Abstract
Although hepatitis C virus E2 protein can bind to human cells by interacting with a putative viral receptor, CD81, the interaction alone is not sufficient to establish permissiveness for hepatitis C virus infection. Using an Epstein-Barr virus-based extrachromosomal replication system, we have screened through a human liver cDNA library and successfully identified a cDNA capable of supporting hepatitis C virus replication in an otherwise nonpermissive cell line. This cDNA encodes a protein exhibiting homology to a group of proteins derived from various evolutionarily distant species, including Oryza sativa submergence-induced protein 2A. The mRNAs encoding this factor are heterogeneous at the 5′ ends and are ubiquitously expressed in multiple tissues, albeit in a very small amount. The longest mRNA contains an in-frame and upstream initiation codon and codes for a larger protein. This 5′-extended form of mRNA was detected in hepatocellular carcinoma, but not in normal liver tissue. Immunofluorescence analysis demonstrated that the hepatic factor was distributed evenly in cells, but occasionally formed aggregations in the peri- or intranuclear areas. In summary, we have identified a hepatic factor capable of supporting hepatitis C virus replication in an otherwise nonpermissive cell line. This factor belongs to a previously uncharacterized protein family. The physiological function of this protein awaits further study.
Hepatitis C virus (HCV) is a major cause of chronic hepatitis worldwide (4). Chronic hepatitis C may lead to severe sequelae, such as liver cirrhosis and hepatocellular carcinoma (18, 19). Although scientists have made important progress in understanding molecular mechanisms for HCV replication, few data are available regarding essential cellular factors required for HCV replication (3, 9, 10). It is believed that as an initial step for viral infection, HCV must bind either directly or indirectly to a viral receptor in order to anchor on the cell membrane. Previously, it was proposed that HCV could associate with low-density lipoprotein (LDL) in the blood and that the complexes interacted with LDL receptor before entering the hepatocytes (1, 13). Recently, it was discovered that HCV E2 protein interacted specifically with CD81 on the cell membrane, which was suggested to be the HCV receptor (15, 20). Although several other groups confirmed the specificity of binding between E2 and CD81, subsequent studies indicated that the interaction alone did not predict susceptibility of cells to HCV infection (2, 5, 7, 12, 14). For example, HCV E2 protein was able to bind CD81 that originated from other species not permissive for HCV infection (2, 12). Structure-function analysis revealed that HCV binding to hepatocytes might not entirely depend on CD81 and that CD81 was an attachment receptor with poor capacity to mediate virus entry (14). These results lead to the argument that other species-specific cellular factors or coreceptors are needed for cell entry and thus replication of HCV. In this study, we have developed a strategy to search for such a molecule.
MATERIALS AND METHODS
Cell lines, transfection, and establishment of transformants.
Human embryonic kidney cells constitutively expressing Epstein-Barr virus nuclear antigen-1 (EBNA-1) protein from Epstein-Barr virus (293EBNA cells; Invitrogen, Carlsbad, Calif.) were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 250 μg of G418 per ml. HepG2 cells were maintained in minimal essential medium containing 10% fetal bovine serum. Huh-7 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. A human liver cDNA library (Clontech Laboratories, Inc., Palo Alto, Calif.) was constructed by inserting the cDNAs into a vector, pDR2, downstream of a Rous sarcoma virus long terminal repeat (LTR) promoter. This plasmid contains Epstein-Barr virus OriP, a gene for hygromycin B selection, and an ampicillin resistance gene. The cDNA clones were first grouped into 200 sets with 100 to 200 cDNA clones per set. Plasmids were then extracted from each set of clones and transfected into 293EBNA cells by the standard CaPO4 precipitation method. Transformants were selected by addition of 150 μg of hygromycin B per ml to the culture medium. A total of 200 transformants were established for the first round of the HCV infection assay.
HCV infection assay and detection of CD81 mRNA.
