Abstract
In a series of papers, we have provided evidence that during its assembly vaccinia virus is enveloped by a membrane cisterna that originates from a specialized, virally modified, smooth-membraned domain of the endoplasmic reticulum (ER). Recently, however, Hollinshead et al. (M. Hollinshead, A. Vanderplasschen, G. I. Smith, and D. J. Vaux, J. Virol. 73:1503–1517, 1999) argued against this hypothesis, based on their interpretations of thin-sectioned material. The present article is the first in a series of papers that describe a comprehensive electron microscopy (EM) analysis of the vaccinia Intracellular Mature Virus (IMV) and the process of its assembly in HeLa cells. In this first study, we analyzed the IMV by on-grid staining, cryo-scanning EM (SEM), and cryo-transmission EM. We focused on the structure of the IMV particle, both after isolation and in the context of viral entry. For the latter, we used high-resolution cryo-SEM combined with cryofixation, as well as a novel approach we developed for investigating vaccinia IMV bound to plasma membrane fragments adsorbed onto EM grids. Our analysis revealed that the IMV is made up of interconnected cisternal and tubular domains that fold upon themselves via a complex topology that includes an S-shaped fold. The viral tubules appear to be eviscerated from the particle during viral infection. Since the structure of the IMV is the result of a complex assembly process, we also provide a working model to explain how a specialized smooth-ER domain can be modulated to form the IMV. We also present theoretical arguments for why it is highly unlikely that the IMV is surrounded by only a single membrane.
Vaccinia virus, the best-characterized member of the family Poxviridae, has a complicated life cycle that includes an elaborate rearrangement of viral membranes to assemble the first of the two infectious forms of the virus, the intracellular mature virus (IMV) (10, 47). Although this process is exceedingly complex, the IMV is usually simplified into a brick-shaped structure with an outer membrane(s) enclosing an inner membrane-like palisade layer that encloses the viral DNA (33). The precursor of the second infectious form, the extracellular enveloped virus (EEV), is initiated by wrapping of the IMV by trans-Golgi network (TGN)-derived viral membranes (45).
The assembly of poxviruses such as vaccinia virus has never ceased to be a controversial topic. Dales has long championed the view that the IMV acquires its membrane(s) via an unprecedented mechanism of de novo viral membrane assembly (10). Our group, in contrast, has consistently argued that the vaccinia virus membranes are derived from the cellular endoplasmic reticulum (ER) and, as a consequence, the viral membranes are necessarily cisternal rather than a single bilayer (29, 43, 47). Recently, controversy has again arisen with a study by Hollinshead et al. (24). These authors return once more to a simple model in which the IMV is surrounded by a single-membrane bilayer. They leave open the difficult question of how a single-membrane bilayer could surround a cytoplasmic virus (see below).
At stages later than 3 to 4 h after infection, one sees the appearance of the large DNA- and protein-rich “factories,” which collectively are similar to the nucleus in size (4). It is from these factory regions that the first membrane structures, the viral crescents, become apparent at 5 to 6 h postinfection (7, 11, 32) (see Fig. 1A in the accompanying paper [20a]). In thin sections observed by electron microscopy (EM), these structures appear to contain one unit membrane, often, but not always, with apparent free ends (or, more literally, what has been interpreted as free ends). Occasionally, two membranes can be directly seen (47), an observation first made by Patrizi and Middlecamp (36). These membranes become more prominent when infected cells are permeabilized in the presence of protease (47). Within the crescent population, a gradation is seen from short, curved elements to more-mature, cup-shaped spheres that in profile can often appear as perfect circles (see Fig. 1A in accompanying paper [20a]). These structures, which are remarkably homogeneous in size, almost spherical, and 280 nm in diameter, are referred to as the immature virus (IV). On the outer aspect of the unit membrane of the IV, a series of ∼6-nm-long spikes, or spicules, are seen that have been described many times (10).
FIG. 1.
Hypothetical models of viral membrane assembly. (A and B) Two possible models for assembly of a single-membraned virus; spicules are shown as a coat. Model A is used by viruses such as rotavirus and bunyaviruses, whereas budding out of a luminal compartment (model B) has not been described for a virus. Model C shows how cisternal assembly (a model which we favor for vaccinia virus) allows the virus to form an entity that eventually “buds” into the cytoplasm. Note that both sides of the crescent expose the cytoplasmic domain to the cytoplasm. L, lumen; C, cytoplasm.
In infected cells, the viral DNA has been directly visualized in different stages of entry into late-stage IV (10, 31). This step, in which the spherical IV undergoes drastic morphological changes, is perhaps the least understood in the assembly process. The DNA is electron dense, shows a distinct periodic spacing (31), and has been visualized as discrete structures attached to the rough ER (15). This process of DNA entry culminates in the assembly of the quasi-brick-shaped IMV particle. In some thin-section profiles this virus always shows two distinct unit membranes (see Fig. 1B in the accompanying paper [20a]). The innermost membrane profile has often been referred to as a protein layer, although it is ultrastructurally indistinguishable from the outer membrane of the IMV and from membrane profiles in general. Moreover, at times, Dales and Kajioka (8) and others quite reasonably referred to this layer as the inner or core membrane. In between the outer and inner membrane profiles is a periodic, presumably linker structure that can be seen by many EM techniques and has been referred to as the palisade layer (10, 34). It is predominantly composed of the core surface protein P39 (A4L) (6). The above description could be considered the textbook description of vaccinia IMV assembly (10, 33).
The controversy started when attempts were made to explain how an apparent single membrane could envelope viral DNA and viral core precursors while allowing the elaboration of the complex, underlying viral core, which acquires an envelope that is indistinguishable from the outer membrane by EM. Moreover, those who favored the single-membrane hypothesis had to explain how a single membrane could engulf viral DNA that is unquestionably synthesized in the cytoplasm (see Fig. 1). Dales and Mosbach (9) proposed the radical notion that this process represented the only known example (then and now) of the assembly of a biological membrane de novo from lipid and protein precursors. We do not take this de novo membrane model seriously since we believe speculations based on examination of plastic sections of vaccinia virus-infected cells are not convincing enough to overturn a fundamental dogma in cell biology, that membranes are always made from preexisting membranes (35). Moreover, we are also far from convinced by claims that lipid analyses have shown any support for this model because the IV has never been isolated in a pure form. We argue that the question of how a membrane was first assembled must be addressed at the level of the evolutionary ancestor of all cells.
We have assumed from the outset that vaccinia virus assembly requires membranes that are derived from cellular membranes, as is the case for all other known membrane viruses. One important trick of such membrane viruses is to allow the cellular machinery to target viral membrane proteins selectively to the specific membrane domain where assembly occurs, most commonly at the plasma membrane (19). Assume, for the sake of argument, that the crescent indeed has only one membrane and that, in the simple model, the IMV is completely surrounded by this crescent-derived membrane (10, 24). If the viral membrane were derived from cellular membranes, insurmountable conceptual barriers to the assembly process would ensue. The first problem is the fact that all biological membranes are always in continuity with themselves, forming continuous, closed systems with no free ends (which would be highly unstable in membrane sheets) (35). As discussed in more detail below, this poses serious problems for the model of a single vaccinia virus membrane. Our idea that the crescent represents a tightly apposed membrane cisterna derived from the smooth ER is much easier to fit into existing cell biological concepts (47). Even Hollinshead et al. (24) conceded that it is likely that viral membranes are acquired from cellular membranes.
When we first started these studies over a decade ago, we considered the possibility that the crescent membranes are indeed single bilayers which would, in that case, be continuous with themselves via tortuous, elusive connections that are perhaps difficult to preserve. If this were true, however, one would be faced with two different scenarios. In scenario A of Fig. 1, a single-membraned virus can form by budding into the lumen of a cellular compartment. In this case, the particle with one membrane is separated from the cytoplasm by both luminal space and the membrane of the compartment. Such a model would be inconsistent with vaccinia IV formation since both sides (i.e., outer and inner surfaces) of the crescent and the IV are completely accessible to gold particles that enter the cytoplasm of permeabilized cells (47), and core proteins are undoubtedly made in the cytoplasm. In scenario B in Fig. 1, the IV buds out of a membrane compartment, in a fashion analogous to the budding of vesicles into the cytoplasmic space, such as clathrin-coated vesicles. This model was also ruled out by the experiments of Sodeik et al. (47) showing that the central region of the IV is a cytoplasmic compartment. Moreover, a cytoplasmically synthesized scaffold protein (P65-D13L, the target of rifampin), as well as core proteins, is on the inner aspect of the crescent (48). Only the cisternal wrapping model shown in Fig. 1C is conceptually compatible with all of our data.
In this, as well as the next series of papers, we provide further support for our model of cisternal wrapping and show that both the assembly and the final structure of vaccinia virus are much more elaborate than previously appreciated. Collectively, our data essentially lay to rest the one-membrane hypothesis as conceived by Hollinshead and colleagues. In this first paper, we focus on the structure of highly purified IMV particles, using many complementary EM techniques.
For a number of experiments, we took advantage of the reducing agent dithiothreitol (DTT) to reduce vaccinia virus disulfide bonds and consequently open up or loosen the tightly packaged IMV (29, 43). Although this treatment has significant effects on the IMV structure, surprisingly, it has no effect on the infectivity of the particle (29). We also used well-defined membrane markers to facilitate the identification of membrane structures. We consider abundant labeling by well-characterized antibodies against viral membrane proteins to be strong evidence for the existence of viral membranes. Finally, the structural studies described here compound the complexity of the IMV by showing that the IMV undergoes a dramatic structural reorganization when the core enters cells. Using cryo-scanning EM (SEM) and a new EM approach we have developed, we took advantage of this entry process to elucidate more details of the structure of the virus as it enters cells. We think that the treatment with DTT simulates many aspects of this process.
We think it will be easier for most readers to follow our subsequent arguments in this paper and those to follow if we first present our working hypothesis (Fig. 2). We argue that the crescent forms (mostly) at the ends of cisternae with the tight zippering of luminal domains. The remainder of this cisterna is continuous with the smooth and rough ER, and a significant fraction of this membrane tucks inside the volume of the IV, a point discussed in more detail in the second paper in this series (20a). We will argue that these loose infoldings of membranes into the IV represent the precursors of distinct domains in the crescent and IVs that will subsequently assemble into the core membrane (cisterna). We therefore explicitly specify that cisternal membrane domains directly connect the core to the outer layer of the IMV. An underlying scaffold enriched in the rifampin-sensitive protein P65 (D13L) has been proposed to facilitate the self-assembly of the IV. The central part of the IV encloses all investigated core proteins (reference 15 and unpublished data). P65 is degraded concomitant with or just prior to IMV closure to make a fully assembled particle (26).
FIG. 2.
Proposed assembly model. For details, see the text. The numbers 1 and 2 indicate the two distinct cytoplasmic compartments. The DNA is shown as a dense structure. In the upper figure, the cisternal organization is drawn, as is the attachment to the rough ER; in the middle and lower figures, the cisterna is drawn as a line.
