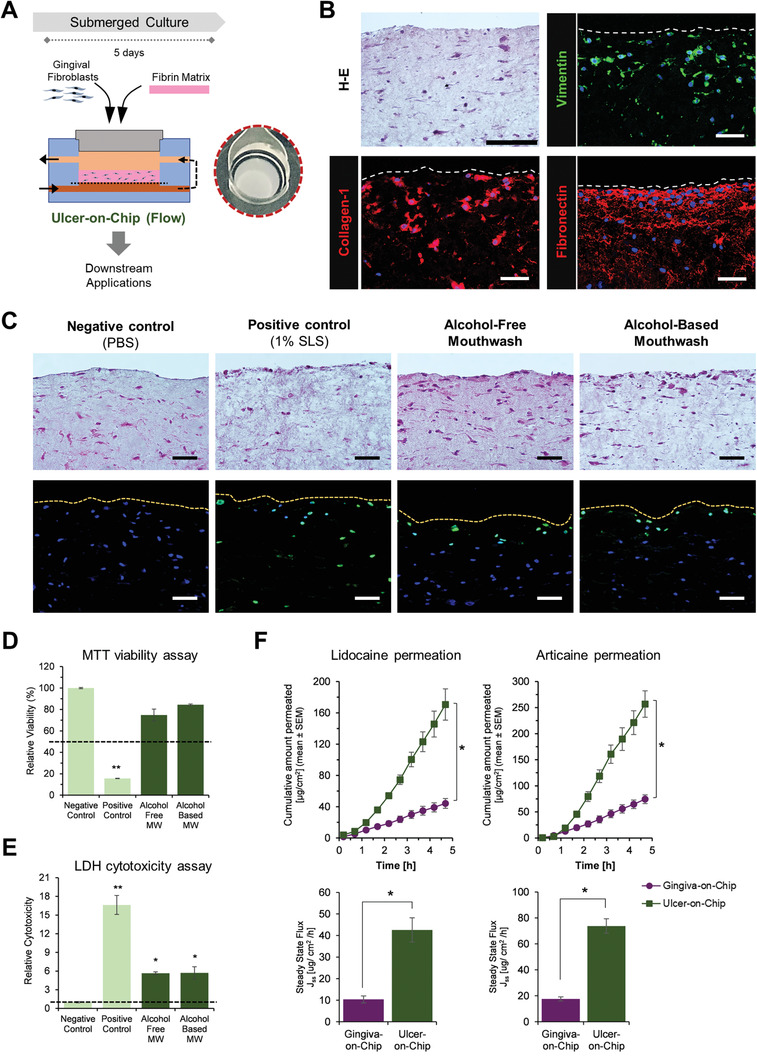
Application of gingiva‐on‐chip for disease modeling and drug permeation studies. A) Schematic representation of the fabrication of ulcer‐on‐chip equivalents under flow conditions, and macroscopic view of its shiny, wet surface. B) Hematoxylin‐eosin and immunostained sections show the expression of vimentin and matrix proteins (collagen‐I and fibronectin). C) Hematoxylin‐eosin and TUNEL‐stained sections, D) relative cell viability, and E) relative LDH release from the ulcer‐on‐chip equivalents after exposure to oral‐care formulations and controls (Scale bar: 50 µm). Dotted lines represent the respective thresholds. F,G) Line and bar graphs show the permeation profiles of lidocaine HCl and articaine HCl permeation through gingiva‐on‐chip and ulcer‐on‐chip equivalents. (n≥3; *p<0.05).
The resulting ulcer‐on‐chip tissues were stable and exhibited a wet and shiny appearance reminiscent of ulcerated tissues (Figure 6A). H‐E stained and immunostained sections demonstrated the presence of spindle‐shaped, vimentin‐positive fibroblasts embedded within the matrix (Figure 6B). The matrix showed the presence of collagen‐I and fibronectin fibers, demonstrating the production of cell‐derived extracellular matrix proteins required for fibroblast adhesion, growth, and migration (Figure 6B). These features of cellular connective tissue without an overlying mucosal epithelium are reminiscent of an ulcerated mucosal tissue.