HCV-positive serum containing 107 copies of HCV RNA per ml, measured by the branched DNA (bDNA) method (Quantiplex HCV-RNA 2.0 assay; Chiron, Emeryville, Calif.), was used for the HCV infection assay. The cells in a 60-mm-diameter petri dish were incubated in medium containing 5 μl of HCV-positive serum for 12 h. The cells were then incubated in fresh medium without HCV-positive serum, and the medium was changed every day. To detect HCV RNA, cells were trypsinized and washed two times with fresh medium by centrifugation. The supernatant of the second wash (as a contamination control) and the washed cells were collected in pairs for RNA extraction and reverse transcription (RT)-PCR. The procedures were described previously (23). The nested primers used were C1 (5′-CGGCAACAGGTAAACTCCAC-3′, antisense, nucleotides [nt] 114 to 95), C2 (5′-CCCTGTGAGGAACTACTGTC-3′, sense, nt −299 to −280), C3 (5′-ACGATCTGACCACCGCCCGG-3′, antisense, nt 92 to 73), and C4 (5′-TTCACGCAGAAAGCGTCTAG-3′, sense, nt −279 to −260). A digoxigenin-labeled probe, flanking by C3 and C4, was used for the subsequent Southern blot analysis. As a control, β-actin mRNAs were detected simultaneously. The primers used were A1 (5′-CACCAACTGGGACGACATGG-3′, sense, nt 301 to 320); A2 (5′-AGGATCTTCATGAGGTAGTC-3′, antisense, nt 651 to 632), A3 (5′-TCTGGCACCACACCTTCTAC-3′, sense, nt 327 to 346), and A4 (5′-GTCAGGTCCCGGCCAGCCAG-3′, antisense, nt 630 to 611). The procedure for minus-strand-specific RT-PCR was also described previously (22, 23). To detect CD81 mRNA, RT was performed with random primers. The primers used for PCR were 5′-CGAGACGCTTGACTGCTGTG-3′ (sense, nt 691 to 710) and 5′-CTCAGTACACGGAGCTGTTC-3′ (antisense, nt 950 to 931). The PCR product was verified by nucleotide sequencing with an automatic DNA sequencer (CEQ 2000; Beckman Instruments, Inc., Fullerton, Calif.).
RACE.
The 5′ rapid amplification of cDNA ends (RACE) experiment was performed with a 5′/3′ RACE kit (Boehringer Mannheim Biochemica, Mannheim, Germany). Total normal liver RNA was used. The primer used for cDNA synthesis was PsipR (5′-CATGAAGATCCGGATCCAC-3′). After being tailed with dATP homopolymer by a terminal deoxynucleotidyl transferase, the tailed cDNA was amplified by PCR with an oligo(dT) anchor primer and P2 (5′-TCCTCCTTGTCCCTCACATC-3′). Finally, a second step of PCR was performed with an anchor primer and P4 (5′-GGATCTCATCGTCCAAGTGC-3′). The details of the experimental procedure and the sequences of oligo(dT) and the anchor primer were described previously (23). The sequences of clones generated by RACE (61.31R1 to 61.31R4) and two artificially created deletion mutants (61.31D1 and 61.31D2) were verified and then inserted into the BamHI-XbaI sites of pDR2 for further transfection. The restriction enzyme sites needed for plasmid construction were generated by PCR-based site-directed mutagenesis (24). Briefly, for insertion of 61.31R1 to 61.31R4 into pDR2, two primers containing engineered BamHI and XbaI sites and complementary to the 5′ and 3′ ends of these clones were designed for amplification. The amplified products were digested with BamHI and XbaI, gel purified, and inserted into pDR2. For generation of the deletion mutants, the downstream primer containing the engineered XbaI site was designed to match the desired positions in 61.31R4 so that the amplified products became truncated at the 3′ portion.
Detection of Sip-L and eSip-L mRNA.
RT was performed with random primers. Sip-L mRNA was detected with P2 and P1 (5′-GGTGCTCTACTGGAAGCTGG-3′). eSip-L mRNA was detected with P4 and P3 (5′-CCGCACTGCGCGTCATGGTG-3′). A digoxigenin-labeled probe, flanked by P1 and P2, was used for the subsequent Southern blot analysis. As a control, β-actin mRNA was also detected simultaneously.
Immunofluorescence analysis.
To perform immunofluorescence analysis, the coding region from clone 61.31 was isolated and inserted in frame with the V5 epitope in pcDNA3.1/V5-His B (Invitrogen, San Diego, Calif.). The plasmid was transfected into 293EBNA cells. The methods of cell fixation and staining were described previously (21). The primary antibody used was mouse anti-V5 monoclonal antibody (Invitrogen). The secondary antibody used was fluorescein isothiocyanate-conjugated goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.).
Establishment of a stable Huh-7 cell line expressing Sip-L.
To establish a stable Huh-7 cell line expressing a high level of Sip-L protein, pSV2neo, which encoded a neomycin-resistant marker, and pCMVEBNA, which encoded EBNA-1 (both from Clontech), were cotransfected into Huh-7 cells. After selection with neomycin, the stable transformant was subsequently transfected with pDR2–61.31R3 and selected with hygromycin B. The stable transformant expressing the highest level of Sip-L mRNA (by Northern analysis) was used for further experimentation.
