Abstract
Bioprinting is considered a key technology for future space missions and is currently being established on the International Space Station (ISS). With the aim to perform bioink production as a critical and resource‐consuming preparatory step already on Earth and transport a bioink cartridge “ready to use” to the ISS, the storability of bioinks is investigated. Hydrogel blends based on alginate and methylcellulose are laden with either green microalgae of the species Chlorella vulgaris or with different human cell lines including immortilized human mesenchymal stem cells, SaOS‐2 and HepG2, as well as with primary human dental pulp stem cells. The bioinks are filled into printing cartridges and stored at 4°C for up to four weeks. Printability of the bioinks is maintained after storage. Viability and function of the cells embedded in constructs bioprinted from the stored bioinks are investigated during subsequent cultivation: The microalgae survive the storage period very well and show no loss of growth and functionality, however a significant decrease is visible for human cells, varying between the different cell types. The study demonstrates that storage of bioinks is in principle possible and is a promising starting point for future research, making complex printing processes more effective and reproducible.
Keywords: bioink storability, bioprinting, cold storages, launch, microalgae
Bioprinting, currently being established for use in space, offers great potential for astronauts to become more independent of medical care from Earth and for generation of tissue models to study the long‐term‐effect of space conditions. The preparation of bioinks is a complex process and thus, preferably done in advance of space missions—this study investigates the storability of bioinks.
1. Introduction
The adaptation of processes established on Earth to microgravity, for example for use in space stations orbiting the Earth or on possible long‐term space missions, requires a complete rethinking of the approach to the overall process and the sub‐processes involved in an applied scientific method. Procedures have to be simplified due to technical, human, or logistical challenges and limitations, adjusted to the local environmental conditions and implemented safety regulations.[ 1 ]
This also applies to 3D bioprinting, in which so‐called bioinks, consisting of cells suspended in a hydrogel‐based ink, are processed into 3D objects of predefined design under computer control[ 2 ] 3D bioprinting is a rapidly developing technology that enables both the patient‐specific production of implants, the development of tissue models for basic research, and, as in the case of green bioprinting, new approaches, and concepts for biotechnological and environmental applications.[ 3 , 4 , 5 , 6 ] Just as the goals of international space travel are moving further and further away from Earth, thereby increasing both the length of the missions and the time frame of a medical retrieval operation, the need to become independent of medical care on Earth is increasing to the same extent. The aforementioned potentials make 3D bioprinting a possible key technology in this endeavor, while in the context of green bioprinting they also offer new technological possibilities for the algae‐containing life support systems established on the International Space Station (ISS).[ 7 , 8 ]
One variant of 3D bioprinting, which, due to its possibility of detailed cell organization, can also be seen as the most suitable for the medical approach in space, is the so‐called extrusion‐based bioprinting, in which bioinks are deposited strand‐wise on top of each other in a layer‐by‐layer approach to form a 3D structure.[ 9 ] Bioinks are predominantly based on hydrogels which must fall within the so‐called biofabrication window, as they have to be dimensionally stable enough for 3D printing of constructs with high shape fidelity and at the same time soft enough to allow for the embedded cells to survive and function.[ 10 ] One ink that fulfills these requirements very well is a blend consisting of 3 wt% alginate and 9 wt% methylcellulose (Alg‐MC) developed by Schuetz et al., as it combines the high cytocompatibility of crosslinked alginate hydrogels with the stabilizing properties of methylcellulose in the printing process, which leads to a higher viscosity, better shear recovery and therefore a better printability of the ink.[ 11 ] During alginate crosslinking and the subsequent cultivation, the methylcellulose is partially released, forming a crosslinked construct of mainly alginate..[ 11 , 12 ] This versatile ink has been successfully applied for bioprinting of various mammalian cell types such as pancreatic islets of Langerhans and chondrocytes, but also for bioprinting of microalgae.[ 13 , 14 , 15 , 16 , 17 , 18 , 19 ]
Based on the promising results in these ground‐based studies, Alg‐MC was the first bioink to be selected by the company OHB for use in space. As part of the ISS mission Cosmic Kiss of the German ESA astronaut Matthias Maurer, it was used as one of two for the mission selected bioinks to generate constructs with the aid of a hand‐held printer, which in combination with dermal fibroblasts should later provide rapid assistance in the wound closure of large‐area skin injuries. Ahlfeld & Cubo et al. were able to modify this ink to support cell adhesion once again in the context of space utilization. For biological functionalization, the biopolymers alginate and methylcellulose were dissolved in human blood plasma instead of PBS; an improvement in cell attachment, spreading, and proliferation could be demonstrated for an immortalized human mesenchymal stem cell line (hTERT‐MSC), primary human dental pulp stem cells (DPSC), human osteoblasts, murine and human fibroblasts as well as the human hepatocellular carcinoma cell line HepG2.[ 20 , 21 , 22 ]
The tests on the ISS with the handheld printer were still carried out without cells in the printed ink. Due to strong limitations regarding the technical equipment, spatial conditions, and crew time, it is extremely difficult to grow large quantities of cells, as necessary for bioink preparation, on board the ISS. Therefore, restrictions in the survival rate of the cells and possibly also in their functionality must be expected, since the conditions and equipment of the cell culture laboratory on the ISS are of course limited compared to a large, fully equipped ground‐based laboratory, that is, a reliable and reproducible cultivation of cells will not be possible to the same extent, and the number of cells will always be limited due to the lack of space in the provided cell culture system. The question is therefore: How can bioprinting in space be made possible with living and functional cells in relevant concentrations, without exceeding the logistical or resource‐specific framework conditions of space travel and research in space?
There are two possible ways to manage that and reduce the workload on board of the space station: (i) Bringing the biomaterial components of the ink in already sufficient quantity and cells as cryo‐preserved stocks separately into space and mix them there with a mixing unit developed recently especially for such use by Dani et al.[ 23 ] This cryo‐preservation is done for a lot of different mammalian cell types for 2D cell culture experiments—where compared to 3D bioprinting much lower cell numbers are needed—on board of the ISS to shield them from negative impacts of long transportation times, vibration and acceleration during rocket launch.[ 24 , 25 ] However, this would again involve time and resource‐consuming cell culture steps for the astronauts on board and increase the risk of contamination. This could be circumvented by (ii) performing all print‐preparing steps already on Earth and flying a bioink cartridge “ready to use” to the ISS. In this case, the astronauts would only have to insert the cartridge into the bioprinter and the experiment could start directly; however, the hurdles that a rocket launch entails would have to be overcome. These include defined storage modules such as the GLACIER, MERLIN, or POLAR system with a possible permanent storage temperature of 4°C as well as the associated storage duration. Even if short rocket loading times and thus short transport times to the ISS are becoming more and more the norm, launch delays can occur, for example, due to weather or technical problems. Therefore, a sample can in a worst case scenario experience a storage period of up to 28 days from loading into the rocket to unpacking and use at the station. The all‐important questions on which this possibility hinges are therefore: What influence does storage of up to 28 days at 4°C have on the printability of bioinks and what influence do these environmental conditions have on the viability and functionality of the cells embedded in the bioinks?
The aim of this study was to answer these questions. The impact of storage at 4°C for up to four weeks on key features of different bioinks based on Alg‐MC was investigated. As key features, rheological properties of the bioinks, as well as viability and activity of the cells within the 3D bioprinted constructs during subsequent cultivation were analyzed. Different cell types were examined: besides green microalgae of the species Chlorella vulgaris, established as a reliable oxygen production source in bioprinted constructs, an immortalized human mesenchymal stem cell line (hTERT‐MSC), HepG2, and SAOS‐2 as common human model cell lines for liver and bone specific cells, respectively, as well as human dental pulp stem cells as an example for primary cells were included in the bioinks. The broad variety of compositions and cell types investigated in this study made it necessary to perform extensive cell expansion as in total about one billion human cells were needed to prepare the respective bioinks.
2. Results
The experimental setup used in the study is shown schematically in Figure 1 . The blends consisting of 3 wt% alginate and 9 wt% methylcellulose (Alg‐MC) were prepared and laden with the different cell types at room temperature; cell densities were chosen based on our previous studies.[ 11 , 12 , 15 , 18 , 20 , 22 , 26 ] One aliquot of each bioink was filled into a cartridge for immediate use for extrusion‐based bioprinting without storage (W0), four aliquots were filled into separate cartridges and the four cartridges were transferred to 4°C for storage. After 1–4 weeks of storage (W1–W4), the cartridges were brought to room temperature and used for bioprinting. The printed cell‐laden scaffolds were ionically crosslinked and incubated under optimal cell culture conditions for up to 28 days. At different time points during cultivation, viability, cell number development, and cell functionality were analyzed.
Figure 1.
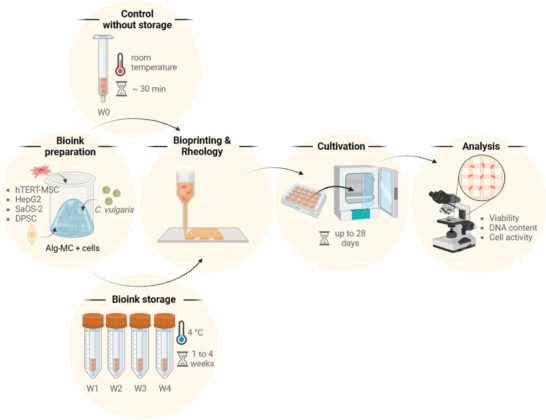
Schematic overview of the experimental setup used in this study. W0: without storage; W1–W4: storage for 1–4 weeks (images created using Biorender.com software).
2.1. Alg‐MC Laden with Microalgae
The blend consisting of 3 wt% alginate and 9 wt% methylcellulose, dissolved in water,[ 17 ] was laden with 1 × 106 C. vulgaris cells per g bioink. One aliquot of the bioink was immediately used (W0) and four aliquots were used after cold storage (W1–W4) for rheological analysis and bioprinting. Over a culture period of 7 days, the impact of cold storage on C. vulgaris‐laden bioinks was investigated in terms of changes in viscosity, cell viability (via live/dead cell imaging), cell number development (via chlorophyll content), and photosynthetic activity.
2.1.1. Rheological Analysis and Printability
The viscosity as key determinant for printability was measured for C. vulgaris‐laden Alg‐MC in fresh state and after storage in two independent experiments. Representative measurements of the viscosity over an increasing shear rate from 1 to 100 s−1 are shown in Figure 2A. Shear thinning properties, which are a crucial requirement for extrusion‐based bioprinting, were observed in fresh as well as in stored bioinks. The viscosity of the fresh bioink was higher at every shear rate compared to the stored bioinks. However, the variable storage time seemed to have no influence on the viscosity. This trend is visible in Figure 2B, where the viscosities of the fresh and stored bioinks at a shear rate of 10 s−1 are compared, whereby values of two individual experiments were combined in this graph. After storage at 4°C for 1 week, the viscosity decreased significantly from 222 ± 22 Pa s in the fresh ink to 147 ± 40 Pa s. After 2–4 weeks of storage, the viscosity did not change significantly compared to the viscosity after 1 week of storage.
Figure 2.
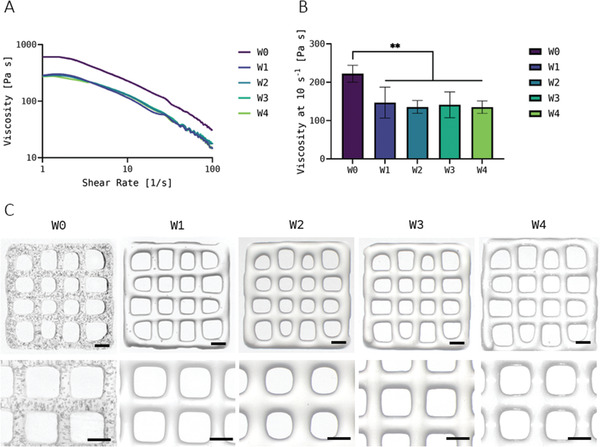
A) Representative measurements of the viscosity of the Chlorella vulgaris‐laden Alg‐MC bioinks immediately after preparation (fresh, W0) and at different time points of bioink storage at 4°C (W1–W4) at an increasing shear rate. The bioinks were brought to room temperature before measurement. B) Viscosity of the bioinks at a shear rate of 10 s−1 (n = 3 of each of the two individual experiments, n = 6 in total; mean ± SD). C) Images of bioprinted scaffolds prepared from fresh (W0) and 1 to 4 weeks stored (W1–W4) bioinks (scale bars = 1 mm). ** p < 0.01.
Despite a decrease in viscosity, the stored bioinks were very well printable: During extrusion, the reduced viscosity was counteracted by an adaptation of the printing parameters pressure and velocity (see Experimental Section). The constructs printed from the fresh and the stored bioinks were comparable with regard to shape fidelity, as shown in Figure 2C. Air bubbles are visible in the scaffolds made from fresh bioinks, which were caused by the mixing of the hydrogel during the production of the bioinks. After a storage time of 1 week, almost all bubbles have leaked out.
