Abstract
Autism spectrum disorder (ASD) is characterized by social and neurocognitive impairments, with mutations of the SHANK3 gene being prominent in patients with monogenic ASD. Using the InsG3680 mouse model with a Shank3 mutation seen in humans, we revealed an unknown role for Shank3 in postsynaptic oligodendrocyte (OL) features, similar to its role in neurons. This was shown by impaired molecular and physiological glutamatergic traits of InsG3680-derived primary OL cultures. In vivo, InsG3680 mice exhibit significant reductions in the expression of key myelination–related transcripts and proteins, along with deficits in myelin ultrastructure, white matter, axonal conductivity, and motor skills. Last, we observed significant impairments, with clinical relevance, in induced pluripotent stem cell–derived OLs from a patient with the InsG3680 mutation. Together, our study provides insight into Shank3’s role in OLs and reveals a mechanism of the crucial connection of myelination to ASD pathology.
Shank3 mutation disrupts glutamate signaling and myelination in ASD mouse models and human iPSCs.
INTRODUCTION
Autism spectrum disorders (ASDs) are a set of neurodevelopmental disorders known for the hallmark symptoms of impaired social behavior and communication, limited interests, and repetitive behavior patterns, among others (1–3). Despite ongoing research, the underlying causes of the various disorders on the spectrum remain unclear, although there is evidence of a strong genetic link for many of these disorders (4–10). These genes include SHANK3, mutations of which are associated with a high risk for monogenic ASD (11–17). While the impact of these mutations has been extensively studied in neurons and at excitatory synapses, its effects on oligodendrocytes (OLs) and myelination are not yet understood. This gap in knowledge is fundamental for our understanding of ASD mechanisms and clinical approaches.
SHANK3 is expressed at the postsynaptic density (PSD) of excitatory neurons (11, 18). Interacting with main PSD proteins to form a complex that connects glutamate receptors to actin filaments, SHANK3 plays an integral role in synaptic transmission at excitatory synapses and in glutamatergic signaling (11, 12, 19). Extensive research has shown how different mutations in Shank3 disturb normal synaptic transmission between neurons due to lower expression of glutamate receptors, reduced numbers of dendritic spines, abnormal cell morphology, altered ionic currents, and decreased glutamate receptor activity (12, 20–23).
OLs undergo a developmental process, starting as OL progenitor cells (OPCs) and differentiating into mature OLs (mOLs) (24–26). OL development and growth depend on various signals, primarily those originating from neurons (26, 27). These signals act at axon-glia synapses, where neurotransmitters are released (28, 29). Such intricate communication is primarily facilitated by glutamate, which is released by neurons and acts as a presynaptic signal (30, 31). OLs have glutamate receptors and express postsynaptic proteins, similarly to dendrites in neurons, and act as postsynaptic receivers of the neuronal signal (32–34). Receptor signal binding leads to calcium and other ions influx, promoting OPC differentiation (35–40). This calcium influx also regulates myelin formation and remodeling of myelin ultrastructure and has a large effect on the expression of key myelination genes (33, 34, 41–46). Disruption of this pathway could thus lead to impaired OL maturation and function (42, 43). Recently, Shank3 was shown to be expressed in OLs (47, 48), although its role at axon-OL synapses and whether Shank3 interacts with other postsynaptic proteins to maintain proper glutamate-mediated interactions in this cell population are unknown.
Neuronal function and neural circuit connectivity rely on proper axonal conductivity, supported by the myelin that wraps axons and maintains action potential propagation (49–53). Myelin, generated by mOLs in the central nervous system, comprises lipids and key myelination proteins, such as Plp1, Mbp, and Mog, responsible for maintaining the unique condensed layer structure (54–56). Interruption of this structure was observed previously in ASDs (57), and there is some evidence of myelination disruption in Shank3-related ASD models (48, 58). However, at present, the roles of Shank3 in OL development and myelination at the molecular, structural, and cellular levels remain unclear.
The InsG3680 mouse model contains an inserted guanine in the Shank3 gene as in the mutated human gene that causes a frameshift and an early stop codon, resulting in marked down-regulation of Shank3 protein, as compared to controls (12). In humans, the corresponding mutation leads to a specific disorder on the autistic spectrum, and therefore, studying this mouse model has a greater potential to understand its underlying mechanisms in humans (12, 13). At the same time, the use of induced pluripotent stem cells (iPSCs) from patients with the human equivalent of the mutation found in InsG3680 mice could provide further insight into the effect of this mutation in human cells. iPSCs are generated from adult somatic cells and can be reprogrammed to differentiate into almost any type of human cell (59). Recent studies found that iPSC-derived neurons from patients with the mutated SHANK3 gene described above exhibit similar defects as found in the InsG3680 mouse model (22, 60, 61). Nevertheless, this mutation’s impact on other cell types was not deeply studied.
Here, to investigate Shank3’s role specifically in OLs and the potential effect of the InsG3680 mutation in Shank3 on OLs properties, we first examined the involvement of Shank3 as a postsynaptic protein and in glutamate signaling in OLs in vitro. Then, to study how dysfunctional OLs affect myelination on the whole organism level, we characterized myelination properties in vivo in this model, to understand the molecular and physiological consequences of the InsG3680 mutation. To corroborate our findings from the mouse model, we also investigated clinically relevant iPSC-derived OPCs obtained from a patient carrying the InsG3680 mutation. Last, we restored SHANK3 levels in primary OPC cultures and explored whether this restoration ameliorates several of the observed deficits.
RESULTS
To perform the following experiments, we used wild-type (WT) littermates, referred to as controls; homozygous InsG3680 mutated mice referred to as InsG3680 mice; heterozygous InsG3680 mutated mice referred to as InsG3680(+/−) mice; iPSC-derived OPCs from a control and a human patient referred to as InsG3680(+/−) patient.
Shank3 mutation results in low expression of key postsynaptic genes in InsG3680 mouse OPCs and affects OL differentiation and morphology
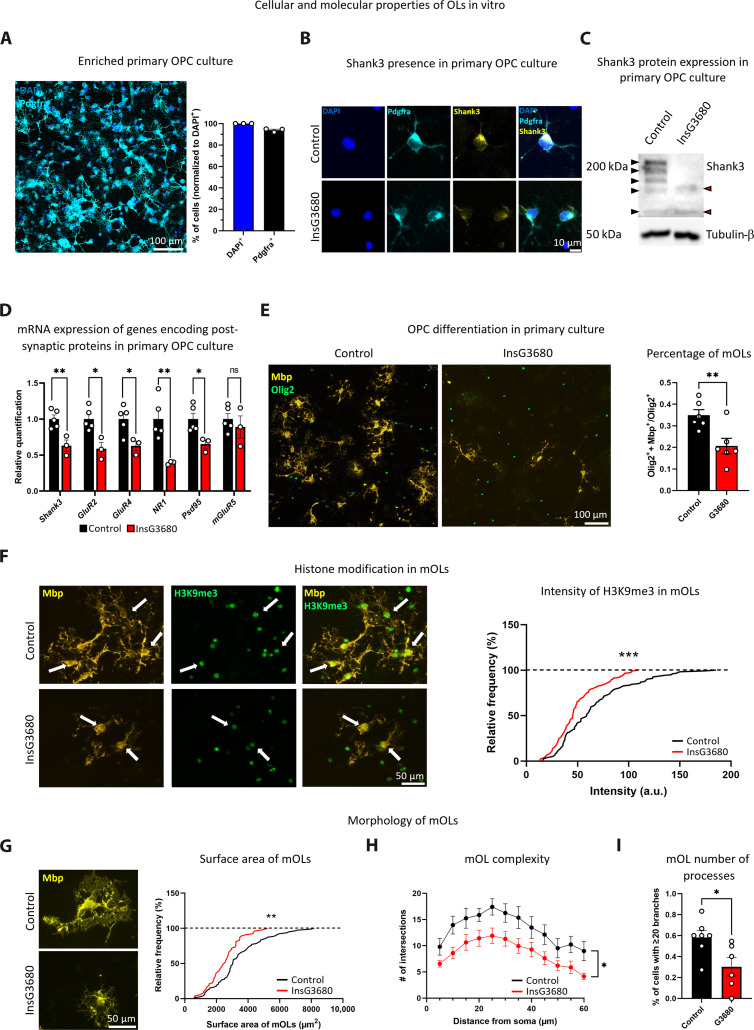
To dissect Shank3 function in OPCs and determine whether InsG3680 mutation could alter the expression of genes encoding postsynaptic proteins in OPCs, we prepared enriched primary OPC cultures from control and InsG3680 mice (Fig. 1A and fig. S1A). We found that Shank3 is expressed in control OPCs but is markedly reduced in InsG3680 mouse OPCs using immunocytochemistry assay (Fig. 1B) and Western blot assay (Fig. 1C). Moreover, we found significant down-regulation in the mRNA expression levels of Shank3 and of genes encoding key postsynaptic proteins and glutamatergic receptors in the OPC culture prepared from the mouse model, as compared to controls (Fig. 1D).
Fig. 1. Reduced expression of transcripts encoding postsynaptic proteins in InsG3680 mouse–derived OPCs and abnormal InsG3680-OL differentiation.
(A) Enriched primary OPC culture (left). Percentage of Pdgfra+ cells out of all cells, as labeled by DAPI (right). (B) Shank3 detection in control Pdgfra+ cells and reduced detection in InsG3680-derived Pdgfra+ cells. (C) Blot showing Shank3 expression in OPC lysate. Black arrowheads indicate control and red arrowheads indicate InsG3680 OPCs Shank3 isoforms. Some of the Shank3 isoforms are expressed by control but not InsG3680-derived OPCs. (D) Significantly reduced mRNA levels of Shank3, GluR2, GluR4, NR1, and Psd95 in InsG3680 mouse–derived OPCs compared to controls. No significant difference in mGluR5 mRNA levels. (E) Immunocytochemistry images showing mOLs (Olig2+ + Mbp+) and OLs (Olig2+) in control and InsG3680 mouse–derived OPCs (left). InsG3680 mice show a significantly lower percentage of mOLs after differentiation (right). (F) H3K9me3 intensity in Mbp+ cells, arrows point to Mbp+ + H3K9me3+ cells, from control and InsG3680 mice (left). H3K9me3 intensity in mOLs from InsG3680 mouse–derived cultures is significantly lower than in controls (right). (G) Control and InsG3680 mOLs (Mbp+ cells) (left). Significantly reduced mOL surface area in InsG3680 mouse–derived cells compared to controls (right). (H) Significantly decreased branching complexity in InsG3680 mouse–derived mOLs compared to controls. (I) A significantly lower percentage of mOLs with 20 or more branches in InsG3680 mouse–derived mOLs compared to controls. ns, nonsignificant. *P < 0.05, **P < 0.01, and ***P < 0.001. Two-tailed t test (D, E, and I), Kolmogorov-Smirnov test (F and G), and repeated measures analysis of variance (ANOVA) (H). Data are shown as means ± SEM (A) n = 3, primary cultures. (D) n = 5, control mice; n = 3 InsG3680 mice. (E) n = 6, control mice; n = 6, InsG3680 mice. (F) n = 122 cells and n = 3, control mice; n = 115 cells and n = 5, InsG3680 mice. (G) n = 79 cells and n = 6, control mice; n = 53 cells and n = 6, InsG3680 mice. (H and I) n = 7, control mice; n = 6, InsG3680 mice. a.u., arbitrary units.
To define whether these alterations in transcription have an effect on cellular properties, we examined the differentiation properties of OPC and mOL morphology in cells from the mouse model and the control. We used common markers to stain distinct populations of OLs; specifically, Olig2 was used to label the entire OL population, Pdgfra to identify OPCs, and Mbp to visualize mOLs. We found in the mouse model–derived cultures a decreased percentage of mOLs, measured by the number of Olig2+ + Mbp+ cells divided by Olig2+ cells, as compared to controls (Fig. 1E) with no significant difference in Olig2+ cells number (fig. S1B). OL differentiation depends on many signals and epigenetic changes, such as histone modification H3K9me3 (62). This histone modification is responsible for heterochromatin formation in certain DNA regions, and it is found in mOLs, where it is important for normal differentiation processes. In keeping with our finding of a decreased percentage of mOLs, we measured lower levels of the H3K9me3 modification in Mbp+ cells (Fig. 1F). Upon addressing whether these changes in the differentiation process led to altered mOL morphology, we found smaller mOL cell surface area in the InsG3680 mouse–derived cultures as compared to controls (Fig. 1G). We also found significantly decreased branching complexity in InsG3680 mOLs, quantified by the number of branches intersecting with concentric circles around the cell body (Fig. 1H). In addition, there was a significantly lower percentage of cells with 20 or more branches (Fig. 1I) in InsG3680 mOLs compared to controls.