Nucleotide sequence accession number.
The GenBank accession no. for Sip-L is AF403478.
RESULTS
Experimental strategy.
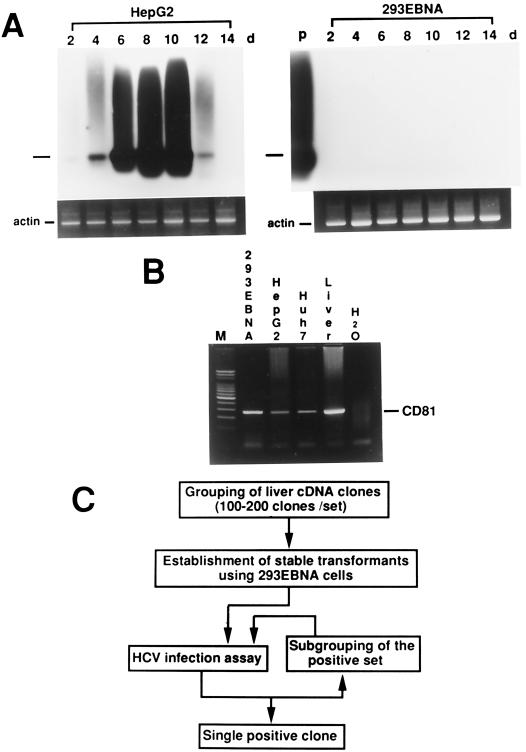
Expression of Epstein-Barr nuclear antigen-1 (EBNA-1) in cells allows extrachromosomal replication of plasmids carrying the Epstein-Barr virus replication origin region (OriP) (8, 11). A cDNA library constructed with this system theoretically expresses a high level of transcripts in EBNA-1-expressing cells. Previous reports showed that HCV could infect HepG2 cells and that the viral RNA could be detected transiently (16). Additionally, HCV replication in the culture cells could be enhanced by expression of EBNA-1 (17). Based on these observations, RT followed by a two-step (nested) PCR was performed serially after inoculation of HepG2 cells with HCV-positive serum. HCV RNA could be detected by Southern blotting, and the signal was strongest on the 8th day after inoculation (Fig. 1A, left panel). The same procedure was repeated with a human embryonic kidney cell line expressing EBNA-1 (293EBNA cells). No HCV RNA signal could be detected (Fig. 1A, right panel). However, CD81 mRNA was readily detected in 293EBNA cells (Fig. 1B). To search for the missing factor, a liver cDNA library equipped with the aforementioned Epstein-Barr virus-based system was first grouped into 200 sets with 100 to 200 clones per set (Fig. 1C). Two hundred sets of 293EBNA transformants, each transfected by one set of mixed cDNA clones, were then established. The HCV infection assay was performed with all 200 transformants to look for the positive set. Once the positive set was identified, the cDNA clones of that set were further subgrouped, and the assay was repeated until a single cDNA clone was obtained.
FIG. 1.
Strategy to clone a cDNA capable of supporting HCV replication in a nonpermissive cell line. (A) HCV infection assay performed with HepG2 cells (left panel) and 293EBNA cells (right panel). Infection assays were performed every 2 days (d) up to 14 days. The experiments were repeated three times. A representative result is shown here. In each panel, the bar to the left indicates the predicted position of the PCR product. As a control, β-actin mRNA was detected simultaneously. P, positive control with HCV-positive serum. (B) Detection of CD81 mRNA by RT-PCR. (C) Flowchart of the cloning strategy.
Identification of a cDNA clone capable of supporting HCV replication in 293EBNA cells.