2.1.2. Viability and Function of Embedded C. vulgaris
Figure 3A shows the influence of bioink storage on the viability of the microalgae in the printed and crosslinked scaffolds which was determined via the ratio of live versus total cell number after 1, 3, and 7 days of cultivation; representative images of live (red autofluorescence of chlorophyll) and dead (green fluorescence after Sytox staining) cells are shown in Figure S1 (Supporting Information). The values measured for day 1 in all five conditions investigated range from 90.8% (W1) to 78.7% (W4). Apart from a small increase from W0 to W1 (89.4% vs 90.8%), the viability of the microalgae embedded in the bioprinted constructs decreased marginally with increasing storage time compared to the freshly mixed bioink. Over the course of the following cultivation, all constructs maintained their high viability independent of storage time, with values continually ranging between 77% (W3, d7) and 90.3% (W0, d7) for all groups investigated. Thus, a significant negative impact of storage at 4°C on the microalgae embedded in the Alg‐MC hydrogel was not observed.
Figure 3.
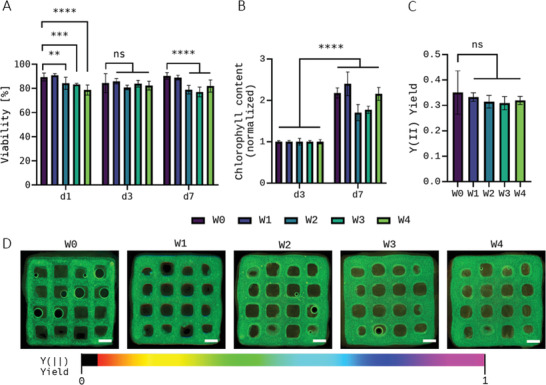
Viability (n = 3 of three individual samples, n = 9 in total; mean ± SD) A) and cell growth (n = 3, mean ± SD) B) after 1, 3, and 7 days of cultivation, and photosynthetic efficiency at day 7 (n = 5 of three individual samples, n = 15 in total, mean ± SD; scale bars = 1 mm) C,D) of Chlorella vulgaris embedded in printed and crosslinked scaffolds. C. vulgaris‐laden Alg‐MC bioinks were used for bioprinting immediately after preparation (W0) and at different time points of bioink storage at 4°C (W1–W4); the bioinks were brought to room temperature before bioprinting. Cell growth was normalized to chlorophyll content of day 3 of the respective unstored control group (W0). ** p < 0.01, *** p < 0.001, **** p < 0.0001, and ns = not significant.
Figure 3B shows the development of the chlorophyll content within the scaffolds from day 3 to 7 in the respective groups. An increased chlorophyll content, observed for all groups, correlates directly with a higher cell number in the scaffold, and is therefore, a reliable parameter for cell growth during cultivation. Day 3 was used as corresponding reference point since on day 1, the chlorophyll content in all groups was still below the detection limit of the assay. The fact that there were enough cells in all groups on day 3 indicates good cell growth from day 1 to 3. Results of the chlorophyll extraction analysis revealed that storage of the microalgae within the bioink did not hamper their proliferation during the subsequent cultivation within the printed scaffolds, regardless of the storage length. Groups W0, W1, and W4 increased their chlorophyll content by > 100%, and W2 and W3 were not far behind with an increase of > 70%. The excellent growth rates were also proven by the fact that on day 7, measurement of the photosynthetic efficiency of the microalgae could be performed in all groups, which is only possible in very densely populated scaffolds.
After seven days of cultivation, pulse–amplitude modulation fluorometry (PAM) was performed to determine the photosynthetic efficiency yield Y(II) of the embedded microalgae. This indicates how much of the light available to the microalgae can actually be absorbed via their photoreceptors, and therefore what proportion can be used for photosynthetic processes inside the cell. This is a good way to test the functionality of the microalgae; the photosynthetic efficiency yield Y(II) also represents an important stress indicator in microalgae biotechnology.[ 27 ] For C. vulgaris embedded in the printed and crosslinked scaffolds, the measurements using the PAM system showed a similar progression to the results of the viability measurements (Figure 3C); representative images of the PAM measurements are shown in Figure 3D. The samples printed from the fresh bioink showed the highest photosynthetic efficiency yield with Y(II) = 0.35, samples of all storage groups were below this value, with Y(II) between 0.33 and 0.31, but the differences were only marginal and not significant. Overall, however, no apparent negative effect of bioink storage on the cell functionality of the microalgae was evident: They could still use the light available to them with similar efficiency as the fresh reference group even after 4 weeks of storage, which also leads to the release of oxygen out of the scaffold, leading to the release of air bubbles, which can be seen in some of the images.
2.2. PBS‐Alg‐MC and Plasma‐Alg‐MC Laden with hTERT‐MSC
The immortalized human mesenchymal stem cell line hTERT‐MSC, which due to its unlimited proliferation capacity easily allows the generation of a sufficiently high number of cells while maintaining their stem cell characteristics,[ 28 ] was used as model cell line to answer the questions (i) if mammalian cells can survive within the cold‐stored Alg‐MC‐based bioinks, and (ii) if integrated proteins, derived from human blood plasma, are stable and able to support the cells within the ink during storage. Thus, the impact of storage on hTERT‐MSC‐laden bioinks based on PBS‐Alg‐MC and on Plasma‐Alg‐MC[ 11 , 20 ] was comparatively investigated regarding changes in viscosity, cell viability, and cell number development via DNA content during cultivation.
The blends consisting of 3 wt% alginate and 9 wt% methylcellulose, dissolved in either PBS or plasma, were laden with 5 × 106 hTERT‐MSC per gram bioink. One aliquot of the bioinks was immediately used (W0) and four aliquots were used after cold storage (W1–W4) for rheological analysis and bioprinting.
2.2.1. Rheological Analysis and Printability
Viscosity was measured for hTERT‐MSC‐laden PBS‐Alg‐MC as well as for hTERT‐MSC‐laden Plasma‐Alg‐MC, both in the fresh state (W0) and after storage (W1–W4). Figure 4A,C shows representative measurements of the viscosity of PBS‐Alg‐MC‐ and Plasma‐Alg‐MC‐based bioinks, respectively. Both bioinks exhibited shear thinning properties in fresh and stored conditions. The viscosity of the PBS‐Alg‐MC bioink decreased slightly from 54 ± 2 Pa s in the fresh state to 45 ± 5 Pa s after 1 week, and significantly to 36 ± 3 Pa s after 2 weeks of storage at a shear rate of 10 s−1 (Figure 4B). The viscosity of the bioinks stored for 3 and 4 weeks, with 55 ± 6 Pa s and 56 ± 5 Pa s at a shear rate of 10 s−1, respectively, was not significantly different from the fresh bioink. Interestingly, the viscosity of the Plasma‐Alg‐MC ink increased from 41 ± 10 Pa s in the fresh state to 57 ± 2 Pa s after 1 week and 56 ± 3 Pa s after 2 weeks of storage (Figure 4D). Measurements of the viscosity after 3 and 4 weeks of bioink storage revealed no significant changes in comparison to the fresh bioinks, with values of 51 ± 1 Pa s and 45 ± 1 Pa s, respectively.
Figure 4.
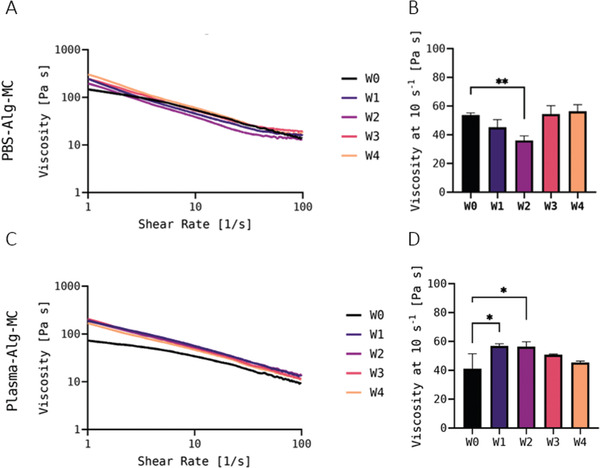
A) Representative measurements of the viscosity of the hTERT‐MSC‐laden PBS‐Alg‐MC (A) and Plasma‐Alg‐MC C) bioinks immediately after preparation (fresh, W0) and at different time points of bioink storage at 4°C (W1–W4) at an increasing shear rate. The bioinks were brought to room temperature before measurement. B,D) Viscosity of the bioinks at a shear rate of 10 s−1 (n = 3; mean ± SD). * p < 0.05 and ** p < 0.01.
Consistent with the maintenance of shear thinning behavior and high viscosity observed for both bioinks during storage at 4°C, the stored bioinks were very well printable.
2.2.2. Viability of Embedded hTERT‐MSC
In the first experiment, viability and cell number development of hTERT‐MSC embedded in PBS‐Alg‐MC or Plasma‐Alg‐MC were analyzed via simultaneous staining of live (calcein) and dead (ethidium homodimer‐1) cells and DNA quantification, respectively, after cultivation of the printed and crosslinked constructs for 1, 7 and 21 days (Figure 5 ). Within the PBS‐Alg‐MC‐based constructs, the viability values were between 55 and 65% on day 1, without significant differences between the fresh control group W0 and the storage groups W1–W4. Over the course of the subsequent cultivation, the values differed between the different experimental groups, but overall remained in a similar viability range (Figure 5A). In contrast, a significant negative influence of storage on the viability was apparent for the cells embedded in Plasma‐Alg‐MC‐based hydrogels: Whereas at day 1 the viability of the fresh control group W0 was > 90%, it was significantly reduced after storage in W1–W4. Surprisingly, the value in W1 was the lowest with ≈ 20% and the value in W4 was closest to W0 with nearly 60%. Over the course of cultivation, viability values decreased in the control group W0 but increased in the storage groups W1–W4 to ≈ 80% until day 21. This indicates recovery of the cells after storage; at day 21, the differences between the five experimental groups were not significant (Figure 5C). For both, the PBS‐Alg‐MC‐ and the Plasma‐Alg‐MC‐based bioinks, results of DNA quantification indicated higher cell numbers in the storage groups W1–W4 compared to the unstored control group W0—except for W4‐samples in Plasma‐Alg‐MC, these differences were significant. In the course of subsequent cultivation, a significantly stronger reduction of the cell number in the storage groups W1–W4 in comparison to the control group W0 was observed on day 7 in PBS‐Alg‐MC but not in Plasma‐Alg‐MC. On day 21, no significant differences between the five experimental groups were observed in both, the PBS‐Alg‐MC‐ and the Plasma‐Alg‐MC‐based bioink (Figure 5B,D).
Figure 5.
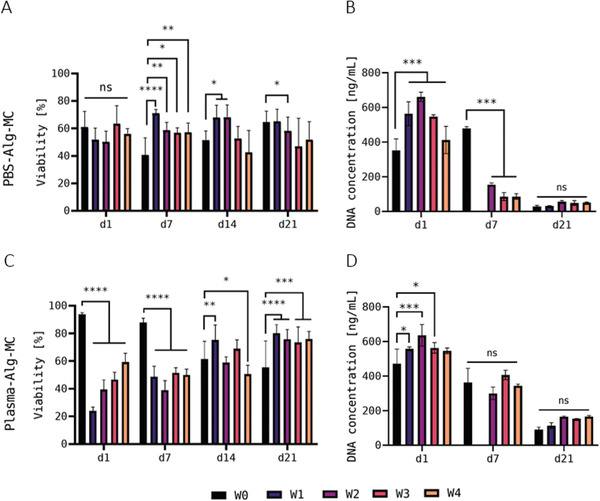
Viability A,C) and DNA content B,D) of hTERT‐MSC in printed and crosslinked scaffolds—first experiment. Viability was determined after 1, 7, 14, and 21 days of cultivation (n = 3 of three individual samples, n = 9 in total; mean ± SD) and DNA content was measured after 1, 7, and 21 days of cultivation (n = 3, mean ± SD). hTERT‐MSC‐laden Plasma‐Alg‐MC bioinks were used for bioprinting immediately after preparation (W0) and at different time points of bioink storage at 4°C (W1–W4); the bioinks were brought to room temperature before bioprinting. Due to a contamination, DNA quantification was not possible for the W1 group for both, PBS‐Alg‐MC and Plasma‐Alg‐MC, at day 7. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, and ns = not significant.
