Abstract
The use of decellularized extracellular matrix (dECM) as a biomaterial has been an important step forward for the development of functional tissue constructs. In addition to tissues and organs, cell cultures are gaining a lot of attention as an alternative source of dECM. In this work, a novel biomimetic hydrogel is developed based on dECM obtained from mesenchymal stem cells (mdECM) for cartilage tissue engineering. To this end, cells are seeded under specific culture conditions to generate an early chondrogenic extracellular matrix (ECM) providing cues and elements necessary for cartilage development. The composition is determined by quantitative, histological, and mass spectrometry techniques. Moreover, the decellularization process is evaluated by measuring the DNA content and compositional analyses, and the hydrogel is formulated at different concentrations (3% and 6% w/v). Results show that mdECM derived hydrogels possess excellent biocompatibility and suitable physicochemical and mechanical properties for their injectability. Furthermore, it is evidenced that this hydrogel is able to induce chondrogenesis of mesenchymal stem cells (MSCs) without supplemental factors and, furthermore, to form hyaline cartilage‐like tissue after in vivo implantation. These findings demonstrate for the first time the potential of this hydrogel based on mdECM for applications in cartilage repair and regeneration.
Keywords: biomaterials, cartilage tissue engineering, decellularized extracellular matrix, hydrogel scaffolds, mesenchymal stem cells, regenerative medicine
A novel biomimetic hydrogel based on biomimetic decellularized matrix derived from mesenchymal stem cells (MSCs) in culture (mdECM) (decellularized extracellular matrix obtained from mesenchymal stem cells, mdECM) is developed for cartilage regeneration. Results show that the mdECM hydrogel possess biocompatibility and suitable physicochemical and mechanical properties for its injectability. Moreover, it is able to induce chondrogenesis of MSCs without supplemental factors and form hyaline cartilage‐like tissue in vivo.
1. Introduction
Articular cartilage (AC) is a specialized connective tissue that develops important functions related with preserving and enabling locomotion.[ 1 ] Due to its avascular structure and physiology, cartilage has a limited intrinsic repair capacity when is damaged, a condition that often results in degenerative diseases such as osteoarthritis.[ 2 , 3 ] To promote cartilage regeneration, tissue engineering (TE) strategies have been an step forward, providing biomaterials as scaffolds in which cells can be placed to locally produce new cartilage‐like tissue.[ 4 , 5 ] Among the scaffolding biomaterials, injectable hydrogels have received a lot of attention not only for their unique biological and physicochemical properties (e.g., cytocompatibility, high water retention, permeability, and tunable mechanical properties) but also for their ability to fill any shaped wound.[ 6 , 7 , 8 , 9 ]
Taking inspiration from the ECM, which plays an important role not only as a mechanical support but also as a key functional regulator for tissue maturation, considerable efforts have been made to develop hydrogels that recreate the composition or properties of native articular cartilage to guide tissue‐specific formation.[ 10 , 11 , 12 , 13 ] Currently, tissue‐derived dECM (t‐dECM) is considered a promising biomaterial since it is a direct way to provide the complex composition of native tissue, which is difficult to reproduce using common biomaterials. Results from studies about cartilage‐derived ECM as hydrogel revealed chondro‐inductivity and potential for supporting new matrix synthesis, without the need for further functionalization.[ 14 , 15 , 16 , 17 ] However, there are still some challenges and problems that need to be addressed. For instance, limited availability of donor tissue when t‐dECM source is autologous, the potential risk of raised immunogenicity and pathogen transmission if it is allogenic or long‐term protocols for its complete decellularization.[ 18 , 19 , 20 , 21 ]
Recently, cell culture has emerged as an alternative source of dECM for TE. The cell‐derived matrix (c‐dECM) is also a functional matrix that contains a complex mixture of macromolecules and signaling factors that may resemble the native tissue microenvironment. In comparison to t‐dECM, this cell‐derived ECM offers some advantages such as its large scale production and readily customization through the use of different cell types or culture conditions, in addition to the excellent biocompatibility and bio‐inductive properties.[ 20 , 22 , 23 , 24 , 25 , 26 , 27 ] Among cell options, MSCs have gained a great attraction for their easy availability, their immunomodulatory activity, the self‐renewal properties that make it an unlimited ECM source, and their ability to differentiate into several cell lineages, which endows flexibility to generate a wide variety of matrices depending on culture conditions.[ 18 , 19 , 20 , 28 , 29 , 30 , 31 , 32 ] Additionally, by controlling the MSCs stepwise differentiation, it is possible to reproduce even better the dynamically changing microenvironment sustained by ECM during tissue development.[ 29 , 30 , 31 , 32 ] Hence, studies that have used matrices from different stage of differentiation showed different effects on tissue formation, being much favorable for inducing tissue‐specific commitment and matrix deposition an ECM at early stage of maturity.[ 33 , 34 , 35 , 36 ]
Since it has been demonstrated that t‐dECM is a promising option as more functional scaffold or cartilaginous graft[ 14 , 37 ] for treatment of cartilage defects, we hypothesized that using the biomimetic hydrogel based on mdECM could promote chondrogenesis and cartilage regeneration without exogenous growth factors, providing for more readily translatable product than xenogeneic tissue‐derived ECM and avoiding invasive surgery. The objective was to develop for first time a new injectable biomimetic mdECM hydrogel for cartilage regeneration. The manuscript describes the synthesis and formulation of a gel based on ECM derived from chondrogenic induced MSCs as well as a detailed characterization in terms of structural composition, biocompatibility, bioactive and rheological properties. Chondro‐inductive potential of this new biomimetic hydrogel was investigated by the analysis of cell differentiation and tissue‐specific formation both in vitro and in vivo.
2. Results
2.1. Production and Characterization of mECM
Isolated MSCs from lipoaspirate of patients were characterized following the established criteria of the International Society for Cellular Therapy (ISCT)[ 38 ] to define multipotent mesenchymal stromal cells (Figure S1, the Supporting Information). In an attempt to produce a hydrogel that better recreates the environment during chondrogenesis, we synthetized an early chondrogenic matrix from MSCs since it contains the biochemical components that are necessary for tissue development. For this, MSCs were cultured in monolayer and in chondrogenic media during different time points (1, 2, 3, and 4 weeks). Quantification of main components of articular cartilage such as Collagen type II and sulfated glycosaminoglycans (GAGs) were performed to investigate the different stages of chondrogenesis. After 1 week in culture, the content of GAGs per µg DNA of MSCs cultured in the chondrogenic medium was at the same level as that cultured in the growth medium (Figure 1A). Nonetheless, the GAGs/DNA ratio increased significantly with increasing the cell culture time. A similar situation was observed for Collagen II, whose levels were appreciated two weeks after MSCs culture under chondrogenic conditions and remarkably increase along time, more specially during the last week (Figure 1B). In view of these results, since the formation of the chondrogenic matrix started after two weeks in culture and this time was enough to induce the initial deposition of chondrogenic components, which characterizes early steps of chondrogenesis, we used this time for the subsequent assays.
Figure 1.
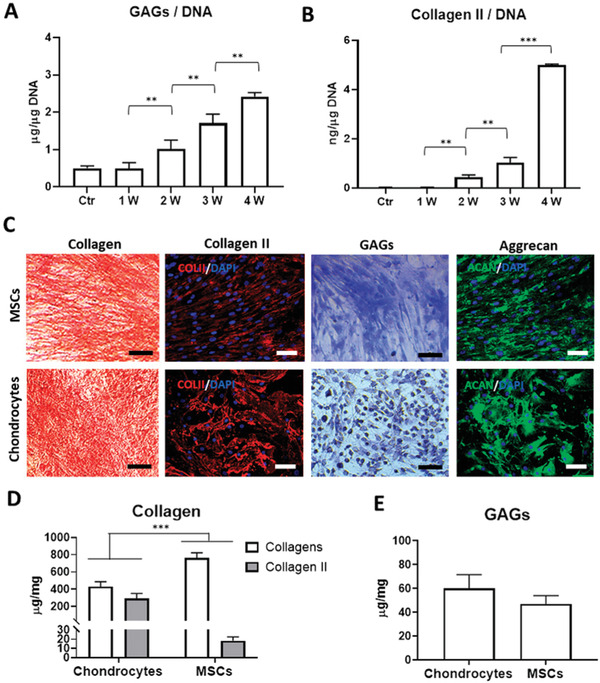
Production and characterization of early chondrogenic mECM. Quantitative analysis of GAGs A) and Collagen type II B) cartilage‐specific components in matrix derived from MSCs, cultured under chondrogenic culture conditions during 1, 2, 3, and 4 weeks, normalized to DNA of cells in culture. Qualitative analysis of main components, collagen and GAGs in matrix derived from MSCs and chondrocytes after 2 weeks in culture by histological staining (Sirius Red, Toluidine O) and immunofluorescence (Collagen type II and Aggrecan) C). Quantitative analysis of collagen, Collagen type II D) and GAGs E) content in matrix derived from MSCs and chondrocytes after 2 weeks in culture. Scale bar: 50 µm. Data represent mean ± S.D. (n = 3). ** p < 0.01; *** p < 0.001, Student's t‐test.
The production of the early chondrogenic matrix was then confirmed by comparing two‐week mECM and mature chondrogenic matrix derived from chondrocytes (cECM) in culture for two weeks. Histology and immunofluorescence staining revealed differential presence and distribution of the main ECM components. Figure 1C shows that both ECMs expressed similar levels of collagen and GAGs, although in the ECM derived from chondrocytes there was a significant higher expression of mature cartilage‐specific components such as Collagen II and Aggrecan. These observations were in accordance with results from quantitative assays of those components, revealing that matrices were rich in collagen (430 µg mg−1 for cECM and 760 µg mg−1 for mECM), being 70% of this content Collagen II in matrix from chondrocytes, in contrast to mECM that was only 2.45% (Figure 1D). Meanwhile, GAGs content expressed values around 60 µg mg−1 in cECM and 47 µg mg−1 in mECM (Figure 1D,E).
In order to obtain more details about the components of mECM we performed a mass spectrometry (MS) characterization (Table 1 ). Results revealed the presence of a complex protein matrix, identifying 100 proteins that included core matrisome and matrisome associated proteins. Core matrisome consisting of structural proteins that confer mechanical properties and cell adhesion to the ECM was the major component. This was rich in collagens, glycoproteins, and proteoglycans. A large variety of collagens were identified, most of them constituents of cartilaginous ECM. There were some members of fibrillar family (Collagen type I, III, and V), fibril associated collagens with interrupted triple (FACIT) helices (Collagens type XII, XIV, and XIV) and network‐forming collagens (Collagen type IV and VIII). Other subtypes of collagen such as Collagen type VI, which is the primary collagen located in the pericellular matrix of cartilage, was also present. Between glycoproteins, it was found some MSC matrix markers like Emilin‐1 but also typical structural proteins of cartilage matrix such as cartilage oligomeric matrix protein (COMP), Tenascin‐C, Lumican (LUM), chondroitin sulfate proteoglycan core protein 2 (CSPG2 or Versican), Prolargin (PRELP), and transforming growth factor‐beta‐induced protein, although the mostly detected was fibronectin. Biglycan, Decorin, and Versican Core Protein, which are also very common in cartilage, were the principal proteoglycans in our mECM. In less abundance were identified other affiliated proteins including regulators, e.g., matrix metalloproteinases (MMP) and their inhibitors (TIMPs), ECM‐associated proteins (i.e., Annexins and Galectins) and signaling ligands (Table 1).
Table 1.