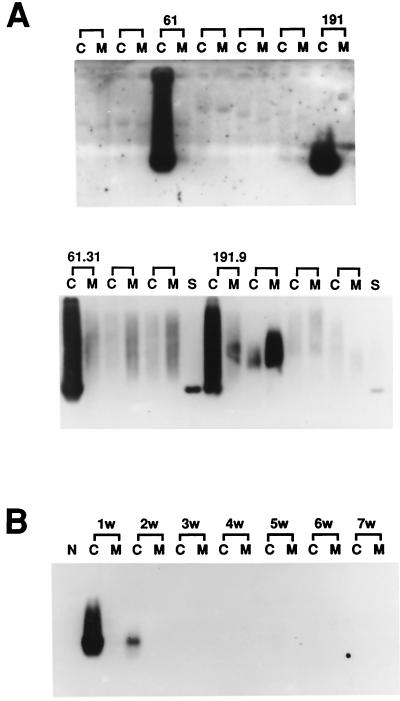
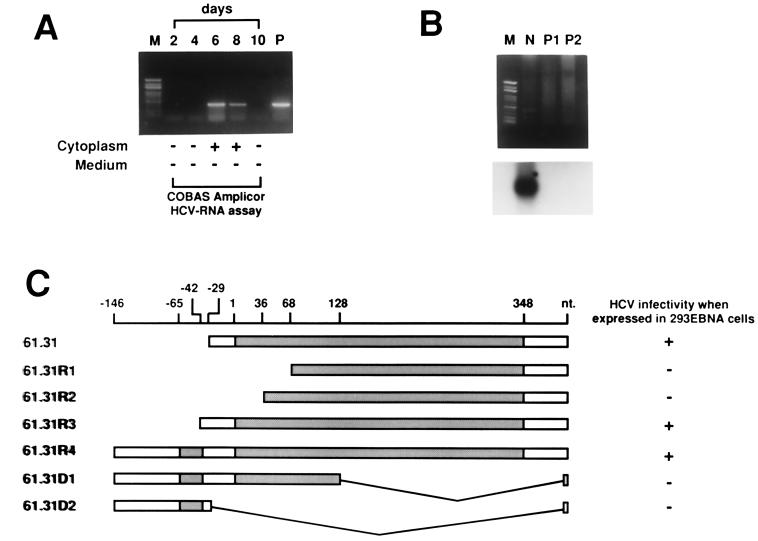
Two sets of transformants, sets 61 and 191, containing mixed cDNA clones were first identified as positive by the HCV infection assay (Fig. 2A, upper panel). After subgrouping and repeating the assays, two cDNA clones from set 61 and one cDNA clone from set 191 were tested as positive by the HCV infection assay (Fig. 2A, lower panel). Nucleotide sequence analysis revealed that all three clones were identical. Clone 61.31 and the corresponding transformant were used for further experiments. In this extrachromosomal replication system, the plasmid containing the cDNA fragment was selected and maintained with hygromycin B. We then cultured the 61.31 293EBNA cells in medium without hygromycin B to allow for the loss of the extrachromosomal plasmid DNA (6). The HCV infection assay was performed serially 1 to 7 weeks after removal of hygromycin B. Cell susceptibility to HCV infection was gradually lost, and the cells became nonpermissive again 3 weeks later (Fig. 2B). To further verify our results, an HCV infection assay was performed with 61.31 293EBNA cells, and serial cell extracts were sent for HCV RNA testing performed by our molecular medicine team. This team routinely performs HCV RNA testing for clinical doctors in this medical center by using a standard assay (COBAS Amplicor HCV-RNA test). Intracellular HCV RNA was positive on days 6 and 8 after inoculation with HCV-positive serum (Fig. 3A). Finally, to confirm the presence of HCV replication in 61.31 293EBNA cells, minus-strand HCV RNA was detected 8 days after inoculation with HCV-positive serum (Fig. 3B). The result showed that minus-strand HCV RNA was indeed present in the infected cells.
FIG. 2.
Results of molecular screening through a liver cDNA library. (A) Sets 61 and 191 of mixed cDNA clones were capable of supporting HCV replication (upper panel). The positive results of the HCV infection assay for two single cDNA clones (61.31 and 191.9) are shown (lower panel). C, cell lysate; M, the second aliquot of fresh medium used to wash the cells; S, HCV-positive serum. (B) Hygromycin B was removed from the medium to allow loss of episomal plasmid DNA. The HCV infection assay was performed every week (w) up to 7 weeks after removal of hygromycin B.
FIG. 3.
Verification of the permissiveness of 61.31 293EBNA cells to HCV infection and determination of the physiological 5′ ends of 61.31 mRNA. (A) HCV-positive serum was used to infect 61.31 293EBNA cells, and HCV RNA was detected every 2 days by RT-nested PCR. The cytoplasmic fractions and the second aliquots of fresh medium used to wash the cells were both subjected to the COBAS Amplicor HCV-RNA assay. (B) Minus-strand-specific RT-PCR for detection of HCV-RNA. M, marker; N, 61.31 293EBNA cells assayed on the 8th day after HCV infection; P1, 50 μl of HCV-positive serum (105 copies/ml); P2, 50 μl of HCV-positive serum (107 copies/ml). The PCR product was verified by Southern blotting (lower panel). (C) Results of the 5′ RACE experiment. Four clones with different 5′ ends are shown (61.31R1 to 61.31R4). The structures of two artificially created deletion mutants are also shown (61.31D1 to 61.31D2). The results of the HCV infection assay for these clones are indicated to the right. Two potential open reading frames, orf-1 (short) and orf-2 (long), are indicated by shaded bars.