A second experiment (Figure 6 ) was conducted to prove the findings of the first experiment in the early cultivation phase, for instance the apparent adjustment of the cells to the storage conditions in the Plasma‐Alg‐MC bioink, reflected by viability values which steadily increased from W1 to W4 at day 1 of cultivation (Figure 5A). However, this was not confirmed in the repeat experiment. In both, the PBS‐Alg‐MC‐ and the Plasma‐Alg‐MC‐based bioink a significantly reduced viability was observed in the storage groups W1–W4 in comparison to the control group W0 (Figure 6A,C; representative images of live (green fluorescence after Calcein staining) and dead (red fluorescence after Ethidium homodimer‐1 staining) cells are shown in Figures S2 and S3, Supporting Information). This is in line with the first experiment for the Plasma‐Alg‐MC‐ but not for the PBS‐Alg‐MC‐based bioinks. DNA quantification indicated a significant reduction in cell number in the storage groups W1–W4 in comparison to the control group W0, which confirmed the findings of the first experiment for day 7 but not for day 1. For the cells in the Plasma‐Alg‐MC bioinks, the results of DNA quantification of the first and the second experiment were slightly different, but in both they do not indicate a strong negative effect of cold storage on cell number development. Altogether, the results of both experiments indicate a negative effect of storage on cell survival that disappeared after bringing the cells back to optimal conditions, during subsequent cultivation. The number of cells in the printed hydrogel constructs was moderately affected by storage.
Figure 6.
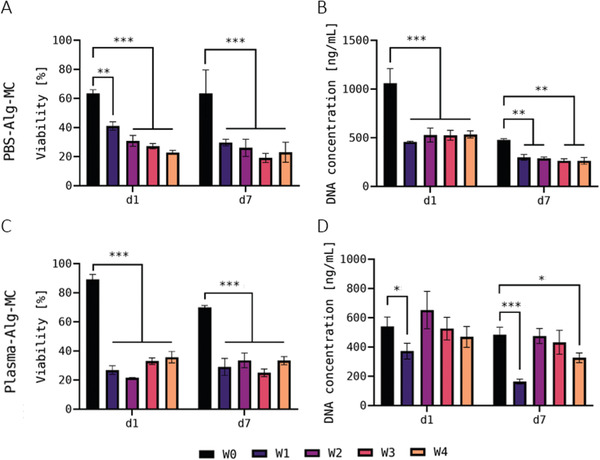
Viability A,C) and DNA content B,D) of hTERT‐MSC in printed and crosslinked scaffolds—second experiment. Viability (n = 3 of three individual samples, n = 9 in total; mean ± SD) and DNA content (n = 3; mean ± SD) were measured after 1 and 7 days of cultivation. hTERT‐MSC‐laden Plasma‐Alg‐MC bioinks were used for bioprinting immediately after preparation (W0) and at different time points of bioink storage at 4°C (W1–W4); the bioinks were brought to room temperature before bioprinting. * p < 0.05, ** p < 0.01, *** p < 0.001, and ns = not significant.
2.3. Plasma‐Alg‐MC Laden with SaOS‐2 and HepG2
The human osteosarcoma cell line SaOS‐2 and the human hepatocellular carcinoma cell line HepG2, commonly applied as model cell lines for osteoblasts and hepatocytes, respectively, were used in this study to investigate whether mammalian cells can also maintain their function after cold storage within the Plasma‐Alg‐MC ink. Therefore, besides viability and cell number development within the stored bioinks, the enzyme activity of lactate dehydrogenase (LDH) was determined as non‐specific metabolic marker, as well as the enzyme activity of alkaline phosphatase (ALP) as specific marker for osteoblasts, and the formation of albumin‐expressing cell clusters as specific phenotype of hepatocytes.
The blend consisting of 3 wt% alginate and 9 wt% methylcellulose, dissolved in plasma, was laden either with 5 × 106 SaOS‐2 or 5 × 106 HepG2 cells per gram bioink. Again, one aliquot of the bioinks was immediately used (W0) and four aliquots were used after cold storage (W1–W4) for bioprinting.
2.3.1. Viability and Function of Embedded SaOS‐2
Figure 7A shows the influence of bioink storage on the viability of SaOS‐2 cells in the printed and crosslinked scaffolds which was again determined via simultaneous staining of live and dead cells after 1, 7, 14, and 21 days of cultivation. The viability rate of SaOS‐2 cells on day 1 after printing without prior storage (W0 control group), averaged nearly 80% of living cells. After storage, the viability rate dropped significantly; however, a high variation was observed between the different storage groups: while it even fell < 20% mark in the groups W1 (12.4%) and W3 (17.8%), the groups W2 and W4 showed a decrease to ≈ 30% and 50%, respectively. After bioprinting, the constructs of all groups were cultivated under optimal conditions for 21 days and over the course of this cultivation, the viability values of all storage groups (W1–W4) recovered. The data indicate an apparent negative influence of storage on cell survival, which, however, was compensated during cultivation of the bioprinted constructs.
Figure 7.
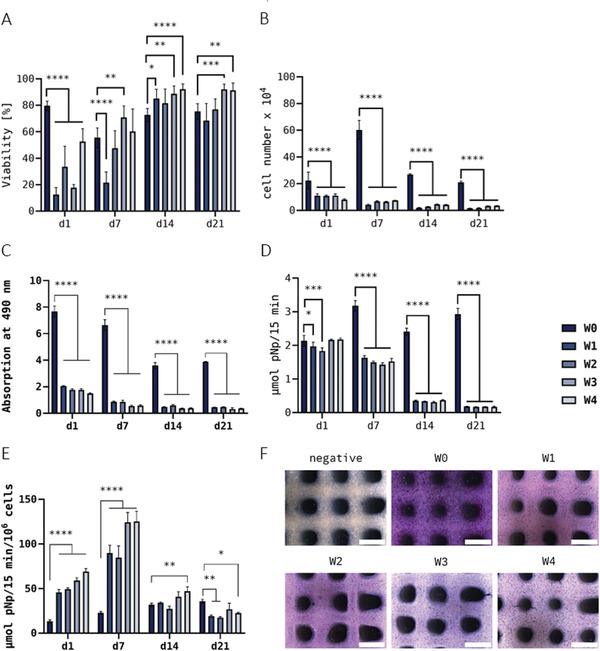
Viability (n = 3 of three individual samples, n = 9 in total; mean ± SD) A), cell growth (n = 3; mean ± SD) B), LDH activity (n = 3; mean ± SD) C), ALP activity per scaffold (n = 3; mean ± SD) D), specific ALP activity (n = 3; mean ± SD) E), and measurement of SaOS‐2 embedded in printed and crosslinked scaffolds after 1, 7, 14, and 21 days of cultivation. Histological ALP‐activity staining F) after 21 days of cultivation (scale bars = 2 mm). SaOS‐2‐laden Plasma‐Alg‐MC bioinks were used for bioprinting immediately after preparation (W0) and at different time points of bioink storage at 4°C (W1–W4); the bioinks were brought to room temperature before bioprinting. * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
The DNA content of the bioprinted constructs was measured and correlated to the cell number (Figure 7B). On day 1 after printing, a significant decrease of the cell number from ≈ 2 × 105 (W0) to 1 × 105 (W1–W3) or 0.7 × 105 (W4) cells per scaffold during storage was observed. Whereas an increase in the cell number during cultivation from day 1 to 7 was detected for the control group W0, the cell numbers in the storage groups W1–W4 decreased further. On day 14 and 21, lower cell numbers were observed for all groups in comparison to those measured on day 7. These data indicate a strong negative effect of storage on growth of SaOS‐2 cells within the hydrogel.
Consistent with these observations, the LDH activity measurements (Figure 7C) revealed a significant decrease of metabolic activity in bioprinted constructs of the storage groups W1–W4 in comparison to the control group W0 on day 1 after printing. A small decrease was also observed from W1 to W4, indicating a marginal influence of storage time. During further cultivation (day 7–21), LDH activity continued to decrease in all groups while the differences between groups W0 and W1–W4 remained. The relation between LDH activity and DNA content (Figure S3, Supporting Information) shows that the relative LDH activity is decreased immediately after printing (day 1), but in the long term the activity evens out during cultivation.
ALP activity, indicating osteogenic differentiation, was detected in all groups whereby the absolute values—that is, the activity per scaffold—of the W0 group increased from day 1 to 7 and remained within the range of these two values during further cultivation, but values in the stored groups W1–W4 decreased from day 1 to 7, and further from day 7 to 14 (Figure 7D). On day 21, the ALP activity per scaffold was much higher in W0 compared to W1–W4 (Figure 7E) which was confirmed by histological staining of ALP activity (Figure 7F). The specific ALP activity—that is, the activity related to the number of metabolically active cells—is shown in Figure 7E: An increase in the values from day 1 to 7, typical for osteogenic differentiation, was observed for all groups, independent of storage. Later on, the specific ALP activity dropped for the stored groups W1–W4 but not for the control group W0. Interestingly, specific ALP activity determined for W1–W4 was significantly higher in comparison to W0. These data indicate maintenance of the osteogenic phenotype after cold storage despite a significantly reduced metabolic activity.
2.3.2. Viability and Function of Embedded HepG2
Figure 8A shows the influence of bioink storage on the viability of Hep‐G2 cells embedded in the printed and crosslinked hydrogels, again determined by simultaneous staining of live and dead cells during the 14 days of cultivation. On day 1 after printing, the viability of the cells in the storage groups W1–W4 decreased significantly by more than half compared to the viability rate of the control group W0 (60.1%), except for W2 (54.5%) which was rather similar to W0. In the course of further cultivation, however, a steady increase in the viability rate was observed in all stored samples, so that all four storage groups were in the range of 76.2% (W2) to 85.5% (W3) living cells on day 14 and thus, even above the viability rate of the control group W0 (57.6%). Again, a negative influence of storage of the bioink on cell survival is indicated, but also a recovery of cell viability during cultivation of the constructs after bioprinting.
Figure 8.

Viability (n = 3 of three individual samples, n = 9 in total; mean ± SD) A), cell growth (n = 3; mean ± SD) B), and LDH activity (n = 3; mean ± SD) C) of Hep‐G2 embedded in printed and crosslinked scaffolds after 1, 7, and 14 days of cultivation. HepG2‐laden Plasma‐Alg‐MC bioinks were used for bioprinting immediately after preparation (W0) and at different time points of bioink storage at 4°C (W1–W4); the bioinks were brought to room temperature before bioprinting. ** p < 0.01, **** p < 0.0001, and ns = not significant.
Looking at the DNA content of the bioprinted constructs and correlating it to the cell number (Figure 8B), the data measured on day 1 after printing show that, in contrast to the SaOS‐2 cells (Figure 7B), apparently no decrease was observed due to storage: The values of the stored groups were in the same range as those of the control group W0 and were independent of the length of storage. However, during further cultivation (day 7 and day 14), the DNA values of stored and unstored samples developed completely differently: Whereas the unstored samples (W0) showed a strong increase from day 1 to 7 (calculated cell numbers: 5.88 × 104 to 2.81 × 105) as well as from day 7 to 14 (7.97 × 105), a significant reduction was observed in the scaffolds bioprinted using stored bioinks (W1–W4; cell numbers: ≈ 6.92 × 104 at day 1 to 1.50 × 104 at day 14).
These findings, indicating a strong negative effect of storage on growth of HepG2, are supported by the measurement of LDH activity which revealed an even stronger effect on the metabolism of the cells: Only very low values were measured at day 1 in the stored samples (W1–W4) and no activity at days 7 and 14; in contrast, the LDH activity in the unstored control group W0 increased significantly during the 14 days of cultivation (Figure 8C).
Fluorescence microscopic analyses were performed to investigate whether the embedded HepG2 was able to form the typical hepatic cell clusters expressing albumin after storage in the bioink. As shown in Figure 9A, cluster formation by proliferating cells was proven for the unstored W0 group. However, in the bioink group stored for 1 week (W1), only very few clusters were observed (left images in Figure 9A for day 7 and 14); the majority of the cells remained in their single state (right images in Figure 9A for day 7 and 14). In addition, compared to the W0 group, the number of proliferating (EdU‐positive) cells in the W1 group was strongly reduced on day 7 and completely disappeared on day 14 of cultivation. In the groups which were stored longer than 1 week (W2–W4), neither cell clusters nor proliferating cells were observed (data not shown). Albumin expression was proven for the control group W0 but not for the storage groups W1–W4 (exemplarily shown in Figure 9B for W1). These findings are in line with the observed decrease in cell number (DNA content) and metabolic activity, indicating a very strong impact of cold storage on HepG2 within the bioink.
Figure 9.
