Abstract
It is often assumed that carbon nanotubes (CNTs) stimulate neuronal differentiation by transferring electrical signals and enhancing neuronal excitability. Given this, CNT–hydrogel composites are regarded as potential materials able to combine high electrical conductivity with biocompatibility, and therefore promote nerve regeneration. However, whether CNT–hydrogel composites actually influence neuronal differentiation and maturation, and how they do so remain elusive. In this study, CNT–hydrogel composites are prepared by in situ polymerization of poly(ethylene glycol) around a preformed CNT meshwork. It is demonstrated that the composites facilitate long‐term survival and differentiation of pheochromocytoma 12 cells. Adult neural stem cells cultured on the composites show an increased neuron‐to‐astrocyte ratio and higher synaptic connectivity. Moreover, primary hippocampal neurons cultured on composites maintain morphological synaptic features as well as their neuronal network activity evaluated by spontaneous calcium oscillations, which are comparable to neurons cultured under control conditions. These results indicate that the composites are promising materials that could indeed facilitate neuronal differentiation while maintaining neuronal homeostasis.
Keywords: carbon nanotubes, composites, hydrogels, neuronal differentiation
A new, versatile method for the preparation of carbon nanotube–poly(ethylene glycol) hydrogel composites is developed. The effects of the composites on neuronal differentiation and activity of pheochromocytoma 12 (PC12) cells, adult neural stem cells, and primary hippocampal neurons are investigated. It is demonstrated that the composites can serve as effective scaffolds promoting neuronal differentiation while maintaining network activity homeostasis.
1. Introduction
Developing biomaterials capable of promoting neuronal differentiation, regrowth, and formation of local and long‐range axonal connections is of great importance for the development of new clinical technologies to tackle neuronal injury.[ 1 ] Conductive scaffolds have been reported to transfer electrical signals from the extracellular matrix to cells and to stimulate neuronal differentiation under electrical stimulation.[ 2 ] In this respect, carbon nanotubes (CNTs) and their analogs with high electrical conductivity are promising materials.[ 3 ] The thin films of CNT meshwork (usually coated on substrates) with organized fractal‐like nanostructures were reported as good substrates for neuronal growth and long‐term survival.[ 4 ] Moreover, there are reports that neurons on CNT meshwork exhibit enhanced signal transmission.[ 5 ] Such an enhancement in network activity was also reported for neurons on graphene (an analog of CNTs),[ 6 ] although it remains controversial.[ 7 ] It is assumed that the contact and interaction between CNTs (or graphene) and the cell membrane can improve neuronal excitability by modifying membrane potential.[ 5 , 6 ] Based on these findings, CNTs and their analogs have rapidly become of interest to the material science community in the design of novel materials in nerve tissue engineering.[ 8 ]
Although CNTs are usually reported as noncytotoxic to neurons, some essential issues need to be carefully addressed before in vivo applications. First, the direct exposure and potential accumulation of CNTs in human tissue, which may cause abnormal immune cell activation and fibroblast proliferation, are of great concern.[ 3b ] These risks can be greatly reduced by incorporating CNTs as fillers into a hydrogel matrix. Some hydrogel‐based materials have very good biocompatibility and exhibit great potential in biomedical fields such as drug delivery,[ 9 ] biosensors,[ 10 ] and tissue engineering.[ 11 ] Therefore, CNT–hydrogel composites are often regarded as potential materials which combine electrical conductivity with biocompatibility promoting nerve regeneration.[ 8 , 12 ] Second, the neuronal excitability boosting caused by CNTs, which is often assumed to promote neuronal regeneration, could actually cause detrimental consequences to the nervous system. For instance, spinal cord injury leads to an increase in the excitability in the motoneurons below the lesion, causing a common debilitating complication, e.g., spasticity.[ 13 ] Excessively increased neuronal excitability will also inhibit axonal elongation and dampen regional brain connectivity.[ 14 ] Moreover, hyper‐excitability of cortical circuits may lead to neuropsychiatric disorders, e.g., epilepsy.[ 15 ] Therefore, the homeostatic properties of neuronal excitability, namely maintaining a target level of electrical activity, are of great significance in processes ranging from memory storage to activity‐dependent neuronal development.[ 16 ]
In this regard, CNT–hydrogel composites where CNTs are embedded within the hydrogel matrix might serve as biocompatible conductive scaffolds while having less effect on modifying intrinsic neuronal excitability due to the reduced contact between CNTs and cell membranes. The CNT–hydrogel composites were reported to stimulate neurite outgrowth of model cell line pheochromocytoma 12 (PC12) cells under electrical stimulation.[ 12a,b ] Moreover, it has been recently reported that the differentiation of PC12 cells,[ 8 , 12 ] dorsal root ganglia,[ 12c ] and stem cells[ 12d ] can be enhanced on CNT–hydrogel composites even without the presence of exogenous electric fields. However, the underlying mechanism remains elusive. Enhanced excitability of electrical activity was often assumed as vital in stimulating neuronal differentiation, although there is no concrete evidence supporting this so far.[ 8a,c,d ] Moreover, higher electrical activity as shown by increased Ca2+ transients during neuronal development may greatly impede neuronal maturation leading to growth cone stalling and axonal retraction.[ 17 ]
Most of the existing studies focus on the expression of cell phenotypes and are largely based on PC12 cells. At the same time, the effect of CNT–hydrogel composites on neurogenesis and neuronal excitability remains largely unexplored. The underlying question is whether the CNTs embedded in hydrogels could indeed affect neurogenesis and neuronal excitability. Moreover, CNT–hydrogel composites are usually prepared in aqueous solutions and CNTs need to be pre‐functionalized to improve dispersion.[ 18 ] In this study, we prepared CNT‐poly(ethylene glycol) (PEG) hydrogel composites using a newly developed simple and versatile method, which does not require the prefunctionalization of CNTs. To gain a deeper understanding on the effect of the composites on neuronal development, we investigated the differentiation of PC12 cells and neurogenesis of adult neural stem cells (NSCs). Moreover, we studied the network activity on composites using primary hippocampal neurons.
2. Results and Discussion
We used an ionic liquid as a medium to prepare CNT‐based hydrogel composites. Ionic liquids, known as “green solvents” with negligible vapor pressure, have been reported to promote the dispersion of carbon nanomaterials in a polymer matrix or solutions even without functionalization.[ 19 ] The main advantage of combing CNTs and ionic liquids is that bucky gels containing CNT meshwork infused with large amounts of ionic liquids can be obtained by simple mixing using mechanical grinding. The entangled CNT bundles are exfoliated to provide finer bundles forming percolation networks in ionic liquids.[ 20 ] We found that such CNT network can be transferred into a hydrogel matrix by in situ polymerizations of suitable monomers in ionic liquids and then turned into CNT–hydrogel composite by solvent replacements. PEG is considered biocompatible, and its use in biomedical applications is well established. Therefore, we selected CNT–PEG hydrogel composites as a model system. Poly(ethylene glycol) diacrylate, which is well dissolvable in ionic liquids, was used as a precursor for synthesizing PEG hydrogel matrix by polymerization and crosslinking under UV illumination.[ 21 ]
To prepare CNT–PEG hydrogels, CNTs (10, 20, 40 mg) of 10–30 µm lengths were mixed with 2 mL 1‐hexyl‐3‐methyl‐imidazolium bis(trifluoromethyl sulfonyl)imide by mechanical grinding for 30 min (Figure 1 ). Then, 0.5 mL poly(ethylene glycol) diacrylate containing 10% (v/v) 2‐hydroxy‐2‐methylpropiophene as a photoinitiator was added to the mixtures by stirring. The final mixtures were stirred for >2 h and used as precursors. 50 µL of the precursors were sandwiched between two glass slides and illuminated by UV light (365 nm, 250 mA) for a specific amount of time (10–240 s, Table S1, Supporting Information) to make a composite film with a thickness of ≈140 µm. The top cover slide was pre‐modified with 3‐(trimethoxysilyl)propyl acrylate to anchor the composite films covalently. The CNT–PEG hydrogel composites with varying CNT loading were obtained by replacing the ionic liquid with acetonitrile and then with water. The composites are denoted as c‐PEG‐x, where x is the initial concentration (w/v) of CNTs in the ionic liquid.
Figure 1.
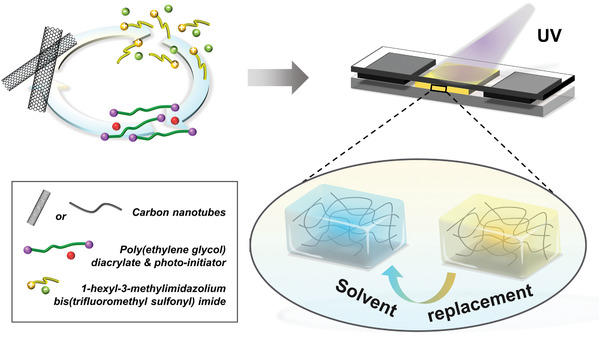
Schematic illustration of the preparation of CNT–PEG hydrogel composites. The CNTs were ground in 1‐hexyl‐3‐methylimidazolium bis(trifluoromethyl sulfonyl) imide. Poly(ethylene glycol) diacrylate with a photoinitiator was added to the mixtures and they were used as precursors. The precursors were sandwiched between two glass slides with two cover slides (140 µm thick) used as spacers. The CNT–PEG hydrogel composites with varying CNT content were prepared by UV‐induced polymerization followed by solvent replacements.
