Abstract
Background
Aberrant accumulation of R-loops leads to DNA damage, genome instability and even cell death. Therefore, the timely removal of harmful R-loops is essential for the maintenance of genome integrity. Nucleolar R-loops occupy up to 50% of cellular R-loops due to the frequent activation of Pol I transcription. However, the mechanisms involved in the nucleolar R-loop resolution remain elusive. The nucleolar acetyltransferase NAT10 harbors a putative RecD helicase domain (RHD), however, if NAT10 acts in the R-loop resolution is still unknown.
Methods
NAT10 knockdown cell lines were constructed using CRISPR/Cas9 technology and short hairpin RNA targeting NAT10 mRNA, respectively. The level of R-loops was detected by immunofluorescent staining combined with RNase H treatment. The helicase activity of NAT10 or DDX21 was determined by in vitro helicase experiment. The interaction between NAT10 and DDX21 was verified by co-immunoprecipitation, immunofluorescent staining and GST pull-down experiments. Acetylation sites of DDX21 by NAT10 were analyzed by mass spectrometry. NAT10 knockdown-induced DNA damage was evaluated by immunofluorescent staining and Western blot detecting γH2AX.
Results
Depletion of NAT10 led to the accumulation of nucleolar R-loops. NAT10 resolves R-loops through an RHD in vitro and in cells. However, Flag-NAT10 ∆RHD mutant still partially reduced R-loop levels in the NAT10-depleted cells, suggesting that NAT10 might resolve R-loops through additional pathways. Further, the acetyltransferase activity of NAT10 is required for the nucleolar R-loop resolution. NAT10 acetylates DDX21 at K236 and K573 to enhance the helicase activity of DDX21 to unwind nucleolar R-loops. The helicase activity of DDX21 significantly decreased by Flag-DDX21 2KR and increased by Flag-DDX21 2KQ in cells and in vitro. Consequently, NAT10 depletion-induced nucleolar R-loop accumulation led to DNA damage, which was rescued by co-expression of Flag-DDX21 2KQ and Flag-NAT10 G641E, demonstrating that NAT10 resolves nucleolar R-loops through bipartite pathways.
Conclusion
We demonstrate that NAT10 is a novel R-loop resolvase and it resolves nucleolar R-loops depending on its helicase activity and acetylation of DDX21. The cooperation of NAT10 and DDX21 provides comprehensive insights into the nucleolar R-loop resolution for maintaining genome stability.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01869-3.
Keywords: NAT10, DDX21, R-loop, Post-translational modification, RNA helicase, Nucleolus
Background
R-loops are unique three-stranded nucleic acid structures that form transiently during transcription. An R-loop structure contains a displaced single-stranded DNA (ssDNA) and an RNA: DNA hybrid formed by the nascently transcribed RNA and the template DNA [1]. Genome-wide sequencing revealed that R-loops occupy approximately 5% of the genome in mammals [2] with an average length ranging between 100 and 450 bp [3]. Under normal conditions, transiently formed R-loops are removed in a short period, with an average half-life of 11 min [4]. Failure to remove harmful R-loops leads to R-loop accumulation, which causes DNA damage through blocking efficient transcription and replication fork progression, and finally threatens genome stability [5, 6]. For instance, R-loop accumulation brings about the recruitment of nucleotide excision repair factors XPG and XPF to cut R-loops at stalled transcription complexes, therefore resulting in double-strand DNA breaks and genome instability [7]. A growing number of studies have shown that aberrations in regulating R-loops are associated with human diseases including autoimmune diseases, neurological disorders and cancer [8–11], highlighting the significance for maintaining R-loop homeostasis.
Although R-loops form potentially all over the transcribed genome, genes with high GC content are prone to R-loop formation due to the high thermodynamic stability of the hybridization of G-rich RNA with C-rich DNA [12]. Ribosomal DNA (rDNA) containing high GC content is the most heavily transcribed gene [13], thus R-loops often form during rDNA transcription. It has been reported that 50% of R-loops are found on the rDNA locus in wild-type yeast cells [14]. Additionally, DNA/RNA immunoprecipitation (DRIP)-seq analysis showed that R-loops are enriched at the 18 S, 5.8 S and 28 S rDNA loci [15], and the length of R-loops on the rDNA locus can reach up to 2kb [3], indicating that rDNA is the primary site of R-loop formation. Importantly, the accumulation of nucleolar R-loops impairs pre-ribosomal RNA (pre-rRNA) transcription and leads to rDNA damage [16], which is tightly associated with rDNA copy number loss and cellular senescence [17–19]. Therefore, timely resolution of nucleolar R-loops is crucial for maintaining rDNA stability and cell viability.
Cells have evolved several mechanisms to maintain nucleolar R-loop homeostasis. Eukaryotic ribonuclease H1 (RNase H1) controls nucleolar R-loop levels through directly and specifically degrading the RNA moiety of the RNA: DNA hybrids [20, 21]. Additionally, RNA helicases such as senataxin, DDX21 and DDX47 have been identified as nucleolar R-loop resolvases in cells and in vitro, while depletion of these RNA helicases leads to nucleolar R-loop accumulation and DNA damage [22–24]. RNA helicases unwind base pairs within the RNA: DNA duplexes locally depending on the ATPase activity, leading to the automatic dissociation of the remaining base pairs [25, 26]. However, how the activity of RNA helicases is regulated remains unclear. In addition, exploring novel RNA helicases would provide scientific evidence for the understanding of nucleolar R-loop resolution.
Nucleolar N-acetyltransferase 10 (NAT10) plays crucial roles in various cellular processes depending on its lysine and/or cytidine acetyltransferase activity. Under normal conditions, NAT10 promotes pre-rRNA transcription and processing through acetylating UBF [27]. Additionally, NAT10 was found to acetylate rRNA, and depletion of NAT10 leads to accumulation of 30 S rRNA and failure of 18 S rRNA processing [28]. NAT10 also acetylates tRNA and mRNA to enhance translation efficiency [29, 30]. Moreover, NAT10 ensures the correct segregation of chromosomes during mitosis through acetylating Eg5 [31]. Upon DNA damage, NAT10 acetylates and stabilizes p53 and PARP1 to promote DNA damage repair [32, 33]. Under energy stress, NAT10 was deacetylated by SIRT1 to promote autophagy [34]. All these data highlight the essential roles of NAT10 in maintaining cellular homeostasis depending on its acetyltransferase activity. A previous study suggested that the homologue of NAT10 in bacteria (TmcA) possesses a RecD helicase domain (RHD), which might remodel tRNA conformation [35]. Additionally, analysis of the protein sequence by Pfam website (http://pfam-legacy.xfam.org/) showed that NAT10 contains a putative RHD (amino acids 282–488) (Supplemental Fig. 1). However, the function of this RHD domain remains undetermined. Since NAT10 activates rDNA transcription, we wondered if NAT10 controls the resolution of nucleolar R-loops by its RHD in situ.
In the present study, we found that depletion of NAT10 led to the accumulation of nucleolar R-loops. NAT10 acts as a novel R-loop resolvase to unwind R-loops in vitro depending on its RHD. Further, the acetyltransferase activity of NAT10 was also required in the nucleolar R-loop resolution. Mechanistically, NAT10 acetylates DDX21 at K236 and K573 to enhance the R-loop unwinding activity of DDX21. Depletion of NAT10 led to DNA damage, which is rescued by co-expression of Flag-NAT10 G641E (the acetyltransferase enzyme-dead mutant) and Flag-DDX21 2KQ, demonstrating that NAT10 resolves nucleolar R-loop through its RNA helicase activity and coordinating with DDX21. These findings highlight the crucial role of NAT10 in maintaining DNA stability through resolving nucleolar R-loops in situ.
Methods
Cell culture and transfection
HeLa and HCT116 cells were maintained in DMEM supplemented with 10% fetal bovine serum. All cell lines were purchased from cell bank of Chinese Academy of Medical Sciences. Cells were routinely tested for Mycoplasma contamination. Cells were transfected with plasmids or siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. In transient transfection experiments, concentrations of plasmids were maintained at a constant level with an empty vector. Sequence of NAT10 siRNAs: UUGCCACGAGUCUCUCUCUUC [36].
Plasmid construction
GFP-NAT10, Flag-NAT10, Flag-NAT10 G641E, GST-NAT10 and its deletion mutant plasmids were generated in our laboratory [31]. Flag-NAT10 ΔRHD, Flag-DDX21 and Flag-DDX21 mutants were cloned into the pCI-neo vector. GST-DDX21 and its deletion mutants were cloned into the pGEX-4T1 vector. His-DDX21 and its mutants was cloned into pET-28b(+) vector. GFP-RNase H1 was cloned into pEGFP-C3 vector. All plasmids cloned with PCR inserts were confirmed by DNA sequencing. Site mutated mutant plasmids including Flag-DDX21 K236R, Flag-DDX21 K406R, Flag-DDX21 K573R, Flag-DDX21 K236R/K573R, Flag-DDX21 K236Q/K573Q, Flag-NAT10 K290A, Flag-NAT10 G641E/K290A, His-DDX21 K236R/K573R, His DDX21 K236Q/K573Q and GFP-RNase H1 D210N were obtained by mutagenesis using the Quick-Change Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol. The presence of mutations in the constructed plasmids were confirmed by DNA sequencing.
Antibodies and reagents
Antibody against NAT10 was generated by our lab previously [32]. Commercial antibodies used were anti-acetyl-lysine (Cell Signaling Technology, 9441), anti-γ-H2AX (Cell Signaling Technology, 9718), anti-β-actin (ABclonal, AC004), anti-α-tubulin (ABclonal, AC012), anti-Flag (TransGen, HT201), anti-GFP (TransGen, HT801), anti-DDX21 (Santa Cruz, sc-376953), anti-Nucleolin (Santa Cruz, sc-13057) and S9.6 antibody (Millipore, MABE1095). Anti-rabbit or anti-mouse secondary antibodies were from LI-COR (926-68071 and 926-32210). Actinomycin D (Act. D, SBR00013) was purchased from Sigma. Remodelin (SD1168) was purchased from Beyotime. RNase H (M0297) was purchased from NEW ENGLAND Biolabs. RNase T1 (R7096) and RNase III (R7086) were purchased from Beyotime.
CRISPR-Cas9-mediated genome editing and stable cell lines construction
The HeLa control sgRNA, HeLa NAT10 sgRNA, HCT116 control sgRNA and HCT116 NAT10 sgRNA cell lines were established by CRISPR–Cas9 genome editing technology in our laboratory previously [31, 32]. The NAT10 stably knockdown and control HeLa cell lines were constructed by short hairpin RNA (shRNA)-mediated genetic knockdown. Briefly, NAT10 shRNA-1, NAT10 shRNA-2 and control shRNA oligos were cloned into the pLKO.1 plasmid to obtain a lentiviral particle, and the HeLa cell lines were infected by the indicated lentivirus. Next, the NAT10 stably knockdown cells were screened by puromycin. NAT10 protein knockdown was confirmed by Western blot. Sequence of NAT10 shRNAs: #1 GAGAUGUAUUCACGGAAUAUG and #2 UUGCCACGAGUCUCUCUCUUC [36].
