Abstract
Iterative Bleaching Extends multipleXity (IBEX) is a versatile method for highly multiplexed imaging of diverse tissues. Based on open science principles, we created the IBEX Knowledge-Base, a resource for reagents, protocols and more, to empower innovation.
The power and pitfalls of multiplexed tissue imaging
Multiplexed tissue imaging is a powerful approach for studying the spatial organization and cellular composition of intact tissues at single cell resolution. The last decade has seen a rapid expansion in the development and commercialization of advanced spatial biology techniques. These methods include technologies that probe RNA molecules using imaging-based approaches or spatial barcoding techniques. In addition, proteins may be targeted with antibodies applied to thin sections as well as thick tissue volumes using a variety of multiplexed antibody-based imaging approaches[1]. These methods vary in the optical resolution, tissue volume, and number and type of targets (RNA, protein, or both) that can be imaged in a single tissue preparation[2]. As with any rapidly evolving field, the technical specifications of a given method are constantly improving, enhancing the value of these approaches. Multiplexed tissue imaging has been especially informative for visualizing and quantifying cell-cell interactions, identifying rare cells, evaluating spatial relationships among cells, and providing new insights into higher level tissue organization. These technologies have been foundational for the construction of single cell atlases and the study of naturally occurring cancers using samples from clinical trials and experimental models of disease. Despite their considerable promise, several challenges prevent the widespread adoption of spatial biology technologies. First and foremost, the majority of these methods require expensive equipment and consumables that may not be available in all research settings. Extensive expertise is also needed to optimize tissue collection, validate reagents, acquire images, and analyze data[1].
IBEX: An open and versatile method for highly multiplexed imaging
To provide a robust and widely usable solution for highly multiplexed imaging, we developed the Iterative Bleaching Extends multipleXity (IBEX) method[3, 4]. This method achieves high parameter imaging (>65 markers) in a single tissue section (5–30 μm) using cyclic rounds of antibody labeling and dye inactivation. Following image acquisition, individual images are registered, pixel-to-pixel, into one composite image using open-source software[5]. Since December 2020[3], IBEX has been adopted by dozens of international scientists from fields as diverse as immunology, developmental biology, comparative anatomy, and cancer biology. Furthermore, IBEX has been used to evaluate tissues obtained from humans, mice, non-human primates, canines, and zebrafish. More importantly, these advances reflect the community’s ability to both adopt and expand the original IBEX method to different applications and laboratory settings. As a result, we have collectively overcome common challenges, developed workflows for automated imaging and immunolabeling, and incorporated new reagents to acquire high quality imaging datasets for a variety of studies.
Motivation and design for IBEX Knowledge-Base
From the beginning, we have strived to share knowledge related to each stage of the multiplexed imaging workflow: sample preparation, antibody selection, antibody validation, panel design, image alignment, image processing, data analysis, and publication of results via open data repositories and scholarly publications. This effort was born out of a desire to reduce the significant time, resources, and expertise required to implement IBEX and other multiplexed imaging techniques[1, 6]. To achieve this aim, we established the IBEX Knowledge-Base, a central resource for reagents, protocols, data, software, and information related to IBEX and other spatial biology methods including non-iterative, standard tissue imaging (Multiplexed 2D Imaging), IBEX imaging with Opal dyes (Opal-plex), thick volume imaging achieved through clearing enhanced 3D (Ce3D)[7], and integration of Ce3D and IBEX (Ce3D-IBEX) to obtain highly multiplexed imaging of thick samples (>300 μm). We anticipate the number of methods supported by the IBEX Imaging Community to grow and include unique extensions of the protocol for the detection of novel chemistries and nucleic acid probes.
The IBEX Knowledge-Base is designed around three facets common to FAIR data and open-source software development: a source/data repository, a static website, and an archive for source data (Yaniv et al., in preparation). The first facet, the IBEX Knowledge-Base GitHub repository, stores source data and scripts used to generate the static website (https://github.com/IBEXImagingCommunity/ibex_imaging_knowledge_base). Furthermore, the GitHub ecosystem provides support for automatic data validation, website creation and hosting, issue reporting, as well as a discussion forum. These latter two utilities provide an open, transparent venue for discussing issues and questions related to the IBEX Knowledge-Base and multiplexed tissue imaging, respectively. There are several benefits to this approach such as increased trust in the data, reduced time in answering the same or similar questions in closed communication channels, and access to a wider pool of expertise. The second facet, the static website (https://ibeximagingcommunity.github.io/ibex_imaging_knowledge_base), is automatically generated with every update to the IBEX Knowledge-Base via a GitHub pull request. The IBEX Imaging Community website was designed to provide a user-friendly platform for browsing the current stage of knowledge and, unlike scientific publications, is constantly evolving with each contribution. The third and final facet of the IBEX Knowledge-Base is publication of an authoritative, citable, archival version through the generalist repository Zenodo[8]. By publishing through Zenodo, the IBEX Knowledge-Base is assigned a persistent digital object identifier, providing a mechanism for members to be rewarded with authorship for their contributions. In contrast to the static website, updates to the Zenodo entry are not continuous but are issued as versioned releases following a significant update.