Before studying neuronal differentiation on the c‐PEG‐x samples, we characterized the physicochemical properties of the composites. Figure 2a shows the frequency (ω) dependence of the real (G′) and imaginary (G″) parts of the complex shear moduli of the precursors as measured by oscillatory shear rheology (see Experimental Section). The frequency response of the precursor of pure PEG hydrogels reveals non‐Newtonian liquid behavior. It is characterized by a typical terminal region with G′(ω) ∼ ω2, G″(ω) ∼ ω1. Upon incorporating the CNTs, a plateau with G′ > G″ is observed at low frequencies in c‐PEG‐5 samples. It indicates the formation of weak CNTs percolation networks. For c‐PEG‐20 samples, G′ > G″ over a wider range of frequency reflects the response of strong CNTs networks. The formation of CNTs percolation networks also resulted in a remarkable increase in the shear viscosity of precursors (Figure S1, Supporting Information). The surface morphology of the composites was inspected by scanning electron microscopy (see Experimental Section). After dehydration, the composites showed some protrusions of CNTs through the top of the surface (Figure 2b, Figure S2, Supporting Information).
Figure 2.
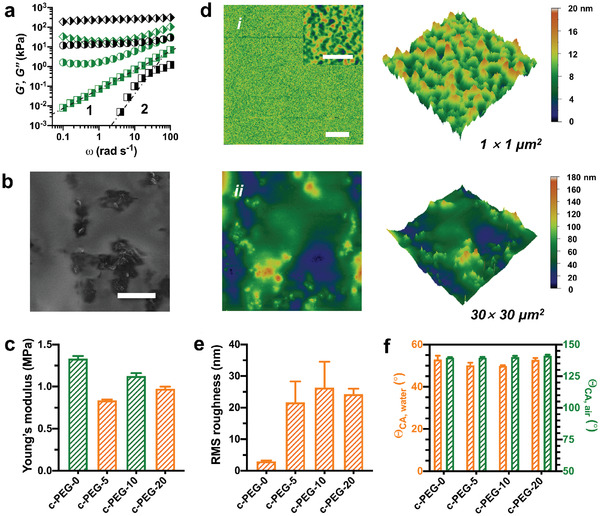
Characterization of CNT–PEG hydrogel composites. a) Storage modulus (G′, black) and loss modulus (G″, green) of the precursors of c‐PEG‐0 (◨), c‐PEG‐5 (◑), c‐PEG‐20 (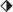 ) samples. b) Scanning electron microscopy image of the surface of c‐PEG‐20 samples. The images were captured after the samples were gradually dehydrated at ambient temperature. Scale bar: 5 µm. c) Young's modulus of the samples obtained from the nanoindentation experiments. d) Surface topographies (2D and 3D height images) of c‐PEG‐0 (i) and c‐PEG‐20 (ii) samples captured in water by atomic force microscopy. Scale bar: 5 µm (inset: 500 nm). e) Root mean square (RMS) roughness of the surface (30 µm × 30 µm) of the samples. f) Apparent contact angles (ΘCA) of water droplets in air (orange) and air bubbles under water (green) on the surface of the samples. Bar graphs show mean ± standard error of the mean (S.E.M.).
) samples. b) Scanning electron microscopy image of the surface of c‐PEG‐20 samples. The images were captured after the samples were gradually dehydrated at ambient temperature. Scale bar: 5 µm. c) Young's modulus of the samples obtained from the nanoindentation experiments. d) Surface topographies (2D and 3D height images) of c‐PEG‐0 (i) and c‐PEG‐20 (ii) samples captured in water by atomic force microscopy. Scale bar: 5 µm (inset: 500 nm). e) Root mean square (RMS) roughness of the surface (30 µm × 30 µm) of the samples. f) Apparent contact angles (ΘCA) of water droplets in air (orange) and air bubbles under water (green) on the surface of the samples. Bar graphs show mean ± standard error of the mean (S.E.M.).
The surface mechanics and topography of materials designed to mimic extracellular matrix have great impact on cell adhesion, migration, and differentiation.[ 22 ] The surface mechanics of the composites in water was investigated by a nanoindenter (see Experimental Section). The elastic modulus of the composites was obtained by fitting indentation curves (Figure S3, Supporting Information) using the Hertz model. The elastic modulus of the composite samples varied from ≈ 0.8 MPa (i.e., c‐PEG‐5) to ≈1.3 MPa (i.e., c‐PEG‐0) (Figure 2c). It should be noted that the precursors with CNTs required longer exposure time to UV illumination for adequate crosslinking and mechanical strength. We attribute the need for prolonged illumination to the reduction of polymerization efficiency resulting from the UV adsorption of CNTs, or the dark color of precursors.[ 23 ] Thus, the UV exposure time was tuned to achieve a comparable elastic modulus of the composite films (Table S1, Supporting Information).
The surface topography of the composites in water was investigated using atomic force microscopy (see Experimental Section). Pure PEG hydrogels show a smooth surface (Figure 2d). The root mean square (RMS) roughness (30 µm × 30 µm) was ≈ 3 nm (Figure 2d). Upon incorporating CNTs, the surface roughness of the composites increased significantly compared to pure hydrogels (Figure 2d, and Figure S4, Supporting Information). The RMS roughness of c‐PEG‐20 samples was ≈24 nm. This indicates the increase of surface microstructures. No remarkable differences in surface roughness among the composite samples could be discerned (Figure 2e). Moreover, no significant difference in the apparent contact angle of water drops in air (or air bubbles under water) was observed (Figure 2f).
The effect of CNT–PEG hydrogel composites on neuronal differentiation was first investigated using PC12 cells. PC12 cells exhibited minimal cell viability after culturing on c‐PEG‐0 samples for 24 h (Figure 3a), which was attributed to the intrinsic cell repellent properties of pure PEG hydrogels due to their minimal protein adsorption (Figure 3b).[ 24 ] However, when compared to pure hydrogels, cell adhesion was increased on the composites. The cell viability increased with higher CNT loading but did not change further above c‐PEG‐10 samples. The results might indicate that CNTs in the composites enhance the adsorption of extracellular matrix proteins, as CNTs are prone to interact with many biomolecules.[ 4 , 25 ] The protein adsorption of the composites was evaluated using bovine serum albumin (see Experimental Section). An increased adsorption of bovine serum albumin was observed when CNTs content was increased (Figure 3b). The cytotoxicity of the composites was evaluated by comparing cell viability (Figure 3c) and apoptosis (Figure 3d) of PC12 cells cultured on poly(l‐lysine) (PLL)‐coated glass substrates (as the positive control) and c‐PEG‐20 samples for different incubation periods (also see Figures S5 and S6, Supporting Information). These results point at minimal cytotoxicity of the composites.
Figure 3.