We next aimed to evaluate the application of the ulcer‐on‐chip platform for in vitro assessment of oral‐care products under flow conditions. H‐E and TUNEL‐stained sections of the ulcer‐on‐chip equivalents postexposure to negative control (PBS), showed no disruption of the mucosal matrix and the fibroblasts displayed a spindle‐shaped morphology reminiscent of their healthy state (Figure 6C). However, the cultures exposed to 1% SLS resulted in fibroblasts with rounded morphology, a reduced number of viable fibroblasts, and the presence of TUNEL‐positive cells throughout the depth of the mucosal matrix (Figure 6C). MTT viability tests on the 1% SLS‐exposed tissues showed viability levels below the 50% threshold, and significantly high amounts of LDH release (Figure 6D,E). The ulcer‐on‐chip cultures exposed to alcohol‐free and alcohol‐based mouthwashes also showed mild disruption, with the fibroblasts on the exposed surface exhibiting a rounded morphology, TUNEL‐positive staining, and release of significant amounts of LDH (Figure 6C,E). Although the tissue viability was more than the 50% threshold, the morphological alterations of the fibroblasts on the exposed surface and LDH release suggest the mild cytotoxicity potential of the mouthwashes when used on oral ulcers.
Next, we used the gingiva‐on‐chip and ulcer‐on‐chip equivalents for assessing the transmucosal permeation of lidocaine HCl and articaine HCl across intact and ulcerated tissues (Figure 6F). Mass balance calculations showed that the recovery of both anesthetics was within the recommended guidelines (90–110%). The ulcer‐on‐chip equivalents presented significantly higher cumulative permeation over the 4.5 h period and ≈4‐fold higher steady‐state flux for lidocaine HCl and articaine HCl when compared to the intact gingiva‐on‐chip equivalents (p<0.01) (Figure 6F, Table 2, Supporting Information).
Maintaining tissue integrity is crucial in avoiding artificial spikes in permeation parameters, which often occur in manually handled permeation studies using diffusion cells. Histological examination of the tissues after permeation experiments showed no apparent disruption in the epithelial and matrix structure, which could be attributed to the integrated culture and downstream permeation studies (Figure S2, Supporting Information).
4. Discussion
Emulating the microenvironment of the gingival tissues in vitro requires the controlled interaction of the gingival epithelium with the cellular and matrix components of the underlying lamina propria and a myriad of extrinsic factors such as saliva, microbes, mechanical forces, food, lifestyle habits, and oral‐care products. In this work, we leveraged on a microfluidic organ‐on‐a‐chip device with vertically stacked design to enable the biofabrication and long‐term culture of full‐thickness gingiva‐on‐chip tissue equivalents under air‐liquid interface and flow conditions. Compared to static conditions, the culture under flow conditions enabled the biofabrication of gingival tissue equivalents with improved epithelial morphogenesis, maturation, and barrier features. Further, the flow feature was utilized to emulate the mechanical action of mouth rinse and integrate the assessment of host‐material interactions and transmucosal permeation of oral‐care formulations.