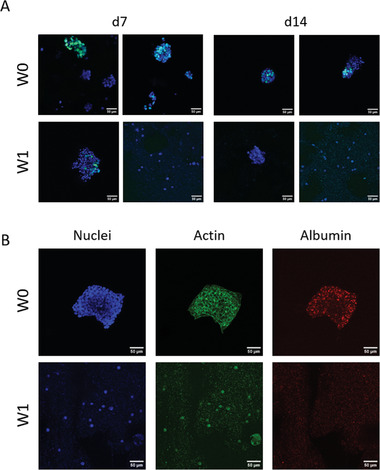
Hep‐G2 in 3D bioprinted scaffolds after bioink storage for 1 week (W1) in comparison to the unstored control group (W0), cultivated over 14 days. A) Proliferating nuclei and cell cluster formation. After 7 and 14 days of cultivation, scaffolds were incubated with EdU for 6 h before fixation. Cells that proliferated in that time frame incorporated EdU. EdU‐positive nuclei: green, counterstaining for all nuclei with DAPI: blue. B) Immunofluorescence staining for nuclei (DAPI, blue), cytoskeletons (actin, green), and albumin (red) after 14 days of cultivation. Scale bars = 50 µm for all.
2.4. Plasma‐Alg‐MC Laden with Human DPSC
Finally, the impact of bioink storage on the viability, cell number development, and metabolic activity of embedded primary, non‐immortalized, and non‐tumorigenic cells was investigated using human dental pulp stem cells (DPSC) obtained from two different donors.
The blend consisting of 3 wt% alginate and 9 wt% methylcellulose, dissolved in plasma, was laden with 5 × 106 DPSC of donor 1 or donor 2 per gram bioink. One aliquot of the bioinks was immediately used (W0) and four aliquots were used after cold storage (W1–W4) for bioprinting. Over a culture period of 28 days, the viability (via live/dead cell staining), cell number development (via DNA content), and metabolic activity (via LDH activity) within the printed and crosslinked constructs were determined.
2.4.1. Viability, Growth, and Metabolic Activity of DPSC
For DPSC of donor 1, the simultaneous staining of live and dead cells at day 1 revealed a decrease in viability of the storage groups (W1–W4) in comparison to the control group W0 (Figure 10A): The percentage of living cells dropped from ≈ 67.5% (W0) to values between 36.9% (W4) and 46.4% (W3). Over the course of further cultivation, however, again a recovery of the viability could be observed. From day 14 on, none of the storage groups were behind the control group (69.1%) in viability, so a negative long‐term influence of the storage at 4°C was not recognizable. Interestingly, viability in the storage groups (W3 and W4, partly also W2 and W1) reached even higher values than the control group W0 (Figure 10A). Samples laden with DPSC of donor 2 behaved very similarly and showed the same trends with regard to viability (Figure 10B): About 50% of the cells survived the bioink preparation and cold storage in almost all samples (from 41.6% in W2 up to 54.3% in W0 on day 1); the differences between stored and unstored bioinks were even lower than in the samples from donor 1. Over the course of the following cultivation, the recovery of cell viability, already observed in other experiments, was evident here as well, with values of over 80% in all groups from day 14 on with slight differences between the individual days of the experiment.
Figure 10.
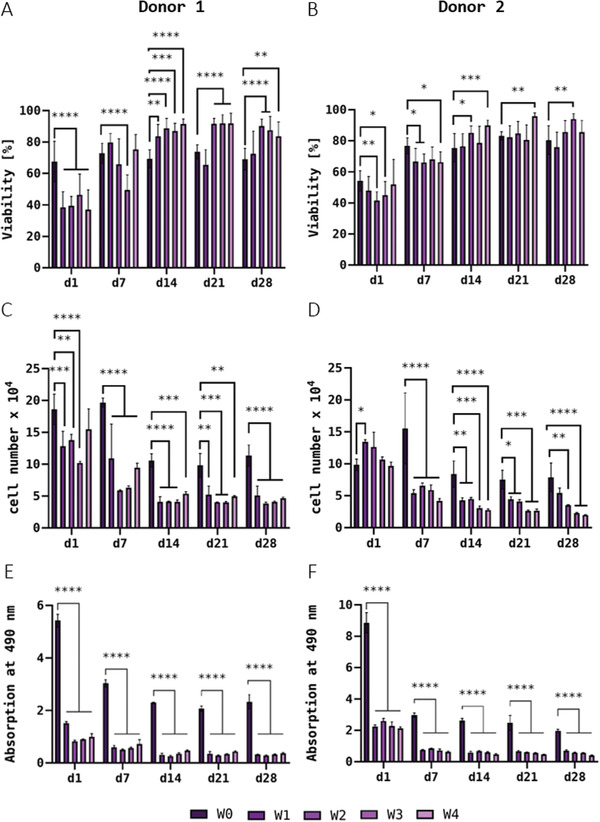
Viability (n = 3 of three individual samples, n = 9 in total; mean ± SD) A,B), cell growth (n = 3; mean ± SD) C,D), and LDH activity (n = 3; mean ± SD) E,F) of DPSC from donor 1 (A,C,E) and donor 2 (B,D,F) embedded in printed and crosslinked scaffolds after 1, 7, 14, 21, and 28 days of cultivation. DPSC‐laden Plasma‐Alg‐MC bioinks were used for bioprinting immediately after preparation (W0) and at different time points of bioink storage at 4°C (W1–W4); the bioinks were brought to room temperature before bioprinting. * p < 0.05, ** p < 0.01, *** p < 0.001, and ****p < 0.0001.
Measurement of the DNA content of bioprinted scaffolds containing DPSC of donor 1 revealed that already on day 1, the scaffolds obtained from the stored bioinks contained significantly fewer cells (≈ 1.29 × 105) than the scaffolds of the control group W0 (1.86 × 105). Again, the length of storage did not seem to have an influence, as the average number of cells per scaffold seemed to remain relatively constant in all groups except for W3. In both the control group and the storage groups, a drop in cell numbers was observed up to day 14; later on, the DNA content and hence the calculated cell number appeared to remain constant in the further course of cultivation (Figure 10C). For bioprinted constructs laden with DPSC of donor 2, DNA quantification revealed no differences between stored and unstored samples on day 1 after printing. During cultivation, however, there was a significant decrease in DNA content up to day 7 in the storage groups (W1–W4), whereas the unstored control group (W0) even showed an increase. Up to day 14, the values decreased further but remained stable at a constant level for all groups from day 14 onward (Figure 10D).
The investigation of the LDH activity in scaffolds laden with DPSC of donor 1 showed a significant reduction in the storage groups (W1–W4) compared to the control group (W0) on day 1. During the further cultivation, the values of all groups decreased until day 14 but remained constant in the further course; the significant difference between the control and the storage groups was observed also on days 7, 14, 21, and 28 (Figure 10E). For constructs containing DPSC of donor 2, the same trend was observed, with significant differences between the control group W0 and the storage groups W1–W4 at all cultivation time points (Figure 10F). The relation between LDH activity and DNA content (Figure S4, Supporting Information) shows that the relative LDH activity is already significantly reduced directly after printing (day 1) and that the activity does not completely even out in the long term over the course of cultivation. Altogether, these data indicate that cold storage has a minor/moderate effect on the viability of DPSC but a stronger effect on their activity.
3. Discussion
General characteristics of the Alg‐MC blend used in this study are a viscosity in ranges ideal for extrusion‐based 3D bioprinting of volumetric structures that retain their shape‐fidelity in air until crosslinking and compatibility with a variety of cell types, such as microalgae, MSC, chondrocytes, and pancreatic islets. In preliminary experiments it also could be demonstrated that the cohesiveness of this blend is so strong that > 30 layers could be printed onto each other upside down and therefore against Earth's gravity[ 7 ] so that it can be expected that extrusion‐based printing will also be feasible under microgravity conditions in space. Depending on the cell type incorporated, the composition of the blend differs slightly, because when microalgae are embedded, Alg is dissolved in water, for mammalian cells in PBS, before MC as dry powder is added.[ 11 , 12 , 13 , 15 , 17 , 18 ] This same blend, with the Alg dissolved in fresh‐frozen human blood plasma[ 29 ] instead of PBS, was demonstrated to exhibit similar excellent shape fidelity; the integration of plasma proteins stimulated proliferation and differentiation of bioprinted human osteoblasts as well as DPSC and increased proliferation and albumin production (an indicator for functionality) in bioprinted HepG2 cells.[ 21 ] With its wide scope of possible applications this blend was therefore chosen as a model material to test the possibility of storage as a bioink containing live cells for extended periods of time in low temperatures. The main two components of a bioink are the biomaterial components and the incorporated cells, both of which can be affected by extended storage and influence each other, so we deemed it necessary to analyze the influence of storage time on rheological properties, as well as cell behavior.
The hydrogel blend is composed of alginate polymer chains, methylcellulose polymer chains, water molecules, and, in case of the PBS‐ and Plasma‐Alg‐MC inks, additional ions, and proteins. Considering these components, the main parameter that is expected to change over time and with temperature is the distribution and interaction of the components—even without further stirring the polymer chains and especially the water molecules slowly change their positions due to ubiquitous molecular movement. This, in turn, can influence the viscosity of the ink as, within this blend, the viscosity is almost exclusively determined by the MC[ 11 , 26 , 30 ]—in our experience neither the alginate nor the presence of cells influence viscosity noticeably.[ 31 ] MC is an inversely thermo‐gelling polymer with high solubility at low and gelation at higher temperatures.[ 32 , 33 , 34 ] In the present study the Alg‐MC blend was prepared in the three different compositions described above, that is, the alginate was dissolved in either water, PBS, or human plasma before dry MC powder was added. With rigorous stirring and a swelling time of >30 min this results in a homogeneous, high‐viscous Alg‐MC paste. The rheological measurements showed a distinct difference between the gel prepared with water and the other two compositions, independent on the cold storage: Overall the viscosity with water was much higher than with either PBS or plasma as solvents (150–200 and 40–60 Pa s, respectively), which has been observed before[ 11 , 20 , 35 ] A direct comparison of PBS‐ and Plasma‐Alg‐MC showed that their viscosities are close in range, with the Plasma‐Alg‐MC showing a slightly lower viscosity. The viscosity values measured in the scope of this study are perfectly in range of those mentioned previously.[ 21 ] This described difference in viscosity was also reflected in different printing pressures used for the bioinks, whereby the Alg‐MC in water required nearly twice as much pressure as the other bioinks, with otherwise completely similar settings.
The solution/gelation mechanism of MC has been the topic of extensive research efforts over the years and has recently been summarized in a comprehensive review, and it is by now widely accepted that during dissolution a hydration of especially the hydrophobic side chains leads to a disruption of intra‐/inter‐chain binding of MC, whereas an increase in temperature increases hydrophobic interactions and thereby results in fibrillation.[ 36 ] It is furthermore undisputed, that this mechanism can be influenced by the presence of ions,[ 37 ] and, to a lesser extent, proteins.[ 38 ] Specifically, a large amount of proteins can reduce the viscosity of an Alg‐MC blend, which might be a reason for the slightly lower viscosity of Plasma‐Alg‐MC compared to PBS‐Alg‐MC;[ 38 ] and different ions can either increase or decrease solution/gelation of MC via “competition” for water molecules. That presence of ions also influences solubility of MC in an alginate solution and is corroborated by our observations that more vigorous and longer stirring is required to achieve a homogeneous paste when dry MC is stirred into alginate dissolved in water, than when it is stirred into PBS‐ or plasma‐alginate. Nevertheless, from reports in the literature the large difference in viscosity between Alg‐MC in water versus PBS and plasma that we observed repeatedly cannot be explained by the ions present, the mechanism of MC hydration, therefore seems to be affected not only by the complex mixture of ions in our inks but also by the presence of alginate, which needs to be further elucidated in a future study.
Storage of Alg‐MC dissolved in water for 1 week at 4°C led to a significant reduction in viscosity, but no further reduction was observed during the following 3 weeks of storage. It can therefore be inferred that storage itself—and thus, the temperature decrease—but not duration of storage influenced viscosity of the bioink prepared with water. On the other hand, the viscosity of PBS‐ and Plasma‐Alg‐MC changed only marginally over the entire duration of storage. That we observed an influence of storage on the ink prepared with water, but not with PBS nor human plasma, again indicated that the hydration of MC in our system is changed by the presence of ions. Altogether, our data indicate that the impact of cold storage on the viscosity of the bioinks is strongly dependent on their composition and, thus, has to be investigated for every specific type of bioink. However, all viscosities measured after cold storage in this study are in ranges suitable for extrusion bioprinting of stable cell‐containing scaffolds with high shape fidelity; the observed differences can be compensated by adjusting the printing pressure.