When examining InsG3680(+/−) mouse–derived cultures, which represent the genotypic but not the phenotypic state of the human condition (12), we found no significant difference in mRNA expression levels between the groups (fig. S2A). However, we observed a trend of decreased percentage of mOLs (fig. S2B) and a significant reduction in mOL surface area in InsG3680(+/−) mice as compared to controls (fig. S2C).
These findings suggest that Shank3 functions as a postsynaptic scaffolding protein in OPCs. Furthermore, the InsG3680 mutation affects the expression of Shank3 isoforms in OPCs and the development of these cells.
Primary OPC cultures from InsG3680 mice show lower calcium activity rate and altered function in response to glutamate and basal conditions
We next investigated whether the altered transcription and the impaired morphological parameters we observed have physiological consequences on OPCs. Since a glutamate signal normally activates OPC glutamate receptors and leads to calcium entry into the cell, we measured calcium flux at the single-cell resolution before and after glutamate administration in primary OPC-enriched cultures (Fig. 2, A to C). Administration of 1 mM glutamate led to a significantly lower rate of calcium flux (calcium event) in InsG3680 mouse–derived OPCs, as compared to their control counterparts (Fig. 2D). Unexpectedly, the amplitude of the first calcium event was significantly higher in the InsG3680 mouse–derived OPC cultures (Fig. 2E), with no significant difference between the two groups in the area under the curve of the whole signal, reflecting the sum of calcium influx throughout the recording (Fig. 2F). This prompted us to examine glutamate signal stability and whether it is similarly maintained in both groups during the course of recording. We further identified that the ratio between the sizes of peaks was significantly lower in the InsG3680 mouse–derived OPCs as compared to controls (Fig. 2G), reflecting how the signal in these OPCs was strong at first and faded as the recording continued. Last, we measured significantly longer interevent intervals in InsG3680 mouse–derived OPCs as compared to controls, throughout the period of recording (Fig. 2H). All these results argue that InsG3680 mouse–derived OPCs that received a glutamate signal showed overall lower activity, with a stronger response at the beginning of the recording window that decayed over time, as compared to controls. Afterward, we studied the calcium response to glutamate in InsG3680(+/−) mouse–derived OPCs and found no significant difference in the calcium signal in all quantified aspects as compared to controls (fig. S2, D to I).
Fig. 2. Impaired calcium signaling in response to glutamate stimulus of InsG3680 mouse–derived OPCs as compared to controls.
(A) The experimental procedure (left) and calcium labeled in OPCs using Fluo-4 (right). An example of the region of interest (ROI) of a cell is circled and pointed in an arrow. (B) Representative traces of the signal generated upon glutamate stimulus in control (black) and InsG3680 mouse–derived OPCs (red). (C) Features that were determined on the basis of the calcium signal. (D) A significantly reduced activity rate of calcium flux was found in InsG3680 mouse–derived OPCs, as compared to controls. (E) A significantly higher amplitude of the first calcium event was measured in InsG3680 mouse–derived OPCs, (F) with no significant differences in the areas under all curves. (G) A significantly smaller ratio between the sizes of peaks was found in InsG3680 mouse–derived OPCs as compared to controls. (H) A significant increase in interevent intervals was measured in InsG3680 mouse–derived OPCs as compared to controls. **P < 0.01 and ***P < 0.001. One sample t test (D to H) and Kolmogorov-Smirnov test (D to H). Data are shown as means ± SEM. (D to F) n = 47 cells from control mice; n = 32 cells from InsG3680 mice. (G) n = 39 events from control OPCs; n = 21 events from InsG3680 OPCs. (H) n = 76 events from control OPCs; n = 32 events from InsG3680 OPCs. (D to H) n = 3, control mice; n = 3, InsG3680 mice (for one-sample t test).
To describe other physiological changes seen in InsG3680 mouse–derived OPC cultures, we measured essential ion currents at single-cell resolution using a patch clamp approach (Fig. 3A). Representative images depicting ionic currents at the highest depolarization step in both control and InsG3680 mouse–derived OPCs (Fig. 3B) and detailed views of the current recordings from control OPCs (Fig. 3C) and InsG3680 mouse–derived OPCs (Fig. 3D) are presented. Sodium currents (between −20 and 70 mV) were significantly reduced in InsG3680 mouse–derived OPCs, as compared to controls (Fig. 3E). Tetrodotoxin (TTX) experiments were performed to demonstrate that sodium currents were recorded (fig. S3, A to D). No significant difference was found in fast potassium currents (between 40 and 70 mV) (Fig. 3F) and slow potassium currents (Fig. 3G) in InsG3680 mouse–derived OPCs as compared to controls. These results suggest that the Shank3 mutation in InsG3680 mouse–derived OPCs affects sodium current conductance in OPCs, as was shown previously in neurons (23).
Fig. 3. Reduced sodium currents measured in OPCs from InsG3680 mice, as compared to OPCs from control mice.
(A) Representative image of cells before patch clamp recordings. (B) A representative trace of ionic currents at a depolarization step in control (black) and InsG3680 mouse–derived OPCs (red). Zoomed-in views of the current recordings from (C) control OPCs and (D) InsG3680 mouse–derived OPCs. (E) The sodium current was significantly reduced in InsG3680 mouse–derived OPCs, as compared to control OPCs. (F) Fast and (G) slow potassium currents were not significantly different between InsG3680 and control mouse–derived OPCs. *P < 0.05. Repeated measures ANOVA (E to G). Data are shown as means ± SEM. (E to G) n = 5, control OPCs; n = 8, InsG3680 OPCs.
Young adult and adult InsG3680 mice show increased OL numbers and reduced myelination gene expression
Having observed reduced mRNA expression levels of genes encoding key postsynaptic proteins and morphological and physiological deficits in InsG3680 mouse–derived OL cultures, we next asked whether similar changes are seen at the organismal level. Specifically, we considered whether Shank3 mutation has a substantial impact on OLs and myelination properties in the mouse model at different developmental stages. As such, we first compared Olig2+ (to identify OLs), Olig2+ + CC1+ (to identify mOLs), Pdgfra+ (to identify OPCs), and Pdgfra+ + Ki67+ (to identify proliferating OPCs) cell numbers in InsG3680 mice and their controls at 30 days postnatal (P30) in the frontal cortex, motor cortex, and striatum (Fig. 4A and fig. S4, A and B). These correspond to regions of interest (ROI) in this mouse model because of Shank3’s relatively high expression and function in these areas (11) and the important role that these areas serve in social, motor, and repetitive behaviors (1, 12, 63).
Fig. 4. Altered myelination-related properties in young adult InsG3680 mice at the cellular and molecular levels.
(A) Representative image of Olig2+, CC1+, Pdgfra+, and Ki67+ cells, with arrows pointing at Olig2+ + CC1+ and Pdgfra+ + Ki67+ colocalized cells. (B) A significantly increased number of Olig2+ cells with no significant difference in the number of Olig2+ + CC1+, Pdgfra+, and Pdgfra+ + Ki67+ cells in the frontal cortex of InsG3680 P30 mice, as compared to controls. (C) A significant increase in the number of Olig2+ cells along with a trend of increase in the number of Olig2+ + CC1+ cells in the striatum was found in P30 InsG3680 mice, as compared to controls. No significant difference was found in Pdgfra+ cell number, although the number of Pdgfra+ + Ki67+ was significantly lower in P30 InsG3680 mice, as compared to controls. Significantly reduced Mbp, Plp1, and Mog mRNA expression levels in both the (D) frontal cortex and (E) striatum of P30 InsG3680 mice were measured, as compared to controls. Significantly reduced Mbp and Plp1 protein expression levels were measured in the crude myelin fraction of P30 InsG3680 mice in the (F) frontal cortex and (G) striatum, as compared to controls. *P < 0.05, **P < 0.01, and ***P < 0.001. Two-tailed t test (B to E), Mann-Whitney (B, Olig2+), and one sample t test (F and G). Data are shown as means ± SEM. (B) n = 14, control; n = 9 InsG3680 mice for Olig2+. (B) n = 14, control; n = 8 InsG3680 mice for Olig2++CC1+ cells. (B) n = 13, control mice; n = 9 InsG3680 mice for Pdgfra+ and Pdgfra++Ki67+. (C) n = 13, control mice; n = 9 InsG3680 mice. (D and E) n = 9, control mice; n = 9, InsG3680 mice. (E) n = 8, control mice, n = 9, InsG3680 mice for Plp1. (F) n = 4, control mice; n = 4, InsG3680 mice, for Mbp. (F) n = 6, control mice; n = 7, InsG3680 mice, for Plp1. (G) n = 5, control mice; n = 5, InsG3680 mice, for Mbp. (G) n = 7, control mice; n = 6, InsG3680 mice, for Plp1.
We found a significantly increased number of OLs with no significant difference in other cell properties in the frontal cortex of P30 InsG3680 mice as compared to controls (Fig. 4B). In the striatum, we also show a significantly increased number of OLs, along with a trend of increased number of mOLs. In addition, we found no significant difference in the number of OPCs between both groups; however, we show a significant decrease in proliferating OPCs in P30 InsG3680 mice as compared to controls (Fig. 4C). No significant difference was found in cellular properties in the motor cortex between both groups (fig. S4C).
To examine whether the Shank3 mutation results in myelination-related abnormalities in vivo, we determined mRNA expression levels of key myelination genes in the frontal cortex and striatum by real-time polymerase chain reaction (RT-PCR). We found a significant reduction in the expression levels of Mbp, Plp1, and Mog in the frontal cortex (Fig. 4D) and striatum (Fig. 4E) of P30 InsG3680 mice, as compared to controls.
To further address the impact of the Shank3 mutation on key myelination proteins in these brain regions, we isolated a crude enriched myelin fraction (64) from P30 mice. We found a significant reduction in Mbp and Plp1 expression levels in the frontal cortex (Fig. 4F) and striatum (Fig. 4G) in P30 InsG3680 mice.
Since this is a model for neurodevelopmental disorders and myelin follows a developmental trajectory that is usually completed after 3 months in mice, we next asked whether the changes seen in young mice (P30) continue into adulthood by addressing the traits considered above in P90 mice. We found a significantly increased number of OLs and mOLs in the motor cortex of P90 InsG3680 mice, as compared to controls (fig. S4D). Unexpectedly, unlike what was shown for P30 InsG3680 mice, there was no significant difference in the number of OLs in the frontal cortex (Fig. 5A) and striatum (Fig. 5B) between the two groups. However, a significant decrease in the number of OPCs was detected in the striatum of P90 InsG3680 mice compared to controls (Fig. 5B), which corresponds with our finding of decreased proliferating OPCs in the striatum of P30 InsG3680 mice. When we examined mRNA expression levels of key myelination genes, we found a significant reduction in the expression levels of Mbp and Plp1 and a similar trend in Mog levels in the frontal cortex of P90 InsG3680 mice, as compared to controls (Fig. 5C). On the contrary, we found no significant difference in the mRNA expression levels of these genes in the striatum between the groups (Fig. 5D).
Fig. 5. Altered myelination-related properties in adult InsG3680 mice at the cellular and molecular levels.
(A) No significant difference between groups in the number of Olig2+, Olig2+ + CC1+, Pdgfra+, and Pdgfra+ + Ki67+ cells in the frontal cortex of P90 mice. (B) No significant difference between groups in the number of Olig2+ and Olig2+ + CC1+ cells in the striatum of P90 mice. A significant decrease in Pdgfra+ cell number in P90 InsG3680 mice, as compared to controls, with no significant difference between groups in the number of Pdgfra+ + Ki67+ cells. (C) Significantly reduced Mbp and Plp1 and a similar trend in Mog mRNA expression levels in the frontal cortex but not in the (D) striatum of P90 InsG3680 mice compared to controls. (E) Significantly reduced protein expression levels of Plp1 but not Mbp in the crude myelin fraction isolated from the frontal cortex of P90 InsG3680 mice, as compared to controls. (F) No significant difference was found between groups in the protein expression levels of Mbp and Plp1 in the crude myelin fraction isolated from the striatum. *P < 0.05 and **P < 0.01. Two-tailed t test (A to D), Mann-Whitney (C, Mbp), and one sample t test (E and F). Data are shown as means ± SEM. (A) n = 11, control mice; n = 11, InsG3680 mice for Olig2+ and Olig2++CC1+. (A) n = 14, control mice; n = 15, InsG3680 mice for Pdgfra+ and Pdgfra++Ki67+. (B) n = 12, control mice; n = 11, InsG3680 mice for Olig2+ and Olig2+ + CC1+. (B) n = 14, control mice; n = 15, InsG3680 mice for Pdgfra+. (B) n = 13, control mice; n = 15, InsG3680 mice for Pdgfra+ + Ki67+. (C) n = 15, control mice; n = 15, InsG3680 mice for Mbp. (C) n = 9, control; n = 11, InsG3680 mice for Plp1. (C) n = 10, control mice; n = 12, InsG3680 mice for Mog. (D) n = 8, control mice; n = 8, InsG3680 mice. (E) n = 6, control mice; n = 6, InsG3680 mice for Mbp. (E) n = 6, control mice; n = 5, InsG3680 mice for Plp1. (F) n = 5, control; n = 5, InsG3680 mice.