Determination of the 5′ end of 61.31 mRNA by RACE.
Four different 5′ ends of 61.31 mRNA were detected in total normal liver RNA by the RACE method. These clones were named 61.31R1 to 61.31R4 (Fig. 3C). Further RACE experiments with a primer located near the 5′ end of 61.31R4 failed to obtain other clones. The 61.31R4 clone contained two open reading frames, orf-1 (short, upstream) and orf-2 (long, downstream). To determine which open reading frame is functional, all four clones obtained from RACE and two artificially deleted mutant clones were transfected into 293EBNA cells to test for HCV infectivity by the same method, as shown in Fig. 3A. Preservation of orf-2 was found to be required for HCV infectivity (Fig. 3C).
Clone 61.31 encoded a protein factor exhibiting homology to Oryza sativa submergence-induced protein 2A.
The amino acid sequence of 61.31R4 orf-2 was compared with existing protein sequences by a BLAST search (National Center for Biotechnology Information [www.ncbi.nlm.nih.gov]). Strikingly, this protein exhibits homology to several proteins derived from various genetically distant species, including Oryza sativa submergence-induced protein 2A (Fig. 4). The amino acid sequence of 61.31R4 orf-2, temporarily named “submergence-induced protein-like factor” (Sip-L), was identical to the carboxyl portion of two other human clones derived from ovarian cancer and placenta choriocarcinoma, respectively. These two sequences, recently deposited in GenBank without formal publication, have an extension of 63 additional amino acid residues at the amino terminus compared with Sip-L. Only 1 amino acid difference was found between the two. They were temporarily named the “amino-terminus-extended form of Sip-L” (eSip-L).
FIG. 4.
Comparison of the open reading frame (orf-2) sequence with various homologs derived from different species. Amino acid residues identical among three or more different species are boxed. Dashes indicate gaps.
Tissue distribution of Sip-L mRNA.
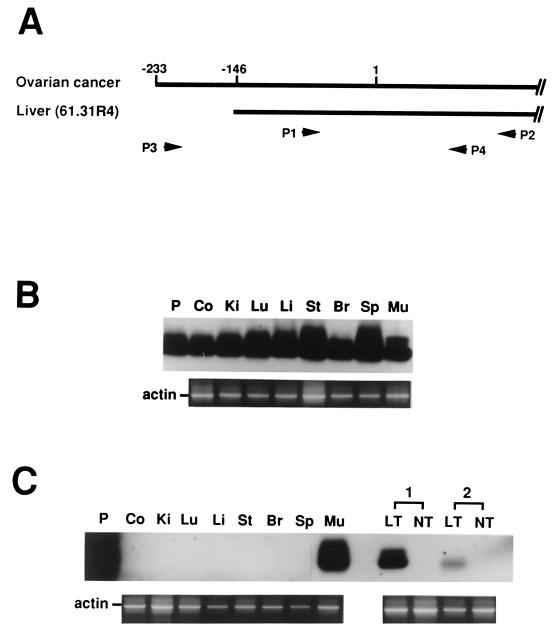
To determine the tissue distribution of Sip-L mRNA, Northern analysis was performed with total RNA obtained from various human organs. This experiment, however, failed to detect any Sip-L mRNA. Thus, RT followed by one-step PCR and Southern analysis was performed. Two sets of primers were designed to detect Sip-L and eSip-L mRNAs (Fig. 5A). The results indicated that Sip-L mRNA was ubiquitously distributed in all kinds of tissues, but eSip-L mRNA was found only in skeletal muscle (Fig. 5B and C). Strikingly, eSip-L mRNA was also detected in two samples derived from hepatocellular carcinoma.
FIG. 5.
Detection of Sip-L and eSip-L mRNA by RT-PCR. (A) To mark the positions of primers (P1 to P4), the first nucleotide of the Sip-L initiation codon was assigned as no. 1. (B) Primers P1 and P2 were used to detect Sip-L mRNA. (C) Primers P3 and P4 were used to detect eSip-L mRNA. The sequence of P, a P1-to-P2 DNA fragment, was verified, and P was then used as a hybridization control. Co, colon; Ki, kidney; Lu, lung; Li, liver; St, stomach; Br, brain; Sp, spleen; Mu, skeletal muscle. Two paired liver tumor tissues, including cancerous (LT) and noncancerous (NT) parts, were also tested. As a control, β-actin mRNA was detected simultaneously for all tissues.