Design features of an organ‐on‐a‐chip system greatly influences its capabilities, potential applications, and limitations. The potential to customize the design features enabled the emulation of the microphysiological environment of tissues and their interface with the internal and external milieu. Dedicated microchannels beneath (in this study, and others[ 17 , 25 ]) or placed adjacent[ 21 , 23 ] to the culture chamber enable continuous replenishment of culture media, removal of metabolic wastes, and collection of biomarkers, mimicking the blood flow. Similarly, microchannels placed on top (in this study, and others[ 17 , 25 ]), or adjacent[ 21 , 23 , 24 , 27 ] to the culture chamber could be utilized to mimic salivary flow, temporally controlled delivery of cells, bacteria, and test substances. This opens avenues to study the impact of blood, tissue fluid, and salivary flow on transmucosal permeation, host‐microbiome, and host‐material interactions.[ 17 , 24 ] In this study, we utilized the upper channel of the microfluidic device to enable air‐liquid interface culture during the tissue maturation phase, and to simulate the swishing motion of a mouth rinse, where the test substances were flown over the surface of gingiva‐on‐chip equivalents using forward and backward flow. Despite the thicker and more mature epithelium in gingiva‐on‐chip cultures, exposure to 1% SLS resulted in a marked epithelial disruption and higher amounts of LDH release. SLS is a synthetic detergent widely used in dentifrices and mouthwashes and often implicated in oral mucosal irritation caused by oral‐care formulations.[ 35 ] Similarly, alcohol‐based mouthwash caused slightly higher LDH release in the gingiva‐on‐chip cultures. These detrimental effects were also evident on ulcer‐on‐chip equivalents, with cell death predominantly on the exposed surface. These findings underscore the importance of considering the mechanical action of oral‐care formulations when assessing their potential for mucosal irritation. The combination of organotypic cultures with microfluidic devices offers a more realistic representation of the swishing motion, enabling a more physiologically relevant assessment of the mucosal irritation potential of oral‐care formulations.
Microfluidic systems designed for the biofabrication of oral mucosa and gingival barrier tissues[ 21 , 22 , 23 , 24 , 25 ] typically feature a configuration where a culture chamber is sandwiched between two channels. These channels can be arranged in either a horizontal or vertical stacking, and each configuration presents unique opportunities and limitations. Horizontally stacked channels in previous studies[ 21 , 23 , 24 ] enabled compartmentalization and real‐time visualization of the cells, matrix, and bacteria and their response to dental materials and microorganisms. However, such a configuration limits the ability to achieve the necessary air‐liquid interface culture required for the stratification and differentiation of keratinocytes, and the attainment of epithelial barrier function.[ 18 ] In contrast, the vertically stacked design combined with open access feature presented in this study, helps mimic the lumen of the oral cavity, and achieve the required air‐liquid interface for efficient differentiation of keratinocytes, and the formation of protective barrier.
Gingiva‐on‐chip equivalents biofabricated under the perfused air‐liquid interface conditions demonstrated the presence of pronounced keratohyalin granules, strong expression of filaggrin and loricrin, and absence of CK13 and CK19 expression. These features are reminiscent of masticatory mucosa that includes gingival and palatal epithelium. Unlike the non‐keratinized oral mucosa (referred to as lining mucosa), the masticatory mucosa expresses terminal differentiation markers and barrier proteins such as filaggrin and loricrin at the interface between granular and cornified layers.[ 4 , 37 ] The strong expression of keratinocyte differentiation markers (CK10 and involucrin), barrier proteins (filaggrin and loricrin), and basement membrane markers (collagen IV) in the gingiva‐on‐chip cultures compared to static conditions highlight the potential role of air‐liquid interface and flow‐induced mechanical stimulation. Besides supporting a continuous nutrient supply and waste removal, dynamic perfusion potentially generates shear stresses and mechano‐transduction mediated induction of fibroblast function,[ 38 , 39 ] keratinocyte maturation and modulation of epithelial barrier function.[ 40 , 41 , 42 ] Owing to the static nature of gingiva‐insert cultures, the nutrient transport to the cells within the 3D matrix is purely diffusive. In contrast, the perfusion of media and moist air through the upper and lower channels respectively, provides a pressure gradient across the gingiva‐on‐chip equivalents, that potentially drives active interstitial media flow through the 3D matrix and keratinocytes in the epithelial layer. Further, the active perfusion of moist air through the upper channel potentially provides shear stresses on the keratinocytes and the external layers of the gingival epithelium. We believe that the shear stress and interstitial fluid flow induced mechano‐transduction cues could have mediated the improved epithelial morphogenesis, maturation, and barrier features in the gingiva‐on‐chip equivalents.
