Abstract
Engineering hierarchical vasculatures is critical for creating implantable functional thick tissues. Current approaches focus on fabricating mesoscale vessels for implantation or hierarchical microvascular in vitro models, but a combined approach is yet to be achieved to create engineered tissue flaps. Here, millimetric vessel‐like scaffolds and 3D bioprinted vascularized tissues interconnect, creating fully engineered hierarchical vascular constructs for implantation. Endothelial and support cells spontaneously form microvascular networks in bioprinted tissues using a human collagen bioink. Sacrificial molds are used to create polymeric vessel‐like scaffolds and endothelial cells seeded in their lumen form native‐like endothelia. Assembling endothelialized scaffolds within vascularizing hydrogels incites the bioprinted vasculature and endothelium to cooperatively create vessels, enabling tissue perfusion through the scaffold lumen. Using a cuffing microsurgery approach, the engineered tissue is directly anastomosed with a rat femoral artery, promoting a rich host vasculature within the implanted tissue. After two weeks in vivo, contrast microcomputer tomography imaging and lectin perfusion of explanted engineered tissues verify the host ingrowth vasculature's functionality. Furthermore, the hierarchical vessel network (VesselNet) supports in vitro functionality of cardiomyocytes. Finally, the proposed approach is expanded to mimic complex structures with native‐like millimetric vessels. This work presents a novel strategy aiming to create fully‐engineered patient‐specific thick tissue flaps.
Keywords: 3D bioprinting, ECM bioink, engineered flap, personalized medicine, tissue engineering, vascularization
Mesoscale tubular scaffolds and 3D bioprinted vascularized hydrogels are combined to create engineered hierarchically vascularized tissues (engineered flap) consisting of rich microvascular networks in communication with an engineered endothelium. The in vitro cultured flaps are directly anastomosed to the host vasculature, which perfuses the implanted tissue. The concept is expanded by 3D printing patient‐specific vascular constructs with complex native architectures.
![]()
1. Introduction
A critical trait shared by most tissues in the body is a complex hierarchical vascular structure to provide nourishment to tissue‐forming cells. Without proper vascularization, cells embedded in a biological environment cannot acquire vital nutrients and gases or remove metabolic waste, leading to cell death and subsequent tissue necrosis.[ 1 ] Therefore, engineered tissues thicker than the nutrient diffusion limit require incorporating a vascular network that facilitates the transport of nutrients and waste.[ 2 , 3 ] A common method to achieve vascularized constructs consists of co‐seeding endothelial cells (ECs) and support cells (SCs) within a suitable environment, such as a scaffold or hydrogel containing extracellular matrix (ECM) proteins, resulting in the spontaneous formation of vascular structures.[ 4 , 5 ] Previous works have shown the importance of including self‐assembling mature microvascular networks into engineered constructs, which improve the overall in vitro tissue development, organization, and functionality[ 4 , 6 , 7 , 8 , 9 , 10 ] and promote the construct‐host microvascular integration upon implantation.[ 11 , 12 , 13 , 14 ] Further efforts demonstrated the feasibility of controlling the engineered microvessels’ development and behavior by applying biochemical[ 15 , 16 , 17 ] and mechanical[ 18 , 19 , 20 ] stimuli or simply changing the cellular microenvironment.[ 11 , 21 , 22 ] Although these studies represent a significant advancement in the microvascular engineering field, they focus on the micrometric scale without addressing how to surgically integrate the engineered tissues directly to the host's vascular tree.[ 23 ] On the other hand, researchers have created implantable artificial vessels at the meso scales (≈1 mm diameter and larger) to replace damaged vessels,[ 24 , 25 ] create artificial bypasses,[ 26 , 27 ] or for repeated cannula insertion during hemodialysis.[ 28 , 29 ] However, many of these studies have not considered the vessels’ interactions with the surrounding tissues, do not use the engineered vessels for in vitro prevascularization of engineered constructs, and their fabrication methods were mainly suitable for creating simple elongated tubular structures.
In this work, we sought to apply 3D bioprinting to combine the microscales and mesoscales in our efforts to engineer functional vascular structures. To achieve this, we bioprinted self‐assembling microvascular networks in an ECM environment, which then connected to the interior of a larger implantable tubular Vascular scaffold (VascFold, for simplification). Thus, the microvessels could receive nutrients from the VascFold and distribute them within the printed hydrogel, resulting in a fully engineered tissue flap, which are tissue units containing a functional hierarchical vascular network. To accomplish this, we designed and 3D printed a water‐soluble mold in which we poured a polymer‐in‐solvent solution. We then lyophilized and washed out the mold to obtain the VascFold, a porous hollow tube with designed radial fenestrations (Figure 1 ‐i).[ 30 ] Separately, we 3D printed alternating layers of recombinant human collagen methacrylate (rhCollMA) containing ECs and SCs together in one layer and a tissue‐specific cell type in the other (Figure 1‐ii). The resulting construct included a circular space to accommodate the VascFold. After we assembled the construct, we cultured it for three days, allowing the printed cells to start the microvascular self‐assembly process (Figure 1‐iii). The cell‐embedded rhCollMA undergoes substantial shrinkage because of this process, which tightly anchored the gel around the VascFold and its fenestrations. Later, we seeded ECs on the inner surface of the VascFold to mimic the natural vascular endothelium. The ECs then proliferated on the inner surface, creating an endothelium‐like structure, and connected to the vascular network located within the outer printed rhCollMA (Figure 1‐iv), enabling nutrient transportation from the VascFold into the surrounding tissue, resulting in the formation of a functional hierarchical vessel network (VesselNet). Finally, using microsurgical techniques, the entire construct was implanted as an engineered tissue flap in a rat model by anastomosing the VascFold lumen directly to the femoral artery (Figure 1‐v).
Figure 1.
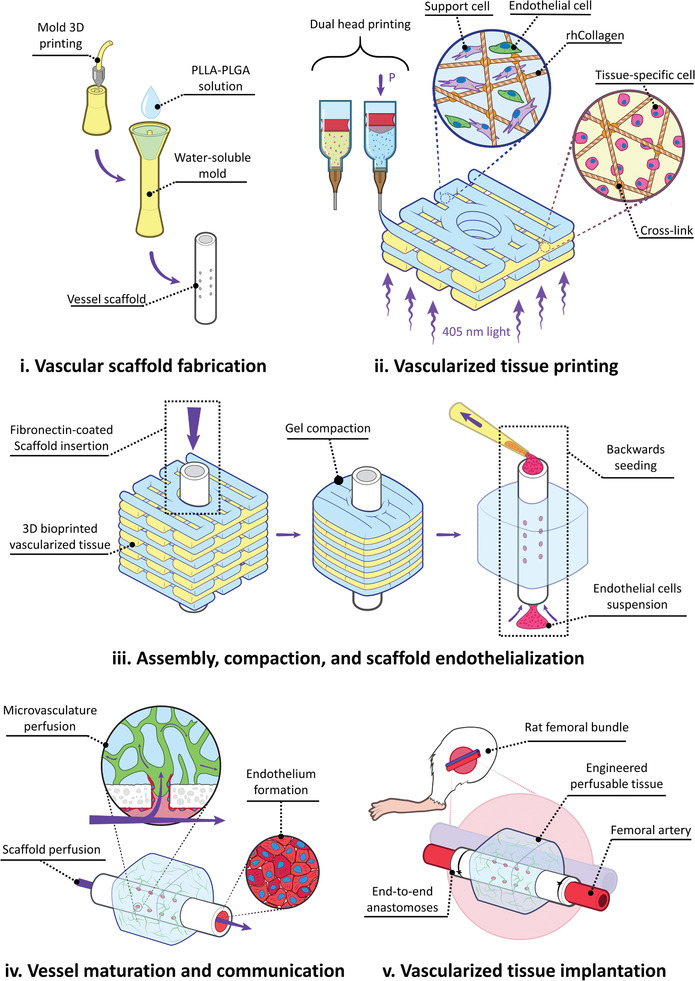
Experiment flow for fabricating and implanting perfusable vascularized tissues. i) Water‐soluble molds are 3D printed, filled with a PLLA‐PLGA polymer solution, lyophilized, and washed away, resulting in tubular fenestrated scaffolds (VascFold). ii) Recombinant human collagen methacrylate (rhCollMA) is used as a bioink to bioprint vascularized tissues. A dual head extrusion system is used to fabricate intercalated rhCollMA layers containing support and endothelial cells or tissue‐specific cells. iii) Immediately after printing, a fibronectin‐coated VascFold is inserted into the printed tissue channel and cultured for two days. The cells in the rhCollMA start organizing into functional tissues, exerting forces that compact the gel. The compaction stabilizes the printed rhCollMA around the scaffold and covers its side fenestrations. Then, endothelial cells are seeded into the VascFold lumen by applying negative pressure. iv) The full construct was cultured for up to 1 week. The spontaneously formed vessels in the hydrogel communicate with the endothelium through the scaffold fenestrations, enabling the microvasculature perfusion. v) After in vitro culture, the constructs are implanted on a rat femoral artery model by performing microsurgery to anastomose the VascFold ends with the femoral artery.
2. Results
2.1. rhCollMA Bioink for 3D Bioprinting Microvascular Networks
The first step in building the engineered vascularized tissue flap was to design a suitable ECM environment. Currently, there is extensive research regarding biological materials that can support the vasculogenesis phenomenon.[ 31 ] We selected recombinant human collagen I methacrylate (rhCollMA) as the ECM environment for its outstanding biological functionality and the additional capabilities of photopolymerization and tunability provided by the methacrylate groups.[ 32 , 33 ] Because we aimed to develop a bioink supporting extensive spontaneously‐formed microvascular networks, we used vascular network parameters as indicators to select the most suitable hydrogel (Figure S1, Supporting Information). We performed all vascularized hydrogel experiments with 1:3 ZsGreen‐ECs:SCs in rhCollMA hydrogels with lithium phenyl‐2,4,6‐trimethylbenzoylphosphinate (LAP) as the photoinitiator. We selected human adipose microvascular endothelial cells (HAMECs) as our ECs and dental pulp stem cells (DPSCs) as our SCs due to their ease of isolation from a patient and their capability of forming rich microvascular networks in‐vitro.[ 22 , 34 , 35 ] We chose the gel concentration by comparing vascular development in hydrogel plugs made with 3, 5, and 8 mg mL−1 rhCollMA. Networks in the 5 mg mL−1 hydrogels extended and developed more than those in the 8 mg mL−1 hydrogels (Figure S2, Supporting Information), whereas 3 mg mL−1 hydrogels drastically shrunk after two days, with no observable vessel networks formations (data not shown). To enhance the rhCollMA vascularization, we examined the effects of mixing a poly(ethylene oxide) (PEO) solution into the hydrogel formulation. The PEO has been shown to function as a porogen by generating pores that improve cell viability in 3D printed gelatin‐methacrylate (GelMA) hydrogels.[ 36 ] SEM images revealed the increased porosity of the rhCollMA hydrogels prepared with 0.8% porogen (+porogen) compared to those with no porogen (‐porogen, Figure 2A). As expected, the networks in +porogen gels presented a higher total vessel length than networks in the ‐porogen hydrogels at days 3 and 7 (Figure 2B). Furthermore, by day 7, the +porogen hydrogel had more interconnected vascular networks than ‐porogen hydrogels, indicated by the significantly fewer vessel endpoints.[ 37 ] After optimizing the rhCollMA bioink formulation, we verified its mechanical properties and suitability for tissue printing. We performed a stress–strain compression testing on these hydrogels (Figure 2C, left). The average elastic modulus of the rhCollMA hydrogel was 32.4 ± 0.9 kPa; a modulus value that is within the range of native human vascularized tissues.[ 38 ] We also performed rheological testing on the rhCollMA hydrogels using an oscillatory strain‐rate controlled rheometer. After performing a strain‐sweep measurement for the hydrogel's linear viscosity regime (Figure 2C, center), we chose a 1% strain to further perform time sweep rheology measurements. Under oscillation, the hydrogel precursor (i.e., pre‐gel) behaved as a viscous fluid until photoinitiation, which facilitated a cross‐linking reaction that culminated when the shear modulus reached its plateau value (Figure 2C, right). The resultant viscoelastic solid exhibited a storage modulus (G′) two orders of magnitude higher than the loss modulus (G″). With these results, we determined that the rhCollMA hydrogel possessed highly suitable biological and physical characteristics for producing engineered vascular tissues.