Detectable proteins in early chondrogenic mECM
| Protein name | Accession number | Score | Sequence coverage | # peptides |
|---|---|---|---|---|
| Collagens | ||||
| Collagen alpha‐1(I) chain OS = Homo sapiens GN = COL1A1 PE = 1 SV = 5 | P02452 | 2713 | 1605 | 16 |
| Collagen alpha‐2(I) chain OS = Homo sapiens GN = COL1A2 PE = 1 SV = 7 | P08123 | 4296 | 2401 | 22 |
| Collagen alpha‐1(IV) chain OS = Homo sapiens GN = COL4A1 PE = 1 SV = 3 | P02462 | 140 | 3 | 4 |
| Collagen alpha‐2(IV) chain OS = Homo sapiens GN = COL4A2 PE = 1 SV = 4 | P08572 | 370 | 7 | 7 |
| Collagen alpha‐3(VI) chain OS = Homo sapiens GN = COL6A3 PE = 1 SV = 5 | P12111 | 32 978 | 60 | 147 |
| Collagen alpha‐1(III) chain OS = Homo sapiens GN = COL3A1 PE = 1 SV = 4 | P02461 | 2018 | 1064 | 11 |
| Collagen alpha‐2(V) chain OS = Homo sapiens GN = COL5A2 PE = 1 SV = 3 | P05997 | 486 | 8 | 8 |
| Collagen alpha‐1(V) chain OS = Homo sapiens GN = COL5A1 PE = 1 SV = 3 | P20908 | 2543 | 15 | 18 |
| Collagen alpha‐1(VI) chain OS = Homo sapiens OX = 9606 GN = COL6A1 PE = 1 SV = 3 | P12109 | 9169 | 2247 | 16 |
| Collagen alpha‐2(VI) chain OS = Homo sapiens OX = 9606 GN = COL6A2 PE = 1 SV = 4 | P12110 | 6068 | 1894 | 14 |
| Collagen alpha‐1(VIII) chain OS = Homo sapiens GN = COL8A1 PE = 1 SV = 2 | P27658 | 64 | 6 | 3 |
| Collagen alpha‐1(XII) chain OS = Homo sapiens OX = 9606 GN = COL12A1 PE = 1 SV = 2 | Q99715 | 9989 | 1877 | 44 |
| Collagen alpha‐1(VII) chain OS = Homo sapiens GN = COL7A1 PE = 1 SV = 2 | Q02388 | 51 | 1 | 1 |
| Collagen alpha‐2(VIII) chain OS = Homo sapiens GN = COL8A2 PE = 1 SV = 2 | P25067 | 16 | 7 | 2 |
| Collagen alpha‐1(XI) chain OS = Homo sapiens GN = COL11A1 PE = 1 SV = 4 | P12107 | 360 | 8 | 11 |
| Collagen alpha‐1(XIV) chain OS = Homo sapiens GN = COL14A1 PE = 1 SV = 3 | Q05707 | 2340 | 42 | 50 |
| Collagen alpha‐1(XVI) chain OS = Homo sapiens GN = COL16A1 PE = 1 SV = 2 | Q07092 | 58 | 2 | 2 |
| Collagen alpha‐1(XXI) chain OS = Homo sapiens GN = COL21A1 PE = 2 SV = 1 | Q96P44 | 93 | 7 | 5 |
| Glycoproteins | ||||
| Fibulin‐1 OS = Homo sapiens GN = FBLN1 PE = 1 SV = 4 | P23142 | 959 | 25 | 13 |
| Fibulin‐2 OS = Homo sapiens GN = FBLN2 PE = 1 SV = 2 | P98095 | 1468 | 29 | 25 |
| Filamin‐A OS = Homo sapiens OX = 9606 GN = FLNA PE = 1 SV = 4 | P21333 | 5443 | 1500 | 28 |
| EMILIN‐1 OS = Homo sapiens OX = 9606 GN = EMILIN1 PE = 1 SV = 3 | Q9Y6C2 | 3200 | 1280 | 9 |
| EMILIN‐2 OS = Homo sapiens GN = EMILIN2 PE = 1 SV = 3 | Q9BXX0 | 316 | 15 | 10 |
| Fibronectin OS = Homo sapiens OX = 9606 GN = FN1 PE = 1 SV = 4 | P02751 | 29406 | 4417 | 65 |
| Fibromodulin OS = Homo sapiens GN = FMOD PE = 1 SV = 2 | Q06828 | 231 | 21 | 5 |
| Cartilage oligomeric matrix protein OS = Homo sapiens GN = COMP PE = 1 SV = 2 | P49747 | 1060 | 29 | 15 |
| Cartilage intermediate layer protein 1 OS = Homo sapiens GN = CILP PE = 1 SV = 4 | O75339 | 893 | 27 | 28 |
| Cartilage intermediate layer protein 2 OS = Homo sapiens GN = CILP2 PE = 2 SV = 2 | Q8IUL8 | 0 | 1 | 1 |
| Chitinase‐3‐like protein 1 OS = Homo sapiens GN = CHI3L1 PE = 1 SV = 2 | P36222 | 88 | 21 | 7 |
| Periostin OS = Homo sapiens GN = POSTN PE = 1 SV = 2 | Q15063 | 7739 | 66 | 48 |
| Thrombospondin‐1 OS = Homo sapiens GN = THBS1 PE = 1 SV = 2 | P07996 | 26 485 | 60 | 64 |
| Thrombospondin‐2 OS = Homo sapiens GN = THBS2 PE = 1 SV = 2 | P35442 | 3219 | 28 | 25 |
| Tenascin OS = Homo sapiens OX = 9606 GN = TNC PE = 1 SV = 3 | P24821 | 9680 | 2690 | 40 |
| Laminin subunit beta‐2 OS = Homo sapiens GN = LAMB2 PE = 1 SV = 2 | P55268 | 159 | 5 | 5 |
| Laminin subunit alpha‐4 OS = Homo sapiens GN = LAMA4 PE = 1 SV = 4 | Q16363 | 0 | 0 | 1 |
| Laminin subunit gamma‐1 OS = Homo sapiens GN = LAMC1 PE = 1 SV = 3 | P11047 | 128 | 2 | 3 |
| SPARC OS = Homo sapiens GN = SPARC PE = 1 SV = 1 | P09486 | 155 | 17 | 4 |
| EGF‐containing fibulin‐like extracellular matrix protein 1 OS = Homo sapiens GN = EFEMP1 PE = 1 SV = 2 | Q12805 | 138 | 15 | 5 |
| EGF‐containing fibulin‐like extracellular matrix protein 2 OS = Homo sapiens GN = EFEMP2 PE = 1 SV = 3 | O95967 | 120 | 12 | 4 |
| Vitronectin OS = Homo sapiens GN = VTN PE = 1 SV = 1 | P04004 | 94 | 3 | 1 |
| Lactadherin OS = Homo sapiens GN = MFGE8 PE = 1 SV = 2 | Q08431 | 28 | 6 | 2 |
| Podocan OS = Homo sapiens GN = PODN PE = 1 SV = 2 | Q7Z5L7 | 0 | 2 | 1 |
| Olfactomedin‐like protein 3 OS = Homo sapiens GN = OLFML3 PE = 2 SV = 1 | Q9NRN5 | 26 | 2 | 1 |
| Secretoglobin family 1D member 2 OS = Homo sapiens GN = SCGB1D2 PE = 2 SV = 1 | O95969 | 22 | 10 | 1 |
| Semaphorin‐3C OS = Homo sapiens GN = SEMA3C PE = 2 SV = 2 | Q99985 | 0 | 1 | 1 |
| Thyroglobulin OS = Homo sapiens GN = TG PE = 1 SV = 5 | P01266 | 0 | 1 | 1 |
| Proteoglycans | ||||
| Basement membrane‐specific heparan sulfate proteoglycan core protein OS = Homo sapiens GN = HSPG2 PE = 1 SV = 4 | P98160 | 6480 | 1018 | 34 |
| Biglycan OS = Homo sapiens OX = 9606 GN = BGN PE = 1 SV = 2 | P21810 | 1954 | 2826 | 8 |
| Versican core protein OS = Homo sapiens OX = 9606 GN = VCAN PE = 1 SV = 3 | P13611 | 1267 | 177 | 5 |
| Decorin OS = Homo sapiens GN = DCN PE = 1 SV = 1 | P07585 | 804 | 31 | 8 |
| Lumican OS = Homo sapiens GN = LUM PE = 1 SV = 2 | P51884 | 533 | 35 | 9 |
| Mimecan OS = Homo sapiens GN = OGN PE = 1 SV = 1 | P20774 | 102 | 17 | 4 |
| Regulators | ||||
| Matrix metalloproteinase‐14 OS = Homo sapiens GN = MMP14 PE = 1 SV = 3 | P50281 | 119 | 13 | 6 |
| A disintegrin and metalloproteinase with thrombospondin motifs 4 OS = Homo sapiens GN = ADAMTS4 PE = 1 SV = 3 | O75173 | 95 | 4 | 2 |
| A disintegrin and metalloproteinase with thrombospondin motifs 3 OS = Homo sapiens GN = ADAMTS3 PE = 2 SV = 4 | O15072 | 41 | 2 | 1 |
| A disintegrin and metalloproteinase with thrombospondin motifs 2 OS = Homo sapiens GN = ADAMTS2 PE = 2 SV = 2 | O95450 | 34 | 3 | 3 |
| Metalloproteinase inhibitor 1 OS = Homo sapiens GN = TIMP1 PE = 1 SV = 1 | P01033 | 30 | 8 | 1 |
| Metalloproteinase inhibitor 3 OS = Homo sapiens GN = TIMP3 PE = 1 SV = 2 | P35625 | 55 | 5 | 1 |
| 72 kDa type IV collagenase OS = Homo sapiens GN = MMP2 PE = 1 SV = 2 | P08253 | 271 | 22 | 10 |
| Interstitial collagenase OS = Homo sapiens GN = MMP1 PE = 1 SV = 3 | P03956 | 48 | 3 | 1 |
| Matrix‐remodeling‐associated protein 5 OS = Homo sapiens GN = MXRA5 PE = 2 SV = 3 | Q9NR99 | 190 | 035 | 1 |
| Matrix‐remodeling‐associated protein 7 OS = Homo sapiens GN = MXRA7 PE = 1 SV = 1 | P84157 | 182 | 539 | 1 |
| Cell migration‐inducing and hyaluronan‐binding protein OS = Homo sapiens GN = CEMIP PE = 1 SV = 2 | Q8WUJ3 | 74 | 4 | 4 |
| 14‐3‐3 protein epsilon OS = Homo sapiens GN = YWHAE PE = 1 SV = 1 | P62258 | 2879 | 6667 | 16 |
| 14‐3‐3 protein zeta/delta OS = Homo sapiens GN = YWHAZ PE = 1 SV = 1 | P63104 | 2158 | 5796 | 12 |
| 14‐3‐3 protein beta/alpha OS = Homo sapiens GN = YWHAB PE = 1 SV = 3 | P31946 | 1340 | 6016 | 8 |
| 14‐3‐3 protein theta OS = Homo sapiens GN = YWHAQ PE = 1 SV = 1 | P27348 | 1200 | 4571 | 6 |
| 14‐3‐3 protein gamma OS = Homo sapiens GN = YWHAG PE = 1 SV = 2 | P61981 | 992 | 4980 | 5 |
| 14‐3‐3 protein eta OS = Homo sapiens GN = YWHAH PE = 1 SV = 4 | Q04917 | 897 | 3902 | 5 |
| Collagen triple helix repeat‐containing protein 1 OS = Homo sapiens GN = CTHRC1 PE = 1 SV = 1 | Q96CG8 | 36 | 3 | 1 |
| Dermcidin OS = Homo sapiens GN = DCD PE = 1 SV = 2 | P81605 | 333 | 2000 | 2 |
| Serine protease HTRA1 OS = Homo sapiens GN = HTRA1 PE = 1 SV = 1 | Q92743 | 857 | 1875 | 5 |
| ECM‐affiliated proteins | ||||
| Latent‐transforming growth factor beta‐binding protein 1 OS = Homo sapiens GN = LTBP1 PE = 1 SV = 4 | Q14766 | 269 | 7 | 8 |
| Prolargin OS = Homo sapiens GN = PRELP PE = 1 SV = 1 | P51888 | 204 | 23 | 7 |
| Microfibrillar‐associated protein 2 OS = Homo sapiens GN = MFAP2 PE = 2 SV = 1 | P55001 | 457 | 27 | 4 |