Western blotting
Cells were harvested and lysed in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 0.5% Nonidet P-40 (NP-40), 1 mM PMSF, 1 mM Na3VO4, 1 mM EDTA, 1 mM NaF and cocktail of protease inhibitors. Following lysate clearance with centrifugation for 30 min at 12,000 rpm and 4℃, protein quantification was performed using Bradford Protein Assay Kit (Beyotime). Proteins were separated on SDS-PAGE and transferred onto 0.45 μm–0.2 μm nitrocellulose blotting membrane (Amersham). Membranes were probed with corresponding primary antibodies after blocking with 5% milk in phosphate-buffered saline (PBS). After extensive washing with PBS/T (0.5% Tween-20 in PBS buffer), the membranes were incubated with IRDye® 800CW or 680RD secondary antibodies (LI-COR). Fluorescence signals were detected using Odyssey® CLx imager. Images were acquired using Image Studio 5.0 (LI-COR).
Co-immunoprecipitation assay
Cell lysates were prepared in Buffer A (25 mM Tris–Cl (pH 7.5), 150 mM NaCl, 1 mM DTT, 2 mM EDTA, 0.5 mM PMSF, 5% glycerol, 0.5% NP-40 and protease inhibitors) and then were sonicated. The supernatant was collected after centrifugation and used for immunoprecipitation. Antibodies were coupled with a 50% suspension of protein A/G-Sepharose beads (GE Healthcare) in IPP500 (500 mM NaCl, 10 mM Tris–Cl (pH 8.0), 0.5% NP-40) for 2 h at 4°C. The coupled beads were incubated with cell lysates overnight at 4°C. After washing, the precipitants were analyzed by Western blot using the indicated antibodies.
DRIP
DRIP experiment was performed as previously reported [37]. Cells were lysed by cell lysis buffer (85 mM KCl, 5 mM PIPES (pH 8.0), 0.5% NP-40 and protease inhibitors) and then were centrifuged. The pellet was washed with cell lysis buffer and lysed by nuclear lysis buffer (10 mM Tris-HCl (pH 7.5), 200 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100, 0.2% sodium deoxycholate, 0.1% SDS, 0.05% sodium lauroyl sarcosinate (SLS) and protease inhibitors). After sonification and centrifugation, the supernatant was collected and diluted with RSBT buffer (10 mM Tris-HCl pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100). Then the diluted lysate was incubated with S9.6-antibody conjugated Protein A/G Sepharose beads overnight at 4℃ in R-loop IP buffer (10 mM Tris-HCl (pH 7.5), 200 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100, 0.05% sodium deoxycholate, 0.025% SDS, 0.0125% SLS). After washing by RSBT buffer four times and RSB buffer (10 mM Tris-HCl (pH 7.5), 200 mM NaCl, 2.5 mM MgCl2) twice, the precipitants were analyzed by Western blot using the indicated antibodies.
Real-time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. Then cDNA was synthesized from 1 µg of total RNA using the Hifair® I 1st Strand cDNA Synthesis SuperMix (YEASEN). The quantitative polymerase chain reaction (qPCR) analysis was performed using the ABI 7500 Real-time PCR System (Applied Biosystems) and Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (YEASEN). The human β-actin was used as an internal control. All real-time PCR data were analyzed by comparative Ct method and normalized to β-actin. The sequences of primers for β-actin are CATGTACGTTGCTATCCAGGC (forward) and CTCCTTAATGTCACGCACGAT (reverse). The sequences of primers for pre-rRNA are GAACGGTGGTGTGTCGTTC (forward) and GCGTCTCGTCTCGTCTCACT (reverse).
Northern blot
Northern blot was performed as described before [27]. Briefly, total RNA was extracted from control sgRNA and NAT10 sgRNA cells. Denatured RNA was separated in 1% agarose gel (with 2.2 mol/L formaldehyde) for 4 h at 60 V (5 V/cm gel). RNA was transferred onto Hybond-N + nylon membrane (Amersham) for 2 h in 20×SSC. Prehybridization was performed for 4 h at 42℃ in 5×SSC, 5×Denhardts’ solution (0.1% (w/v), Ficoll 400, 0.1% (w/v) BSA, 0.1% (w/v) polyvinylpyrrolidone-40), 1% SDS, 100 µg/ml salmon sperm DNA and 50 mM Na2HPO4. Biotin-labeled DNA probes was added into prehybridization buffer, and hybridization was carried out at 42℃ for 16 h. The membrane was washed in 0.5×SSC and 0.5% SDS for 10 min for 3 times at 65℃. Finally, the membrane was incubated with HRP-labeled streptavidin and analyzed by chemiluminescence. The sequence of the biotin-labeled DNA probe is 5’-CCTCTCCAGCGACAGGTCGCCAGAGGACAGCGTGTCAGC-3’.
Immunofluorescence staining
HeLa or HCT116 cells were fixed with ice-cold 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature (RT). After permeabilization by 0.5% Triton X-100 for 10 min at RT, cells were blocked with 10% goat serum in PBS for 45 min at 37℃ and then incubated with appropriate primary antibodies at 4℃ overnight. After washing with PBS, cells were incubated with secondary antibodies conjugated with DyLight 488 or DyLight 594 (1:100) (Earthox) for 45 min at 37℃. The nuclei were stained with DAPI (Beyotime) for 10 min at RT and the coverslips were mounted. Images were processed and quantified using ImageJ software. The mean of intensity of S9.6 signal was calculated by dividing the total S9.6 fluorescence intensity of a cell nuclei/nucleoli by the area of the cell nuclei/nucleoli. Mean intensity of nuclear γH2AX signal was calculated as the same.
In-cell acetylation experiment
Cells were incubated with 1 μm trichostatin A (TSA) and 5 mM nicotinamide (NAM) for 6 h before harvested. Cell extracts were prepared in whole cell lysis buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM PMSF, 0.1% NP-40, 2 mM EDTA, 1 mM DTT and 5% glycerol, protease inhibitors, 10 μm TSA and 5 mM NAM) and then were sonicated. Proteins were incubated with anti-acetyl-lysine antibody-conjugated protein A/G-Sepharose beads overnight at 4°C. The beads were washed five times with wash buffer (10 mM Tris-HCl (pH 7.5), 150mM NaCl, 5mM MgCl2, 0.1% NP-40). The immunoprecipitates and total proteins were subjected to Western blot using the indicated antibodies.
Protein purification
Flag-NAT10-His was purified in sf9 cells by our lab previously [32]. GST-NAT10 mutants, GST-DDX21 and its mutants, His-DDX21, His-DDX21 2KR, His-DDX21 2KQ, His-NAT10 and His-NAT10 ΔRHD were purified in E. coli strain BL21 or Rosetta (TransGen). Plasmids were transformed into BL21 or Rosetta, and isopropy-β-D-thiogalactoside (IPTG) was added at the concentration of 0.1 mM to induce protein expression for 4 h at 37℃. Cells were resuspended in GST lysis buffer (500 mM NaCl, 20% glycerol, 50 mM Tris-HCl (pH 7.5), 0.2% NP-40 and protease inhibitors) or His lysis buffer (20 mM Tris-Cl (pH 7.5), 500 mM KCl, 10% glycerol, 0.1% Triton X-100, 5 mM imidazole and protease inhibitor cocktail), followed by sonication and centrifugation. GST fused proteins were purified using the Glutathione Sepharose 4B (GE Healthcare) and His-fused proteins were purified using Ni-NTA agarose (Qiagen).
In vitro acetylation experiment
Purified His-DDX21 and Flag-NAT10-His were incubated in reaction buffer (50 mM Tris-HCl (pH 7.5), 10% glycerol, 0.1 mM EDTA, 1 mM PMSF, 10 mM sodium butyrate, 10 µM acetyl‐CoA) at 30°C for 1 h. Reaction mixtures were separated by SDS-PAGE and analyzed by Western blot using the indicated antibodies.
GST pull-down experiment
GST fusion proteins were prepared following standard protocol. For in vitro binding assays, GST fusion proteins bound to the Glutathione Sepharose 4B (GE Healthcare) were incubated with His-tagged purified proteins in GST binding buffer (100 mM NaCl, 2 mM EDTA, 20 mM Tris-HCl (pH 7.5), 2 mM DTT, 5% glycerol, 1% NP-40). After washing, the bound proteins were separated by SDS–PAGE and immunoblotted with indicated antibodies.
Identification of acetylation site in DDX21 by LC-MS/MS analysis
In vitro acetylation experiment was performed using purified His-DDX21 and Flag-NAT10-His proteins. The reaction mixture was resolved by SDS-PAGE and visualized by Coomassie blue staining. The bands of DDX21 were cut from SDS-PAGE gel, fully trypsinized, and analyzed by Extractive liquid chromatography-tandem mass spectrometry (LC-MS/MS) using mass spectrometer Orbitrap Velos Pro (Thermo). Mass spectrometry was carried out at the Key Laboratory of Proteomics of Peking University Health Science Center, and data were processed using the Proteome Discoverer software (Version 1.4).
In vitro helicase experiment
The R-loop substrates were generated by annealing 5’ 6-carboxyfluorescein (FAM)-labeled reverse DNA oligonucleotides (GACGCTGCCGAATTCTACCATGTGAAATTTCGCGCGATGAGACTGCCTACAGGTTCACCC), forward DNA oligonucleotides (GGGTGAACCTGTAGGCAGTCGTGCATTAATGGCTGGTTAGTGGTAGAATTCGGCAGCGTC) and RNA oligonucleotides (UCAUCGCGCGAAAUUUCACA) at a molar ratio of 1:2:4. The annealing reaction was carried out by heating the mixture for 5 min at 95℃ followed by slow cooling to 25℃ in annealing buffer (25 mM Tris-HCl (pH 7.5), 3.5 mM MgCl2, 100 mM NaCl, 2 mM DTT). The helicase experiment was performed by incubating the 10 nM R-loop substrates with indicated recombinant proteins in 20 µL of reaction buffer (25 mM Tris-HCl (pH 7.5), 3.5 mM MgCl2, 5 mM ATP, 2 mM DTT, 50 mM NaCl) at 37℃ for 30 min. ATP was lastly added to start the reaction. Reactions were terminated by adding 3 µL of loading buffer containing 0.25% (w/v) bromophenol blue and xylene cyanol in 30% (v/v) glycerol and incubated on ice for 5 min. The products were separated by native PAGE (12%, 1× TBE). Gel images were acquired by Amersham Typhoon (GE Healthcare).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.0.0 for Windows, GraphPad Software, San Diego, California USA, www.graphpad.com. One-way ANOVA followed by Tukey’s multiple comparisons test or Newman-Keuls multiple comparisons test was used to analyze the differences among more than two groups. The two groups were analyzed using t-test. The group size is the minimum number that is sufficient to define a functional parameter and the variance is similar between the groups. All data met the assumptions of the tests and represented in the figures with the error of the mean (mean ± SEM or SD). P < 0.05, P < 0.01, P < 0.001 and P < 0.0001 were considered statistically significant.