Although Alg‐MC dissolved in water exhibited the highest viscosities of the three bioinks tested in this study and therefore the highest printing pressures were needed, it did not seem to have a negative influence on the viability of the embedded microalga C. vulgaris. Thakare et al. studied the effect of different pressures (300, 500 and 700 kPa) during extrusion‐based bioprinting on the subsequent growth of microalgae (Chlamydomonas reinhardtii) and showed the highest cell concentrations after 6 days of culture at a printing pressure of 300 kPa.[ 39 ] That is even higher than the highest pressure used in the present study, 270 kPa for the fresh (W0) Alg‐MC bioink which exhibited the highest viscosity. The high viability (≈ 90% observed for W0) and the growth of embedded C. vulgaris after bioprinting in Alg‐MC is consistent with the findings for other bioprinted microalgae such as C. reinhardtii, Chlorella sorokiniana and Scenedesmus sp.[ 17 , 18 , 40 ]
The storage at 4°C can be expected to have a stronger impact on the living part of bioinks, the cells. For the microalgae, only a marginal decrease of viability throughout the 7 days of cultivation, due to cold storage of the bioink was detected. Since the chlorophyll concentration increased similarly during cultivation in the scaffolds prepared from fresh as well as stored bioinks, the cells not only remained viable but were also able to grow in the printed scaffolds after storage. To determine the functionality of the microalgae, photosynthetic efficiency was measured using PAM fluorometry: It was approximately the same in constructs bioprinted from fresh (W0) and stored bioinks at all measured storage times (W1–W4), indicating that the embedded microalgae did not exhibit a stress response after cold storage. The photosynthetic activity is also an indicator for oxygen production, a potentially relevant function of microalgae, for example, in life support systems or tissue engineering concepts, as the light‐dependent reaction converts light energy into chemical energy, producing oxygen as a by‐product.[ 18 , 41 , 42 ] C. vulgaris is distributed worldwide in freshwater, marine, and terrestrial environments and has a high photosynthetic capacity under a variety of environmental conditions.[ 43 ] Both photosynthetic and respiratory processes are highly temperature dependent and are reduced in suboptimal temperature ranges, < 15°C and > 35°C.[ 44 ] Microalgae adapt to these conditions through physiological changes, such as an increase in fatty acids and cellular accumulation of amino acids and their derivatives, which may contribute to cell tolerance to low temperatures.[ 43 ] The results of this study show that C. vulgaris adapt to the suboptimal conditions, indicated by full recovery of their viability, growth ability, and functionality in terms of photosynthetic activity, making them suitable organisms to be stored in hydrogels at 4°C for several weeks prior to use.
Immortalized MSC (hTERT‐MSC) were used to gain a first insight into how well mammalian cells—whose physiological conditions lie in a much narrower window—tolerate the bioink storage at 4°C; viability and DNA content in PBS‐ and Plasma‐Alg‐MC were determined in two independent experiments. Viability of hTERT‐MSC in the unstored bioinks (W0) on day 1 was ≈ 60% and 90% in PBS‐Alg‐MC and Plasma‐Alg‐MC, respectively. These ranges fit to previous reports for cell viability of hTERT‐MSC in these gels[ 20 , 23 ] With respect to the influence of storage of bioinks, while there was some variability between the experiments, we observed an overall trend to lower viability which, however, recovered completely over the cultivation period of 21 days. The drop in viability between W0, d1 and W1, d1 appeared more pronounced in Plasma‐Alg‐MC than in PBS‐Alg‐MC; however, in Plasma‐Alg‐MC the initial viability (W0, d1–d7) was much higher. While viability reflects the ratio of living versus total cells, the DNA content is a measure of the number of cells present in the bioprinted constructs. The DNA content showed a clear trend toward reduction as a result of storage in PBS‐Alg‐MC but not in Plasma‐Alg‐MC. All in all, these data indicate, that cold storage, mainly the beginning of it (differences W0–W1 > W1–W4), influences the viability and cell number of hTERT‐MSC within Alg‐MC‐based bioinks, but also, that even mammalian cells can survive such unfavorable conditions to a considerable degree in these inks.
The effects of hypothermic conditions on mammalian cells is of interest to many fields of research and medicine, not least for the transport of organs for transplantation and with respect to the use of cells for tissue engineering outside of specialized laboratories. These effects were, for example, comprehensively summarized by Rubinsky et al.: In short, exposure to hypothermia has a beneficial effect—it results in a strongly reduced cell metabolism accompanied by reduced energy consumption and reduced oxygen demand, which results in a relatively robust reaction to short‐term hypothermia exposure—but also a number of harmful effects, the most prominent of which are the cell membrane becoming “leaky” due to a lipid phase transition and a reduced activity of ionic pumps, resulting in an osmotic shock.[ 45 , 46 ] It has been hypothesized previously that this osmotic shock can be alleviated by membrane stabilization and reduced ion diffusion via encapsulation of cells in protective hydrogel microcapsules: adipose tissue‐derived MSC (AT‐MSC) encapsulated in alginate were able to keep their differentiation potential after 2 weeks at 15°C[ 47 ] and human umbilical vein endothelial cells encapsulated in alginate with carboxymethyl chitosan showed an 80% viability after 1 week at 4°C[ 48 ] In these studies the cells were encapsulated in crosslinked hydrogels, and previous studies investigating cold‐storage of cells concentrated on a timeframe of up to 2 weeks. In a very recent study published only this year,[ 49 ] the authors also reported that cell‐containing alginate beads can be dissolved and used for bioprinting after storage for 1 week at 15°C with a very high viability of the embedded AT‐MSC in printed constructs over 14 days; during the culture time they observed a drastic drop in cell number though. This shows that the storage of cells in crosslinked hydrogel beads with subsequent bioprinting is in principle possible, it involved a number of intermediate steps though and the choice of materials is limited. With respect to bioprinting in space, we instead aimed at an uncrosslinked ready‐to‐use bioink stored at 4°C, which is the controlled temperature for which ESA offers the largest storage space, and for up to 4 weeks, the worst‐case timeframe until launch. Based on our results for hTERT‐MSC in PBS‐Alg‐MC we hypothesize that the stabilizing effect of the surrounding matrix is also partially present when MSC are stored in uncrosslinked gels.
We were furthermore interested in whether the beneficial effects on MSC we see in the Alg‐MC blend when we supplement it with human plasma, would also be present after storage at 4°C. We assume that the cells show a generally higher metabolic activity in the Plasma‐containing Alg‐MC and a higher tendency for growth due to the high amount of supportive factors they are surrounded by.[ 20 ] While this leads to a higher viability in freshly prepared Plasma‐Alg‐MC compared to PBS‐Alg‐MC (W0 groups), in stored inks (especially W1 groups) it might have the opposite effect. Exactly this higher metabolic activity and proliferation could lead to a slower reduction of metabolic activity in response to the cold, and a large drop in viability, especially compared to the very high viability in unstored samples. Additionally, the lipid transition can result in an increase in intracellular calcium, which could lead to a stronger activation of Ca2+‐dependent proteases and phospholipases with subsequent damage to the cytoskeleton.[ 45 ] As human plasma contains ≈ 2.5 mm calcium, the PBS we used was calcium‐free though, this might also explain the stronger reduction in viability. In contrast to this, at 15°C supplementation of alginate with platelet lysate has been reported to have a beneficial effect on the viability of AT‐MSC in microbeads.[ 50 ] Nevertheless, the observed recovery in viability over the storage time indicates that after re‐warming the plasma still maintained its beneficial effect on MSC. This was also reflected in the DNA‐content of the bioprinted samples: Comparing the storage groups with the control group W0, a strong reduction in PBS‐Alg‐MC was observed whereas it remained at a similar level in Plasma‐Alg‐MC. The DNA‐measurements coupled with the viability analysis indicate a sizable number of remaining cells even after 4 weeks of storage and an additional 4 weeks of culture and thereby indicate a beneficial effect of supplementation with plasma, despite the initially higher drop in viability. The overall decrease of the DNA content with longer cultivation times (independent of storage time and observed for PBS‐Alg‐MC and–to a less extent–for Plasma‐Alg‐MC) is in line with previous findings and can likely be attributed to a lack of cellular attachment sites in the alginate itself, and to the fact that MSC generally seem to prefer stiffer surfaces to the soft hydrogel.[ 20 , 26 , 51 ] The latter is indicated by a migration of MSC from Plasma‐Alg‐MC onto calcium–phosphate cement (CPC) in biphasic scaffolds—hybrid scaffolds composed of alternating bioink and CPC strands—where the cells immediately spread out followed by a high proliferation.[ 20 ]
Considering the literature reports and our own observations in this study, so far the following factors influencing the sensitivity of mammalian cells to cold storage in hydrogels can be hypothesized: (1) the state/stiffness of the gel matrix depending on crosslinking, (2) the state of the cells prior to bringing them into the cold (attached vs suspended,[ 52 ] actively proliferating versus quiescent[ 53 ]), (3) the presence of cell‐supportive molecules (e.g., signaling factors and/or components of the extracellular matrix), and (4), considering other reports in the literature, the cell type (permanent/immortal cell lines versus primary cells and/or tissue‐specificity). We therefore additionally analyzed the influence of bioink storage specifically on the activity and function of embedded mammalian cells by using two different human tissue‐specific model cell lines: SaOS‐2 for osteoblasts and HepG2 for hepatocytes—both cell lines are derived from tumors and therefore exhibit a high metabolic activity and high proliferative capacity—as well as primary DPSC. Based on our previous studies with osteoblastic cells (human osteoblasts and osteogenically induced DPSC) and HepG2, Plasma‐Alg‐MC was chosen as ink for these experiments.[ 20 , 21 , 22 ]
Similarly, to the hTERT‐MSC, the viability of bioprinted SaOS‐2 was significantly lower in the storage groups (W1–W4) compared to the unstored control group (W0) on day 1 after printing and recovered over the cultivation period, reaching values similar to and even slightly surpassing the control W0 after 14 and 21 days of culture. However, in contrast to the hTERT‐MSC the DNA content of the SaOS‐2‐laden constructs showed a strong decrease as result of the cold storage—again, the storage (at 4°C) per se, but not the storage duration, was crucial. This strong reduction in the cell number was accompanied by a strong drop in the metabolic activity, indicated by cytosolic LDH activity measurements. When the measured LDH activity is compared with the quantified DNA content, it is evident that the relative activity of the non‐stored cells is higher than that of the stored cells. Despite this negative effect of cold storage, ALP activity was detected—using biochemical measurements as well as histological staining—which indicates maintenance of cell function of SaOS‐2 after storage. The absolute values, that is, ALP activity per scaffold, in the storage groups W1–W4 decreased over the cultivation period and were significantly lower at days 7, 14, and 21 than the absolute values in the control group W0. This can be attributed to the significantly lower amount of metabolically active cells in the cold‐stored bioinks and was confirmed by differences observed in the intensity of ALP staining in the scaffolds between W0 and W1–W4 after 21 days of cultivation. The specific ALP activity values, that is, ALP activity related to the number of metabolically active cells, also showed differences in the amount as well as in the course of cultivation time between the control group W0 and the storage groups W1–W4. Surprisingly, the values were significantly higher in the storage groups than in the control group at early time points of cultivation (days 1 and 7), but dropped already between days 7 and 14 that is rather early compared to our experience in former studies.[ 54 , 55 ] Although we cannot completely exclude that technical reasons could have had a certain influence—for instance the lysis of cells embedded in a 3D hydrogel construct is more challenging compared to cells attached on the surface of a stiff substrate—potential reasons for this observation might be the strongly differing cell density within the hydrogel constructs and the fact that the conditions during/after storage did not support cell proliferation and thus, favor cell differentiation. However, more detailed investigations, for example, using gene expression analysis, are necessary to better understand these observations.
According to their natural microenvironment in bone tissue, osteoblastic cells prefer stiff substrates—as observed in our previous study by migration of the cells out of the Plasma‐Alg‐MC hydrogel strands toward the surface of adjacent CPC strands in printed biphasic scaffolds.[ 20 ] This is probably the reason why even in the control group W0, the SaOS‐2 cells did not increase in cell number over the cultivation time of 21 days and also why a decrease in the metabolic activity from day 1 to 14 was observed. In contrast, for the HepG2‐laden constructs of the control group W0, a strong increase of both, the DNA content and the LDH activity, was observed over the cultivation period of 14 days, indicating cell proliferation and high metabolic activity of the cells within the Plasma‐Alg‐MC. This is in line with our previous study[ 22 ] and can be attributed to the fact that hepatic cells prefer and are better adapted to soft hydrogels[ 56 ] due to their natural microenvironment.[ 22 , 56 ] Interestingly, despite the good performance of the HepG2 in the control group W0, the most dramatic effect of cold storage was observed for these cells: Whereas on day 1 after printing, the cell numbers in the storage groups W1–W4 were comparable to the control group W0, very low cell numbers were detected on day 7 and 14 of cultivation. The LDH activity as metabolic marker was strongly reduced in all storage groups in comparison to the control group already at day 1 after printing and at the detection limit later on. Fluorescence microscopic analysis of cell functionality completed this picture: HepG2 formed albumin‐expressing clusters by cell proliferation, as described previously,[ 22 ] in the control group W0, but nearly no cluster formation, strongly reduced proliferation and no albumin expression was detected in the storage groups, indicating that the cells were not functional anymore after storage. Reports on the storage of isolated rat hepatocytes in suspension also indicate a strong negative effect of hypothermia, with a large drop in viability and a loss of the ability to attach even after brief periods in the cold.[ 57 ] Altogether, these data suggest that metabolically very active cells are much more affected by cold storage—at least in the Plasma‐Alg‐MC bioink—than more quiescent cells such as MSC, and that the stabilizing effect of the surrounding alginate‐based matrix is not sufficient to protect these cells.