In parallel, we argue that the DNA associates with ER membranes (15) and subsequently matures into an apparently regular, roughly brick-shaped structure that probably represents one unit of the genome. This ∼60-μm-long, double-stranded DNA of one virion is aligned into long parallel fibrils (16, 31, 32). Electron diffraction of EM images of these structures reveals a 2.5-nm repeat (V. Guenebaut, K. Leonard, and G. Griffiths, unpublished observations; see also references 28 and 31). This value strongly suggests that the DNA is free, rather than bound to protein, and that perhaps it is arranged as a liquid crystal as it enters the IV. We argue that the DNA is partially bound to these ER membranes as it enters the IMV. The final model proposes a tubular cisternal structure that packages the DNA by enveloping it via complex, highly interwoven folds. A unique consequence of this model is that the IMV contains two distinct cytoplasmic compartments that are physically separated (Fig. 2).
In our model, the inner-membrane (core) cisternal domain is always in direct continuity with the crescent cisterna, as mentioned above. However, we also argue that this connection between the core membrane cisterna and the outer cisterna must be broken when the core enters the cell during viral infection, using a (proposed) nonfusion mechanism (see below). In agreement with this view, the most abundant DNA-binding protein, P11 (F17R), remains outside the cell with the remains of the viral outer membranes when the core enters cells. This protein localizes to the IMV subcompartment that does not contain the DNA (37) (Fig. 2). In contrast, the second, abundant DNA-binding protein P25 (L4R) colocalizes with the DNA (compartment 1 in Fig. 2) and enters the cell as part of the core during the infection process (37). While the details of this model will be elaborated from thin-section analyses in the second paper (20a), we focus here on the use of different techniques to analyze intact or DTT-disrupted particles, as well as particles in the process of entering cells.
A detailed analysis of all the images we have examined using all the different approaches has led us to conclude that the virus is a highly asymmetric structure, although when viewed from the most commonly seen perspective it is roughly brick shaped with rounded corners. However, this apparent simplicity is highly misleading because all of these corners are differently organized, as are right from left, front from back, and top from bottom. This will be our terminology for describing the IMV: we arbitrarily define the top of the IMV, and where possible we have tried to align the particles such that the front of the “brick” faces up on the page, the back faces down, and (when viewed from above) the left and right lobes are shown accordingly. Now, if this particle is simply turned over 180° (exposing the bottom upward), what is defined as left is now seen on the right side of the particle, whereas front and back keep their original orientations. We have attempted to define the different faces of the virus in the EM images. Where possible, the top, bottom, front, back, right side, and left side are indicated on all micrographs. We emphasise that our attempts to identify these different faces of the virus are not always clear-cut, due to its complexity, so our assignments should be considered only as guides. It should be noted that in on-grid stained and dried preparations, the IMV particles collapse to different extents, depending greatly on the orientation of the particle that is facing the viewer. Because of the complexity of the particle, we cannot be sure whether all particles assemble identically or whether, for example, left and right forms exist.
MATERIALS AND METHODS
Cells, virus, and antibodies.
HeLa cells were grown as described previously (47). Vaccinia virus strain WR was grown in HeLa cells and semipurified as described by Pedersen et al. (37). Antibodies to P16 (A14L) and P21 (A17L) have been described before (37, 44).
Grid-tap method.
HeLa cells were grown on small pieces of glass coverslips. Purified vaccinia virus was added to these cells in serum-free medium at a density of 100 PFU/cell. After 15 or 20 min, the coverglass pieces were held by forceps with one hand while a 400-mesh grid (Formvar and carbon coated) was held by a second forceps. The grid was touched to the surface of the cells for 1s and then separated. Following two brief rinses in distilled water (a few seconds each), the grids were subjected to the Tokuyasu procedure (see below). A variation of this method, used to obtain the data shown in Fig. 10, involved sandwiching ∼1 μl of virus between two grids. After 5 min, the grids were separated and both were stained with uranyl acetate-methyl cellulose (see below). At least 100 images were prepared by this method in four different experiments.
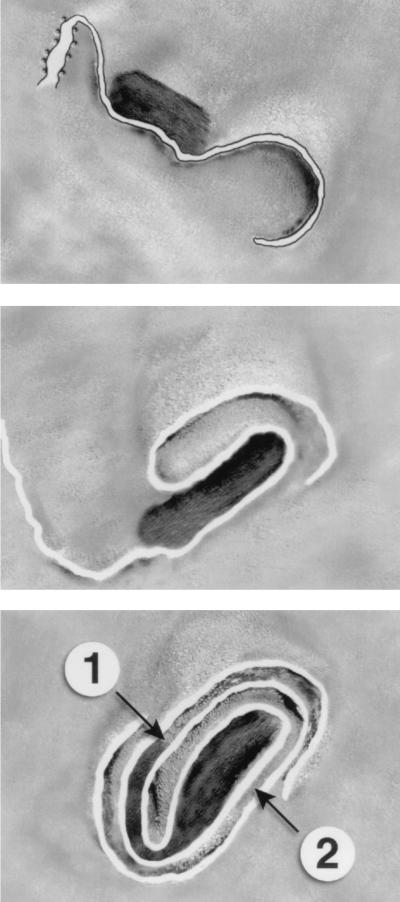
FIG. 10.
IMV particles adsorbed to two grids and mechanically separated. This approach results in opening up of some particles. (A) Bottom (B) view of a particle that has two displaced membrane domains (star), one of which is attached to the particle by a rigid membrane tubule (double arrow). The large arrowheads indicate two of the ∼30-nm-diameter tubules that are connected to different parts of the IMV. The small arrow shows a bulbous tip on the upper tubule that also appears kinked, presumably effecting tight curvature at this site. f, front. (B) A single tubule is seen to emanate from the right (R) side of the particle seen in the top view. Again, the tip of this tubule is bulbous (arrow). (C and D) The same particle, tilted 50° relative to each other. The arrow indicates the bulbous tip of a tubule on one side that has extended from the IMV. The top core (c) domain is identifiable by its roughly rectangular shape (with one more regular 90° corner [cf. FIG. 9A]). This is highly interconnected with membrane tubules. (E and F) Two tilted (25°) views of a particle in which a large membrane fold (star) has been pulled away from the IMV, revealing the inside of the core. The still-intact connection between the two structures is indicated by the arrow. The arrowhead shows membrane folds with the core. We suggest that this image resembles the process by which the core enters cells; the side of the arrow would then be separated between the intracellular core and the extracellular cisternal remnants. (G) What we interpret as the lower part of the IMV that has been pulled out of an IMV (cf. FIG. 6E). Two tubules are evident, one with the characteristic bulbous top (arrow) and the other with a curved end (arrowhead). Bars, 50 nm.
EM preparation methods for intact particles. i. Tokuyasu's method.
Formvar- and carbon-coated 400-mesh grids were glow discharged and incubated for 5 min on a drop of IMV suspension. After brief (a few seconds) rinses in H2O, the section was embedded, using a ∼3- to 3.5-mm-diameter loop, with a mixture of 8 parts methyl cellulose (25 centipoise; 2%) and 2 parts uranyl acetate (3%). The grids were looped out of the solution, and excess liquid was removed by contact with filter paper. The methyl cellulose solution was made by mixing the powder with cold water and stirring for at least 24 h in a cold (4°C) room. The solution was then centrifuged at 100,000 × g for 1 h at 4°C and then left undisturbed in the refrigerator. The solution was changed every 2 months. This method reduces the extent of air-drying artifacts compared to conventional negative staining and gives a range of positive and negative contrast (see references 18, 21, and 49). Many hundreds of images have been prepared using this approach.
ii. Conventional negative staining.
Negative staining was carried out as described above except that the grid was incubated with 2% uranyl acetate or 2% ammonium molybdate in H2O for 20 s and air dried after being blotted briefly with filter paper (22). Since conventional negative stains are acidic (e.g., uranyl acetate solutions are pH 4) and this low pH may induce membrane artifacts, we also used neutral-pH uranyl acetate-oxalate for some experiments. This solution was prepared as follows: two stock solutions, 2% uranyl acetate and 24 mM oxalic acid, were mixed at a 1:1 ratio, and the pH was adjusted by addition of 25% ammonia to a pH of ∼6.8. To avoid precipitation, the solution was kept on ice and used within 1 h after preparation. Over 200 images were obtained by these different approaches.
iii. Specimen preparation for cryo-EM.
The thin-film vitrification method of Adrian et al. (1) has been described in detail by Roos and Morgan (42) and Dubochet et al. (14). A 3- to 5-μl droplet of suspension was placed on a grid supporting a perforated carbon film with hole diameters of between 2 and 5 μm. The drop was left on the grid for 1 to 3 min in the case of IMV and for up to 10 min in the case of EEV. Blotting paper (Whatman no. 1) was then firmly applied to one side of the grid for about 1 s. The plunger holding the grid was immediately released, and within about 0.1 s the grid was plunged into a liquid-ethane slush cooled with liquid nitrogen, thereby vitrifying the thin liquid film. The vitrified samples were either stored in liquid nitrogen or observed immediately. Transfer and observations were performed with side-entry, cryo-transfer holders (Gatan [Gatan, Varrendale, Pa.] 626 and Oxford Instruments (Oxford, United Kingdom] CT-3500) in a Jeol-2000EX cryo-electron microscope. Observations were made under minimum-irradiation conditions at magnifications ranging from 3,000× to 40,000× and at 3.5 to 10.5 μm underfocus; low-dose micrographs were recorded on Kodak SO-163 electron microscopy film and developed in full-strength D-19 developer (Kodak) for 12 min (speed, ca. 2.2 μm2/electron × optical density unit). Magnification was calibrated with a cross-grating replica, with an estimated error of 2%. In the course of this study, we collected many hundreds of images by this approach.
iv. Cryo-SEM.
Specimens were prepared for cryo-SEM as described by Krijnse Locker et al. (30) and Ritter et al. (41). For more details on the background to this approach, see reference 23. Over 200 images were obtained from at least five different experiments.
v. Tomography.
A grid of IMV was prepared according to Tokuyasu's method. At a relatively low primary magnification (10,000×), fields of viruses were selected such that about 10 to 20 viruses would be seen on each negative. Using the eucentric tilt stage of the Phillips 400T EM, we then took images every 10° from −57° to +57° (the maximum possible). Over a dozen complete series were prepared, as well as many hundreds of less-extensive tilt series. For more details, see references 3 and 27.
vi. Immunolabeling.
IMVs, or IMVs pretreated for 30 min with freshly prepared DTT (20 mM; 30 min at 37°C), were adsorbed onto grids. In some cases, the DTT was applied after viruses had already adsorbed onto grids. The grids were blocked with 1% fish skin gelatin plus 0.3% bovine serum albumin for 5 min and incubated with anti-P16 (A14L) or anti-P21 (A17L) antibodies (37, 44) for 15 min. After being rinsed in phosphate-buffered saline (PBS) for 10 min, the grids were incubated with protein A-gold (10 nm diameter). After being rinsed in PBS (20 min) and weaked with H2O, the grids were embedded in methyl cellulose as described above. For double labeling, we followed the general sequential regime used for labeling of cryosections (46).
RESULTS
Methyl cellulose-uranyl acetate: untreated particles.