Figure 2.
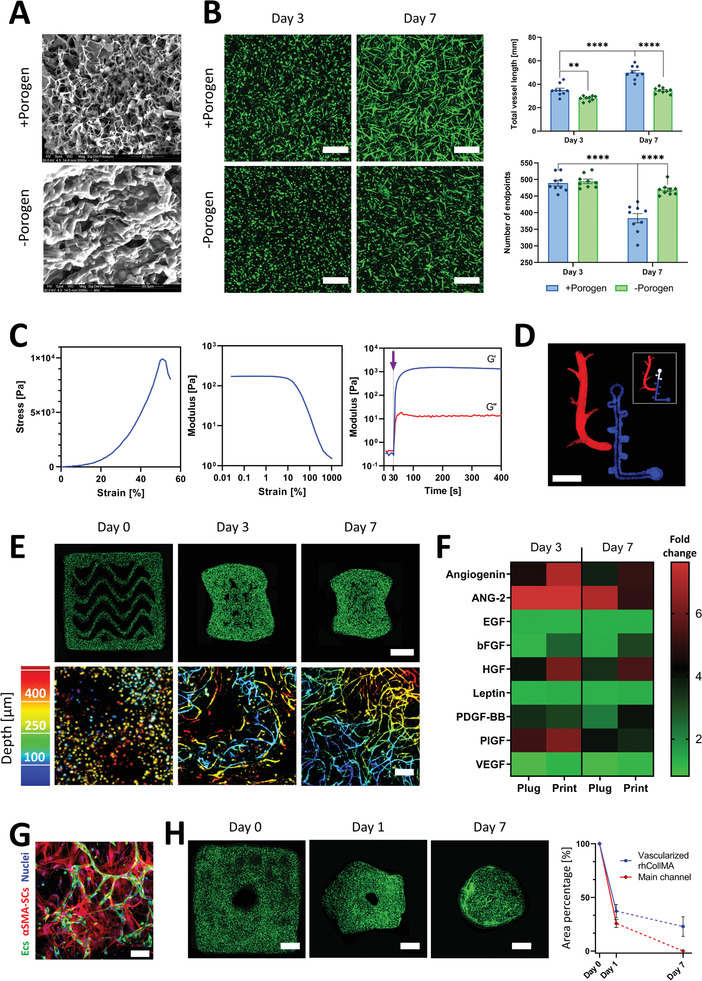
Bioink calibration and characterization for 3D bioprinting of functional self‐assembling vascular networks. A) Representative SEM images of rhCollMA with (top) and without (bottom) PEO porogen. B) Left, vessel spontaneous formation in ‐porogen and +porogen rhCollMA plugs at days 3 and 7 (scale bar: 200 µm). Right, total vessel length and number of vessel endpoints quantification for –porogen and +porogen plugs for days 3 and 7. C) Rheological and mechanical characterization for the bioink formulation: left, characteristic stress–strain curve during compression tests of a rhCollMA plug; center, linear viscoelastic range assessment using an amplitude sweep test; right, storage modulus G′ and loss modulus G″ curves for the bioink before and after cross‐linking. The purple arrow indicates the time of initialization of the photopolymerization process. (n > 3) D) Confocal image of a bioprinted Levenberg Lab logo using a multi‐material rhCollMA bioprinter with two fluorescent polystyrene microsphere colors; the insert shows the actual logo design (scale bar: 5 mm). E) Confocal images of vessel network formation in 3D bioprinted constructs at days 0, 3, and 7. The top row shows the full construct in which the ZsGreen‐ECs (green) organize and form vessels (scale bar: 1 mm). The bottom row shows higher magnification confocal images of the engineered vessel networks using a depth color‐coded projection (scale bar: 200 µm). F) Heatmap comparing secreted cytokines during the vascularization process by vessels forming within bioink plugs and 3D bioprinted structures. Values are calculated as fold change from day 1. G) Representative confocal image of formed vessels (ZsGreen‐ECs, green) and αSMA‐expressing SCs (red) at day 7, evidencing SCs recruitment and differentiation, an indication of vessel network maturation and stabilization. H) Bioprinted tissues present significant compaction during vessel formation. Left, confocal images of a bioprinted vascularized construct presenting a designed main channel and the subsequent gel shrinkage during one week of culture. Right, quantification of the total printed area and main channel area shrinkage as a percentage of their original total area. By day 7, the main channel was not discernable (scale bar: 1 mm). The dashed lines represent a non‐quantified progression. For all experiments: n > 3, **p < 0.01, ****p < 0.0001.
To verify the bioink printability, we added fluorescent polystyrene microspheres to the pregel solution. Using two print heads with different microsphere colors, we printed the Levenberg Lab logo into a gelatin support bath,[ 39 ] which was removed after photocrosslinking the printed material (Figure 2D). The next step was to observe the vasculogenesis process after bioprinting ZsG‐ECs and non‐labeled SCs in rhCollMA hydrogels. We printed 4×4×1 mm structures with a layer thickness of 150 µm and a wavy pattern infill, and took low and high magnification confocal images at days 0, 3, and 7 (Figure 2E). As expected, the ECs showed a circular morphology immediately after printing (day 0), and the construct shape was unchanged from its design. After 3 days, the vessel formation process was evident, with sparse vessels resembling those present in bioink plugs. The overall construct shape showed a significant shrinkage, and the inner pattern was almost indistinguishable. By day 7, the pattern stripes merged almost completely, and the vessels formed rich interconnected networks that reached up to 550 µm deep from the hydrogel surface, over 2.5 times the diffusion limit (Figure 2E, bottom right). It is critical to clarify that the stated 550 µm depth is only related to the maximum achievable depth using our confocal microscopy setup, and it does not represent the actual coverage of the vascular network within the depth of the construct. As we confirmed the feasibility of printing vascular constructs, we studied the biochemical differences and similarities between printed vascular networks and their non‐printed counterparts (i.e., plug constructs). Therefore, we performed an angiogenic cytokine analysis for vessels in printed constructs and hydrogel plugs at days 3 and 7, which we quantified as the x‐fold changes from day 1 (Figure 2F). The results displayed a similar behavior for both systems, with no significant differences between any of the cytokine secretion when we compared both systems for the same time points, e.g., plug at day 3 versus printed at day 3 (Figure S3, Supporting Information). The lack of significant differences confirmed that vascular networks in printed constructs behave similarly to those in hydrogel plugs, which is a known experimental configuration for in vitro and in vivo angiogenic models (e.g., aortic ring assay).[ 40 , 41 , 42 ] We expected to see SCs expressing α‐smooth muscle actin (αSMA), indicating vascular maturation and stabilization.[ 43 ] On day 7, we observed the presence of αSMA‐expressing SCs (αSMA‐SCs) surrounding the spontaneously‐formed microvessel (Figure 2G). SCs in general, and αSMA‐SCs in particular, are associated with a high contractile activity,[ 44 ] which would explain why the vascularized printed constructs significantly shrink during culture, as shown in Figure 2E. Taking this shrinkage into consideration, we sought to test the feasibility of bioprinting prevascularized tissues with main perfusable channels that will preserve their lumen during culture. For this, we printed 6×6×2 mm structures with 1.5 mm diameter main channels and tracked the total construct area and main channel area for one week (Figure 2H). By day 1, the printed area was reduced to 37% of the original area, whereas the main channel area shrunk to 25%. By day 7, the printed area shrunk to 23% of the original area, and the main channel was indiscernible. These results indicated that creating vascularized hydrogel constructs from rhCollMA with perfusable channels that would retain their shape was unlikely. Furthermore, it would be virtually impossible to implant them in direct anastomosis to a host vessel due to the hydrogel deformations during culture. Due to these critical shortcomings, we focused on preserving a perfusable lumen in the vascularized hydrogels by including a vessel‐mimicking scaffold that would provide mechanical support for the tissue.
2.2. Polymeric Tubular Scaffold as an Engineered Mesoscale Vessel
Figure 3A shows the vessel‐like scaffold's fabrication process (VascFold) using a soluble negative mold approach.[ 30 ] We printed the mold using the water‐soluble material butanediol vinyl alcohol copolymer (BVOH; Figure 3A‐i). We designed the mold (Figure 3A‐ii) in the shape of a tube (2 mm inner diameter) with a concentric rod (900 µm diameter), communicated by perpendicular thinner rods (220 µm diameter), and a usable length of 18 mm (Figure S4, Supporting Information). We filled the mold's funnel using a 7% PLLA‐PLGA in dioxane solution and centrifuged it for 1 min at 2000 g to force the viscous solution into the narrower areas (Figure 3A‐iii). After filling the mold, we froze it at −80°C and lyophilized it overnight, removing the solvent from the frozen solution and only leaving a porous polymeric structure (Figure 3A‐iv). After lyophilization, we dissolved the BVOH mold in water (Figure 3A‐v), resulting in a polymeric tubular scaffold with side fenestrations (Figure 3A‐vi). Figure S5 (Supporting Information) shows a BVOH mold and a finished VascFold next to a 1 NIS coin for size comparison. To characterize the resulting scaffold, we scanned it using microcomputed tomography imaging (µCT) and scanning electron microscopy (SEM) imaging. Figure 3B shows the resemblance between macroscope photographs and µCT images of the scaffold side and inner structure, although the µCT images provided greater detail of the scaffold walls. Noticeably, the outer wall presented a ridged repeating pattern, and the fenestrations were not circular; these discrepancies between design and scaffold arose from inherent 3D printing accuracy limitations. SEM images revealed the VascFold central lumen and side fenestrations in greater detail (Figure 3C‐i,iii), and the high wall porosity observed from the outer surface, and a transversal cut plane (Figure 3C‐ii,iv). The wall bulk had a high mean porosity (94.62 ± 0.97%) and represented 56% of the scaffold's void volume, whereas the side fenestrations and the main lumen represented 27% and 17%, respectively (Figure 3D). These results indicate a sizeable free volume within the scaffold walls, potentially enhancing cell infiltration, nutrient diffusion, tissue regeneration, and remodeling.[ 45 , 46 ] Additionally, the VascFold longitudinal wall stiffness was 4.5 ± 0.8 kPa (Figure 3E), indicating a relatively soft and flexible scaffold, which was advantageous for scaffold manipulations during microsurgical implantation.
Figure 3.
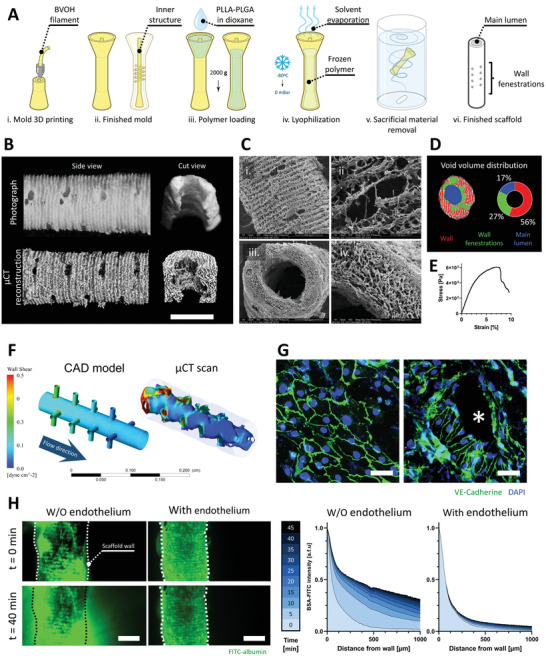
VascFold fabrication, characterization, and endothelialization for creating mesoscale vessel‐like constructs. A) Schematic explaining the VascFold fabrication process: i) A mold is printed using water‐soluble BVOH material, resulting in ii) a hollow cylindrical shape with a central rod connected to the outer wall by several perpendicular rods. iii) Using a centrifuge, the mold is filled with a 7% PLLA‐PLGA in dioxane solution, iv) frozen at −80 °C and then lyophilized, which removes the solvent from the solution, leaving a porous polymeric scaffold within the mold. v) The construct is then washed in DDW, vi) resulting in the finished scaffold with a central lumen and wall fenestrations. B) Representative side view and longitudinal cut view photographs and µCT reconstructions of a scaffold segment (scale bar: 1 mm). C) SEM images showing (i) the side fenestrations, (ii) the outer wall porosity, (iii) the main lumen, and (iv) the wall bulk porosity. D) Left, representative µCT pore segmentation of a scaffold transversal section containing wall fenestrations. Right, VascFold void volume distribution. E) Representative stress‐strain curve for the porous scaffold wall under axial stretching. F) Wall shear stress simulations of 50 µL min−1 medium flow within the CAD model and the actual inner VascFold surface using µCT imaging reconstruction (the outer wall can be seen grayed out). G) Under low flow (50 µL min−1), ECs form an endothelium‐like organization on the VascFold lumen wall), observable by the intercellular adhesion protein VE‐Cad (green; nuclei in blue). On areas with low wall shear stress, ECs show a cobblestone organization (left), while in areas next to the fenestrations (denoted by an asterisk), the ECs align in the direction of the flow (right; scale bar: 50 µm). H) Left, confocal images of FITC‐albumin diffusing freely through VascFold walls without endothelium over time but remaining entrapped in the lumen of endothelialized VascFolds (scale bar: 1 mm). Right, quantification of FITC‐albumin fluorescent intensity as a function of the distance from the scaffold wall. Darker shades of blue indicate later time steps after the FITC‐albumin perfusion started (n > 3).