Results
Depletion of NAT10 leads to nucleolar R-loop accumulation
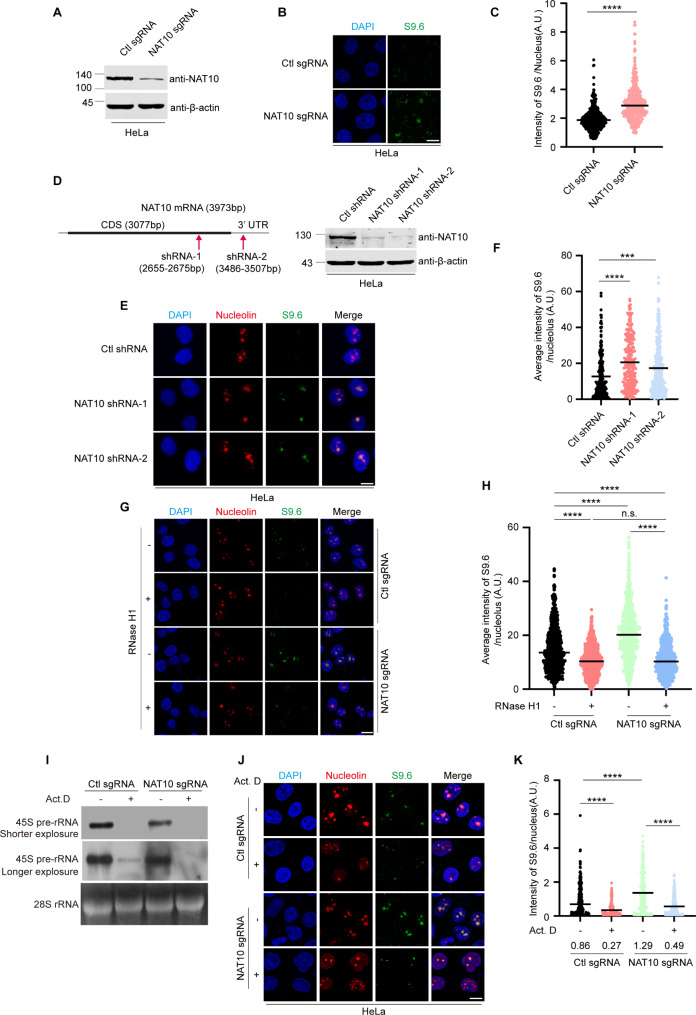
To determine the function of NAT10 in resolving R-loops, we generated NAT10 sgRNA HeLa cell lines using CRISPR-Cas9 technology, and the genetic background for NAT10 in control sgRNA cells and NAT10 sgRNA cells was verified by DNA sequencing (Supplemental Fig. 2). The level of R-loops was evaluated through immunofluorescent staining using a S9.6 antibody, which recognizes RNA: DNA hybrids [38]. The nuclear intensity of S9.6 significantly increased (Fig. 1A-C) in the NAT10 sgRNA cells, suggesting that NAT10 is required for R-loop resolution in the nucleus. The role of NAT10 in R-loop resolution was confirmed in the HCT116 NAT10 sgRNA cell lines (Supplemental Fig. 3A-3C). Since NAT10 is a nucleolar protein, we examined the subcellular localization of NAT10-associated R-loops. The intensity of S9.6 signal in each nucleolus was measured in the NAT10 shRNA cells and control shRNA cells. The nucleolar R-loop levels significantly increased in the NAT10 shRNA cells (Fig. 1D-F), indicating that NAT10 is required for the resolution of nucleolar R-loops. It is reported that S9.6 antibody might recognize rRNAs, thus S9.6 IF might bring about artifacts [39, 40]. To exclude the off-targets of the S9.6 antibody, we have treated control sgRNA cells and NAT10 sgRNA cells with RNase H1, RNase T1 or RNase III, and performed immunofluorescent staining to evaluate the level of nucleolar S9.6 intensity. The results show that upon treatment with RNase H1, which specifically digests DNA: RNA hybrid, the nucleolar S9.6 intensity decreased in both control sgRNA and NAT10 sgRNA cells with no significant difference (Fig. 1G, H), confirming that NAT10 depletion increases accumulation of nucleolar R-loop. Upon treatment with RNase T1, which digests single-stranded RNA, the nucleolar intensity of S9.6 in NAT10 sgRNA cells was still significantly higher than that in the control sgRNA cells, though it decreased in both NAT10 sgRNA cells and control cells. This result further confirmed that depletion of NAT10 leads to accumulation of R-loop. Additionally, it indicated that single-stranded RNAs are also recognized by the S9.6 antibody as previously reported [40]. Upon treatment with RNase III, which digests double-stranded RNA, the nucleolar S9.6 intensity remained unchanged in both control sgRNA and NAT10 sgRNA cells (Supplemental Fig. 4). To further validate that NAT10 knockdown induces R-loop accumulation, we transfected control sgRNA and NAT10 sgRNA cells with the enzyme-dead mutant of RNase H1-GFP-RNase H1 D210N. RNase H1 D210N still binds to DNA: RNA hybrid but is unable to hydrolyze the RNA, thus has been used to detect R-loops [41]. The nucleolar GFP intensity elevated in NAT10 sgRNA cells (Supplemental Fig. 5). Taken together, these results demonstrate that knockdown of NAT10 causes accumulation of R-loops in the nucleolus.
Fig. 1.
Depletion of NAT10 leads to nucleolar R-loop accumulation (A) NAT10 was knocked down using CRISPR-Cas9 technology. Proteins of NAT10 sgRNA and control sgRNA HeLa cells were extracted and subjected to Western blot and probed with indicated antibodies. (B) Cells described in A were fixed and immunofluorescent staining was performed with S9.6 antibody. Nucleus was stained with DAPI. Scale bar represents 10 μm. (C) Quantitative comparison of average intensity of nuclear S9.6 signal between NAT10 sgRNA cells and Ctl sgRNA cells is shown (n > 300 cells). P value was calculated using t-test. ****P < 0.0001. A.U. denotes arbitrary unit. (D) NAT10 knockdown HeLa cells were generated using shRNAs targeting NAT10 mRNA. The target sites of shRNAs are shown in the schematic diagram (left panel). Proteins in cell lysates of indicated cells were subjected to Western blot to evaluate the level of NAT10 (right panel). (E) NAT10 shRNA and Ctl shRNA HeLa cells were fixed and immunostained with indicated antibodies. Nucleus was stained with DAPI. The nucleolar area was determined by Nucleolin staining. Scale bar represents 10 μm. (F) Average intensity of nucleolar S9.6 fluorescence of cells described in E was calculated (n > 200 nucleoli). (G) Ctl sgRNA cells and NAT10 sgRNA cells were treated with 0.3 U/µL RNase H1 for 30 min. Cells were fixed and immunofluorescent staining was performed using the indicated antibodies. Nucleus was stained with DAPI. The nucleolar area was determined by Nucleolin staining. Scale bar, 10 μm. (H) Average intensity of nucleolar S9.6 fluorescence of cells described in G was quantified (n > 200 nucleoli). P values were calculated using one-way ANOVA. ****P < 0.0001. n.s. denotes no significance, A.U. denotes arbitrary unit. (I)-(K) Ctl sgRNA and NAT10 sgRNA HeLa cells were treated with 8 nM Act. D or DMSO for 8 h. (I) Total RNAs were extracted and subjected to Northern blot probed with indicated probes. 28 S rRNA was used as a loading control. (J) Ctl sgRNA and NAT10 sgRNA cells were fixed and stained with indicated antibodies. Scale bar represents 10 μm. (K) The average intensity of nuclear S9.6 signal of cells described in J was calculated (n > 200 cells). P values in F,H and K were calculated by one-way ANOVA. *P < 0.05, ***P < 0.001, ****P < 0.0001, A.U. denotes arbitrary unit
Since the formation of nucleolar R-loops is closely associated with the rDNA transcription [21], we treated cells with a low dose of actinomycin D (Act. D) to specifically inhibit Pol I transcription [42] (Fig. 1I). The levels of nucleolar R-loops were significantly reduced after Act. D treatment, indicating that the nucleolar R-loops form during Pol I transcription (Fig. 1I-K). Although NAT10 depletion inhibited Pol I transcription, NAT10 knockdown-induced R-loop accumulation was 1.5-fold (1.29/0.86) more than that in the control sgRNA cells (Fig. 1J, K), demonstrating that NAT10 plays essential roles in the nucleolar R-loop resolution.
NAT10 resolves R-loops depending on the RHD in vitro
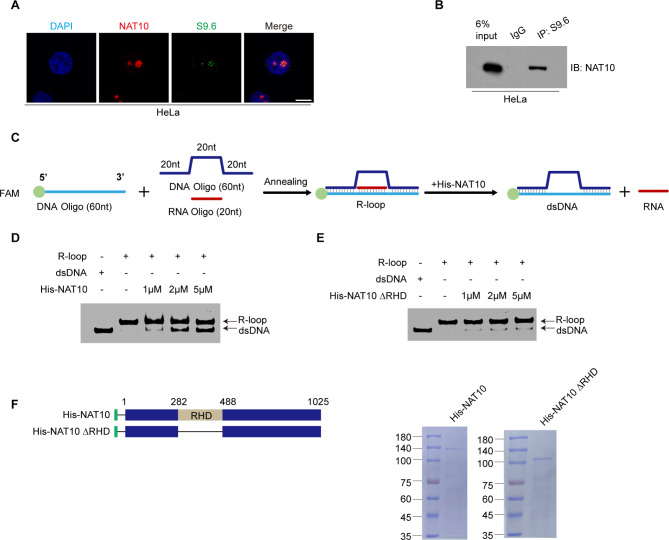
To unravel the mechanism by which NAT10 resolves nucleolar R-loops, we firstly verified the interaction between NAT10 and R-loops. NAT10 colocalizes with S9.6 signals in the nucleolus (Fig. 2A). Meanwhile, DRIP experiment confirmed that NAT10 binds to RNA: DNA hybrids (Fig. 2B), suggesting that NAT10 might resolve nucleolar R-loops directly. To verify if NAT10 resolves R-loops directly, we performed the in vitro helicase experiment as previously described [22], in which a FAM-labeled DNA oligo was annealed with a 20 nt complementary RNA oligo, and a DNA oligo flanking the FAM-labeled RNA: DNA hybrid (Fig. 2C). His-NAT10 and its helicase domain deletion mutant (His-NAT10 ΔRHD) were purified from E. coli and used in the in vitro helicase experiment with the annealed R-loop products. His-NAT10 unwinds the R-loops in vitro (Fig. 2D, F), while His-NAT10 ΔRHD failed to do so (Fig. 2E, F). We thus identify NAT10 as an R-loop resolvase which directly resolves R-loops in vitro depending on its RHD.
Fig. 2.