In the crude myelin fraction isolated from the frontal cortex, we found a significant decrease in the protein expression levels of Plp1, but not in Mbp proteins in InsG3680 mice, as compared to controls (Fig. 5E). In addition, we found no significant difference in the expression levels of either protein in the striatum of InsG3680 and control mice (Fig. 5F). Together, these findings highlight OL deficits in mutated mice, resulting in myelin production deficits which are most prominent during early adulthood and become moderated later in development.
Postsynaptic and myelination protein expression is reduced in the myelin fraction of InsG3680 mouse brains, alongside deficits in myelin ultrastructure
To dissect whether the reduced levels of key myelination proteins seen in the selected brain regions described above is a wider phenomenon in this mouse model, affecting other brain regions as well, we isolated a pure myelin fraction (64) from the whole brain of P90 mice. Given how this moiety is generated by OLs, the purification of myelin allowed us to examine changes in this aspect of OL biology and, in doing so, increase the resolution of the myelin studies described above.
Since the pure myelin fraction is highly enriched and contains almost no axons or other neuronal fractions (fig. S5), we were able to consider those postsynaptic proteins that are found in myelin and which may play an important role in neuro-OL synapse function. Accordingly, we first assessed the expression levels of Shank3, the postsynaptic protein Psd95, and the glutamate N-methyl-d-aspartate receptor (NMDAR) subunit NR1 and found significantly reduced amounts of all three proteins in InsG3680 mice, as compared to controls (Fig. 6A). When we examined crude myelin samples, which also contain neuronal cells compartments, we found different isoforms of Shank3 in the pure myelin fraction, all extracted from control mice (Fig. 6B). Next, to increase the resolution of our earlier findings in the frontal cortex and striatum, we assessed the expression levels of key myelination proteins in the pure myelin fraction isolated from whole brain. Here, we found a significant reduction in both Mbp and Plp1 expression levels in this fraction prepared from InsG3680 mice, as compared to controls (Fig. 6C).
Fig. 6. Reduced expression levels of synaptic and key myelination proteins in a pure myelin fraction, along with myelin ultrastructure deficits in adult InsG3680 mice.
(A) Significant reduction in the protein expression levels of Shank3, Psd95, and NR1 in a pure myelin fraction from the whole brain of P90 InsG3680 mice, as compared to controls. (B) Different Shank3 isoforms (marked in arrowheads) are detected in pure and crude myelin fractions from whole brain of P90 control mice. (C) Significant reduction in the protein expression levels of Mbp and Plp1 proteins in a pure myelin fraction from the whole brain of P90 InsG3680 mice, as compared to controls. (D) Representative images of myelin ultrastructure from both genotypes (left). Significantly reduced g-ratio [axon diameter/(axon diameter + myelin thickness)] (right) and (E) decreased axon diameter in P90 InsG3680 mice, as compared to controls. (F) Representative images of impaired ultrastructure (red) in InsG3680 mice compared to controls (left). Significantly increased percentage area of myelin ultrastructure abnormalities in the myelin sheaths of P90 InsG3680 mice, as compared to controls (right). *P < 0.05, **P < 0.01, and ****P < 0.0001. One sampled t test (A and C), linear regression (D), two-tailed t test (D to F), and Kolmogorov-Smirnov (E). Data are shown as means ± SEM. (A and C) n = 6, control mice; n = 6, InsG3680 mice. (A) n = 5, control mice; n = 5, InsG3680 mice for Psd95. (D and E) n = 679 axons, control mice; n = 513 axons, InsG3680 mice. (D to F) n = 6, control mice; n = 3, InsG3680 mice.
As these proteins are crucial for the unique condensed layered structure of myelin (65–68), we subsequently analyzed the ultrastructure of myelin sheaths in the cortico-striatal circuitry of InsG3680 and control mice. We specifically chose the cortico-striatal circuitry based on prior research that identified cortico-striatal synaptic dysfunction in InsG3680 mice (12) and since concomitant deficits in these regions are associated with autistic-like behaviors (69). We found a significantly decreased g-ratio (Fig. 6D), a parameter often used to assess axonal myelination, and lower axon diameter (Fig. 6E) in InsG3680 mice, as compared to controls. The decreased g-ratio, together with the reduced expression of proteins essential for myelin compaction, prompted us to examine the myelin ultrastructure specifically and found a significantly higher percentage of ultrastructural abnormalities in the myelin of the fibers in the cortico-striatal circuitry of P90 InsG3680 mice, as compared to controls (Fig. 6F). These data demonstrate that the InsG3680 mutation disrupts both postsynaptic proteins and essential myelination proteins, leading to abnormal myelin ultrastructure properties.
InsG3680 mice exhibit reduced connectivity between brain regions, abnormal axonal conductivity in the corpus callosum, and motor skill deficits
We next investigated how the observed myelin deficits affected wide connectivity, physiological aspects, and behavior. For this, we first performed magnetic resonance imaging (MRI) diffusion tensor imaging (DTI) scans, to analyze whole-brain tractography. We found a significant reduction in fractional anisotropy (FA) values, a common MRI-DTI–generated parameter indicative of the degree of myelination, and in the number of tracts, in numerous brain regions in InsG3680 mice, as compared to controls (Fig. 7A). Only a few areas showed a significant increase in FA values, with no areas displaying a significant increase in the number of tracts in InsG3680 mice compared to controls (fig. S6). To dissect whether these changes also appear in the corpus callosum (CC), corresponding to the largest white matter (WM) tract, we performed tractography in this region and noted a significantly reduced number of tracts in InsG3680 mice, as compared to controls (Fig. 7B).
Fig. 7. Reduced brain-wide connectivity, abnormal CC axonal conductivity, and motor skill deficits in InsG3680 mice.
(A) Connectogram depicting the significant reduction in FA values of tracts (left) in numerous brain regions and in the number of tracts (right) in P90 InsG3680 mice, as compared to controls. The scale bar indicates the level of significance, ranging from light to dark lines. Light lines represent significance levels of P < 0.05 and close to this threshold, while dark lines indicate higher levels of significance. (B) P90 InsG3680 mice exhibit a significantly lower number of tracts in the CC than do controls. (C) Schematic representation of CAP recordings in the CC, with representative CAP traces obtained from control (black) and InsG3680 (red) mice. (D) No significant difference in the N1 amplitude of InsG3680 mice and controls. (E) A significantly higher N2 amplitude was measured in InsG3680 mice, as compared to controls. (F) Significant reduction in latency to fall in the rotarod test, along with (G) significantly reduced grip strength in the forelimbs (H) but not in the hindlimbs of InsG3680 mice, as compared to controls. *P < 0.05 and **P < 0.01. Two-tailed t test (A, B, and D to H) and repeated measures ANOVA (D and E). Data are shown as means ± SEM. (A) n = 19, control mice; n = 19, InsG3680 mice. (B) n = 18, control mice; n = 19, InsG3680 mice. (D and E) n = 4, control mice; n = 4, InsG3680 mice. (F and G) n = 15, control mice; n = 14, InsG3680 mice. (H) n = 15, control mice; n = 15, InsG3680 mice.
We next considered physiological aspects of the CC and how such aspects were affected by the reduced connectivity we found in this area. Accordingly, we performed compound action potential (CAP) electrophysiological recordings in which a stimulus was applied to one side of the CC and the response on the other side was recorded (Fig. 7C). No significant differences were observed in N1 amplitude, a value which reflects myelinated axon properties or larger axon diameter, between the two groups (Fig. 7D). Nevertheless, a significantly increased signal was measured in the N2 amplitude in InsG3680 mice, reflective of a higher percentage of unmyelinated axons and smaller diameter axons (Fig. 7E). These data match our MRI-DTI and electron microscopy (EM) findings, although the EM results were collected from cortico-striatal tracts while the electrophysiological recordings were obtained from the CC.
Because myelin and connectivity deficits could cause impaired motor behavior, we analyzed the mouse motor skills in two different tests, namely, the rotarod test, which examines coordination, and the grip strength measurement (GSM) test, which is correlated to the activity properties of motor neurons, connected to both the front and rear limbs. As shown before (12, 70), InsG3680 mice had significantly shorter latency until they fell off the rotarod, as compared to controls (Fig. 7F). In addition, we found significantly lower grip strength in the forelimbs (Fig. 7G) but not in the hindlimbs (Fig. 7H) of InsG3680 mice, as compared to controls. These findings demonstrate that the InsG3680 mutation has widespread effects throughout the body and the brain. The altered structure of WM tracts in the mutant mouse brain leads to physiological and behavioral consequences.
Human iPSC-derived OPCs from a patient with the InsG3680 SHANK3 mutation show lower calcium activity rate and altered function in response to glutamate
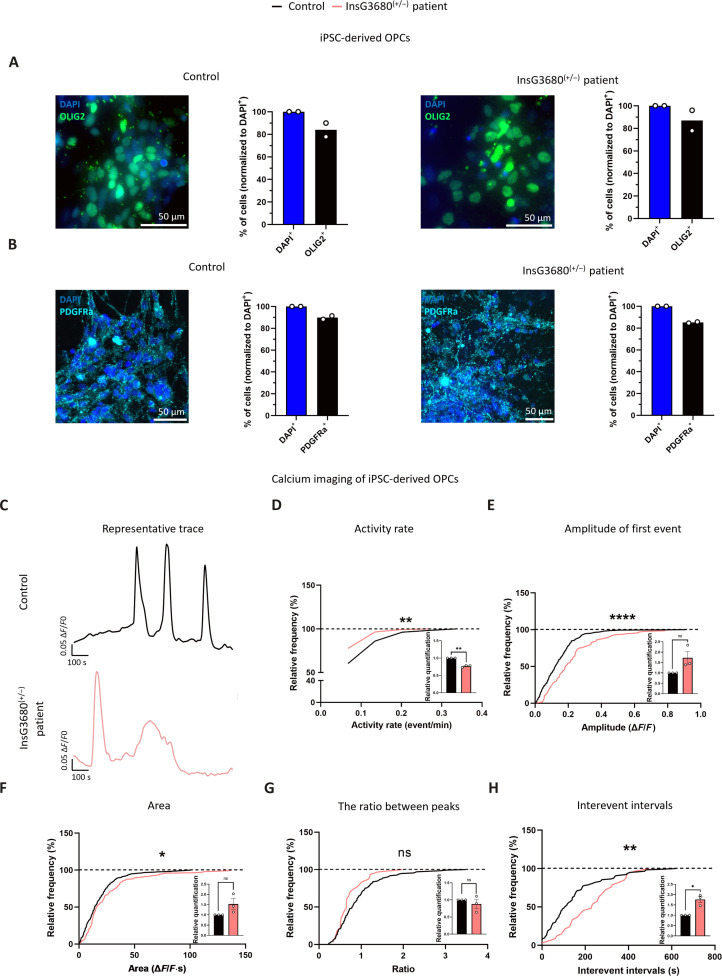
As mentioned above, the InsG3680 mutation also exists in humans, causing monogenic ASD. Therefore, we addressed iPSC-derived OPCs from a patient with the counterpart of the mutation presented by InsG3680 mice to validate some of our findings describing physiological deficits in InsG3680 mouse OPCs. Calcium imaging after glutamate administration was carried out on iPSC-derived OPCs prepared from a control and a patient with the equivalent Shank3 mutation (71) as found in InsG3680 mice, at week 8 postdifferentiation. To confirm the accurate differentiation of iPSCs into OPCs, we conducted staining using the OL marker OLIG2 (Fig. 8A and fig. S7, A and B) and the OPC marker platelet-derived growth factor receptor alpha (PDGFRA) (Fig. 8B and fig. S7, A and B) and observed a substantial percentage of OLIG2+ and PDGFRA+ cells in both cell lines. When we examined whether some of the cells differentiated into neurons using the NEUN marker, we found a considerably low percentage of this cell population in both cell lines (fig. S7, C to E). Representative calcium traces from both control and InsG3680(+/−) patient iPSC–derived OPCs are shown in Fig. 8C. We found a significantly lower activity rate of calcium events in iPSC-derived OPCs from a InsG3680(+/−) patient, as compared to the control, in response to glutamate administration (Fig. 8D). The amplitude of the first calcium event was significantly higher in the InsG3680(+/−) patient iPSC–derived OPC cultures (Fig. 8E), accompanied by a significantly larger area under the curve of the entire signal, as compared to controls (Fig. 8F). Unlike our findings in mice, the ratio between the sizes of peaks remained consistent between both cell lines (Fig. 8G). In addition, we measured significantly longer interevent intervals in InsG3680(+/−) patient iPSC–derived OPCs compared to controls throughout the recording period (Fig. 8H). These data align with our findings in mice, as both the mouse and human results demonstrated substantial physiological deficits in OPC processing in response to glutamate due to the SHANK3 mutation.