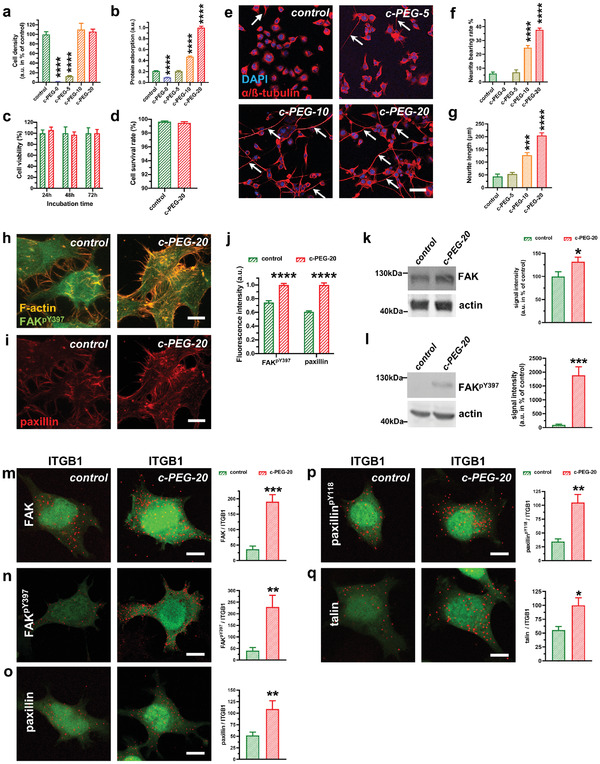
Proliferation and differentiation of PC12 cells on poly(l‐lysine) (PLL)‐coated glass and CNT–PEG substrates. a) Cell density of PC12 cells after incubation on PLL‐coated glass substrates (control) and different CNT–PEG composite samples (n = 7 for control and c‐PEG‐20, 3 for c‐PEG‐0, c‐PEG‐5 and c‐PEG‐10, 7 for c‐PEG‐20, one‐way analysis of variance (ANOVA)). b) Normalized protein adsorption of PLL‐coated glass substrates and different CNT–PEG composite samples (n = 6 for all conditions), one‐way ANOVA. c) Cell viability of PC12 cells cultured on PLL‐coated glass substrates (control, green) and c‐PEG‐20 samples (red) at different culture time points following plating (n = 7 for 24 h, 6 for 48 h, and 5 for 72 h control and 7 for all time points for c‐PEG‐20 samples, two‐way ANOVA). d) Cell survival rate of PC12 cells after 24 h on PLL‐coated glass substrates and on c‐PEG‐20 samples (n = 6 for controls and 5 for c‐PEG‐20 samples). e) Immunostaining images of PC12 cells after differentiation for 12 d on PLL‐coated glass substrates (control), c‐PEG‐5, c‐PEG‐10 and c‐PEG‐20 samples. Cells were stained with α/β‐tubulin antibody (red) and the nuclei were stained with DAPI (blue). Arrows point to neurites. Scale bar: 50 µm. f) Neurite analysis of PC12 cells cultured on different substrates. Cells with a protrusion length of more than 5 µm were assigned as neurite‐bearing cells (n = 5 for PLL‐coated glass substrates, 3 for c‐PEG‐0, 3 for c‐PEG‐5, 8 for c‐PEG‐10 and 16 for c‐PEG‐20, one‐way ANOVA). g) Neurite length measurement of PC12 cells cultured on different substrates. (n = 21 cells for control, 67 cells for c‐PEG‐5, 80 cells for c‐PEG‐10 and 110 cells for c‐PEG‐20, one‐way ANOVA). h,i) Immunofluorescent detection of phosphorylated focal adhesion kinase (FAKpY397) and of paxillin in PC12 cells cultured on PLL‐coated glass substrates (left) or c‐PEG‐20 samples (right). Note that the fluorescence intensity for PLL‐coated glass substrates (left) was adjusted for brightness/contrast to allow for better visualization. F‐actin (orange) displays cell protrusions and shows presence of FAKpY397 in cell filopodia, which was especially visible when PC12 cells were cultured on c‐PEG‐20 samples. j) Quantitative assessment of fluorescence intensity revealed a significant increase of FAKpY397 and paxillin in PC12 cells cultured on c‐PEG‐20 samples (n = 6 cells for each condition, two‐sided t‐test). k,l) Western blot of the total FAK (k) and of the active, phosphorylated FAK (FAKpY397, l) show a significant increase of FAK and of FAKpY397 when PC12 cells were cultured on c‐PEG‐20 samples (n = 6 for all conditions, two‐sided unpaired t‐test). m–q) Images of proximity ligation assay (PLA) pointing protein‐protein interaction of integrin beta 1 (ITGB1) with FAK (m), FAKpY397 (n), paxillin (o), paxillinpY118 (p) and talin (q). Note the higher protein‐protein interaction signals (depicted by red signals) on c‐PEG‐20 samples when compared to PLL‐coated glass substrates (controls). Quantitative analysis of signals pointing to protein/protein interaction revealed significant higher interaction for FAK/ITGB1 (m), FAKpY397/ITGB1 (n), paxillin/ITGB1 (o), paxillinpY118/ITGB1 (p), talin/ITGB1 (q). (n = 5 cells on control glass substrates and 6 cells on c‐PEG‐20 samples for FAK/ITGB1; 5 cells on control substrate and 5 cells on c‐PEG‐20 samples for FAKpY397/ITGB1; 6 cells on control substrate and 6 cells on c‐PEG‐20 samples for paxillin/ITGB1; 6 cells on control substrate and 6 cells on c‐PEG‐20 samples for paxillinpY118/ITGB1; 6 cells on control substrate and 6 cells on c‐PEG‐20 samples for talin/ITGB1; unpaired two‐tailed t‐test was used for normal distributed data and Mann‐Whitney test was used for non‐parametric data) Bar graphs show mean ± S.E.M. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The differentiation of PC12 cells was investigated by culturing them in a low‐serum medium (containing 2.5% fetal bovine serum) where proliferation was inhibited while no exogenous neurotrophic factors were supplemented. After 12 d, PC12 cells could hardly be observed on pure hydrogels while cells on PLL‐coated glass substrates differentiated to a minimal degree (Figure 3e,f). The differentiation of PC12 cells was enhanced with increasing CNT content in the composites. For instance, cells cultured on c‐PEG‐20 samples exhibited a maximum 38% proportion of cells bearing neurites (Figure 3f). No significant changes in the neurite bearing rate of PC12 cells were observed when cells were cultured on a sample with even higher CNT loading (Figure S7, Supporting Information). As discussed above, the protein adsorption was enhanced on the composites (Figure 3b). This points to the possibility that the adsorption of growth factors (i.e., diffusible signaling proteins that stimulate cell growth, differentiation, etc.) secreted by the differentiated PC12 cells was enhanced when PC12 cells were cultured on the composites, a fact, which might contribute to long‐term survival and differentiation.[ 4 , 26 ]
We therefore analyzed the potential mechanism for triggering the differentiation of PC12 cells on the composites and focused on the focal adhesion‐mediated signaling. The surface microstructures have been reported to promote cell differentiation by triggering intracellular signal transduction pathways via the integrin‐mediated focal adhesions.[ 2a ] Focal adhesions are macromolecular complexes consisting mainly of integrins, focal adhesion kinases (FAK), paxillin, talin, which form mechanical links between intracellular actin bundles and the extracellular matrix or substrates. The integrin activation results in various intracellular signaling alterations related to cell motility, survival, proliferation, and differentiation and was shown to be important for neuronal adhesion as well as for neuronal maturation, where it plays an important role in the formation of synaptic contacts, morphologically visible as dendritic spines.[ 27 ] The phosphorylation of FAK at tyrosine‐397 (FAKpY397) is regarded as the most critical event for neuronal differentiation since FAKpY397 functions as an activated form and a “switch” for multiple signaling outputs.[ 28 ] One of the most important downstream targets of FAK is paxillin. The phosphorylation of paxillin (paxillinpY118) acts as a platform for the downstream signaling.
PC12 cells cultured on c‐PEG‐20 samples for 3 d developed more neurites as shown by filamentous actin (F‐actin), which reflects the neuronal polarization during the early differentiation stage (Figure 3h). The fluorescence intensity of immunostaining indicates a higher expression level of FAKpY397 and paxillin of PC12 cells cultured on c‐PEG‐20 samples compared to the control (Figure 3i,j, and Figure S8, Supporting Information). In order to quantify the upregulation of FAK and of its active form, FAKpY397, we performed fluorescence Western blotting, which allows for linear signal quantification. Here, we detected higher total FAK levels as well as increased levels of the active FAK form (FAKpY397) in cells cultured on c‐PEG‐20 pointing indeed to increased formation of focal adhesions (Figure 3k,l). To further evaluate whether the composites are able to promote focal adhesions formation, we performed proximity ligation assay (PLA), which is a robust tool to detect protein complexes and an accepted method to detect protein–protein interactions in situ (see Experimental Section). Our data showed significantly increased numbers of FAK/integrin β1(ITGB1), paxillin/ITGB1 and talin/ITGB1 complexes in PC12 cells cultured on c‐PEG‐20 samples when compared to control cultures (Figure 3g,h), suggesting an increase of focal adhesion formation induced by the composites. Moreover, PC12 cells on c‐PEG‐20 samples displayed more FAKpY397/ITGB1 and paxillinpY118/ITGB1 complexes, supporting the surmise that CNT–PEG hydrogel composite induces differentiation‐supporting intracellular signal transduction. Taken together, these results indicate that the surface microstructures promote focal adhesion formation and downstream pro‐differentiation signaling activation.
While PC12 cells are a commonly used model system to investigate cell viability, adhesion, and differentiation, they are derived from the peripheral nerve system and do not form synapses and are a difficult model to assess regenerative properties of substrates meant to be used for therapy of central nervous system lesions. Neural stem cells (NSCs) have been shown to be suitable for restoring connectivity after spinal cord injury.[ 29 ] Thus, after observing an enhanced differentiation of PC12 cells, the differentiation of adult murine NSCs on the composites was further investigated (Figure 4 ). NSCs are self‐renewing, multipotent cells capable of differentiation into neurons and astrocytes.[ 30 ] After differentiation for 35 d, both NSCs cultured on PLL‐coated glass substrates (control) and c‐PEG‐20 samples differentiated into neurons (Figure 4a) and developed into neuron‐astrocyte networks (Figure 4b). No differences in the gross morphology of networks were discerned between PLL‐coated glass substrates and c‐PEG‐20 samples. However, the astrocytes‐to‐neurons ratio was observed to be lower in c‐PEG‐20 samples compared to glass substrates (Figure 4c). Further analysis revealed that the neuronal cell density on the c‐PEG‐20 samples was comparable to that on the glass substrates, while the cell density of astrocytes was significantly reduced (Figure S9, Supporting Information). These results suggest that c‐PEG‐20 coated samples were likely to inhibit the proliferation or survival of the NSCs differentiated astrocytes.