| Microfibrillar‐associated protein 5 OS = Homo sapiens GN = MFAP5 PE = 1 SV = 1 | Q13361 | 157 | 18 | 3 |
| Transforming growth factor‐beta‐induced protein ig‐h3 OS = Homo sapiens GN = TGFBI PE = 1 SV = 1 | Q15582 | 4751 | 4890 | 24 |
| Cartilage‐associated protein OS = Homo sapiens GN = CRTAP PE = 1 SV = 1 | O75718 | 1332 | 2195 | 7 |
| Procollagen C‐endopeptidase enhancer 1 OS = Homo sapiens GN = PCOLCE PE = 1 SV = 2 | Q15113 | 633 | 1782 | 5 |
| Annexin A6 OS = Homo sapiens GN = ANXA6 PE = 1 SV = 3 | P08133 | 7169 | 6300 | 35 |
| Annexin A2 OS = Homo sapiens GN = ANXA2 PE = 1 SV = 2 | P07355 | 5203 | 7286 | 34 |
| Annexin A5 OS = Homo sapiens GN = ANXA5 PE = 1 SV = 2 | P08758 | 4748 | 7000 | 27 |
| Annexin A1 OS = Homo sapiens GN = ANXA1 PE = 1 SV = 2 | P04083 | 3612 | 6098 | 18 |
| Annexin A4 OS = Homo sapiens GN = ANXA4 PE = 1 SV = 4 | P09525 | 2243 | 4765 | 12 |
| Annexin A7 OS = Homo sapiens GN = ANXA7 PE = 1 SV = 3 | P20073 | 1654 | 2070 | 9 |
| Annexin A11 OS = Homo sapiens GN = ANXA11 PE = 1 SV = 1 | P50995 | 1033 | 1465 | 6 |
| Galectin‐1 OS = Homo sapiens GN = LGALS1 PE = 1 SV = 2 | P09382 | 1445 | 7037 | 8 |
| Galectin‐3 OS = Homo sapiens GN = LGALS3 PE = 1 SV = 5 | P17931 | 188 | 1400 | 3 |
| Protein CYR61 OS = Homo sapiens GN = CYR61 PE = 1 SV = 1 | O00622 | 213 | 341 | 1 |
| Secreted factors | ||||
| Myeloid‐derived growth factor OS = Homo sapiens GN = MYDGF PE = 1 SV = 1 | Q969H8 | 1146 | 3757 | 6 |
| Insulin‐like growth factor 2 mRNA‐binding protein 2 OS = Homo sapiens GN = IGF2BP2 PE = 1 SV = 2 | Q9Y6M1 | 652 | 1085 | 5 |
| Hepatoma‐derived growth factor OS = Homo sapiens GN = HDGF PE = 1 SV = 1 | P51858 | 336 | 1792 | 3 |
| Connective tissue growth factor OS = Homo sapiens GN = CTGF PE = 1 SV = 2 | P29279 | 268 | 630 | 2 |
| Guanylate‐binding protein 1 OS = Homo sapiens GN = GBP1 PE = 1 SV = 2 | P32455 | 219 | 507 | 2 |
| Pentraxin‐related protein PTX3 OS = Homo sapiens GN = PTX3 PE = 1 SV = 3 | P26022 | 569 | 1339 | 4 |
| Leukocyte elastase inhibitor OS = Homo sapiens GN = SERPINB1 PE = 1 SV = 1 | P30740 | 482 | 792 | 3 |
| Mesencephalic astrocyte‐derived neurotrophic factor OS = Homo sapiens GN = MANF PE = 1 SV = 3 | P55145 | 319 | 2143 | 3 |
| Spondin‐1 OS = Homo sapiens GN = SPON1 PE = 1 SV = 2 | Q9HCB6 | 30 | 1 | 1 |
| Sushi repeat‐containing protein SRPX OS = Homo sapiens GN = SRPX PE = 1 SV = 1 | P78539 | 90 | 8 | 3 |
| Sushi repeat‐containing protein SRPX2 OS = Homo sapiens GN = SRPX2 PE = 1 SV = 1 | O60687 | 23 | 3 | 1 |
2.2. Formulation of mdECM Hydrogel
The development of ECM‐based hydrogel involves two steps, including i) decellularization of mECM, and ii) preparation of mdMEC gel. First, cell removal from early chondrogenic matrix was performed to maximize the elimination of cellular material, while minimizing ECM loss and damage. Treatment containing Triton X‐100 and ammonium hydroxide (NH4OH) successfully eliminated the cellular content as demonstrated the absence of nuclei on phase‐contrast images and hematoxylin and eosin (H&E)‐stained sections of decellularized matrix (Figure 2A), as well as the reduction in ≈98% in DNA content (Figure 2B). In parallel, it was proved that ECM composition was properly preserved. Comparative information about protein content by mass spectrometry revealed similar patterns before and after decellularizing treatment (Table 2 ). Similarly, histological analysis using Masson's Trichrome and Toluidine O staining and quantitative biochemistry assays of collagen and GAGs respectively, also showed the non‐significant differences in remaining content between mECM and mdECM (Figure 2C,D). Taken together, these results indicate an efficient decellularization of mECM without influencing ECM integrity.
Figure 2.
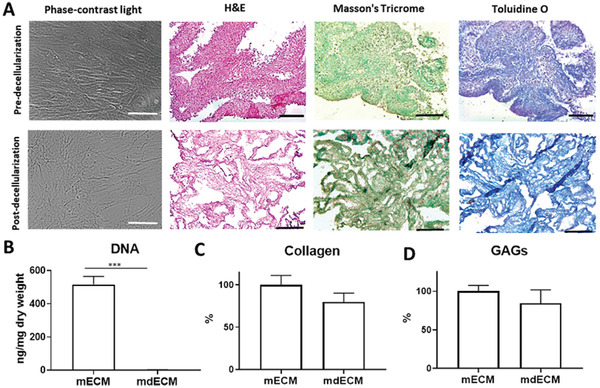
Decellularization of early chondrogenic matrix derived from MSCs. A) Qualitative analysis by optical microscope image (phase‐contrast light) and histological staining: H&E (cellular content), Masson's Trichrome (collagen) and Toluidine O (GAGs) staining. Quantitative analysis of DNA B), collagen C), and GAGs D) content. Scale bar: 50 µm (phase‐contrast) and 200 µm (histological staining). Data represent mean ± S.D. (n = 3). *** p < 0.001, Student's t‐test.
Table 2.
Detectable proteins in mECM previously and after decellularization process
| Protein name | mECM | mdECM |
|---|---|---|
| Collagens | ||
| Collagen alpha‐1(I) chain OS = Homo sapiens GN = COL1A1 PE = 1 SV = 5 | X | X |
| Collagen alpha‐2(I) chain OS = Homo sapiens GN = COL1A2 PE = 1 SV = 7 | X | X |
| Collagen alpha‐1(IV) chain OS = Homo sapiens GN = COL4A1 PE = 1 SV = 3 | X | X |
| Collagen alpha‐2(IV) chain OS = Homo sapiens GN = COL4A2 PE = 1 SV = 4 | X | X |
| Collagen alpha‐3(VI) chain OS = Homo sapiens GN = COL6A3 PE = 1 SV = 5 | X | X |
| Collagen alpha‐1(III) chain OS = Homo sapiens GN = COL3A1 PE = 1 SV = 4 | X | X |
| Collagen alpha‐2(V) chain OS = Homo sapiens GN = COL5A2 PE = 1 SV = 3 | X | X |
| Collagen alpha‐1(V) chain OS = Homo sapiens GN = COL5A1 PE = 1 SV = 3 | X | X |
| Collagen alpha‐1(VI) chain OS = Homo sapiens OX = 9606 GN = COL6A1 PE = 1 SV = 3 | X | X |
| Collagen alpha‐2(VI) chain OS = Homo sapiens OX = 9606 GN = COL6A2 PE = 1 SV = 4 | X | X |
| Collagen alpha‐1(VIII) chain OS = Homo sapiens GN = COL8A1 PE = 1 SV = 2 | X | X |
| Collagen alpha‐1(XII) chain OS = Homo sapiens OX = 9606 GN = COL12A1 PE = 1 SV = 2 | X | X |
| Collagen alpha‐1(VII) chain OS = Homo sapiens GN = COL7A1 PE = 1 SV = 2 | X | X |
| Collagen alpha‐2(VIII) chain OS = Homo sapiens GN = COL8A2 PE = 1 SV = 2 | X | |
| Collagen alpha‐1(XI) chain OS = Homo sapiens GN = COL11A1 PE = 1 SV = 4 | X | X |
| Collagen alpha‐1(XIV) chain OS = Homo sapiens GN = COL14A1 PE = 1 SV = 3 | X | X |
| Collagen alpha‐1(XVI) chain OS = Homo sapiens GN = COL16A1 PE = 1 SV = 2 | X | X |
| Collagen alpha‐1(XXI) chain OS = Homo sapiens GN = COL21A1 PE = 2 SV = 1 | X | |
| Glycoproteins | ||
| Fibulin‐1 OS = Homo sapiens GN = FBLN1 PE = 1 SV = 4 | X | X |
| Fibulin‐2 OS = Homo sapiens GN = FBLN2 PE = 1 SV = 2 | X | X |
| Filamin‐A OS = Homo sapiens OX = 9606 GN = FLNA PE = 1 SV = 4 | X | X |
| EMILIN‐1 OS = Homo sapiens OX = 9606 GN = EMILIN1 PE = 1 SV = 3 | X | X |
| EMILIN‐2 OS = Homo sapiens GN = EMILIN2 PE = 1 SV = 3 | X | X |
| Fibronectin OS = Homo sapiens OX = 9606 GN = FN1 PE = 1 SV = 4 | X | X |
| Fibromodulin OS = Homo sapiens GN = FMOD PE = 1 SV = 2 | X | X |
| Cartilage oligomeric matrix protein OS = Homo sapiens GN = COMP PE = 1 SV = 2 | X | X |
| Cartilage intermediate layer protein 1 OS = Homo sapiens GN = CILP PE = 1 SV = 4 | X | X |
| Cartilage intermediate layer protein 2 OS = Homo sapiens GN = CILP2 PE = 2 SV = 2 | X | X |
| Chitinase‐3‐like protein 1 OS = Homo sapiens GN = CHI3L1 PE = 1 SV = 2 | X | X |
| Periostin OS = Homo sapiens GN = POSTN PE = 1 SV = 2 | X | X |
| Thrombospondin‐1 OS = Homo sapiens GN = THBS1 PE = 1 SV = 2 | X | X |
| Thrombospondin‐2 OS = Homo sapiens GN = THBS2 PE = 1 SV = 2 | X | X |
| Tenascin OS = Homo sapiens OX = 9606 GN = TNC PE = 1 SV = 3 | X | X |
| Laminin subunit beta‐2 OS = Homo sapiens GN = LAMB2 PE = 1 SV = 2 | X | X |
| Laminin subunit alpha‐4 OS = Homo sapiens GN = LAMA4 PE = 1 SV = 4 | X | X |
| Laminin subunit gamma‐1 OS = Homo sapiens GN = LAMC1 PE = 1 SV = 3 | X | X |
| SPARC OS = Homo sapiens GN = SPARC PE = 1 SV = 1 | X | X |
| EGF‐containing fibulin‐like extracellular matrix protein 1 OS = Homo sapiens GN = EFEMP1 PE = 1 SV = 2 | X | X |
| EGF‐containing fibulin‐like extracellular matrix protein 2 OS = Homo sapiens GN = EFEMP2 PE = 1 SV = 3 | X | X |
| Vitronectin OS = Homo sapiens GN = VTN PE = 1 SV = 1 | X | X |
| Lactadherin OS = Homo sapiens GN = MFGE8 PE = 1 SV = 2 | X | X |