The first EM method we describe in detail for investigating IMV structure is methyl cellulose-uranyl acetate embedding. This method, introduced by Tokuysasu (18, 49) for use with thawed cryosections, can also be used for particle staining and gives a complex mixture of negative contrast and positive staining due to stain absorption. It offers a number of advantages over conventional negative staining because the structures collapse much less upon drying; cryosections generally collapse ∼20% with methyl cellulose versus 70 to 80% in its absence (20). However, very thick layers of negative stain (which are highly beam sensitive) can also help to reduce the collapse phenomenon (49). In Fig. 3 we show an array of our purest, highest-titer preparation of virus prepared by using the methyl cellulose recipe that we developed earlier (21). The overall appearance and dimensions of these 10 particles (all on the same negative) are evident at different angles of tilt, from −57 to +57. These images, which provide remarkable information at higher magnification, are available online (http://www.emblheidelberg.de/wendt.vaccinia.html).
FIG. 3.
Tomographic representation of 10 IMV particles from the same EM negative following preparation using methyl cellulose-uranyl acetate. These images were tilted in a series at −57° to +57°C from the horizontal and in 10 increments. Thus, each particle is revealed as 13 different tilt images. For reference, the particles in rows 1 and 4 (0° tilt) show a “top” view of the IMV, whereas the particles in rows 8 and 10 (0°) show “bottom” views. These images can be downloaded from the website http://www.embl-heidelberg.de/wendt.vaccinia.html.
As one rotates the particle systematically in these images (Fig. 3), the increasing complexity of the other parts of the IMV becomes evident. In these preparations, the reference top domain of the virus usually resembles the flattened head of a snake whose body is coiled into the structure below (see, e.g., column 4, −17° in Fig. 3). When these images are examined at a higher magnification, it becomes evident that this top domain is connected via loops into an extended S-shaped structure (as will become more clear below). A striking feature evident in Fig. 3 is the tortuous coiling and bending of different lobes of the virus; boundary regions between different domains are generally more densely stained. Also, many tubules can be seen to emanate from the lobed regions (see, e.g., column 2, −7°, and column 4, −57). The images at −17° to +20° in column 9 suggest that on the underside of the IMV, three or more lobes come together to form a mouth-like structure, like the petals of a flower, that can close tightly. Overall, these images reveal the IMV to be a highly asymmetric particle that is only ∼110 nm in one of its dimensions (height).
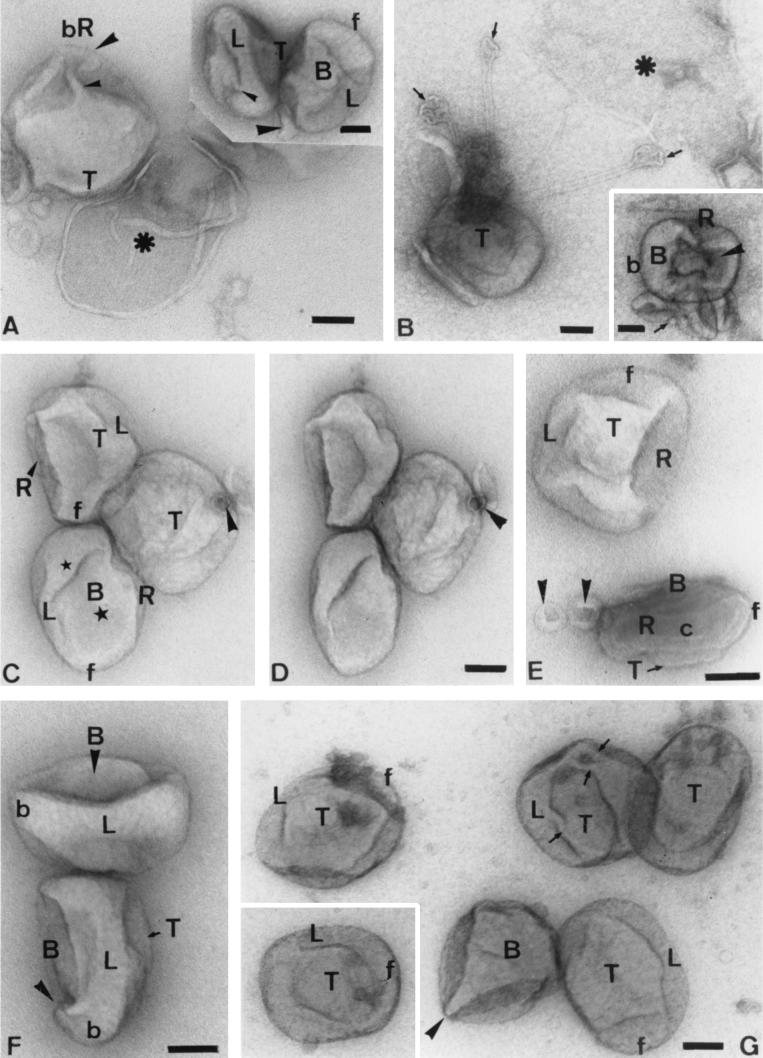
In preparations with a relatively thick layer of methyl cellulose, many particles show no detectable collapse and appear almost completely smooth, especially when viewed from above (Fig. 4A and B). As the film thickness decreases, the particles start to collapse, revealing “creases” (Fig. 4C to F). These creases are zones where two or more different tubulo-cisternal folds impinge on each other. The pattern of these creases usually allows us to identify the top and bottom of the flattened brick-shaped particle (Fig. 4C to F). The top of the particle has a spherical “cap” of folded membrane domains (the “snakehead”) that tends to collapse less than the surrounding parts of the IMV, and consequently, in top views, this cap is usually raised above the remaining parts. This top domain is seen to be connected to the rest of the IMV by tubules. In contrast, the bottom of the particle usually shows longitudinal creases (but depending on orientation) (Fig. 4C and F). Figure 4C shows an IMV conventionally negative stained with neutral-pH uranyl acetate, which we recommend as a general method (if methyl cellulose is to be avoided). Here, two large central domains of membrane can be seen to be surrounded by interconnected tubules. The topological relationship between these tubules is more evident in Fig. 4F, in which the particle has visibly collapsed.
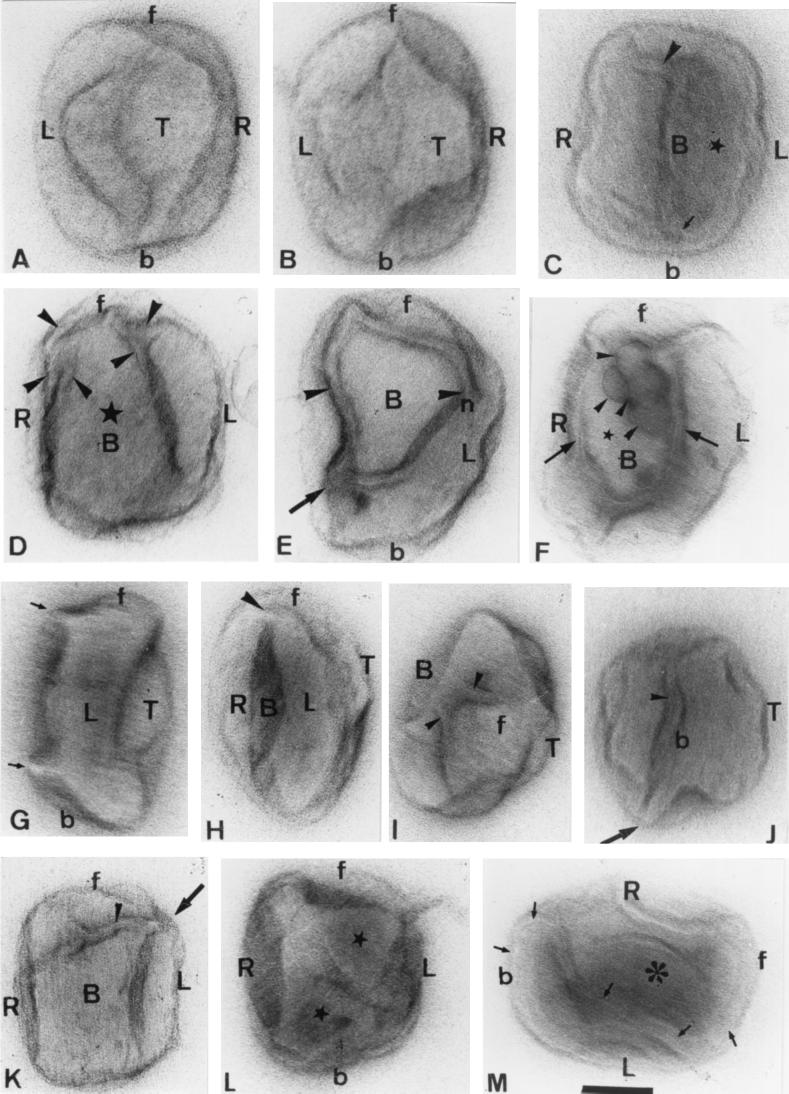
FIG. 4.
On-grid staining of untreated particles with uranyl acetate-methyl cellulose (except panel C, which shows a particle negatively stained with neutral-pH uranyl acetate). In all images, the top (T) domain is arbitrarily selected as the brick- or oval-shaped view that is most commonly seen. Relative to this are indicated the bottom (B) and two side domains (right [R] and left [L]), while f indicates the front and b indicates the back of the particle. When the layer of methyl cellulose is relatively thick, the particle appears almost smooth from top views (A and B). With thinner methyl cellulose layers, the particle starts to collapse and contrast increases. (D) A top view with the characteristic square to oval top fold. The left (L) domain is recognizable by a fold that extends the whole length on one side of the particle whereas the opposite (R) side is interrupted by a crease. (E) The particle is rotated (relative to panel D) such that more of the highly tubular right side is seen. (C and F) Different views of the bottom (B) of the particle. The arrowheads show tubular projections; the latter particle is especially flattened. (G) An image of the left side of the IMV. (H and I) Views of the right side of the IMV, showing a long, thin, branched tubular domain that extends the whole length of the right side of the particle. The complexity of this right side is evidently much greater than that of the left side. The arrow indicates the tip of a highly curved tubule. (J) Left side, with its connection to the top domains. Whereas the top domain is curved and devoid of significant interaction with the stain, the bottom aspect often gives the impression of being cut open and stains heavily. (K and L) Examples of regular particles that are seen in untreated IMV preparations but appear more often after treatment with DTT. These particles appear indistinguishable from some images seen in thin sections (cf. the particle in FIG. 1B of the accompanying paper [20a] indicated by a star) showing two membrane profiles. We believe that this particle has lost its top domain membranes and shows the top of the core (Tc). The rather homogeneous appearance of the core surface is deceptive; in panel L is seen a similar particle that is attached to the grid on one edge (i.e., is tilted relative to the particle in panel K). This shows an apparent connection between a part of the core and the outer membrane (arrowhead). Bar, 50 nm (for all panels).
Figures 4G and J show the left side of the virus. This is an extensive flap of membranes that extends continuously from front to back while connected laterally to other tubular domains on the right side of the particle. These domains are evident in Fig. 4H and I, which reveal the complex right side of the virus. This is the first clear hint of a labyrinth of interconnected membrane tubules (Fig. 4F, H, and I); the same information can be seen in Fig. 3 at a higher magnification. The curved end of one tubule is evident at one corner in Fig. 4H. In Fig. 4I, this tubule is compacted into the particle, which in the normal virus is probably still more closely packed—in effect, sealed. This figure also shows how the top domain is connected at its front and back aspects to tubular structures. Figure 4J shows how in one view the virus can look highly asymmetric, the top domain is rounded, while the bottom looks as if it had been cut with a knife.