Figure 4.
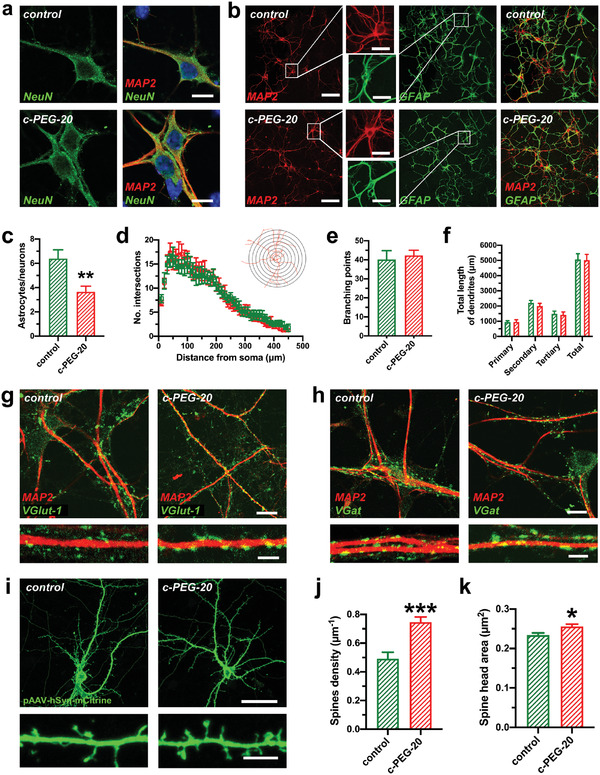
Differentiation of adult mouse neural stem cells (NSCs) on PLL‐coated glass substrates and c‐PEG‐20 samples. a) Expression of the neuronal marker NeuN was comparable in neurons cultivated on PLL‐coated glass substrates and c‐PEG‐20 samples. Scale bar: 10 µm. b) Immunostaining images of networks derived from NSCs differentiation on PLL‐coated glass substrates and c‐PEG‐20 samples for 5 weeks. Neurons and astrocytes are stained with MAP2 and GFAP‐antibodies, respectively. Scale bar: 50 µm (inset: 20 µm). c) Astrocytes‐to‐neurons ratio of NSC‐differentiated on glass substrates (control, green) and c‐PEG‐20 samples (red) (n = 8 control and 7 c‐PEG‐20, two‐sided unpaired t‐test). d) Sholl analysis plots comparing dendritic complexity of neurons derived from NSC differentiation on glass substrates (green) and c‐PEG‐20 samples (red). (n = 10 neurons cultured on PLL‐coated glass substrates and 10 neurons on c‐PEG‐20 samples, two‐way ANOVA). The inset shows a conceptual example of Sholl analysis. e) Branching points and f) total length of neuron dendrites derived from NSCs on glass substrates (green) and c‐PEG‐20 samples (red) (n = 10 neurons per condition, e) two‐sided t‐test and f) two‐way ANOVA). g) Assessment of glutamatergic input revealed comparable VGlut‐1 punctae along MAP‐2 stained dendrites on neurons cultivated on control glass substrates and on c‐PEG‐20 samples. h) Immunostaining revealed similar distribution of inhibitory synapses (shown by VGat staining) on MAP‐2 stained dendrites of neurons cultivated on glass substrates and on c‐PEG‐20 samples. i) Immunostaining showing typical NSC‐differentiated neurons displaying complex dendritic trees and an axon cultivated on PLL‐coated glass substrates (control) and c‐PEG‐20 samples. Neurons were transfected with the pAAV‐hSyn‐IRES‐mCitrine plasmid and stained with GFP‐antibody to visualize the dendritic spines. Higher detail below shows dendritic spines of neurons from NSCs differentiated on control glass substrates or on c‐PEG‐20 samples. Scale bar: 30 µm, 5 µm. j) Spine density of neurons derived from NSCs show significant higher spine density when cultivated on c‐PEG‐20 samples. (n = 333 spines from 11 fields of view on glass substrates and n = 612 spines from 14 fields of view on c‐PEG‐20 samples, unpaired t‐test). k) Spine head area was significantly increased when neurons were cultivated on c‐PEG‐20 samples when compared to control substrates (n = 737 spines on neurons on control glass substrates and 875 spines from neurons cultivated on c‐PEG‐20 samples, two‐sided t‐test). Bar graphs show mean ± S.E.M. *p < 0.05, **p < 0.01, ***p < 0.001.
The dendritic development of neurons was evaluated by a semiautomatic Sholl analysis (see Experimental Section). The analysis is conceptually achieved by drawing concentric circles around the soma in a distant‐dependent manner. The number of intersections at a varying distance (step = 10 µm) from the soma reflects dendritic complexity (Figure 4d). No distinct differences were observed between glass substrates and c‐PEG‐20 samples. The branching and total length of the dendrites were further investigated (Figure 4e,f). The total length of primary, secondary, and tertiary dendrites was thus classified. The results show no significant differences between samples on glass substrates and c‐PEG‐20 samples. We next assessed connectivity patterns of the NSC‐derived neurons and interrogated excitatory and inhibitory synaptic inputs. Immunostaining displayed well‐developed glutamatergic synapses (depicted by the presynaptic excitatory marker VGlut1) and inhibitory, GABAergic synapses in the neuronal network, which were clearly visible along the MAP‐2 stained dendritic structures (Figure 4g,h). Detailed dendritic investigation (lower image in Figure 4g,h) pointed to bigger and denser synaptic inputs on dendrites of NSC‐derived neurons cultured on c‐PEG‐20 samples.
To quantify synaptic connectivity patterns, we performed detailed morphological analyses (Figure 4i–k, and Figure S10, Supporting Information) finding that neurons cultivated on c‐PEG‐20 samples showed increased density of dendritic spines and a larger spine head area (Figure 4i–k). The dendritic spines are specific microstructures for neuronal communication and are morphological correlates of functional synapses.[ 31 ] The spine density corresponds to the density of synapses, while the spine head area reflects synaptic strength.[ 32 ] The mean spine density of neurons on PLL‐coated glass substrates and c‐PEG‐20 samples was 0.49 µm–1, and 0.74 µm–1, respectively. These results indicate a higher synaptic connectivity of the neurons cultivated on c‐PEG‐20 samples when compared to neurons cultivated on PLL‐coated glass substrates. As discussed above for PC12 cells, the enhanced protein adsorption and surface microstructures mediated focal adhesion formation might well contribute to the development and maturation of the neurons differentiated from NSCs, which is consistent with the previous report that integrin activation promotes spine formation.[ 27 ]
After investigating the differentiation of NSCs, the maturation and the network activity of primary hippocampal neurons were further evaluated (Figure 5 ). Primary hippocampal neurons were prepared from the embryonic (day 17) mouse hippocampal anlage and represent an intermediate time point in the development of this postmitotic neuronal population,[ 33 ] which makes them especially suitable for analysis of effects of different substrates on neuronal differentiation and maturation. Cultures of primary neurons form synaptic connections and are therefore used as in vitro models to study neuronal excitability regulation. Primary hippocampal neurons exhibited similar gross morphology and network densities on PLL‐coated glass substrates (control) and c‐PEG‐20 samples after 16 d in vitro (DIV 16) (Figure 5a,b). In order to assess neuronal network excitation/inhibition balance, we first performed immunofluorescent staining of excitatory, glutamatergic and GABAergic, inhibitory inputs finding similar distribution of excitatory and inhibitory synapses along the MAP‐2 stained dendrites (Figure 5c,d). The maturity of neurons was then analyzed by evaluating dendritic spine densities, measurement of the spine head area, spine head length and by assessment of the different spine categories. The mean spine densities of neurons on glass substrates and c‐PEG‐20 samples were 0.94 and 0.97 µm–1, respectively (Figure 5f). Our data suggest that primary hippocampal neurons grown on c‐PEG‐20 substrate displayed a comparable spine head area, spine head length and a spine maturation as shown by analysis of the different spine categories by careful assessment of spine morphology (Figure 5g‐i). These results indicate that c‐PEG‐20 samples are optimally suited for development of primary immature neurons which exhibit a robust innate differentiation program, a finding which is consistent with previous studies.[ 5 , 6 ] Therefore, primary hippocampal neurons serve as a good model for investigating the properties of network activity on the composites.
Figure 5.
