Abstract
The family Cystoviridae comprises several bacteriophages with double-stranded RNA (dsRNA) genomes. We have previously purified the catalytic polymerase subunit (Pol) of one of the Cystoviridae members, bacteriophage φ6, and shown that the protein can catalyze RNA synthesis in vitro. In this reaction, both bacteriophage-specific and heterologous RNAs can serve as templates, but those containing 3′ termini from the φ6 minus strands are favored. This provides a molecular basis for the observation that only plus strands, not minus strands, are transcribed from φ6 dsRNA segments in vivo. To test whether such a regulatory mechanism is also found in other dsRNA viruses, we purified recombinant Pol subunits from the φ6-related bacteriophages φ8 and φ13 and assayed their polymerase activities in vitro. The enzymes catalyze template-dependent RNA synthesis using both single-stranded-RNA (ssRNA) and dsRNA templates. However, they differ from each other as well as from φ6 Pol in certain biochemical properties. Notably, each polymerase demonstrates a distinct preference for ssRNAs bearing short 3′-terminal sequences from the virus-specific minus strands. This suggests that, in addition to other factors, RNA transcription in Cystoviridae is controlled by the template specificity of the polymerase subunit.
Bacteriophage φ6, infecting the plant-pathogenic bacterium Pseudomonas syringae, is one of the best-characterized double-stranded RNA (dsRNA) viruses (15, 16). Its genome consists of three linear dsRNA segments: small (S), medium (M), and large (L). These are brought into the host cell inside a transcriptionally active virus core particle. In the course of transcription, plus-sense single-stranded RNAs (ssRNAs) s+, m+, and l+ are produced and extruded into the cytoplasm, where they are translated by the cellular protein synthesis machinery. Early φ6 proteins P1, P2, P4, and P7 assemble into the polymerase complex, or procapsid (PC). This is followed by packaging of s+, m+, and l+ into the PC and their subsequent replication to form S, M, and L, respectively. We have previously demonstrated that purified P2 protein, the Pol subunit of the φ6 PC, can act as a replicase and transcriptase in vitro (11, 13). Although it accepts RNA templates of both φ6 and heterologous origin, φ6 Pol shows a clear preference for templates containing φ6-specific plus-strand initiation signals at their 3′ ends. This suggests that the enzyme may play an active role in selection between plus- and minus-strand initiation signals, thus favoring the synthesis of plus strands during transcription in vivo (11).
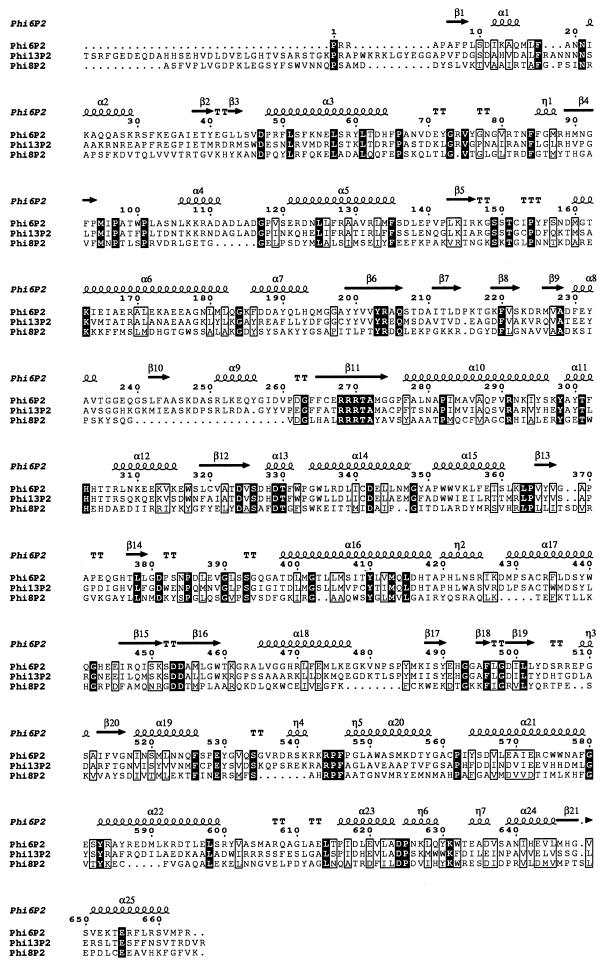
φ6 has long been the only known species in the family Cystoviridae (20). Recently, the family has been updated with several newly isolated bacteriophages (17). Like φ6, they contain tripartite dsRNA genomes and a lipid membrane as a structural element. Genomes of two of these bacteriophages, φ8 and φ13, have been sequenced completely, thus enabling their phylogenic analysis (8, 18). φ13 shows limited sequence homology with φ6, whereas φ8 differs from φ6 and φ13 dramatically. At the amino acid level, the polymerase complex proteins P1, P2, P4, and P7 are homologous between φ6 and φ13. However, the only protein that shows detectable similarity across all three bacteriophages is the P2 polymerase, referred to here as Pol (Fig. 1).
FIG. 1.