Occasionally, particles such as that shown in Fig. 4K are seen even in our purest IMV preparations; these become much more common after DTT treatment. These particles are remarkably regular and are indistinguishable from one profile of an IMV particle shown in a thin section (see Fig. 1B in the accompanying paper [20a]). We believe that this is a top view of the core surface, exposed after artifactual loss of the top membrane domain. Tilting views of this particle reveal that its upper surface is dome shaped, and we suspect that a part of the original crescent sphere is responsible for maintaining this dome-shaped top structure. The deceptiveness of the appearance of symmetry is also evident in a particle that has attached to the grid via a different side (i.e., the image was not manually tilted [Fig. 4L]); here, the core folds are evident, as are inward invaginations of outer membranes. Thin tubular extensions of the core start to become evident in this image. The average dimensions of the virus after the methyl cellulose-uranyl acetate treatment were a length of 309 nm (standard deviation [SD], 31 nm) a width of 237 nm (SD, 27 nm), and a width of 137 nm (SD, 18 nm). These data are in excellent agreement with cryo-EM estimates (see below).
Use of DTT to open up the virus.
The analysis of intact IMV made it clear that a vast complexity lies buried in the interior of the particle. Further progress necessitated a means of loosening up the tight organization of the virus. For this, we used the reducing agent DTT and also took advantage of the dramatic rearrangements in the structure that occur upon viral entry into cells (see below). DTT reduces the viral cytoplasmically disposed disulfide bonds and induces a partial unfolding of the virus (29, 43). Remarkably, this treatment has no effect on viral infectivity (29), which makes us confident that this treatment does not destroy the robust structure of the IMV.
For most studies, IMV was treated in suspension with 20 mM DTT for 30 min at 37°C and then adsorbed onto grids and stained as described above. The same features that we saw with untreated IMV were readily apparent; however, the details usually became more distinct after DTT treatment. Figure 5 shows a gallery of IMV particles that had been titled manually in the electron microscope until interesting detail was revealed. A detailed description of these images is given in the legend to Fig. 5. It becomes even more apparent from this panel that we are facing a topological problem of tremendous complexity if we are to understand the structure of the IMV. We believe that all these views are representative of the real structure, although different particles are probably distorted to different extents by the drying process.
FIG. 5.
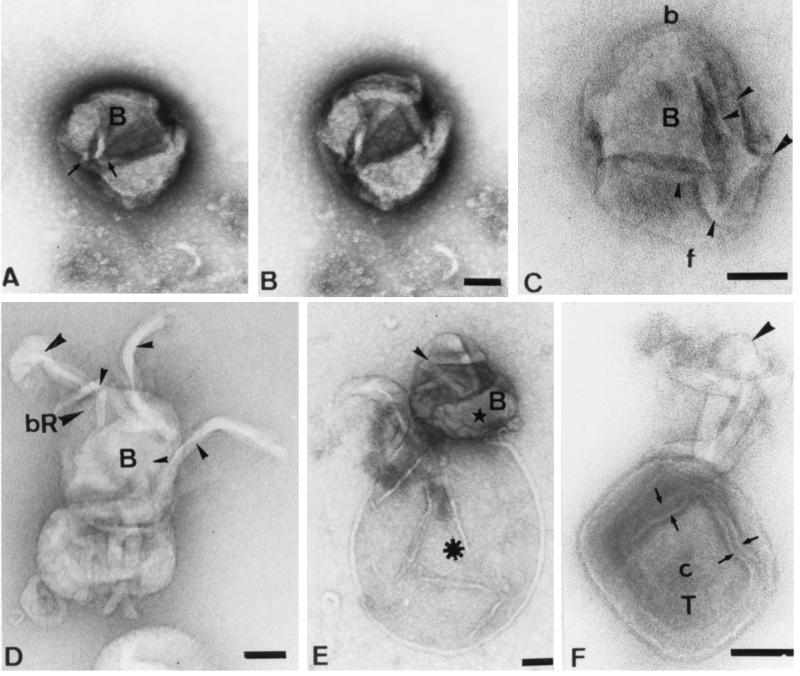
Selected IMV particles treated with DTT. Many of these particles were tilted in the electron microscope until an interesting view was observed. As in FIG. 4, the arbitrarily selected top (T), the bottom (B) right (R) and left (L) sides, and the front (f) and back (b) are indicated. (A and B) Particles aligned similarly but distorted to different extents. (C) Particle viewed from below. In this relatively flattened particle, the core (star) is on the right side of the image (adjacent to the left domain); the arrow shows an inward-curving membrane connection from the outer membranes. A tubular extension is indicated by the arrowhead. (D, E, F, and K) Different views of the bottom of the particle. (D) The large bottom flap (star) is only slightly distorted, revealing the interconnected underlying tubules that show branching (arrowheads). (E) Particle is more collapsed, revealing a highly curved, tubular structure (arrowheads) that coalesces into a protrusion at one site (arrow). (F) We believe that the membrane flap that covered the lower surface of the core either has been lost or has collapsed inward (star), revealing the underlying shape of the core (arrows) and the four spherical structures (arrowheads) that we consider to be highly curved tubules. (G) Side view of the left side of the particle, revealing curved, elbow-like tubular projections (arrows). (H) A Close to en face view of the back end of the IMV. The arrowhead indicates the connection between the left and right lobes. (I and J) Highly tilted, mostly corner images of the IMV. Tubules are indicated by arrowheads, and the arrow in panel J indicates the projecting kink or elbow (cf. panel G). (K) A different view of the bottom of the particle and the interconnected lobes (arrowhead). The arrow shows the convergence of tubules at one corner; the same structure is indicated by the arrows in panel G. (L) Stars show two quasi-triangular lobes of the core in this collapsed bottom-view image of the IMV. (M) Particle viewed from the right side is strongly collapsed, revealing the interior of the core. The arrows trace the infolding of the outer membranes that extend around the core domain (asterisk). Bar, 100 nm (for all panels).
The basic organizational principle that becomes even more evident from these images is that of highly extended and folded membrane cisternae of various shapes and long, tubular extensions. Since the fully assembled IMV is obviously a distinct entity, we believe that the overall organization of the structure is a single labyrinth of interconnected cisternal sheets and tubules. Figures 5A and B show similar but not identical orientations of the “top” of the particle. The particles in Fig. 5C to F and K to and M show the opposite, bottom aspect of the virus. We believe that these images appear different because of often subtle differences in the parts of the virus that adsorbed onto the grid, as well as in how the structure was distorted during drying. As a result of both of these phenomena, the virus can appear quite different from one particle to the next. We believe that one of the lobes that overlies the core in Fig. 5F has been distorted and pushed aside, revealing the underlying core structure, one part of which has four bulbous structures that probably represent highly curved membrane tubules. It seems likely that the high stain density of these structures is due to the bound viral DNA. In relatively rare cross sections through the IMV, these “balls” (which we believe to be highly curved tubules) can also be seen in thin sections (see Fig. 9B in the accompanying paper [20a]). In remarkable images obtained by Peters and Müller (40), these structures were beautifully shown using both negative staining of intact particles and thin sections (see also reference 34).
FIG. 9.
(A to C) Conventional uranyl acetate negative-stained IMV. (A and B) Bottom (B) view. In both of these particles, the back left (L) corner of the core is seen as a 90° bend whereas the other (right [R]) side of the IMV is asymmetrical- f, front; b, back. In panel A, two distinct, stain-filled cavities are seen (arrowheads) (cf. Fig. 2). Since the particles in panels A and B are similarly orientated, it is evident that the front right corner of the particle has been pushed outward in B relative to panel A. The arrow shows one of the bulbous, curved tubules in one of these cavities (cf. FIG. 6F). In panel C the top (T) of the particle has been removed (possibly indicated by the arrowhead), revealing the core. Note that from this view the core (c) appears relatively symmetrical. (D and E) Negative staining of DTT-treated IMV with ammonium molybdate. The arrowhead in panel D indicates a tubular projection with a bulbous tip, while the arrows show two of the four interconnected tubules that lie free on the grid. (E) One intact particle is seen (above) next to a particle that is evidently uncoating; the arrow indicates a tubule, while the arrowhead shows its tip. Bars, 100 nm.
Views of the particles as they enter cells: cryo-SEM.
We next used cryo-SEM to investigate the structure of IMV particles that are involved in the process of viral entry. By this approach we could view the virus and outer surface of plasma membrane projections, but of course we could not be certain that all of the particles shown were really in the process of entry. However, we believe that the images shown can be interpreted in the context of the overall IMV structure that is obtained by the different approaches. An important aspect of the cryo-SEM approach we used is that the specimen, seen in a natural setting, is physically fixed by vitrification, which prevents potential artifacts of chemical fixation; the latter are likely to be more of a problem when the virus uncoats.
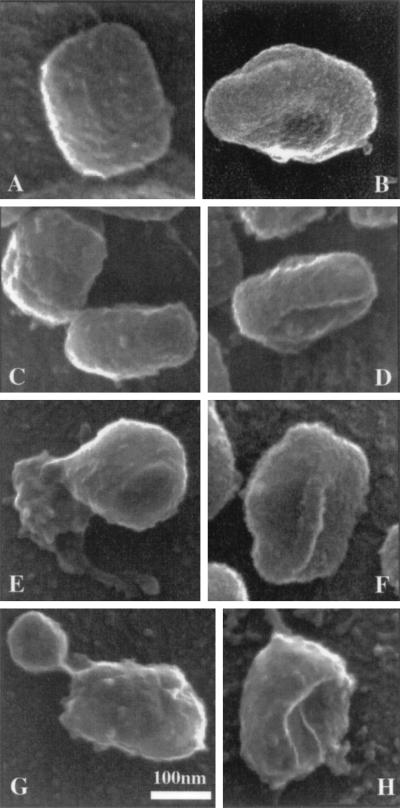
Although the metal coating obscures many details, the overall shape of the particle and its appendages seen by SEM and by on-grid staining look similar (Fig. 6). The image in Fig. 6A shows the classical view of the top of the brick-shaped particle. One corner (top left of this image) of this particle is more indented; the same feature is evident by cryo-transmission EM (13, 43) (see below); from the uranyl acetate-stained preparations, we can identify this corner as the back right of the particle. The overall dimensions of these particles are 299 (± 18) by 237 (± 13) by 106 (+ 1 to 10) nm; the side or height (thickness is evident in Fig. 6B to D; range, 90 to 123 nm). However, it can be appreciated in these images, as with grid staining methods, that estimates of the side or height depend greatly on the angle of tilt and on the part of the structure that is measured. Figure 6F shows the underside of the particle, revealing a protruding ridge along its length.
FIG. 6.