Subcellular localization of Sip-L protein in 293EBNA cells.
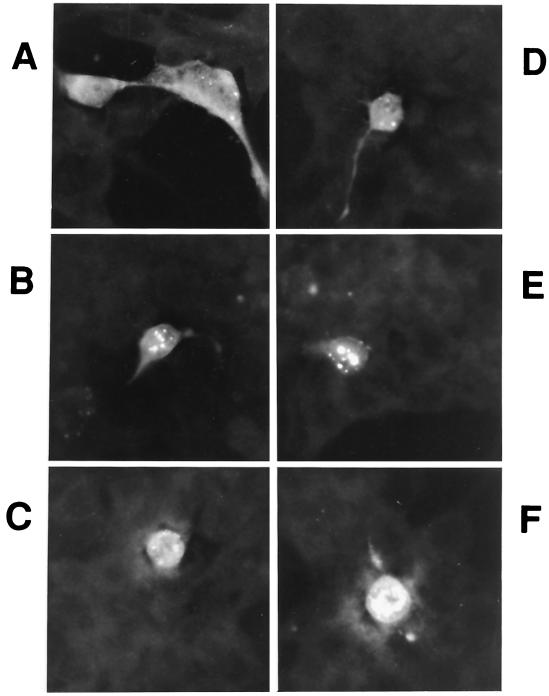
To gather clues for the possible physiological function of Sip-L, we have examined the subcellular localization of Sip-L by immunofluorescence analysis. Sip-L was tagged with a paramyxovirus SV5 epitope for detection with anti-V5 antibody. It was found that this protein is distributed evenly in both cytoplasm and nucleus in the majority of cells. In 60% of cells, various numbers and sizes of Sip-L protein aggregations were found in either the peri- or intranuclear areas (Fig. 6A to F). In a few cells, the protein was heavily clustered in the nucleus (Fig. 6F). The cause or nature of these aggregations was unknown.
FIG. 6.
Immunofluorescence analysis of Sip-L protein. (A to F) Sip-L was tagged with a V5 epitope for immunofluorescence staining in 293EBNA cells.
Infectivity of HCV in Huh-7 cells stably expressing Sip-L.
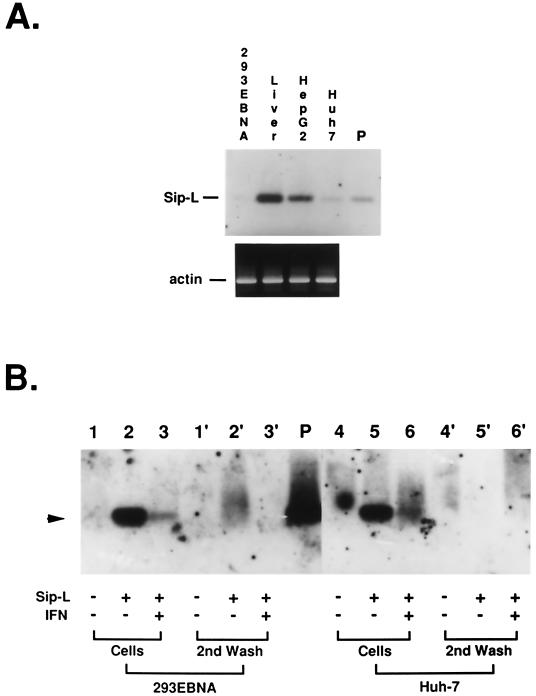
The presence of Sip-L mRNA was determined in 293EBNA cells, HepG2 cells, Huh-7 cells, and normal liver tissue by RT-PCR (Fig. 7A). The amount of Sip-L mRNA in HepG2 cells was relatively smaller than that in normal liver tissue. Only a trace of Sip-L mRNA was found in Huh-7 cells. No Sip-L mRNA was detected in 293EBNA cells. An Huh-7 cell line stably expressing a high level of Sip-L mRNA was thus established (see Materials and Methods). It was found that this higher-expression line was permissive for HCV infection (Fig. 7B). Furthermore, addition of 5,000 U of alpha interferon (Schering-Plough Corp., Kenilworth, N.J.) per ml to the medium from the 4th day of the infection assay resulted in a significant decrease in intracellular HCV RNA on the 7th day. Similar results were observed in 293EBNA cells.
FIG. 7.