Comparision of three recombinant Pol proteins (P2 proteins) from φ6, φ8, and φ13. The protein sequences can be accessed at http://www.ncbi.nlm.nih.gov (phi6P2, AAA32355; phi8P2 AAF63300; phi13P2, AAG00444). Amino acid sequence alignment is based on structure alignment with ClustalW (http://www.ebi.ac.uk/clustalw/) and ESPript 1.9 (http://www-pgm1.ipbs.fr:8080/ESPript/). Strictly conserved residues are highlighted, and similar residues are boxed. Numbers correspond to the φ6 Pol sequence. Secondary-structure elements are from the high-resolution crystal structure of φ6 Pol (3), courtesy of S. J. Butcher.
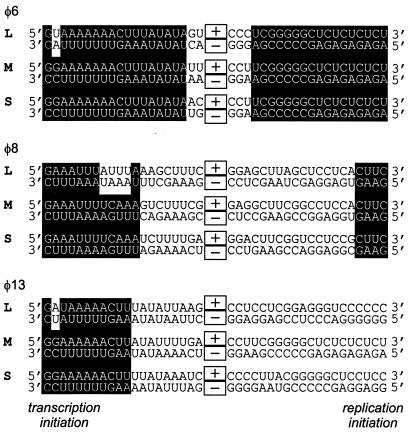
Replication and transcription initiation sites vary considerably between φ6, φ8, and φ13 (Fig. 2). It is obvious, however, that the 3′ ends of both minus and plus strands, used in the initiation of plus- and minus-strand syntheses, respectively, are pyrimidine rich, with an invariable C at the 3′-most position of all minus strands. Within each bacteriophage genome, the 3′-proximal sequences of l−, m−, and s− are generally more conserved than those of l+, m+, and s+. φ13 is the best example of this asymmetry. Interestingly, the 3′ end of l− does not match the m−/s− consensus at one (φ6 and φ13) or several (φ8) positions. In the case of φ6, this single-nucleotide substitution is known to be the reason for the lower transcription efficiency of L compared to M or S (6, 11).
FIG. 2.
Terminal homologies among dsRNA genomic segments S, M, and L in φ6, φ8, and φ13. Twenty nucleotides (each) of the left- and right-hand termini are shown. Boxed plus and minus signs represent the rest of the plus- and minus-strand sequences, respectively. Nucleotides conserved within each genome are highlighted. Positions of transcription (left-hand) and replication (right-hand) initiation sites are indicated.
Since φ6 Pol has been suggested to control the bacteriophage RNA metabolism through selective initiation at the transcriptional promoter, the question arises whether a similar regulatory function could also be assigned to the polymerases of φ8 and φ13. To this end, we purified the two recombinant polymerase subunits and assayed their activities in vitro. Like their φ6 counterpart, both polymerases catalyze RNA-dependent RNA synthesis in vitro. However, they differ from each other and from φ6 Pol in several biochemical properties. Most interestingly, each enzyme has a distinct template specificity, with a clear bias for ssRNAs containing 3′ ends similar to those found in bacteriophage-specific minus strands. These data suggest a possible scenario in which polymerase subunit specificity has coevolved with the minus-strand 3′-proximal sequence to ensure efficient transcription initiation. On the other hand, the results with genomic RNAs indicate that some additional factors, such as the melting temperature of the initiation site, are likely to be involved in the regulation of RNA synthesis from dsRNA templates.
MATERIALS AND METHODS
Plasmids and strains.
φ6 polymerase was produced using Escherichia coli strain BL21(DE3/pEM2) (13). The φ8 Pol expression strain was prepared as follows. The p2 gene of φ8 was first PCR amplified from plasmid pLM2424 (8) using the primer pair p2phi8up and p2phi8down primer pair (Table 1) and Pfu DNA polymerase (Stratagene). The PCR fragment was digested with NdeI and BamHI (underlined sites in primer sequences) and ligated with the similarly cut vector pMG60, a derivative of pMG59 (7). E. coli BL21 was then transformed with the resultant plasmid pHY1 to produce BL21(pHY1). To construct the φ13 Pol expression strain, the p2 gene of φ13 was amplified from pLM2200 (18) using primers p2phi13up and p2phi13down. The PCR fragment was inserted into pET32b(+) (Novagen) at NdeI-HindIII sites to produce plasmid pHY2. This was introduced into BL21(DE3) (Novagen), yielding BL21(DE3/pHY2).
TABLE 1.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| p2phi8up | 5′ CGAGCCGTACATATGGCATCGTTCGT |
| p2phi8down | 5′ GGAGTTGGATCCTGGTGTAACTTTCGT |
| p2phi13up | 5′ CAGGCGCTGACATATGACTTCCCGCT |
| p2phi13down | 5′ GCCGAAGCTTATCTGACATCCCTCGT |
| T7-1 | 5′ CGCGTAATACGACTCACTATAG |
| 3′end | 5′ AGAGAGAGAGCCCCCGA |
| 3′end1 | 5′ AAGAGAGAGAGCCCCCGA |
| 3′end2 | 5′ CAGAGAGAGAGCCCCCGA |
| 3′end3 | 5′ GAGAGAGAGAGCCCCCGA |
| 3′end4 | 5′ TAGAGAGAGAGCCCCCGA |
| pT7-3′end | 5′ TAAGCTTGGGCTGCAGGT |
| pT7-3′end1 | 5′ ATAAGCTTGGGCTGCAGGT |
| pT7-3′end2 | 5′ CTAAGCTTGGGCTGCAGGT |
| pT7-3′end3 | 5′ GTAAGCTTGGGCTGCAGGT |
| pT7-3′end4 | 5′ TTAAGCTTGGGCTGCAGGT |
| pT7-3′end5 | 5′ GGTAAGCTTGGGCTGCAGGT |
| pT7-3′end9 | 5′ AGAGAGAGATAAGCTTGGGCTGCAGGT |
| pT7-3′end11 | 5′ GATAAGCTTGGGCTGCAGGT |
| pT7-3′end12 | 5′ GAAATTTTCTAAGCTTGGGCTGCAGGT |
| pT7-3′end13 | 5′ GGAAAAAACTAAGCTTGGGCTGCAGGT |
| pT7-3′end14 | 5′ GGGTAAGCTTGGGCTGCAGGT |
Restriction sites used for cloning are underlined.