Fig. 8. Impaired calcium signaling in response to glutamate stimulus of human iPSC-derived OPCs from an individual presenting a heterozygous InsG3680 SHANK3 mutation.
Enriched human iPSC-derived OPC cultures. (A) Representative images of OLIG2+ cells in iPSC-derived OPC cultures. Percentage of OLIG2+ out of all cells in control cultures (left) and InsG3680(+/−) patient cultures (right). (B) Representative images of PDGFRa+ cells in iPSC-derived OPC cultures. Percentage of PDGFRa+ cells out of all cells in control cultures (left) and InsG3680(+/−) patient cultures (right). (C) Representative traces of the signal generated upon glutamate stimulus in control (black) and InsG3680(+/−) patient–derived OPCs (pink). (D) A significantly reduced activity rate of calcium flux was found in InsG3680(+/−) patient–derived OPCs, as compared to controls. (E) A significantly higher amplitude of the first calcium event was measured in InsG3680(+/−) patient–derived OPCs, (F) with a significantly larger area under all curves, compared to the control. (G) No significant difference was found in the ratio between the sizes of peaks between the two groups. (H) A significant increase in interevent intervals was measured in InsG3680(+/−) patient–derived OPCs compared to controls. *P < 0.05, **P < 0.01, and ****P < 0.0001. Kolmogorov-Smirnov test (D to H) and one sample t test (D to H). Data are shown as means ± SEM. (D to F) n = 345 cells from control; n = 143 cells from InsG3680(+/−) patient OPCs. (G) n = 136 events from control; n = 31 events from InsG3680(+/−) patient OPCs. (H) n = 118 events from control; n = 33 events from InsG3680(+/−) patient OPCs. (D to H) n = 3, control; n = 3 InsG3680(+/−) patient OPCs (for one-sample t test).
SHANK3 restoration in InsG3680 mouse–derived OPCs results in increased expression of postsynaptic proteins
To examine whether SHANK3 restoration could rescue some of the deficits in OPCs, we transfected InsG3680 mouse–derived OPCs with a plasmid containing the human sequence of SHANK3 full-length isoform. Ninety-six hours after transfection, we examined the expression of SHANK3 (Fig. 9A) and the postsynaptic proteins Psd95 and NR1 in transfected OPCs compared to untransfected OPCs.
Fig. 9. Increased expression of Psd95 and NR1 following SHANK3 restoration in InsG3680 primary OPC cultures.
(A) Representative images of SHANK3 expression in transfected InsG3680 primary OPC culture. (B) Representative images of SHANK3 and Psd95 expression (left). (a) A magnified view of the dashed square shows colocalization between SHANK3 and Psd95, with the Pearson’s coefficient (r) indicating the correlation between the two and the Manders’ coefficient (M1) indicating the overlap of Psd95 with SHANK3. (b) A binary image showing colocalized regions of both proteins (white). Line scan graphs showing the immunofluorescence intensity along the arrow of SHANK3 and Psd95 (right). (C) A significant correlation was found between SHANK3 and Psd95 expression levels. (D) A significant increase in Psd95 expression levels in transfected cells in comparison to untransfected cells. (E) Representative image of SHANK3 and NR1 expression (left). Line scan graphs showing the immunofluorescence intensity along the arrow of SHANK3 and NR1 (right). (F) A significant correlation was found between SHANK3 and NR1 expression levels. (G) A significant increase in NR1 expression levels in transfected cells in comparison to untransfected cells. *P < 0.05. One-sample t test (D and G) and Spearman’s correlation test (C and F). Data are shown as means ± SEM. (C) n = 1378 pixels of SHANK3+ + Psd95+ image. (D) n = 16 nontransfected cells; n = 5 transfected cells. (F) n = 1315 pixels of SHANK3+ + NR1+ image. (G) n = 15 nontransfected cells; n = 3 transfected cells.
SHANK3 was prominently localized in both the soma and processes, and its restoration led to Psd95 expression in proximity to SHANK3 localization (Fig. 9B). However, Psd95 expression was not observed in several SHANK3-restored regions. Moreover, Psd95 expression levels increased in correlation with SHANK3 levels (Fig. 9C), leading to significant overall up-regulation of Psd95 expression in transfected OPCs compared to untransfected controls (Fig. 9D). Similar effects were observed for NR1 (Fig. 9, E to G). These findings indicate a causal relationship between SHANK3 and other postsynaptic proteins, suggesting that SHANK3 restoration is pivotal in reestablishing Psd95 within postsynaptic regions.
In summary, our findings reveal that the InsG3680 mutation in Shank3 profoundly disrupts the function of the postsynaptic region in OPCs and their response to glutamate. Moreover, it impairs the differentiation properties of these cells, resulting in deficits in OLs and compromised myelination properties. These physiological and behavioral deficits are evident as a consequence of the mutation. We validated these findings by studying human iPSC-derived OPCs extracted from an individual with a heterozygous InsG3680 mutation, which also displayed glutamate-related functional deficits in line with our observations. Clinically relevant, we found that the restoration of SHANK3 levels in InsG3680 OPC primary cultures leads to up-regulation in the expression of other postsynaptic proteins. This suggests that increasing SHANK3 expression may help ameliorate synaptic deficits by enhancing the overall postsynaptic structure, potentially providing therapeutic benefits for conditions associated with SHANK3 mutations.
DISCUSSION
Our study identified a previously unknown role of Shank3 as a postsynaptic protein in OLs. Furthermore, we found that the Shank3 human-based mutation presented by InsG3680 model mice leads to major deficits in WM and myelination properties. We examined the role of Shank3 in OLs by exploring the effect of the InsG3680 mutation on this cell population and found a marked reduction in postsynaptic-related mRNA and protein levels, morphological abnormalities, and major physiological deficits in response to glutamate stimulus. This was followed by in vivo findings showing how OLs and myelination are affected by other cell signals, in the whole organism. We found increased OL numbers, a major reduction in myelin-related gene transcript and protein levels, abnormal myelin ultrastructure, and myelin-related physiological and behavioral deficits. These results were further supported by the similar physiological deficits seen in iPSC-derived OPCs from a human patient with the same mutation in SHANK3 seen in InsG3680 mice. Last, the restoration of SHANK3 expression in InsG3680 OPCs resulted in an up-regulation in the expression of both the NMDAR subunit NR1 and the postsynaptic protein Psd95. This indicates that SHANK3 restoration can positively influence the synaptic protein environment in OPCs, potentially leading to improved synaptic function and connectivity. These data suggest that Shank3 is crucial for OL function and axon-OL synapses and that the InsG3680 mutation in mice, along with its human equivalent, may lead to abnormal OL function and significant myelination deficits.
While Shank3’s role is well-known in the PSD of neuronal synapses (11, 12, 18), its part in OLs remains unclear. Glutamate receptors are essential not only for neuronal communication but also for axon-OL signaling (27, 30, 31, 36), mediated by calcium and other ions entering OLs via these receptors (37, 41, 42). This influx was shown to affect OPC development and regulate myelination processes (35, 36, 38). As mentioned earlier, Shank3 is a part of the key complex used for binding glutamate receptors to the cytoskeleton and for maintaining proper synaptic transmission (11). Our findings suggest that Shank3 plays a similar role in OLs. We showed that not only is Shank3 present in OPCs and in myelin-containing fractions but also that major isoforms of Shank3 are not found in InsG3680 mice OLs. We have also observed distinct expressions of Shank3’s primary isoforms in both pure and crude myelin fractions. Notably, the crude myelin fraction includes axons as well. This indicates that specific isoforms may play varying roles in different cell types, underscoring the need for further exploration in future research. Moreover, the reduced expression levels of glutamate receptors and the postsynaptic protein Psd95, as well as the impaired calcium flux in response to a glutamate signal, expand our understanding of SHANK3-linked ASD and likely contribute to some of the in vivo myelin deficits we observed. In a prior study, researchers demonstrated a positive correlation between high-frequency and low-amplitude calcium activity and the elongation of myelin sheaths (72). While their investigation focused on mOLs and ours on OPCs, these findings reinforce our discovery of impaired calcium activity in InsG3680 OPCs compared to controls. Such alterations in the physiological function could potentially lead to deficits in proper differentiation and growth of OLs (73).
To explore a causal relationship between SHANK3 and other postsynaptic proteins in OLs and a potential future treatment, we engineered a plasmid containing the human sequence of SHANK3. Our results revealed that mouse primary cells recognized the human sequence and expressed the protein in both soma and processes. We also found that restoring SHANK3 in mutated OPCs resulted in increased levels of the glutamate NMDAR subunit NR1 and the postsynaptic protein Psd95, supporting our hypothesis of SHANK3’s important role as a postsynaptic protein in OPCs.
PSD95 is an essential protein for the structure of the PSD, interacting with SHANK3 through SAP90/PSD95-associated proteins to connect NMDARs to SHANK3, as previously shown in neurons (11). Therefore, the expression of Psd95 in the proximity of the restored SHANK3 protein in our study indicates a reconstruction of postsynaptic regions in SHANK3-restored OLs. Nonetheless, further investigation into the connections between Psd95 and Shank3 in OLs, as well as the overall state of the synapse, is needed for a more comprehensive understanding. It is also important to mention that the challenges associated with transfecting mouse OPCs, coupled with the large size of the SHANK3 gene plasmid, restricted our analysis to a small number of cells.
This study relied on the InsG3680 mouse model and iPSC-derived OPCs from a patient with the same mutation. We should note that in clinical conditions, the InsG3680 SHANK3 mutation is heterozygous, whereas, in the current study, we used homozygous mutant mice to get a clear understanding of the physiological role of the Shank3 gene and the underlying functional consequences of its disruption. Nevertheless, the findings from iPSC-derived OPCs bearing the InsG3680 SHANK3 mutation indicate that similar deficits may also occur in human OPCs due to this mutation.
We also studied OPCs derived from heterozygous InsG3680 mice to analyze the genetic condition analogous to that observed in humans. Our observations revealed only a minor phenotype characterized by impaired differentiation of OLs and no change in glutamate response. These outcomes might be attributable to the robust compensatory mechanisms present in mice, which are less pronounced in humans. Previous research has indicated that the heterozygous mutation in mice exhibits far less severity compared to humans, whereas the homozygous mutation in mice better mirrors the human disorder (12).
The variance between our in vivo and in vitro results in cellular properties suggests a potential autonomous impact of the InsG3680 mutation on OPCs. In vivo, the differentiation and proliferation of OPCs could be influenced by other cell types, primarily neurons. Conversely, our in vitro observations allow us to specifically delineate the mutation’s effects on this cell type. It is important to note that our in vivo experiments were conducted in specific brain regions, potentially not representing the entire OPC and OL population across the brain. In addition, we observed these effects in adult mice, unlike the developmental stage of pups in our in vitro experiments. Although we noted an increased number of OLs in certain areas of P30 InsG3680 mice, it appears that their maturation process is disrupted. This is evidenced by impaired histone modification and reduced surface area of mOLs in vitro, as well as decreased expression of key myelination proteins and defects in myelin ultrastructure in vivo.
The diminished myelination seen in InsG3680 mice in contrast to controls can primarily be ascribed to a decrease in axon numbers and fiber tract density. Nevertheless, additional observations propose that cellular changes within OLs and abnormalities in myelin properties play a major role in the pathology. Specifically, compromised OL differentiation, aberrant histone modifications, and morphological anomalies, as evidenced in vitro, imply intrinsic cellular defects, independent of neuronal influences.
In summary, this study unveiled an unknown role of Shank3 in OLs through the InsG3680 mutation. In addition, we identified myelin-related deficits in InsG3680 mice, providing crucial insights into the clinical condition in humans. As the mechanisms underlying ASD symptoms remain elusive, our findings hold promise for innovative treatment approaches targeting the gene mutation–linked ASD. Focusing on rescuing improper myelination and supporting OPC development may offer potential therapeutic avenues.
MATERIALS AND METHODS
Animal work statement
Breeding
Breeding was performed with InsG3680(+/−) C57 B6/S129 Sv mixed background mice. The newborn mice were either homozygous for the mutation (InsG3680(+/+)), heterozygous for the mutation (InsG3680(+/−)), or did not carry the mutation (WT). Only male mice were used for the in vivo experiments in this study, while mice from both sexes were used for in vitro experiments. The mutation occurs in all the cells of the body.
Housing
Mice of identical gender were housed in cages alongside two to four littermates of random genotypes. The housing environment maintained stable conditions with temperatures ranging from 20° to 24°C and a consistent 12-hour light/dark cycle (lights on from 07:00 to 19:00). Food and water were provided ad libitum throughout the study. All experimental protocols adhered to the guidelines set by the Institutional Animal Care and Use Committee of Tel Aviv University, Tel Aviv, Israel (2409-139-2). Every effort was made to reduce animal discomfort and distress while minimizing the number of animals used.