Producing a tube‐like structure is only the first step to mimicking the native vessels, which consists of three concentric layers: tunica adventitia, tunica media, and tunica intima. The adventitia and media compositions vary, while the intima is similar for all vessels, with a basal membrane and an endothelium. The endothelium is a continuous single‐ECs layer that lines all vessels in the body. Blood circulating through vessels exert wall shear stress forces (WSS) on the native endothelium, affecting the ECs’ behavior.[ 47 ] Hence, studying these forces can provide a further understanding of the endothelium within the engineered vessel. For this, we used the finite element analysis (FEA) to observe the WSS distribution in the scaffold lumen during medium perfusion. Figure 3F shows the WSS for the VascFold µCT reconstruction when perfused with medium at 50 µL min−1. As expected, the highest WSS were present at the scaffold entrance and side fenestrations. Interestingly, when performing the same FEA to the CAD model used to design the scaffold, the WSS had a similar distribution to the µCT model, suggesting its potential use as an estimation tool for the WSS in the lumen. Next, we seeded ECs on the lumen wall (Figure S6, Supporting Information) and cultured the scaffolds under flow conditions for two days. Following the FEA results, the cells formed an endothelium with a cobblestone organization in low WSS areas (Figure 3F, left) and aligned with the flow direction next to fenestrations (Figure 3F, right), where the WSS was higher. This engineered endothelium response to WSS is in accordance with the literature on engineered and native endothelia.[ 48 ] After creating the VascFold endothelium, we sought to test its functionality. Because the native endothelium does not allow the passive diffusion of blood proteins to the tissues, we assessed the VascFold endothelium functionality by observing the outward diffusion of FITC‐albumin perfused into the scaffold lumen over time (Figure 3G, left; Movie S1: Supporting Information). Perfused VascFolds without endothelium revealed a steady accumulation of FITC‐albumin over time in the scaffold periphery, whereas endothelialized VascFolds did not show signs of passive albumin diffusion through the scaffold's walls (Figure 3G, right), suggesting that the engineered endothelium can mimic the function of its native counterpart. It is essential to clarify that the diffused albumin values are not necessarily physiological, and the experimental setup might limit its measurement, but the barrier function achieved by endothelializing the VascFold has an evident effect on the albumin diffusion. With this in mind, we then sought to combine the VascFold with 3D bioprinted capillaries to create a hierarchical vascular network.
2.3. Bioprinted Microvasculature and Scaffold Endothelium Cooperatively form Hierarchical Vascular Networks (VesselNet)
As we previously explained (Figure 1D), we combined 3D‐printed vascularized structures with fenestrated vessel‐like scaffolds to create a vascular hierarchy. First, we printed a vascularized construct with a size of 6×6×5 mm, a layer thickness of 150 µm, a grid pattern infill, and a central circular channel with a 2.5 mm diameter (Figure S7, Supporting Information). We then inserted a fenestrated scaffold within the printed channel (Figure S8, Supporting Information) and cultured it for two days. As expected, the printed gel contracted and constricted the VascFold, creating a mechanically stable assembly (Movie S2, Supporting Information). After gel compaction, we seeded the ECs in the scaffold lumen to create the endothelium. We used ZsGreen‐expressing ECs (green) for the printed structure and dTomato‐expressing ECs (red) for the endothelium to discern the cells’ original location after culturing for one week. Figure 4A‐i shows a side view of a cultured assembly, with the vascularized gel tightly attached to the endothelialized scaffold. The gel compaction is evidenced by its uneven geometry and the meniscus formations adjacent to the VascFold, marked by the arrowheads. The dashed line marks where we divided the assembly to gain access to the cut plane shown in Figure 4A‐ii, in which we can clearly observe the printed vascular networks in green (ZsGreen‐ECs) and the endothelium ECs in red (dTomato‐ECs), which also lined the fenestrations (marked by the arrowheads). Close‐up confocal images of an area outside the fenestrations (Figure 4A‐iii) reveal the integration between the printed and endothelium ECs, which formed microvessels in cooperation.
Figure 4.
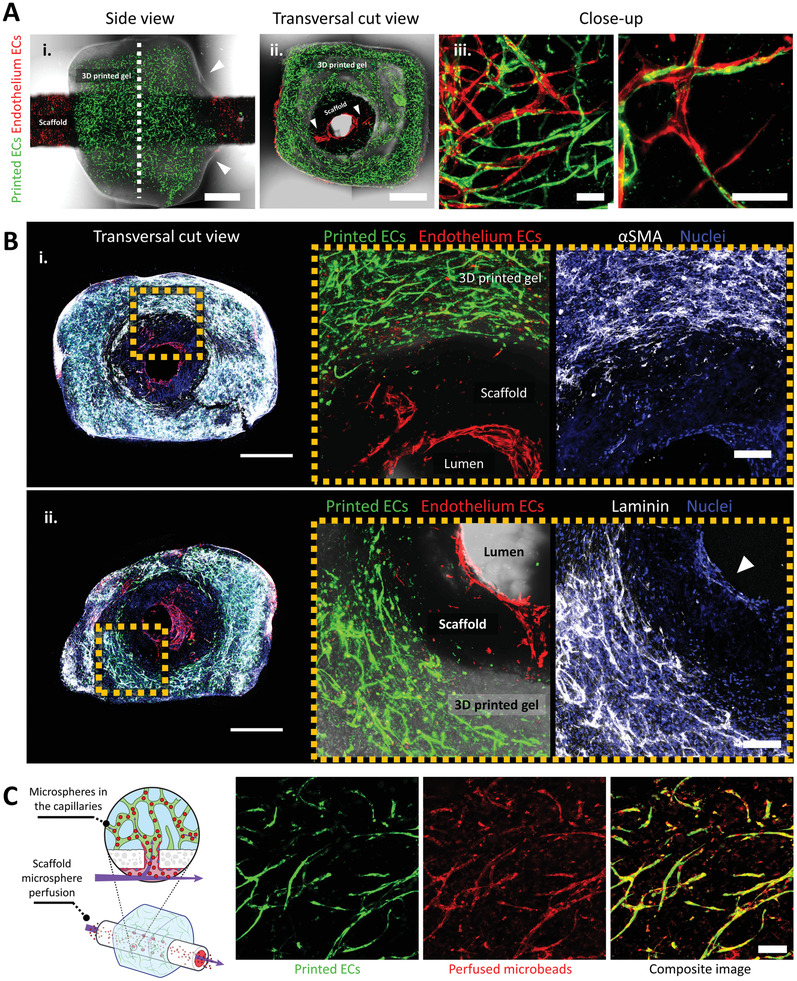
In vitro characterization of VesselNets. A) Representative confocal images of assembled vascular constructs with endothelium ECs (dTomato‐ECs, red) and printed microvascular ECs (ZsGreen‐ECs, green) after one week in culture. (i) Side view of a full construct; the arrowheads mark the hydrogel meniscus shape, indicating the tight gel constriction around the VascFold, and the dashed line marks where the construct was cut to observe the transversal planes. (ii) Transversal cut view of the construct; the arrowheads denote the endothelium ECs lining the wall fenestrations. (iii) Endothelium and printed ECs cooperate to form vascular networks which communicate the scaffold lumen with the surrounding hydrogel (scale bar (i, ii): 1 mm; scale bar (iii): 200 µm). B) Representative wide confocal images (left) and close‐up images (right) of characteristic vascular markers in cultured constructs; the dashed yellow squares indicate the close‐up region from the full construct images. (i) αSMA‐SCs (white) provide support to the printed microvasculature (ZsGreen‐ECs, green) but do not reach the endothelium ECs (dTomato‐ECs, red). (ii) The ECM protein laminin (white) is deposited by the printed microvasculature and the endothelium ECs (arrowhead; scale bar(left) 1 mm, scale bar(right): 200 µm). C) The printed microvasculature and VascFold form a hierarchical vascular construct after one week in vitro. Fluorescent microspheres (red) injected into the scaffold lumen appear in the hydrogel microvasculature (ZsGreen‐ECs, green), indicating the vascular continuity between the scaffold lumen and the surrounding vascular networks (scale bar: 100 µm).
Further investigation of the constructs revealed remarkable aspects of vascularization development (Figure 4B). First, the constructs showed interconnected vascular networks covering the whole printed gel thickness, from the scaffold external wall to the outer gel surface, with distances reaching over 700 µm (Figure S9, Supporting Information), three and a half times the diffusion limit.[ 49 ] The networks also showed vessel maturation within the construct, represented by αSMA‐SCs located in the microvessels vicinity (Figure 4B‐i).[ 43 ] The gel microvasculature and the endothelium exhibited laminin deposition (Figure 4B‐ii), a basal membrane glycoprotein present in all vascular tissues, which provides vascular stability, affects cell proliferation and migration, and contributes to the endothelium barrier function.[ 50 ] The presence of +αSMA‐SCs and laminin deposition surrounding the engineered microvessels are positive indications of native‐like networks. However, these biological phenomena are only some characteristics of a functional vasculature. A crucial aspect of assessing vascular functionality within our VesselNet construct was the feasibility of perfusing the microvasculature through the VascFold lumen. Thus, we injected a fluorescent microspheres solution into the VascFold lumen of fixed constructs and retrieved a portion of the printed hydrogel for imaging (Figure 4C). Confocal images of the perfused printed gel revealed microspheres trapped within the mature microvessels, giving unequivocal evidence of a functional vascular hierarchy.
2.4. Implanted Engineered Tissue Flaps Mimic Vascular Hierarchy In Vivo
An ideal engineered tissue should be vascularized so that the tissue can receive blood perfusion immediately upon implantation.[ 51 ] Hence, we designed our vascularized tissues to allow direct anastomosis with a host vessel, which would enable immediate blood perfusion into the tissue through the VesselNet. We sought to assess our approach's in vivo potential using a rat femoral artery model due to its similar diameter to the scaffold lumen diameter. Figure S10 (Supporting Information) provides a schematic explanation of the microsurgery process used for all in vivo experiments. Briefly, using a cuff microsurgery technique, we resected a femoral artery segment and encased the artery stumps with polyimide cuffs. Then, we introduced the cuffed stumps into the scaffold lumen and secured the attachment with circumferential sutures. In this way, the scaffold and stumps shared a lumen.
First, we wanted to investigate the VascFold capability to maintain normal blood flow over time with no surrounding printed gel. For this, we implanted endothelialized non‐fenestrated VascFolds (to avoid blood leaking to the scaffold periphery) and compared their blood flow to non‐endothelialized scaffolds and artery ligatures (Figure 5A). Since ligatures interrupt all flow through the artery, we included the non‐endothelialized scaffolds as a perfused control. After surgery, we measured the rats’ treated limb (right) blood flow and compared it to the healthy limb (left) on days 1, 4, 7, 10, and 14. Figure 5A shows representative laser speckle contrast imaging (LSCI) of rats from the three groups on day 14 on the left and blood flow quantification for the whole experiment on the right. Over time, the ligature group displayed a decreased blood perfusion, with maximum flows slightly over 50% of the healthy leg. Although the non‐endothelialized group appears to present higher values for the first week, it is important to note that two of the three animals did not survive after day 4. We speculate that the scaffold's lack of endothelium caused thrombus formations in its lumen (Figure S11, Supporting Information) that were later dislodged and traveled through the bloodstream, causing organ infarction.[ 52 ] On the other hand, the endothelialized scaffolds showed an average flow above 65% for all time points, reaching 85% after two weeks, evidencing the VascFold's capability to maintain blood flow over time.