Expression and purification of recombinant polymerase subunits.
Recombinant polymerases from φ8 and φ13 were expressed by the procedure described for φ6 Pol (4, 13). Briefly, starter cultures were grown in Luria-Bertani medium containing 150 mg of ampicillin/ml at 37°C to an optical density at 540 nm (OD540) of 0.6. Cultures were then diluted 50-fold, and incubation was continued until the OD540 reached 1.0. Expression of the recombinant polymerase was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After addition of IPTG, bacterial cultures were shaken for 15 h at 23°C [for BL21(pHY1)] or 18 h at 19°C [for BL21(DE3/pHY2)]. Protein purification was carried out at +4°C. Throughout purification, the pH value was 7.4 for φ8 Pol and 8.0 for φ13 Pol. Bacterial pellets were resuspended in 35 ml of 100 mM NaCl–50 mM Tris-HCl–1 mM EDTA. Suspensions were passed three times through a precooled French pressure cell at ∼105 MPa. Phenylmethylsulfonyl fluoride was added to 1 mM after the first passage. Lysates were cleared at 120,000 × g for 2.5 h. φ6 Pol and φ13 Pol were purified successively on Cibacron Blue 3GA agarose (Sigma), heparin agarose (Sigma), and a HiTrap Q Sepharose column (Pharmacia) as described elsewhere (13). For the φ8 Pol purification, Reactive Brown 10 agarose (Sigma) and a Superdex 75 gel filtration column (Pharmacia) were used. Protein concentrations were determined by absorbance at 280 nm in 6 M guanidine hydrochloride based on an optical density (OD) of 1.21 OU for 1 mg of φ8 Pol/ml and an OD of 1.37 for 1 mg of φ13 Pol/ml (5). Purified proteins were stored at +4°C.
Preparation of RNA substrates.
ssRNA substrates were produced by in vitro transcription with T7 RNA polymerase as described previously (13, 14). Templates for the transcription were prepared either by cutting recombinant plasmid DNA with restriction endonucleases or by PCR amplification with Pfu DNA polymerase. For PCR, oligonucleotide T7-1, containing the T7 polymerase promoter sequence, was used as an upstream primer. Oligonucleotides 3′end to 3′end4 were used as downstream primers for amplification of the Δs+ fragment from pEM15 (11), whereas pT7-3′end to pT7-3′end14 served as downstream primers to amplify the luciferase gene from pT7luc (9). Genomic dsRNA was extracted from purified bacteriophage particles with phenol-chloroform, precipitated with ethanol, and dissolved in sterile water.
Polymerase activity assay.
Polymerase activity was assayed in 10-μl reaction mixtures. In the initial experiments, we used the conditions reported previously for the φ6 polymerase (11, 13). These were further optimized to 50 mM HEPES-KOH (pH 7.4 to 7.8; see Results for details), 20 mM ammonium acetate (NH4OAc), 6% (wt/vol) polyethylene glycol 4000, 5 mM MgCl2, 2 mM MnCl2, 0.1 mM EDTA, 0.1% Triton X-100, 1 mM (each) ATP and GTP, 0.2 mM (each) CTP and UTP, 0.8 U of RNasin/μl, and 0.25 mCi of [α-32P]UTP (Amersham; 3,000 Ci/mmol)/ml. The final concentration of RNA substrates was 50 to 200 μg/ml. Reactions were started by adding one of the three polymerases up to 0.02 to 0.04 mg/ml, and reaction mixtures were further incubated at 30°C for 1 h. Reaction products were separated by standard agarose gel electrophoresis (13). Gels were dried and exposed to Fuji Super RX film or analyzed with a Fuji BAS1500 phosphorimager.
RESULTS
Expression and purification of recombinant Pol proteins.