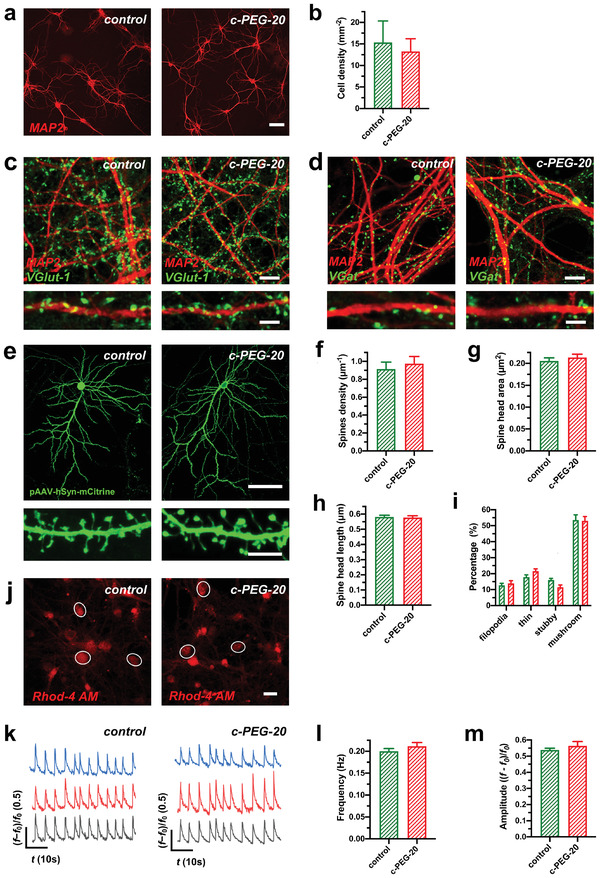
Differentiation and neuronal activity of primary hippocampal neurons cultured on PLL‐coated glass substrates and c‐PEG‐20 samples. a,b) Immunostaining of dendritic processes (stained with MAP‐2) of neuronal networks of primary hippocampal neuron cultures grown on PLL‐coated glass substrates (control) and c‐PEG‐20 samples for 16 d in vitro (DIV16). Quantitative assessment of cell densities of neurons cultured on the different substrates did not reveal significant differences (n = 3 cultures on control and 4 cultures on c‐PEG‐20 samples for cell density, two‐tailed t‐test). Scale bar: 50 µm. c) Glutamatergic input revealed comparable VGlut‐1 puncta along MAP‐2 stained dendrites on primary hippocampal neurons cultivated on control glass substrates and on c‐PEG‐20 samples. d) Inhibitory synapses on MAP‐2 stained dendrites of neurons cultivated on glass substrates displayed a similar distribution when compared to neurons cultivated on c‐PEG‐20 samples. Scale bars: 10 µm for (c,d) and 5 µm for detail images in (c,d). e) Exemplary images of GFP‐transfected primary hippocampal neurons and their dendritic spines (below) from neuronal cultures grown on PLL–glass substrates and c‐PEG‐20 samples for 16 d in vitro (DIV16). For visualization, neurons were transfected with the pAAV‐hSyn‐IRES‐mCitrine plasmid and stained with GFP antibody for visualizing dendritic spines (shown below). Scale bars: 50 µm and 5 µm for detail. f) Dendritic spines densities, g) spine head area, h) spine head length and quantitative analysis of spine categories (i) were comparable when primary hippocampal neurons were cultured on PLL‐coated glass substrates (control, green) or c‐PEG‐20 samples (red) (n = 614 spines from neurons cultivated on control and 534 spines from neurons cultivated on c‐PEG‐20 samples (f), n = 520 spines from neurons cultivated on control and 570 spines from neurons cultivated on c‐PEG‐20 samples (g), n = 520 spines from neurons cultivated on control and 570 spines from neurons cultivated on c‐PEG‐20 samples (h), n = 614 spines analyzed from neurons cultivated on control glass substrates and 534 spines from neurons cultivated on c‐PEG‐20 samples (i), two tailed t‐test or Mann‐Whitney test for nonparametric data). j) Overview on rhodamine‐4 AM loaded neurons cultivated on control and c‐PEG‐20 samples. Circled neurons were assessed for Ca2+‐oscillations as exemplary depicted by original traces shown in (k). Scale bar: 20 µm. l,m) Frequencies and amplitudes of synchronized calcium oscillations did not show significant differences for primary hippocampal neurons when grown on PLL‐coated glass substrates or c‐PEG‐20 samples (n = 13 recordings on control and 11 recordings on c‐PEG‐20 samples for frequency analysis and n = 36 measurements for amplitudes from neurons cultivated on control glass substrates and 33 measurements for amplitudes from neurons plated on c‐PEG‐20 samples, two sided t‐test). Bar graphs show mean ± S.E.M.
The network activity of primary hippocampal neurons was next investigated by calcium imaging, which is a reliable method used to track the activity of neuronal populations.[ 34 ] The primary hippocampal neurons cultured in vitro for 14 d exhibited spontaneous synchronized calcium oscillations driven by synaptic activity.[ 35 ] Using a calcium indicator (Rhod‐4 AM) and confocal microscopy (see Experimental Section), we recorded the oscillations of free cytosolic calcium in primary cultures of hippocampal neurons (Figure 5j,k, and Movie S1, Supporting Information). The frequency of calcium oscillations, which is dependent on the periodic changes of the membrane potential driven by action potentials, reflects neuronal network excitability.[ 35 , 36 ] The amplitude reflects the efficacy of action potential in evoking postsynaptic calcium current through glutamatergic receptors and voltage‐gated calcium channels.[ 35 , 37 ] Here, we compared frequencies and amplitudes of calcium oscillations in the neurons cultured on PLL‐coated glass substrates and c‐PEG‐20 samples (Figure 5l,m). These results indicate that the CNT–PEG hydrogel composites are able to maintain the homeostasis of neuronal network activity.
These results differ from earlier studies performed on CNT‐coated substrates where an increased neuronal excitability was reported.[ 5b,c ] The main difference between CNT‐coated substrates and CNT–PEG hydrogel composites is that in the latter case, the CNTs were embedded in the hydrogel and thus their interactions with the neurons might have been partially disrupted. Thus, the encapsulation of the CNT network into the hydrogel matrix could indeed play a critical role in maintaining the homeostasis of neuronal network activity. We therefore conclude that the encapsulation of the CNT network into the hydrogel matrix did not change the developmental balance between excitatory synapses and inhibitory synapses, thereby preserving intrinsic neuronal activity and neuronal network homeostasis.
3. Conclusions
We have described and characterized a novel and versatile method for preparing CNT–hydrogel composites. The effects of CNT–PEG hydrogel composites on neuronal differentiation and electrical activity were investigated using PC12 cells, adult NSCs which were differentiated into neurons and astrocytes and primary hippocampal neurons. We demonstrated that the composites were beneficial for the long‐term survival and differentiation of PC12 cells and NSCs. The results show an enhanced differentiation of PC12 cells and an increased neuron‐to‐astrocyte ratio of NSCs on the composites compared to the control conditions of glass substrates. Furthermore, the spine density of neurons derived from NSCs was increased. The enhanced differentiation of PC12 cells and NSCs may be ascribed to the enhanced surface microstructures and protein adsorption. Moreover, network activity of primary cultures of hippocampal neurons cultured on the composites was maintained within a physiological range and was comparable to the neuronal activity of primary neurons from the same preparation cultured on standard control conditions. In addition, c‐PEG‐20 composites allowed for optimal neuronal differentiation up to the level of dendritic spines. To sum up, our data suggest that the CNT–PEG hydrogel composites are novel versatile substrates for culturing neurons, suitable for the development of lesion interpolates for potential use in vivo. These substrates display several advantages for neuronal differentiation while maintaining homeostatic properties of neuronal networks which is an important prerequisite when intended to be used for repair of CNS lesions, which are often associated with pathological hyperexcitability as present, e.g., in the case of ischemic stroke.
4. Experimental Section
Materials and Sample Preparation
Poly(ethylene glycol) diacrylate (average M n = 575 Da), 2‐hydroxy‐2‐methylpropiophene (D1173, 97%), 3‐(trimethoxysilyl)propyl acrylate (92%), and 1‐hexyl‐3‐methyl‐imidazolium bis(trifluoromethyl sulfonyl)imide (T g = −9 °C) were purchased from Sigma‐Aldrich. CNTs (multiwalled, outside diameter = 18–28 nm, length = 10–30 µm, >96%, 1.7% —OH, 98 S cm−1) were purchased from Nanografi Nano Technology. Microscope glass slides (No. 1.5H) were purchased from Thermo Fisher Scientific. The cover slides were pre‐modified by immersing them into a solution of 2% (v/v) 3‐(trimethoxysilyl)propyl acrylate in ethanol overnight, followed by rinsing with ethanol and drying with nitrogen gas. Poly(l‐lysine) (PLL)‐coated glass substrates were used as the control. All the samples for cell plating were pre‐treated by immersing them into an aqueous solution of 0.1 wt% PLL (Sigma‐Aldrich) at 4 °C overnight.