NAT10 resolves R-loops depending on the RHD in vitro. (A) Immunofluorescent staining was performed using indicated antibodies after HeLa cells were fixed. Nuclei were stained with DAPI. Scale bar represents 10 μm. (B) Nuclear fractionation of HeLa cells were extracted and immunoprecipitated with control IgG and S9.6 antibody. The immunoprecipitants were resolved by SDS-PAGE and probed with anti-NAT10 antibody. (C) Schematic diagram of in vitro helicase experiment procedure. (D) In vitro helicase experiment was performed with purified His-NAT10. The reaction products were analyzed by gel electrophoresis and fluorescence imaging. The bands of R-loops and dsDNA are indicated by corresponding arrows. (E) In vitro helicase experiment was performed with His-NAT10 ΔRHD as described in D. (F) His-NAT10 and His-NAT10 ΔRHD proteins were purified from E. coli. Schematic diagram represents the constructs of His-NAT10 and His-NAT10 ΔRHD (left panel). Purified proteins were resolved by SDS-PAGE and stained with Coomassie blue R-250 (right panel)
The acetyltransferase activity of NAT10 is also required for the nucleolar R-loop resolution
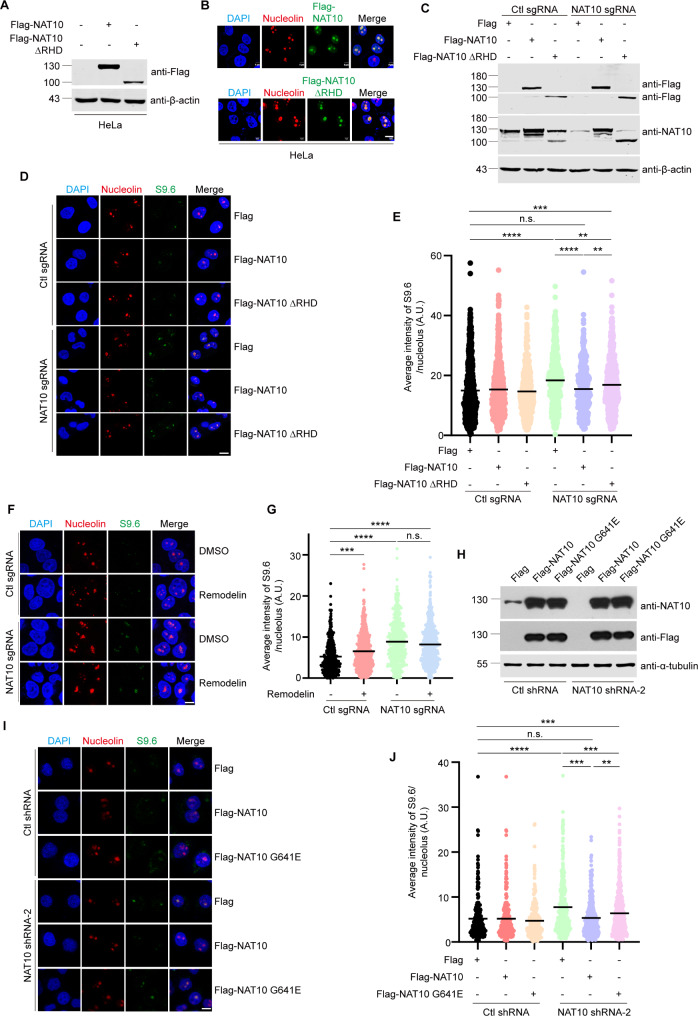
Next, we determined if NAT10 resolves R-loops depending on its RHD in cells. Flag-NAT10 ΔRHD localizes in the nucleolus (Fig. 3A, B), and was still capable of partially reducing the NAT10 knockdown-induced R-loop accumulation (Fig. 3C-E). Considering that depletion of RHD might lead to disruption in NAT10’s structure and function, we tried to find out the site responsible for the helicase catalytic activity in NAT10 RHD domain. In the RHD of NAT10, we have found a conserved “RGRGKS” Walker A motif (286–291 a.a.), which might be responsible for ATP hydrolysis [35]. Previous studies proved that mutation of lysine to alanine within the Walker A motif abolishes the helicase activity of RNA helicases [43, 44]. We thus constructed Flag-NAT10 K290A and evaluated the effect of this NAT10 mutant. Ectopic Flag-NAT10 K290A only partially reduced the nucleolar R-loop accumulation in NAT10 sgRNA cells compared with Flag-NAT10 WT (Supplemental Fig. 6), demonstrating that Flag-NAT10 K290A is the helicase catalytic-dead mutant of NAT10. These results further indicate that NAT10 might resolve nucleolar R-loops depending on other functions. Since NAT10 participates in multiple cellular processes through acetylating various substrates, we wanted to know if NAT10 resolves R-loops through its lysine acetyltransferase activity. We found that the nucleolar R-loops accumulated significantly when the acetyltransferase activity of NAT10 was specifically inhibited by Remodelin [45] (Fig. 3F, G), while Remodelin has no significant effects on NAT10 sgRNA cells. This result indicates that the acetyltransferase activity of NAT10 is required for the nucleolar R-loop resolution. Additionally, Flag-NAT10 G641E, which abolishes the acetyltransferase activity of NAT10, was not capable of reducing the R-loop accumulation as efficiently as Flag-NAT10 did (Fig. 3H-J), confirming that the acetyltransferase activity of NAT10 is also required for the nucleolar R-loop resolution.
Fig. 3.
The acetyltransferase activity of NAT10 is also required for the nucleolar R-loop resolution. (A) HeLa cells were transfected with indicated plasmids. Cells were harvested after 24 h. Proteins in cell lysates were subjected to Western blot probed with indicated antibodies. Beta-actin was used as a loading control. (B) HeLa cells were transfected with Flag-NAT10 or Flag-NAT10 ΔRHD. Cells were fixed and immunostained with indicated antibodies. (C)-(E) Ctl sgRNA and NAT10 sgRNA cells were transfected with indicated plasmids. (C) Cells were lysed, and proteins were subjected to Western blot and probed with indicated antibodies. Beta-actin was used as a loading control. (D) Cells were fixed and stained with indicated antibodies. (E) Quantification of the average intensity of nucleolar S9.6 signal in cells described in D (n > 600 nucleoli). (F) Ctl sgRNA cells and NAT10 sgRNA cells were treated with DMSO or 5µM Remodelin for 48 h. Cells were fixed and immunofluorescent staining was performed using indicated antibodies. (G) The average intensity of nucleolar S9.6 signal of cells described in F was calculated (n > 600 nucleoli). (H)-(J) NAT10 shRNA HeLa cells were transfected with indicated plasmids. (H) Cells were harvested, and proteins extracted from cell lysates were subjected to Western blot and probed with indicated antibodies. Tubulin was used as a loading control. (I) Cells were fixed and immunostained with indicated antibodies. (J) The plots represent the average intensity of nucleolar S9.6 signal of cells described in I (n > 250 nucleoli). In immunofluorescent staining experiments, nucleus was stained with DAPI and the nucleolar area was determined by Nucleolin staining. Scale bar represents 10 μm. All P values were calculated using one-way ANOVA. *P < 0.05, ***P < 0.001, ****P < 0.0001, n.s. denotes no significance, A.U. denotes arbitrary unit
The RNA helicase DDX21 is identified as a NAT10 partner in the nucleolar R-loop resolution
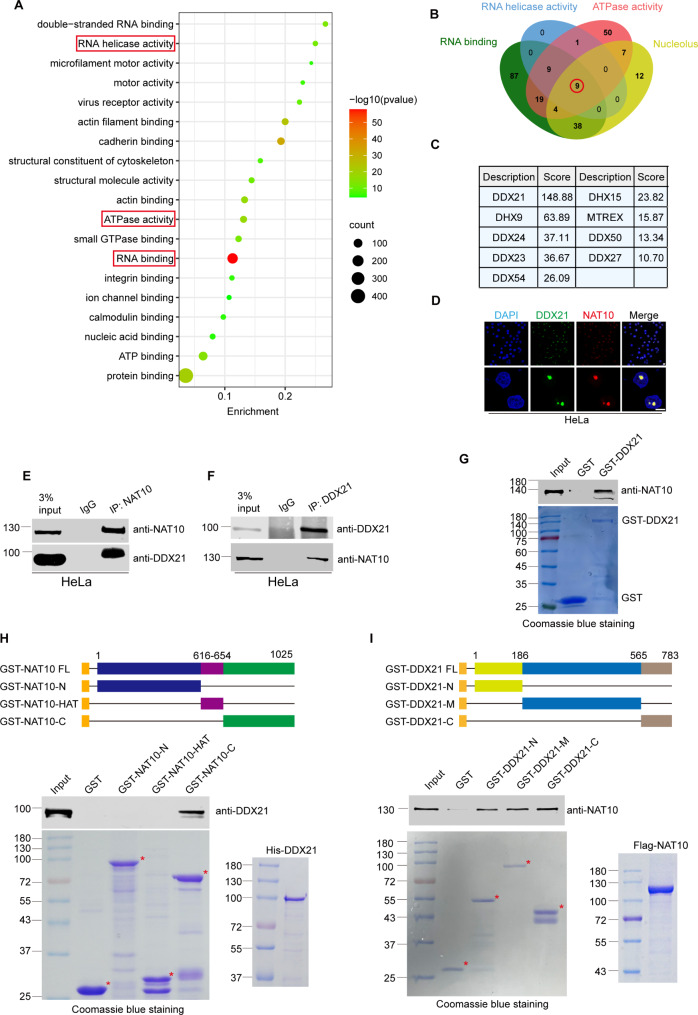
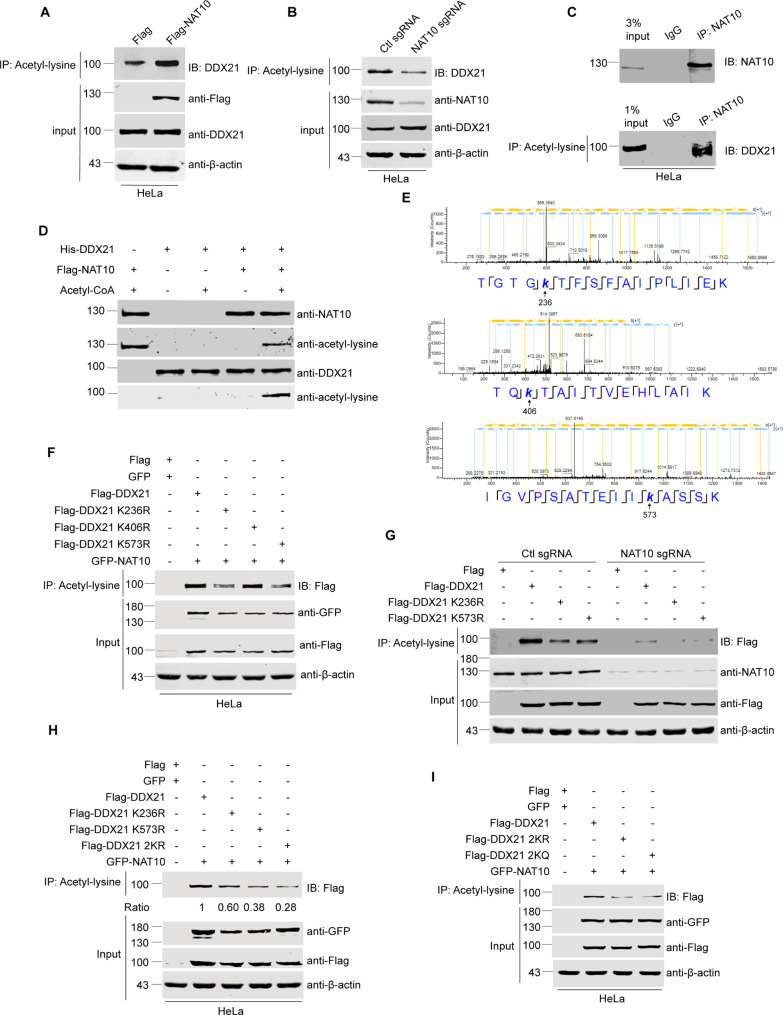
To identify the acetylated substrate of NAT10 in the R-loop resolution, we classified the Flag-NAT10-binding proteins previously identified by mass spectrometric analysis after immunoprecipitation [31] using gene ontology analysis (https://david.ncifcrf.gov/) (Fig. 4A and Supplemental Table. S1). The NAT10-binding proteins are most significantly enriched in the RNA helicase activity (with a high enrichment score) and RNA binding (with the lowest P value), suggesting that the critical NAT10-binding partner might be a helicase with potential RNA binding ability. Additionally, ATPase activity is also one of the critical properties of NAT10-interacting proteins. All these evidences point to RNA helicases as NAT10 partners in the R-loop resolution. To further identify the predominant RNA helicase that collaborates with NAT10 in the nucleolar R-loop resolution, we comprehensively analyzed the protein datasets with the properties of RNA binding, RNA helicase activity, ATPase activity and nucleolar localization, and 9 common proteins are shared in these datasets (Fig. 4B). Among these 9 proteins, the nucleolar RNA helicase DDX21 hosts the highest score (Fig. 4C). To validate the interaction between DDX21 and NAT10, immunofluorescent staining was performed and DDX21 co-localizes with NAT10 in the nucleolus (Fig. 4D). Furthermore, co-immunoprecipitation experiments showed that endogenous DDX21 interacts with NAT10 in cells (Fig. 4E, F). To determine if NAT10 directly binds DDX21, we performed in vitro GST pull-down experiments using GST-DDX21 and Flag-NAT10 purified from sf9 insect cells (Fig. 4G). The result shows that GST-DDX21 directly binds to Flag-NAT10. We further determined the minimal DDX21-interacting domain in NAT10. GST pull-down experiments were performed using GST-NAT10 and its deletion mutants, and His-DDX21 purified from E. coli. His-DDX21 directly binds GST-NAT10-C (Fig. 4H). We thereafter determined the minimal NAT10-interacting domain in DDX21. Flag-NAT10 directly binds GST-DDX21-N, GST-DDX21-M and GST-DDX21-C (Fig. 4I). These data demonstrate that DDX21 interacts with NAT10 in cells and in vitro, and might collaborate with NAT10 in the nucleolar R-loop resolution.