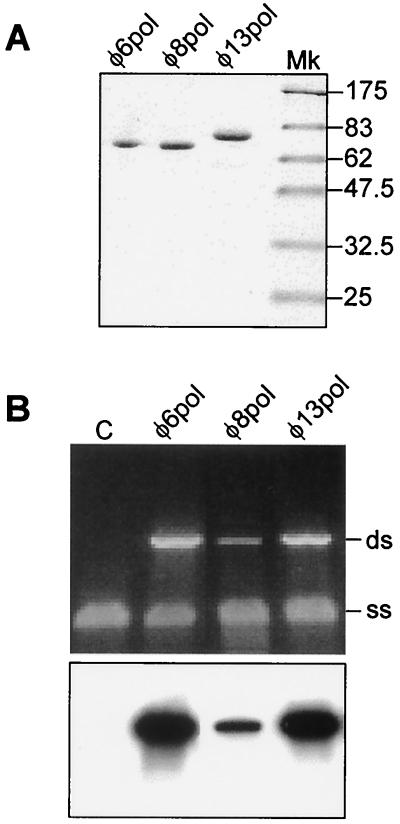
To construct plasmids for φ8 Pol and φ13 Pol expression, full-length polymerase genes were cloned under the control of inducible promoters and strong Shine-Dalgarno sequences. Soluble polymerases from bacteriophages φ6, φ8, and φ13 were expressed by incubating the corresponding E. coli strains at 16 to 23°C in the presence of IPTG. Expression at higher temperatures (28 to 37°C) led to substantial increases in overall polymerase production, but most of the protein was insoluble. The φ6 Pol purification protocol, employing a combination of Cibacron Blue 3GA agarose and heparin agarose and an anion-exchange column, gave satisfactory results for φ13 Pol (Fig. 3A). However, φ8 Pol failed to bind to the blue agarose, leading us to test other dye affinity resins (Sigma kit no. RDL-9). Reactive Brown 10 agarose bound φ8 Pol well, without retarding most of the contaminating proteins (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA). Bound φ8 Pol was eluted with 50 mM Tris-HCl (pH 7.4)–500 mM NaCl– 1 mM EDTA and was further purified to near-homogeneity using a Superdex 75 gel filtration column equilibrated with 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 0.1 mM EDTA (Fig. 3A). The relative electrophoretic mobilities of the three purified enzymes correlated with their calculated molecular masses (Table 2): φ13 Pol is larger, and φ8 Pol is smaller, than φ6 Pol. The yields of purified proteins in milligrams per liter of bacterial culture are given in Table 2.
FIG. 3.
Purified polymerase subunits catalyze RNA replication in vitro. (A) SDS-PAGE analysis of purified recombinant Pol proteins from φ6, φ8, and φ13. Lane Mk, protein marker. Molecular masses (in kilodaltons) are given on the right. (B) Agarose electrophoresis of reaction mixtures containing s+13 RNA and one of the three purified polymerases: φ6 Pol, φ8 Pol, or φ13 Pol. Lane C, control without polymerase. The upper panel shows an ethidium bromide-stained gel, with the ssRNA (ss) and dsRNA (ds) marked on the right; the lower panel is the corresponding autoradiogram.
TABLE 2.
Properties of the Pol subunits from φ6, φ8, and φ13
| Protein name | Length (aa)a | Molecular size (kDa) | Predicted pI | Yield (mg/liter) |
|---|---|---|---|---|
| φ6 Pol | 664 | 74.8 | 6.8 | 6 |
| φ8 Pol | 635 | 71.5 | 8.7 | 18 |
| φ13 Pol | 712 | 79.6 | 6.0 | 3.5 |
Without the first formylmethionyl residue.
φ8 Pol and φ13 Pol catalyze RNA replication in vitro. RNA replication activity of the purified polymerases was assayed as described earlier for the φ6 enzyme (13). The mixtures containing 50 mM Tris-HCl (pH 8.9), 80 mM NH4OAc, 5 mM MgCl2, 1 mM MnCl2, 6% polyethylene glycol 4000, 0.1 mM EDTA, and the φ6-specific ssRNA template s+13 (s+ segment with the extension CTAGAGGATCCCC 3′) were incubated for 1 h at 30°C. Both φ8 Pol and φ13 Pol were enzymatically active, producing full-length dsRNA forms indistinguishable from the φ6 Pol dsRNA product (Fig. 3B). φ13 Pol was nearly as active as φ6 Pol, whereas the specific activity of φ8 Pol was lower under the conditions employed. No radioactive bands were detected when polymerase or the RNA substrate was omitted from the reaction mixture (Fig. 3B, lane 1; also not shown). In addition, labeled products did not appear in the absence of the four unlabeled nucleoside triphosphates (NTPs). These results unequivocally demonstrate that the newly isolated proteins φ8 Pol and φ13 Pol possess RNA-dependent replicase activity in vitro.
Effects of reaction conditions on the polymerase activity.
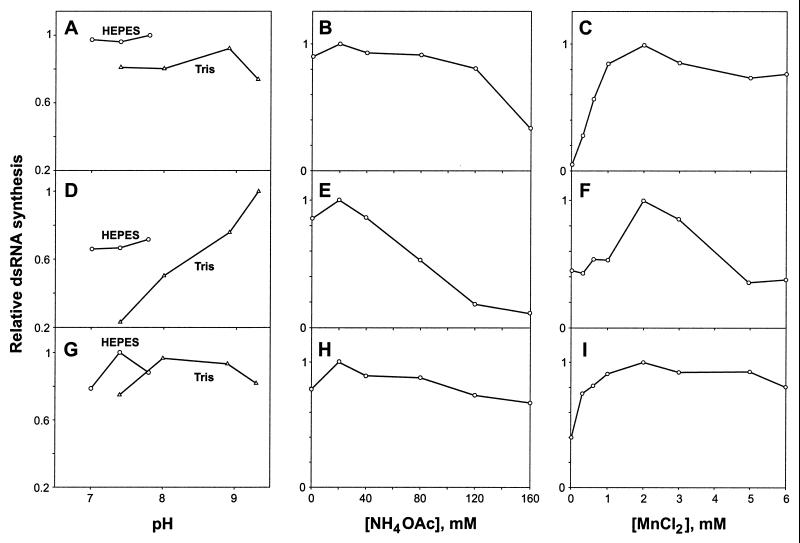
The above assay was utilized to study the effects of several parameters on the activities of the three polymerases. All three enzymes synthesized dsRNA over a wide pH range, with the Tris-HCl pH optima increasing in the order φ13 Pol–φ6 Pol–φ8 Pol. Interestingly, the enzyme activity in HEPES-KOH was always higher than that in Tris-HCl at the same pH (Fig. 4A, D, and G).