Viscoelastic Properties
The viscoelastic properties of the precursors were measured by a DHR‐3 rheometer (TA Instruments). A 1° cone– plate geometry with a diameter of 40 mm was used for the measurements. The sample thickness was around 1 mm. A strain amplitude sweep was performed first, in order to define the linear viscoelastic zone. The storage modulus, loss modulus, and complex viscosity were obtained by a frequency sweep carried out at room temperature within the range of 0.1–100 rad s−1.
Surface Morphology
The samples were gradually dehydrated and dried at ambient temperature. The surface morphology of the samples was captured by scanning electron microscopy (Zeiss, LEO Gemini 1530) with an accelerating voltage of 3 kV.
Surface Modulus
The samples were placed in a Petri dish which was filled with Milli‐Q water (18.2 MΩ cm). The surface modulus was measured using an Asylum MFP‐3D nanoindenter (Oxford instruments). A spherical indenter made of sapphire was used with a radius of 0.5 mm. The indentation was done under force control. After touching the sample surface with a threshold force of 5–10 µN, the force was ramped up to 200 µN within 5 s, then held at 200 µN for 5 s, and then reduced to 0 µN within 5 s again. Indentation curves were fitted using the Hertz model, assuming a sample Poisson ratio of 0.5 for the polymer.
Surface Topography
The samples were placed in a Petri dish which was filled with Milli‐Q water (18.2 MΩ cm). The surface topography was captured using atomic force microscopy (JPK NW III, QI mode) under water. The cantilever with a CONT‐W tip (NanoWorld) at the very end was used. The resonant frequency of the cantilever is 13 kHz (9–17 kHz). The spring constant of the cantilever is 0.2 N m−1 (0.07–0.4 N m−1). The images and RMS surface roughness were analyzed by free software, Gwyddion (v2.56).
Cell Viability and Cytotoxicity, and Focal Adhesion
PC12 cells (6 × 104 cells cm–2) were plated and maintained in DMEM (Invitrogen) supplemented with 10% FBS, 0.1 × 10−3 m minimum essential medium nonessential amino acids (MEM‐NEAA; Invitrogen), 2 × 10−3 m l‐glutamine (Gibco), 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin. For cell viability, cells were fixed and stained with α/β‐Tubulin Antibody (Cell signaling, #2148, 1:2000) after culturing for 24, 48, and 72 h, and the cell number was calculated. For cell apoptosis, the cells (48 h) were stained with cleaved Caspase3 (a marker for dead cells). Cleaved Caspase3 positive cells were counted. For analysis of focal adhesion, the medium was replaced with 2.5% FBS culture medium after 24 h and maintained for another 48 h. Primary antibodies were used for immunostaining as following: Phospho‐FAK (pY397) antibody (Cell signaling, #3283S, 1:2000); Paxillin antibody (Thermo scientific, MA5‐13356, 1:1000). To visualize F‐actin, cells were further stained with Alexa Fluor 568 Phalloidin (Thermo Scientific, A12380, 1:1000).
Protein Adsorption
Samples with a diameter of 2 cm were placed in a six‐well plate, and each well was filled with 2 mL of 1 wt% bovine serum albumin (BSA) solution in phosphate‐buffered saline (PBS, pH 7.4, Sigma). The samples were left to allow for protein adsorption at ambient temperature (22 °C) for 1 h while stirring in an orbital shaker at 90 rpm. Then, the samples were rinsed 3 times with PBS and placed into 1.5 mL of 1 wt% SDS solution for 24 h to thoroughly extract BSA from the samples. The BSA concentration in the SDS solution was analyzed with Micro BCA Protein‐Assay‐Kit (Thermo Scientific, 23235).
Differentiation of PC12 Cells
PC12 cells were plated at a low density (1 × 104 cells cm−2) in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 0.1 × 10−3 m minimum essential medium non‐essential amino acids (MEM‐NEAA; Invitrogen), 2 × 10−3 m l‐glutamine (Gibco), 100 U mL−1 penicillin and 100 µg mL−1 streptomycin. The medium was replaced with DMEM (Invitrogen) supplemented with 2.5% FBS, 0.1 × 10−3 m minimum essential medium nonessential amino acids (MEM‐NEAA; Invitrogen), 2 × 10−3 m l‐glutamine (Gibco), 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin after 16 h. Cells were retained for 12 d for subsequent analysis.
Proximity Ligation Assay (PLA)
PC12 cells were plated on the control and c‐PEG‐20 samples in DMEM medium containing 10% FBS at about 40% confluence. 6 h later, cells were transfected with CAG‐integrin β 1‐HA‐ires‐GFP plasmid, in which an HA tag was added to the intracellular N‐terminal of integrin β 1. 24 h later, the medium was replaced with 2.5% FBS culture medium and the cells were cultured for another 24 h. Cells were fixed with 4% PFA followed by blocking with blocking solution and were incubated with primary antibodies at 4 °C for overnight. Then the incubations with the PLA probes (antimouse and antirabbit IgG antibodies conjugated with oligonucleotides), ligation, and amplification according to the user manual of Duolink Proximity Ligation Assay (Sigma). The following primary antibodies were used for PLA: HA‐Tag (Cell signaling, 6E2, #23671:1000), FAK Antibody (Cell signaling, #3285, 1:1000) and Phospho‐FAK (Tyr397) Antibody (Cell signaling, #3283, 1:1000), Paxillin Antibody (Cell signaling, #2542, 1:1000), Phospho‐Paxillin (Tyr118) Antibody (Cell signaling, #2541, 1:1000) and Talin‐1 Antibody (Cell signaling, C45F1, #4021, 1:1000). Imaging was obtained on a Leica SP8 confocal laser scanning microscope. Quantitative analysis was performed by counting PLA‐positive dots per cell.
Differentiation of Adult Neural Stem Cells
Adult neural stem cells culture was prepared from the lateral wall of the subventricular zone (SVZ) of young adult (8–12 weeks) C57/Bl6 mice. All experiments were conducted in accordance with the national laws for the use of animals in research. Prior to commencing the culture, plates were coated with 0.1% (w/w) poly(d‐lysine) (Sigma‐Aldrich) and laminin (5 µg mL−1, Sigma) at 4 °C overnight. After anesthetizing the mice according to the appropriate institutional guidelines and performing cervical dislocation, the mouse brains were quickly removed and transferred into ice‐cold Hank's balanced salt solution (HBSS, Gibco) medium. The thin layer of tissue surrounding the ventricle was carefully dissected and stored in HBSS medium on ice. Thereafter, the tissue was digested in pre‐warmed 0.05% Trypsin–EDTA in HBSS for 10 min in a water bath at 37 °C, followed by centrifugation at 300g for 5 min, discarding the supernatant. Then, the tissue was washed twice with growth medium, composed of Neural Basal Medium A (Invitrogen) with 2% B27 (Invitrogen), 1× GlutaMAX (Invitrogen). The tissue was suspended again in 2 mL of growth medium and dissociated by gently pipetting up and down approximately 10× using a P1000 pipette, followed by passing the cell suspension through a 40 µm sieve to remove debris and un‐dissociated tissue clumps.
The resulting cell suspension was plated onto the poly(d‐lysine)/laminin coated glass plates, maintained in growth medium supplemented with 20 ng mL−1 purified mouse receptor‐grade epidermal growth factor (EGF, Sigma) and 20 ng mL−1 recombinant bovine fibroblast growth factor (FGF‐2, Sigma). At this stage, the yield of adult neural stem cells was very low. EGF and FGF‐2 stimulate stem cell proliferation and yield enough adult neuronal stem cells for subsequent experiments. For the differentiation, equal amounts of stem cells were plated on the samples and maintained in growth medium with EGF and FGF‐2. When the cells reached approximately 80% confluency, the growth medium was replaced with a medium containing 5 ng mL−1 FGF‐2 and no EGF. Following 2 d in 5 ng mL−1 FGF‐2, the medium was replaced again with a growth medium in the absence of both EGF and FGF‐2. Cells were retained for ≈5 weeks for subsequent analysis. Primary antibodies were used for immunostaining as following: MAP2 antibody (Sigma, #HPA012828, 1:2000) and NeuN antibody (Chemicon, MAB377, 1:1000) for neurons, GFAP antibody (Sigma, #G6171, 1:2000) for astrocytes.
Primary Culture of Hippocampal Neurons
Hippocampi from C57BL/6J mouse embryos at embryonic day 17 (E17) were dissected and incubated in HBSS supplemented with 0.05% trypsin for 15 min at 37 °C. All experiments were conducted in accordance with the national laws for the use of animals in research. Then the hippocampi were washed twice and dissociated in a plating medium (MEM, Invitrogen) supplemented with 10% horse serum, 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin (Gibco), 0.6% glucose (Fresenius Kabi) using Pasteur pipettes with polished tips. Neurons (1 × 105 cells cm−2) were seeded on the samples and kept at 37 °C and 5% carbon dioxide (CO2). Cells were gradually washed with PBS and incubated in a neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen), 100 U mL−1 penicillin, 100 µg mL−1 streptomycin (Gibco), and 0.5 × 10−3 m l‐glutamine (Gibco) after 3 h. The cells were retained for ≈2 weeks and ≈3 weeks for analysis and calcium imaging, respectively.