Fig. 4.
The RNA helicase DDX21 is identified as a NAT10 partner in the nucleolar R-loop resolution. (A) Proteins immunoprecipitated by Flag-NAT10 were analyzed by gene ontology analysis (https://david.ncifcrf.gov/). Bubble chart was plotted to show the 15 clusters of lowest P values. The depth of bubble color represents P value. The size of each bubble represents number of proteins in indicated datasets. (B) Venn diagram of 4 datasets (“RNA binding”, “RNA helicase activity”, “ATPase activity” and “Nucleolus”) is shown. Nine proteins are shared in 4 datasets. (C) Details and mass spectrum score of the 9 proteins in B. (D) HeLa cells were fixed and immunofluorescent staining was performed using anti-DDX21 and anti-NAT10 antibodies. Nucleus was stained with DAPI. Scale bar represents 10 μm. (E)-(F) Proteins of HeLa cells were extracted and immunoprecipitated by anti-NAT10 antibody or anti-DDX21 antibody. Immunoprecipitants were resolved by SDS-PAGE and probed with indicated antibodies. (G) GST-DDX21 was purified in E. coli. GST pull-down assay was performed with purified GST-DDX21 and Flag-NAT10 proteins. The amounts of GST fusion proteins used in the GST pull-down assay are shown in the lower panel. (H) The schematic diagram represents the constructs of GST-NAT10 deletion mutants (upper panel). GST pull-down assay was performed using purified GST-NAT10 deletion mutants and His-DDX21. The purified GST protein or GST-NAT10 deletion mutants and His-DDX21 were resolved by SDS-PAGE and stained by coomassie blue staining (lower panel). (I) The schematic diagram represents the constructs of GST-DDX21 deletion mutants (upper panel). GST pull-down assay was performed using purified GST-DDX21 deletion mutants and Flag-NAT10. The purified GST protein or GST-DDX21 deletion mutants and Flag-NAT10 were resolved by SDS-PAGE and stained by coomassie blue staining (lower panel)
NAT10 acetylates DDX21 at lysine 236 and lysine 573
We further explored if NAT10 acetylates DDX21. The acetylation level of DDX21 increased upon ectopic expression of Flag-NAT10 (Fig. 5A) and decreased in NAT10 sgRNA cells (Fig. 5B), while the protein level and the localization of DDX21 were not affected by NAT10 depletion (Supplemental Fig. 7A and 7B). These data illustrate that NAT10 acetylates DDX21 in cells without changing the expression level and localization of DDX21. To determine if the acetylated form of endogenous DDX21 binds to NAT10, we firstly performed immunoprecipitation with the anti-NAT10 antibody, then eluted the precipitants and performed immunoprecipitation with the anti-pan-acetyl-lysine antibody. We show that endogenous acetylated DDX21 binds to NAT10 (Fig. 5C).
Fig. 5.
NAT10 acetylates DDX21 at K236 and K573. (A)-(B) HeLa cells were transfected with indicated plasmids and harvested after 24 h (A) NAT10 sgRNA cells and Ctl sgRNA cells were harvested (B) Proteins from whole cell lysates were immunoprecipitated by anti-pan-acetyl-lysine antibody. Immunoprecipitants were resolved by SDS-PAGE and probed with anti-DDX21 antibody. (C) HeLa cells were harvested and proteins from whole cell lysates were firstly immunoprecipitated by anti-NAT10 antibody. Immunoprecipitants were resolved by SDS-PAGE and probed with anti-NAT10 antibody (upper panel), or eluted. Eluted proteins were immunoprecipitated by anti-pan-acetyl-lysine antibody, and the immunoprecipitants were resolved by SDS-PAGE probed with anti-DDX21 antibody (lower panel). (D) In vitro acetylation experiment was performed using His-DDX21 and Flag-NAT10. Proteins were resolved by SDS-PAGE and probed with anti-pan-acetyl-lysine antibody. (E) In vitro acetylation experiment was performed as described in D. The reaction products were resolved by SDS-PAGE. The band of acetylated DDX21 was cut, fully trypsinized and analyzed by LC-MS/MS using mass spectrometer (Thermo). Mass spectrometry data were processed using the Proteome Discoverer software (Version 1.4). Three acetylation sites (K236, K406 and K573) of DDX21 were suggested. (F)-(I) HeLa cells, Ctl sgRNA or NAT10 sgRNA cells were transfected with indicated plasmids and harvested after 24 h. Proteins in cell lysates were immunoprecipitated by anti-pan-acetyl-lysine antibody. Immunoprecipitants were resolved by SDS-PAGE and probed with anti-Flag antibody. Proteins in whole cell lysates were subjected to Western blot and probed with indicated antibodies. Beta-actin was used as a loading control
To confirm the direct acetylation of DDX21 by NAT10, we performed in vitro acetylation experiments using Flag-NAT10 purified from sf9 insect cells and His-DDX21 purified from E. coli. NAT10 acetylates DDX21 in vitro (Fig. 5D), in addition to acetylating itself in the presence of acetyl-CoA as previously reported [46]. To specify the acetylated residues of DDX21 catalyzed by NAT10, mass spectrometry was applied to analyze the acetylated sites of DDX21 after in vitro acetylation experiments performed with purified His-DDX21 and Flag-NAT10. DDX21 might be acetylated at K236, K406 and K573 by NAT10 (Fig. 5E). To validate these acetylation sites of DDX21, we ectopically expressed site-specific Flag-DDX21 mutants and evaluated the acetylation level of these mutants. Both the acetylation level of Flag-DDX21 K236R and Flag-DDX21 K573R decreased in the presence of GFP-NAT10, while Flag-DDX21 K406R remained unchanged compared with that of Flag-DDX21 (Fig. 5F), indicating that NAT10 acetylates DDX21 at K236 and/or K573. To further confirm the acetylation of DDX21 at K236 and K573 residues is reliant on NAT10 in cells, we determined the acetylation level of ectopic Flag-DDX21 and its mutants in NAT10 sgRNA cells (Fig. 5G). In control shRNA cells, the acetylation level of Flag-DDX21 K236R and DDX21 K573R is lower than that of Flag-DDX21. In NAT10 sgRNA cells, the acetylation levels of Flag-DDX21 and its mutants drastically decreased accordingly. This result proves that acetylation of DDX21 K236 and K573 is reliant on NAT10. Further, we constructed Flag-DDX21 2KR (K236R/K573R, hypo-acetylation mimic) and Flag-DDX21 2KQ (K236Q/K573Q, hyper-acetylation mimic) plasmids. The acetylation level of Flag-DDX21 2KR and Flag-DDX21 2KQ was lower than Flag-DDX21 K236R or Flag-DDX21 K573R, demonstrating that NAT10 acetylates DDX21 at K236 and K573 simultaneously (Fig. 5H, I).