OPC-enriched culture preparation and differentiation to mOLs
OPC-enriched cultures were prepared from 0- to 3-day-old control or InsG3680 mouse pups. The cortex was isolated and minced with scissors and digested in a digestion solution [Hanks’ balanced salt solution (HBSS), Hepes, CaCl2, EDTA, deoxyribonuclease, cysteine, and papaine]. Digestion was stopped by adding mixed glia medium containing Dulbecco’s modified Eagle’s medium (DMEM) F-12 (Sigma-Aldrich, catalog no. D6421), glutamine supplement (Sigma-Aldrich, catalog no. G7513), 10% fetal bovine serum, and 1% penicillin-streptomycin (PS). The cell suspension was centrifuged for 5 min at 1000 rpm, and the pellet was mechanically dissociated 10 times in mixed glia medium using a glass Pasteur pipette. The dissociated cells were strained through a 100-μm cell strainer and centrifuged again for 5 min at 1000 rpm. Cells were seeded in mixed glia medium in T25 flasks with poly-d-lysine (PDL) coating and grown for 8 to 9 days at 37°C in a 5% CO2 atmosphere, with fresh medium exchange every 2 to 3 days. On the fifth day, insulin (5 μg/ml; Sigma-Aldrich, catalog no. I0516) was added to the medium.
Next, OPCs and microglia were separated by manually shaking for 15 times. To discard microglia, the cells were seeded onto uncoated petri dishes for 45 min. Medium with OPCs was collected from the petri dishes and centrifuged for 6 min at 1000 rpm. The cell suspension was diluted with OPC medium (DMEM F-12, 20% SM1, and 1% PS) and counted using a cell countess and methylene blue. Cells were seeded on PDL at a density of 104 cells/cm2 and grown for 3 days at 37°C in a 5% CO2 atmosphere. For differentiation of OPCs into mOLs, cells were grown for 8 days with the same medium. For RNA and protein extraction from isolated OPCs, these cells were taken immediately after isolation without seeding. Two samples were pooled together for protein extraction.
Immunocytochemistry
Enriched OPC cell cultures were fixed after 3 days postseeding for 10 min and then washed three times with phosphate-buffered saline (PBS). Differentiated mOLs were fixed after 8 days. After fixation, the cells were blocked with 2% normal goat serum (NGS) and 0.3% Triton X-100 and induced with primary antibodies for an hour. After three washes with PBS, secondary antibodies were added for an hour. Three washes were then performed on each sample. Last, for mounting onto glass slides, we used VECTASHIELD Hardset Antifade Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI; catalog no. H-1500-10, Vector Laboratories). Images were captured using a light microscope (IX-83, Olympus, Tokyo, Japan), with the experimenter blind to the genotype. For quantification of cellular properties, images were taken at ×10, ×20, or ×40 magnification, according to the type of staining and analysis. Cell number and intensity were quantified manually using the ImageJ program. Intensity was calculated by the mean of the gray value divided by the cell area. The sholl analysis method was used to evaluate the extent of branching of mOLs’ processes (74). This methodology involves quantifying the intersections of these processes with concentric circles of increasing radii centered around the cell soma, providing insights into their branching patterns and complexity. Commercial antibodies used were as follows: anti-Olig2 (diluted 1:1000; catalog no. AB9610, Sigma-Aldrich), anti-Pdgfrα (CD140a) (diluted 1:700; catalog no. 14-1401-82, Invitrogen), anti-Mbp (diluted 1:500; catalog no. MAB386, Sigma-Aldrich), anti-Shank3 (diluted 1:400; catalog no. 64555, Cell Signaling), anti-H3K9me3 (diluted 1:400; catalog no. ab8898, Abcam), and secondary antibodies Alexa Fluor 488 (diluted 1:1000; catalog nos. ab150077 and ab15016, Abcam) or Alexa Fluor 555 (diluted 1:1000; catalog nos. A32732 and A21434, Thermo Fisher Scientific).
Calcium imaging of OPCs
OPCs were incubated in a custom “well-chip” to ensure high-quality imaging and higher throughput. The well-chip, specifically designed to create a modular system for imaging (75), was created from a polydimethylsiloxane (PDMS) sheet prepared from Sylgard 184 (Sigma-Aldrich, 761028) mixed with a curing agent (1, 10), followed by curing at 60°C for at least 4 hours. The PDMS sheets were cut into rings and attached to a 13-mm cover glass (internal diameter, 8 mm) by mechanical pressure. The “well chips” were sterilized by ethanol washes and ultraviolet light for 30 min, followed by coating with PDL before OPC seeding.
OPCs were incubated for 30 min with 5 μM Fuo-4 AM calcium dye (Abcam ab241082-10-B) at 37°C in a 5% CO2 atmosphere. Excess dye was removed by washing with calcium buffer containing 20 mM Hepes (catalog no. 03-025-1B, Sartorius), 10 mM glucose (catalog no. G8769, Sigma-Aldrich), and 2 mM CaCl2 in HBSS (catalog no. 02-018-1A, Biological Industries). Calcium flux was followed using an Olympus IX83 confocal microscope, with images being acquired at 1.1-s intervals over 15 min, using an UCPLFLN20X Universal plan semi-apochromat objective, numerical aperture of 0.7. Glutamate (1 mM) was added using a pipette after 2 min of baseline signal acquisition (movie S1).
Calcium imaging data analysis of OPC primary cultures
Image analysis was performed with Fiji software to define ROIs, corresponding to the soma of an OPC. First, frames of each record were stacked using average intensity projection, and automated segmentation of ROIs was performed using a binary mask to minimize bias in ROI selection. Then, raw calcium fluorescence values were extracted from each ROI by averaging all data points of an ROI in one-time series. Using a designated MATLAB script, calcium traces were baseline-corrected with the average basal calcium fluorescence intensity before glutamate administration (F0). To analyze this, glutamate was added 120 s after the initiation of each recording session. The exact time of glutamate addition was verified using recorded video footage. During data analysis in MATLAB, we aligned all events to this specific moment of glutamate addition. This alignment enabled us to establish the baseline of each calcium trace using the calcium levels preceding the glutamate addition, and we performed subsequent analyses on the calcium trace following this event. To measure ΔF/F0 we used the following formula: ΔF/F0 = F(t) − F(0)/F(0) where F(t) is the fluorescence intensity in the ROI in which the calcium transient was observed at time t. Preprocessed calcium traces were classified into oscillatory and non-oscillatory signals to ensure that only ROIs showing calcium-related activity were retained. ROIs with calcium fluctuations (calcium events) were defined as those with at least one prominent local maximum that meets the threshold criteria detailed in fig. S8. For each oscillatory calcium trace, the rate of calcium activity was calculated as the number of events per minute. For each event, the amplitude was calculated as an average of three data points around the maxima of the event. The peak was measured as the vertical distance between the peak and its lowest contour line (the prominence of the peak), and a critical threshold was set at 30% of the peak. The onset of an event was detected when the signal crossed the critical threshold before reaching amplitude value, and the offset of the event was detected similarly after reaching the amplitude value. The event area was measured as the area under the curve bounded in time by the onset and offset of the event. Interevent intervals were set as the duration between the offset of an event and the onset of the following event.
Whole-cell patch clamp recording
Whole-cell patch clamp recordings were performed on mouse OPC cultures. Thirteen-millimeter cover glass were placed inside a recording chamber filled with Hepes-based artificial cerebrospinal fluid (aCSF) containing 152 mM NaCl, 10 mM Hepes, 2.8 mM KCl, 2 mM CaCl2, and 10 mM d-glucose [pH 7.3, adjusted using NaOH (1 M), osmolarity adjusted to 320 to 330 mOsm] at room temperature. The recording micropipettes (tip resistance of 10 to 15 megohm) were filled with an internal solution containing 134 mM k-gluconate, 2 mM MgCl2, 10 mM Na-Hepes, 10 mM Na-Pi creatine, 2 mM Mg2–adenosine triphosphate, and 0.3 mM Na3–guanosine triphosphate. The pH was maintained using KOH (1 M). To confirm the recording of sodium currents, we selectively blocked voltage-gated sodium channels pharmacologically with 1 μM TTX. Data were recorded at room temperature using Clampex v11.1 software, with a sampling rate of 20 kHz.
Analysis of sodium and fast and slow potassium currents
Current measurements for sodium and potassium were taken in voltage-clamp mode, recorded from the soma of the OPC, as previously described (76). Voltage steps of 400 ms in the −90- to 80-mV range were produced while holding the cells at a voltage of −60 mV. Cell capacitance was used to normalize currents. Cell capacitance was acquired through a membrane test of the Clampex SW software. The maximal outward current that occurs right after a depolarization step, usually within a time range of a few milliseconds, was used to calculate the fast potassium current. The slow potassium current was measured as the current at the end of the 400-ms depolarization phase. The sodium current was computed by subtracting the sodium current after stabilization from the lowest value of the inward sodium current. The fast and slow potassium currents were analyzed between 40 and 70 mV, while the sodium current was analyzed between −20 and 70 mV. In TTX experiments, we observed a notable reduction in the measured current, confirming that it is a sodium current.
Immunohistochemistry
Brain tissue extraction
Mice underwent deep anesthesia with isoflurane, with consciousness being assessed by their response to a foot pinch. Tissue samples were obtained from one ear for genetic verification. Subsequently, the mice were subjected to perfusion using 15 ml of ice-cold PBS solution, followed by 15 ml of ice-cold 4% paraformaldehyde (PFA). The brain was then extracted. The collected samples were immersed in 4% PFA for 24 hours and subsequently transferred to PBS solution.
Staining was done as previously described (70). Briefly, brains were extracted and sectioned into 100-μm-thick slices using a vibratome. Sections from the motor and frontal cortices were stained using the free-floating method. After washing and permeabilization, the sections were blocked and incubated overnight with primary antibodies. Following additional washes, the sections were incubated with secondary antibodies. Last, the sections were mounted onto glass slides using a mounting medium. Images were captured at a ×10 magnification using a light microscope (IX-83, Olympus), and cellular properties in the motor cortex, frontal cortex, and striatum were quantified manually using ImageJ software. Commercial antibodies used were as follows: anti–adenomatous polyposis coli (APC) (Ab-7, CC1) (diluted 1:500; catalog no. OP80, Calbiochem), anti-Olig2 (diluted 1:1000; catalog no. AB9610, Sigma-Aldrich), and secondary antibody Alexa Fluor 488 or 555 (diluted 1:1000; catalog nos. ab150077 and ab150118, Abcam).
RNA extraction, cDNA preparation, and RT-PCR
These procedures were performed as previously described (70).
Brain tissue extraction
The mice were euthanized by cervical dislocation, and the frontal cortex and striatum were collected for analysis into RNA-later (catalog no. AB-AM7020, Thermo Fisher Scientific) for 24 hours and then frozen in −80°C.
RNA extraction
Samples were homogenized in safe-lock tubes with stainless steel beads and cold TRIzol reagent. After incubation and addition of chloroform, the samples were centrifuged to separate the RNA layer. The RNA was then mixed with isopropanol, centrifuged, washed with ethanol, and dried. Diethyl pyrocarbonate–treated water was added, followed by heating and pipetting to attain homogeneity. RNA concentration was measured using a NanoDrop One device. Samples were stored at −80°C.
cDNA preparation
Extracted RNA was diluted to a concentration of 20 ng/μl based on the original concentration of a sample. Reverse transcription was carried out using random primers and the High-Capacity cDNA Reverse Transcription Kit from Thermo Fisher Scientific. The following protocol was used with the C1000 Touch thermal cycler (Bio-Rad): 10 min at 25°C, 120 min at 37°C, 5 min at 85°C, and a final step at 4°C until storage. The resulting cDNA was then stored at −20°C.
Real-time polymerase chain reaction
mRNA levels were measured by RT-PCR with Fast SYBR Green PCR Master Mix (Thermo Fisher Scientific) and the CFX Connect Real-Time PCR Detection System (Bio-Rad). The protocol included an initial denaturation step, 40 amplification cycles, and melt curve analysis. mRNA levels were determined using the comparative cycle threshold (Ct) method. Glyceraldehyde-3-phosphate dehydrogenase mRNA was used for normalization. Primers were custom-designed and obtained from Hy Laboratories (Rehovot, Israel). Detailed primer sequences are provided in table S1.
Myelin isolation
Myelin was isolated as previously described (64), with slight adjustments. Mice were euthanized by cervical dislocation, and the frontal cortex, striatum, or whole brain was dissected and rapidly frozen with liquid nitrogen before storing at −80°C. For the frontal cortex and striatum, samples from two mice with the same genotype were pooled in a single tube. Next, the tissue was homogenized in a 0.32 M sucrose solution containing a protease inhibitor (catalog no. 539131, Sigma-Aldrich). The homogenized samples were layered onto 0.85 M sucrose solution and subjected to ultracentrifugation using an Optima XPN80 ultracentrifuge with an SW41-Ti rotor (Beckman Coulter). This process resulted in the crude myelin fraction being visible at the interphase of the two sucrose concentrations. To purify the myelin fraction, two washing and osmotic shock steps with filtered double-distilled water were performed, followed by centrifugation on a second sucrose step gradient (0.85 M at the bottom and 0.32 M at the top) and another washing step, while for the crude myelin fraction, washes were performed and the pellet was collected.
