ABSTRACT
Two phages belonging to Arthrobacter phage cluster AK were isolated from soil samples collected in Newburgh, NY in 2021. Both are lytic with a genome organization typical of siphoviruses except for two genes encoding minor tail proteins with pyocin-knob domains found early in the genome, before the terminase gene.
KEYWORDS: bacteriophage assembly, Arthrobacter, annotation
ANNOUNCEMENT
To gain a better understanding of Actinophage diversity and the networks through which horizontal gene transfer may occur in these phages, the SEA-PHAGES program has been isolating phages on diverse Actinobacteria, including Arthrobacter species (1–3). Although found in many environments, Arthrobacter is a genus of predominantly soil bacteria, with some species relevant to bioremediation (4). Here we report on two Arthrobacter phages belonging to cluster AK (2, 3).
Soil samples were collected in 2021 in Newburgh, NY, USA (GPS coordinates in Table 1). From these samples, two phages were isolated on Arthrobacter sp. ATCC 21022 following enrichment protocol in the SEA-PHAGES Discovery Guide (https://seaphagesphagediscoveryguide.helpdocsonline.com/home). Briefly, soil samples were incubated in peptone yeast calcium media enriched with the bacterial host at 30°C and shaken at 250 rpm. Individual plaques were isolated and triple-plaque purified on the lawns of the isolation host. High titer lysates (HTL) were prepared by collecting phage buffers from flooded webbed plates of plaques.
TABLE 1.
Genome characteristics and sequencing data of cluster AK Arthrobacter phages BrotherBlo and Misaenga
| Phage | BrotherBlo | Misaeng |
|---|---|---|
| Genome length (bp) | 43,650 | 43,817 |
| Number of ORFs | 62 | 61 |
| GC content (%) | 61.1 | 60.6 |
| Isolation GPS | 41.510556 N, 74.0125 W | 41.50941 N, 74.0139 W |
| Coverage | 2911 | 2359 |
| Accession number | PP725413 | PP725418 |
| SRA | SRR28773367 | SRR28773353 |
| Number of reads | 880,402 | 716,031 |
ORF = open reading frame.
For each phage, DNA was extracted from HTL using the Promega Wizard DNA CleanUp System (A7280) and sequenced at the Pittsburgh Bacteriophage Institute. DNA libraries were prepared with the NEB Ultra II DNA Library Kit and sequenced on an Illumina MiSeq system (MiSeq reagent kit v3), using a v3 150 SE flow cell. Reads (150 bp single-end) assembled into one contig with Newbler v2.9 and were quality controlled with Consed v29.0 (5, 6). Genome termini were identified as cohesive ends (3′ ends of 13 bases: GGTAACCGTGATA) from read overrepresentations (5).
Finished sequences were imported into DNAMaster v5.23.6 (http://cobamide2.bio.pitt.edu/computer.htm). Both Glimmer v3.0 and GeneMark v2.5 algorithms were used to call putative genes (7, 8). NCBI BLASTp (searched on the non-redundant protein sequence database) v.2.13 and HHPred (searched on PDB_mmCIF70, SCOPe70, Pfam-A, and NCBI_Conserved_Domains) were used to predict putative protein functions (9, 10). For BLASTp matches, an E-value below 10−5 was required to assign a function. For HHpred matches, a high probability (>85%), substantial coverage (>50%), and low E-value (<10−5) were required. The absence of tRNA genes was confirmed with Aragorn (11). Default settings were used in all programs. Genome annotations have been submitted to NCBI (accession and SRA numbers in Table 1).
Genome maps were generated by importing annotations into the Phamerator database (12). Proteins were grouped into phage families, or “phams,” using the PhaMMseqs pipeline (13). Phages that share 35% or more of their phams are classified as belonging to the same cluster (14, 15).
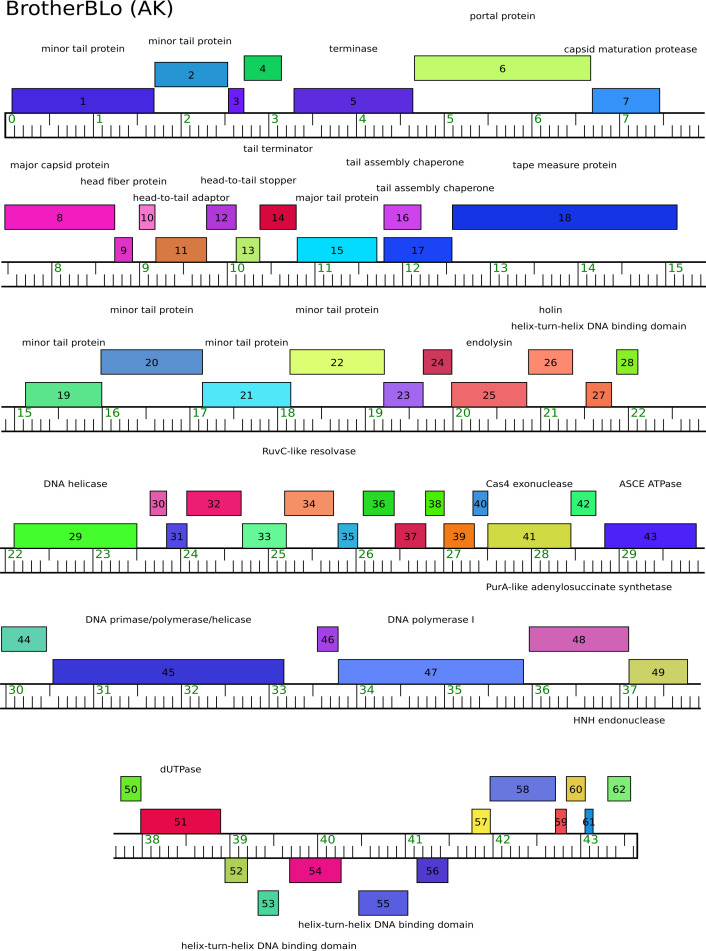
Both phages are similar in genome length, number of genes, and GC content (Table 1), have a high nucleotide identity to each other (79.9%; 16), and belong to cluster AK (Duplodnaviria, Heunggongvirae, Uroviricota, Caudoviricetes, Korravirus) (2, 3). Like other AK phages, they are lytic (confirmed by clear plaque morphology; figures available at phagesdb.org), have a siphovirus morphology and genome organization, except for the presence of two genes encoding minor tail proteins at the beginning of their genome (Fig. 1). Based on NCBI_Conserved_Domains, these gene products contain a pyocin-knob domain (17). In addition, both phages have genes that putatively encode proteins involved in DNA metabolism (Fig. 1).