FIG. 4.
Effects of pH (A, D, and G) and of ammonium acetate (B, E, and H) and Mn2+ (C, F, and I) concentrations on the activities of the three polymerases. Reaction mixtures containing s+13 RNA and φ6 Pol (A, B, and C), φ8 Pol (D, E, and F) or φ13 Pol (G, H, and I) were incubated at 30°C for 1 h and analyzed by agarose gel electrophoresis. Radioactivity in the bands of the newly produced dsRNA was quantified with a phosphorimager. Tris-HCl (Tris) or HEPES-KOH (HEPES) buffers were used in panelsA, D, and G, whereas in the other panels pH was buffered with HEPES-KOH (pH 7.8 for φ6 Pol and φ8 Pol, and pH 7.4 for φ13 Pol). Graphs are normalized so that the highest observed value within each panel is set to 1.
To determine the monovalent cation optimum, reaction mixtures were supplemented with different concentrations of NH4OAc. Although all three enzymes were active without NH4OAc, low salt concentrations (20 mM) reproducibly stimulated RNA synthesis. Further increases in NH4OAc concentration were inhibitory, particularly for φ8 Pol (Fig. 4B, E, and H).
Manganese (Mn2+) has been reported to stimulate φ6 Pol (11, 13). Here, we systematically studied the effect of this ion on the three RNA polymerases. Regardless of the enzyme origin, the optimal Mn2+ concentration was 2 mM. However, there were differences in concentration dependence (Fig. 4C, F, and I). Added at concentrations up to 1 to 2 mM, Mn2+ increased the polymerase activity of φ6 Pol more than 10-fold, whereas this stimulatory effect was relatively modest in the case of φ8 Pol and φ13 Pol (∼2.5-fold). Furthermore, higher concentrations of manganese (5 to 6 mM) inhibited φ8 Pol but were well tolerated by φ6 Pol and φ13 Pol.
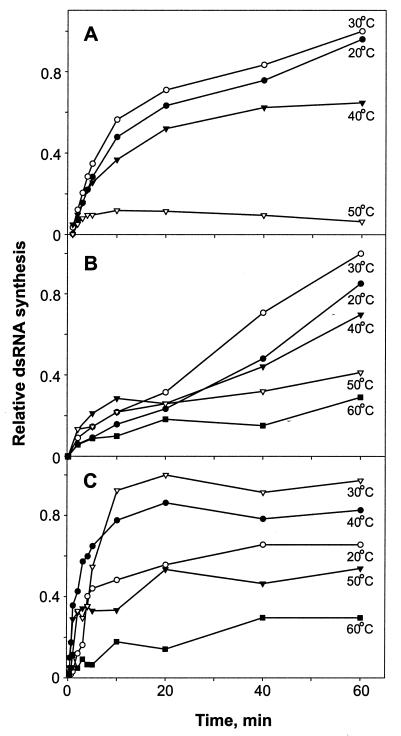
To test the effect of incubation temperature, RNA-synthesis was carried out in the optimized buffer containing 50 mM HEPES-KOH (pH 7.8 for φ6 Pol and φ8 Pol and pH 7.4 for φ13 Pol), 20 mM NH4OAc, and 2 mM MnCl2, in addition to other components listed in Materials and Methods. Reaction mixtures were incubated at 20 to 60°C for 1 h, and aliquots were sampled at different time points. For all the polymerases, the highest dsRNA synthesis was observed at 30°C (Fig. 5). Increasing the temperature to 40°C accelerated the initial RNA synthesis, at least for φ8 Pol and φ13 Pol (notice the difference in the slopes at 30 and 40°C in Fig. 5B and C), but compromised the final yield. Furthermore, φ8 Pol and φ13 Pol retained a substantial fraction of their activity at 50 and even 60°C, whereas φ6 Pol was almost completely inactivated at ≥50°C.
FIG. 5.
Effects of temperature on replication. Replication mixtures containing s+13 RNA and φ6 Pol (A), φ8 Pol (B), or φ13 Pol (C) were incubated at 20 to 60°C, as indicated. Aliquots were withdrawn at different time points and analyzed by agarose gel-electrophoresis followed by phosphorimager analysis. The highest observed value within each panel is set to 1.
φ8 and φ13 polymerases catalyze dsRNA transcription in vitro.
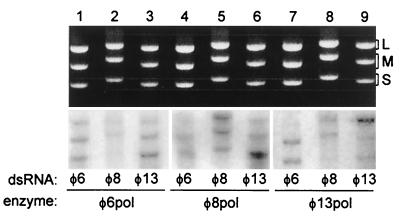
In addition to its replication activity, purified φ6 Pol can catalyze dsRNA transcription in vitro employing a semiconservative (strand displacement) mechanism (11). To test the transcriptional activities of φ8 Pol and φ13 Pol, the two polymerases, along with φ6 Pol, were assayed with genomic dsRNAs extracted from bacteriophage particles. In all three cases, radioactive products that migrated at the position of the input dsRNA species were found, consistent with the idea of semiconservative transcription (Fig. 6). However, the product patterns varied significantly depending on the polymerase and the dsRNA source.