Microfluidic design also enables miniaturization, reducing the sample requirements, lower limits of detection, and higher sensitivity owing to reduced dilution of the permeated compounds.[ 19 ] Human gingival tissues are typically scarce and small, leading to the common use of fresh or frozen porcine tissues for drug permeation studies. Gingiva‐on‐chip serves as an effective alternative to conventional Franz diffusion cells (that demand large tissue size) and provides a physiologically relevant alternative to human and animal‐derived tissues. The in situ reconstruction of gingival tissues within the microfluidic device and integrated downstream mucosal irritation and permeation assays avoid the need for manual handling of fragile tissues, which could potentially impact tissue interaction and drug permeation studies.[ 43 ] As demonstrated in this study and by others on skin tissues,[ 17 , 25 , 44 ] the vertical‐stacked configuration also allows the integration of drug permeation studies through the barrier tissues. Most permeation studies focus on the epithelium alone and typically employ heat or chemical‐based methods to separate the oral epithelium from the underlying lamina propria.[ 45 , 46 , 47 ] However, this in turn can have detrimental effects on the cells and affect the barrier properties of the test tissue.[ 46 ] Further, as the methods and conditions of tissue storage affect the permeation characteristics,[ 48 , 49 ] the integration of culture and drug permeation studies within the microfluidic device offers the potential advantage of performing experiments on fresh tissues. Furthermore, the potential to fabricate lamina propria alone or ulcer‐on‐chip equivalents avoids the need for physical or chemical separation methods and associated tissue damage. Though the epithelial tissue forms the primary permeability barrier,[ 50 ] the connective tissue component also has significant barrier properties depending on the physico‐chemical characteristics of the test compound.[ 47 ] Therefore, information on the permeation through lamina propria or ulcerated tissue is crucial, especially for topical formulations and/or actives with anesthetic and wound healing properties.
In conclusion, the gingiva‐on‐chip presented in this study models the microphysiological features of the gingival tissue in healthy and diseased states, and their interface with the external milieu. The potential to model healthy and diseased states provides a valuable opportunity to investigate gingival and oral mucosal biology, aid reduction and/or replacement of animal models for toxicity and biocompatibility evaluation of dental materials and oral‐care products under near‐physiologic conditions. Moving forward, the gingiva‐on‐chip could be incorporated with salivary flow and microbial colonization to understand gingival and periodontal disease, development, and assessment of novel periodontal therapeutics. Further, inclusion of endothelial cells would enable the recapitulation of microvasculature and incorporation of immune cells.[ 51 ] Similarly, for the ulcer‐on‐chip, inclusion of keratinocytes and a wound can enable closer recapitulation of oral ulcers and its application for wound healing studies. These improvisations could then pave way to study the impact of the microbiome and external agents on the innate immune response of the gingival tissues and its modulation using oral‐care products, as well as of effective transmucosal drug and nutraceutical delivery. Furthermore, integrating with other organ‐on‐a‐chip models such as the skin, gut, liver, and/or cardiac tissues could enable the potential to study mucocutaneous drug interactions, metabolic and systemic impact of periodontal disease on diabetes and cardiovascular diseases.
Conflict of Interest
The authors would also like to disclose that two of the authors, M.A. and G.S., are co‐founders of Revivo Biosystems Pte Ltd., and the microfluidic device used in this study is based on a patent application (WO2018030958A1) licensed to the company.
Author Contributions
G.M. designed methodology, performed validation, formal analysis, investigation, and visualization, and wrote and reviewed the original draft. R.H.S.T. and Y.D. designed methodology and performed investigation and formal analysis. R.W. performed supervision, funding acquisition, resource acquisition, and wrote, reviewed, and edited the manuscript. M.A. performed conceptualization, design of methodology, formal analysis, writing, reviewing, and editing of the manuscript, supervision, funding acquisition, and resource acquisition. G.S. performed conceptualization, design of methodology, formal analysis, visualization, writing, reviewing, and editing of the manuscript, supervision, project administration, funding acquisition, and resource acquisition. All authors contributed to the revision and approval of the final version of the manuscript.