Cryo-SEM images of IMV on the surfaces of HeLA cells after a 5-min infection. Panels A and C (upper particle) show top views of seemingly normal particles. The top left corner of the particle in panel A reveals the slightly indented (back right) corner of the particle. (B to D) Different side views of the IMV. In panel C, the lower particle is quite uniform. When tilted slightly (evident in panel D), the end folding of the left side membrane domain around the particle can be seen. (F) Bottom view in which a long ridge is seen to bisect the particle. (E) An irregularly shaped membrane mass seems to have ejected out of the bottom of the particle; this domain is continuous with two tubular projections, one of which has a bulbous tip. (G) A spherically shaped projection has separated from the particle. The highly curved top domain is indicated. (H) Particle from which the loose internal domains on the underside of the particle have presumably been lost, leaving a cavity. Note that the rest of the particle has kept its normal shape.
We believe that the process of viral uncoating that occurs upon contacting and activating cells (30, 37) can explain the images shown in Fig. 6E, G, and H. Evidently, a large subdomain of the virus has been ejected from the main body of the particle, a significant mass of membrane seems to have been displaced (Fig. 6E). This structure often showed projections: in this image, two possible tubular extensions of ∼300 nm have bulbous ends (cf. Fig. 6E, Fig. 7B, and 8D). Figures 6G and H show the ejection of an oval-shaped lobe. These delicate structural intermediates are seemingly quite sensitive to preparation, and we had the impression that many are artifactually lost during preparation. These images suggest that the entry process leads to a significant unfolding and ejection of the bottom part of the virus. When these domains have been lost from the particle, a cavity is evident in one side of the particle (Fig. 6H). It seems likely that what is left after this unfolding process is predominantly the viral core that during infection can traverse the plasma membrane barrier.
FIG. 7.
Visualization of IMV during cell entry using the grid tap rip-off method and methyl cellulose-uranyl acetate. As for Fig. 4 and 5, the top (T), bottom (B) right (R) and left (L), and front (f) and back (b) are indicated. We emphasize that these viruses are not treated with DTT; the structural changes observed here are related to the entry process. (A) IMV attached to a cellular membrane vesicle (asterisk). From one of the large lobes at the top left side of the IMV projects at least one tubule (small arrowhead). The latter bends abruptly at its point of convergence with other highly curved tubules (large arrowhead). The inset shows views of the side (left particle) and bottom (right particle) aspects of the IMV. The small arrowhead indicates a tubular connection between two larger lobes on the bottom side of the virus. The large arrowhead indicates the curved tubule at the bottom right corner of the virus. (B) Three 30 to 40-nm-diameter tubules that emanate from one corner of an IMV. The ends of these tubules are bulbous or are plug or sucker like (arrows). One of these (on the right) is attached to a cellular, ripped-off cell vesicle (asterisk). The inset in panel B shows a bottom-aspect image of an IMV particle that is evidently eviscerating viral tubules (arrow) that are seen outside the particle, leaving a cavity (arrowhead) in the IMV. At the opposite side of this particle, a stain-filled gap is seen between two lobes; that is an opening into the core interior. (C and D) Stereo pair (16° tilt) view of three IMV particles; the top particle show clearly the planar view of the right side of the IMV. The middle particle shows the same side in contact with the particle above it. The arrowhead indicates a ring-like structure that is probably an en face view of the “sucker” shown in panel B. The lower particle shows a bottom (B) view with continuity between the larger flap-like lobe (large star) and the thinner lamellar (left) domain that curves around one end of the particle (small star). (E) Top (T) view of a particle (upper) in which the left (L) and right (stained) domains are evident. The lower particle shows an en face view of the right (R) side in which the core (c) structure is evident next to the top domains. The arrowheads indicate two curved tubules at one end of the IMV. (F) Two particles in which the distinction between the curved top (T) domain and the flattened and more opened bottom (B) domain is seen. The arrowhead shows one of the kinked corner domains. (G) Major domain organization of the IMV. In one particle (second from right upper row), the extreme flattening of the particle has allowed the top domain to separate from the connected domains, and their topographical relationships can be clearly seen (arrows indicate tubular connections). The arrowhead in this panel indicates an extreme edge-on projection of the virus in which the large triangular domain converges into the thinner tubule at the back right corner projections (cf. FIG. 5G). Bars, 100 nm.
FIG. 8.
More examples of the “rip-off” method for visualizing IMV entry intermediates. (A and B) Stereo pair (15° tilt) showing the impression of twofold symmetry on the bottom (B) of the virus, as well as the interconnectivities between the larger viral lobes and the tubules (arrows). (C) Convergence of many tubules (small arrowheads) into a kink on the right side is indicated by the large arrowhead. Up to four of these tubules can be seen extending from this site. (D) Unfolding of three tubules at the back and bottom (B) side and one from the front of the IMV is evident (arrowheads). bR, back right. The end of one of these tubes is more bulbous (larger arrowhead). (E) Bottom view of an IMV attached by a large membrane domain (star) to a membrane vesicle (asterisk). Note the apparent unfolding of the membrane tubules. (F) Top layer of the IMV has presumably been displaced (arrowhead), exposing the top (T) of the core (c). In projection a cisternal structure is evident (arrows). Bars, 100 nm.
The grid-tap method.
We realized that a more-detailed analysis of the viral structure as the IMV enters cells required a different approach. We were aware that within minutes of adding IMV to HeLa cells, the cells respond with signaling events that induce the assembly of prominent microvillar projections to which IMV readily attach (30). We therefore reasoned that it might be possible to attach such membrane projections with attached virions simply by touching the cell surfaces with a carbon- and Formvar-coated EM grid. For this, a high titer of IMV was added to HeLa cells (grown on glass coverslips) for 30 min, a period more than sufficient to induce massive cell surface projections (30). The coverslips were allowed to contact EM grids for ∼1 s, and following brief rinses in water, the grids were stained and dried with uranyl acetate and methyl cellulose. We believe that the resulting images show snapshots of interesting viral intermediates likely involved in entry.
In Fig. 7 and 8, one sees the same unusual features of the virus as were seen with the other approaches, especially after DTT treatment, although additional features become evident here. The particle shown in Fig. 7A is a flattened view of the particle attached to a plasma membrane-derived vesicle that allows one to clearly appreciate three-dimensional continuity between the curved, tongue-like cisternal domains and the connecting tubules. Moreover, the ends of two different tubules seem to converge and twist into a spherical structure, like the end of a corkscrew spiral, at the back right corner of the particle. These curved tubular rings are also evident in Fig. 7C to E.
The inset to Fig. 7A shows highly informative views of the sides and bottom (right particle) of the IMV. The particle on the left provides a clear illustration of the so-called lateral bodies, evident as highly interconnected tubular lobes on the top and bottom that tuck into the side of the particle. Having seen this image, these structures can also be appreciated in Fig. 4H and I. The particle on the left in Fig. 7A (inset) shows the long left-side domain that curves around both ends of the particle. A tubular projection from the lateral body (top domain) can be seen to either connect to or be tucked beneath the long side lobe. The bottom view (right particle) shows the tight packing of large domains connected by tubular connections. One of these domains appears to be contacting a highly curved tube. The latter, as before, emerges from the right back part of the IMV, evidenced as an indentation of the particle at this site.
Some of the structures shown in Fig. 7B are putative vesicles derived from the plasma membrane, most likely from microvillar or filipodial extensions, to which the virus had attached. The main part of this panel is a rare image in which three highly regular tubules with bulbous tips are seen extending from the virus. We believe that these are the same structures as those shown in Fig. 4H and 6E and just before ejection by the tubes seen in Fig. 7A. One of these tips in Fig. 7B seems to be attached to the plasma membrane vesicle. At a higher magnification, these 30 to 40-nm-diameter tubes exhibit a periodic structure (not shown) that is highly reminiscent of the tubes that form when IMV is treated with NP-40 and mercaptoethanol (50). By comparison to this figure it seems likely that the same tips or plugs can be seen (presumably more flattened) in Fig. 7C to E.
Figure 7C and D show a stereo pair in which the striking sidedness of the IMV again becomes apparent (top particle); whereas the right side of this particle is highly curved and seemingly completely enclosed, the “right” side shows a rather straight edge, again as if the particle had been sliced by a knife (see also the two particles in Fig. 7F). It is by facing this “open” right side that one sees the side-view details of the core structure (Fig. 7E, bottom). In Fig. 7F, it also becomes clear that the two side projections of the IMV (shown also in Fig. 5G) at the bottom of this image consist of curved tubules that project downward like two “elbows”. Because of a slightly different projection, these “elbows” are not seen in the upper particle of this figure. A different view of this right aspect of the IMV is seen in the top particle of Fig. 7E. A general feature seen in these images is that the right side of the particle is always rather planar whereas the left side is more hemispherical after partial drying of the particle.
The interconnections of many of the viral domains are clearly evident in Fig. 7G. In this figure, one particle (second from right, top row) has flattened severely, which has the effect of accentuating the tubular connections between the larger membrane lobes.
In Fig. 8A and B is shown a stereo pair of the bottom of a particle, which gives the impression of (pseudo-) twofold symmetry around the central cavity. This image gives a detailed view of the folding and interconnections of the tubules, which are evident at higher magnifications in Fig. 8C. It is evident that this particle has significantly loosened its organization, but all of the membranes still appear within the particle. Figure 8D shows early stages of unfolding of up to four distinct tubules, one in the front and three in the back. The length of the longest extension exceeds the largest dimension of the IMV. In Fig. 8E, an IMV is seen to be attached to a cell membrane-derived vesicle. This image suggests that the membranes are ejected on the side away from these putative membrane attachment sites.
Figure 8F shows a particle in which the top surface of the core is evidently enclosed by a partially rectangular tubule in which stain may have entered the lumen of cisternae but more likely reveal two fine tubules in projection. In this particle, the continuity of the outer domains with domains deeper inside the particle is quite evident.
Conventional negative staining.
To relate our data to earlier published data, we also show images of DTT-treated IMVs that were conventionally negatively stained with uranyl acetate (Fig. 9A to C). In these images it is clear that a significant amount of stain has had access to the interior regions. We presume that the stained part is predominantly the (positive stained) DNA-containing structures while the stain-excluding parts are domains of cisternae and tubules. In Fig. 9A and B, the shape of the core is evident as an L- or bookend-like structure, having an almost 90° edge at the back left corner of these two particles. One of the curved tubular structures is evident as a stain-excluding structure in Fig. 9A. On the opposite sides of these particles one can see the infolding of viral membrane domains that are intact and exclude stain. Although the preservation of these preparations is clearly inferior to that with the Tokuyasu procedure, the crude outlines of the shape of the core resemble what was seen with the methyl cellulose procedure. In Fig. 9D and E, negative staining with ammonium molybdate was used with DTT-treated virus. In Fig. 9D, one particle shows a single tubule with a bulbous tip. To the left of this image is a branched tubular structure that we believe has separated from an IMV (cf. Fig. 7B and 11D). In Fig. 9E, the upper particle appears intact whereas the bottom particle seems to be uncoating; stain has entered the core region, and a single, highly curved tubule is seen to be unraveling itself at the back right corner of this virus.