| Podocan OS = Homo sapiens GN = PODN PE = 1 SV = 2 | X | X |
| Olfactomedin‐like protein 3 OS = Homo sapiens GN = OLFML3 PE = 2 SV = 1 | X | X |
| Secretoglobin family 1D member 2 OS = Homo sapiens GN = SCGB1D2 PE = 2 SV = 1 | X | X |
| Semaphorin‐3C OS = Homo sapiens GN = SEMA3C PE = 2 SV = 2 | X | X |
| Thyroglobulin OS = Homo sapiens GN = TG PE = 1 SV = 5 | X | X |
| Proteoglycans | ||
| Basement membrane‐specific heparan sulfate proteoglycan core protein OS = Homo sapiens GN = HSPG2 PE = 1 SV = 4 | X | X |
| Biglycan OS = Homo sapiens OX = 9606 GN = BGN PE = 1 SV = 2 | X | X |
| Versican core protein OS = Homo sapiens OX = 9606 GN = VCAN PE = 1 SV = 3 | X | X |
| Decorin OS = Homo sapiens GN = DCN PE = 1 SV = 1 | X | X |
| Lumican OS = Homo sapiens GN = LUM PE = 1 SV = 2 | X | X |
| Mimecan OS = Homo sapiens GN = OGN PE = 1 SV = 1 | X | X |
| Regulators | ||
| Matrix metalloproteinase‐14 OS = Homo sapiens GN = MMP14 PE = 1 SV = 3 | X | X |
| A disintegrin and metalloproteinase with thrombospondin motifs 4 OS = Homo sapiens GN = ADAMTS4 PE = 1 SV = 3 | X | X |
| A disintegrin and metalloproteinase with thrombospondin motifs 3 OS = Homo sapiens GN = ADAMTS3 PE = 2 SV = 4 | X | X |
| A disintegrin and metalloproteinase with thrombospondin motifs 2 OS = Homo sapiens GN = ADAMTS2 PE = 2 SV = 2 | X | X |
| Metalloproteinase inhibitor 1 OS = Homo sapiens GN = TIMP1 PE = 1 SV = 1 | X | X |
| Metalloproteinase inhibitor 3 OS = Homo sapiens GN = TIMP3 PE = 1 SV = 2 | X | X |
| 72 kDa type IV collagenase OS = Homo sapiens GN = MMP2 PE = 1 SV = 2 | X | X |
| Interstitial collagenase OS = Homo sapiens GN = MMP1 PE = 1 SV = 3 | X | X |
| Matrix‐remodeling‐associated protein 5 OS = Homo sapiens GN = MXRA5 PE = 2 SV = 3 | X | X |
| Matrix‐remodeling‐associated protein 7 OS = Homo sapiens GN = MXRA7 PE = 1 SV = 1 | X | |
| Cell migration‐inducing and hyaluronan‐binding protein OS = Homo sapiens GN = CEMIP PE = 1 SV = 2 | X | X |
| 14‐3‐3 protein epsilon OS = Homo sapiens GN = YWHAE PE = 1 SV = 1 | X | X |
| 14‐3‐3 protein zeta/delta OS = Homo sapiens GN = YWHAZ PE = 1 SV = 1 | X | X |
| 14‐3‐3 protein beta/alpha OS = Homo sapiens GN = YWHAB PE = 1 SV = 3 | X | X |
| 14‐3‐3 protein theta OS = Homo sapiens GN = YWHAQ PE = 1 SV = 1 | X | X |
| 14‐3‐3 protein gamma OS = Homo sapiens GN = YWHAG PE = 1 SV = 2 | X | X |
| 14‐3‐3 protein eta OS = Homo sapiens GN = YWHAH PE = 1 SV = 4 | X | X |
| Collagen triple helix repeat‐containing protein 1 OS = Homo sapiens GN = CTHRC1 PE = 1 SV = 1 | X | X |
| Dermcidin OS = Homo sapiens GN = DCD PE = 1 SV = 2 | X | X |
| Serine protease HTRA1 OS = Homo sapiens GN = HTRA1 PE = 1 SV = 1 | X | |
| ECM‐affiliated proteins | ||
| Latent‐transforming growth factor beta‐binding protein 1 OS = Homo sapiens GN = LTBP1 PE = 1 SV = 4 | X | X |
| Prolargin OS = Homo sapiens GN = PRELP PE = 1 SV = 1 | X | X |
| Microfibrillar‐associated protein 2 OS = Homo sapiens GN = MFAP2 PE = 2 SV = 1 | X | X |
| Microfibrillar‐associated protein 5 OS = Homo sapiens GN = MFAP5 PE = 1 SV = 1 | X | X |
| Transforming growth factor‐beta‐induced protein ig‐h3 OS = Homo sapiens GN = TGFBI PE = 1 SV = 1 | X | X |
| Cartilage‐associated protein OS = Homo sapiens GN = CRTAP PE = 1 SV = 1 | X | X |
| Procollagen C‐endopeptidase enhancer 1 OS = Homo sapiens GN = PCOLCE PE = 1 SV = 2 | X | X |
| Annexin A6 OS = Homo sapiens GN = ANXA6 PE = 1 SV = 3 | X | X |
| Annexin A2 OS = Homo sapiens GN = ANXA2 PE = 1 SV = 2 | X | X |
| Annexin A5 OS = Homo sapiens GN = ANXA5 PE = 1 SV = 2 | X | X |
| Annexin A1 OS = Homo sapiens GN = ANXA1 PE = 1 SV = 2 | X | X |
| Annexin A4 OS = Homo sapiens GN = ANXA4 PE = 1 SV = 4 | X | |
| Annexin A7 OS = Homo sapiens GN = ANXA7 PE = 1 SV = 3 | X | |
| Annexin A11 OS = Homo sapiens GN = ANXA11 PE = 1 SV = 1 | X | |
| Galectin‐1 OS = Homo sapiens GN = LGALS1 PE = 1 SV = 2 | X | X |
| Galectin‐3 OS = Homo sapiens GN = LGALS3 PE = 1 SV = 5 | X | X |
| Protein CYR61 OS = Homo sapiens GN = CYR61 PE = 1 SV = 1 | X | X |
| Secreted factors | ||
| Myeloid‐derived growth factor OS = Homo sapiens GN = MYDGF PE = 1 SV = 1 | X | |
| Insulin‐like growth factor 2 mRNA‐binding protein 2 OS = Homo sapiens GN = IGF2BP2 PE = 1 SV = 2 | X | X |
| Hepatoma‐derived growth factor OS = Homo sapiens GN = HDGF PE = 1 SV = 1 | X | |
| Connective tissue growth factor OS = Homo sapiens GN = CTGF PE = 1 SV = 2 | X | |
| Guanylate‐binding protein 1 OS = Homo sapiens GN = GBP1 PE = 1 SV = 2 | X | |
| Pentraxin‐related protein PTX3 OS = Homo sapiens GN = PTX3 PE = 1 SV = 3 | X | X |
| Leukocyte elastase inhibitor OS = Homo sapiens GN = SERPINB1 PE = 1 SV = 1 | X | |
| Mesencephalic astrocyte‐derived neurotrophic factor OS = Homo sapiens GN = MANF PE = 1 SV = 3 | X | |
| Spondin‐1 OS = Homo sapiens GN = SPON1 PE = 1 SV = 2 | X | |
| Sushi repeat‐containing protein SRPX OS = Homo sapiens GN = SRPX PE = 1 SV = 1 | X | X |
| Sushi repeat‐containing protein SRPX2 OS = Homo sapiens GN = SRPX2 PE = 1 SV = 1 | X | X |
After decellularization, the early chondrogenic mdECM was lyophilized, milled into a fine white powder and then solubilized with pepsin to liquefy it at final concentrations of 3% and 6% w/v (Figure 3A). The resulting homogeneous acidic mdECM solutions were adjusted to physiological pH using basic solution (NaOH) before encapsulating cells. Neutralized mdECM solutions exhibited a viscous‐like appearance with a high opacity that increased with the concentration. These solutions were thermoresponsive; behaving as a viscous liquid at low temperatures (<15 °C) and transforming into a gel when incubating for 20 min at 37 °C.
Figure 3.
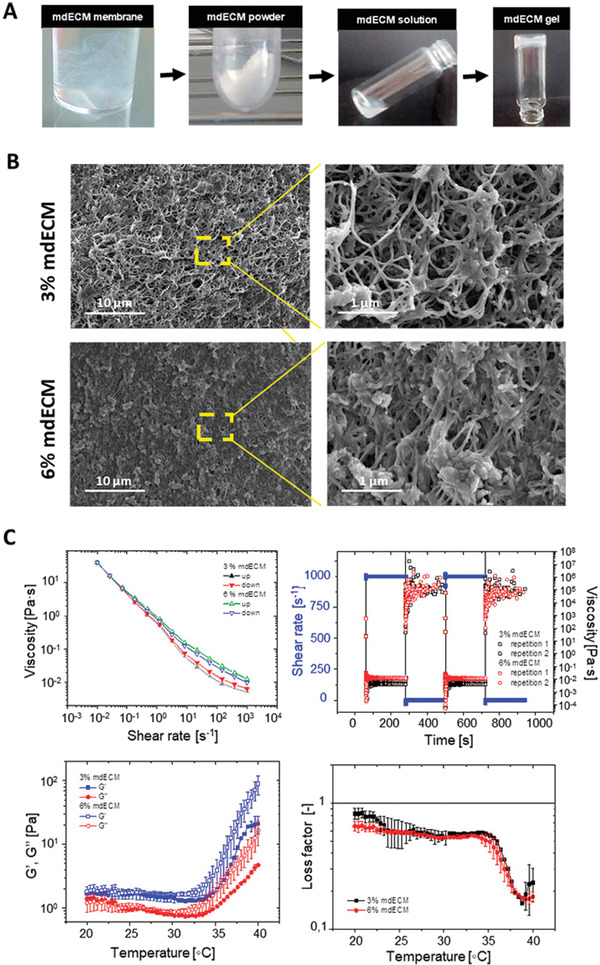
Formulation and characterization of mdECM hydrogel. A) Schematic elucidating preparation of hydrogel based on early chondrogenic mdECM. B) Scanning electron micrographs (SEM) of crosslinked mdECM hydrogel. Magnifications of 10 000× (at left) and 40 000× (at right). C) Steady shear flow behavior of the hydrogel under an isothermal up and down shear rate ramp at 15 °C. D) Transient shear recovery behavior of the hydrogel under isothermal stepwise changes in the shear rate at 15 °C. Thermal‐induced gelation of the hydrogel as determined by linear viscoelasticity tests E) and by the loss factor (G′′/G′) F) of the hydrogels from 20 to 40 °C. Data represent mean ± S.D. (n = 3).
2.3. Characterization of mdECM Hydrogel
We used scanning electron microscopy (SEM) to examine the ultrastructure of the mdECM hydrogel at 37 °C. Micrographs showed that the generated gel presented a 3D sponge‐like appearance with a randomly oriented nanofibrillar structure, typical of decellularized ECM‐based gels[ 39 ] (Figure 3B). 3% and 6% mdECM hydrogels appeared visually similar with a homogenous microporous system, although fibrillar density and interconnectivity was higher with increasing concentration (Figure 3E). Porous of 2.72 ± 1.54 µm in diameter were formed inside the 3% mdECM and 1.35 ± 0.77 µm in 6% mdECM.
The injectability and gelling abilities of the hydrogel, at 3% and 6% mdECM, were analyzed using steady shear, intermittent flows, and dynamic oscillation experiments. Figure 3C shows the viscosity curves for the mdECM hydrogels at 15 °C. As observed, the hydrogels exhibit a very strong shear thinning behavior. This is evidenced by the decrease of the viscosity with the shear rate, ranging values from 39.65 and 40.1 Pa s at 0.01 s–1 shear rate to 0.0057 and 0.0113 Pa s at 1000 s–1 shear rate for 3% and 6% mdECM, respectively. As expected, the higher the matrix concentration, the larger the viscosity. Also important is to note that the curves overlap when performing up‐and‐down shear rate sweeps. This is an indication that the sample is thixotropic.