Figure 5.
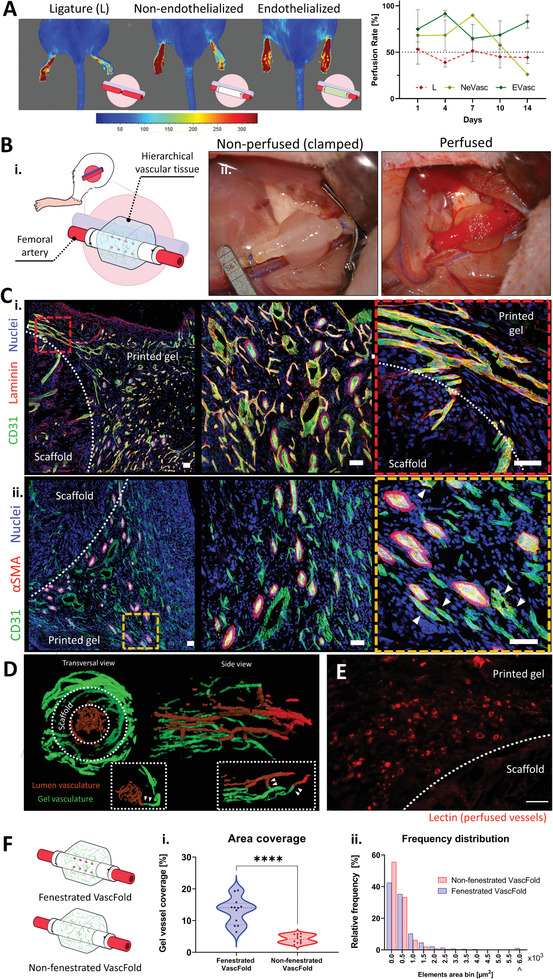
Engineered constructs promote host functional vasculature ingrowth upon anastomosis with rats’ femoral artery. A) Left, characteristic LSCI images of blood flow in hind limbs of rats implanted with endothelialized Vascfolds (Evasc), non‐endothelialized (NeVasc) VascFolds, and with ligated femoral arteries (L). The left hind limb remained untreated for all animals. Right, quantification of average blood flow over two weeks for all treatments (n = 3); the untreated left hind limb was used as a positive control (100% flow). B) Engineered flap anastomosis with rat femoral artery. i) Schematic of the implanted engineered flap. ii) Microsurgery photographs show the implanted construct before blood perfusion (left) and after blood perfusion (right). Blood can be seen filling the VascFold immediately after the clamps are released. C) Two weeks after implantation, explanted tissues are highly vascularized and present characteristic vascular network markers (all scale bars: 50 µm). The dashed white lines indicate the limit between the explanted scaffolds and hydrogels, and the red and orange dashed squares indicate the close‐up regions. i) The basal membrane protein laminin (red) is present in all vessels’ close vicinity within the printed gel, marked by the endothelial cell marker CD31 (green). Close‐up images show a variety of vascular phenotypes and well‐defined vessel lumina. ii) αSMA sheaths surround several vessels (CD31, green) but not all the vascular networks. Close‐up images reveal the presence of +αSMA cells next to non‐sheathed vessels (arrowheads). D) Transversal and side µCT images of perfused vascular networks in explanted engineered tissues. Vascular structures in the scaffold lumen (brown) communicate with vessels located in the surrounding hydrogel (green). The dashed line inserts show regions with vessel segments crossing the scaffold wall and communicating the luminal and external vasculatures. E) Representative confocal images of cryosections showing vascular networks stained with lectins (red), which were perfused before tissue explantation. The white dashed line indicates the limit between the scaffold and the hydrogel (scale bar: 50 µm). F) After two weeks in vivo, constructs with fenestrated VascFold display i) a higher hydrogel vascularized area percentage and ii) an element area distribution skewed toward larger areas, when compared with non‐fenestrated VascFold constructs (n > 3, ****p < 0,0001).
The next step was to implant the engineered flaps using an animal model (Figure 5B‐i). As part of the microsurgery procedure, we clamped both ends of the artery next to the stumps, completely stopping the implantation region's blood flow (Figure 5B‐ii, left). After successfully anastomosing artery and scaffold, we released the clamps, resulting in the implanted construct's instant perfusion, evidenced by the blood traveling through the scaffold and the red hue on the printed gel (Figure 5B‐ii, right). Since this work's focus was studying the implanted gel integration occurring through the VesselNet, we isolated all implanted constructs with 0.2 µm polycarbonate filters, preventing vascular ingrowth from surrounding tissues.[ 53 ] After two weeks in vivo, we retrieved the constructs and studied the vascularization within the implanted printed gel using cryosection, histological (H&E), and immunohistofluorescence techniques (Figure 5C and Figure S13: Supporting Information). The histological staining revealed structures resembling vessels (both axially and transversally cut), including several that seemed to contain red blood cells in their interior (Figure S13: Supporting Information, right). Furthermore, using confocal microscopy, we investigated the vascularization within the printed gel by staining the sections for host CD31, nuclei, and laminin (Figure 5C‐i) or αSMA (Figure 5C‐ii). Confocal images revealed a considerable host vasculature presence in the implanted flap, with vessels presenting diverse morphologies and sizes, including different diameters, circularities, and variable +αSMA sheaths; regardless, most vessels showed clear lumen evidencing their tube‐like structure (Figure S14, Supporting Information). To test the vasculature's functionality within the gel, we injected a contrast agent into the host bloodstream, explanted the engineered tissue, and imaged it using µCT imaging. The µCT scans allowed us to create a 3D visualization of the contrast‐filled vessels, which revealed an array of parallel vessels inside the VascFold lumen (brown) and a complex and interconnected vascular plexus within the printed gel (green; Figure 5D). Moreover, we found vessel segments connecting both vasculatures across the VascFold wall (white arrowheads). We hypothesize that these vessels were able to cross through the VascFold's fenestrations, a phenomenon we also observed in stained cryosections (Figure S12, Supporting Information). We successfully detected functional microvessels by injecting a lectin solution into the host bloodstream before tissue explantation. Figure 5E shows a confocal image of a tissue section with lectin‐stained capillaries (red). The presence of the injected materials in the explanted flap strongly evidenced the host's bloodstream perfusion of the engineered tissue. Furthermore, the perfused vascular networks within the explanted tissue were not localized only close to the fenestrations but were present in the whole explanted area (Figure S15, Supporting Information).
As mentioned above, we observed large vessels crossing from the scaffold lumen to the surrounding gel through the engineered fenestrations. Because current mesoscale scaffolds do not include designed fenestrations, we sought to investigate their effect on the host vascular ingrowth into the printed gel. We implanted assembled constructs with fenestrated (FS) and non‐fenestrated (nFS) scaffolds and, after 14 days in vivo, we compared the vascularized area percentage and the vessel size distribution in the surrounding engineered tissue of both groups (Figure 5G). FS constructs had 13.12 ± 4.22% of the surrounding tissue covered by vessels, while for nFS constructs, the vessel coverage was 4.16 ± 1.27%. Interestingly, capillaries (0–1250 µm2) accounted for 95.2% of the nFS vessels and 88.1% of the FS vessels. Larger vessels (1250–5750 µm2) represented 4.5% and 10.8% of the nFS and FS vessels, respectively. The number of largest vessels (>6000 µm2) was almost four times higher for the FS constructs (1.1%) compared to the nFS constructs (0.3%). These results evidence the positive impact the fenestrations have on the host vascular ingrowth and the implanted tissue's vasculature hierarchization.
2.5. Hierarchical Vascular Constructs Support iPSCs‐Derived Cardiomyocytes In Vitro
The vascular system's primary role is circulating blood to provide nutrients and gases to the body's organs.[ 1 ] Hence, a robust engineered vascular construct should be capable of nourishing tissue‐specific cells.[ 2 ] To achieve this, we combined the VesselNet with human cardiomyocytes and investigated their beating activity and response to a chronotropic drug. First, we differentiated GCaMP‐expressing induced pluripotent stem cells (iPSCs) into cardiomyocytes (GCaMP‐CM) following the monolayer‐based directed differentiation protocol published by Burridge et al.[ 54 ] We selected these transfected iPSCs because GCaMP is a calcium indicator that changes its fluorescence intensity in the presence of calcium ions (Ca+2) and, when expressed in the GCaMP‐CMs, it enables real‐time imaging of cardiac calcium transients.[ 55 ] Once the GCaMP‐CM started spontaneously contracting (10 to 14 days after starting the differentiation protocol), we suspended them in the bioink and bioprinted grid structures to assess the cardiomyocytes’ functionality following printing. After three days in culture, the 3D printed cardiomyocytes clustered in locally organized bundles as evidenced by the positive staining for cardiac sarcomeric marker troponin I (cTnI, Figure 6A). Movie S3 (Supporting Information) shows a contrast‐enhanced time‐lapse microscopy visualization of the GCaMP‐CMs contracting the gel bulk. A closer look of the printed cardiomyocytes (Movie S4, Supporting Information) revealed the GCaMP‐CMs continuous contractions and spreading calcium transients (green), showing the established electrophysiological connections between GCaMP‐CMs within the bioink after 3D printing (Figure 6B). After assessing the bioprinted GCaMP‐CMs sustained functionality, we created cardiac vascularized hierarchical constructs, following the steps described in Figure 1a–f. Briefly, we 3D printed alternating rhCollMA layers with vascularizing cells or GCaMP‐CMs and assembled them with VascFolds (Figure S16, Supporting Information). The printed geometry was a 6 × 6 × 5 mm prism, with a 2.5 mm diameter inner channel, a grid pattern, and a layer thickness of 150 µm. We cultured the assembly for two days until the gel shrunk and then seeded the endothelium ECs, resulting in engineered tissues as described in Figure 6C‐i. One week after printing, confocal images of the construct (Figure 6C‐ii) revealed developed vascular networks, with several vessels migrating toward the GCaMP‐CMs clusters, an indication of integration between the layers. The next step was to verify that the cardiomyocytes in the assembled construct were functional. A widespread method to test cardiomyocyte functionality is by observing the CM response to isoproterenol, a ß‐adrenergic agonist used to treat bradycardia.[ 56 , 57 ] To test the vascularized GCaMP‐CMs, we supplemented the assembled constructs’ media with 1 × 10−6 m isoproterenol and compared these (Iso) and non‐treated constructs (control) chronotropic responses. We used confocal line‐scan imaging of the GCaMP‐CMs’ calcium transients to quantify the cells beating rate and calcium transient decay time within the hierarchically vascularized structures (Figure S17 and Movie S5: Supporting Information). Figure 6D‐i shows representative Ca+2 transients of control and isoproterenol‐treated constructs, exhibiting an increased beating rate after isoproterenol stimulation. The average beating frequency for control constructs was 46.9 ± 16.5 beats‐per‐minute (BPM), while for isoproterenol‐treated constructs was 90.5 ± 20.0 BPM (Figure 6D‐ii). The decay time for control and iso constructs was 277.3 ± 96.3 and 215 ± 36.9 ms, respectively. These results reveal the proper functionality of the vascularized GCaMP‐CMs, which presented a chronotropic response similar to the native cardiac tissue.[ 58 ]
Figure 6.