Infectivity of HCV in Huh-7 cells stably expressing a higher level of Sip-L mRNA. (A) Sip-L mRNA was detected by RT-PCR and Southern blotting in 293EBNA cells, normal liver tissue, HepG2 cells, and Huh-7 cells. As a control, β-actin mRNA was detected simultaneously. (B) The HCV infection assay was performed with 293EBNA and Huh-7 cells. No HCV RNA can be detected by RT-PCR (arrowhead) in the nontransfected cells (lanes 1 and 4), whereas HCV RNA can be detected in cells expressing Sip-L (lanes 2 and 5). The signals were significantly decreased when 5,000 U of alpha interferon per ml was added to the medium from the 4th day of the infection assay (lanes 3 and 6). The second aliquots of medium used to wash the infected cells were used as contamination controls (lanes 1′ to 3′ and 4′ to 6′).
DISCUSSION
Binding of viruses to receptors on cell membrane is considered to be the first step for cell entry. However, successful binding by itself does not ensure cell entry and replication of the virus. In our pilot study, we washed the cells on the culture dishes three times daily after inoculation of HCV serum and trypsinized the cells from the dishes every 2 days. HCV RNA was not detectable in the third aliquots of medium used to wash the cells after the 2nd day. However, HCV-RNA was detectable by one-step RT-PCR in the supernatant after trypsinization of the cells (if no further wash was performed) up to the 4th day. We subsequently demonstrated that 293EBNA, HepG2, and Huh-7 cells all expressed CD81, thereby allowing docking of HCV on the cell membrane. To achieve a valid infection assay, we further washed the trypsinized cells two times by centrifugation and used the second aliquot of washing medium as a contamination control. By this method, it was shown that HCV could not replicate in either Huh-7 or 293EBNA cells, but could replicate in HepG2 cells, albeit transiently. By using a molecular screening strategy, a molecule (Sip-L) was identified that was capable of supporting HCV replication in 293EBNA cells. This finding confirms the concept that additional cellular factors are required for essential viral replication.
Our results, however, did not solve the problem of tissue tropism in HCV infection. Sip-L mRNA is ubiquitously expressed in all tissue examined, albeit in a very small amount. Thus, Sip-L does not account for the tissue tropism of HCV. Several possibilities can be postulated. For example, Sip-L may activate another factor that is essential for HCV infection, and that factor may be expressed only in susceptible cells. Tissue-specific negative regulatory factors may suppress the effect of Sip-L, or Sip-L is unevenly expressed in the same tissue (such as hepatocytes or mononuclear cells), and only a small number of hepatocytes or mononuclear cells express a sufficient amount of Sip-L to allow HCV replication. Additionally, since Sip-L supports HCV replication only transiently, it is possible that many organs are in fact permissive to transient but silent HCV replication, while other tissue-specific factors are required to achieve persistent viral replication. Presumably, Sip-L is merely one of a number of factors required for essential HCV replication.
Although it is not known how the Sip-L mRNA level is regulated, current data indicate that heterogeneities at the 5′ end may be involved. An intriguing fact is that eSip-L mRNA can be detected in cancerous tissues, including hepatocellular carcinoma, ovarian cancer, and placenta choriocarcinoma. Presumably, transcription from farther upstream of this gene to generate eSip-L mRNA requires factors expressed in cancerous cells. Alternatively, a suppression factor may be produced in noncancerous cells to inhibit its transcription. It is unclear why skeletal muscle also expresses eSip-L mRNA.
The physiological function of Sip-L or eSip-L is unknown. This protein is so evolutionarily conserved even prokaryotes harbor a homologous protein. The function of this protein, therefore, should be essential to life. In plants, this protein is induced during submergence in water, a hypoxic situation, suggesting its involvement in a novel stress-related pathway. At the subcellular level, this protein forms aggregations in the peri- or intranuclear areas, suggesting its association with nucleus-related activities. These clues may help us to design experiments for further studies.
In summary, by using a molecular screening strategy, we have identified a cellular factor, Sip-L, capable of supporting HCV replication in an otherwise nonpermissive cell line. This factor shares extensive amino acid homology with a group of proteins derived from various evolutionarily distant species. The molecular mechanism of Sip-L-supported HCV replication is unknown. The physiological function of this protein requires further study.
ACKNOWLEDGMENTS
We thank C. Y. Peng for helpful discussions.
This work was supported partly by a 3-year grant (CMRP 752) and partly by a continuous grant for pilot projects (CMRP 800) from the Chang Gung Medical Research Council.