Figure 3D shows the viscosity transients under sudden changes in the shear rate at 15 °C. Results show that the transients are extraordinarily short, and the samples respond very quickly (relaxation times ≈1 s) to the excitation; both at large and small shear rates. This is in agreement with the fact that up and down steady shear curves superimposed in Figure 3C.
The thermal‐driven gelation of the suspensions is shown in Figure 3E. This Figure represents the linear viscoelastic moduli (storage modulus G′ and loss modulus G′′) as a function of the temperature in the interval ranging from 20 to 40 °C. Independent on the concentration and the temperature, the storage modulus remains always larger than the loss modulus evidencing the existence of a gel‐like structure in the range of temperatures investigated. However, at a particular temperature of ≈35 °C the viscoelastic moduli increase further due to self‐assemble of main components of the matrix, such as collagen fibers or fibronectin, among others.
Figure 3F shows the loss factor (G′′/G′) of the hydrogels as a function of the temperature. The loss factor is a measure of the relative importance of viscous dissipation over elastic recovery of the samples. Independently on the concentration, the loss factor is always smaller than one as expected from the formation of a gel (G′ > G′′). The loss factor dramatically decreases for temperatures above 35 °C. It is worth to remark that contrary to the viscoelastic moduli, the loss factor is independent on the concentration.
2.4. Biocompatibility of mdECM Hydrogel
We evaluated cell viability and proliferation of MSCs encapsulated within the mdECM hydrogels at 3% and 6%, respectively for 28 days in culture. Cell survival was tested using a Live/Dead staining using confocal microscopy. Images evidenced that cells were homogenously distributed throughout the hydrogel and maintained the rounded morphology in both concentrations of mdECM and throughout the culture period (Figure 4A). Results showed that more than 90% of embedded MSCs within the hydrogels at 3% and 6% mdECM were viable (day 1), without remarkable differences between them. This high viability was maintained over time in 6% mdECM hydrogel, meanwhile in 3% mdECM it was observed a lower number of living cells by the end of the culture period, although this value (83.3%) was also indicative of a high cell viability (Figure 4B).
Figure 4.
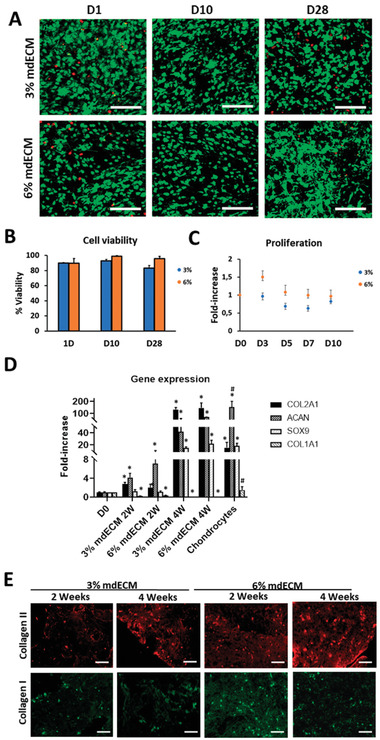
Biological assessments in vitro of mdECM hydrogel. A) Representative confocal images of cell viability using Live/Dead assay at 1, 10, and 28 days (green for live cells, red for dead cells). B) Cell viability (%) in the mdECM hydrogel after 1, 10, and 28 days. C) Proliferation rate of MSCs cultured in mdECM hydrogel in comparison to day 0 of culture. D) Gene expression analysis of specific (COL2A1, ACAN, SOX9) and nonspecific chondrogenic (COL1A1) genes of MSCs embedded in 3% and 6% mdECM over time in culture (2, 4 weeks) and chondrocytes in culture for 2 weeks (control +). E) Immunostaining analysis for Collagen type II and Collagen type I. Scale bar 100 µm. Data are expressed as mean ± S.D. (n = 3); *, # p < 0.01, Student's t‐test. (*)Value significantly different in comparison to values from MSCs cultured at day 0; (#) Value significantly different in comparison to values from MSCs in 3% and 6% mdECM hydrogels after 4 weeks in culture.
On the other hand, Alamar Blue (AB) assay revealed a tendency to maintain cell proliferation over culture time in both concentrations. In 3% mdECM this tendency was produced from the beginning, showing a slight reduction in cell proliferation between day 5 and day 7. Meanwhile, the proliferation rate in 6% mdECM was maintained after three days in culture a decreasing up to day 14 at similar level than 3% mdECM (Figure 4C). Altogether, these results indicate that the new biomimetic hydrogel based on mdECM provided a cell‐friendly environment, supporting high cell viability at different concentrations of mdECM.
2.5. Chondrogenic Differentiation of MSCs‐Loaded mdECM Biomimetic Hydrogel
Chondro‐inductivity of mdECM hydrogel was investigated by gene expression analysis of embedded MSCs during 1 month in culture without any inductive factor in the media (Figure 4D). qRT‐PCR results evidenced a constant increment in the expression of chondrogenic markers and decrease on non‐chondrogenic markers over time. High levels in Collagen type II (COL2A1) and Aggrecan (ACAN) genes were observed by day 14 in MSCs embedded in 3% and 6% mdECM hydrogels, without significant differences between concentrations, in comparison to these cells at day 0 of culture. At day 28, the increase in gene expression of these chondrogenic markers were even more evident. The chondrogenic inductor factor gene Sox‐transcription factor 9 (SOX9) and COL2A1 raised values that were similar or even higher to the positive control chondrocytes cultured during 2 weeks in vitro, respectively. However, ACAN expression in chondrocytes were significantly higher than MSCs in mdECM hydrogels by day 28. Although this upregulation seemed to be in a concentration dependent manner, differences in expression levels among 3% and 6% mdECM were not statistically significant. A decrease in fibrotic marker gene, Collagen type I (COL1A1) was also observed from day 14, and much more significant after 28 days in both hydrogels in comparison to day 0, whereas the expression of Collagen type X (COL10A1) hypertrophic marker gene was undetectable from the beginning.
Also, to confirm results from gene expression, we evaluated chondrogenesis at protein level using immunofluorescence techniques (Figure 4E). Immunofluorescence staining revealed the presence of Collagen type II and Collagen type I after 14 days in culture. In accordance with the qRT‐PCR results previously described, it was observed a good evolution of the hyaline cartilaginous matrix over time, with more positive staining for Collagen type II and less for Collagen Type I by day 28, without appreciable differences between concentrations. Altogether, these results indicated that mdECM environment promoted chondrogenic commitment of MSCs and tissue‐specific formation despite the concentration.
2.6. In Vivo Biocompatibility and Cartilage‐Like Tissue Development of mdECM Hydrogel
To evaluate whether mdECM would have biocompatibility and good integration in vivo, hydrogels were subcutaneously implanted at both concentrations into dorsal regions of immunocompetent CD1 mice along 4 weeks. Figure 5A shows good integration of acellular mdECM hydrogels with no red or swelling appearance. DAPI staining also revealed infiltration of surrounding cells (Figure 5B). The increase in number of nucleus over time indicated that cells were able to attach, penetrate, and grow into the 3D structure of the hydrogels.
Figure 5.
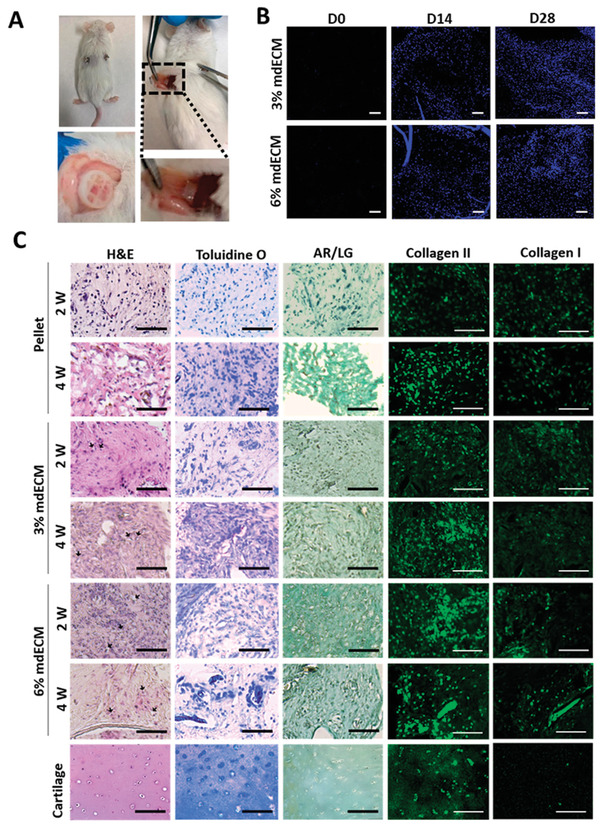
Biological assessments in vivo of mdECM hydrogel. A) Images at 4 weeks of post‐implantation in CD1 mice. B) DAPI staining showing cells infiltrating the acellular mdECM gels at 3% and 6% at day 0, 14, and 28 after their subcutaneously implantation in CD1 mice. C) Histological analysis of mdECM gels at 3% and 6% concentrations, pellets of MSCs predifferentiated in vitro with chondrogenic media after subcutaneous implantation in NSG mice at 2 and 4 weeks, and articular cartilage (control +). Arrows indicate cells embedded in lacunas. Scale bar 100 µm.
Tissue formation was determined in immunodeficient NOD SCID gamma mice by histological analysis of 3% and 6% MSCs‐loaded mdECM hydrogels in comparison to MSCs predifferentiated towards chondrogenic lineage in pellets (Figure 5C). H&E staining showed that cells in mdECM hydrogel acquired the typical shape of chondrocytes embedded in lacunas after 2 weeks in vivo, being more evident in the highest concentration. Using Toluidine O staining, it was evidenced an increase in the deposition of GAGs, showing an intense staining in four‐week implanted 3% and 6% mdECM gels, that was higher than that observed in control pellet. By immunostaining, it was determined high levels of Collagen type II and low levels of Collagen type I in both concentrations of mdECM over time. Both in histological and immunostaining assays, staining levels were higher in mdECM gels than control pellet. Alizarin Red/Light green (AR/LG) staining also revealed no presence of mineralization in any condition. These results indicate the formation of a mature cartilaginous matrix in mdECM hydrogels, especially in 6% mdECM, whose matrix was more similar to cartilage tissue, and confirm the ability of mdECM hydrogels to induce cartilage‐like tissue in vivo.
3. Discussion
Hydrogels are ideal scaffolds for cartilage repair and regeneration.[ 7 , 8 , 9 ] Cell‐derived ECM offers an alternative option to produce biomimetic dECM hydrogels as it contains a complex assembly of macromolecules, including fibrillar proteins and GAGs, and it is easy to obtain on a larger scale by simple monolayer culture. Especially, mECM can be customized depending on culture conditions. In this work, we developed a new biomimetic hydrogel based on an early chondrogenic matrix derived from MSCs in culture, with suitable physicochemical and biological properties to be applied to promote cartilage tissue‐specific regeneration. In particular, we proved that mdECM hydrogel could successfully induce chondrogenesis in vitro and further form cartilage‐like tissue cartilage after in vivo implantation.
During tissue development, including chondrogenesis, the composition of extracellular microenvironments is constantly changing in order to regulate the process and provide appropriated signals at each development state.[ 29 , 30 , 31 , 32 ] In this line, there are studies that tested c‐dECM as coating culture substrate at different stage of maturity, demonstrating different effects on cells, including gene expression profile or metabolic activity that regulate cell phenotype, ECM synthesis or proliferation rate, among others.[ 22 , 34 , 40 , 41 ] Particularly, it has been demonstrated that the differentiation of MSCs towards chondrogenic,[ 42 ] osteogenic,[ 33 ] or adipogenic lineage[ 43 ] occurred more rapidly on early matrices than in later stage matrices. These results were related to biochemical composition, as early stage matrix contains elements and factors that are not present in the mature one. In addition, to provide more favorable microenvironment for tissue specific formation, the use of matrices from early stages of differentiation supposes a reduction in time required to generate biomimetic hydrogels for clinical applications.