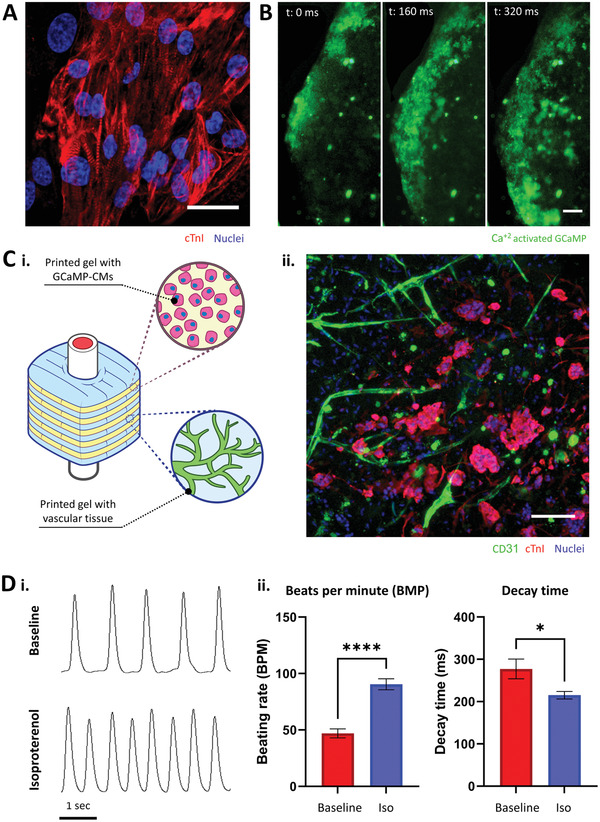
Hierarchical vasculature supports iPSCs‐derived cardiomyocytes. A) iPSC‐derived cardiomyocytes printed in rhCollMA cluster and display characteristic sarcomere striated patterns (cTnI, red; scale bar: 20 µm). B) Bioprinted GCaMP‐CMs spontaneously beat and develop electromechanical communications, evidenced by calcium transients (green) traveling distances over 1 mm (scale bar: 200 µm). C‐i) Schematic describing the assembled and cultured vascularized cardiomyocytes constructs. ii) Confocal image showing the presence of vascular networks (CD31, green) and cardiomyocytes (cTnI, red) after seven days in vitro (scale bar: 100 µm). D) Cardiomyocytes chronotropic behavior responses to isoproterenol. i) Representative line‐scan images showing the GCaMP‐CMs intracellular Ca+2 transients before and after isoproterenol was added to the culture medium. ii) Comparison of the average beats per minute and Ca+2 transients decay time between vascularized cardiac constructs in regular culture medium (baseline) and isoproterenol‐supplemented medium (iso). N > 4, *p < 0.05, ****p < 0.0001.
2.6. Creating Patient‐Specific VesselNets
One of the major drawbacks of current vascular scaffold fabrication approaches at the mesoscale is their geometry limitation. Most used techniques are limited to fabricating simple tubular hollow scaffolds with no designed fenestrations.[ 3 ] The tubular structure limits the engineered vessels’ applicability when more complex structures are required, e.g., replacing a bifurcated vessel. With this in mind, we sought to verify our novel approach's potential to fabricate patient‐specific complex geometries. Figure 7A describes the adapted workflow for creating personalized VesselNets: i) we obtained a right coronary artery 3D model created from actual CT contrast images,[ 59 ] ii) we used this model to fabricate a water‐soluble mold, iii) we created the bifurcated scaffold by filling the mold with a polymeric solution, lyophilizing the filled mold, and dissolving the water‐soluble material and, iv) we 3D printed a 40% w/v Pluronic F‐127 bioink around the scaffold regardless of its complex shape. It is critical to clarify that for this experiment, we used Pluronic F‐127 as our bioink due to several technical aspects: Pluronic is an easily printable hydrogel at high temperatures and liquid at low temperatures, making its polymerization reversible and the material reusable for several attempts; we could increase the visibility of the printed details by adding color to the bioink without interfering with its cross‐linking capabilities, as it would happen with the light‐based cross‐linking of CollMA; and, because this experiment is meant as a proof‐of‐concept with no printed cells, the selected bioink does not have to be biocompatible nor biodegradable. The resulting coronary VascFold, like the 3D model, was hollow, had an inlet, two outlets, and different curvatures for each branch, evidencing the complexity of the native vasculature (Figure 7B). To encase the scaffold in a hydrogel, we designed and printed a base following the coronary artery geometry, which could hold the VascFold in place. Then, we placed the VascFold on top of the base and printed the hydrogel over it with a specific design based on the coronary artery model shape (Movie S6, Supporting Information). As a result, we obtained a complex patient‐specific vascular scaffold entirely encased by a 3D bioprinted hydrogel (Figure 7C). Furthermore, Figure S18 (Supporting Information) reveals the possibility of creating the same patient‐specific coronary flap, albeit on a smaller scale, using our biocompatible CollMA bioink formulation. This experiment reveals the high versatility and potential of our proposed technique and represents an important step toward creating personalized implantable vascularized engineered tissues.
Figure 7.
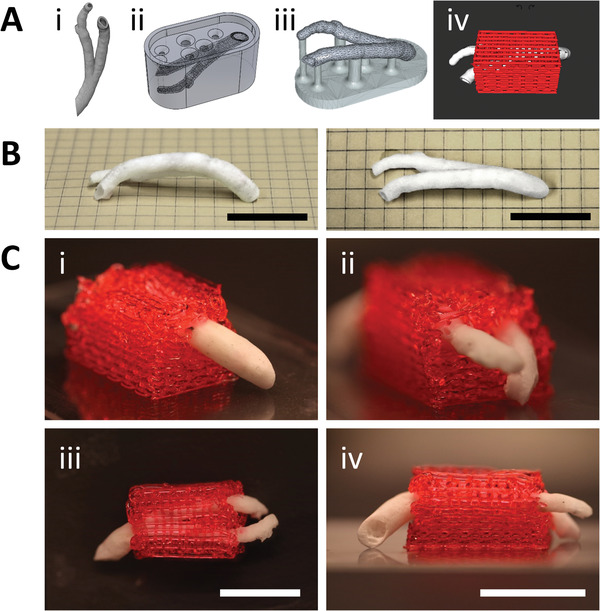
Coronary VascFold fabrication and integration with a 3D printed surrounding hydrogel structure. A) Diagram of the fabrication and integration processes. i) A medical CT image of a human right coronary artery was reconstructed as a CAD solid object. ii) The object was used to create a water‐soluble mold. iii) The mold was filled with the PLLA‐PLGA solution, lyophilized, and washed away, resulting in a hollow VascFold representation of the native coronary artery. iv) A 3D hydrogel was printed around the scaffold, surrounding the sidewalls and leaving the open ends accessible for perfusion. B) Side and top photographs of the coronary artery VascFold. C) Photographs showing the resulting hydrogel and native‐mimicking VascFold construct from i) the single‐entry side, ii) the dual‐exit side, iii) the top, and iv) the front. The pictures show the hydrogel fully encasing the scaffold (scale bar: 2 mm).
3. Discussion
The cardiovascular system is a fundamental biological trait that appeared over 600 million years ago and is still present in modern vertebrates and invertebrates alike.[ 60 ] Its tree‐like structure helps distribute gases and nutrients to tissues, allows cell waste removal, and plays a central role in the immune system.[ 1 ] Thus, engineering hierarchical vascularized tissues has been one of the main foci since the field's beginnings, and despite many recent advances, it remains an unsolved challenge to this day.[ 2 ] Current techniques for in vitro engineering of implantable vascularized tissues center on creating self‐assembled microvascular networks[ 9 , 11 , 13 , 22 , 61 , 62 ] or fabricating mesoscale vessel replacements,[ 24 , 25 , 26 , 27 ] but recreating the system's hierarchical structure was beyond these works’ scope. Ideally, a vascularized construct should be implanted in direct communication with the host's vascular tree, immediately perfusing an associated engineered microvascular network.[ 5 ]
In this work, we developed a novel approach that takes advantage of state‐of‐the‐art tissue engineering techniques and bridges between the previously separated micro‐ and mesoscale vasculatures. For this purpose, we combined 3D bioprinted microvessels with a vessel‐mimicking scaffold, resulting in fully vascularized implantable tissues.
3D bioprinting is a versatile technology that has gained broad attention for the advantages that it offers over traditional tissue engineering techniques. The key features responsible of moving increased efforts into 3D bioprinting is the possibility to deposit cells or biological inks (bioinks) in a controlled and automated fashion, the wide range of bioinks that can be used to create 3D structures (including the use of multiple bioinks simultaneously),[ 63 ] its capability to create complex organs with intricate microarchitectures (e.g., liver, cardiac tissue, kidney), and the technology's high replicability and repeatability.[ 64 , 65 ] A key aspect of bioprinting is selecting a bioink, a hydrogel consisting of polysaccharides or ECM proteins that can be tuned to support the target tissue's development and organization.[ 66 ] Among the ECM proteins, type I collagen stands out for being the most abundant in the human body (the main building block of most tissues in the body), providing anchorage sites for cells, and interacting with other ECM molecules and growth factors.[ 67 ] Moreover, recently developed recombinant techniques enable fabricating human type I collagen (e.g., human type I collagen production in tobacco plants),[ 33 ] which can then be processed to further its applications by, for example, adding methacrylate groups for photopolymerization.[ 32 ] In contrast, other widely used biomaterials for vascular tissue engineering (e.g., fibrin) lack mechanical stability[ 68 ] and must be extracted from animal or human sources.[ 69 ] However, the bioink's main material is not the only aspect to consider when creating vascularized tissues. Other factors, such as gel concentration (Figure S2, Supporting Information), significantly impact vascular development. We formulated the rhCollMA bioink to maximize vascular coverage and development, including a gel porogen that drastically increased network development (Figure 1B); despite its positive impact, gel porosity is often overlooked in the literature.[ 36 ] The rhCollMA bioink displayed suitable mechanical properties (Figure 1C), good printability (Figure 1D), and vascular networks in bioink constructs displayed an biochemical profile similar to those grown in gel plugs, a method used for angiogenic analysis (e.g., Matrigel plug angiogenesis assay, aortic ring assay)[ 40 , 41 , 42 ] (Figure 1F and Figure S3: Supporting Information). A prominent characteristic of vascularized hydrogels seldomly addressed in the literature is their tendency to shrink due to cell exerted forces (Figure 1E,H). In vascularized tissues, support cells (Figure 1D) exert strong forces that compact the gel they reside in, significantly affecting the surrounding cells, drastically deforming the gel's original shape and impeding the constructs’ implantability.[ 22 , 70 , 71 ] This is particularly relevant because the most common approach for engineering hierarchical vascular structures is creating vascularized gel bulks with large channels in lieu of perfusable vessels.[ 15 , 72 , 73 ] However, since shrinkage is not a fully predictable phenomenon, we believe studies analyzing the 3D printed hydrogel shrinkage due to cell‐exerted forces (either by computational modelling or real‐time microscopy imaging) could shed light on this fascinating matter.
It is worth mentioning that despite the cell‐exerted forces compacting the construct until no pattern is detected (resembling a non‐porous structure), 3D bioprinted hydrogels with designed porosity present higher cell viability and differentiation within the construct over other techniques such as casting hydrogel in molds or using thick gel plugs,[ 74 ] which usually create non‐porous structures that do not allow an easy flow of nutrients to the innermost areas of the construct. Some molding techniques may also provide porosity but achieving higher structure complexity and construct handling are more challenging than with bioprinting technologies.
We created highly porous vascular scaffolds (VascFolds) using 3D‐printed water‐soluble molds to provide mechanical support for the hydrogels during culture and facilitate the flap implantation (Figure 3A).[ 30 ] We printed the molds using a hydrophilic material, BVOH, and a standard plastic extrusion 3D printer, resulting in a cost‐effective technique with immense versatility. Furthermore, BVOH does not react with organic solvents, enabling the use of a wide variety of polymer solutions. This approach's main downside is the resolution limitations inherent to the printing technique,[ 75 ] resulting in micrometric discrepancies between the CAD model and the final scaffold (Figure 3B–E). Nonetheless, because 3D printing is evolving rapidly, we speculate that this issue will be solved shortly, either by improving extrusion printing accuracy or developing hydrophilic materials for more accurate technologies (e.g., stereolithography 3D printing). A higher print fidelity will also enable more accurate FEAs of relevant biological phenomena such as WSS and flow dynamics (Figure 3E), eliminate the need to fabricate and image the scaffold to obtain a CAD model, and facilitate the iterative design.[ 76 ] However, it is important stating that cells are capable of remodeling polymeric scaffolds (e.g., by exerting mechanical forces or excreting enzymes that degrade the polymer),[ 77 ] which might affect the significance of the mentioned fabrication fidelity. The complexity of remodeling is an important issue and should be furtherly studied in following works.