Western blot
Samples were homogenized in solubilization buffer [50 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA (pH 8), 1 mM EGTA (pH 8), 1.5 mM MgCl2, and 200 μM Na3VO4], with protease inhibitor cocktail 1 diluted 1:100 (Merck). Protein levels were determined using Quick Start Bradford 1X Dye reagent (catalog no. 5000205, Bio-Rad). Equal amounts of protein were separated by 4 to 20% SDS–polyacrylamide gel electrophoresis. The proteins were then transferred onto a nitrocellulose membrane using a transfer buffer composed of 25 mM tris-HCl (pH 8.5), 190 mM glycine, and 20% methanol. Following transfer, the membranes were blocked with 6% skimmed milk in TBST buffer [0.05 M tris-HCl (pH 7.5), 0.15 M NaCl, and 0.1% Tween 20] for 45 min at room temperature. After three consecutive washes with TBST, the membranes were incubated overnight at 4°C with primary antibodies, with gentle shaking. The next day, the membranes were washed three times for 15 min each in TBST and subsequently incubated with horseradish peroxidase–conjugated secondary antibodies for 45 min at room temperature, with gentle shaking. Following another three washes in TBST, the immunoreactive bands were visualized using the SuperSignal West Pico PLUS Chemiluminescent Substrate (catalog no. 34577, Thermo Fisher Scientific). Images were captured under specific exposure parameters for each antibody and protein load. Antibody-labeled/housekeeping protein ratios were determined using ImageJ software by an experimenter blind to the origin of the samples.
Commercial antibodies used were as follows: anti-Mbp (diluted 1:1000; catalog no. MAB386, Merck), anti-tubulin (diluted 1:5000; catalog no. AB108342, Abcam), anti-tubulin β4 (diluted 1:1000; catalog no. AB179509, Abcam), anti-Plp1 (diluted 1:12000; catalog no. AB28486, Abcam), anti-Shank3 (diluted 1:1000; catalog no. 64555, Cell Signaling), anti-Psd95 (diluted 1:1000; catalog no. CST-3450S, Cell Signaling), and anti-NR1 (diluted 1:1000; catalog no. AGC-001, Alomone Labs). Secondary antibodies included goat anti-rabbit (diluted 1:10,000; catalog no. AP132P, Merck) and goat anti-rat (diluted 1:10,000; catalog no. AP136P, Merck) antibodies.
Electron microscopy
Mice were transcardially perfused with a modified fixation solution containing 2% PFA, 2.5% glutaraldehyde, and 0.1 M sodium cacodylate buffer (pH 7.4). Brain tissue from the dorsolateral striatum between the fornix and the CC was dissected into ~1-mm cubes. The samples were incubated overnight in the fixation solution, washed, and transferred into 0.1 M sodium cacodylate buffer for storage at 4°C for 2 days at the most. Ultrathin sections were prepared by the EM unit at Tel Aviv University. Images were captured using a transmission electron microscope (JEM-1400Plus) at various magnifications. Approximately 50 to 200 axons per mouse were analyzed for g-ratio quantification. The g-ratio values were determined by measuring the ratio of axon diameter divided by the myelinated axon diameter, with the measurements being performed by an experimenter blind to the genotype. For ultrastructure abnormality analysis, ImageJ was used to automatically mark the area of myelin and the area of damaged myelin (size of particles: 0.0003 to 0.3 μm).
MRI-DTI scanning and analysis
A 7 T MRI scanner (Bruker, Billerica, MA) with a 30-cm bore and gradient strength up to 400 mT/m was used for MRI scanning. The diffusion imaging protocol involved acquiring 18 slices, each with a thickness of 0.6 mm, at a resolution of 0.175 mm × 0.175 mm (matrix size, 128 × 96). A diffusion-weighted spin-echo echo-planar imaging pulse sequence was used, with a repetition time of 3000 ms, an echo time of 25 ms, and Δ/δ values of 10/2.5 ms. The protocol included four echo-planar imaging segments, 15 noncollinear gradient directions, and a single b value shell at 1000 s/mm2, along with three b0 images. Analysis of the diffusion MRI data was carried out using ExploreDTI. This encompassed motion and distortion correction, transformation into atlas space, whole-brain tractography, and quantification of the reconstructed number of tracts and FA values for subsequent statistical analysis between groups.
Electrophysiological recordings from brain slices
Acute brain slice preparation
Acute brain slices were prepared as previously described (77) from four control and four InsG3680 male mice at 6 months of age. Briefly, mice were anesthetized with avertine (tribromoethanol; 20 mg/ml) via intraperitoneal injection at a dosage of 0.5 mg/g body weight. Mice were then intracardially perfused with a modified N-methyl-d-glucamine aCSF cutting solution (NMDG-aCSF) (78) [30 mM NaHCO3, 25 mM glucose, 20 mM Hepes, 18 mM NMDG, 10 mM MgSO4, 2.5 mM KCl, 5 mM Na-ascorbate, 2 mM thiourea, 3 mM Na-pyruvate, 1.2 mM NaH2PO4, 0.5 mM CaCl2 (all reagents from Sigma-Aldrich)], 300 mOsm (pH 7.3 to 7.4), saturated with carbogen (95% O2 and 5% CO2). Coronal brain slices (400-μm thick) were prepared in carbogenated NMDG-aCSF, using a vibratome (Leica, VS1000) at a speed of 0.10 mm/s, a frequency of 100 Hz, and an amplitude of 0.6 mm. Slices containing the CC (0.8 to −0.8 mm from bregma) were collected and allowed to recover at 34°C for 10 min in carbogenated NMDG-aCSF. Slices were then transferred into aCSF saturated with carbogen (95% O2 and 5% CO2) at room temperature and allowed to recover for 1 hour before recordings. aCSF composition was as follows: 119 mM NaCl, 24 mM NaHCO3, 12.5 mM glucose, 2.5 mM KCl, 2.0 mM CaCl2, 2.0 mM MgSO4, and 1.2 mM NaH2PO4 (all reagents from Sigma-Aldrich), 300 mOsm (pH 7.3 to 7.4).
CAP recordings
Three slices per animal were used to record CAP as previously described (79). Briefly, each slice was individually placed in a recording chamber continuously perfused (3 ml/min) with carbogenated aCSF at room temperature and visualized using an Olympus BX51WI microscope. Afferents were stimulated with a 0.1-ms stimulation step (ISO-Flex, A.M.P.I.), delivered at 0.05-Hz frequency using a platinum iridium concentric bipolar electrode (FHC, 30213) placed in the CC (0.5 mm from the midline). CAP responses were recorded using borosilicate-glass capillaries (GB150F-8P, Science Products), pulled on a P1000 horizontal puller (Sutter Instruments), backfilled with 3 M NaCl solution, and placed in the CC of the contralateral hemisphere at a distance of approximately 1.0 mm from the stimulating electrode (0.5 mm from the midline). Optimal signal was usually obtained at ~150- to 200-μm depth from the slice surface. After placing the electrodes and finding optimal signals, the electrodes were left in place 5 min to accommodate before recordings. Triplicate measurements were obtained for all CAP recordings and averaged to determine peak amplitude and latency. Signals were amplified using a Multiclamp 700B (Axon Instruments), acquired at a sampling rate of 10 kHz with Digidata 1440A (Axon Instruments), and analyzed offline with Clampfit 10.2 software (Axon Instruments/Molecular Devices).
Behavioral tests
Behavioral tests were conducted with the experimenter blinded to genotype. Before the tests, mice underwent a 1-hour habituation period in the test room. Each group of mice participated in two behavioral tests, with at least a 3-day interval between tests. The mice were tested at 3 months of age.
GSM test
The grip strength of mice was evaluated using a grip strength meter (TSE Systems) following the manufacturer’s guidelines. Three consecutive trials were conducted for each mouse to obtain the average force (in grams).
Rotarod test
The mice were positioned on the apparatus of an accelerating rotarod to assess motor coordination, and their latency to fall off the apparatus was recorded. Each mouse underwent three trials, with a 30-min gap between each test.
Differentiation of iPSCs into OPCs
Human iPSCs generated from a SHANK3 mutant patient and a healthy control were grown until ~80% confluence (71). Using a published protocol (80), the iPSCs were differentiated into OPCs. To generate embryoid bodies (EBs), iPSCs were grown to confluency and dissociated using Accutase (STEMCELL Technologies) at 37°C for 5 min. Once the colonies were between 100 and 250 μM in diameter, EBs were cultured in neural induction medium [mTeSR custom medium containing 10 μM SB431542, 250 nM LDN193189, and 100 nM retinoic acid (RA)] added freshly on the day of use for 8 days. Thereafter, the media was replaced by N2 medium [DMEM/F-12 with GlutaMAX, 1% antibiotic-antimycotic, 0.1 mM 2-mercaptoethanol, 1% nonessential amino acids (NEAA), N2 supplement, 100 nM all-trans-RA, and 1 μM smoothened agonist (SAG)] for 4 days.
On day 12, cells were mechanically dissociated to prepare EBs which were grown in N2B27 medium [DMEM/F-12 with GlutaMAX, 1% antibiotic-antimycotic, 0.1 mM 2-mercaptoethanol, 1% NEAA, N2 supplement, 100 nM RA, 1 μM SAG, B27 supplement, and insulin (25 μg/ml)] for 9 days. On day 20, two-thirds of the N2B27 medium was replaced with PDGF medium [DMEM/F-12 with GlutaMAX, 1% antibiotic-antimycotic, 0.1 mM 2-mercaptoethanol, 1% NEAA, 100× N2 supplement, 50× B27 supplement, insulin (25 μg/ml), PDGFaa (10 ng/ml), insulin-like growth factor 1 (10 ng/ml), hepatocyte growth factor (5 ng/ml), NT3 (10 ng/ml), T3 (60 ng/ml), biotin (100 ng/ml), and 1 μM cyclic adenosine 3′,5′-monophosphate (cAMP)] for 10 days.
On day 30, EBs that are round and golden brown in the center with a diameter of 300 to 800 μm were picked and plated onto laminin/poly-ornithine–coated six-well plates. The adherent cells were fed with two-thirds of PDGF medium every other day until day 50 postdifferentiation. After day 50, iPSCs were transitioned into OPCs using glial medium [DMEM/F-12 with GlutaMAX, 1% antibiotic-antimycotic, 0.1 mM 2-mercaptoethanol, 1% NEAA, N2 supplement, B27 supplement, insulin (25 μg/ml), T3 (60 ng/ml), biotin (100 ng/ml), 1 μM cAMP, 10 mM Hepes, and ascorbic acid (20 μg/ml)]. The use of these iPSCs was approved by the Institutional Ethics Committee (223/23), and written informed consent was obtained from the patient’s guardians.
Calcium imaging experiments and analyses from iPSC-derived OPCs
Coverslips containing OPC cultures derived from iPSCs of a patient with a SHANK3 mutation and a healthy control 8 weeks following differentiation were transferred into a 24-well plate before the recording experiments. Coverslips were incubated for 45 min with 5 μM Fluo-4 AM calcium dye (Abcam, ab241082-10-B) at 37°C. Excess dye was removed by washing with calcium buffer containing 20 mM Hepes (catalog no. 03-025-1B, Sartorius), 10 mM glucose (catalog no. G8769, Sigma-Aldrich), and 2 mM CaCl2 in HBSS (catalog no. 02-018-1A, Biological Industries). Image acquisition was performed using a Leica THUNDER imager, and the obtained images were subsequently processed and analyzed as described above for primary OPC cultures.
Immunocytochemistry of iPSC-derived OPCs
Cells were fixed in 4% PFA for 15 min, followed by three washes of PBS. Then, cells were blocked and permeabilized in 2% NGS and 0.3% Triton X-100 for 1 hour. The coverslips were incubated with primary antibodies in the blocking solution overnight at 4°C. On the next day, they were washed in PBS and incubated with the corresponding secondary antibodies for 60 min at room temperature. Then, coverslips were washed three times, mounted on glass slides using VECTASHIELD Hardset Antifade Mounting Medium with DAPI (catalog no. H-1500-10, Vector Laboratories), and dried overnight while protected from light. Fluorescence signals were detected using a Leica TCS SP5 Olympus confocal microscope and a light microscope (IX-83, Olympus, Tokyo, Japan), with the experimenter blind to the genotype. For quantification of culture purity, images were taken at ×20 or ×40 magnification. Cell numbers were quantified manually using the ImageJ program. Commercial antibodies used were as follows: anti-Olig2 (diluted 1:1000; catalog no. AB9610, Sigma-Aldrich), anti-Pdgfrα (CD140a) (diluted 1:700; catalog no. 14-1401-82, Invitrogen), anti-NeuN (diluted 1:1000; catalog no. MAB377, Sigma-Aldrich), and secondary antibody Alexa Fluor 555 (diluted 1:1000; catalog nos. A32732 and A21434, Thermo Fisher Scientific) or Alexa Fluor 647 (diluted 1:1000; catalog no. A32728, Thermo Fisher Scientific).