In our work, we differentiated MSCs during two weeks in culture to generate an early chondrogenic matrix consisting in the initial expression of major components of native tissue, Collagen type II and GAGs, as well as proteins that have been linked to the first steps of chondrogenesis such as Fibronectin, Collagen type I, Versican or Tenascin‐C.[ 31 ] In addition to those markers, mass spectrometry also showed the identification of other components that constitute this rich and complex matrix, including structural proteins such as fibrillar family and associated collagens (type III, IV,V,VIII,XII), proteoglycans (Lumican, Biglycan, Decorin) and glycoproteins (Emilins, COMP), that confer mechanical properties and cell adhesion,[ 31 , 44 ] as well as affiliated proteins that contribute to ECM function and dynamics such as Annexins, Galectins, MMPs or TIMPs,[ 44 , 45 ] and signaling factors such as IGF2, HDGF, or CTGF, which are known to stimulate chondrocyte differentiation, deposition of fibrillary extracellular matrix and cell proliferation.[ 46 , 47 , 48 , 49 , 50 ]
The formulation of the c‐dECM hydrogel, as in the case of t‐dECM, involves a process that includes decellularization and solubilization of matrix. The first step was the most important, since it must ensure that no cellular material remains while preserving the most ECM components, which provide matrix functionality. There are several protocols that have been applied to decellularize ECM from monolayer culture.[ 51 ] Among these, the most frequently used has been the combination of Triton X‐100 (mild detergent) and ammonium hydroxide (alkaline reagent), to break down cellular and nuclear membranes, followed by enzymatic treatment based on nucleases to disrupt cytoplasmic content and break down remnant DNA and RNA.[ 52 , 53 ] Using this method, researchers have demonstrated decellularization efficiency without influencing ECM integrity. Here, we also effectively decellularized the early chondrogenic matrix derived from MSCs. The absence of nuclei and DNA content, which was lower than the upper limit content considered (<50 ng mg−1 DNA),[ 54 ] confirmed the complete removal of cellular components while similar patterns in terms of composition indicated minimal subtraction of ECM elements after the decellularization process. A biomimetic hydrogel based on mdECM was finally attained after solubilization and neutralization of the decellularized matrix, at different concentrations (3% and 6% w/v). The hydrogel behaved as a shear thinning, nonthixotropic material with a very short relaxation time. It also exhibited a thermoresponsive character as evidenced from SAOS tests. In the temperature range investigated, the storage modulus was always larger than the loss modulus. However, above 35 °C the viscoelastic moduli increased. This was expected due to the presence of self‐assembling molecules responsible for the gelation such as collagens, laminins, and proteoglycans, and it would assure stability of extruded gel for long time.[ 55 , 56 , 57 ] The fact that the loss factor is independent on the concentration suggests a similar scaling of G′ and G′′ with the concentration. Both steady and unsteady rheological tests demonstrated that mdECM hydrogel at both concentrations have suitable properties to be used as an injectable hydrogel not only for local cartilage repair but also for development of cartilage‐like biomimetic substitutes by 3D bioprinting,[ 58 ] which would provide more advantages structurally and mechanically. On the one hand, it exhibited a strong shear‐thinning behavior, which is essential to assure good extrusion and cell protection during the pass through the nozzle. On the other hand, the mdECM hydrogel demonstrated a very quick stress recovery (i.e., a small transient upon stepwise changes in shear rate), which would ensure the filling of the defect and gel retention after extrusion.
Besides biochemical composition and mechanical properties, other important requirements for hydrogels are biocompatibility and bioactivity.[ 57 , 59 ] Here, we showed that mdECM hydrogel provided a cell‐friendly environment, supporting high cell viability at different concentrations of 3% and 6%. These results are in agreement with other studies reporting the benefits of c‐dECM as biomaterial in TE, including improvement of cell maintenance, reduction of apoptosis and intracellular reactive oxygen species activity decrease.[ 23 , 24 , 26 , 60 ] In our study, SEM results showed that biocompatibility was also provided by its porous ultrastructure, which allows diffusion of nutrients and oxygen, even in the highest matrix concentration, while preserving the rounded shape of cells, which is essential to promote the chondrogenic phenotype.[ 61 , 62 ] In terms of functionality, the mdECM hydrogel was capable to induce chondrogenic differentiation of MSCs and cartilage tissue formation without stimulating supplements. These effects were observed by an increase in cartilage‐specific markers, both at genetic (COL2A1, ACAN, and SOX9 genes) and protein (Collagen type II and GAGs) levels using qRT‐PCR and histological techniques, respectively. These results highlight the role that ECM plays on cell behavior and, therefore, the importance of its structural composition that it is known to promote tissue‐specific differentiation of progenitor cells. The fact that there were no significant differences between mdECM concentrations would be a great benefit for the generation of a functional hydrogel using reduced amount of mdECM. Accordingly, many studies have evidenced the capacity of specific ECMs, from tissue or cells in culture, to direct stem cell fate toward particular cell lineages.[ 25 , 33 , 63 , 64 , 65 , 66 ] In this line, interesting results have also been reported by using stepwise‐mimicking matrices derived from stem cells, as it presents a composition that reproduce better the environment and contain adequate factors necessaries for tissue development.[ 22 , 33 ] Here, cell differentiation could also explain the minimal contraction observed in mdECM hydrogels, slightly more noticeable in the higher concentration (data not shown). In fact, it has been reported in literature that cell aggregation is a crucial step during chondrogenesis process and it might involve the contraction of hydrogel where cells are embedded.[ 67 ] In addition, MSCs differentiation would explain the maintenance of proliferation that was observed along the culture period in both concentrations, although 6% showed an increase during the first days. This fact could be related to a higher content in fibronectin, which is involved in the promotion of cell survival and proliferation during the first stage of chondrogenesis[ 31 ] and/or by restriction of cell migration due to the high density of the fibrillar structures.
As a follow‐up step, we evaluated the potential of mdECM hydrogels in vivo by their subcutaneous implantation in a mice model. Similar to the in vitro study, it was demonstrated the biocompatibility by the high cell infiltration and no immune reaction or inflammation in immunocompetent mice. These results could be explained by the effective decellularization of the mECM and the presence of immunomodulatory factors, secreted by MSC during matrix production and retained in after decellularization.[ 68 , 69 ] In addition, after implantation of MSCs‐loaded mdECM in immunodeficient mice, the histological analysis also revealed that mdECM hydrogels support hyaline cartilage development, showing an increased deposition of cartilage components (GAGs and Collagen type II) and no presence of hypertrophic markers over 4 weeks. Furthermore, the neo‐cartilage tissue formed in both concentrations of mdECM gels was better than control pellet, which is the current methodology used to demonstrate chondrogenesis, and acquired a maturity that could resemble cartilaginous matrix of native tissue, in the case of mdECM hydrogel at the higher concentration (6% mdECM). In accordance with our results, there are other in vivo studies that have reported cartilage formation using lyophilized c‐dECM scaffolds.[ 70 , 71 ] In particular, these studied used chondrocyte‐derived ECM or stem cell‐derived ECM with encapsulated chondrocytes for in vivo neo‐cartilage formation, obtaining better results than current strategies such as Autologous Chondrocyte Implantation (ACI) or autologous cultured chondrocytes on porcine collagen membrane (MACI).[ 70 , 72 , 73 ] However, unlike these studies, this is the first time that it has been evidenced the generation of articular cartilage using a MSCs‐loaded hydrogel based on chondrogenic ECM derived from MSCs in culture. The advantage of this mdECM is the preservation of biological activity, which is mediated by specific biochemical and physical signaling motifs contained in the matrix, which guides chondrogenic differentiation, without the limitation on donor cartilage tissue availability.
4. Conclusions
We successfully demonstrated that MSCs are an attractive source of ECM useful to produce biomimetic hydrogels. Their stem cell properties, such as self‐renewal and capacity to commit towards several cell lineages, allowed the generation of a biomimetic cartilaginous ECM easily scalable and with low cost. We have formulated a novel hydrogel based on an early chondrogenic matrix, that mimic the environment established during chondrogenesis, with appropriate rheological properties. Moreover, the mdECM hydrogel exhibits biocompatibility and capability to induce chondrogenesis and further hyaline cartilage formation without stimulating supplements. Although it has been addressed for cartilage regeneration, our results encourage applying this strategy to produce different biomimetic hydrogels for repair and regeneration other tissues and organs through tissue engineering, including 3D bioprinting.
5. Experimental Section
Isolation, Culture, and Characterization of MSCs
Human MSCs were isolated from adipose tissue of patients that had liposuction procedure after informed consent and authorization from the Ethics Committee of Clinical University Hospital of Málaga, Spain. Isolation characterization of MSCs were previously reported.[ 74 ] Cells were seeded in growth media (high‐glucose Dulbecco's modified Eagle's medium (DMEM; Sigma‐Aldrich) supplemented with 10% fetal bovine serum (FBS; Sigma‐Aldrich), 100 U mL−1 penicillin, and 100 mg mL−1 streptomycin (Invitrogen Inc.)) at 37 °C in a humidified atmosphere with 5% CO2. Medium was changed every 3 days and when cells reached 80% of confluence, they were subcultured. For all the experiments MSCs were used between passages 4 and 6.
Phenotype and differentiation potential of isolated cells were characterized, as previously described.[ 38 ] To examine their immunophenotype, cells were trypsinized, washed, and resuspended in phosphate‐buffered saline (PBS) with 1% bovine serum albumin (BSA; Sigma‐Aldrich). A total of 2 × 105 cells was incubated in the dark for 30 min with fluorochrome‐conjugated monoclonal antibodies [CD34, CD45, CD90, CD73, CD105, and CD133 (Miltenyi Biotec, Auburn, CA, USA)], washed in PBS and analyzed by flow cytometry in a FACSCanto II cytometer (BD Biosciences). For the differentiation assays, MSCs were plated at 1 × 105 cells cm−2 in DMEM‐FBS into 6‐well culture plates. After 48 h, the culture medium was replaced with specific differentiation‐inductive medium. For adipogenic, osteogenic, and chondrogenic differentiation, cells were cultured for 2 weeks in MSC Adipogenic Differentiation BulletKit (Lonza), MSC Osteogenic Differentiation BulletKit (Lonza) and StemMACS ChondroDiff Medium (Miltenyi Biotec), respectively. Differentiated cell cultures were stained with oil red O (Amresco, Solon, OH, USA) for adipogenic differentiation, alizarin red (Lonza) for osteogenic differentiation, or toluidine blue (Sigma‐Aldrich) for chondrogenic differentiation.
Isolation and Culture of Chondrocytes
Chondrocytes were isolated from articular cartilage of patients with knee osteoarthritis (described in detail in the Supporting Information), as previously reported.[ 38 ] Cells were seeded in chondrocytes medium (growth medium supplemented with 1% ITS (Insulin‐Transferrin‐Selenium, Gibco), 50 µg µL−1 of ascorbic acid (Sigma), 40 µg µL−1 of proline (Sigma) and 100 U mL−1 penicillin and 100 µg mL−1 streptomycin), at 37 °C in a humidified atmosphere with 5% CO2. At 80% of confluency cells were subcultured.
Production of the Early Chondrogenic mECM
At complete confluence, MSCs were released with TriPLE (Invitrogen) and subcultured in Petri dishes at the cell density of 1 × 105 cells cm−2. To allow the formation of ECM membrane, they were cultured in monolayer during 1, 2, 3, or 4 weeks in chondrocytes media supplemented with dexamethasone (100 × 10−9 m) and TGF‐β3 (10 ng mL−1) at 37 °C humidified atmosphere containing 5% CO2. The medium was refreshed every 2 or 3 days. As a control, ECM was generated from chondrocytes in culture in the same way, but without using supplements.
Decellularization of the Early Chondrogenic mECM
At the completion of culture period, cells were removed from the underlying matrix by exposition to a decellularization treatment, selected from a variety of methodologies that have been previously reported to prepare cultured c‐dECM scaffolds.[ 35 ] Briefly, the chemical procedure consisted on incubation in a detergent solution containing 0.25% Triton X‐100 and 10 × 10−3 m NH4OH at 37 °C for 5 min, followed by treatment with 50 units mL−1 deoxyribonuclease (DNase) I and 50 mg mL−1 ribonuclease (RNase) A (Invitrogen) for 2 h. After several washes with PBS, culture plates were examined by optical microscopy to ensure that all cellular material were removed. Finally, the supernatant was removed and matrix remaining on the plate was scraped and lyophilized.