Within the VascFold's lumen, the seeded ECs organized into an endothelium‐like structure under flow conditions (50 µL min−1; Figure 3G). The ECs localized on the areas displaying the lowest values of WSS (Figure 3F,G) presented a cobblestone‐like organization, while ECs localized on areas with higher WSS (i.e., surrounding the wall fenestrations) adopted an elongated configuration corresponding with the flow direction. However, the computed WSS values (≈0.3–0.5 dyne cm−2) that promoted the ECs directionality within the VascFold are lower to the minimum needed WSS values reported by the bibliography (>3 dyne cm−2).[ 78 , 79 ] This should be furtherly explored to clarify the difference between the reported values and those obtained in our model.
The VascFold's engineered endothelium displayed a native‐like barrier function by preventing a FITC‐albumin solution from diffusing through the scaffold walls passively (Figure 3F). Furthermore, when we assembled the vascularized printed tissue and scaffold, the endothelium ECs sprouted into the gel and formed microcapillaries in cooperation with the printed ECs (Figure 4A). We speculate that this happens due to the hydrogel compaction and subsequent endothelium seeding, which lined the compacted gel covering the fenestrations, and the endothelium ECs sprouted into the gel. Many works have presented very similar occurrences in microfluidic devices,[ 80 , 81 ] gel channels,[ 61 , 82 ] or perforated scaffolds,[ 83 ] but, to our knowledge, this is the first time this phenomenon was observed in a system built using 3D printing technologies. Additionally, we successfully perfused the gel's vascular network by injecting a fluorescent microsphere solution into the scaffold lumen (Figure 4C), furtherly evidencing the communication between endothelium and microvasculature.
As a first step to assess the VascFold's capability to work as an artificial vessel, we surgically anastomosed endothelialized and non‐endothelialized VascFolds (EVasc and nEVasc, respectively; Figure 5A) to the femoral artery in a rat model using a microsurgery cuff technique[ 84 ] (Figure S9, Supporting Information), facilitated by the VascFold's mechanical stability. Throughout the experiment, the treated rats received heparin to reduce the risk of clot formation. Interestingly, the implanted EVascs remained patent for two weeks (the experiment's endpoint), while the nEVascs showed clot formation (Figure S11, Supporting Information) regardless of the heparin injections. This phenomenon might be explained by the findings described by Trindade et al.[ 85 ] suggesting that heparin binds to the endothelial cells’ ECM and promotes the synthesis and subsequent secretion of the antithrombogenic molecule heparan sulfate (HS) by the ECs. A later work by Onishi et al.[ 86 ] explains that the activity of the antithrombin (AT) is highly enhanced in the presence of heparin or HS. AT is a small protein that inhibits coagulation and binds to HS or heparan sulfate proteoglycans (HSPG) on the endothelial cells’ glycocalyx.
After proving the EVascs’ potential as an artificial vessel, we fabricated and cultured VesselNet constructs for a week and then anastomosed them with the femoral artery. Two weeks after implantation, the explanted printed hydrogel presented a rich host vasculature that grew from the scaffold lumen (Figure S12, Supporting Information). Interestingly, αSMA and laminin staining revealed three distinct vessel groups within the gel (Figure 5C): small capillary‐like vessels (< 40 µm diameter) with thin laminin layers, medium tubular vessels (40–60 µm diameter) with thick laminin and αSMA sheaths (arteriole‐like), and large vessels (> 60 µm diameter) with thin laminin layer, no αSMA sheath and amorphous lumen (venule‐like). The sprouting vessel variety raises the question of how sprouts from an artery reveal an apparent venous phenotype (a thin layer of αSMA‐SCs surrounding the vessel wall and a large lumen with low circularity)[ 87 ] and if it is similar to the vein arterialization process occurring during cardiac embryogenesis;[ 88 ] a specialized study could shed light on this matter.
We successfully verified the communication between the hydrogel vasculature and the host bloodstream by perfusing the rats’ tail vein with a µCT contrast material and lectin solution before construct explantation, which we then detected in the explanted tissue's microvasculature (Figure 5D,E). Although the µCT scan provided an excellent 3D visualization of the vasculature, the images did not show the microvasculature we observed in the cryosections, likely related to the contrast agent's high viscosity.[ 89 ] However, the water‐based lectin solution reached and stained the smallest capillaries, revealing blood perfusion regardless the vessel diameter. These combined results suggest the existence of a proper vessel perfusion from the host vascular tree into the vessels within the engineered flap, and no blood leakage through the fenestrations. In case of blood leakage, the µCT agent contrasts would not display well‐defined vessel structures but amorphous blobs of material, while the lectin solution would not be able to bind to the ECs glycocalyx[ 90 ] (resulting in no specific staining), since the vessel walls are impermeable to blood.
Additionally, we confirmed the positive vessel ingrowth effect of the scaffold wall fenestrations, which enhanced the hydrogel vascularized area and allowed larger vessels to infiltrate into the gel (Figure 5F). Based on our information, this is the first work to show the in vivo vascularization of a fully engineered flap directly anastomosed to a host artery.
The heart is the organ responsible for pumping blood through the cardiovascular system. As a biological pump, it contracts its muscle walls between 60 and 100 times per minute. The cardiac muscle contractions consume a high amount of energy that requires a constant blood supply to the active cardiomyocytes, provided by a rich vasculature.[ 1 ] The native heart heterogenous population consists of cardiomyocytes, fibroblasts, support cells, and endothelial cells, with varying ratios depending on the heart region.[ 91 ] Recently, 3D bioprinting gained attention for its potential to recreate the heart's complex structure.[ 39 , 73 , 92 ] Thus, we created VesselNets with iPSCs‐derived cardiomyocytes as an in vitro proof‐of‐concept to verify the engineered vasculature's capability to support tissue‐specific cells (Figure 6C‐i). Vascularized iPSC‐CMs displayed the characteristic sarcomere striated pattern and spontaneous beating (Figure 6C‐ii and Movies S4 and S5: Supporting Information). Moreover, when we supplied the tissues with the chronotropic drug isoproterenol, the cardiomyocytes showed a native‐like response,[ 56 , 58 ] displaying increased beating rates and faster calcium transients decay times (Figure 6D), demonstrating our approach's potential for creating vascularized cardiac tissues.
However, cell composition and functionality are just among the many complexities of recreating the native heart. The cardiac vasculature consists of a network of vessels with intricate bifurcating geometries. The most explored mesoscale scaffold configuration is a straight tube, which is useful for specific applications (e.g., creating artificial bypass vessels[ 25 , 26 ]) but cannot recreate native‐like geometries, reducing its application in tissue engineering. The water‐soluble molding technique allowed us to create coronary‐mimicking scaffolds based on actual medical CT images (Figure 7A,B). Due to the shape of the coronary scaffold, we could not assemble the construct as we did previously, so we adapted our approach to overcoming this shortcoming. For this, we assembled the construct by designing an ad hoc hydrogel base in which we placed the scaffold and then continued printing on top of it, successfully encasing the VascFold within the printed bioink (Figure 7C and Movie S6: Supporting Information). In this work, we have shown the feasibility of creating native‐like structures for implantation; nonetheless, we believe that furtherly researching some aspects of the model could improve the concept and make it even more versatile and robust. One of these aspects is the simultaneous fabrication of the polymeric vessel within the 3D‐printed hydrogel. Unfortunately, the fabrication process of the VascFold cannot be combined with living cells due to the use of organic solvents and the need for lyophilization. However, printing the main vessel using reinforced or highly stiff hydrogels that would not deform under cells‐exerted forces could allow the simultaneous fabrication of the main vessel and the surrounding vascularized tissue, eliminating the need to fabricate the VascFold separately and subsequently assemble the scaffold into the hydrogel.
We believe that this work represents a step forward in the personalized medicine field as a combination of human collagen bioinks, VascFolds designed from medical images, and iPSC‐derived cells has the potential to create patient‐specific implantable vascularized tissues. Future works will focus on assessing the scalability and translatability of the proposed approach by adapting the VesselNet constructs to be implanted in larger animal models (e.g., pigs).
4. Conclusion
The current work presents a novel and versatile technique for creating hierarchical vascular tissues that can be implanted as tissue flaps. We successfully perfused a 3D bioprinted microvascular network through a mesoscale vessel scaffold, recreating the vasculature's tree‐like organization. After construct implantation in a rat femoral artery model, the host vasculature covered the hydrogel with functional microvessels. The proposed approach is a versatile and adaptable technique that may cement a new path toward fully lab‐grown patient‐specific tissues.
5. Experimental Section
Cell Culture
Human adipose microvascular endothelial cells (HAMECS or ECs; ScienceCell), ZsGreen‐ECs, and dTomato‐ECs were grown in ECM bullet kit (ScienceCell). Dental pulp stem cells (DPSCs or SCs; Lonza) were cultured in low‐glucose Dulbecco's modified Eagle medium (DMEM, Gibco), supplemented with 10% fetal bovine serum (Hyclone), 1% non‐essential amino acids (Gibco), 1% GlutaMAX (Gibco), and 1% penicillin‐streptomycin‐nystatin solution (Biological Industries).
rhCollMA Bioink Preparation
An acidic rhCollMA solution (10 × 10−3 m HCl, >90% degree of functionalization) was generously provided by CollPlant (Rehovot, Israel). A 10X phosphate buffer (PB) was prepared by adding 5.495 g Na2HPO4, 1.55 g NaH2PO4, and 30 mg NaCl in 50 mL DDW; a 1X PB was obtained by diluting 1:9 the 10X PB in DDW. A collagen solution was prepared by neutralizing the stock solution with 10% 10X PB of the desired final volume and 1X PB until reaching 10 mg mL−1. The porogen‐photoinitiator solution (LAP‐PEO) was prepared by mixing 0.2% lithium phenyl‐2,4,6‐trimethylbenzoylphosphinate (LAP, Sigma‐Aldrich) and 1.6% polyethylene oxide (8000 kDa PEO, Polysciences) in phenol red‐free DMEM (Gibco). To obtain a 5 mg mL−1 rhCollMA bioink, the collagen stock solution and LAP‐PEO solution were mixed in a 1:1 ratio. rhCollMA with no porogen were prepared in the same way but with no PEO added.
Sacrificial Mold 3D Printing
The sacrificial mold was designed in SolidWorks 2019 (Dassault Systèmes SE), as shown in Figure S4 (Supporting Information). The mold STL file was sliced using Prusa Slic3r MK3 and printed with a 3D printer Prusa i3 MK3S (Prusa) with a 250 µm nozzle. Molds were printed using butanediol vinyl alcohol copolymer (BVOH, Verbatim), and, after printing, they were kept in a vacuum desiccator until use to avoid moisture.
Vascular Scaffold Fabrication
A 7% w/v PLLA‐PLGA solution was prepared by dissolving 3.5 g of PLLA (Polysciences) and 3.5 g of PLGA (Boehringer Ingelheim) solution in 100 mL dioxane under constant stirring until homogeneous. The solution was then added to the molds’ funnel, which then were placed in 2 mL eppendorfs and centrifuged for 3 min at 2000 g. The filled molds were put at −80°C for a minimum of an hour and lyophilized overnight for solvent removal. After lyophilization, the molds were washed in warm water until no traces of BVOH were observed on the scaffolds. The scaffolds were then frozen and lyophilized again (to remove moisture) and kept in a vacuum desiccator until use.
Bioprinting of Cellular and Acellular rhCollMA Constructs
2·106 ZsGreen‐ECs and 6·106 were suspended in 5 mg mL−1 bioink and pipetted into a 10 mL printing cartridge (Nordson) with a 27 G needle (CML Supply). For acellular printing, only the bioink was loaded into the cartridge. The cartridge was loaded into an LT‐head of a 3D‐Bioplotter Manufacturer Series bioprinter (EnvisionTEC GmbH). The CAD object was created in Solidworks 2019, sliced using Perfactory (EnvisionTEC GmbH) and converted into a print file using the VisualMachines software (EnvisionTEC GmbH), which was also used to control the bioprinter. The chosen printing parameters were 0.1 bar extrusion pressure, 15 mm s−1 extruder movement speed, 10°C stage temperature, and a layer height of 150 µm. All rhCollMA bioprinting was performed using the FRESH technique using LifeSupport sacrificial bath (FluidForm), which was prepared per the indications of the manufacturer. After printing, the printed rhCollMA in the support bath was cross‐linked by illuminating for 30 s using a 405 nm light source with an intensity of 3 mW cm−2. After cross‐linking, the rhCollMA in the support bath was placed at 37°C and 5% CO2 until the bath was dissolved. Then, the printed constructs were either cultured in EC medium or assembled with VascFolds. When dual‐head printing was performed, the same process was used for both printing heads.