FIG. 6.
Polymerase subunits from φ6, φ8, and φ13 catalyze dsRNA transcription in vitro. Reaction mixtures contained φ6 Pol (lanes 1 to 3), φ8 Pol (lanes 4 to 6) or φ13 Pol (lanes 7 to 9) and dsRNA extracted from φ6 (lanes 1, 4, and 7), φ8 (lanes 2, 5, and 8) or φ13 (lanes 3, 6, and 9). The upper panel shows an ethidium bromide-stained gel, and the lower panel is the corresponding autoradiogram. The positions of dsRNA segments L, M, and S are marked on the right.
Effect of the template 3′ end on the efficiency of RNA synthesis.
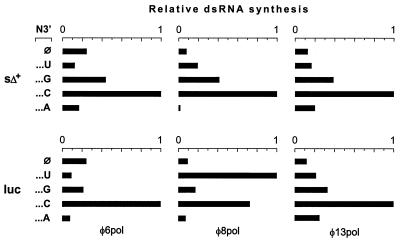
To further explore the template preferences of φ6 Pol, φ8 Pol, and φ13 Pol, we compared replication efficiencies with five variants of sΔ+ ssRNA (φ6 s+ segment with an extensive internal deletion [11]). One of the variants, sΔ+(φ), had the natural φ6 terminus CUCUCUCUCU3′ (also present in the φ13 m+; see Fig. 2); the other four templates contained different one-nucleotide additions: A3′, C3′, G3′, and U3′. For all three polymerases, replication efficiency increased considerably when C3′ was used as a terminal template nucleotide. G3′ was the second-best addition (Fig. 7, sΔ+). The effects of the other two terminal bases were polymerase dependent. Both A3′ and U3′ reduced φ6 Pol replication, whereas for φ13 Pol they were neutral or somewhat stimulatory. φ8 Pol was substantially inhibited by A3′ and was activated by U3′. A similar result was also obtained for the set of five firefly luciferase mRNAs: luc(φ), containing CCCAAGCUUA3′, and its one-nucleotide-longer derivatives luc(A3′), luc(C3′), luc(G3′), and luc(U3′). However, in this case, U3′ enhanced φ8 Pol replication by an order of magnitude (Fig. 7, luc).
FIG. 7.
Effect of the 3′-terminal nucleotide of the template on replication efficiency. Two sets of ssRNA templates, luc and sΔ+, were assayed with φ6 Pol, φ8 Pol, or φ13 Pol at 30°C for 1 h. Each of the sets contained five RNA species with different 3′ ends (N3′): either without modifications (φ) or extended with one additional 3′-terminal nucleotide (U, G, C or A). Reaction products were separated by agarose gel-electrophoresis, and radioactivity in the dsRNA product bands was quantified with a phosphorimager. The graphs are normalized so that the highest value within each panel is set to 1.
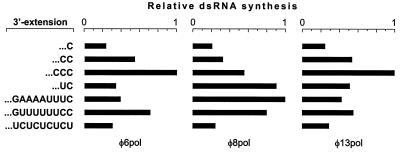
We also assayed luciferase mRNAs containing longer 3′-terminal extensions (Fig. 8). Compared to C3′, CC3′ doubled replication efficiency for all three polymerases. The enhancement was even stronger when the CCC3′ terminus was used. Nine terminal bases of φ13 m− or s− (GUUUUUUCC 3′; also similar in φ6 m− and s−, as shown in Fig. 2) added to the luc RNA caused approximately the same effect as CC3′ for φ6 Pol and φ13 Pol. This sequence, however, stimulated φ8 Pol replication more significantly. Similarly, UC3′ was a somewhat weaker enhancer than CC3′ for φ6 Pol and φ13 Pol, whereas it was clearly superior to CC3′ for φ8 Pol. The φ8-specific minus-strand 3′ terminus GAAAAUUUC3′ (Fig. 2) was the best initiation signal for φ8 Pol but not for φ6 Pol and φ13 Pol. Finally, luc RNA with a UCUCUCUCU 3′ terminus, found in all three plus strands of φ6 as well as in m+ of φ13 (Fig. 2), was a relatively inefficient template for all three polymerases.
FIG. 8.
Replication efficiency of luc ssRNAs containing different 3′-terminal extensions. Reaction mixtures containing one of the seven luciferase ssRNA templates with the different 3′ extensions and φ6 Pol, φ8 Pol, or φ13 Pol were incubated at 30°C for 1 h and analyzed as for Fig. 7. The highest value within each panel is set to 1.
DISCUSSION
Previous studies have shown that the polymerase subunit of bacteriophage φ6 can catalyze RNA synthesis without the assistance of other proteins (11, 13). In addition, the high-resolution crystal structures of the φ6 Pol apoenzyme, as well as its template and NTP complexes, have recently been solved (3). We now demonstrate RNA-dependent RNA polymerization activity of two polymerase subunits isolated from the φ6-related bacteriophages φ8 and φ13.