Preparation of Hydrogel Based on Early Chondrogenic mdECM
Lyophilized mdECM was crushed into powder, using liquid N2. Required amount of mdECM powder was digested in pepsin solution (1 mg mL−1 in 0.1 n HCl) for 48 h. It was prepared at two concentrations: 3% and 6% w/v mdECM. After solubilizing mdECM, the pH was neutralized with dropwise addition of 1 m NaOH and 10× PBS (to 1× final dilution), at low temperatures (below 15 °C) to avoid gelation of the mdECM. The resulting pre‐gel mdECM was mixed with MSCs at a concentration of 5 × 106 cells mL−1 for further biological studies. After gelation at 37 °C, mdECM hydrogels with embedded cells were maintained in growth medium for specific time, defined in each experiment.
Rheological Assays
The rheological properties of the mdECM hydrogels (3% and 6% w/v) were investigated with a torsional rheometer (MCR301, Anton Paar, Austria) using a cone‐plate geometry (20 mm diameter and 2° angle). Steady shear tests were carried out to determine the shear viscosity at 15 °C. These tests consisted in four steps. In the first one the sample was presheared at a constant shear rate (500 s–1) during 30 s. In the second step the sample was allowed to rest during 30 s in the absence of shear. In the third step the shear rate was logarithmically increased from 0.1 to 1000 s–1. Finally, in the fourth step the shear rate was decreased from 1000 to 0.1 s–1 to explore whether the sample is thixotropic or not.
To get a better insight on the thixotropic behavior of the samples, intermittent flow tests were also carried out using stepwise changes in shear rate with a logarithmic measuring point duration from 0.01 to 10 s at 15 °C. The protocol consisted in six steps. In the first step the sample was presheared at a constant shear rate (500 s–1) during 30 s. In the second step the sample was allowed to rest during 30 s in the absence of shear. The third step consisted in a start‐up test at 1000 s–1 while the fourth step consisted in suddenly reducing the shear rate to 0 s–1. Steps three and four were repeated in intervals five and six for consistency in order to measure the viscosity transients.
The thermal driven gelation of the samples was investigated using small amplitude oscillatory shear (SAOS) tests at a strain amplitude of 1% and a frequency of 1 Hz. The protocol consisted in two steps. In the first step, the samples were equilibrated at 20 °C for 5 min. Finally, in the second step SAOS tests were performed while the temperature increased from 20 to 40 °C at a rate of 1 °C min−1.
Cell Viability
The Live/Dead kit assay (Thermo Fisher Scientific) was used to evaluate cell viability, following manufacturer's instructions. Images from the samples were observed using a confocal microscope (Nikon Eclipse Ti‐E) and analyzed with Image J software (v. 1.52i, USA). The percentage of viable cells were obtained by counting six regions of each sample (n = 3).
Cell Proliferation
The proliferation rate of cells embedded in mdECM hydrogels (n = 3) were assessed by colorimetric AB assay (Thermo Fisher Scientific) at different time points, following manufacturer's instructions. It was normalized to the appropriate control without cells. The absorbance data were represented as fold increase to day 0.
Gene Expression Analysis
Total messenger RNA (mRNA) of MSCs embedded in mdECM hydrogels (n = 3) at different time points was isolated using TriReagent (Sigma) and reverse‐transcribed into cDNA using the Reverse Transcription System kit (Promega), according to manufacturer's instructions. Quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed using a SYBR green master mix (Promega), following the manufacturer's instructions. Gene expression levels for (COL2A1, ACAN, SOX‐9, and COL1A1 were normalized to the housekeeping gene Glyceraldehyde 3‐phosfate dehydrogenase (GADPH) and showed as fold change relative to the value of embedded MSCs at day 0.
Histology
mdECM hydrogels, pellets, and articular cartilage tissue were fixed in 4% paraformaldehyde (PFA), serially dehydrated, embedded in paraffin, and sectioned (4 µm thickness) with a microtome (Leica RM2255, Leica Biosystems, USA). Samples were rehydrated and stained with H&E, Toluidine O, Masson's Tricrome and AR/LG. For monolayer staining, fixed cells were directly stained using Toluidine O and Sirius Red. All samples were photographed using Leica DM 5500B microscope under bright field and analyzed using Image J software.
Quantitative Biochemistry Assays
Lyophilized mECM and mdECM were papain digested and assayed for GAG and DNA quantification. GAGs content was estimated by dimethyl methylene blue (DMMB) colorimetric assay. DNA content was quantified by fluorometric assay using DAPI staining. For collagen quantification, lyophilized samples were firstly solubilized in pepsin solution (2 mg mL−1 in acetic acid 0.5 m). Total collagen content was measured by Picrosirius red staining. Collagen type II content was determined using a commercially available Collagen type II ELISA kit (Chondrex). The quantification assays are described in detail in the Supporting Information.
Immunostaining Assay
Cell monolayers, mdECM hydrogels, pellets, and articular cartilage tissue were fixed with 4% PFA in PBS for 20 min at RT. For analysis by immunofluorescence, mdECM hydrogels, pellets, and articular cartilage tissue were embedded in optimal cutting temperature (OCT) compound and sectioned using cryotome in 10 mm thickness. Sections and monolayers were treated with a primary antibody against Collagen type I (Santa Cruz Biotechnology, 1:100), Collagen type II (Santa Cruz Biotechnology, 1:100), and Aggrecan (Sigma, 1:100). Sections were then incubated with AlexaFluor 488 or 594‐conjugated secondary antibodies (ThermoFisher, 1:500) and counterstained with DAPI. Images were obtained using a Leica DM 5500B microscope and analyzed with Image J software.
Analysis by MS
The full characterization of protein content in mECM was performed by tandem mass spectroscopy (LC/MS). Samples were resuspended in 50 × 10−3 m NH4HCO3 (pH 8.5) and a small part (1/20) was digested. The dried‐down peptide mixtures were analyzed in a nanoAcquity liquid chromatographer (Waters) coupled to a LTQ‐Orbitrap Velos (Thermo Scientific) mass spectrometer. The data were utilized to search against a modified version of the public database SwissProt human. Database search was performed with Sequest HT search engine using Thermo Proteome Discover (v.1.4.1.14). The results were observed in Proteome Discoverer (v.1.4.1.14) and the lists of identified proteins was exported as Excel file. More detailed information about the sample preparation and the analysis is found in the Supporting Information.
SEM Analysis
The internal microstructure of hydrogel was examined using SEM. mdECM hydrogel at different concentrations were fixed in cold 2.5% glutaraldehyde. After several washes with PBS, samples were dehydrated using a graded series of ethanol (30–100%). These were critically point dried in an Emscope CPD 750 critical point dryer, attached to aluminum SEM specimen mounting stubs and sputter coated with a gold palladium alloy, by using a Sputter Coater 108 Auto. Images from samples were obtained using a SEM (Quanta 400 (FEI), at a 10 000× and 40 000× magnification.
In Vivo Experiments
In vivo assays were performed according to approved guidelines of University of Granada and following institutional and international standards for animal welfare and experimental procedure. All experiments were approved by the Research Ethics Committee of the University of Granada (sol438‐CEEA‐OH‐2020).
In vivo biocompatibility was assessed in immunocompetent CD1 (ICR) mice. Acellular gels were introduced in PCL scaffolds (a porous cylinder‐type structure, 8 × 5 mm; 3 mm pore size) for easy localization, and transplanted into the back subcutaneous tissue of mice anesthetized by isoflurane inhalation (n = 6).
For in vivo evaluation of tissue formation and cartilage maturation, 3% and 6% mdECM hydrogels loaded with cells (5 × 106 cells mL−1), previously cultured in growth media during 2 weeks in vitro, were subcutaneously injected in immunodeficient NOD SCID gamma (NOD.Cg‐Prkdcscid Il2rgtm1Wjl/SzJ, NSG) mice (n = 8) following the same methodology as in CD1 mice experiment. As a positive control (n = 6) predifferentiated MSCs toward chondrogenic lineage in pellet system were used during 2 weeks in vitro. For this, 3 × 105 MSCs were centrifuged (1500 rpm, 5 min) in a 15 mL conical tube and incubated with loosened tops at 37 °C and 5% CO2. Chondrogenic media was changed every other day for 2 weeks and the tubes were gently shaken to avoid the adherence of the pellet to the plastic walls. Animals were maintained in a microventilated cage system with food and water ad libitum. Mice were manipulated in a laminar air‐flow to maintain sterile conditions. Two and four weeks later, mice were sacrificed via an overdose injection of anesthetics, and hydrogels were excised for further histologic and immunofluorescence analysis.
Statistical Analysis
All graphed data represent the mean +/−SD from at least three experiments. To determine differences between conditions, the two‐tailed Student's t‐test was used for quantitative biochemistry analysis to determine differences between conditions and two‐way ANOVA test for gene expression analysis using GraphPad Prism 8.3.0 software. Assumptions of homoscedasticity and normality of data were tested and assured by using transformed data sets [log(dependent variable value + 1)] when necessary. p‐values <0.001 (***), < 0.01 (**) and < 0.05 (*) were considered statistically significant in all cases.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Acknowledgements
Ana Santos, Mohamed Tassi, Jose Manuel Entrena, Gustavo Ortiz, and Isabel Sánchez from the C.I.C. (University of Granada) are acknowledged for excellent technical assistance. C.A. acknowledges the predoctoral fellowship from the Spanish Ministry of Education, Culture and Sports (BOE‐A‐2014–13539). This research was supported by the Ministerio de Economía, Industria y Competitividad (ERDF funds, project RTC‐2016‐5451‐1), Fundación Mutua Madrileña (project FMM‐AP17196‐2019), Consejería de Economía, Conocimiento, Empresas y Universidad de la Junta de Andalucía (ERDF funds, projects B‐CTS‐230‐UGR18, PY18‐2470, SOMM17‐6109, and P18‐FR‐2465), and the Instituto de Salud Carlos III, ERDF funds (DTS19/00145).