Sholl Analysis, and Spine Analysis
The neurons for Sholl analysis and spine analysis were transfected with the pAAV‐hSyn‐IRES‐mCitrine plasmid to visualize cell morphology (see DNA transfection in Supporting Information). Cells were fixed and stained with a GFP antibody 2 d after transfection. Fluorescent images were captured by confocal laser scanning microscopy (Leica SP8, and Zeiss LSM880). The images were quantified using ImageJ (Fiji). For Sholl analysis, a 20× objective (NA = 0.8) was used and Z‐series stacks with a step size of 1 µm were recorded. Z‐projected (Max intensity) was performed to convert the stacks into 2D images. The dendrites of neurons were traced with the semi‐automated Simple Neurite Tracer plugin.[ 38 ] The tracers were saved as SWC (or Traces) files. The Sholl analysis was performed by the Sholl Analysis plugin using the SWC files with a step distance of 10 µm.[ 39 ] For spine analysis, a 63× oil objective (NA = 1.4) was used and Z‐series stacks with a step of 0.2 µm were recorded. Spines were defined as dendritic protrusions from 0.5 µm up to 5 µm in length and were manually counted along a selected dendritic segment. For each dendritic segment, the number of spines per µm dendrite was calculated.
Calcium Imaging
Primary hippocampal neurons were incubated with 1 × 10−6 m Rhod‐4 AM (Abcam, #ab142780) for 30 min at 37 °C in 5% CO2. The samples were washed and conditioned in 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES)‐buffered Krebs–Ringer solution (Avantor, J67795.AP, containing 120 × 10−3 m sodium chloride, 5 × 10−3 m potassium chloride, 2 × 10−3 m calcium chloride, 1 × 10−3 m magnesium chloride, 25 × 10−3 m sodium bicarbonate, 5.5 × 10−3 m HEPES, and 1 × 10−3 m d‐glucose, pH 7.2) for 30 min at 37 °C. The calcium imaging was then performed by confocal laser scanning microscopy (Zeiss LSM880) equipped with an incubator (the temperature was equilibrated at 37°C). A 20× Plan‐Apochromat objective (NA = 0.8) was used. The excitation was done with a HeNe laser operating at 543 nm and the emitted fluorescence was detected in the range from 560 to 700 nm. The fluorescent fluctuation of the regions of interest (soma) was recorded and analyzed by the ZEN v2.3 software (Zeiss). Changes in cytosolic calcium are shown as (f – f 0)/f 0, where f is the fluorescence intensity at soma over time, f 0 is the fluorescence intensity at the rest state.
Details of DNA transfection, western blotting, and immunostaining are available in Supporting Information.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
L.Y. and H.J. contributed equally to this work. K.K., J.V., and H.‐J.B. are equally contributing senior authors. L.Y. conceived the idea of this project. L.Y. and H.J. designed and performed the main experiments. K.K., J.V., and H.‐J.B. supervised the project. J.L., C.‐H.T., and M.K. participated in sample characterizations and analyses. L.Y., H.J., K.K., J.V., and H.‐J.B. wrote the manuscript. All the authors reviewed the manuscript and approved the final version.
Supporting information
Supporting Information
Supplemental Movie 1
Acknowledgements
The authors gratefully acknowledge the fruitful discussions they had with Dr. Rüdiger Berger at the Max Planck Institute for Polymer Research (MPIP), and the technical support given by Andreas Hanewald, Uwe Rietzler, and Andreas Best at the MPIP, and Dr. Stefan Wagner at University of Cologne. L.Y. acknowledges the participation of Heng Zhang, Chao Zhu at the MPIP, and Heiko Endle at the University Medical Center (Mainz) in further productive discussions. K.K. gratefully acknowledges the funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)–Projektnummer 213555243. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 1451) and by Germany's Excellence Strategy – EXC 2030 – 390661388 to J.V.
Open access funding enabled and organized by Projekt DEAL.
Ye L., Ji H., Liu J., Tu C.‐H., Kappl M., Koynov K., Vogt J., Butt H.‐J., Carbon Nanotube–Hydrogel Composites Facilitate Neuronal Differentiation While Maintaining Homeostasis of Network Activity. Adv. Mater. 2021, 33, 2102981. 10.1002/adma.202102981
Contributor Information
Kaloian Koynov, Email: koynov@mpip-mainz.mpg.de.
Johannes Vogt, Email: johannes.vogt@uk-koeln.de.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.a) Houshyar S., Bhattacharyya A., Shanks R., ACS Chem. Neurosci. 2019, 10, 3349; [DOI] [PubMed] [Google Scholar]; b) Steel E. M., Sundararaghavan H. G. in Neural Engineering: From Advanced Biomaterials to 3D Fabrication Techniques (Eds: Zhang L. G., Kaplan D. L.), Springer International Publishing, Cham, Switzerland: 2016, pp. 145–179; [Google Scholar]; c) Farrukh A., Zhao S., del Campo A., Front. Mater. 2018, 5, 62. [Google Scholar]
- 2.a) Eftekhari B. S., Eskandari M., Janmey P. A., Samadikuchaksaraei A., Gholipourmalekabadi M., Adv. Funct. Mater. 2020, 30, 1907792; [Google Scholar]; b) Burnstine‐Townley A., Eshel Y., Amdursky N., Adv. Funct. Mater. 2020, 30, 1901369. [Google Scholar]
- 3.a) Fabbro A., Prato M., Ballerini L., Adv. Drug Delivery Rev. 2013, 65, 2034; [DOI] [PubMed] [Google Scholar]; b) Oprych K. M., Whitby R. L., Mikhalovsky S. V., Tomlins P., Adu J., Adv. Healthcare Mater. 2016, 5, 1253. [DOI] [PubMed] [Google Scholar]
- 4.a) Mattson M. P., Haddon R. C., Rao A. M., J. Mol. Neurosci. 2000, 14, 175; [DOI] [PubMed] [Google Scholar]; b) Hu H., Ni Y., Montana V., Haddon R. C., Parpura V., Nano Lett. 2004, 4, 507; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chao T. I., Xiang S., Lipstate J. F., Wang C., Lu J., Adv. Mater. 2010, 22, 3542; [DOI] [PubMed] [Google Scholar]; d) Chen Y. S., Hsiue G. H., Biomaterials 2013, 34, 4936. [DOI] [PubMed] [Google Scholar]
- 5.a) Cellot G., Cilia E., Cipollone S., Rancic V., Sucapane A., Giordani S., Gambazzi L., Markram H., Grandolfo M., Scaini D., Gelain F., Casalis L., Prato M., Giugliano M., Ballerini L., Nat. Nanotechnol. 2009, 4, 126; [DOI] [PubMed] [Google Scholar]; b) Mazzatenta A., Giugliano M., Campidelli S., Gambazzi L., Businaro L., Markram H., Prato M., Ballerini L., J. Neurosci. 2007, 27, 6931; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Lovat V., Pantarotto D., Lagostena L., Cacciari B., Grandolfo M., Righi M., Spalluto G., Prato M., Ballerini L., Nano Lett. 2005, 5, 1107. [DOI] [PubMed] [Google Scholar]
- 6.a) Pampaloni N. P., Lottner M., Giugliano M., Matruglio A., D'Amico F., Prato M., Garrido J. A., Ballerini L., Scaini D., Nat. Nanotechnol. 2018, 13, 755; [DOI] [PubMed] [Google Scholar]; b) Tang M., Song Q., Li N., Jiang Z., Huang R., Cheng G., Biomaterials 2013, 34, 6402. [DOI] [PubMed] [Google Scholar]
- 7. Capasso A., Rodrigues J., Moschetta M., Buonocore F., Faggio G., Messina G., Kim M. J., Kwon J., Placidi E., Benfenati F., Bramini M., Lee G.‐H., Lisi N., Adv. Funct. Mater. 2021, 31, 2005300. [Google Scholar]