Guiding principles
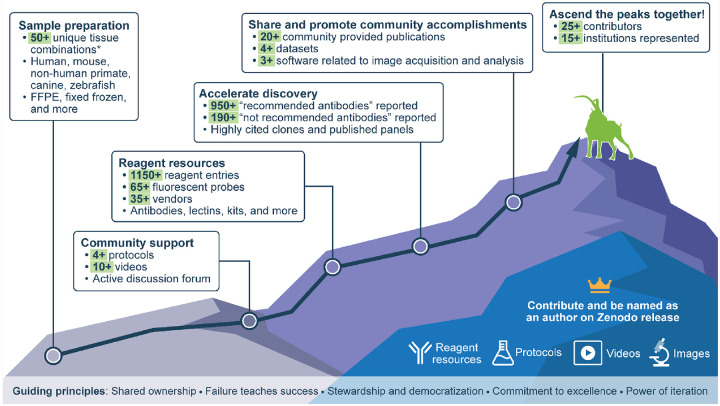
The IBEX Knowledge-Base was founded on five guiding principles (Figure 1). First, we are better together and, importantly, achieve more together by adopting a mindset of shared ownership. For this reason, everyone who contributes knowledge in the form of a reagent resource, antibody validation image, protocol created expressly for the community, or video tutorial is named as an author on the Zenodo dataset and static website. Our second principle is failure teaches success. Unlike publications in which only successful work is described, the goal of the IBEX Knowledge-Base is to document both successful and failed work. By sharing failures, we advance science at a faster pace, reduce financial costs, and instill confidence in the resulting data. While it’s good to learn from our own failures, it’s better to learn from our failures and the failures of others. Accordingly, our discussion forum provides an opportunity for community members to learn from others. Our third principle, stewardship and democratization, is rooted in the open science principles of data sharing, equity, and inclusion. Beyond sharing recommended reagents, we actively encourage the communication of unsatisfactory reagents to prevent other researchers from wasting time and resources. Through stewardship we make science more equitable, reduce the significant cost of validating antibodies[1, 6], and empower scientists from around the world to perform multiplexed imaging. Fourth, members of the IBEX Knowledge-Base are distinguished by a commitment to excellence. To achieve this goal, we collect metadata critical for the performance of a reagent including the target species, tissue preservation method, antigen retrieval conditions, detergent used in blocking and immunolabeling buffers, and information on the best conjugate or antibody clone. We also designed a mechanism for self-correction whereby members of the community can “agree” or “disagree” with reagent entries using their Open Researcher and Contributor ID (ORCID). Like PLos Biology[9], we believe in the importance of being second because reproducible science is good science. Our most current state of knowledge, found on the static website, reports sixteen reagent entries replicated by two independent authors and one reagent entry replicated by three experts. More than a year after its launch, we celebrated our first disagreement regarding an antibody that labels anticipated cell types as well as unusual cell types in the mouse lymph node. We welcome you to join the conversation in the post titled, “Our first disagreement” in the discussion forum. This contribution exemplifies our fifth and final principle by demonstrating the power of iteration, particularly as it applies to correcting and refining our collective state of knowledge. With each addition to the IBEX Knowledge-Base, our knowledge about sample preparation, reagents, and many other aspects of multiplexed imaging and analysis grows (Figure 1).
Figure 1. The IBEX Knowledge-Base is a central portal for IBEX and related multiplexed tissue imaging techniques.
The IBEX Knowledge-Base is an open, global repository providing information related to IBEX and other spatial biology methods. The evolving state of knowledge is reflected by the plus signs associated with the information found here. The 50+ unique tissue combinations are calculated using details related to the target species, tissue preservation method, target tissue, and tissue state, e.g., infected with a particular pathogen. The crown denotes contributions leading to authorship on the Zenodo release (some restrictions apply). The ibex (goat) climbing the mountain is symbolic of the method’s namesake.