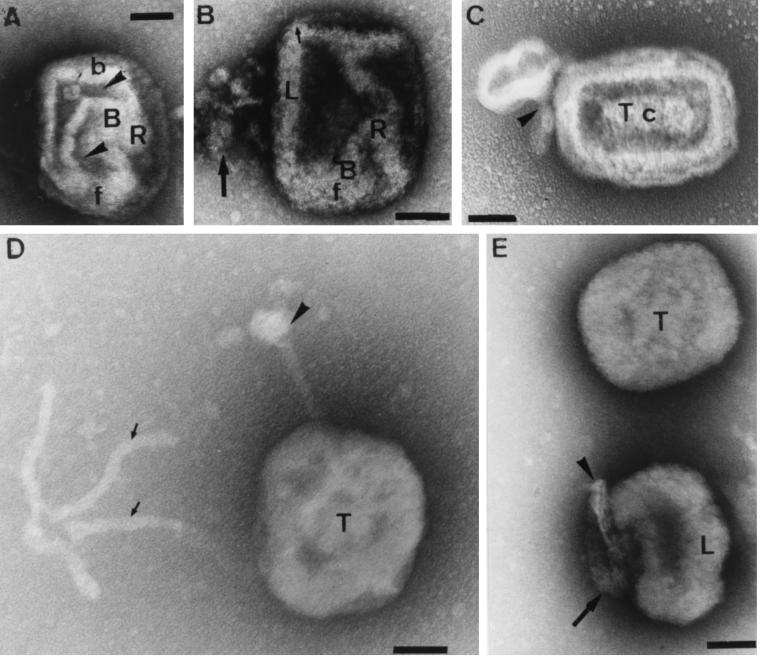
FIG. 11.
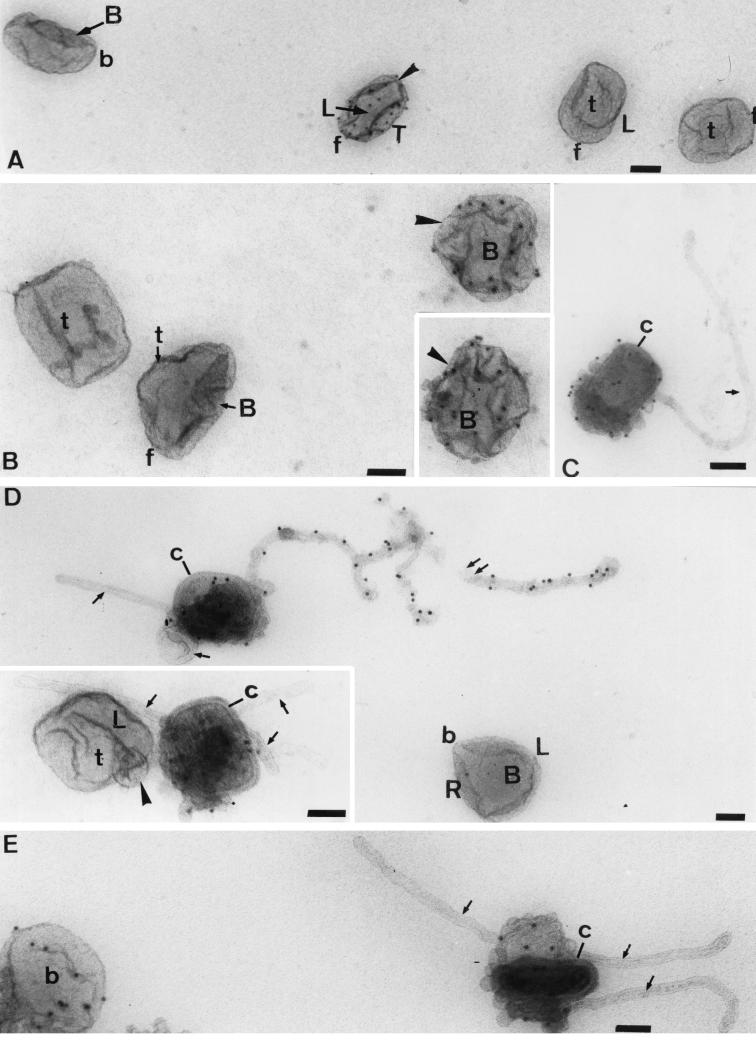
Effects of DTT and P16 immunolabeling of IMV in vitro. The particles were treated for 15 min with 20 mM DTT before adsorption onto grids and labeling with anti-P16 (cytoplasmic domain) and protein A-gold. In untreated particles, the P16 is hardly accessible to antibodies, but accessibility increases with increasing DTT treatment. In these images, unlabeled particles can be seen adjacent to strongly labeled ones (arrowheads in panels A and B). In panels C to E, the increased accessibility to anti-P16 coincides with a peeling off of the outer membranes, revealing the underlying core (c) structure. Note also the extensive, ∼30-nm-diameter tubules emanating from the disrupted particles (arrows). The long, branched tubule in panel D is strongly labeled for P16, but the second tubule, as well as those in the inset to panel D and in panel C and E, are unlabeled. In panel D, one of the unlabeled tubules can be seen to curve into a spherical structure, while in the inset this structure is more compressed into a ball (arrowhead). As for previous figure, where possible the top (T), bottom (B) and (L) sides, and front (f) and back (b) of the IMV are indicated. Bars, 100 nm.
Grid separation method.
As an alternative method of physically unraveling the viral structure, in a few experiments we applied a 1-μl volume of virus suspension between two grids and then separated the grids. In a few cases we seemed to have indeed pulled out some structures. As shown in Fig. 10A and B, the above-described tubules with the bulbous ends could be considerably extended. In Fig. 10A it is (again) evident that the two tubular extensions are extended from two different corners of the virus. Also, membrane folds that presumably covered the particle have been opened up. In Fig. 10C and D, two tilted (50°) views are shown of a corner at the bottom of a particle in which the bulbous tubular extension has only partially opened. The shape of the underlying core is evident, as is its extensive connections to the tubular labyrinth. A different view of these structures, again with two extensions, is seen in Fig. 10G. In Fig. 10E and F, the outer membrane has been pulled out as a sheet but is still clearly connected to the underlying core. By comparing these two images, one gains an impression of the folding of the unstained membrane tubes within the core. We suggest that this image resembles the separation of the core from the outer membranes that occurs during viral entry.
P16 Immunolabeling after DTT treatment.
As mentioned above, treatment of the IMV with DTT loosens up the structure without affecting infectivity. In Fig. 11 we show images of DTT-treated virus (in suspension) that was adsorbed onto an EM grid, fixed, and immunolabeled for the membrane protein P16 by using a highly specific antibody against this protein's cytoplasmic domain (AI4L). These micrographs show different stages of DTT-induced uncoating. In Fig. 11A, three particles are classified as intact. These particles are almost completely devoid of label for P16, in agreement with earlier data showing that very little of this protein is exposed on the outside of the intact IMV (44).
As shown in Fig. 11A and B, under conditions in which some particles are completely devoid of P16 labeling and after a 15-min incubation with DTT, a few particles became strongly labeled. There is a clear hint that when the particle is clearly labeled, some domains label whereas others appear to be unlabeled. In Fig. 11C to E, tubular extensions are evident in some particles after 30 min of DTT treatment. Strikingly, a significant amount of the C terminus of P16 is exposed on some of these tubules (Fig. 11D and E), whereas others are completely devoid of labeling (Fig. 11 C to E). In many of these particles, the structure of the underlying core is revealed (Fig. 11C to E). This was confirmed by their being labeled with an antibody against the vaccinia virus core (surface) (results not shown). In these images, between one and three tubes can be seen to project from different sites on the virus.
These data can most rationally be explained by assuming that in the intact particle, the bulk of P16 is on membranes that are mostly buried within the IMV (44). Upon breakage of the cytoplasmically enriched disulfide bonds of vaccinia virus with DTT (29, 43), the structure opens out, exposing tubular and cisternal domains that were previously hidden. In our cisternal-tubular model, such an outward movement of P16 can easily be envisaged as a lateral diffusion of antigen from one cisternal domain to another and by the labeled tubular domains being hidden deep within the untreated particle. While we believe that a number of tubules are an integral part of the virus structure, it also seems possible that with prolonged DTT treatment, cisternal domains artifactually tubularize. We also cannot be certain that some of these tubules do not artifactually form during air drying. Indeed, we argue that such a process induces the surface tubular elements seen with classical negative-staining approaches. Nevertheless, the differential labeling with P16 in our preparations indicates clearly that these tubes are not identical.
Cryo-EM.
In a previous paper (43) we described an analysis of the IMV by cryo-EM using the bare-grid method of Adrian et al. (1). In this method, the particles are suspended in a drop of aqueous medium, blotted to make a thin film, and then directly vitrified in liquid ethane and observed at a temperature below −160°C. The studies by Roos et al. (43) and Krijnse Locker and Griffiths (29) showed that DTT treatment reduced the (mostly cytoplasmic) IMV disulfide bonds, leading to a loosening of the outer envelope from the particle and therefore allowing heavy-metal stains to gain access to the underlying viral core. As mentioned, this treatment with DTT has no quantitative effect on the viral infection process (29). This observation is one of many we have made that is inconsistent with the well-accepted notion that the IMV enters cells by fusing with the plasma membrane (2, 5, 17) (see Discussion). If the IMV were surrounded by only a single membrane that lysed and separated even partially from the core, it is quite difficult to imagine how such a virus could fuse and still insert the core intracellularly.
A great advantage of the cryo-EM approach is that, unlike with the negative-staining methods, information is gained via projection of electrons through the specimen (14), allowing the whole three-dimensional structure to be visualized, at least in principle. It must be emphasized, however, that the relatively large size of the IMV particle (0.3 μm in the largest dimension) is at the limit for being useful for cryo-EM analysis, and extensive overprojection of an exceedingly asymmetric and convoluted membrane structure makes it difficult to approach a higher-resolution analysis without an enormous investment in this approach. Following the procedure of Roos et al.(43), we decided to focus exclusively on the use of a low concentration of uranyl acetate stain just prior to vitrification, since this additional contrast highlights more structure (but may also induce more artifacts).
In Fig. 12 we show untreated IMV prepared by this method. In Fig. 12A, both top-up and bottom-up orientations are seen. When the particles lie free on the air-water interface, the top aspect can usually be ascertained by the fact that the indented corner is at the back right side of the particle, whereas in bottom-up particles it is on the back left corner (Fig. 12C). The two membrane layers and the layer of spicules are clearly evident in these images. In Fig. 12B, the particle has attached to the side of the supporting film, revealing the “right” side view into the projected core that appears deceptively regular. This view shows the classical dumbbell-shaped image that has been widely described from thin sections (see Fig. 1B in the accompanying paper [20a]). The structures referred to as the lateral bodies (10) are also seen faintly in projection. Figure 12C shows that a preponderance of the IMV align similarly, giving the impression of ovoid bricks. Branching tubules reminiscent of those described above (possibly swollen by the pure water used for vitrification) are evident in the background. The dimensions of these particles prepared by cryo-EM (300 by 250 by 150 nm [43]) are very similar to the estimates of the stained particles, as well as to the SEM measurements (see above).
FIG. 12.