REFERENCES
- 1.Agnello V, Abel G, Elfahal M, Knight G B, Zhang Q X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander T, Forns X, Emerson S U, Purcell R H, Bukh J. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology. 2000;277:358–367. doi: 10.1006/viro.2000.0617. [DOI] [PubMed] [Google Scholar]
- 3.Blight K J, Kolykhalov A A, Rice C M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 4.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 5.Flint M, Maidens C, Loomis-Price L D, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating J A. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hambor J E, Hauer C A, Shu H K, Groger R K, Kaplan D R, Tykocinski M L. Use of an Epstein-Barr virus episomal replicon for anti-sense RNA-mediated gene inhibition in a human cytotoxic T-cell clone. Proc Natl Acad Sci USA. 1988;85:4010–4014. doi: 10.1073/pnas.85.11.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higginbottom A, Quinn E R, Kuo C-C, Flint M, Wilson L H, Bianchi E, Nicosia A, Monk P N, McKeating J A, Levy S. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J Virol. 2000;74:3642–3649. doi: 10.1128/jvi.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung S C, Kang M S, Kieff E. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci USA. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 10.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 11.Margolskee R F, Kavathas P, Berg P. Epstein-Barr virus shuttle vector for stable episomal replication of cDNA expression libraries in human cells. Mol Cell Biol. 1988;8:2837–2847. doi: 10.1128/mcb.8.7.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meola A, Sbardellati A, Bruni Ercole B, Cerretani M, Pezzanera M, Ceccacci A, Vitelli A, Levy S, Nicosia A, Traboni C, McKeating J, Scarselli E. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J Virol. 2000;74:5933–5938. doi: 10.1128/jvi.74.13.5933-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monazahian M, Bohme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J Med Virol. 1999;57:223–229. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Petracca R, Falugi F, Galli G, Norais N, Rosa D, Campagnoli S, Burgio V, Di Stasio E, Giardina B, Houghton M, Abrignani S, Grandi G. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J Virol. 2000;74:4824–4830. doi: 10.1128/jvi.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 16.Seipp S, Mueller H M, Pfaff E, Stremmel W, Theilmann L, Goeser T. Establishment of persistent hepatitis C virus infection and replication in vitro. J Gen Virol. 1997;78:2467–2476. doi: 10.1099/0022-1317-78-10-2467. [DOI] [PubMed] [Google Scholar]
- 17.Sugawara Y, Makuuchi M, Kato N, Shimotohno K, Takada K. Enhancement of hepatitis C virus replication by Epstein-Barr virus-encoded nuclear antigen 1. EMBO J. 1999;18:5755–5760. doi: 10.1093/emboj/18.20.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomimatsu M, Ishiguro N, Taniai M, Okuda H, Saito A, Obata H, Yamamoto M, Takasaki K, Nakano M. Hepatitis C virus antibody in patients with primary liver cancer (hepatocellular carcinoma, cholangiocarcinoma, and combined hepatocellular-cholangiocarcinoma) in Japan. Cancer. 1993;72:683–688. doi: 10.1002/1097-0142(19930801)72:3<683::aid-cncr2820720310>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 19.Tremolada F, Casarin C, Alberti A, Drago C, Tagger A, Ribero M L, Realdi G. Long-term follow-up of non-A, non-B (type C) post-transfusion hepatitis. J Hepatol. 1992;16:273–281. doi: 10.1016/s0168-8278(05)80657-9. [DOI] [PubMed] [Google Scholar]
- 20.Wünschmann S, Medh J D, Klinzmann D, Schmidt W N, Stapleton J T. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J Virol. 2000;74:10055–10062. doi: 10.1128/jvi.74.21.10055-10062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh C-T, Liaw Y-F, Ou J-H. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J Virol. 1990;64:6141–6147. doi: 10.1128/jvi.64.12.6141-6147.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh C-T, Chu C-M, Liaw Y-F. Distinct composition of viral quasispecies between ascites and serum samples from patients with late stage chronic hepatitis C. Biochem Biophys Res Commun. 1996;227:524–529. doi: 10.1006/bbrc.1996.1540. [DOI] [PubMed] [Google Scholar]
- 23.Yeh C-T, Lu S-C, Chu C-M, Liaw Y-F. Molecular cloning of a defective hepatitis C virus genome from the ascitic fluid of a patient with hepatocellular carcinoma. J Gen Virol. 1997;78:2761–2770. doi: 10.1099/0022-1317-78-11-2761. [DOI] [PubMed] [Google Scholar]
- 24.Yeh C-T, Chien R-N, Chu C-M, Liaw Y-F. Clearance of the original hepatitis B virus YMDD-motif mutants with emergence of distinct lamivudine-resistant mutants during prolonged lamivudine therapy. Hepatology. 2000;31:1318–1326. doi: 10.1053/jhep.2000.7296. [DOI] [PubMed] [Google Scholar]