The investigation of human DPSC‐laden bioinks based on Plasma‐Alg‐MC demonstrated that cold storage is in principle also possible with primary cells which are known to be more sensitive and less proliferative than cell lines and may be more responsive to sub‐optimal conditions in their microenvironment. On day 1 after printing, the DPSC of both donors exhibited somewhat lower viability in Plasma‐Alg‐MC in the control group W0 compared to the hTERT‐MSC and the SaOS‐2 (≈ 55 and 70% vs ≈ 90 and 80%); however, the decrease in cell viability after storage (groups W1–W4) was moderate and even less pronounced compared to the other cell types. Over the cultivation time of 28 days, the cell viability recovered completely and partially even surpassed viability in unstored samples—similar to SaOS‐2 and hTERT‐MSC. The DNA content measured for DPSC‐laden constructs of both donors was not significantly reduced due to storage (W0 vs W1–W4) on day 1 after printing, but at later time points of cultivation, significant differences were observed. Thus, DNA measurements indicate that the DPSC seemed to be more sensitive to cold storage than the hTERT‐MSC, but similar to the SaOS‐2 cell line. That is also reflected by the significantly reduced metabolic activity of DPSC after cold storage; the LDH activity measured for DPSC of both donors was in the same range as those of the SaOS‐2 cells in the control and the storage groups. Again, when the measured LDH activity is compared with the quantified DNA content, it is evident that the relative activity of the non‐stored cells is higher than that of the stored cells. These results suggest that bioinks laden with primary cells also have the potential to be stored prior to bioprinting, enabling the realization of patient‐specific approaches in space missions.
Both the cultivation of sufficient quantities of cells and the production of the hydrogel blends used for bioprinting require the completion of numerous steps of varying complexity and can therefore be assessed as very time‐intensive, especially for operations in space such as on the ISS. Due to the small number of astronauts simultaneously present on the station and the large number of different projects and experiments, which are often outside the astronauts' area of expertise, crew time is a very valuable and limited resource in space science and the experimental steps should be kept as simple as possible. Successful prior ground‐based mixing of the bioinks by experienced researchers would not only significantly reduce the workload of the crew on the station, but also make the experiments safer and less error‐prone. There would be more crew time available for working on a careful execution of the actual experimental process—the 3D bioprinting itself. In addition to the fact that this would generally lead to a lower vulnerability to errors, a prior preparation of the bioinks on Earth would transfer process steps with a possible risk of contamination from the ISS to ground, where a safer execution in fully equipped laboratories ideally set up for the processes is possible. All these factors in themselves already contribute to a more successful experiment and at the same time increase the probability of approval for flight experiments through the advantages mentioned above as well as simultaneously simplifying the proposal that needs to be submitted for every experiment through the reduction of process steps that are relevant for the space agencies in terms of safety. The results of the present study can be seen as a first and promising step toward this goal.
Future research on bioink storage is needed for a more in‐depth characterization of the metabolic activity and state of the cells during, as well as survival after, storage in dependence of different storage temperatures (Investigation of the behavior at different temperature points in the range between the storage temperature 4°C of the study and the 37°C normal cultivation temperature). Moreover, the effect of the bioink components (alg, MC, plasma) on cell metabolism and survival has to be figured out in detail by comparing storage in these (single) components versus normal cell culture medium. Further aspects to be investigated concern the impact of other biomaterial components (for instance gelatin which forms a gel < 37°C and thus, provide a matrix with cell adhesion sites) or bioink supplements (such as growth factors or nutrients) on cell survival in the stored bioink. Furthermore, the oxygen and nutrient content in the cartridge should be monitored over the storage period—for example, by using optical sensor nanoparticles for oxygen imaging, mixed into the bioink as described[ 58 ] or by determining the glucose content. Another question to be answered is the cell density in the cartridge which could have an influence on the cell survival, too. It might also be possible that cells survive better when they form spheroids, having contact to other cells. For a magnetic levitational bioassembly process at ISS, Parfenov et al. have already shown that a cell‐protective transport of living tissue spheroids embedded in a thermoreversible hydrogel is possible to bring them safely and vital to the station.[ 59 ] Even if it is unlikely due to the trouble‐free centrifugation without loss of function of the hydrogels in the lab, it remains also to be seen how the general influence of vibrations and acceleration during the rocket launch on the bioinks is. It also still has to be observed whether microgravity could possibly also have an influence on the cold‐stored cells, which would require adjustments in the storage process.
Cryopreservation of cell suspensions for storage is commonly done by cooling to temperatures far < 0°C. In this case, dimethylsulfoxide (DMSO) is required to prevent the formation of ice crystals. However, DMSO is highly toxic to active cells at 37°C, and therefore, fast removal or dilution of DMSO is an important issue than taking the cells in culture. Nevertheless, cryopreservation of bioinks prepared on ground and transported to ISS should not completely be excluded from consideration: The general possibility of using DMSO‐containing bioinks has been demonstrated very recently by Zhang and coworkers who developed a cryobioprinting strategy: Gelatin methacryloyl‐based bioinks containing DMSO and the trisaccharide melezitose as cryoprotective agents were extrusion bioprinted on a freezing plate; the incorporated cells (C2C12 cell line, human MSC, murine NIH/3T3 fibroblast cell line, Human Umbilical Vein Endothelial Cells) maintained their viability and functionality after thawing and crosslinking.[ 60 ] The applicability of temperatures < 0°C for bioink storage strongly depends on the thawing process and the temperature which is needed for bioprinting—the faster the bioink is used after defrosting, the higher the cell survival rate will be.
Another interesting field of research from which suggestions for storage of cell‐laden bioinks could be derived is the preservation of human tissues before replantation. Several studies describe improved storage conditions for, for example, teeth, or limbs like fingers, hands or even arms that can become functional again after replantation.[ 61 ] Besides perfusion of the tissues with specific protective solutions also static storage conditions are described which might be most interesting to find parallels to bioink storage. Again, the storage temperature seems to be of utmost importance here.[ 62 ]
4. Conclusion
The data presented in this work suggest that cold storage of cell‐laden bioinks over a period of several weeks is in principle possible. The composition of the biomaterial component of a bioink is a crucial determinant of the influence of cold storage on the rheological properties. However, for the Alg‐MC‐based bioinks, we have demonstrated that they maintained their excellent printability, even if the rheological properties changed due to cold storage. On the cell component of a bioink, storage at 4°C had a significant impact, however, the extent of which was strongly dependent on the cell type. The microalgae C. vulgaris, which can naturally adapt to a variety of environmental conditions, survived the storage period very well and showed no loss of growth and functionality during cultivation in the bioprinted constructs. Thus, microalgae‐laden bioinks can be excellently used to address any potential solutions such as a life support system application. As expected, mammalian cells, naturally occurring in a very specific microenvironment and more demanding, are more affected by cold storage. The observed differences in the impact of cold storage on the four mammalian cell types utilized in this study suggest that metabolically very active cells are much more influenced than more quiescent cells. Nevertheless, survival of cells was proven for all cell types investigated herein. In addition, the data suggest that the storage at low temperature per se is the critical factor in the effect of storage on the bioink properties and that the storage duration (1–4 weeks) is of less importance. Therefore, even longer storage periods could be applicable. This study is a promising starting point for further research of bioink storage. Storable bioinks would not only be advantageous for bioprinting in space but also interesting for bioprinting on Earth, making complex printing processes more effective and reproducible.
5. Experimental Section
Cell Expansion of Microalgae
C. vulgaris CCALA 269 was purchased from the Culture Collection of Autotrophic Organisms (CCALA) (Třeboň, Czec Republic). The microalgal strain was inoculated in 50 mL modified trisphosphate (TP) medium (according to[ 63 ]); the ammonium chloride (NH4Cl; Merck KGaA, Germany) originally present in the medium was substituted with 0.75 g L−1 sodium nitrate (NaNO3, Merck KGaA). Cell expansion was performed in 250 mL Erlenmeyer shake flasks (ROTILABO, Carl Roth GmbH + Co. KG) at 25°C and an illumination of 180 µmol m−2 s−1 using cool white fluorescent lamps for seven days, followed by sub‐cultivation of 5 mL into 45 mL fresh TP medium.
Cell Expansion of Human Cells
A human mesenchymal stem cell line expressing telomerase reverse transcriptase (hTERT‐MSC),[ 28 ] was kindly provided by Prof. Matthias Schieker (Laboratory of Experimental Surgery and Regenerative Medicine, Ludwig Maximilian University, Munich, Germany). The cells were cultivated in Dulbecco's modified Eagle's medium (DMEM; Gibco, Life Technologies, Germany) with 10% fetal calf serum (FCS; Corning Inc., NY, USA), 100 U mL−1 penicillin and 100 µg mL−1 streptomycin (P/S; both Gibco, Life Technologies). The human cell line SaOS‐2 (ACC‐243, DSMZ Braunschweig, Germany) was cultivated in DMEM containing 15% fetal calf serum, 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin. The human hepatocellular carcinoma cell line (HepG2), purchased from DSMZ (Braunschweig), was cultivated in DMEM containing 10% FCS, 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin (P/S; both Gibco, Life Technologies). Human primary dental pulp stem cells (DPSC), isolated from deciduous teeth as previously published,[ 64 ] were provided by the Department of Maxillofacial Surgery at University Hospital Carl Gustav Carus Dresden. The ethics commission of TU Dresden approved the application of DPSC for experiments (EK 106042010). DPSC were cultivated in DMEM containing 20% fetal calf serum, 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin. Cells in passage 5 were used for bioink preparation. All cell types were expanded in monolayer culture in T175 cell culture flasks (Sarstedt) at 37°C and 5% CO2 with change of medium twice per week.
Bioink Preparation
For the bioprinting of C. vulgaris, an alginate‐methylcellulose bioink (Alg‐MC) was prepared as described.[ 17 , 18 ] Briefly, 3 w/v% alginate (Alg; Sigma Aldrich, USA), were dissolved in double distilled water (ddH2O) and autoclaved at 121°C for 20 min; 9 w/v% methylcellulose (MC; Sigma Aldrich, USA) were autoclaved as powder at 121°C for 20 min and mixed manually with a sterile spatula to a homogeneous paste. The mixture was kept at room temperature for 2 h, allowing the MC to swell; following that 100 µL algae suspension in TP medium per gram bioink was added and homogenously manually mixed into the ink, so that the final C. vulgaris concentration was equal to 106 cells per gram bioink.
The PBS‐Alg‐MC ink was prepared by dissolving 3 w/v% alginate in phosphate buffered saline (PBS; Gibco by ThermoScientific, USA) and autoclaving this solution at 121°C for 20 min, before adding 9 w/v% of autoclaved (121°C, 20 min) methylcellulose powder and mixing it manually into the Alg‐PBS solution with a sterile spatula.[ 11 ] The blend was set to swell at room temperature for 2 h. 100 µL cell (hTERT‐MSC) suspension per gram bioink was added to the blend and mixed manually until homogeneous. The final cell concentration was 5 × 106 cells per gram bioink.
The Plasma‐Alg‐MC ink was prepared as follows:[ 20 ] alginate and methylcellulose were sterilized by autoclaving the powders separately for 20 min at 121°C. Alginate powder (3 w/v%) was freshly dissolved in fresh frozen plasma (i.e., platelet‐poor plasma prepared within 6 h of blood donation and frozen immediately for long‐term storage at −20°C) acquired from German Red Cross (BSD Ost, Dresden, Germany). To minimize the influence of donor variations, plasma of 5 different donors was pooled for all the experiments presented in this study. After dissolution, 0.7 mg of tranexamic acid (Amchafibrin, Rottapharm, Barcelona, Spain) per mg fibrinogen (2.26 mg per mL plasma) was added to prevent fibrinolysis. Afterward, 9 w/v% methylcellulose powder was added, manually mixed into the plasma‐alginate solution with a spatula until homogeneous, and stored for 2 h at room temperature, allowing the methylcellulose to swell. Further, 100 µL of a cell suspension (hTERT‐MSC, SaOS‐2, HepG2 or DPSC) per gram bioink were added and mixed into the blend with a spatula. The final cell concentration was 5 × 106 cells per gram bioink.
Bioink Storage
After the 10 mL polypropylene printing cartridges (Nordson EFD, Oberhaching, Germany) were filled with 5 g of the bioinks, they were transferred into sterile 50 mL tubes (Eppendorf, Hamburg, Germany) and stored at 4°C in a dark cold room. Between taking the cartridges out of the cold room and the bioprinting and rheological characterization of the inks, the cartridges were acclimatized to room temperature for 1 h.