Transfection of primary OPCs
Twenty-four hours after seeding, primary OPCs were transfected with a plasmid containing the SHANK3 main isoform human sequence (2 μg of DNA) using calcium phosphate transfection. A full media change was performed 24 hours after transfection. The cells were fixed 96 hours posttransfection to allow sufficient time for gene expression. Fluorescence signals were detected using a Leica TCS SP5 Olympus confocal microscope and a light microscope (IX-83, Olympus, Tokyo, Japan). Protein intensity was quantified manually using the ImageJ program. The intensity was calculated by the mean of gray value divided by the cell area, in transfected cells in comparison to untransfected cells in the field of view of the same image. Pearson’s coefficient and Manders’ coefficient were calculated using JaCoP plugin in ImageJ program. Commercial antibodies used were as follows: anti-Pdgfrα (CD140a) (diluted 1:700; catalog no. 14-1401-82, Invitrogen), anti-SHANK3 (diluted 1:400; catalog no. 64555, Cell Signaling), anti-Psd95 (diluted 1:1000; catalog no. ab192757, Abcam), anti-NR1 (diluted 1:100; catalog no. ab134308, Abcam), and secondary antibody Alexa Fluor 555 (diluted 1:1000; catalog nos. A32732 and A21434, Thermo Fisher Scientific) or Alexa Fluor 647 (diluted 1:1000; catalog no. A32728, Thermo Fisher Scientific) or Alexa Fluor 405 (diluted 1:500; catalog no. ab175671, Abcam).
Statistical analysis
Data are presented as the mean ± SEM, as calculated by GraphPad Prism 9.4.1 for Windows (GraphPad Software, San Diego, CA). P values were calculated using Student’s t test, one sample t test, Kolmogorov-Smirnov, Mann-Whitney test, linear regression, repeated measures analysis of variance (ANOVA), and Spearman’s correlation test, with P < 0.05 being considered significant (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). The normality of distributions and equality of variances were checked and addressed accordingly using the appropriate statistical analysis. Outliers were determined via the extreme studentized deviate method (*P < 0.05).
Acknowledgments
We gratefully acknowledge the patient with the genetic condition for her contribution of the iPSCs, past and current members of the Barak laboratory, and E. Peles’ laboratory for technical support with the OPC primary culture protocol. S.St. is supported by the Zuckerman STEM leadership program.
Funding: This work was supported by grants awarded to B.B. from Fritz Thyssen Stiftung, Israel Science Foundation (grant no. 2305/20), Federation of European Biochemical Societies (FEBS), and the National Institute of Psychobiology in Israel. B.B. was the recipient of The Alon Fellowship for Outstanding Young Researchers awarded by the Israeli Council for Higher Education.
Author contributions: I.F. and B.B. conceptualized the study, designed the experiments, and wrote the manuscript. I.F. and B.B. also led the design, collection, analysis, and interpretation across in vitro OL cultures, in vivo assays, iPSC-derived OPC experiments, and transfection studies, including the SHANK3 plasmid design. S.Sh. was involved in the conceptualization, design, writing, collection, analysis, and interpretation of calcium imaging in OPC primary cultures and iPSC-derived OPCs, as well as RT-PCR, Western blot, and myelin isolation in both in vitro and in vivo experiments. Y.L.-B. was involved in the conceptualization, writing, design, collection, and analysis of calcium imaging from OPC primary cultures, supported by design and analysis by A.So. R.N. contributed to the design and collection of iPSC-derived OPC differentiation and calcium imaging experiments. G.W. handled the design and analysis of OL primary cultures, in vivo studies, and analysis of immunofluorescence of iPSC-derived OPC cultures. I.R. led patch clamp studies, supported by data collection by A.Sh. and data collection and analysis by U.T. M.R. managed data collection and analysis of in vivo immunofluorescence, myelin isolation, and behavioral tests. A.C.C. and P.M. were involved in electrophysiological studies of the CC, involving conceptualization, design, data collection, analysis, interpretation, and writing methods and results. G.C. and A.Sc. designed and cloned SHANK3 plasmid for transfection experiments. Y.A. designed and interpreted MRI-DTI experiments. G.E.-S. and S.S.-T. conceptualized, designed, and collected data and interpreted in vitro OPC primary cultures, in vivo Western blot, myelin isolation, with G.E.-S. also leading transfection experiments. E.B. designed, collected, analyzed, and interpreted in vitro OL primary culture molecular properties. Y.H. collected, analyzed, and interpreted basal activity in iPSC-derived OPC experiments and wrote the methods for iPSC-derived OPCs. S.St. conceptualized, designed, collected, analyzed, interpreted, and wrote the results and methods for patch clamp and iPSC-derived OPC experiments. B.M.M. conceptualized, designed, collected, analyzed, interpreted, and wrote the methods for calcium imaging experiments from OPC primary cultures. S.Sh., Y.L.-B., A.So., and B.M.M. were involved in writing software for calcium imaging analysis, with S.Sh. leading this aspect. I.F., Y.A., and B.B. handled software development for DTI analysis. Y.H. and S.St. wrote software for basal activity in calcium imaging analysis, with S.St. also responsible for patch clamp analysis software. All authors have read and agreed to the published version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
The PDF file includes:
Tables S1 and S2
Figs. S1 to S7
Uncropped Western blots
Legend for movie S1
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Barak B., Feng G., Neurobiology of social behavior abnormalities in autism and Williams syndrome. Nat. Neurosci. 19, 647–655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral D. G., Schumann C. M., Nordahl C. W., Neuroanatomy of autism. Trends Neurosci. 31, 137–145 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Lord C., Cook E. H., Leventhal B. L., Amaral D. G., Autism spectrum disorders. Neuron 28, 355–363 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Huguet G., Ey E., Bourgeron T., The genetic landscapes of autism spectrum disorders. Annu. Rev. Genom. Hum. Genet. 14, 191–213 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Satterstrom F. K., Kosmicki J. A., Wang J., Breen M. S., De Rubeis S., An J. Y., Peng M., Collins R., Grove J., Klei L., Stevens C., Reichert J., Mulhern M. S., Artomov M., Gerges S., Sheppard B., Xu X., Bhaduri A., Norman U., Brand H., Schwartz G., Nguyen R., Guerrero E. E., Dias C., Betancur C., Cook E. H., Gallagher L., Gill M., Sutcliffe J. S., Thurm A., Zwick M. E., Børglum A. D., State M. W., Cicek A. E., Talkowski M. E., Cutler D. J., Devlin B., Sanders S. J., Roeder K., Daly M. J., Buxbaum J. D., Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willsey H. R., Willsey A. J., Wang B., State M. W., Genomics, convergent neuroscience and progress in understanding autism spectrum disorder. Nat. Rev. Neurosci. 23, 323–341 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willsey A. J., State M. W., Autism spectrum disorders: From genes to neurobiology. Curr. Opin. Neurobiol. 30, 92–99 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan B. N., Bohlen J. F., Davis B. A., Ye Z., Chen H.-Y., Mayfield B., Sripathy S. R., Cerceo Page S., Campbell M. N., Smith H. L., Gallop D., Kim H., Thaxton C. L., Simon J. M., Burke E. E., Shin J. H., Kennedy A. J., Sweatt J. D., Philpot B. D., Jaffe A. E., Maher B. J., A myelin-related transcriptomic profile is shared by Pitt–Hopkins syndrome models and human autism spectrum disorder. Nat. Neurosci. 23, 375–385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rubeis S., Buxbaum J. D., Genetics and genomics of autism spectrum disorder: Embracing complexity. Hum. Mol. Genet. 24, R24–R31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta A. R., State M. W., Recent advances in the genetics of autism. Biol. Psychiatry 61, 429–437 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Monteiro P., Feng G., SHANK proteins: Roles at the synapse and in autism spectrum disorder. Nat. Rev. Neurosci. 18, 147–157 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y., Kaiser T., Monteiro P., Zhang X., Van der Goes M. S., Wang D., Barak B., Zeng M., Li C., Lu C., Wells M., Amaya A., Nguyen S., Lewis M., Sanjana N., Zhou Y., Zhang M., Zhang F., Fu Z., Feng G., Mice with Shank3 mutations associated with ASD and schizophrenia display both shared and distinct defects. Neuron 89, 147–162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand C. M., Betancur C., Boeckers T. M., Bockmann J., Chaste P., Fauchereau F., Nygren G., Rastam M., Gillberg I. C., Anckarsäter H., Sponheim E., Goubran-Botros H., Delorme R., Chabane N., Mouren-Simeoni M.-C., de Mas P., Bieth E., Rogé B., Héron D., Burglen L., Gillberg C., Leboyer M., Bourgeron T., Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 39, 25–27 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harony-Nicolas H., De Rubeis S., Kolevzon A., Buxbaum J. D., Phelan McDermid syndrome: From genetic discoveries to animal models and treatment. J. Child Neurol. 30, 1861–1870 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verpelli C., Montani C., Vicidomini C., Heise C., Sala C., Mutations of the synapse genes and intellectual disability syndromes. Eur. J. Pharmacol. 719, 112–116 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Buxbaum J. D., Multiple rare variants in the etiology of autism spectrum disorders. Dialogues Clin. Neurosci. 11, 35–43 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Rubeis S., He X., Goldberg A. P., Poultney C. S., Samocha K., Cicek A. E., Kou Y., Liu L., Fromer M., Walker S., Singh T., Klei L., Kosmicki J., Fu S.-C., Aleksic B., Biscaldi M., Bolton P. F., Brownfeld J. M., Cai J., Campbell N. G., Carracedo A., Chahrour M. H., Chiocchetti A. G., Coon H., Crawford E. L., Crooks L., Curran S. R., Dawson G., Duketis E., Fernandez B. A., Gallagher L., Geller E., Guter S. J., Hill R. S., Ionita-Laza I., Gonzalez P. J., Kilpinen H., Klauck S. M., Kolevzon A., Lee I., Lei J., Lehtimäki T., Lin C.-F., Ma’ayan A., Marshall C. R., McInnes A. L., Neale B., Owen M. J., Ozaki N., Parellada M., Parr J. R., Purcell S., Puura K., Rajagopalan D., Rehnström K., Reichenberg A., Sabo A., Sachse M., Sanders S. J., Schafer C., Schulte-Rüther M., Skuse D., Stevens C., Szatmari P., Tammimies K., Valladares O., Voran A., Wang L.-S., Weiss L. A., Willsey A. J., Yu T. W., Yuen R. K. C.; The DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium; The Autism Sequencing Consortium, Cook E. H., Freitag C. M., Gill M., Hultman C. M., Lehner T., Palotie A., Schellenberg G. D., Sklar P., State M. W., Sutcliffe J. S., Walsh C. A., Scherer S. W., Zwick M. E., Barrett J. C., Cutler D. J., Roeder K., Devlin B., Daly M. J., Buxbaum J. D., Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sala C., Piëch V., Wilson N. R., Passafaro M., Liu G., Sheng M., Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31, 115–130 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Sala C., Segal M., Dendritic spines: The locus of structural and functional plasticity. Physiol. Rev. 94, 141–188 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Gauthier J., Champagne N., Lafrenière R. G., Xiong L., Spiegelman D., Brustein E., Lapointe M., Peng H., Côté M., Noreau A., Hamdan F. F., Addington A. M., Rapoport J. L., DeLisi L. E., Krebs M.-O., Joober R., Fathalli F., Mouaffak F., Haghighi A. P., Néri C., Dubé M.-P., Samuels M. E., Marineau C., Stone E. A., Awadalla P., Barker P. A., Carbonetto S., Drapeau P., Rouleau G. A., Team T. S. D., Daignault K., Diallo O., Duguay J., Drits M., Henrion E., Jolivet P., Kuku F., Lachapelle K., Laliberté G., Laurent S., Liao M., Marino C., Piton A., Raymond A., Reynolds A., Rochefort D., St-Onge J., Thibodeau P., Tsurudome K., Yang Y., Leroy S., Ossian K., Chayet M., Gourion D., De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 107, 7863–7868 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi F., Danko T., Botelho S. C., Patzke C., Pak C., Wernig M., Südhof T. C., Autism-associated SHANK3 haploinsufficiency causes channelopathy in human neurons. Science 352, aaf2669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kathuria A., Nowosiad P., Jagasia R., Aigner S., Taylor R. D., Andreae L. C., Gatford N. J. F., Lucchesi W., Srivastava D. P., Price J., Stem cell-derived neurons from autistic individuals with SHANK3 mutation show morphogenetic abnormalities during early development. Mol. Psychiatry 23, 735–746 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussein Y., Tripathi U., Choudhary A., Nayak R., Peles D., Rosh I., Rabinski T., Djamus J., Vatine G. D., Spiegel R., Garin-Shkolnik T., Stern S., Early maturation and hyperexcitability is a shared phenotype of cortical neurons derived from different ASD-associated mutations. Transl. Psychiatry 13, 246 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulanger J. J., Messier C., From precursors to myelinating oligodendrocytes: Contribution of intrinsic and extrinsic factors to white matter plasticity in the adult brain. Neuroscience 269, 343–366 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Nishiyama A., Komitova M., Suzuki R., Zhu X., Polydendrocytes (NG2 cells): Multifunctional cells with lineage plasticity. Nat. Rev. Neurosci. 10, 9–22 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Bozzali M., Wrabetz L., Axonal signals and oligodendrocyte differentiation. Neurochem. Res. 29, 979–988 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Osso L. A., Chan J. R., Architecting the myelin landscape. Curr. Opin. Neurobiol. 47, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Shen X., Zhang Y., Zheng X., Cepeda C., Wang Y., Duan S., Tong X., Interactions of glial cells with neuronal synapses, from astrocytes to microglia and oligodendrocyte lineage cells. Glia 71, 1383–1401 (2023). [DOI] [PubMed] [Google Scholar]