Fig 1.
Genome map for Cluster AK Arthrobacter phage BrotherBlo. The ruler shows genome length (in kilobases) with forward and reverse genes shown above and below the ruler, respectively. Function or putative functions are listed. Map created using Phamerator (12). Note a number of genes that putatively encode proteins involved in DNA metabolism: a DNA primase/polymerase/helicase, a PurA-like adenylosuccinate synthetase, a RuvC-like resolvase, an ASCE ATPase, and a deoxyuridine triphosphatase.
ACKNOWLEDGMENTS
This work was conducted as part of the HHMI-supported SEA-PHAGES program. We thank Graham F. Hatfull for the SEA-PHAGES program and members of his lab, particularly Deborah Jacobs-Sera and Dan Russell, for logistical support. Funding for publication was provided by a gift to the Biology Department at Gettysburg College from Gail Seygal Biology '67.
Contributor Information
Véronique A. Delesalle, Email: delesall@gettysburg.edu.
John J. Dennehy, Department of Biology, Queens College, Queens, New York, USA
DATA AVAILABILITY
All sequencing and annotation data related to these phages are publicly available and accession numbers for both the assembly and raw reads are provided in Table 1.
REFERENCES
- 1. Hatfull GF. 2020. Actinobacteriophages: genomics, dynamics, and applications. Annu Rev Virol 7:37–61. doi: 10.1146/annurev-virology-122019-070009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klyczek KK, Bonilla JA, Jacobs-Sera D, Adair TL, Afram P, Allen KG, Archambault ML, Aziz RM, Bagnasco FG, Ball SL, et al. 2017. Tales of diversity: genomic and morphological characteristics of forty-six Arthrobacter phages. PLoS ONE 12:e0180517. doi: 10.1371/journal.pone.0180517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klyczek KK, Jacobs-Sera D, Adair TL, Adams SD, Ball SL, Benjamin RC, Bonilla JA, Breitenberger CA, Daniels CJ, Gaffney BL, et al. 2018. Complete genome sequences of 44 Arthrobacter phages. Genome Announc 6:e01474–17. doi: 10.1128/genomeA.01474-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gobbetti M, Rizzello CG. 2014. Arthrobacter, p 69–76. In Carl A Batt, Mary Lou Tortoello (ed), Encyclopedia of food microbiology (Second edition). Academic Press. [Google Scholar]
- 5. Russell DA. 2018. Sequencing, assembling, and finishing complete bacteriophage23genomes. Methods Mol Biol 1681:109–125. doi: 10.1007/978-1-4939-7343-249_9 [DOI] [PubMed] [Google Scholar]
- 6. Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29:2936–2937. doi: 10.1093/bioinformatics/btt515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res 33:W451–4. doi: 10.1093/nar/gki487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct 7:12. doi: 10.1186/1745-6150-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gabler F, Nam SZ, Till S. 2020. Protein sequence analysis using the MPI6 bioinformatics toolkit. Curr Protoc Bioinformatics 72:e108. doi: 10.1002/cpbi.108.820 [DOI] [PubMed] [Google Scholar]
- 11. Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA9genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for 12 comparative bacteriophage genomics. BMC Bioinformatics 12:395. doi: 10.1186/1471-2105-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gauthier CH, Cresawn SG, Hatfull GF. 2022. PhaMMseqs: a new pipeline for 15 constructing phage gene phamilies using MMseqs2. G3 (Bethesda) 12:jkac233. doi: 10.1093/g3journal/jkac233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pope WH, Bowman CA, Russell DA, et al. 2015. Whole genome comparison of a large 18 collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife 4:e06416. doi: 10.7554/eLife.06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pope WH, Mavrich TN, Garlena RA, Guerrero-Bustamante CA, Jacobs-Sera D, Montgomery MT, Russell DA, Warner MH, Hatfull GF, Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) . 2017. Bacteriophages of Gordonia spp. display a spectrum of diversity and genetic relationships. mBio 8:e01069-17. doi: 10.1128/mBio.01069-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoon SH, Ha SM, Lim J, Kwon S, Chun J. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. doi: 10.1007/s10482-017-0844-4 [DOI] [PubMed] [Google Scholar]
- 17. Buth SA, Shneider MM, Scholl D, Leiman PG. 2018. Structure and analysis of R1 and R2 pyocin receptor-binding fibers. Viruses 10:427. doi: 10.3390/v10080427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequencing and annotation data related to these phages are publicly available and accession numbers for both the assembly and raw reads are provided in Table 1.