Supporting information
Supporting Information
Acknowledgements
Donor‐derived primary gingival fibroblasts were isolated from the gingival tissues attached to healthy, non‐carious impacted third molars after institutional review board approval (DSRB No. 2018/00256). This work was partially supported by grants to G.S.: from National University of Singapore and Singapore Ministry of Education (A‐0002944‐00‐00; A‐0002944‐01‐00), Fellowship for Young Professor (FLY) Programme (AMNUS‐2021‐008) and NUHS Internal Grant Funding under its funding scheme (NUHS RO Project No. NUHSRO/2021/107/RO5+6/Seed‐Sep/10; A‐8000164‐00‐00). This project is also partially supported by grants to M.A.: 16th SMART Innovation Grant from Singapore‐MIT Alliance for Research and Technology Centre (ING‐000158 BIO) and Gap Fund from Accelerate, Agency for Science, Technology and Research (ACCL‐19‐GAP001‐R20H). G.M. is supported by NUS Research Scholarship.
Muniraj G., Tan R. H. S., Dai Y., Wu R., Alberti M., Sriram G., Microphysiological Modeling of Gingival Tissues and Host‐Material Interactions Using Gingiva‐on‐Chip. Adv. Healthcare Mater. 2023, 12, 2301472. 10.1002/adhm.202301472
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Klausner M., Ayehunie S., Breyfogle B. A., Wertz P. W., Bacca L., Kubilus J., Toxicol. In Vitro 2007, 21, 938. [DOI] [PubMed] [Google Scholar]
- 2. Bierbaumer L., Schwarze U. Y., Gruber R., Neuhaus W., Tissue Barriers 2018, 6, 1479568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Viñuela‐Prieto J. M., Sánchez‐Quevedo M. C., Alfonso‐Rodríguez C. A., Oliveira A. C., Scionti G., Martín‐Piedra M. A., Moreu G., Campos A., Alaminos M., Garzón I., J. Periodontal Res. 2015, 50, 658. [DOI] [PubMed] [Google Scholar]
- 4. Presland R. B., Dale B. A., Crit. Rev. Oral Biol. Med. 2000, 11, 383. [DOI] [PubMed] [Google Scholar]
- 5. Ibrahim M. S., El‐Wassefy N. A., Farahat D. S., Biomaterials for Oral and Dental Tissue Engineering, Woodhead Publishing, Cambridge: 2017. [Google Scholar]
- 6. Thurnheer T., Belibasakis G. N., Bostanci N., Arch. Oral Biol. 2014, 59, 977. [DOI] [PubMed] [Google Scholar]
- 7. Bostanci N., Bao K., Wahlander A., Grossmann J., Thurnheer T., Belibasakis G. N., Mol. Oral Microbiol. 2015, 30, 323. [DOI] [PubMed] [Google Scholar]
- 8. Zanetti F., Sewer A., Mathis C., Iskandar A. R., Kostadinova R., Schlage W. K., Leroy P., Majeed S., Guedj E., Trivedi K., Martin F., Elamin A., Merg C., Ivanov N. V., Frentzel S., Peitsch M. C., Hoeng J., Chem. Res. Toxicol. 2016, 29, 1252. [DOI] [PubMed] [Google Scholar]
- 9. Makkar H., Atkuru S., Tang Y. L., Sethi T., Lim C. T., Tan K. S., Sriram G., J. Tissue Eng. 2022, 13, 204173142211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ingendoh‐Tsakmakidis A., Mikolai C., Winkel A., Szafranski S. P., Falk C. S., Rossi A., Walles H., Stiesch M., Cell. Microbiol. 2019, 21, e13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buskermolen J. K., Janus M. M., Roffel S., Krom B. P., Gibbs S., J. Dent. Res. 2018, 97, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shang L., Deng D., Buskermolen J. K., Janus M. M., Krom B. P., Roffel S., Waaijman T., Van Loveren C., Crielaard W., Gibbs S., Sci. Rep. 2018, 8, 16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moharamzadeh K., Brook I. M., Scutt A. M., Thornhill M. H., Van Noort R., J. Dent. 2008, 36, 331. [DOI] [PubMed] [Google Scholar]