Rheological Characterisation
The fresh and stored bioinks were evaluated regarding their rheological characteristics using a rheometer (Rheotest RN 4, Medingen, Germany) with a plate‐plate configuration with a distance of 0.1 mm at a constant temperature of 25°C. The dynamic viscosity was measured by applying increasing shear rates from 0 to 100 s−1 over 600 s. All measurements were performed in triplicates.
3D Bioprinting
Scaffolds were fabricated using a pneumatic‐driven multichannel extrusion 3D printer (Bioscaffolder 3.1, GeSiM mbH, Radeberg, Germany). The bioinks were extruded through gamma‐sterilized dosing needles (d = 410 µm; Globaco, Rödermark, Germany) connected to the cartridges. Scaffolds were fabricated by printing four layers with a layer‐to‐layer orientation of 90° and a strand distance of 2 mm. For bioprinting the algae‐laden scaffolds, the printing pressure, and speed used were 230–270 kPa and 4 mm s−1 with the fresh bioink, and 200230 kPa and 5–6 mm s−1 with the stored bioink. For mammalian‐cell‐laden PBS‐AlgMC, the printing parameters were 100 kPa and 7 mm s−1, and for mammalian‐cell‐laden Plasma‐AlgMC, 60 kPa and 7 mm s−1 were used. After printing, the scaffolds were crosslinked for 10 min with 1.5 mL of a 100 mm CaCl2 solution and subsequently cultured in 12‐well‐plates. Scaffolds containing the different cell types were cultured in 1.5 mL of the respective media, as described above in “cell expansion”, until further analysis. For cultivation of SaOS‐2‐containing scaffolds, the medium was supplemented with 10−7 m dexamethasone, 50 µm ascorbic acide‐2‐phosphate and 10 mm β‐glycerophosphate (all from Sigma–Aldrich, Germany) to induce the osteogenic phenotype.
Assessment of Cell Viability
The cell viability of C. vulgaris in bioprinted scaffolds was analyzed using fluorescence microscopy as previously described.[ 17 ] Briefly, dead cells were stained with SYTOX™ Green nucleic acid stain (Thermo Fisher Scientific, USA) and live cells were imaged via exploiting chlorophyll autofluorescence for visualization. Fluorescence microscopy using a BZ‐X800 fluorescent light microscope (Keyence, Japan) followed by semi‐automatic area determination of living and dead cells using ImageJ (Fiji, Version 1.52p)[ 65 ] to assess quantitative cell viability. Viability was defined as the ratio of the area of living cells divided by the area of all cells. As per the analyzed state, three scaffolds were imaged at three different positions (n = 9).
The cell viability of mammalian cells was investigated using simultaneous staining of live and dead cells with CalceinAM (BD bioscience, Heidelberg, Germany) and ethidium homodimer1 (Biotium, Fremont, USA), respectively, (LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells, Thermo Fisher Scientific) following the manufacturer's instructions. The scaffolds were imaged using a BZ‐X800 fluorescent light microscope (Keyence) and image data were analyzed using ImageJ as described above. As per the analyzed state, three scaffolds were imaged at three different positions (n = 9).
Cell Number Quantification
Chlorophyll Quantification of Microalgae: To analyze the growth of bioprinted microalgae, the chlorophyll content was determined and correlated with the cell concentration. For sampling, whole bioprinted constructs were stored at −80°C until further analysis. After thawing, 3 mL of 100 mm sodium citrate solution was added to dissolve the constructs at 4°C overnight. Dissolved samples were centrifuged at 12 000 rpm for 15 min and the supernatant was removed. The microalgal pellet was resuspended in 250 µL dimethyl sulphoxide (DMSO, Sigma) and transferred to a Precelllys tube (Peqlab, Erlangen, Germany). The suspension was frozen at −80°C for 20 min, and after thawing, 1 mL of DMSO and three ceramic Precellys beads (Peqlab) were added. In the cell homogenizer (Precelllys 24 system, Peqlab), the tube was shaken three times for 30 s at 5000 rpm. 100 µL of lysate was transferred to a transparent 96‐well plate and the optical density was measured at 435 nm using a microplate reader (Infinite M200 pro, Tecan, Switzerland). All measurements were performed in triplicates.
DNA Quantification of Human Cells: To quantify the mammalian cells, the DNA content was analyzed and correlated with the cell number. The scaffolds were transferred to falcon tubes and frozen at −80°C for at least 24 h at desired time points of cultivation. To lyse the cells in the hTERT‐MSC‐laden scaffolds, 3 mL of 100 mm sodium citrate was added per falcon tube and the scaffolds were put in a water bath for 3–4 h at 37°C. Afterward, the falcon tubes were placed into a 60°C water bath overnight to lyse the cells, followed by a sonication for 10 min on ice and a second freezing of the solutions. To lyse HepG2, SaOS‐2, and DPSC, 450 µL of Hanks Balanced Salt Solution (HBSS) was added to the frozen, cell‐laden scaffolds and transferred to the cell homogenizer (Precelllys 24 system), the tubes were shaken 2 times for 10 s at 5900 rpm. Afterward, 50 µL of a 10 v/v% Triton‐X 100 solution was added to each sample and incubated on a shaker at room temperature for 50 min. Samples were centrifuged for 5 min at 1400 rpm (Centrifuge 5415D, Eppendorf), and the supernatant was collected and stored in 1.5 mL tubes on ice. DNA quantification in the lysates was performed with the QuantiFluor dsDNA system (Promega Corporation, USA) as per the manufacturer's instructions. Briefly, QuantiFluor was diluted 1:800 with TE buffer and 190 µL of the diluted solution was added to 10 µL of cell lysate in black 96‐well‐plates. Samples were incubated for 5 min on a shaker in the dark. Fluorescence was measured at excitation and emission wavelengths of 485 and 535 nm, respectively, using a microplate reader (Infinite M200 pro). All measurements were performed in triplicates. DNA content of SaOS‐2, HepG2, and DPSC was correlated with the cell number using a calibration line.
Cell Functionality
Evaluation of Photosynthetic Activity of Microalgae: The photosynthetic efficiency of microalgae (suspended and bioprinted) was determined using pulse‐amplitude modulated fluorometry (PAM) utilizing the stationary measurement device MINI‐PAM blue (Heinz Walz GmbH, Effeltrich, Germany, equipped with a blue LED), and the control software ImagingWinGigE (Heinz Walz GmbH). The principle of the measurement was extensively described by Dani and Windisch.[ 18 ] Briefly, the microalgae‐laden scaffolds were first incubated for 5 min in the dark, followed by a treatment with saturation light pulses repeatedly for 300 s in 20 s intervals and measurement of the responsive fluorescent radiation emitted. From these measurements, the Effective Photosystem II Quantum yield (Y(II), photosynthetic activity) was calculated. Y(II) illustrates the efficiency of quantum utilization in photosystem II (PS II), a higher Y(II) value indicates that a higher percentage of the photons absorbed by PS II has been converted into chemically fixed energy usable by the cell. For comparison of photosynthetic activity within this study, the recorded Y(II) values under saturation light were averaged over a measurement and depicted as bar graphs instead of presenting the complete induction kinetic curve. In order to factor in possible variations in different areas of the analyzed scaffolds, five Areas of Interest (AOI) were set for each sample on defined positions and averaged for calculation. All measurements were performed in triplicates.
Measurement of LDH Activity in Human Cells: To evaluate the metabolic activity of SaOS‐2, HepG2, and DPSC cells, the lactate dehydrogenase (LDH) activity was determined. The cell‐laden scaffolds were lysed as previously described in “DNA quantification of human cells”. 50 µL of each lysate was transferred to a 96‐well‐plate and 50 µL of CytoTox96‐Kit substrate (Promega) was added, followed by an incubation for 30 min on a shaker at room temperature in the dark. Afterward, the reaction was stopped with 50 µL of acetic acid from the CytoTox96‐Kit, and the absorbance was measured at 490 nm, using the microplate reader (Infinite M200 pro, Tecan). All measurements were performed in triplicates.
Measurement and Staining of ALP Activity in SaOS‐2: The functionality of the SaOS‐2 cells in the bioprinted scaffolds was determined by analyzing alkaline phosphatase (ALP) activity, using a colorimetric assay to quantify the reaction of colorless p‐nitrophenylphosphate (pNpp) to yellow p‐nitrophenolate (pNp) mediated by ALP. First, SaOS‐2‐laden scaffolds were lysed as previously described in “DNA quantification of human cells”. 20 µL of each lysate was incubated with 80 µL of substrate solution (1 mg mL−1 4‐nitrophenylphosphate in 0.1 m diethanolamine, 0.1% Triton X‐100, 1 mm MgCl2, pH 9.8 − all from Sigma) for 15 min at 37°C. The enzymatic reaction was stopped with 1 m NaOH, and the absorbance was measured at 405 nm with a microplate reader (Infinite M200 pro, Tecan). A calibration line was established with different concentrations of 4‐nitrophenol. All measurements were performed in triplicates. The total ALP activity was calculated in relation to the number of metabolically active cells, obtained from LDH activity measurement of the individual lysates, to obtain the specific ALP activity.
Histological ALP staining within the scaffolds was performed on day 21 of culture using ALP staining Kit from Sigma–Aldrich (86R‐1KT) according to manufacturer's instruction but without performing the counterstain step. Briefly, samples were fixed with Citrate–Acetone–Formaldehyde and stained for 15 min with alkaline‐dye mixture containing Naphtol AS–BI phosphate, fast red violet LB base, and sodium nitrite. In between, the samples were washed with deionized water.
Fluorescence Microscopic Analysis of Proliferation and Albumin Expression in HepG2: Newly formed cells were detected through incorporation of EdU. Staining was performed using the Click‐iT EdU Cell Proliferation Kit (ThermoFisher Scientific, Germany). Briefly, the modified thymidine analogue EdU was diluted in fresh culture medium at a concentration of 1 µL mL−1. At given time points, scaffolds were incubated with the prepared solution at 37°C, 5% CO2 for 17 h. Afterward, the constructs were fixed with 4% formaldehyde (FA) in HBSS for 30 min and stored in 1% FA at 4°C until further analysis. The scaffolds were then washed with HBSS, stained for EdU with the reaction cocktail from the kit prepared according to the manufacturer's instructions, and stained for nuclei using DAPI (1:2000; Roche, Switzerland). Nuclei stained with Alexa Fluor 488 dye corresponded to newly formed cells, indicating proliferation, while DAPI stained all nuclei, though EdU‐positive nuclei only weakly. Imaging via confocal laser scanning microscopy (cLSM) was performed using a Leica TCS SP5.
Fluorescence immunostainings were performed for morphological and functional cell evaluation. At desired time points, at least two samples of cultured HepG2‐laden scaffolds were washed twice with HBSS, fixed with 4% FA for 30 min, and stored in 1% FA at 4°C until staining. The cells were then permeabilized with 1v/v% TritonX‐100 in HBSS for 15 min and blocked with 3 w/v% bovine serum albumin (BSA; Albumin Fraction V, Roth, Germany) in HBSS for 1 h at room temperature. Scaffolds were incubated overnight at 4°C with rabbit anti‐human serum albumin primary antibody diluted 1:100 in 0.5% BSA and 0.2 v/v% TritonX‐100 The secondary antibody, Alexa 594‐tagged anti‐rabbit IgG (1:200), was added together with Phalloidin iFluor 488 reagent to stain cytoskeletal F‐actin filaments (1:1000) and DAPI to stain the nuclei (1:1000), diluted in 1 wt/v% BSA in HBSS. Scaffolds were incubated with the secondary antibody for 1 h. Between all steps of the staining protocol, scaffolds were washed 3x with HBSS. Imaging via cLSM was performed using a Leica TCS SP5.
Statistics
Data analysis and statistical evaluation were carried out with the software GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). One‐way analysis of variance (ANOVA) with Dunnett's comparison test was performed to analyze the statistical significance (p < 0.05) of the viscosity of the bioinks (Figures 2B and 4B,D) and the photosynthetic efficiency (Figure 3C). Statistical significance of all other data was determined by performing a two‐way ANOVA with Dunnett's comparison test considering a 95% confidence interval.
Ethics Approval Statement
Human DPSC, isolated from wisdom teeth, were used for the experiments after informed consent of patients, as approved by the ethics committee of Technische Universität Dresden (EK 106042010).
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Acknowledgements
This study was motivated by intense discussions with several experts of the European Space Research and Technology Centre (ESTEC) of the European Space Agency (ESA), especially Leonardo Surdo and Dr. Angelique Van Ombergen, as well as by discussions within the ESA Topical Team on 3D Bioprinting of living tissue for utilization in space exploration and extraterrestrial human settlements. The authors thank Carola Petto (Department of Oral and Maxillofacial Surgery, University Hospital Dresden) for providing the DPSC as well as Anna–Maria Placht for excellent technical assistance. The authors were grateful for the support regarding the expertise and equipment of the Core Facility Cellular Imaging (CFCI) of the Faculty of Medicine Carl Gustav Carus at TU Dresden. The authors were grateful to the German Space Agency at DLR for the financial support of the project (grant number 50WB2032).