Cryo-EM of normal IMV using uranyl acetate positive staining. (A) Two particles are seen attached to the side of the support film. Two layers are evident (arrows) that are separated by the spike or spicule layer (arrowhead). (B) The particle has attached to the grid support in a manner such that a classical side view is evident. The two membrane layers are indicated with large arrows, and the spikes are indicated by an arrowhead. The star indicates the projection of one of the lateral bodies that we believe is due to the overlapping of the peripheral lobes of the virus (top and bottom). The small arrow indicates an inward groove in the membrane. (C) Note the rather uniform appearance of the IMV due to their similar orientation at the air-water interface, albeit with significant differences in density across the particles. The more-rounded corner of the particle (back/right) is indicated by arrowheads. This can be used to determine whether a particle is exposing its top or the bottom (in which this corner would appear at back right and back left, respectively). The arrow indicates branched tubular structures that are probably derived from disrupted particles. The particle indicated by the star shows the brick-shaped underlying core structure as a higher-density region. Bars, 100 nm (A and B, same magnification).
A particular advantage of DTT treatment, especially with incubations longer than 5 min, was that it led to particle swelling, which tended to make the particles more spherical (Fig. 13A and B). As a result, the IMVs no longer all aligned along one axis but were free to rotate on the air-water interface prior to freezing. Even in a single image one can see many different projections through the IMV that are concentrated on the air-water interface.
FIG. 13.
Cryo-EM of DTT-treated IMV using positive staining. twenty four different particles are evident in these images. Note that only a few particles now appear normal (e.g., particle 6 in panel A); most are more oval or spherical in appearance. The underlying core is seen in projection in many different views. In some particles (e.g., inset 2 or particle 6 in panel A), the core (c) appears roughly brick shaped. However, as indicated by the arrowhead in inset 1 of panel B), the flat core lobe curves and extends into an S-shaped organization. The connectivities of two different lobes can be appreciated in particle 5 of panel B. In side views, the core can appear completely spherical (inset 2, panel B) or like eyeglasses (inset 1 of panel A or particle 9 of panel B). In inset 1 of panel A, the “top” of the virus projects upwards—in other words, the top domain (the top of the “eyeglasses”) represents the flat, brick-like domain of the core. In some images (e.g., particle 4, panel A) the core appears as an extended U-shaped structure (arrowheads). The arrows in all figures indicate the positions of the lateral bodies, i.e., projections of lobes at the “top” and “bottom” of the IMV. Bars, 100nm.
The lobed extensions on the two sides of the particle that are responsible for the lateral bodies are evident in these images (Fig. 13A; particles 3 and 5; Fig. 13B, particles 3 and 4). The fact that these structures are generally closely apposed to the outer membrane layers supports our contention that they are sites of attachment of core membrane domains to the outer membranes. In these images, the complexity of the core structure is even more evident than before. When viewed from above, it appears in projection as a quasi-rectangle (see, e.g., Fig. 13A, particle 6). However, in different views it can appear bilobed (eyeglass like; see inset 1 of Fig. 13A). More-regular bilobes are evident in particles 2 and 4 of Fig. 13A and in particles 5, 6, and 8 of Fig. 13B. Other images reveal that these lobes are connected via an S-shaped configuration that is rotated along one axis (e.g., particle 1 in Fig. 13B).
Because of the complexity of the core organization and the significant overlap of information at any one point in the structure, we have not yet been able to provide a detailed reconstruction that would reveal the three-dimensional organization of the core or of the whole virus. Nevertheless, we point out that the S-shaped folding of the core seen in these images of the IMV (which were not treated with traditional EM fixatives) is in agreement with our theoretical model (Fig. 2) and with the images of dehydrated virus that we have shown above.
DISCUSSION
In Fig. 2 we introduced our working model that shows the essential organizational framework of the vaccinia IMV. The key features of this model are a roughly S-shaped configuration of an ER-derived cisterna that leads to a particle with two different cytoplasmic compartments. It must be emphasized, however, that the model shown in Fig. 2 is only a crude caricature of the real IMV structure, as revealed in this study. The assembly process leads to a particle that is highly asymmetrical, with differences between the top and bottom, between the right and left, and between the front and back of the particle (arbitrarily defined). Topologically, the virus is an interconnected labyrinth of membrane cisternae and tubules. Moreover, up to four tubules seem to be able to extend from different sites at the front and lower back of the particle.
Our data strongly suggest that during the infection process the virus eviscerates inner membrane contents that resemble intestines. Although further, more definitive studies are needed, our data strongly suggest that these “intestines” represent a lamellar fold connected to tubules that can extend 0.5 μm or more from the virus.
The data collectively suggest that the “top” of the particle is covered by cisternal flaps from which up to four tubules emanate. At one end of this domain, an extended, tongue-like long domain seems to continue around one corner and ends up running along the entire left side of the virus. The top cisternal fold seems to be tightly apposed to the underlying core structure. This dome-shaped top domain was already described by Peters in 1956 (38), although he concluded that the particle was otherwise a rather symmetrical “brick.” His model fairly accurately conveyed the rather flattened shape of the particle (though it was probably flattened even more by the specimen preparation methods available at that time). Peters also accurately described a section profile perpendicular to the top/bottom axis. However, the complex features on the bottom and inside of the particle were not revealed, and perhaps such a clear, and for its time excellent, model helped to reinforce the idea that vaccinia virus has a rather symmetrical, brick-shaped structure. In later papers (39, 40) this author revealed unexpected complexity in the structure of a triad of bulbous elements that seemed to be made up of highly coiled tubules. This was also the interpretation of Hohenberg, who made a detailed artistic drawing of the virus (see reference 34). Our data are consistent with this model with respect to these spherical structures; we argue that these highly curved tubes exhibit continuity with the rest of the labyrinth. Because of the propensity of these structures surfaces to stain heavily with uranyl acetate, we suggest that they are structural membrane scaffolds for the viral DNA. Although one never sees more than three of these spherical structures in cross-sections through the virus (see Fig. 9B in the accompanying paper [20a], up to four can be seen in whole mounts (Fig. 5F).
Our data show clearly that all models to date have greatly underestimated the complexity of vaccinia virus structure. In particular, the bottom and inner aspects of the virus had not previously been revealed in this tightly packed structure. A major reason why we could visualize the “guts” of the virus was the availability of two different methods for uncoating the virus. The first, the use of the reducing agent DTT, reduces the mostly cytoplasmic disulfide bonds in vaccinia virus and loosens its structure. The second is the natural process of uncoating, which seems to accompany core entry.
The top and left side of the particle are, at first glance, rather regular, and these domains tend to be organized at ∼90° angles to one another. This organization is probably a consequence of the fact that the top and left side of the underlying core have the same regular structure as the same faces of the IMV. In contrast, the bottom and right side of the particle are more irregular, and again this lack of symmetry is reflected in the structure of the underlying core, especially when viewed from below. We expect that a more complete model of the three-dimensional organization of the virus will emerge from a detailed tomographic analysis which is now in progress.
During the presumed uncoating response, at least two, and possibly four, tubules seem to be ejected from the inner cavity of the IMV. Under normal conditions, these tubes are tightly packed in the inner parts of the virus. However, the extremities of these tubules are highly curved and occasionally exposed at the front and back right corners. Even in untreated virus particles, these tubes are easily displaced and can extend various distances from the IMV particle. We noted that on contacting cell projections up to three such tubules could be seen, which extended 500 nm or more. In many cases, the ends of these tubes were bulbous and contrasted with the seeming rigidity of the bulk of the tubes. The latter are usually nonrigid when the IMVs have been treated with DTT, but after contact with cell membrane projections they often appear quite rigid and show a structure highly reminiscent of the rigid tubules isolated from IMV by Wilton et al. (50). We speculate that these tubular extensions may be either cell sensors or attachment sites that could facilitate the infection process by initiating the rapid and extensive membrane signaling events that occur in response to IMV entry (30).
In the one-membrane hypothesis of Hollinshead et al. (24), the only conceivable mechanism of entry which fits known principles is membrane fusion, either at the plasma membrane or in an endocytic (or phagocytic) compartment. Indeed, membrane fusion has been proposed to mediate entry of IMV, as well as the EEV, at the plasma membrane (2, 5, 12, 24). Our model of the IMV (Fig. 2) is theoretically inconsistent with a mechanism of membrane fusion since it would leave the core outside of the cell. In fact, our data in this and in other studies (30, 37) are completely at odds with the idea of a fusion event facilitating core entry, both for the IMV and the EEV. The following arguments summarize the main evidence against typical fusion events (we emphasize these points because the question of how the virus enters cells is intimately linked to the organization of its structure).
(i) Extensive immunogold labeling of thin sections of cells infected for short periods with IMV or EEV shows that neither IMV nor EEV membrane antigens are significantly incorporated into the host cell plasma membrane but are left outside, as membrane fragments (30). It is possible that these viral cisternal remnant structures subsequently flatten on top of the cell membrane, giving the impression of fusion, or perhaps represent a real but secondary fusion event that may serve to clear viral membranes from the plasma membrane (2, 5, 17). We point out that the most convincing images of the references 5 and 17 were apparently prepared by centrifuging the virus at 200 × g onto the surface of the spin culture cells. This process may well have flattened cisternal fragments on top of the plasma membrane.
(ii) Since the EEV, relative to the IMV, has one additional membrane, which undoubtedly surrounds the particle completely, a fusion process would be expected to reveal the complete IMV free in the cytoplasm. This has never been observed in extensive analysis. For both IMV and EEV, the only identifiable structure in the cytoplasm is the viral core, which is indistinguishable after both infections.
(iii) DTT greatly disrupts the organization of the particle but has no effect on infectivity. This observation is difficult to fit into a conventional fusion model. Moreover, if the IMV had only one, tight-fitting membrane envelope, DTT's effect could only be easily interpreted as being due to membrane lysis. If a conventional fusion mechanism was indeed operative, a lysed virus would be unlikely to enter cells as efficiently as an intact particle.
(iv) In separate studies, we showed that the two DNA-binding proteins of the viral core, P11 (F17R) and P25 (L4R), are in different compartments (30, 37) (Fig. 3). P25 colocalizes with the viral DNA, whereas P11 does not. Remarkably, while P25 enters the infected cell as an integral and necessary component of the core, P11 (which is essential not for infection but rather is required for assembly) is left outside the cell, along with the membrane fragments (37). While we still lack a molecular description of the process, this observation is incompatible with a viral entry process that follows expected rules of viral fusion.
Our model predicts that during infection, the additional membranes that are concentrated in the cavity of the virus are left behind while the core must traverse the membrane barrier (perhaps by local activation of a viral or plasma membrane lipase). It is possible that the whole structure is under some tension and that a force is provided that helps to “inject” the core. This model also predicts the presence of transient direct membrane connections, between the core cisterna and the outer membranes, that remain extracellular. A local lysis mechanism would conceivably be required to sever the connection between the core and the outer cisternae, perhaps facilitated by a mechanical force obtained during unpacking of the tightly knit virus structure. What such an intermediate might look like is evident in Fig. 10E and F, in which the virus has been pulled apart mechanically.
The lateral bodies have long been a prominent feature of the IMV, as seen in thin sections through one particular plane through the virus (10). Even when the virus is dissociated by chemical treatment, these structures retain at least a part of their organization (25). Dubochet et al. (13) concluded from cryo-EM observations of the IMV that the lateral bodies are artifactually induced by chemical fixation, since they failed to see them unless fixatives were added. We disagree with this view and argue that those interpretations were based on examination of preparations in which the great majority of the IMV aligned top or bottom up on the air-water interface. Occasional viewing of particles attached via their left sides to the sides of the holey film revealed lateral bodies (seen from a “right” perspective in Fig. 13B). While we cannot yet give a complete three-dimensional description of the lateral bodies, we suggest that they represent end projections of cisternal domains of the core that attach, like adhesive plaques, to the outer cisternae on opposite (top and bottom) sides of the particle.