- 14. Moharamzadeh K., Franklin K. L., Brook I. M., Van Noort R., J. Periodontol. 2009, 80, 769. [DOI] [PubMed] [Google Scholar]
- 15. Hu S., Muniraj G., Mishra A., Hong K., Lum J. L., Hong C. H. L., Rosa V., Sriram G., Dent. Mater. 2022, 38, 1385. [DOI] [PubMed] [Google Scholar]
- 16. Ahadian S., Civitarese R., Bannerman D., Mohammadi M. H., Lu R., Wang E., Davenport‐Huyer L., Lai B., Zhang B., Zhao Y., Mandla S., Korolj A., Radisic M., Adv. Healthcare Mater. 2018, 7, 1700506. [DOI] [PubMed] [Google Scholar]
- 17. Sriram G., Alberti M., Dancik Y., Wu B., Wu R., Feng Z., Ramasamy S., Bigliardi P. L., Bigliardi‐Qi M., Wang Z., Mater. Today 2018, 21, 326. [Google Scholar]
- 18. Huang C., Sanaei F., Verdurmen W. P. R., Yang F., Ji W., Walboomers X. F., J. Dent. Res. 2023, 102, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alberti M., Dancik Y., Sriram G., Wu B., Teo Y. L., Feng Z., Bigliardi‐Qi M., Wu R. G., Wang Z. P., Bigliardi P. L., Lab Chip 2017, 17, 1625. [DOI] [PubMed] [Google Scholar]
- 20. Koning J. J., Rodrigues Neves C. T., Schimek K., Thon M., Spiekstra S. W., Waaijman T., de Gruijl T. D., Gibbs S., Front Toxicol 2021, 3, 824825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ly K. L., Luo X., Raub C. B., Biofabrication 2022, 15, 015007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ly K. L., Rooholghodos S. A., Rahimi C., Rahimi B., Bienek D. R., Kaufman G., Raub C. B., Luo X., Biomed. Microdevices 2021, 23, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rahimi C., Rahimi B., Padova D., Rooholghodos S. A., Bienek D. R., Luo X., Kaufman G., Raub C. B., Biomicrofluidics 2018, 12, 054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Makkar H., Zhou Y., Tan K. S., Lim C. T., Sriram G., Adv. Healthcare Mater. 2023, 12, e2202376. [DOI] [PubMed] [Google Scholar]
- 25. Jin L., Kou N., An F., Gao Z., Tian T., Hui J., Chen C., Ma G., Mao H., Liu H., Biosensors 2022, 12, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Franca C. M., Tahayeri A., Rodrigues N. S., Ferdosian S., Puppin Rontani R. M., Sereda G., Ferracane J. L., Bertassoni L. E., Lab Chip 2020, 20, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodrigues N. S., Franca C. M., Tahayeri A., Ren Z., Saboia V. P. A., Smith A. J., Ferracane J. L., Koo H., Bertassoni L. E., J. Dent. Res. 2021, 100, 1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bao K., Belibasakis G. N., Selevsek N., Grossmann J., Bostanci N., Sci. Rep. 2015, 5, 15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bao K., Papadimitropoulos A., Akgül B., Belibasakis G. N., Bostanci N., Virulence 2015, 6, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee E.‐J., Kim Y., Salipante P., Kotula A. P., Lipshutz S., Graves D. T., Alimperti S., Bioengineering 2023, 10, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheung J. W. C., Rose E. E., Paul Santerre J., Acta Biomater. 2013, 9, 6867. [DOI] [PubMed] [Google Scholar]