Open access funding enabled and organized by Projekt DEAL.
Windisch J., Reinhardt O., Duin S., Schütz K., Rodriguez N. J. N., Liu S., Lode A., Gelinsky M., Bioinks for Space Missions: The Influence of Long‐Term Storage of Alginate‐Methylcellulose‐Based Bioinks on Printability as well as Cell Viability and Function. Adv. Healthcare Mater. 2023, 12, 2300436. 10.1002/adhm.202300436
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Inokuchi H., Fukui K., Kogure K., Takaoki M., Kinoshita K., Izumi R., Fujimori Y., Planning Guide for Space Experiment Research, Japan Aerospace Exploration Agency, Japan Space Forum, 2007.
- 2. Groll J., Burdick J. A., Cho D.‐W., Derby B., Gelinsky M., Heilshorn S. C., Jüngst T., Malda J., Mironov V. A., Nakayama K., Ovsianikov A., Sun W., Takeuchi S., Yoo J. J., Woodfield T. B. F., Biofabrication 2018, 11, 013001. [DOI] [PubMed] [Google Scholar]
- 3. Kilian D., Sembdner P., Bretschneider H., Ahlfeld T., Mika L., Lützner J., Holtzhausen S., Lode A., Stelzer R., Gelinsky M., Bio‐Des. Manuf. 2021, 4, 818. [Google Scholar]
- 4. Santoni S., Gugliandolo S. G., Sponchioni M., Moscatelli D., Colosimo B. M., Bio‐Des. Manuf. 2022, 5, 14. [Google Scholar]
- 5. Mobaraki M., Ghaffari M., Yazdanpanah A., Luo Y., Mills D. K., Bioprinting 2020, 18, e00080. [Google Scholar]
- 6. Krujatz F., Dani S., Windisch J., Emmermacher J., Hahn F., Mosshammer M., Murthy S., Steingröwer J., Walther T., Kühl M., Gelinsky M., Lode A., Biotechnol. Adv. 2022, 58, 107930. [DOI] [PubMed] [Google Scholar]
- 7. Cubo‐Mateo N., Podhajsky S., Knickmann D., Slenzka K., Ghidini T., Gelinsky M., Biofabrication 2020, 12, 043001. [DOI] [PubMed] [Google Scholar]
- 8. Ghidini T., J. Thorac. Dis. 2018, 10, S2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Placone J. K., Engler A. J., Adv. Healthcare Mater. 2018, 7, 1701161. [Google Scholar]
- 10. Malda J., Visser J., Melchels F. P., Jüngst T., Hennink W. E., Dhert W. J. A., Groll J., Hutmacher D. W., Adv. Mater. 2013, 25, 5011. [DOI] [PubMed] [Google Scholar]
- 11. Schütz K., Placht A.‐M., Paul B., Brüggemeier S., Gelinsky M., Lode A., J. Tissue Eng. Regener. Med. 2017, 11, 1574. [DOI] [PubMed] [Google Scholar]
- 12. Hodder E., Duin S., Kilian D., Ahlfeld T., Seidel J., Nachtigall C., Bush P., Covill D., Gelinsky M., Lode A., J. Mater. Sci.: Mater. Med. 2019, 30, 10. [DOI] [PubMed] [Google Scholar]
- 13. Duin S., Schütz K., Ahlfeld T., Lehmann S., Lode A., Ludwig B., Gelinsky M., Adv. Healthcare Mater. 2019, 8, 1801631. [DOI] [PubMed] [Google Scholar]
- 14. Duin S., Bhandarkar S., Lehmann S., Kemter E., Wolf E., Gelinsky M., Ludwig B., Lode A., Biomedicines 2022, 10, 1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kilian D., Ahlfeld T., Akkineni A. R., Bernhardt A., Gelinsky M., Lode A., Sci. Rep. 2020, 10, 8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kilian D., Cometta S., Bernhardt A., Taymour R., Golde J., Ahlfeld T., Emmermacher J., Gelinsky M., Lode A., Biofabrication 2022, 14, 014108. [DOI] [PubMed] [Google Scholar]
- 17. Lode A., Krujatz F., Brüggemeier S., Quade M., Schütz K., Knaack S., Weber J., Bley T., Gelinsky M., Eng. Life Sci. 2015, 15, 177. [Google Scholar]
- 18. Dani S., Windisch J., Valencia Guerrero X. M., Bernhardt A., Gelinsky M., Krujatz F., Lode A., Front. Bioeng. Biotechnol. 2022, 10, 994134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu S., Kilian D., Ahlfeld T., Hu Q., Gelinsky M., Biofabrication 2023, 15, 025013. [DOI] [PubMed] [Google Scholar]
- 20. Ahlfeld T., Cubo‐Mateo N., Cometta S., Guduric V., Vater C., Bernhardt A., Akkineni A. R., Lode A., Gelinsky M., ACS Appl. Mater. Interfaces 2020, 12, 12557. [DOI] [PubMed] [Google Scholar]
- 21. Taymour R., Kilian D., Ahlfeld T., Gelinsky M., Lode A., Sci. Rep. 2021, 11, 5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taymour R., Chicaiza‐Cabezas N. A., Gelinsky M., Lode A., Biofabrication 2022, 14, 045019. [DOI] [PubMed] [Google Scholar]
- 23. Dani S., Ahlfeld T., Albrecht F., Duin S., Kluger P., Lode A., Gelinsky M., Gels 2021, 7, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kidder L. S., Zea L., Countryman S. M., Stodieck L. S., Hammer B. E., Gravit. Space Res. 2020, 8, 25. [Google Scholar]
- 25. Kacena M. A., Todd P., Landis W. J., In Vitro Cell. Dev. Biol. Anim. 2003, 39, 454. [DOI] [PubMed] [Google Scholar]
- 26. Ahlfeld T., Cidonio G., Kilian D., Duin S., Akkineni A. R., Dawson J. I., Yang S., Lode A., Oreffo R. O. C., Gelinsky M., Biofabrication 2017, 9, 034103. [DOI] [PubMed] [Google Scholar]
- 27. Klughammer C., Schreiber U., J. PAM Applikation Notes 2008, 1, 27. [Google Scholar]
- 28. Böcker W., Yin Z., Drosse I., Haasters F., Rossmann O., Wierer M., Popov C., Locher M., Mutschler W., Docheva D., Schieker M., J. Cell. Mol. Med. 2008, 12, 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cubo N., Garcia M., del Cañizo J. F., Velasco D., Jorcano J. L., Biofabrication 2016, 9, 015006. [DOI] [PubMed] [Google Scholar]
- 30. Li H., Tan Y. J., Leong K. F., Li L., ACS Appl. Mater. Interfaces 2017, 9, 20086. [DOI] [PubMed] [Google Scholar]
- 31. Emmermacher J., Spura D., Cziommer J., Kilian D., Wollborn T., Fritsching U., Steingroewer J., Walther T., Gelinsky M., Lode A., Biofabrication 2020, 12, 025022. [DOI] [PubMed] [Google Scholar]
- 32. Sannino A., Demitri C., Madaghiele M., Materials 2009, 2, 353. [Google Scholar]
- 33. Nasatto P. L., Pignon F., Silveira J. L. M., Duarte M. E. R., Noseda M. D., Rinaudo M., Polymers 2015, 7, 77. [Google Scholar]
- 34. Ahlfeld T., Guduric V., Duin S., Akkineni A. R., Schütz K., Kilian D., Emmermacher J., Cubo‐Mateo N., Dani S., von Witzleben M., Spangenberg J., Abdelgaber R., Richter R. F., Lode A., Gelinsky M., Biomater. Sci. 2020, 8, 2102. [DOI] [PubMed] [Google Scholar]
- 35. Ahlfeld T., Köhler T., Czichy C., Lode A., Gelinsky M., Gels 2018, 4, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coughlin M. L., Liberman L., Ertem S. P., Edmund J., Bates F. S., Lodge T. P., Prog. Polym. Sci. 2021, 112, 101324. [Google Scholar]
- 37. Heymann E., Trans. Faraday Soc. 1935, 31, 846. [Google Scholar]
- 38. Eskens O., Villani G., Amin S., Cosmetics 2021, 8, 3. [Google Scholar]
- 39. Thakare K., Jerpseth L., Qin H., Pei Z., J. Manuf. Sci. Eng. 2020, 143, 014501. [Google Scholar]
- 40. Krujatz F., Lode A., Brüggemeier S., Schütz K., Kramer J., Bley T., Gelinsky M., Weber J., Eng. Life Sci. 2015, 15, 678. [Google Scholar]
- 41. Revellame E. D., Aguda R., Chistoserdov A., Fortela D. L., Hernandez R. A., Zappi M. E., Algal Res. 2021, 55, 102258. [Google Scholar]
- 42. Maharjan S., Alva J., Cámara C., Rubio A. G., Hernández D., Delavaux C., Correa E., Romo M. D., Bonilla D., Santiago M. L., Matter 2021, 4, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Richmond A., Handbook of Microalgal Culture: Biotechnology and Applied Phycology, Blackwell, Oxford, OX, UK : 2004. [Google Scholar]
- 44. de Morais M. G., Vaz B. d. S., de Morais E. G., Costa J. A. V., Biomed Res. Int. 2015, 2015, e835761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rubinsky B., Heart Failure Rev. 2003, 8, 277. [DOI] [PubMed] [Google Scholar]
- 46. Roobol A., Carden M. J., Newsam R. J., Smales C. M., FEBS J. 2009, 276, 286. [DOI] [PubMed] [Google Scholar]
- 47. Swioklo S., Constantinescu A., Connon C. J., Stem Cells Transl. Med. 2016, 5, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X., Cao Y., Zhao G., Biopreserv. Biobanking 2020, 18, 305. [DOI] [PubMed] [Google Scholar]
- 49. Kostenko A., Connon C. J., Swioklo S., Bioengineering 2022, 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Branco A., Tiago A. L., Laranjeira P., Carreira M. C., Milhano J. C., dos Santos F., Cabral J. M. S., Paiva A., da Silva C. L., Fernandes‐Platzgummer A., Bioengineering 2022, 9, 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Andersen T., Auk‐Emblem P., Dornish M., Microarrays 2015, 4, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hunt L., Hacker D. L., Grosjean F., De Jesus M., Uebersax L., Jordan M., Wurm F. M., Biotechnol. Bioeng. 2005, 89, 157. [DOI] [PubMed] [Google Scholar]
- 53. Nelson R. J., Kruuv J., Exp. Cell Res. 1972, 70, 417. [DOI] [PubMed] [Google Scholar]
- 54. Bernhardt A., Lode A., Peters F., Gelinsky M., Clin. Oral Implants Res. 2013, 24, 441. [DOI] [PubMed] [Google Scholar]
- 55. Bernhardt A., Dittrich R., Lode A., Despang F., Gelinsky M., J. Mater. Sci.: Mater. Med. 2013, 24, 1755. [DOI] [PubMed] [Google Scholar]
- 56. Aparnathi M. K., Patel J. S., Biosci. Biotech. Res. Comm. 2016, 9, 463. [Google Scholar]
- 57. Pless‐Petig G., Singer B. B., Rauen U., PLoS One 2012, 7, e40444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trampe E., Koren K., Akkineni A. R., Senwitz C., Krujatz F., Lode A., Gelinsky M., Kühl M., Adv. Funct. Mater. 2018, 28, 1804411. [Google Scholar]
- 59. Parfenov V., Petrov S., Pereira F., Levin A., Koudan E., Nezhurina E., Karalkin P., Vasiliev M., Petrov O., Komlev V., Khesuani Y. D., Mironov V., Int. J. Bioprint. 2020, 6, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ravanbakhsh H., Luo Z., Zhang X., Maharjan S., Mirkarimi H. S., Tang G., Chávez‐Madero C., Mongeau L., Zhang Y. S., Matter 2022, 5, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ozer K., J. Hand Surg. 2020, 45, 626. [DOI] [PubMed] [Google Scholar]
- 62. Lin C.‐H., Aydyn N., Lin Y.‐T., Hsu C.‐T., Lin C.‐H., Yeh J.‐T., Ann. Plast. Surg. 2010, 64, 286. [DOI] [PubMed] [Google Scholar]
- 63. Hoober J. K., Science 1989, 246, 1503. [DOI] [PubMed] [Google Scholar]
- 64. Neunzehn J., Weber M.‐T., Wittenburg G., Lauer G., Hannig C., Wiesmann H.‐P., Head Face Med. 2014, 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schindelin J., Arganda‐Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.‐Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Nat. Methods 2012, 9, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