Antich C., Jiménez G., de J. Vicente, López‐Ruiz E., Chocarro‐Wrona C., Griñán‐Lisón C., Carrillo E., Montañez E., Marchal J. A., Development of a Biomimetic Hydrogel Based on Predifferentiated Mesenchymal Stem‐Cell‐Derived ECM for Cartilage Tissue Engineering. Adv. Healthcare Mater. 2021, 10, 2001847. 10.1002/adhm.202001847
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Heinegård D., Paulsson M., Methods Enzymol. 1987, 145, 336. [DOI] [PubMed] [Google Scholar]
- 2. Ryu J., Treadwell B. V., Mankin H. J., Arthritis Rheum. 1984, 27, 49. [DOI] [PubMed] [Google Scholar]
- 3. Poole A. R., Kojima T., Yasuda T., Mwale F., Kobayashi M., Laverty S., Clin. Orthop. Relat. Res. 2001, 391, S26. [DOI] [PubMed] [Google Scholar]
- 4. Zylińska B., Silmanowicz P., Sobczyńska‐Rak A., Jarosz Ł., Szponder T., In Vivo 2018, 32, 1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armiento A. R., Stoddart M. J., Alini M., Eglin D., Acta Biomater. 2018, 65, 1. [DOI] [PubMed] [Google Scholar]
- 6. Van Tomme S. R., Storm G., Hennink W. E., Int. J. Pharm. 2008, 355, 1. [DOI] [PubMed] [Google Scholar]
- 7. Vilela C. A., Correia C., Oliveira J. M., Sousa R. A., Espregueira‐Mendes J., Reis R. L., ACS Biomater. Sci. Eng. 2015, 1, 726. [DOI] [PubMed] [Google Scholar]
- 8. Pascual‐Garrido C., Aisenbrey E. A., Rodriguez‐Fontan F., Payne K. A., Bryant S. J., Goodrich L. R., Am. J. Sports Med. 2019, 47, 212. [DOI] [PubMed] [Google Scholar]
- 9. Jooybar E., Abdekhodaie M. J., Alvi M., Mousavi A., Karperien M., Dijkstra P. J., Acta Biomater. 2019, 83, 233. [DOI] [PubMed] [Google Scholar]
- 10. Roseti L., Cavallo C., Desando G., Parisi V., Petretta M., Bartolotti I., Grigolo B., Materials 2018, 11, 1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gungor‐Ozkerim P. S., Inci I., Zhang Y. S., Khademhosseini A., Dokmeci M. R., Biomater. Sci. 2018, 6, 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo Y., Wei X., Huang P., J. Biomed. Mater. Res., Part B 2019, 107, 1695. [DOI] [PubMed] [Google Scholar]
- 13. Jessop Z. M., Gao N., Manivannan S., Al‐Sabah A., Whitaker I. S., 3D Bioprinting for Reconstructive Surgery—Techniques and Applications, Elsevier Inc., Amsterdam: 2018, pp. 277–304. [Google Scholar]
- 14. Burnsed O. A., Schwartz Z., Marchand K. O., Hyzy S. L., Olivares‐Navarrete R., Boyan B. D., Acta Biomater. 2016, 43, 139. [DOI] [PubMed] [Google Scholar]
- 15. Beck E. C., Barragan M., Tadros M. H., Kiyotake E. A., Acosta F. M., Kieweg S. L., Detamore M. S., Ann. Biomed. Eng. 2016, 44, 1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beck E. C., Barragan M., Tadros M. H., Gehrke S. H., Detamore M. S., Acta Biomater. 2016, 38, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bordbar S., Lotfi Bakhshaiesh N., Khanmohammadi M., Sayahpour F. A., Alini M., Baghaban Eslaminejad M., J. Biomed. Mater. Res., Part A 2020, 108, 938. [DOI] [PubMed] [Google Scholar]
- 18. Fitzpatrick L. E., McDevitt T. C., Biomater. Sci. 2015, 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang W., Zhu Y., Li J., Guo Q., Peng J., Liu S., Yang J., Wang Y., Tissue Eng., Part B 2016, 22, 193. [DOI] [PubMed] [Google Scholar]
- 20. Lu H., Hoshiba T., Kawazoe N., Chen G., Biomaterials 2011, 32, 2489. [DOI] [PubMed] [Google Scholar]
- 21. Hoshiba T., J. Mater. Chem. B 2017, 5, 4322. [DOI] [PubMed] [Google Scholar]
- 22. Cai R., Nakamoto T., Kawazoe N., Chen G., Biomaterials 2015, 52, 199. [DOI] [PubMed] [Google Scholar]
- 23. Hidalgo‐Bastida L. A., Cartmell S. H., Tissue Eng., Part B 2010, 16, 405. [DOI] [PubMed] [Google Scholar]
- 24. Li J., Hansen K. C., Zhang Y., Dong C., Dinu C. Z., Dzieciatkowska M., Pei M., Biomaterials 2014, 35, 642. [DOI] [PubMed] [Google Scholar]
- 25. Tang C., Jin C., Xu Y., Wei B., Wang L., Tissue Eng., Part A 2016, 22, 222. [DOI] [PubMed] [Google Scholar]
- 26. Liu X., Zhou L., Chen X., Liu T., Pan G., Cui W., Li M., Luo Z. P., Pei M., Yang H., Gong Y., He F., Mater. Sci. Eng., C 2016, 61, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang C., Xu Y., Jin C., Min B.‐H., Li Z., Pei X., Wang L., Artif. Organs 2013, 37, E179. [DOI] [PubMed] [Google Scholar]
- 28. Prewitz M. C., Seib F. P., Von Bonin M., Friedrichs J., Stißel A., Niehage C., Müller K., Anastassiadis K., Waskow C., Hoflack B., Bornhäuser M., Werner C., Nat. Methods 2013, 10, 788. [DOI] [PubMed] [Google Scholar]
- 29. Daley W. P., Peters S. B., Larsen M., J. Cell Sci. 2008, 121, 255. [DOI] [PubMed] [Google Scholar]
- 30. Nishimura R., Hata K., Ikeda F., Ichida F., Shimoyama A., Matsubara T., Wada M., Amano K., Yoneda T., J. Bone Miner. Metab. 2008, 26, 203. [DOI] [PubMed] [Google Scholar]
- 31. Singh P., Schwarzbauer J. E., J. Cell Sci. 2012, 125, 3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Assis‐Ribas T., Forni M. F., Winnischofer S. M. B., Sogayar M. C., Trombetta‐Lima M., Dev. Biol. 2018, 437, 63. [DOI] [PubMed] [Google Scholar]
- 33. Hoshiba T., Kawazoe N., Tateishi T., Chen G., J. Biol. Chem. 2009, 284, 31164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoshiba T., Kawazoe N., Chen G., Biomaterials 2012, 33, 2025. [DOI] [PubMed] [Google Scholar]
- 35. Hoshiba T., Lu H., Yamada T., Kawazoe N., Tateishi T., Chen G., Biotechnol. Prog. 2011, 27, 788. [DOI] [PubMed] [Google Scholar]
- 36. Cai R., Nakamoto T., Hoshiba T., Kawazoe N., Chen G., Acta Biomater. 2016, 35, 185. [DOI] [PubMed] [Google Scholar]
- 37. Almeida H. V., Cunniffe G. M., Vinardell T., Buckley C. T., O'Brien F. J., Kelly D. J., Adv. Healthcare Mater. 2015, 4, 1043. [DOI] [PubMed] [Google Scholar]
- 38. Dominici M., Blanc K. L. e., Mueller I., Slaper‐Cortenbach I., Marini F. C., Krause D. S., Deans R. J., Keating A., Prockop D. J., Horwitz E. M., Cytotherapy 2006, 8, 315. [DOI] [PubMed] [Google Scholar]
- 39. White L. J., Saldin L. T., Keane T. J., Cramer M. C., Shakesheff K. M., Badylak S. F., Compr. Biomater. II 2017, 8, 532. [Google Scholar]
- 40. Hoshiba T., Yamada T., Lu H., Kawazoe N., Chen G., J. Biomed. Mater. Res., Part A 2012, 100 A, 694. [DOI] [PubMed] [Google Scholar]
- 41. Lai Y., Sun Y., Skinner C. M., Son E. L., Lu Z., Tuan R. S., Jilka R. L., Ling J., Chen X. D., Stem Cells Dev. 2010, 19, 1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cai R., Kawazoe N., Chen G., Colloids Surf., B 2015, 126, 381. [DOI] [PubMed] [Google Scholar]
- 43. Hoshiba T., Kawazoe N., Tateishi T., Chen G., Adv. Mater. 2010, 22, 3042. [DOI] [PubMed] [Google Scholar]
- 44. Hynes R. O., Naba A., Cold Spring Harbor Perspect. Biol. 2012, 4, a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anasiz Y., Ozgul R. K., Uckan‐Cetinkaya D., Stem Cell Rev. Rep. 2017, 13, 587. [DOI] [PubMed] [Google Scholar]
- 46. Shi‐Wen X., Leask A., Abraham D., Cytokine Growth Factor Rev. 2008, 19, 133. [DOI] [PubMed] [Google Scholar]
- 47. Hu T. H., Huang C. C., Liu L. F., Lin P. R., Liu S. Y., Chang H. W., Changchien C. S., Lee C. M., Chuang J. H., Tai M. H., Cancer 2003, 98, 1444. [DOI] [PubMed] [Google Scholar]
- 48. Kao Y. H., Chen C. L., Jawan B., Chung Y. H., Sun C. K., Kuo S. M., Hu T. H., Lin Y. C., Chan H. H., Cheng K. H., Cheng K. H., Wu D.‐C., Goto S., Cheng Y.‐F., Chao D., Tai M. H., J. Hepatol. 2010, 52, 96. [DOI] [PubMed] [Google Scholar]
- 49. Kang H., Jeong Y. D., Verma P., Roh S., Tissue Eng. Regener. Med. 2013, 10, 380. [Google Scholar]
- 50. Hamamura K., Zhang P., Yokota H., Cell Biol. Int. 2008, 32, 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheng C. W., Solorio L. D., Alsberg E., Biotechnol. Adv. 2014, 32, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shakouri‐Motlagh A., O'Connor A. J., Brennecke S. P., Kalionis B., Heath D. E., Acta Biomater. 2017, 55, 1. [DOI] [PubMed] [Google Scholar]
- 53. Keane T. J., Swinehart I. T., Badylak S. F., Methods 2015, 84, 25. [DOI] [PubMed] [Google Scholar]
- 54. Crapo P. M., Gilbert T. W., Badylak S. F., Biomaterials 2011, 32, 3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Choudhury D., Tun H. W., Wang T., Naing M. W., Trends Biotechnol. 2018, 36, 787. [DOI] [PubMed] [Google Scholar]
- 56. Pati F., Jang J., Ha D. H., Won Kim S., Rhie J. W., Shim J. H., Kim D. H., Cho D. W., Nat. Commun. 2014, 5, 3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim B. S., Kim H., Gao G., Jang J., Cho D. W., Biofabrication 2017, 9, 034104. [DOI] [PubMed] [Google Scholar]
- 58. Nam S. Y., Park S. H., Adv. Exp. Med. Biol. 2018, 1064, 335. [DOI] [PubMed] [Google Scholar]
- 59. Gopinathan J., Noh I., Biomater. Res. 2018, 22, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takashi H., Katsumi M., Toshihiro A., Biochem. Biophys. Res. Commun. 2007, 359, 151. [DOI] [PubMed] [Google Scholar]
- 61. Ma H. L., Hung S. C., Lin S. Y., Chen Y. L., Lo W. H., J. Biomed. Mater. Res., Part A 2003, 64, 273. [DOI] [PubMed] [Google Scholar]
- 62. Herlofsen S. R., Küchler A. M., Melvik J. E., Brinchmann J. E., Tissue Eng., Part A 2011, 17, 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang X., Chen G., Huang C., Tu H., Zou J., Yan J., Oncotarget 2017, 8, 98336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mao Y., Hoffman T., Wu A., Goyal R., Kohn J., J. Mater. Sci.: Mater. Med. 2017, 28, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Santhakumar R., Vidyasekar P., Verma R. S., PLoS One 2014, 9, e114697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cheng H. W., Tsui Y. K., Cheung K. M. C., Chan D., Chan B. P., Tissue Eng., Part C 2009, 15, 697. [DOI] [PubMed] [Google Scholar]
- 67. Shu K., Thatte H., Spector M., IEEE 35th Annu. Northeast Bioengineering Conf., Piscataway, NJ: 2009. [Google Scholar]
- 68. Ryan J. M., Barry F. P., Murphy J. M., Mahon B. P., J. Inflammation 2005, 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Aggarwal S., Pittenger M. F., Blood 2005, 105, 1815. [DOI] [PubMed] [Google Scholar]
- 70. Tang C., Xu Y., Jin C., Min B. H., Li Z., Pei X., Wang L., Artif. Organs 2013, 37, e179. [DOI] [PubMed] [Google Scholar]
- 71. Yang J., Zhang Y. S., Yue K., Khademhosseini A., Acta Biomater. 2017, 57, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jin C. Z., Choi B. H., Park S. R., Min B. H., J. Biomed. Mater. Res., Part A 2010, 92, 1567. [DOI] [PubMed] [Google Scholar]
- 73. Yang Y., Lin H., Shen H., Wang B., Lei G., Tuan R. S., Acta Biomater. 2018, 69, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. López‐Ruiz E., Perán M., Cobo‐Molinos J., Jiménez G., Picón M., Bustamante M., Arrebola F., Hernández‐Lamas M. C., Delgado‐Martínez A. D., Montañez E., Marchal J. A., Osteoarthritis Cartilage 2013, 21, 246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