- 8.a) Park J., Jeon J., Kim B., Lee M. S., Park S., Lim J., Yi J., Lee H., Yang H. S., Lee J. Y., Adv. Funct. Mater. 2020, 2003759; [Google Scholar]; b) Hu X., Wang X., Xu Y., Li L., Liu J., He Y., Zou Y., Yu L., Qiu X., Guo J., Adv. Healthcare Mater. 2020, 9, 1901570; [DOI] [PubMed] [Google Scholar]; c) Liu X., Miller A. L., Park S., Waletzki B. E., Zhou Z., Terzic A., Lu L., ACS Appl. Mater. Interfaces 2017, 9, 14677; [DOI] [PubMed] [Google Scholar]; d) Liedtke W., Yeo M., Zhang H., Wang Y., Gignac M., Miller S., Berglund K., Liu J., Small 2013, 9, 1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Li J., Mooney D. J., Nat. Rev. Mater. 2016, 1, 16071; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sankaran S., Becker J., Wittmann C., del Campo A., Small 2019, 15, 1804717. [DOI] [PubMed] [Google Scholar]
- 10. Jung I. Y., Kim J. S., Choi B. R., Lee K., Lee H., Adv. Healthcare Mater. 2017, 6, 1601475. [Google Scholar]
- 11.a) Villiou M., Paez J. I., del Campo A., ACS Appl. Mater. Interfaces 2020, 12, 37862; [DOI] [PubMed] [Google Scholar]; b) Jia Z., Gong J., Zeng Y., Ran J., Liu J., Wang K., Xie C., Lu X., Wang J., Adv. Funct. Mater. 2021, 31, 2010461. [Google Scholar]
- 12.a) Imaninezhad M., Pemberton K., Xu F. L., Kalinowski K., Bera R., Zustiak S. P., J. Neural Eng. 2018, 15, 056034; [DOI] [PubMed] [Google Scholar]; b) Koppes A. N., Keating K. W., McGregor A. L., Koppes R. A., Kearns K. R., Ziemba A. M., McKay C. A., Zuidema J. M., Rivet C. J., Gilbert R. J., Thompson D. M., Acta Biomater. 2016, 39, 34; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang L., Wu Y., Hu T., Ma P. X., Guo B., Acta Biomater. 2019, 96, 175; [DOI] [PubMed] [Google Scholar]; d) Hasanzadeh E., Ebrahimi‐Barough S., Mirzaei E., Azami M., Tavangar S. M., Mahmoodi N., Basiri A., Ai J., J. Biomed. Mater. Res., Part A 2019, 107, 802. [DOI] [PubMed] [Google Scholar]
- 13. Ramer L. M., Ramer M. S., Bradbury E. J., Lancet Neurol. 2014, 13, 1241. [DOI] [PubMed] [Google Scholar]
- 14. Vogt J., Kirischuk S., Unichenko P., Schlüter L., Pelosi A., Endle H., Yang J.‐W., Schmarowski N., Cheng J., Thalman C., Strauss U., Prokudin A., Bharati B. S., Aoki J., Chun J., Lutz B., Luhmann H. J., Nitsch R., Cereb. Cortex 2017, 27, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Winkelmann A., Maggio N., Eller J., Caliskan G., Semtner M., Häussler U., Jüttner R., Dugladze T., Smolinsky B., Kowalczyk S., Chronowska E., Schwarz G., Rathjen F. G., Rechavi G., Haas C. A., Kulik A., Gloveli T., Heinemann U., Meier J. C., J. Clin. Invest. 2014, 124, 696; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nelson S. B., Valakh V., Neuron 2015, 87, 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turrigiano G. G., Nelson S. B., Nat. Rev. Neurosci. 2004, 5, 97. [DOI] [PubMed] [Google Scholar]
- 17. Gomez T. M., Spitzer N. C., Nature 1999, 397, 350. [DOI] [PubMed] [Google Scholar]
- 18. Mehrali M., Thakur A., Pennisi C. P., Talebian S., Arpanaei A., Nikkhah M., Dolatshahi‐Pirouz A., Adv. Mater. 2017, 29, 1603612. [DOI] [PubMed] [Google Scholar]
- 19.a) Zhao L., Li Y., Liu Z., Shimizu H., Chem. Mater. 2010, 22, 5949; [Google Scholar]; b) Zhao L., Li Y., Cao X., You J., Dong W., Nanotechnology 2012, 23, 255702; [DOI] [PubMed] [Google Scholar]; c) Guan J., Xing C., Wang Y., Li Y., Li J., Compos. Sci. Technol. 2017, 138, 98. [Google Scholar]
- 20.a) Fukushima T., Kosaka A., Ishimura Y., Yamamoto T., Takigawa T., Ishii N., Aida T., Science 2003, 300, 2072; [DOI] [PubMed] [Google Scholar]; b) Lee J., Aida T., Chem. Commun. 2011, 47, 6757. [DOI] [PubMed] [Google Scholar]
- 21. Fdz De Anastro A., Porcarelli L., Hilder M., Berlanga C., Galceran M., Howlett P., Forsyth M., Mecerreyes D., ACS Appl. Energy Mater. 2019, 2, 6960. [Google Scholar]
- 22.a) Chaudhuri O., Cooper‐White J., Janmey P. A., Mooney D. J., Shenoy V. B., Nature 2020, 584, 535; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Guilak F., Cohen D. M., Estes B. T., Gimble J. M., Liedtke W., Chen C. S., Cell Stem Cell 2009, 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shin S. R., Bae H., Cha J. M., Mun J. Y., Chen Y.‐C., Tekin H., Shin H., Farshchi S., Dokmeci M. R., Tang S., Khademhosseini A., ACS Nano 2012, 6, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khademhosseini A., Yeh J., Jon S., Eng G., Suh K. Y., Burdick J. A., Langer R., Lab Chip 2004, 4, 425. [DOI] [PubMed] [Google Scholar]
- 25. Shao H., Li T., Zhu R., Xu X., Yu J., Chen S., Song L., Ramakrishna S., Lei Z., Ruan Y., He L., Biomaterials 2018, 175, 93. [DOI] [PubMed] [Google Scholar]
- 26. Convertino D., Fabbri F., Mishra N., Mainardi M., Cappello V., Testa G., Capsoni S., Albertazzi L., Luin S., Marchetti L., Coletti C., Nano Lett. 2020, 20, 3633. [DOI] [PubMed] [Google Scholar]
- 27. Liu X., Huai J., Endle H., Schlüter L., Fan W., Li Y., Richers S., Yurugi H., Rajalingam K., Ji H., Cheng H., Rister B., Horta G., Baumgart J., Berger H., Laube G., Schmitt U., Schmeisser M. J., Boeckers T. M., Tenzer S., Vlachos A., Deller T., Nitsch R., Vogt J., Dev. Cell 2016, 38, 275. [DOI] [PubMed] [Google Scholar]
- 28.a) Teo B. K. K., Wong S. T., Lim C. K., Kung T. Y. S., Yap C. H., Ramagopal Y., Romer L. H., Yim E. K. F., ACS Nano 2013, 7, 4785; [DOI] [PubMed] [Google Scholar]; b) Ivankovic‐Dikic I., Grönroos E., Blaukat A., Barth B.‐U., Dikic I., Nat. Cell Biol. 2000, 2, 574. [DOI] [PubMed] [Google Scholar]
- 29.a) Ceto S., Sekiguchi K. J., Takashima Y., Nimmerjahn A., Tuszynski M. H., Cell Stem Cell 2020, 27, 430.e435; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fischer I., Dulin J. N., Lane M. A., Nat. Rev. Neurosci. 2020, 21, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.a) Obernier K., Alvarez‐Buylla A., Development 2019, 146; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lim D. A., Alvarez‐Buylla A., Cold Spring Harbor Perspect. Biol. 2016, 8, a018820; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Clarke D. L., Johansson C. B., Wilbertz J., Veress B., Nilsson E., Karlström H., Lendahl U., Frisén J., Science 2000, 288, 1660. [DOI] [PubMed] [Google Scholar]
- 31. Alvarez V. A., Sabatini B. L., Annu. Rev. Neurosci. 2007, 30, 79. [DOI] [PubMed] [Google Scholar]
- 32.a) Kasai H., Fukuda M., Watanabe S., Hayashi‐Takagi A., Noguchi J., Trends Neurosci. 2010, 33, 121; [DOI] [PubMed] [Google Scholar]; b) Matsuzaki M., Honkura N., Ellis‐Davies G. C., Kasai H., Nature 2004, 429, 761; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) McKinney R. A., J. Physiol. 2010, 588, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a) Altman J., Bayer S. A., J. Comp. Neurol. 1990, 301, 343; [DOI] [PubMed] [Google Scholar]; b) Altman J., Bayer S. A., J. Comp. Neurol. 1990, 301, 325. [DOI] [PubMed] [Google Scholar]
- 34. Grienberger C., Konnerth A., Neuron 2012, 73, 862. [DOI] [PubMed] [Google Scholar]
- 35. Bacci A., Verderio C., Pravettoni E., Matteoli M., Eur. J. Neurosci. 1999, 11, 389. [DOI] [PubMed] [Google Scholar]
- 36. Murphy T. H., Blatter L. A., Wier W. G., Baraban J. M., J. Neurosci. 1992, 12, 4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang Y.‐M., Wang L.‐Y., J. Neurosci. 2006, 26, 5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Longair M. H., Baker D. A., Armstrong J. D., Bioinformatics 2011, 27, 2453. [DOI] [PubMed] [Google Scholar]
- 39. Ferreira T. A., Blackman A. V., Oyrer J., Jayabal S., Chung A. J., Watt A. J., Sjöström P. J., van Meyel D. J., Nat. Methods 2014, 11, 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supplemental Movie 1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