Vascular Scaffold Endothelialization
Before use, scaffolds were disinfected in 70% ethanol for a minimum of 30 min and washed in PBS. The scaffolds were coated with a 50 µg mL−1 human fibronectin solution in PBS (Sigma‐Aldrich) and cultured at 37°C for 1 h. dTomato‐ECs were trypsinized and suspended in ECs medium with a concentration of 107 cells mL−1. For each scaffold, a 20 µL droplet of the ECs suspension was placed on a hydrophobic surface, and the scaffold was gently held in direct contact with the droplet using forceps. Then, a pipette tip was placed on the opposing end of the scaffold, and the droplet was aspirated through the scaffold, filling its lumen (Figure S6, Supporting Information). The seeded scaffold was placed in 1.5 mL eppendorfs and cultured under axial rotation for 1 h at 37°C and 5% CO2. Then, the endothelialized scaffolds were placed in 12‐well plates with ECs medium for two days.
Hierarchical Vascular Structures)VesselNet) Fabrication
rhCollMA vascular tissues were printed as explained above. Immediately after bioprinting, a fibronectin‐coated scaffold was placed in the main channel of the printed structure, and the whole construct was cultured for two days in ECs medium. After two days, the cells compacted the gel, firmly anchoring it to the scaffold. Then, the scaffold lumen was endothelialized using dTomato‐ECs, as explained above. The constructs were cultured in EC medium for a week.
Animal Experiments
All animal procedures below were approved and conducted under the supervision of the Technion Pre‐Clinical Research Authority (PCRA Technion, approval ethics no. 058‐05‐20). Endothelialized VascFolds, acellular VascFolds, and VascFold (with or without perforations) cultured with rhCollMA were used for the implantation experiments. Three days prior to implantation, male Sprague‐Dawley rats (275–350 g) received a daily dose of cyclosporine administered subcutaneously (10 mg kg−1 body weight; Novartis). On the day of surgery, rats were anesthetized using 3% isoflurane inhalation according to the institution's SOP, and the analgesic buprenorphine was administered subcutaneously (0.03 mg kg−1 body weight), as well as the prophylactic anticoagulation drug heparin (200 IU Kg−1). All procedures involving vascular anastomosis were performed using a microsurgical dissection microscope (Tivato700, Zeiss). A cuff technique was used to facilitate micro‐surgical anastomosis between VascFold scaffolds and native vessels as previously described[ 93 ] with some modifications (Figure S9, Supporting Information). First, the right hind limb of each animal was thoroughly cleaned using septal scrub and prepared for surgery. An oblique 2 cm long incision was made along the concavity between the abdominal wall and the leg. Next, the inguinal fat pad was dissected while preserving its vascular stalk and laterally reflected. The common femoral vessels were exposed with the artery dissected out of its vascular sheath, extending from the inguinal ligament proximally and up to the epigastric vessel's origin distally. Any arterial branches were cauterized and dissected. The artery was clamped, ligated, and cut at the approximate midpoint, and two 1.25 mm × 1.25 mm 25 G polyimide cuffs were applied to both vessel stumps. The artery ends were everted over the cuffs and stabilized using 8‐0 silk sutures. The cuffed vessel ends were then inserted into the VascFolds’ lumen openings and fixed in place using 6‐0 polypropylene sutures. At this point, a 0.2 µm polystyrene membrane filter was wrapped around the anastomosed VascFold to isolate the implanted construct from adjacent soft tissues. Then, the clamps were removed, restoring blood perfusion, and once hemostasis was verified, soft tissues were sutured layer by layer. For the control group, both artery ends were ligated and left without implantation. In all procedures, the overlying fat was sutured using 6‐0 polypropylene sutures, and the skin was sutured using 5‐0 PGA absorbable sutures. Animals recovered on a heating pad in an oxygenated cage and were monitored daily for 14 days. For postoperative pain and animal management, rats received tramadol (0.1% in drinking water) for three days after the surgery. Anticoagulation therapy with heparin was maintained for the remainder of the experiment (200 IU Kg−1 daily subcutaneous injection).
Statistical Analysis
Quantitative results were obtained from a minimum of four independent samples for in vitro studies and a minimum of three samples (rats) for in vivo experiments. Statistical analyses were performed using Prism 9. Two‐group comparisons were made using’ Student's t‐test, and multiple‐group comparisons were made using one‐way or two‐way analysis of variance (ANOVA) with ‘Tukey's test for posthoc analysis. All data is presented as mean ± SEM, and significance levels are as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Additional methods can be found in the Supporting Information (SI).
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Supplemental Video 1
Supplemental Video 2
Supplemental Video 3
Supplemental Video 4
Supplemental Video 5
Supplemental Video 6
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Programme (grant agreement no. 818808. This work was supported by The Czech Science Foundation through Project No. GA18‐05510S. The authors thank the Technion's Pre‐clinical Research Authority for the assistance with animal care, Janette Zavin for histological processing, and Galia Ben David for her constant invaluable assistance.
Szklanny A. A., Machour M., Redenski I., Chochola V., Goldfracht I., Kaplan B., Epshtein M., Simaan H. Yameen, Merdler U., Feinberg A., Seliktar D., Korin N., Jaroš J., Levenberg S., 3D Bioprinting of Engineered Tissue Flaps with Hierarchical Vessel Networks (VesselNet) for Direct Host‐To‐Implant Perfusion. Adv. Mater. 2021, 33, 2102661. 10.1002/adma.202102661
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Levick J. R., in An Introduction to Cardiovascular Physiology (Ed: Levick J. R.), Elsevier, London: 1991, p. 1. [Google Scholar]
- 2. Novosel E. C., Kleinhans C., Kluger P. J., Adv. Drug Delivery Rev. 2011, 63, 300. [DOI] [PubMed] [Google Scholar]
- 3. Song H. H. G., Rumma R. T., Ozaki C. K., Edelman E. R., Chen C. S., Cell Stem Cell 2018, 22, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levenberg S., Rouwkema J., Macdonald M., Garfein E. S., Kohane D. S., Darland D. C., Marini R., van Blitterswijk C. A., Mulligan R. C., D'Amore P. A., Langer R., Nat. Biotechnol. 2005, 23, 879. [DOI] [PubMed] [Google Scholar]
- 5. Rouwkema J., Khademhosseini A., Rivron N. C., van Blitterswijk C. A., Khademhosseini A., Trends Biotechnol. 2016, 34, 733.27032730 [Google Scholar]
- 6. Caspi O., Lesman A., Basevitch Y., Gepstein A., Arbel G., Habib I. H. M., Gepstein L., Levenberg S., Circ. Res. 2007, 100, 263. [DOI] [PubMed] [Google Scholar]
- 7. Osaki T., Sivathanu V., Kamm R. D., Sci. Rep. 2018, 8, 5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sobrino A., Phan D. T. T., Datta R., Wang X., Hachey S. J., Romero‐López M., Gratton E., Lee A. P., George S. C., Hughes C. C. W., Sci. Rep. 2016, 6, 31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kutys M. L., Polacheck W. J., Welch M. K., Gagnon K. A., Koorman T., Kim S., Li L., McClatchey A. I., Chen C. S., Nat. Commun. 2020, 11, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palikuqi B., Nguyen D.‐H. T., Li G., Schreiner R., Pellegata A. F., Liu Y., Redmond D., Geng F., Lin Y., Gómez‐Salinero J. M., Yokoyama M., Zumbo P., Zhang T., Kunar B., Witherspoon M., Han T., Tedeschi A. M., Scottoni F., Lipkin S. M., Dow L., Elemento O., Xiang J. Z., Shido K., Spence J. R., Zhou Q. J., Schwartz R. E., De Coppi P., Rabbany S. Y., Rafii S., Nature 2020, 585, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Landau S., Szklanny A. A., Yeo G. C., Shandalov Y., Kosobrodova E., Weiss A. S., Levenberg S., Biomaterials 2017, 122, 72. [DOI] [PubMed] [Google Scholar]
- 12. Ben‐Shaul S., Landau S., Merdler U., Levenberg S., Proc. Natl. Acad. Sci. USA 2019, 116, 2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baranski J. D., Chaturvedi R. R., Stevens K. R., Eyckmans J., Carvalho B., Solorzano R. D., Yang M. T., Miller J. S., Bhatia S. N., Chen C. S., Proc. Natl. Acad. Sci. USA 2013, 110, 7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mirabella T., Macarthur J. W., Cheng D., Ozaki C. K., Woo Y. J., Yang M. T., Chen C. S., Nat. Biomed. Eng. 2017, 1, 0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nguyen D.‐H. T., Stapleton S. C., Yang M. T., Cha S. S., Choi C. K., Galie P. A., Chen C. S., Proc. Natl. Acad. Sci. USA 2013, 110, 6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X., Phan D. T. T., Sobrino A., George S. C., Hughes C. C. W., Lee A. P., Lab Chip 2016, 16, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S., Lee H., Chung M., Jeon N. L., Lab Chip 2013, 13, 1489. [DOI] [PubMed] [Google Scholar]
- 18. Rosenfeld D., Landau S., Shandalov Y., Raindel N., Freiman A., Shor E., Blinder Y., Vandenburgh H. H., Mooney D. J., Levenberg S., Proc. Natl. Acad. Sci. USA 2016, 113, 3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landau S., Ben‐Shaul S., Levenberg S., Adv. Sci. 2018, 5, 1800506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S., Chung M., Ahn J., Lee S., Jeon N. L., Lab Chip 2016, 16, 4189. [DOI] [PubMed] [Google Scholar]
- 21. Raghavan S., Nelson C. M., Baranski J. D., Lim E., Chen C. S., Tissue Eng., Part A 2010, 16, 2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szklanny A. A. A., Debbi L., Merdler U., Neale D., Muñiz A., Kaplan B., Guo S., Lahann J., Levenberg S., Adv. Funct. Mater. 2019, 30, 1901335. [Google Scholar]
- 23. Gui L., Niklason L. E., Curr. Opin. Chem. Eng. 2014, 3, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dahl S. L. M., Kypson A. P., Lawson J. H., Blum J. L., Strader J. T., Li Y., Manson R. J., Tente W. E., DiBernardo L., Hensley M. T., Carter R., Williams T. P., Prichard H. L., Dey M. S., Begelman K. G., Niklason L. E., Sci. Transl. Med. 2011, 3, 68ra9. [DOI] [PubMed] [Google Scholar]
- 25. Luo J., Qin L., Zhao L., Gui L., Ellis M. W., Huang Y., Kural M. H., Clark J. A., Ono S., Wang J., Yuan Y., Zhang S. M., Cong X., Li G., Riaz M., Lopez C., Hotta A., Campbell S., Tellides G., Dardik A., Niklason L. E., Qyang Y., Cell Stem Cell 2020, 26, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gutowski P., Gage S. M., Guziewicz M., Ilzecki M., Kazimierczak A., Kirkton R. D., Niklason L. E., Pilgrim A., Prichard H. L., Przywara S., Samad R., Tente B., Turek J., Witkiewicz W., Zapotoczny N., Zubilewicz T., Lawson J. H., in Journal of Vascular Surgery, Vol. 72, Mosby Inc., Maryland Heights: 2020, p. 1247. [DOI] [PubMed] [Google Scholar]
- 27. Jiang Y. C., Jiang L., Huang A., Wang X. F., Li Q., Turng L. S., Mater. Sci. Eng., C 2017. [Google Scholar]