Acetylation of DDX21 significantly enhances its helicase activity in cells and in vitro
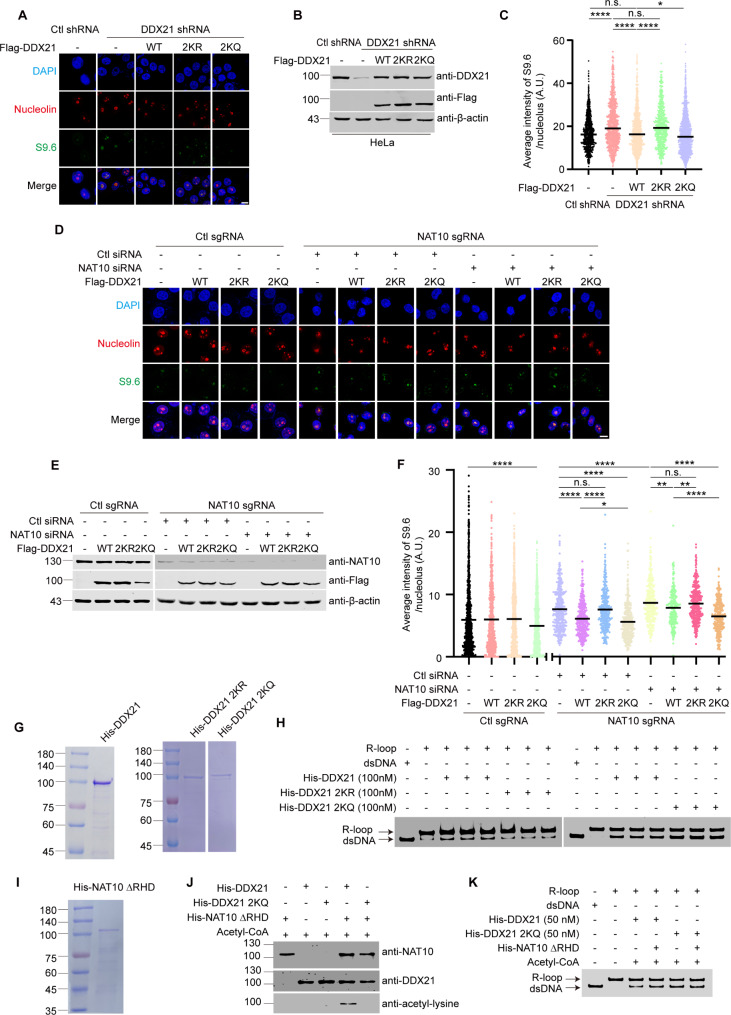
We further determined the role of NAT10-mediated acetylation of DDX21 in the nucleolar R-loop resolution. As previously reported [22], DDX21 depletion induced R-loop accumulation, and Flag-DDX21 rescued nucleolar accumulation of R-loops in DDX21-depleted cells (Fig. 6A-C). Flag-DDX21 2KQ reduced nucleolar R-loops levels more efficiently than Flag-DDX21 did, while Flag-DDX21 2KR failed to reduce R-loop levels. Both Flag-DDX21 2KR and Flag-DDX21 2KQ localized in the nucleolus (Supplemental Fig. 8A and 8B). These data demonstrate that acetylation of DDX21 by NAT10 enhances its capability of resolving nucleolar R-loops in cells. To determine if the acetylation of DDX21 could rescue the NAT10-depletion-caused R-loop accumulation, we ectopically expressed Flag-DDX21 and its mutants in the NAT10 sgRNA cells. To minimize the impact of remaining NAT10 in NAT10 sgRNA cells, we transfected NAT10 sgRNA cells with a NAT10 siRNA and determined the effect of ectopic Flag-DDX21 and its mutants on R-loop resolution. The NAT10 knockdown-induced accumulation of nucleolar R-loops was reduced by Flag-DDX21 and Flag-DDX21 2KQ but not by Flag-DDX21 2KR (Fig. 6D-F, Supplemental Fig. 8C and 8D). In the NAT10 sgRNA cells transfected with control siRNA, Flag-DDX21 shows slightly reduced helicase activity to resolve nucleolar R-loops compared with Flag-DDX21-2KQ. However, when NAT10 was further silenced in NAT10 sgRNA cells by siRNA, Flag-DDX21 2KQ reduced nucleolar R-loops much more effectively than Flag-DDX21 did. Notably, the level of the nucleolar R-loops was reduced by Flag-DDX21 2KQ in control sgRNA cells, while it remained unchanged upon ectopic expression of Flag-DDX21 or Flag-DDX21 2KR (Fig. 6D-F, Supplementary Fig. 8E). These data further confirm that DDX21 acetylation at K236 and K573 enhances the activity of DDX21 in the R-loop resolution.
Fig. 6.
Acetylation of DDX21 significantly enhances its helicase activity in cell and in vitro. (A)-(C) Ctl shRNA and DDX21 shRNA HeLa cells were transfected with indicated plasmids for 48 h. (A) Cells were fixed and immunofluorescent staining was performed using indicated antibodies. Nucleus was stained with DAPI and the nucleolar area was determined by Nucleolin staining. Scale bar represents 10 μm. (B) Cells were harvested and proteins extracted from cell lysates were subjected to Western blot and probed with indicated antibodies. Beta-actin was used as a loading control. (C) Quantitative comparison of average intensity of nucleolar S9.6 signal of cells described in A (n > 600 nucleoli). (D)-(F) Ctl sgRNA were transfected with indicated plasmids. NAT10 sgRNA HeLa cells were transfected with control and NAT10 siRNAs, and then cells were transfected with indicated plasmids 48 h later. (D) Cells were fixed and immunofluorescent staining was performed using indicated antibodies. Nucleus was stained with DAPI and the nucleolar area was determined by Nucleolin staining. Scale bar represents 10 μm. (E) Cells were harvested and proteins extracted from cell lysates were subjected to Western blot and probed with indicated antibodies. Beta-actin was used as a loading control. (F) Quantitative comparison of average intensity of nucleolar S9.6 signal of cells described in D (n > 300 nucleoli). P values in C and F were calculated using one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. denotes no significance, A.U. denotes arbitrary unit. (G) Purified His-DDX21, His-DDX21 2KR and His-DDX21 2KQ were resolved by SDS-PAGE and the gel was stained by coomassie blue. (H) In vitro helicase experiment was performed using R-loops and His-DDX21, His-DDX21 2KR or His-DDX21 2KQ. The bands of R-loop and dsDNA are indicated by corresponding arrows. (I) Purified His-NAT10 ΔRHD was resolved by SDS-PAGE and the gel was stained by coomassie blue. (J) In vitro acetylation experiment was performed using His-NAT10 ΔRHD and His-DDX21 or His-DDX21 2KQ. Proteins were resolved by SDS-PAGE and probed with indicated antibodies. (K) In vitro acetylation experiment was performed as described in J, and the products were used in the following in vitro helicase experiment. The bands of R-loop and dsDNA are indicated by corresponding arrows
As NAT10 resolves R-loops by its RHD, we wonder if the double mutation of NAT10 (K290A/G641E), which loses both acetyltransferase and helicase activities, fully abolishes NAT10’s function in R-loop resolution. Ectopic Flag-NAT10 G641E/K290A failed to reduce the level of nucleolar R-loops in NAT10 sgRNA cells (Supplemental Fig. 9), demonstrating that NAT10 resolves R-loops by both of its RHD and acetyltransferase activity. To further verify the effect of the DDX21 acetylation on the R-loop unwinding activity, the in vitro helicase experiment was performed using purified His-DDX21, His-DDX21 2KR and His-DDX21 2KQ (Fig. 6G). The helicase activity of DDX21 was reduced by His-DDX21 2KR and increased by His-DDX21 2KQ (Fig. 6H). In addition, we found that the helicase-deficient mutant His-NAT10 ΔRHD enhanced helicase activity of His-DDX21 but not that of His-DDX21 2KQ in vitro (Fig. 6I-K). These results demonstrate that NAT10 acetylates DDX21 at K236 and K573 to enhance its helicase activity to resolve R-loops in cells and in vitro.
NAT10-delpetion-induced DNA damage is rescued by Flag-DDX21 2KQ and Flag-NAT10 G641E
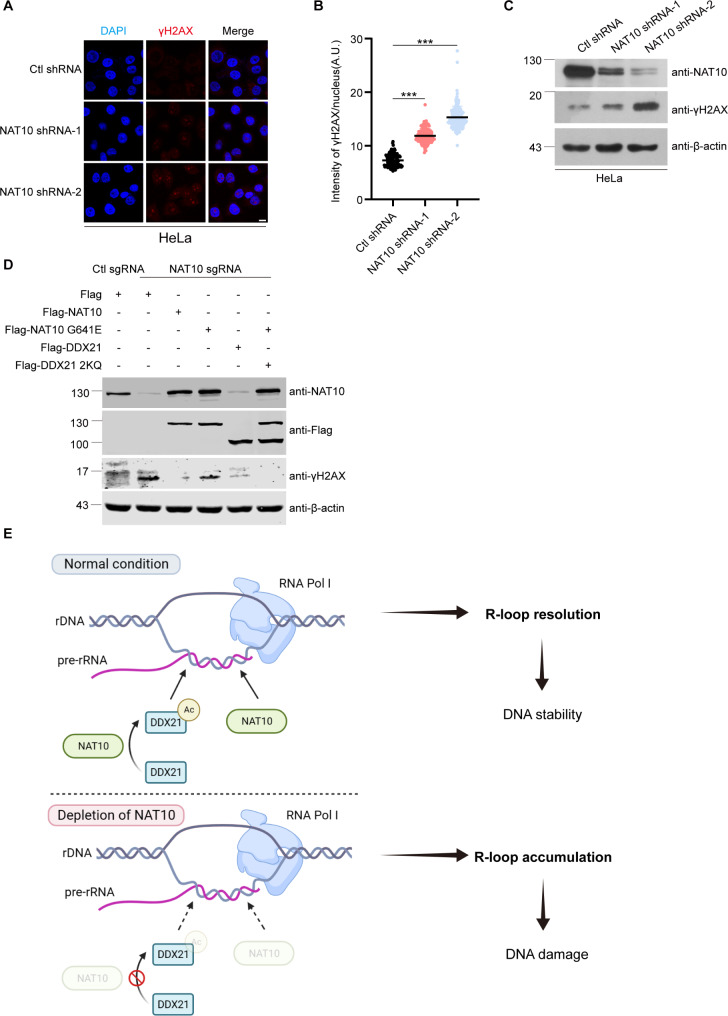
We further evaluated the phosphorylation on serine 139 of histone H2AX (γH2AX) to determine if NAT10 depletion-caused R-loop accumulation results in DNA damage. Immunofluorescence and Western blot showed that γH2AX increased in the NAT10-depleted cells (Fig. 7A-C), indicating that depletion of NAT10 causes DNA damage. To determine if the acetylation of DDX21 by NAT10 is required for maintaining genome stability, we ectopically expressed Flag-NAT10, Flag-NAT10 G641E, Flag-DDX21 and Flag-DDX21 2KQ in NAT10 sgRNA cells. The elevated γH2AX in NAT10 sgRNA cells was reduced by Flag-NAT10, and partially by Flag-NAT10 G641E or Flag-DDX21, indicating that the RHD of NAT10 acts in the prevention of DNA damage through resolving nucleolar R-loops. Additionally, co-expression of Flag-NAT10 G641E with Flag-DDX21 2KQ fully reduced the elevated γH2AX in NAT10 sgRNA cells (Fig. 7D), demonstrating that NAT10 maintains genome stability through its resolvase activity and acetylation of DDX21. Furthermore, we found that depletion of NAT10 leads to rDNA copy number loss and cellular senescence (Supplemental Fig. 10). These data indicate that NAT10 resolves nucleolar R-loops to protect cells from DNA damage.
Fig. 7.