- 32. Navarro F. A., Mizuno S., Huertas J. C., Glowacki J., Orgill D. P., Wound Repair Regen. 2001, 9, 507. [DOI] [PubMed] [Google Scholar]
- 33. Sriram G., Sudhaharan T., Wright G. D., Methods Mol. Biol. 2020, 2150, 195. [DOI] [PubMed] [Google Scholar]
- 34. Shahzad Y., Waters L. J., Barber C., Colloids Surf. A 2014, 458, 96. [Google Scholar]
- 35. Herlofson B. B., Barkvoll P., Eur. J. Oral Sci. 1996, 104, 21. [DOI] [PubMed] [Google Scholar]
- 36. Moharamzadeh K., Brook I. M., Van Noort R., Scutt A. M., Smith K. G., Thornhill M. H., J. Mater. Sci. Mater. Med. 2008, 19, 1793. [DOI] [PubMed] [Google Scholar]
- 37. Mackenzie I. C., Rittman G., Gao Z., Leigh I., Lane E. B., J. Periodontal Res. 1991, 26, 468. [DOI] [PubMed] [Google Scholar]
- 38. Dan L., Chua C. K., Leong K. F., Biotechnol. Bioeng. 2010, 107, 1. [DOI] [PubMed] [Google Scholar]
- 39. Van Der Pauw M. T. M., Klein‐Nulend J., Van Den Bos T., Burger E. H., Everts V., Beertsen W., J. Periodontal Res. 2000, 35, 335. [DOI] [PubMed] [Google Scholar]
- 40. O'neill A. T., Monteiro‐Riviere N. A., Walker G. M., Cytotechnology 2008, 56, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sidhaye V. K., Schweitzer K. S., Caterina M. J., Shimoda L., King L. S., Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Q., Wang C., Li X., Zhang J., Zhang Z., Yang K., Ouyang J., Zha S., Sha L., Ge J., Chen Z., Gu Z., J. Tissue Eng. 2023, 14, 204173142311685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lestari M. L. A. D., Nicolazzo J. A., Finnin B. C., J. Pharm. Sci. 2009, 98, 4577. [DOI] [PubMed] [Google Scholar]
- 44. Mori N., Morimoto Y., Takeuchi S., Biomaterials 2017, 116, 48. [DOI] [PubMed] [Google Scholar]
- 45. Pinto S., Pintado M. E., Sarmento B., Expert Opin. Drug Deliv. 2020, 17, 33. [DOI] [PubMed] [Google Scholar]
- 46. Castro P., Madureira R., Sarmento B., Pintado M., Concepts and Models for Drug Permeability Studies, Woodhead Publishing, Cambridge, 2016, pp. 189–202. [Google Scholar]
- 47. Kulkarni U., Mahalingam R., Pather S. I., Li X., Jasti B., J. Pharm. Sci. 2009, 98, 471. [DOI] [PubMed] [Google Scholar]
- 48. Franz‐Montan M., Serpe L., Martinelli C. C. M., Da Silva C. B., Santos C. P. D., Novaes P. D., Volpato M. C., De Paula E., Lopez R. F. V., Groppo F. C., Eur. J. Pharm. Sci. 2016, 81, 52. [DOI] [PubMed] [Google Scholar]
- 49. Marxen E., Axelsen M. C., Pedersen A. M. L., Jacobsen J., Int. J. Pharm. 2016, 511, 599. [DOI] [PubMed] [Google Scholar]
- 50. Squier C. A., Hall B. K., Arch. Oral Biol. 1985, 30, 485. [DOI] [PubMed] [Google Scholar]
- 51. Makkar H., Lim C. T., Tan K. S., Sriram G., Biofabrication 2023, 15, 045008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