- 28. Kirkton R. D., Santiago‐Maysonet M., Lawson J. H., Tente W. E., Dahl S. L. M., Niklason L. E., Prichard H. L., Sci. Transl. Med. 2019, 11, eaau6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lawson J. H., Glickman M. H., Ilzecki M., Jakimowicz T., Jaroszynski A., Peden E. K., Pilgrim A. J., Prichard H. L., Guziewicz M., Przywara S., Szmidt J., Turek J., Witkiewicz W., Zapotoczny N., Zubilewicz T., Niklason L. E., Lancet 2016, 387, 2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaplan B., Merdler U., Szklanny A. A., Redenski I., Guo S., Bar‐Mucha Z., Michael N., Levenberg S., Biomaterials 2020, 251, 120062. [DOI] [PubMed] [Google Scholar]
- 31. Bittner K. R., Jiménez J. M., Peyton S. R., Adv. Healthcare Mater. 2020, 9, 1901459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stein H., Wilensky M., Tsafrir Y., Rosenthal M., Amir R., Avraham T., Ofir K., Dgany O., Yayon A., Shoseyov O., Biomacromolecules 2009, 10, 2640. [DOI] [PubMed] [Google Scholar]
- 33. Shoseyov O., Posen Y., Grynspan F., Tissue Eng., Part A 2013, 19, 1527. [DOI] [PubMed] [Google Scholar]
- 34. Guo S., Redenski I., Landau S., Szklanny A., Merdler U., Levenberg S., Adv. Healthcare Mater. 2020, 9, 2000974. [DOI] [PubMed] [Google Scholar]
- 35. Redenski I., Guo S., Machour M., Szklanny A., Landau S., Kaplan B., Lock R. I., Gabet Y., Egozi D., Vunjak‐Novakovic G., Levenberg S., Adv. Funct. Mater. 2021, 2008687, 10.1002/adfm.202008687. [DOI] [Google Scholar]
- 36. Ying G. L., Jiang N., Maharjan S., Yin Y. X., Chai R. R., Cao X., Yang J. Z., Miri A. K., Hassan S., Zhang Y. S., Adv. Mater. 2018, 30, 1805460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blinder Y. J., Freiman A., Raindel N., Mooney D. J., Levenberg S., Sci. Rep. 2015, 5, 17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guimarães C. F., Gasperini L., Marques A. P., Reis R. L., Nat. Rev. Mater. 2020, 5, 351. [Google Scholar]
- 39. Lee A., Hudson A. R., Shiwarski D. J., Tashman J. W., Hinton T. J., Yerneni S., Bliley J. M., Campbell P. G., Feinberg A. W., Science 2019, 365, 482. [DOI] [PubMed] [Google Scholar]
- 40. Mikirova N. A., Casciari J. J., Riordan N. H., J. Angiog. Res. 2010, 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Iqbal F., Szaraz P., Librach M., Gauthier‐Fisher A., Librach C. L., Stem Cell Res. Ther. 2017, 8, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Malinda K. M., Methods Mol. Biol. 2009, 467, 287. [DOI] [PubMed] [Google Scholar]
- 43. Potente M., Gerhardt H., Carmeliet P., Cell 2011, 146, 873. [DOI] [PubMed] [Google Scholar]
- 44. Talele N. P., Fradette J., Davies J. E., Kapus A., Hinz B., Stem Cell Rep. 2015, 4, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Battiston K. G., Cheung J. W. C., Jain D., Santerre J. P., Biomaterials 2014, 35, 4465. [DOI] [PubMed] [Google Scholar]
- 46. Nam J., Huang Y., Agarwal S., Lannutti J., Tissue Eng. 2007, 13, 2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buchanan C. F., Verbridge S. S., Vlachos P. P., Rylander M. N., Cell Adhes. Migr. 2014, 8, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Levesque M. J., Nerem R. M., J. Biomech. Eng. 1985, 107, 341. [DOI] [PubMed] [Google Scholar]
- 49. Jain R. K., Au P., Tam J., Duda D. G., Fukumura D., Nat. Biotechnol. 2005, 23, 821. [DOI] [PubMed] [Google Scholar]
- 50. Hallmann R., Horn N., Selg M., Wendler O., Pausch F., Sorokin L. M., Physiol. Rev. 2005, 85, 979. [DOI] [PubMed] [Google Scholar]
- 51. Yang G., Mahadik B., Choi J. Y., Fisher J. P., Prog. Biomed. Eng. 2020, 2, 012002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Opitz F., Schenke‐Layland K., Cohnert T. U., Starcher B., Halbhuber K. J., Martin D. P., Stock U. A., Cardiovasc. Res. 2004, 63, 719. [DOI] [PubMed] [Google Scholar]
- 53. van Gastel N., Stegen S., Eelen G., Schoors S., Carlier A., Daniëls V. W., Baryawno N., Przybylski D., Depypere M., Stiers P. J., Lambrechts D., Van Looveren R., Torrekens S., Sharda A., Agostinis P., Lambrechts D., Maes F., Swinnen J. V., Geris L., Van Oosterwyck H., Thienpont B., Carmeliet P., Scadden D. T., Carmeliet G., Nature 2020, 579, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Burridge P. W., Matsa E., Shukla P., Lin Z. C., Churko J. M., Ebert A. D., Lan F., Diecke S., Huber B., Mordwinkin N. M., Plews J. R., Abilez O. J., Cui B., Gold J. D., Wu J. C., Nat. Methods 2014, 11, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tallini Y. M., Ohkura M., Choi B. R., Ji G., Imoto K., Doran R., Lee J., Plan P., Wilson J., Xin H. B., Sanbe A., Gulick J., Mathai J., Robbins J., Salama G., Nakai J., Kotlikoff M. I., Proc. Natl. Acad. Sci. USA 2006, 103, 4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zeng H., Wang J., Clouse H., Lagrutta A., Appl. In Vitro Toxicol. 2020, 6. [Google Scholar]
- 57. Ronaldson‐Bouchard K., Ma S. P., Yeager K., Chen T., Song L., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak‐Novakovic G., Nature 2018, 556, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goldfracht I., Efraim Y., Shinnawi R., Kovalev E., Huber I., Gepstein A., Arbel G., Shaheen N., Tiburcy M., Zimmermann W. H., Machluf M., Gepstein L., Acta Biomater. 2019, 92, 145. [DOI] [PubMed] [Google Scholar]
- 59.NIH, Phantom Coronary Artery Models | NIH 3D Print Exchange. 2019.
- 60. Monahan‐Earley R., Dvorak A. M., Aird W. C., J. Thromb. Haemostasis. 2013, 11, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Osaki T., Sivathanu V., Kamm R. D., Biomaterials 2018, 156, 65. [DOI] [PubMed] [Google Scholar]
- 62. Song W., Chiu A., Wang L.‐H., Schwartz R. E., Li B., Bouklas N., Bowers D. T., An D., Cheong S. H., Flanders J. A., Pardo Y., Liu Q., Wang X., Lee V. K., Dai G., Ma M., Nat. Commun. 2019, 10, 4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gopinathan J., Noh I., Biomater. Res. 2018, 22, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chia H. N., Wu B. M., J. Biol. Eng. 2015, 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Murphy S. V., Atala A., Nat. Biotechnol. 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- 66. Skardal A., Devarasetty M., Kang H. W., Mead I., Bishop C., Shupe T., Lee S. J., Jackson J., Yoo J., Soker S., Atala A., Acta Biomater. 2015, 25, 24. [DOI] [PubMed] [Google Scholar]
- 67. Di Lullo G. A., Sweeney S. M., Körkkö J., Ala‐Kokko L., San Antonio J. D., J. Biol. Chem. 2002, 277, 4223. [DOI] [PubMed] [Google Scholar]
- 68. Lesman A., Koffler J., Atlas R., Blinder Y. J., Kam Z., Levenberg S., Biomaterials 2011, 32, 7856. [DOI] [PubMed] [Google Scholar]
- 69. Watts D. C., in Biomaterials Science: An Introduction to Materials: Third Edition, Elsevier Inc., New York: 2013, p. 889. [Google Scholar]
- 70. Rivron N. C., Vrij E. J., Rouwkema J., Le Gac S., van den Berg A., Truckenmuller R. K., van Blitterswijk C. A., Proc. Natl. Acad. Sci. USA 2012, 109, 6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pagan‐Diaz G. J., Ramos‐Cruz K. P., Sam R., Kandel M. E., Aydin O., Saif T. M. A., Popescu G., Bashir R., Proc. Natl. Acad. Sci. USA 2019, 116, 25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kolesky D. B., Homan K. A., Skylar‐Scott M. A., Lewis J. A., Proc. Natl. Acad. Sci. USA 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Noor N., Shapira A., Edri R., Gal I., Wertheim L., Dvir T., Adv. Sci. 2019, 6, 1900344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fedorovich N. E., Kuipers E., Gawlitta D., Dhert W. J. A., Alblas J., Tissue Eng., Part A 2011, 17, 2473. [DOI] [PubMed] [Google Scholar]
- 75. Karakurt I., Lin L., Curr. Opin. Chem. Eng. 2020, 28, 134. [Google Scholar]
- 76. Zahedmanesh H., Lally C., Biomech. Model. Mechanobiol. 2012, 11, 363. [DOI] [PubMed] [Google Scholar]
- 77. Dhandayuthapani B., Yoshida Y., Maekawa T., Kumar D. S., Int. J. Polym. Sci. 2011, 2011, 1389. [Google Scholar]
- 78. Vion A. C., Perovic T., Petit C., Hollfinger I., Bartels‐Klein E., Frampton E., Gordon E., Claesson‐Welsh L., Gerhardt H., Front. Physiol. 2021, 11, 1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sonmez U. M., Cheng Y.a‐W., Watkins S. C., Roman B. L., Davidson L. A., Lab Chip 2020, 20, 4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wong K. H. K., Chan J. M., Kamm R. D., Tien J., Annu. Rev. Biomed. Eng. 2012, 14, 205. [DOI] [PubMed] [Google Scholar]
- 81. Fong A. H., Romero‐López M., Heylman C. M., Keating M., Tran D., Sobrino A., Tran A. Q., Pham H. H., Fimbres C., Gershon P. D., Botvinick E. L., George S. C., Hughes C. C. W., Tissue Eng., Part A 2016, 22, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Song H. G., Lammers A., Sundaram S., Rubio L., Chen A. X., Li L., Eyckmans J., Bhatia S. N., Chen C. S., Adv. Funct. Mater. 2020, 30, 2003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Debbi L., Zohar B., Shandalov Y., Levenberg S., bioRxiv 2020, 2020.07.07.190900. [Google Scholar]
- 84. Zhang B., Montgomery M., Chamberlain M. D., Ogawa S., Korolj A., Pahnke A., Wells L. A., Masse S., Kim J., Reis L., Momen A., Nunes S. S., Wheeler A. R., Nanthakumar K., Keller G., Sefton M. V., Radisic M., Nat. Mater. 2016, 15, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Trindade E. S., Oliver C., Jamur M. C., Rocha H. A. O., Franco C. R. C., Bouças R. I., Jarrouge T. R., Pinhal M. A. S., Tersariol I. L. S., Gouvêa T. C., Dietrich C. P., Nader H. B., J. Cell. Physiol. 2008, 217, 328. [DOI] [PubMed] [Google Scholar]
- 86. Onishi A., St Ange K., Dordick J. S., Linhardt R. J., Front. Biosci. 2016, 21, 1372. [DOI] [PubMed] [Google Scholar]
- 87. Ovalle W. K., Nahirney P. C., Netter's Essential Histology, 3rd ed. (Ed: Netter F. H.), Elsevier, New York: 2008. [Google Scholar]
- 88. Su T., Stanley G., Sinha R., D'Amato G., Das S., Rhee S., Chang A. H., Poduri A., Raftrey B., Dinh T. T., Roper W. A., Li G., Quinn K. E., Caron K. M., Wu S., Miquerol L., Butcher E. C., Weissman I., Quake S., Red‐Horse K., Nature 2018, 559, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ghanavati S., Yu L. X., Lerch J. P., Sled J. G., J. Neurosci. Methods 2014, 221, 70. [DOI] [PubMed] [Google Scholar]
- 90. Beyer S., Blocki A., Cheung M. C. Y., Wan Z. H. Y., Mehrjou B., Kamm R. D., Life 2021, 11, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lim G. B., Nat. Rev. Cardiol. 2020, 17, 759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lee S., Sani E. S., Spencer A. R., Guan Y., Weiss A. S., Annabi N., Adv. Mater. 2020, 32, 2003915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schleimer K., Grommes J., Greiner A., Jalaie H., Kalder J., Langer S., Koeppel T. A., Jacobs M., Kokozidou M., J. Visualized Exp. 2012, e4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supplemental Video 1
Supplemental Video 2
Supplemental Video 3
Supplemental Video 4
Supplemental Video 5
Supplemental Video 6
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