NAT10-delpetion induced DNA damage is rescued by Flag-DDX21 2KQ and Flag-NAT10 G641E. (A) Ctl shRNA and NAT10 shRNA HeLa cells were fixed and immunofluorescent staining was performed with anti-γH2AX antibody. Nucleus was stained with DAPI. Scale bar represents 10 μm. (B) Nuclear average intensity of γH2AX of cells described in A was calculated (n > 100 cells). P values were calculated using t-test. ***P < 0.001, A.U. denotes arbitrary unit. (C) Western blot analysis of indicated protein levels in Ctl shRNA and NAT10 shRNA HeLa cells. (D) Ctl sgRNA and NAT10 sgRNA HeLa cells were transfected with indicated plasmids for 48 h. Proteins in cell lysates were subjected to Western blot and probed with indicated antibodies. Beta-actin was used as a loading control. (E) Summary of this study. NAT10 mediates nucleolar R-loop resolution through its helicase activity and acetylating DDX21 at K236 and K573, guaranteeing DNA stability. When NAT10 is knockout, nucleolar R-loops accumulate, leading to DNA damage and DNA instability. This working model is created with BioRender.com
Discussion
RNase H and several RNA helicases including senataxin, DDX21 and DDX47 have been found to remove harmful nucleolar R-loops [20, 22–24, 47]. However, the resolution of nucleolar R-loops is much more complicated than anticipated as the adequate level of nucleolar R-loops plays essential roles in the maintenance of nucleolar functions. In human cells, rDNA is organized into ten clusters of multiple tandem units on the 5 chromosomes and each rDNA unit consists of a 13 kb-long ribosomal gene and a 30 kb-long intergenic spacer (IGS) [48]. The nucleolar R-loops within the 13 kb-long ribosomal gene maintain the location of the rDNA locus since the rRNAs in rRNA: rDNA hybrid bind the ribonucleoproteins (RNPs) to keep the rDNA locus in the fibrillar center [49]. Meanwhile, the R-loops in the IGSs formed by anti-sense intergenic non-coding RNAs (asincRNAs) regulate the nucleolar organization and rDNA transcription, through inhibiting the transcription of the sense intergenic transcripts, which might disrupt nucleolar structure and impair rDNA transcription [50]. Therefore, nucleolar R-loops should be precisely controlled. In the present study, we demonstrate that NAT10 is required for the nucleolar R-loop resolution and depletion of NAT10 results in DNA damage. R-loops form during transcription, and inhibition of Pol I reduces R-loops levels. However, R-loops still accumulated when Pol I was inhibited in NAT10 sgRNA cells, demonstrating that NAT10 plays critical roles in the nucleolar R-loop resolution.
Aberrant accumulation of nucleolar R-loops leads to rDNA damage [16]. To repair rDNA damage, the damaged rDNA locus moves to the nucleolar cap region to commit non-homologous end joining (NHEJ) or homologous repair (HR) [51–53]. However, NHEJ might result in rDNA mutagenesis, leading to rDNA instability [54], and HR might lead to rDNA copy number loss [19]. Therefore, accumulated nucleolar R-loops must be timely removed to maintain the rDNA stability, and identification of novel nucleolar helicases will help us to better understand the maintenance of rDNA homeostasis. In the present study, we identified NAT10 as a novel RNA helicase with a RecD helicase domain. RecD helicase was first identified in E. coli as a subunit of the RecBCD complex to unwind DNA: DNA hybrid [55]. Subsequent research showed that Pif1, the human homologue of E. coli RecD unwinds RNA: DNA hybrid in vitro [56]. However, if RecD helicase resolves R-loops in mammalian cells remains unknown. Here, we demonstrate that NAT10 unwinds R-loops through its RHD in vitro, and NAT10 resolves R-loops in cells partially depending on its RHD, we thus provide evidence for RNA helicase activity of the RecD domain.
Given that NAT10 resolves R-loops in vitro depending on the RecD domain, deletion of the RHD might abolish the R-loop-resolving activity of NAT10 in cells. However, the Flag-NAT10 ΔRHD mutant still partially resolved nucleolar R-loops. In addition, we uncovered that Flag-NAT10 K290A is the helicase catalytic-dead mutant of NAT10 and it still partially resolved nucleolar R-loops. This point mutant of NAT10 avoids unanticipated disruptions in NAT10 structure and function by deleting RHD. It confirmed that NAT10 resolves nucleolar R-loops through other functions in addition to its helicase activity. We further showed that the acetyltransferase activity of NAT10 is also required for nucleolar R-loop resolution. We thus uncover that NAT10 participates in R-loop resolution depending on both the helicase activity and acetyltransferase activity. NAT10 functions in multiple cellular processes through acetylating various substrates including proteins and RNAs [27, 28, 30–32]. Here, DDX21 was further identified as a novel substrate of NAT10. It has been recently reported that both NAT10 and DDX21 interact with RNase H1 and assist in increasing the potency and safety of phosphorothioate oligonucleotides [57], yet the regulation between NAT10 and DDX21 is unknown. In the present study, we found that NAT10 acetylates DDX21 in cells and in vitro, and two novel acetylation sites of DDX21 at K236 and K573 are identified. Acetylation of these two sites by NAT10 strengthens the function of DDX21 in the nucleolar R-loop resolution. Our findings provide new insights into the role of DDX21 acetylation in nucleolar R-loop resolution. Notably, K236 is among the ATP-binding motif of DDX21 and is thought to be associated with ATP binding [58]. However, if the acetylation of DDX21 at K236 regulates the ATP binding capability needs further study. As lysine573 is within the dimerization domain of DDX21, and DDX21 dimerization is essential for its helicase activity [59], thus acetylation at K573 might facilitate its dimerization to enhance the helicase activity of DDX21.
DDX21 is a member of the DEAD box RNA helicase family, playing essential roles in the nucleolus. DDX21 promotes pre-rRNA transcription and processing through interacting with snoRNAs, and also promotes elongation of Pol II transcription through facilitating the release of P-TEFb from 7SK snRNP in the nucleoplasm [60]. Importantly, DDX21 has been found to resolve R-loop in cells and in vitro, and deletion of DDX21 leads to R-loop accumulation and spontaneous DNA damage, highlighting its importance to maintain genome stability. The R-loop unwinding activity of DDX21 is inhibited by CBP-dependent acetylation at K18, K137 and K600, while Sirt7 counteracts with CBP by de-acetylating DDX21 to maintain its helicase activity [22]. Here, we found that acetylation of DDX21 at K236 and K573 enhances its helicase activity, suggesting that acetylation exerts dual effects on helicase activity of DDX21. Our study unravels a novel mechanism for the regulation of DDX21 by NAT10, highlighting the role of the cooperation of two nucleolar proteins in maintaining genome stability.
In addition to lysine acetylation, NAT10 modifies various types of RNAs through cytidine acetylation. NAT10 acetylates tRNA to assist in the correct codon reading through stabilizing the ribose 3’ endo conformation [29]. Additionally, NAT10 promotes rRNA processing through acetylating 18 S rRNA, and NAT10 enhances the stability and translation efficiency of mRNA through acetylating mRNA [28, 30]. However, if NAT10 acetylates RNAs in R-loops remains unknown. Modifications of RNAs in R-loops including methyl-5-cytidine (m5C), methyl-3-cytidine (m3C) and methyl-6-adenosine (m6A) are important for the maintenance of R-loop stability and functions, while m3C modification promotes tumorigenesis [61–64]. However, if cytidine acetylation regulates R-loops needs further exploration.
Collectively, we demonstrate that NAT10 resolves R-loops formed during Pol I transcription, in addition to activating rDNA transcription during cell proliferation [27, 28], to protect cells from DNA damage. It was reported that sustained rDNA damage leads to the loss of rDNA copy number, which in turn inhibits ribosome biogenesis and triggers cellular senescence and cell death [19, 65–67]. Therefore, the function of NAT10 in R-loop resolution aligns with its role in maintaining cellular homeostasis. When NAT10’s functions including its helicase activity and acetyltransferase activity are inhibited, nucleolar R-loops are accumulated, resulting in genome instability and potentially triggering cellular senescence or cell death (Fig. 8E and Supplemental Fig. 10). Considering NAT10 is upregulated in multiple types of cancers and promotes cancer progression [68–70], inhibiting the helicase activity and NAT10-mediated acetylation of DDX21 might be an alternative strategy for cancer therapy.
Conclusion
In summary, our findings uncover the novel function of the nucleolar protein NAT10 to serve as an RNA helicase to resolve nucleolar R-loops. In addition, NAT10 acetylates classical RNA helicase DDX21 to enhance its ability to resolve R-loops. The cooperation of these two nucleolar proteins maintains genome instability and cellular homeostasis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Xiajuan Zou for assistance in the mass spectrometric analysis. We thank Dr. Bo Zhang for providing us with the anti-NAT10 antibody.
Abbreviations
- Act. D
Actinomycin D
- DDX21
DExD-box helicase 21
- DRIP
DNA/RNA immunoprecipitation
- GST
Glutathione-S-transferase
- NAT10
N-acetyltransferase 10
- Pol I
RNA polymerase I
- Pol II
RNA polymerase II
- rDNA
Ribosomal DNA
- rRNA
Ribosomal RNA
- RHD
RecD helicase domain
- RNase H1
Ribonuclease H1
- γH2AX
Gamma H2AX, phosphorylation on serine 139 of histone H2AX
Author contributions
K.S. designed and performed experiments, analyzed the data and wrote the manuscript. Z.Z. helped with many experiments including Northern blot, Western blot and immunofluorescent staining. Y.W. helped with construction of NAT10 knockdown cell lines. S.S. provided much experiment help. X.L. provided purified Flag-NAT10-His. C.Z. helped with cell culture and purchased reagents. Y.J. edited the English in this manuscript, helped with the design of the study and helped with experiments. X.D. designed and acquired the funding for the study, analyzed the data and revised the manuscript.
Funding
This work is supported by grant from the National Natural Science Foundation of China (No. 82372631 and 82173024), and grant from the Beijing Natural Science Foundation (Grant No. 7212061).
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yang Jiang, Email: 1410305107@bjmu.edu.cn.
Xiaojuan Du, Email: duxiaojuan100@bjmu.edu.cn.