The three polymerases are homologous to each other, with an overall amino acid identity of 11% and a mean similarity of 41% (Fig. 1). Pairwise sequence alignments reveal ∼50 and 20% identity of φ6 Pol to φ13 Pol and φ8 Pol, respectively (8, 18). All three proteins contain the (G/S)DD motif (Fig. 1, turn between β15 and β16) critical for catalysis in all RNA-dependent RNA polymerases (10). Also strictly conserved is the positively charged RRRTA sequence (β11), which has been shown to interact with the phosphate groups of the incoming NTP substrate in φ6 Pol (3). We also notice the conservation in the C-terminal domain (α23 to α24), implicated in stabilizing the initiation complex via stacking interactions with the first two nucleotides of the nascent strand (3). More-reliable molecular comparisons, however, require knowledge of the atomic structures for φ8 Pol and φ13 Pol. Crystallization experiments are under way.
The polymerases are shown to catalyze replication and transcription in vitro using, respectively, ssRNA and dsRNA templates (Fig. 3 and 6). Replication is at least 1 order of magnitude more efficient than transcription for all three enzymes (data not shown), in agreement with earlier data on φ6 Pol (11).
The three enzymes can be distinguished biochemically. In Tris-HCl buffer, φ6 Pol is the most active at pH 8.9, with φ13 Pol having a lower and φ8 Pol a higher pH optimum (Fig. 4A, D, G). This distribution is mirrored by the predicted isoelectric points (Table 2) and is likely to be a function of protein surface charge at different pH values. When Mn2+ is added to a reaction mixture containing an excess of Mg2+, it increases the activities of all three enzymes, albeit to different extents (Fig. 4C, F, and I). Manganese has been reported to affect the activities of several RNA-dependent RNA polymerases, including those from φ6 (11, 13), hepatitis C virus (1, 21), brome mosaic virus (19), bacteriophage Qβ (2), and some other viruses. Recent structural data for φ6 Pol suggest that Mn2+ could serve to stabilize the polymerase molecule in a compact initiatory conformation (3). Intriguingly, φ8 Pol and φ13 Pol are stimulated by Mn2+ to a lesser extent than φ6 Pol and, at the same time, show a higher thermostability (Fig. 5).
Most importantly, the purified enzymes have different template specificities, for both ssRNAs and dsRNAs (Fig. 6 to 8). Several generalizations are appropriate in this respect. Both φ6 Pol and φ13 Pol prefer ssRNA templates with the 3′-terminal cytosine; CC3′ and CCC3′ are even better initiation signals (Fig. 7 and 8). While φ8 Pol is also increasingly stimulated with C3′, CC3′, and CCC3′, it initiates more efficiently at the termini containing a 3′-proximal uridine(s), such as GAAAAUUUC 3′, GUUUUUUCC 3′, UC 3′, or U 3′. This is further corroborated by the observation that s+13 RNA (ending in … CCCC 3′) is a better template for φ6 Pol and φ13 Pol than for φ8 Pol (Fig. 3B). Although φ13 Pol shows a somewhat higher affinity for the U-containing 3′-terminal extensions than φ6 Pol (Fig. 7 and 8), the two polymerases are obviously closer to each other than to φ8 Pol in terms of their template specificities, exactly as one would expect from the sequence comparison.
The polymerases from the Cystoviridae do not require a primer to commence RNA synthesis (12, 13). As shown for φ6 Pol, the polymerase forms a quaternary initiation complex where the template 3′ terminus is engaged in Watson-Crick base pairing with two cognate NTPs (3). The NTPs are additionally stabilized by stacking interactions to each other and to a special C-terminal “platform” in the polymerase. Since purine bases possess a very high stacking propensity, it is not surprising that the cystoviral polymerases show a bias for pyrimidine-rich template termini. In fact, many other RNA- and DNA-dependent RNA polymerases also initiate de novo and prefer purine nucleotides at the 5′ end of the nascent RNA, thus indicating that all these enzymes might utilize protein-NTP stacking to facilitate the initiation step.
There is a clear correlation between the terminal preferences of the polymerase subunits and the transcription initiation sequences from the corresponding bacteriophages (Fig. 2). Indeed, all three φ8 minus strands end with UC 3′, which is shifted to (C/U)C 3′ in φ13 and (C/A)C 3′ in φ6, consistent with the Pol specificity profiles. As the transcription in dsRNA viruses is asymmetric, we suggest that the polymerase subunit and the plus sense initiation sites have become mutually fit as a result of coevolution. The replication promoters seem to experience a lower evolutionary pressure, consistent with weaker conservation of the right-hand termini in φ8 and φ13 (Fig. 2). Indeed, both φ6 Pol and φ13 Pol prefer the minus-strand 3′-terminal sequence to that of the plus strands (compare GUUUUUUCC 3′ and UCUCUCUCU 3′ in Fig. 8).
In addition to the 3′-terminal preferences of the polymerase subunits, plus-strand synthesis in the Cystoviridae is also likely to be controlled by the melting temperature of the dsRNA termini. The GC content of the transcription initiation sites is noticeably lower than that of the replication promoters in all three bacteriophages (Fig. 2), which should facilitate access of the polymerase to the 3′ termini of the minus strands. This is consistent with the fact that the template tunnel inside the polymerase molecule can accommodate only ssRNA, not dsRNA (3). The efficiency of the dsRNA terminal unzipping could explain, for example, why minus strands of φ6 and φ13 do not end in CCC 3′, although it is preferred by φ6 Pol and φ13 Pol for ssRNA (Fig. 8). Furthermore, different accessibilities of initiation sites in a single-stranded form might explain the results shown in Fig. 6, where φ8 Pol utilized the φ13 S segment more efficiently than homologous templates, and φ13 Pol preferred φ13 L to M and S (Fig. 6).