In the accompanying paper (20a), we extend these observations by focusing on the still more complex situation using thin-section analysis, the approach upon which all of the existing models of IMV assembly and structure have been almost exclusively based.
REFERENCES
- 1.Adrian M, Dubochet J, Lepault J, McDowell A W. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong J A, Metz D H, Young M R. The mode of entry of vaccinia virus into L cells. J Gen Virol. 1973;21:533–537. doi: 10.1099/0022-1317-21-3-533. [DOI] [PubMed] [Google Scholar]
- 3.Baumeister W, Grimm R, Walz J. Electron tomography of molecules and cells. Trends Cell Biol. 1999;9:99–104. doi: 10.1016/s0962-8924(98)01423-8. [DOI] [PubMed] [Google Scholar]
- 4.Cairns H J F. The initiation of vaccinia infection. Virology. 1960;11:603–623. doi: 10.1016/0042-6822(60)90103-3. [DOI] [PubMed] [Google Scholar]
- 5.Chang A, Metz D H. Further investigations on the mode of entry of vaccinia virus into cells. J Gen Virol. 1976;32:275–282. doi: 10.1099/0022-1317-32-2-275. [DOI] [PubMed] [Google Scholar]
- 6.Cudmore S, Blasco R, Vincentelli R, Esteban M, Sodeik B, Griffiths G, Krijnse Locker J. A vaccinia virus core protein, p39, is membrane associated. J Virol. 1996;70:6909–6921. doi: 10.1128/jvi.70.10.6909-6921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dales S. The uptake and development of vaccinia virus in strain L cells followed with labeled viral deoxyribonucleic acid. J Cell Biol. 1963;18:51–72. doi: 10.1083/jcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dales S, Kajioka R. The cycle of multiplication of vaccinia virus in Earle's strain L cells. 1. Uptake and penetration. Virology. 1964;24:278–294. doi: 10.1016/0042-6822(64)90167-9. [DOI] [PubMed] [Google Scholar]
- 9.Dales S, Mosbach S. Vaccinia as a model for membrane biogenesis. Virology. 1968;35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- 10.Dales S, Pogo B G T. Biology of poxviruses. Virol Monogr. 1981;1981:1–156. doi: 10.1007/978-3-7091-8625-1. [DOI] [PubMed] [Google Scholar]
- 11.Dales S, Siminovitch L. The development of vaccinia virus in Earl's L strain cells as examined by electron microscopy. J Biophys Biochem Cytol. 1961;10:475–503. doi: 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doms R W, Blumenthal R, Moss B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J Virol. 1990;64:4884–4892. doi: 10.1128/jvi.64.10.4884-4892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubochet J, Adrian M, Richter K, Garces J, Wittek R. Structure of intracellular mature virus observed by cryoelectron microscopy. J Virol. 1994;68:1935–1941. doi: 10.1128/jvi.68.3.1935-1941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubochet J, Adrian M, Chang J J, Homo J C, Lepault J, McDowell A W, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 15.Ericsson M, Cudmore S, Shuman S, Condit R C, Griffiths G, Krijnse Locker J. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J Virol. 1995;69:7072–7086. doi: 10.1128/jvi.69.11.7072-7086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteban M, Flores L, Holowczak J A. Topography of vaccinia virus DNA. Virology. 1977;82:163–181. doi: 10.1016/0042-6822(77)90040-x. [DOI] [PubMed] [Google Scholar]
- 17.Granados R R. Entry of an insect poxvirus by fusion of the virus envelope with the host cell membrane. Virology. 1973;52:305–309. doi: 10.1016/0042-6822(73)90422-4. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths G. Fine structure immunocytochemistry. Heidelberg, Germany: Springer-Verlag; 1993. [Google Scholar]
- 19.Griffiths G, Rottier P J M. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Cell Biol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths G, McDowell A W, Back R, Dubochet J. On the preparation of cryosections for immunochemistry. J Ultrastruct Res. 1984;89:65–78. doi: 10.1016/s0022-5320(84)80024-6. [DOI] [PubMed] [Google Scholar]
- 20a.Griffiths G, Roos N, Schleich S, Krijnse-Locker J. Structure and assembly of intracellular mature vaccinia virus: thin-section analysis. J Virol. 2001;75:11056–11070. doi: 10.1128/JVI.75.22.11056-11070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths G, Simons K, Warren G, Tokuyasu K T. immunoelectron microscopy using thin, frozen sections: application to studies of the intracellular transport of Semliki Forest virus spike glycoproteins. Methods Enzymol. 1983;96:466–485. doi: 10.1016/s0076-6879(83)96041-x. [DOI] [PubMed] [Google Scholar]
- 22.Harris R J, Horne R J. Negative staining: a brief assessment of current technical benefits, limitations and future possibilities. Micron. 1994;24:5–13. [Google Scholar]
- 23.Hermann R, Walther P, Mueller M. Double layer coating for high resolution scanning electron microscopy of frozen-hydrated cells. J Microsc. 1995;7:343–350. doi: 10.1111/j.1365-2818.1995.tb03635.x. [DOI] [PubMed] [Google Scholar]
- 24.Hollinshead M, Vanderplasschen A, Smith G L, Vaux D J. Vaccinia virus intracellular mature virions contain only one lipid membrane. J Virol. 1999;73:1503–1517. doi: 10.1128/jvi.73.2.1503-1517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichihashi Y, Oie M, Tsuruhara T. Location of DNA-binding proteins and disulphide-linked proteins in vaccinia virus structural elements. J Virol. 1984;50:929–938. doi: 10.1128/jvi.50.3.929-938.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen O N, Houthaeve T, Shevchenko A, Cudmore S, Mann M, Griffiths G, Krijnse Locker J. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J Virol. 1996;70:7485–7497. doi: 10.1128/jvi.70.11.7485-7497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koster A, Grimm R, Typke A, Hegerl R, Stoschek A, Walz J, Baumeister W. Perspectives of molecular and cellular electron tomography. J Struct Biol. 1997;120:276–308. doi: 10.1006/jsbi.1997.3933. [DOI] [PubMed] [Google Scholar]
- 28.Krajioka R, Siminovich L, Dales S. The cycle of multiplication of vaccinia virus in Earle's strain L cells. Virology. 1964;24:295–308. doi: 10.1016/0042-6822(64)90168-0. [DOI] [PubMed] [Google Scholar]
- 29.Krijnse Locker J, Griffiths G. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J Cell Biol. 1999;144:267–279. doi: 10.1083/jcb.144.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krijnse Locker J, Kuehn A, Rutter G, Hohenberg H, Wepf R, Griffiths G. Vaccinia virus entry at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol Biol Cell. 2000;11:2497–2511. doi: 10.1091/mbc.11.7.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan C. Vaccinia virus reexamined: development and release. Virology. 1976;73:43–58. doi: 10.1016/0042-6822(76)90059-3. [DOI] [PubMed] [Google Scholar]
- 32.Morgan C, Ellison S A, Rose H M, Moore D H. Structure and development of viruses observed in the electron microscope. II. Vaccinia and fowl pox virus. J Exp Med. 1955;100:301–311. doi: 10.1084/jem.100.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moss B. Poxviridae and their replication. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Fields virology. New York, N.Y: Raven Press; 1990. pp. 661–684. [Google Scholar]
- 34.Mueller G, Williamson J D. Poxviridae, p. In: Nermut M, Stevens A, editors. 421–433. Animal virus structure. Amsterdam, The Netherlands: Elsevier; 1987. [Google Scholar]
- 35.Palade G E. Problems in intracellular membrane traffic. Ciba Found Symp. 1982;12:1–4. doi: 10.1002/9780470720745.ch1. [DOI] [PubMed] [Google Scholar]
- 36.Patrizi G, Middlecamp J N. Immature forms of vaccinia virus: morphological observations from thin sections of infected human skin. Virology. 1968;34:189–192. doi: 10.1016/0042-6822(68)90029-9. [DOI] [PubMed] [Google Scholar]
- 37.Pedersen K, Snijder E J, Schleich S, Roos N, Griffiths G, Krijnse Locker J. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J Virol. 2000;74:3525–3536. doi: 10.1128/jvi.74.8.3525-3536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters D. Morphology of resting vaccinia virus. Nature. 1956;178:1453–1455. doi: 10.1038/1781453a0. [DOI] [PubMed] [Google Scholar]
- 39.Peters D. Strukturaufklaerung am Elementarkoerper des Vaccinia-virus durch Abbau mit Trypsin. In: Wurzelbacher P, editor. European regional conference on electron microscopy vol. 1960. II. Amsterdam, The Netherlands: Elsevier; 1961. pp. 694–698. [Google Scholar]
- 40.Peters D, Mueller G. The fine structure of the DNA-containing core of the vaccinia virus. Virology. 1963;21:267–269. doi: 10.1016/0042-6822(63)90267-8. [DOI] [PubMed] [Google Scholar]
- 41.Ritter M, Henry D, Wiesener S, Pfeiffer S, Wepf R. A versatile high-vacuum cryo-transfer for cryo-FESEM, cryo-SPM and other imaging techniques. Microsc Microanal. 1999;5(Suppl.):1–5. [Google Scholar]
- 42.Roos N, Morgan A J. Microscopy handbooks of the Royal Microscopical Society. 21. Cryopreparation of thin biological specimens for electron microscopy: methods and applications. Oxford, United Kingdom: Oxford University Press; 1990. [Google Scholar]
- 43.Roos N, Cyrklaff M, Cudmore S, Blasco R, Krijnse Locker J, Griffiths G. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996;15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 44.Salmons T, Kuhn A, Wylie F, Schleich S, Rodriguez J R, Rodriguez D, Esteban M, Griffiths G, Krijnse Locker J. Vaccinia virus membrane proteins p8 and p16 are co-translationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J Virol. 1997;71:7404–7420. doi: 10.1128/jvi.71.10.7404-7420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmelz M, Sodeik B, Ericsson M, Wolffe E, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slot J W, Geuze H J, Gigengack S, Lienhard G E, James D E. Immuno-localization of the insulin-regulatable glucose transporter in brown adipose tissues of the rat. J Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sodeik B, Griffiths G, Ericsson M, Moss B, Doms R W. Assembly of vaccinia virus: effects of rifampicin on the intracellular distribution of viral protein p65. J Virol. 1994;68:1103–1114. doi: 10.1128/jvi.68.2.1103-1114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tokuyasu K T. A study of positive staining of ultrathin frozen sections. J Ultrastruct Res. 1978;63:287–307. doi: 10.1016/s0022-5320(78)80053-7. [DOI] [PubMed] [Google Scholar]
- 50.Wilton S, Mohandas A R, Dales S. Organization of the vaccinia envelope and relationship to the structure of intracellular mature virions. Virology. 1995;214:503–511. doi: 10.1006/viro.1995.0061. [DOI] [PubMed] [Google Scholar]