References
- 1.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. [DOI] [PubMed] [Google Scholar]
- 2.Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X, Chedin F. Prevalent, dynamic, and conserved R-Loop structures associate with specific epigenomic signatures in mammals. Mol Cell. 2016;63:167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malig M, Hartono SR, Giafaglione JM, Sanz LA, Chedin F. Ultra-deep Coverage single-molecule R-loop footprinting reveals principles of R-loop formation. J Mol Biol. 2020;432:2271–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crossley MP, Bocek MJ, Hamperl S, Swigut T, Cimprich KA. qDRIP: a method to quantitatively assess RNA-DNA hybrid formation genome-wide. Nucleic Acids Res 2020, 48. [DOI] [PMC free article] [PubMed]
- 5.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-replication conflict orientation modulates R-Loop levels and activates distinct DNA damage responses. Cell. 2017;170:774–e786719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Rubio M, Aguilera P, Lafuente-Barquero J, Ruiz JF, Simon MN, Geli V, Rondon AG, Aguilera A. Yra1-bound RNA-DNA hybrids cause orientation-independent transcription-replication collisions and telomere instability. Genes Dev. 2018;32:965–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim YW, Sanz LA, Xu XQ, Hartono SR, Chédin F. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutieres syndrome. Elife 2015, 4. [DOI] [PMC free article] [PubMed]
- 9.Lam FC, Kong YW, Yaffe MB. Inducing DNA damage through R-loops to kill cancer cells. Mol Cell Oncol 2021, 8. [DOI] [PMC free article] [PubMed]
- 10.Park K, Ryoo J, Jeong H, Kim M, Lee S, Hwang SY, Ahn J, Kim D, Moon HC, Baek D et al. Aicardi-Goutieres syndrome-associated gene SAMHD1 preserves genome integrity by preventing R-loop formation at transcription-replication conflict regions. PLoS Genet 2021, 17. [DOI] [PMC free article] [PubMed]
- 11.Petermann E, Lan L, Zou L. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat Rev Mol Cell Biol. 2022;23:521–40. [DOI] [PubMed] [Google Scholar]
- 12.Ginno PA, Lim YW, Lott PL, Korf I, Chédin F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Financsek I, Mizumoto K, Mishima Y, Muramatsu M. Human Ribosomal-Rna Gene - Nucleotide-Sequence of the Transcription Initiation Region and Comparison of 3 mammalian genes. Proc Natl Acad Sci United States America-Biological Sci. 1982;79:3092–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30:1327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadel J, Athanasiadou R, Lemetre C, Wijetunga NA, Sato POB, Zhang H, Jeddeloh Z, Montagna J, Golden C. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin. 2015;8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Hage A, French SL, Beyer AL, Tollervey D. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komseli ES, Pateras IS, Krejsgaard T, Stawiski K, Rizou SV, Polyzos A, Roumelioti FM, Chiourea M, Mourkioti I, Paparouna E, et al. A prototypical non-malignant epithelial model to study genome dynamics and concurrently monitor micro-RNAs and proteins in situ during oncogene-induced senescence. BMC Genomics. 2018;19:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanotti S, Vanhauwaert S, Van Neste C, Olexiouk V, Van Laere J, Verschuuren M, Van der Meulen J, Mus LM, Durinck K, Tilleman L, et al. MYCN-induced nucleolar stress drives an early senescence-like transcriptional program in hTERT-immortalized RPE cells. Sci Rep. 2021;11:14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warmerdam DO, van den Berg J, Medema RH. Breaks in the 45S rDNA lead to recombination-mediated loss of repeats. Cell Rep. 2016;14:2519–27. [DOI] [PubMed] [Google Scholar]
- 20.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen W, Sun H, De Hoyos CL, Bailey JK, Liang XH, Crooke ST. Dynamic nucleoplasmic and nucleolar localization of mammalian RNase H1 in response to RNAP I transcriptional R-loops. Nucleic Acids Res. 2017;45:10672–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song C, Hotz-Wagenblatt A, Voit R, Grummt I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017;31:1370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasanova Z, Klapstova V, Porrua O, Stefl R, Sebesta M. Human senataxin is a bona fide R-loop resolving enzyme and transcription termination factor. Nucleic Acids Res. 2023;51:2818–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchena-Cruz E, Camino LP, Bhandari J, Silva S, Marqueta-Gracia JJ, Amdeen SA, Guillen-Mendoza C, Garcia-Rubio ML, Calderon-Montano JM, Xue X, et al. DDX47, MeCP2, and other functionally heterogeneous factors protect cells from harmful R loops. Cell Rep. 2023;42:112148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–63. [DOI] [PubMed] [Google Scholar]
- 26.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong R, Zhang L, Hu L, Peng Q, Han W, Du X, Ke Y. hALP, a novel transcriptional U three protein (t-UTP), activates RNA polymerase I transcription by binding and acetylating the upstream binding factor (UBF). J Biol Chem. 2011;286:7139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, Suzuki T. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem. 2014;289:35724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai G, Hashizume T, Miyazawa T, McCloskey JA, Yokoyama S. Conformational characteristics of 4-acetylcytidine found in tRNA. Nucleic Acids Symp Ser 1989:61–2. [PubMed]
- 30.Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, et al. Acetylation of Cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1872–e18861824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng J, Tan Y, Liu X, Zhang C, Su K, Jiang Y, Luo J, Li L, Du X. NAT10 regulates mitotic cell fate by acetylating Eg5 to control bipolar spindle assembly and chromosome segregation. Cell Death Differ. 2022;29:846–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, Deng H, Luo J, Ke Y, Du X. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17:349–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Li DQ. MORC2 regulates DNA damage response through a PARP1-dependent pathway. Nucleic Acids Res. 2019;47:8502–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Cai S, Zhang C, Liu Z, Luo J, Xing B, Du X. Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Res. 2018;46:9601–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chimnaronk S, Suzuki T, Manita T, Ikeuchi Y, Yao M, Suzuki T, Tanaka I. RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. EMBO J. 2009;28:1362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Su K, Wang C, Deng T, Liu X, Sun S, Jiang Y, Zhang C, Xing B, Du X. Chemotherapy-induced acetylation of ACLY by NAT10 promotes its nuclear accumulation and acetyl-CoA production to drive chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2024;15:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beghe C, Gromak N. R-Loop immunoprecipitation: a method to detect R-Loop interacting factors. Methods Mol Biol. 2022;2528:215–37. [DOI] [PubMed] [Google Scholar]
- 38.Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA. RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89:123–30. [DOI] [PubMed] [Google Scholar]
- 39.Chedin F, Hartono SR, Sanz LA, Vanoosthuyse V. Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J. 2021;40:e106394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smolka JA, Sanz LA, Hartono SR, Chedin F. Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J Cell Biol 2021, 220. [DOI] [PMC free article] [PubMed]
- 41.Cerritelli SM, Sakhuja K, Crouch RJ. RNase H1, the Gold Standard for R-Loop detection. Methods Mol Biol. 2022;2528:91–114. [DOI] [PubMed] [Google Scholar]
- 42.Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol. 1970;76:127–39. [DOI] [PubMed] [Google Scholar]
- 43.Yuan W, Al-Hadid Q, Wang Z, Shen L, Cho H, Wu X, Yang Y. TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. Nucleic Acids Res. 2021;49:8573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boleslavska B, Oravetzova A, Shukla K, Nascakova Z, Ibini ON, Hasanova Z, Andrs M, Kanagaraj R, Dobrovolna J, Janscak P. DDX17 helicase promotes resolution of R-loop-mediated transcription-replication conflicts in human cells. Nucleic Acids Res. 2022;50:12274–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai SY, Liu XF, Zhang CF, Xing BC, Du XJ. Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem Biophys Res Commun. 2017;483:624–9. [DOI] [PubMed] [Google Scholar]
- 47.Cargill M, Venkataraman R, Lee S. DEAD-Box RNA helicases and Genome Stability. Genes (Basel) 2021, 12. [DOI] [PMC free article] [PubMed]
- 48.Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A. 1972;69:3394–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrimore J, Kolbin D, Stanton J, Khan M, de Larminat SC, Lawrimore C, Yeh E, Bloom K. The rDNA is biomolecular condensate formed by polymer-polymer phase separation and is sequestered in the nucleolus by transcription and R-loops. Nucleic Acids Res. 2021;49:4586–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abraham KJ, Khosraviani N, Chan JNY, Gorthi A, Samman A, Zhao DRY, Wang ML, Bokros M, Vidya E, Ostrowski LA, et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature. 2020;585:298–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korsholm LM, Gal Z, Nieto B, Quevedo O, Boukoura S, Lund CC, Larsen DH. Recent advances in the nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2020;48:9449–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harding SM, Boiarsky JA, Greenberg RA. ATM Dependent silencing links nucleolar chromatin reorganization to DNA damage recognition. Cell Rep. 2015;13:251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Sluis M, McStay B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015;29:1151–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–93. [DOI] [PubMed] [Google Scholar]
- 56.Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006;34:1393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Bernardo KD, Vickers TA, Tian J, Liang XH, Crooke ST. NAT10 and DDX21 proteins interact with RNase H1 and affect the performance of Phosphorothioate oligonucleotides. Nucleic Acid Ther. 2022;32:280–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Baysac KC, Yee LF, Saporita AJ, Weber JD. Elevated DDX21 regulates c-Jun activity and rRNA processing in human breast cancers. Breast Cancer Res. 2014;16:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcaida MJ, Kauzlaric A, Duperrex A, Sulzle J, Moncrieffe MC, Adebajo D, Manley S, Trono D. Dal Peraro M: The Human RNA Helicase DDX21 Presents a Dimerization Interface Necessary for Helicase Activity. iScience 2020, 23:101811. [DOI] [PMC free article] [PubMed]
- 60.Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015;518:249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang H, Lachtara EM, Ran X, Hopkins J, Patel PS, Zhu X, Xiao Y, Phoon L, Gao B, Zou L, et al. The RNA m5C modification in R-loops as an off switch of Alt-NHEJ. Nat Commun. 2023;14:6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen L, Zhang C, Ma W, Huang J, Zhao Y, Liu H. METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability. Nucleic Acids Res. 2022;50:11619–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abakir A, Giles TC, Cristini A, Foster JM, Dai N, Starczak M, Rubio-Roldan A, Li M, Eleftheriou M, Crutchley J, et al. N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat Genet. 2020;52:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang LH, Zhang XY, Hu T, Chen XY, Li JJ, Raida M, Sun N, Luo Y, Gao X. The SUMOylated METTL8 induces R-loop and tumorigenesis via m3C. iScience. 2020;23:100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warmerdam DO, Wolthuis RMF. Keeping ribosomal DNA intact: a repeating challenge. Chromosome Res. 2019;27:57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Payea MJ, Anerillas C, Tharakan R, Gorospe M. Translational control during Cellular Senescence. Mol Cell Biol 2021, 41. [DOI] [PMC free article] [PubMed]
- 67.Negi SS, Brown P. Transient rRNA synthesis inhibition with CX-5461 is sufficient to elicit growth arrest and cell death in acute lymphoblastic leukemia cells. Oncotarget. 2015;6:34846–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu HY, Liu YY, Yang F, Zhang L, Zhang FL, Hu X, Shao ZM, Li DQ. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48:3638–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Jing Y, Wang Y, Tang J, Zhu X, Jin WL, Wang Y, Yuan W, Li X, Li X. NAT10 promotes gastric cancer metastasis via N4-acetylated COL5A1. Signal Transduct Target Ther. 2021;6:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie R, Cheng L, Huang M, Huang L, Chen Z, Zhang Q, Li H, Lu J, Wang H, Zhou Q, et al. NAT10 drives cisplatin chemoresistance by enhancing ac4C-Associated DNA repair in bladder Cancer. Cancer Res. 2023;83:1666–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information.