In summary, biochemical comparison of three Pol subunits from Cystoviridae reveals, in addition to the expected similarity, a substantial degree of functional divergence. Further experiments with the purified polymerases will address the molecular basis for the difference in template specificity and will provide more-detailed insights into the regulation of the viral RNA metabolism.
ACKNOWLEDGMENTS
We thank Sarah Butcher for providing Fig. 1 and for critical reading of the manuscript. Leonard Mindich and A. Marika Grahn are thanked for donating materials used in this study. Purified φ8 and φ13 virions were kindly provided by Aušra Gaidelyte. The expert technical assistance of Riitta Tarkiainen and Marja-Leena Perälä is greatly appreciated.
This work was supported by the Academy of Finland (“Finnish Centre of Excellence Program 2000–2005” grants 162993 and 164298) and the European Union (grant BIO4-CT97-2364).
REFERENCES
- 1.Alaoui-Ismaili M H, Hamel M, L'Heureux L, Nicolas O, Bilimoria D, Labonte P, Mounir S, Rando R F. The hepatitis C virus NS5B RNA-dependent RNA polymerase activity and susceptibility to inhibitors is modulated by metal cations. J Hum Virol. 2000;3:306–316. [PubMed] [Google Scholar]
- 2.Blumenthal T. Qβ replicase template specificity: different templates require different GTP concentrations for initiation. Proc Natl Acad Sci USA. 1980;77:2601–2605. doi: 10.1073/pnas.77.5.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butcher S J, Grimes J M, Makeyev E V, Bamford D H, Stuart D I. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 4.Butcher S J, Makeyev E V, Grimes J M, Stuart D I, Bamford D H. Crystallization and preliminary X-ray crystallographic studies on the bacteriophage φ6 RNA-dependent RNA polymerase. Acta Crystallogr. 2000;D56:1473–1475. doi: 10.1107/s0907444900010702. [DOI] [PubMed] [Google Scholar]
- 5.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 6.Frilander M, Poranen M, Bamford D H. The large genome segment of dsRNA bacteriophage φ6 is the key regulator in the in vitro minus and plus strand synthesis. RNA. 1995;1:510–518. [PMC free article] [PubMed] [Google Scholar]
- 7.Grahn A M, Caldentey J, Bamford J K, Bamford D H. Stable packaging of phage PRD1 DNA requires adsorption protein P2, which binds to the IncP plasmid-encoded conjugative transfer complex. J Bacteriol. 1999;181:6689–6696. doi: 10.1128/jb.181.21.6689-6696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoogstraten D, Qiao X, Sun Y, Hu A, Onodera S, Mindich L. Characterization of φ8, a bacteriophage containing three double-stranded RNA genomic segments and distantly related to φ6. Virology. 2000;272:218–224. doi: 10.1006/viro.2000.0374. [DOI] [PubMed] [Google Scholar]
- 9.Kolb V A, Makeyev E V, Spirin A S. Co-translational folding of a eukaryotic multidomain protein in a prokaryotic translation system. J Biol Chem. 2000;275:16597–16601. doi: 10.1074/jbc.M002030200. [DOI] [PubMed] [Google Scholar]
- 10.Koonin E V, Gorbalenya A E, Chumakov K M. Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases. FEBS Lett. 1989;252:42–46. doi: 10.1016/0014-5793(89)80886-5. [DOI] [PubMed] [Google Scholar]
- 11.Makeyev E V, Bamford D H. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 2000;19:6275–6284. doi: 10.1093/emboj/19.22.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makeyev E V, Bamford D H. Primer-independent RNA sequencing with bacteriophage φ6 RNA polymerase and chain terminators. RNA. 2001;7:774–781. doi: 10.1017/s1355838201002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makeyev E V, Bamford D H. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage φ6. EMBO J. 2000;19:124–133. doi: 10.1093/emboj/19.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makeyev E V, Kolb V A, Spirin A S. Enzymatic activity of the ribosome-bound nascent polypeptide. FEBS Lett. 1996;378:166–170. doi: 10.1016/0014-5793(95)01438-1. [DOI] [PubMed] [Google Scholar]
- 15.Mindich L. Precise packaging of the three genomic segments of the double-stranded RNA bacteriophage φ6. Microbiol Mol Biol Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mindich L. Reverse genetics of dsRNA bacteriophage φ6. Adv Virus Res. 1999;53:341–353. doi: 10.1016/s0065-3527(08)60355-3. [DOI] [PubMed] [Google Scholar]
- 17.Mindich L, Qiao X, Qiao J, Onodera S, Romantschuk M, Hoogstraten D. Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J Bacteriol. 1999;181:4505–4508. doi: 10.1128/jb.181.15.4505-4508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao X, Qiao J, Onodera S, Mindich L. Characterization of φ13, a bacteriophage related to φ6 and containing three dsRNA genomic segments. Virology. 2000;275:218–224. doi: 10.1006/viro.2000.0501. [DOI] [PubMed] [Google Scholar]
- 19.Sun J H, Adkins S, Faurote G, Kao C C. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 20.Vidaver A K, Koski R K, Van Etten J L. Bacteriophage φ6: a lipid-containing virus of Pseudomonas phaseolicola. J Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong W, Uss A S, Ferrari E, Lau J Y, Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]