An open invitation to use and contribute
The evolving state of knowledge, reflected by the plus signs associated with the number of sample preparations, protocols, videos, reagents, etc. (Figure 1), is made possible by contributors like you! To date, more than 25 contributors from Brazil, Canada, Germany, Mexico, Switzerland, the United Kingdom, and the United States have shared their expertise with the community. The IBEX Knowledge-Base operates under the Creative Commons Attribution 4.0 license which allows anyone to use the resources collected here with attribution. Before embarking on multiplexed tissue imaging, we invite you to use the Knowledge-Base to identify the best way to prepare your samples based on protocols, videos, publications, and support offered via the discussion forum. There are several ways to use the “Reagent Resources” tab on the IBEX Imaging Community website to find suitable reagents for your study. The most common approach is to use the filter function to find community-validated antibodies that are “recommended Yes” for your target species, tissue preservation method (formalin-fixed paraffin embedded (FFPE), fixed frozen, etc.), and antigen retrieval conditions. Another option is to use the “Reagent Resources” tab and community provided publications to identify what other members are examining in the same or similar tissues. Lastly, the extensive list of vendors (35+) may help investigators find where to purchase antibodies for non-traditional experimental animal model systems, e.g., Zebrafish International Resource Center (ZIRC). Finally, in accordance with our guiding principles we encourage you to return to the IBEX Knowledge-Base and add your failures and successes, celebrate your accomplishments, and share your knowledge with others. Your success is our success!
Footnotes
Competing interests
SAT is a remunerated Scientific Advisory Board member of Qiagen, Foresite Labs, OMass Therapeutics, and a consultant and equity holder of TransitionBio and EnsoCell, and a non-executive board director of 10x Genomics, as well as part-time employee of GlaxoSmithKline. JC is an employee and stakeholder of BioLegend (revvity inc.). All other authors declare no competing interests.
References
- 1.Hickey JW, Neumann EK, Radtke AJ, Camarillo JM, Beuschel RT, Albanese A, et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nature Methods. 2022;19(3):284–95. doi: 10.1038/s41592-021-01316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radtke AJ, Roschewski M. The follicular lymphoma tumor microenvironment at single-cell and spatial resolution. Blood. 2024;143(12):1069–79. doi: 10.1182/blood.2023020999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radtke AJ, Kandov E, Lowekamp B, Speranza E, Chu CJ, Gola A, et al. IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc Natl Acad Sci USA. 2020;117(52):33455–65. doi: 10.1073/pnas.2018488117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radtke AJ, Chu CJ, Yaniv Z, Yao L, Marr J, Beuschel RT, et al. IBEX: an iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues. Nat Protoc. 2022;17(2):378–401. doi: 10.1038/s41596-021-00644-9. [DOI] [PubMed] [Google Scholar]
- 5.Yaniv Z. Lowekamp B. SimpleITK Imaris Extensions. Zenodo. 2023. doi: 10.5281/zenodo.7854019. [DOI] [Google Scholar]
- 6.Quardokus EM, Saunders DC, McDonough E, Hickey JW, Werlein C, Surrette C, et al. Organ Mapping Antibody Panels: a community resource for standardized multiplexed tissue imaging. Nat Methods. 2023;20(8):1174–8. doi: 10.1038/s41592-023-01846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Germain RN, Gerner MY. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (C(e)3D). Proc Natl Acad Sci U S A. 2017;114(35):E7321–e30. doi: 10.1073/pnas.1708981114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yaniv Z, Anidi I., Arakkal L., Arroyo-Mejias A., Beuschel R. T., Börner K., Chu C. J., Clatworthy M., Colautti J., Croteau J., Dutra W., Fullam S., Gerner M., Gola A., Gollob K. J., Ichise H., Jonigk D., Kandov E., Kastenmüller W., … Radtke A. J. Iterative Bleaching Extends Multiplexity (IBEX) Knowledge-Base. Zenodo. 2023. doi: 10.5281/zenodo.7693279. [DOI] [Google Scholar]
- 9.The PBSE. The importance of being second. PLOS Biology. 2018;16(1):e2005203. doi: 10.1371/journal.pbio.2005203. [DOI] [PMC free article] [PubMed] [Google Scholar]