- 29.Moura D., Brennan E. J., Brock R., Cocas L. A., Neuron to oligodendrocyte precursor cell synapses: Protagonists in oligodendrocyte development and myelination, and targets for therapeutics. Front. Neurosci. 15, 779125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fannon J., Tarmier W., Fulton D., Neuronal activity and AMPA-type glutamate receptor activation regulates the morphological development of oligodendrocyte precursor cells. Glia 63, 1021–1035 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Spitzer S., Volbracht K., Lundgaard I., Káradóttir R. T., Glutamate signalling: A multifaceted modulator of oligodendrocyte lineage cells in health and disease. Neuropharmacology 110, 574–585 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Hughes A. N., Appel B., Oligodendrocytes express synaptic proteins that modulate myelin sheath formation. Nat. Commun. 10, 4125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wake H., Lee P. R., Fields R. D., Control of local protein synthesis and initial events in myelination by action potentials. Science 333, 1647–1651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Miramontes T. G., Czopka T., Monk K. R., Synaptic input and Ca2+ activity in zebrafish oligodendrocyte precursor cells contribute to myelin sheath formation. Nat. Neurosci. 27, 219–231 (2024). [DOI] [PubMed] [Google Scholar]
- 35.Cavaliere F., Urra O., Alberdi E., Matute C., Oligodendrocyte differentiation from adult multipotent stem cells is modulated by glutamate. Cell Death Dis. 3, e268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergles D. E., Roberts J. D. B., Somogyi P., Jahr C. E., Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature 405, 187–191 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Káradóttir R., Hamilton N. B., Bakiri Y., Attwell D., Spiking and nonspiking classes of oligodendrocyte precursor glia in CNS white matter. Nat. Neurosci. 11, 450–456 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pease-Raissi S. E., Chan J. R., Building a (w)rapport between neurons and oligodendroglia: Reciprocal interactions underlying adaptive myelination. Neuron 109, 1258–1273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen T. J., Kula B., Nagy B., Barzan R., Gall A., Ehrlich I., Kukley M., In vivo regulation of oligodendrocyte precursor cell proliferation and differentiation by the AMPA-receptor subunit GluA2. Cell Rep. 25, 852–861.e57 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Li C., Xiao L., Liu X., Yang W., Shen W., Hu C., Yang G., He C., A functional role of NMDA receptor in regulating the differentiation of oligodendrocyte precursor cells and remyelination. Glia 61, 732–749 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Butt A. M., Neurotransmitter-mediated calcium signalling in oligodendrocyte physiology and pathology. Glia 54, 666–675 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Soliven B., Calcium signalling in cells of oligodendroglial lineage. Microsc. Res. Tech. 52, 672–679 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Gautier H. O. B., Evans K. A., Volbracht K., James R., Sitnikov S., Lundgaard I., James F., Lao-Peregrin C., Reynolds R., Franklin R. J. M., Káradóttir R. T., Neuronal activity regulates remyelination via glutamate signalling to oligodendrocyte progenitors. Nat. Commun. 6, 8518 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q., Brus-Ramer M., Martin J. H., McDonald J. W., Electrical stimulation of the medullary pyramid promotes proliferation and differentiation of oligodendrocyte progenitor cells in the corticospinal tract of the adult rat. Neurosci. Lett. 479, 128–133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wake H., Ortiz F. C., Woo D. H., Lee P. R., Angulo M. C., Fields R. D., Nonsynaptic junctions on myelinating glia promote preferential myelination of electrically active axons. Nat. Commun. 6, 7844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marisca R., Hoche T., Agirre E., Hoodless L. J., Barkey W., Auer F., Castelo-Branco G., Czopka T., Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat. Neurosci. 23, 363–374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakurela S., Garding A., Jung R. B., Müller C., Goebbels S., White R., Werner H. B., Tiwari V. K., The transcriptome of mouse central nervous system myelin. Sci. Rep. 6, 25828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malara M., Lutz A.-K., Incearap B., Bauer H. F., Cursano S., Volbracht K., Lerner J. J., Pandey R., Delling J. P., Ioannidis V., SHANK3 deficiency leads to myelin defects in the central and peripheral nervous system. Cell. Mol. Life Sci. 79, 371 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saab A. S., Tzvetanova I. D., Nave K.-A., The role of myelin and oligodendrocytes in axonal energy metabolism. Curr. Opin. Neurobiol. 23, 1065–1072 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Pajevic S., Basser P. J., Fields R. D., Role of myelin plasticity in oscillations and synchrony of neuronal activity. Neuroscience 276, 135–147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nave K. A., Werner H. B., Myelination of the nervous system: Mechanisms and functions. Annu. Rev. Cell Dev. Biol. 30, 503–533 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Nave K. A., Werner H. B., Ensheathment and myelination of axons: Evolution of glial functions. Annu. Rev. Neurosci. 44, 197–219 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Pajevic S., Plenz D., Basser P. J., Fields R. D., Oligodendrocyte-mediated myelin plasticity and its role in neural synchronization. eLife 12, e81982 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhn S., Gritti L., Crooks D., Dombrowski Y., Oligodendrocytes in development myelin generation and beyond. Cells 8, 1424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenfield S., Brostoff S., Eylar E., Morell P., Protein composition of myelin of the peripheral nervous system. J. Neurochem. 20, 1207–1216 (1973). [DOI] [PubMed] [Google Scholar]
- 56.Jahn O., Siems S. B., Kusch K., Hesse D., Jung R. B., Liepold T., Uecker M., Sun T., Werner H. B., The CNS myelin proteome: Deep profile and persistence after post-mortem delay. Front. Cell. Neurosci. 14, 239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen B., Linke A., Olson L., Kohli J., Kinnear M., Sereno M., Müller R.-A., Carper R., Fishman I., Cortical myelination in toddlers and preschoolers with autism spectrum disorder. Dev. Neurobiol. 82, 261–274 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jesse S., Müller H. P., Schoen M., Asoglu H., Bockmann J., Huppertz H. J., Rasche V., Ludolph A. C., Boeckers T. M., Kassubek J., Severe white matter damage in SHANK3 deficiency: A human and translational study. Ann. Clin. Transl. Neurol. 7, 46–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou H., Wu S., Joo J. Y., Zhu S., Han D. W., Lin T., Trauger S., Bien G., Yao S., Zhu Y., Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4, 381–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gouder L., Vitrac A., Goubran-Botros H., Danckaert A., Tinevez J.-Y., André-Leroux G., Atanasova E., Lemière N., Biton A., Leblond C. S., Poulet A., Boland A., Deleuze J.-F., Benchoua A., Delorme R., Bourgeron T., Cloëz-Tayarani I., Altered spinogenesis in iPSC-derived cortical neurons from patients with autism carrying de novo SHANK3 mutations. Sci. Rep. 9, 94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang G., Chen S., Chen X., Zheng J., Xu Z., Doostparast Torshizi A., Gong S., Chen Q., Ma X., Yu J., Zhou L., Qiu S., Wang K., Shi L., Uncovering the functional link between SHANK3 deletions and deficiency in neurodevelopment using IPSC-derived human neurons. Front. Neuroanat. 13, 23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu J., Magri L., Zhang F., Marsh N. O., Albrecht S., Huynh J. L., Kaur J., Kuhlmann T., Zhang W., Slesinger P. A., Casaccia P., Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation. J. Neurosci. 35, 352–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sestan N., State M. W., Lost in translation: Traversing the complex path from genomics to therapeutics in autism spectrum disorder. Neuron 100, 406–423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erwig M. S., Hesse D., Jung R. B., Uecker M., Kusch K., Tenzer S., Jahn O., Werner H. B., Myelin: Methods for purification and proteome analysis. Methods Mol. Biol. 1936, 37–63 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Aggarwal S., Snaidero N., Pähler G., Frey S., Sánchez P., Zweckstetter M., Janshoff A., Schneider A., Weil M.-T., Schaap I. A., Myelin membrane assembly is driven by a phase transition of myelin basic proteins into a cohesive protein meshwork. PLOS Biol. 11, e1001577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Inouye H., Kirschner D. A., Evolution of myelin ultrastructure and the major structural myelin proteins. Brain Res. 1641, 43–63 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Greer J. M., Lees M. B., Myelin proteolipid protein—The first 50 years. Int. J. Biochem. Cell Biol. 34, 211–215 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Ruskamo S., Raasakka A., Pedersen J. S., Martel A., Škubník K., Darwish T., Porcar L., Kursula P., Human myelin proteolipid protein structure and lipid bilayer stacking. Cell. Mol. Life Sci. 79, 419 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mei Y., Monteiro P., Zhou Y., Kim J.-A., Gao X., Fu Z., Feng G., Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature 530, 481–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fischer I., Shohat S., Levy G., Bar E., Trangle S. S., Efrati S., Barak B., Hyperbaric oxygen therapy alleviates social behavior dysfunction and neuroinflammation in a mouse model for autism spectrum disorders. Int. J. Mol. Sci. 23, 11077 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nayak R., Rosh I., Rabinski T., Falik D., Mendel Percia M., Stern S., Generation and characterization of iPSC lines (UOHi003-A, UOHi002-A) from a patient with SHANK3 mutation and her healthy mother. Stem Cell Res. 64, 102899 (2022). [DOI] [PubMed] [Google Scholar]
- 72.Baraban M., Koudelka S., Lyons D. A., Ca2+ activity signatures of myelin sheath formation and growth in vivo. Nat. Neurosci. 21, 19–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iyer M., Kantarci H., Cooper M. H., Ambiel N., Novak S. W., Andrade L. R., Lam M., Jones G., Münch A. E., Yu X., Khakh B. S., Manor U., Zuchero J. B., Oligodendrocyte calcium signaling promotes actin-dependent myelin sheath extension. Nat. Commun. 15, 265 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cristobal C. D., Wang C. Y., Zuo Z., Smith J. A., Lindeke-Myers A., Bellen H. J., Lee H. K., Daam2 regulates myelin structure and the oligodendrocyte actin cytoskeleton through Rac1 and gelsolin. J. Neurosci. 42, 1679–1691 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rauti R., Ess A., Roi B. L., Kreinin Y., Epshtein M., Korin N., Maoz B. M., Transforming a well into a chip: A modular 3D-printed microfluidic chip. APL Bioeng. 5, 026103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brant B., Stern T., Shekhidem H. A., Mizrahi L., Rosh I., Stern Y., Ofer P., Asleh A., Umanah G. K. E., Jada R., Levy N. S., Levy A. P., Stern S., IQSEC2 mutation associated with epilepsy, intellectual disability, and autism results in hyperexcitability of patient-derived neurons and deficient synaptic transmission. Mol. Psychiatry 26, 7498–7508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodrigues D., Jacinto L., Falcão M., Castro A. C., Cruz A., Santa C., Manadas B., Marques F., Sousa N., Monteiro P., Chronic stress causes striatal disinhibition mediated by SOM-interneurons in male mice. Nat. Commun. 13, 7355 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ting J. T., Daigle T. L., Chen Q., Feng G., Acute brain slice methods for adult and aging animals: Application of targeted patch clamp analysis and optogenetics. Methods Mol. Biol. 1183, 221–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bokobza C., Jacquens A., Guenoun D., Bianco B., Galland A., Pispisa M., Cruz A., Zinni M., Faivre V., Roumier A., Lebon S., Vitalis T., Csaba Z., Le Charpentier T., Schwendimann L., Young-Ten P., Degos V., Monteiro P., Dournaud P., Gressens P., Van Steenwinckel J., Targeting the brain 5-HT7 receptor to prevent hypomyelination in a rodent model of perinatal white matter injuries. J. Neural Transm. 130, 281–297 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Douvaras P., Fossati V., Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 1143–1154 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2
Figs. S1 to S7
Uncropped Western blots
Legend for movie S1
Movie S1