Abstract
Aleutian mink disease parvovirus (ADV) causes a persistent infection associated with circulating immune complexes, immune complex disease, hypergammaglobulinemia, and high levels of antiviral antibody. Although antibody can neutralize ADV infectivity in Crandell feline kidney cells in vitro, virus is not cleared in vivo, and capsid-based vaccines have proven uniformly ineffective. Antiviral antibody also enables ADV to infect macrophages, the target cells for persistent infection, by Fc-receptor-mediated antibody-dependent enhancement (ADE). The antibodies involved in these unique aspects of ADV pathogenesis may have specific targets on the ADV capsid. Prominent differences exist between the structure of ADV and other, more-typical parvoviruses, which can be accounted for by short peptide sequences in the flexible loop regions of the capsid proteins. In order to determine whether these short sequences are targets for antibodies involved in ADV pathogenesis, we studied heterologous antibodies against several peptides present in the major capsid protein, VP2. Of these antibodies, a polyclonal rabbit antibody to peptide VP2:428-446 was the most interesting. The anti-VP2:428-446 antibody aggregated virus particles into immune complexes, mediated ADE, and neutralized virus infectivity in vitro. Thus, antibody against this short peptide can be implicated in key facets of ADV pathogenesis. Structural modeling suggested that surface-exposed residues of VP2:428-446 are readily accessible for antibody binding. The observation that antibodies against a single target peptide in the ADV capsid can mediate both neutralization and ADE may explain the failure of capsid-based vaccines.
The interactions between virus and antiviral antibodies play a crucial role in the pathogenesis of Aleutian mink disease parvovirus (ADV) infections (4, 15, 18, 51). Adult mink infected with pathogenic isolates of ADV develop a persistent infection associated with high levels of antiviral antibodies and hypergammaglobulinemia (4, 15, 17, 18, 51). In spite of this robust immune response, virus is not eliminated in vivo (15, 30, 33, 49) and severe immune complex disease and vasculitis develop (51, 53). In fact, complexes containing infectious virus have been demonstrated, denoting the direct involvement of antiviral antibody in this syndrome (50). Furthermore, antiviral antibody enables ADV to infect cells such as macrophages or the monocytic cell line, K562, via an Fc-receptor-dependent mechanism termed antibody-dependent enhancement (ADE) of infection (29, 35). Macrophages are the target cells for persistent ADV infection in vivo, and their infection may play a role in the genesis of the immune disorder (15, 34, 36, 42). Finally, as might be anticipated from these observations, vaccination of mink or the presence of preexisting antiviral antibody does not protect adult mink from ADV infection but, rather, leads to an accelerated form of disease upon challenge (1, 52).
Antiviral antibodies in some circumstances can also play a beneficial role in ADV infections. For example, antibody is able to neutralize ADV infectivity for Crandell feline kidney (CrFK) cells in vitro (1, 35, 59). In addition, antiviral antibody has a mitigating effect on ADV infection in mink kits (2, 10, 11), where presence of natural or passively administered antibody prevents the fulminant, fatal pneumonitis associated with the permissive infection of type II alveolar cells by ADV (9, 10, 15). The mechanism for this effect is unclear, although at the level of the individual cell, the antibody converts permissive infection into a restricted infection (10, 11).
ADV infections stand in sharp contrast to infections of mink with another nondefective parvovirus, mink enteritis virus (MEV), which is a viral host range variant of feline panleukopenia virus (46, 47, 48). Capsid-based vaccines against MEV promptly induce neutralizing antibody and prevent infection and disease (22, 39). In addition, persistent infections do not develop. Consequently, the atypical picture observed during ADV infections cannot be ascribed simply to a generic response of mink to parvoviruses.
The ADV capsid consists of 60 individual capsid proteins. In native capsids from ADV-infected cells, ca. 90% is the 647-amino-acid major capsid protein, VP2 (8, 23). The minor capsid protein, VP1, contains the entire VP2 sequence but has 43 additional unique residues at the N terminus (8, 23, 24, 63). When VP2 from the ADV-G isolate is expressed in either recombinant vaccinia viruses (24) or baculoviruses (23, 63), the proteins assemble into empty capsids. Recent work with prokaryotic expression vectors has localized immunodominant targets for the antibody response to specific regions of the VP2 capsid protein (16, 28). The most immunoreactive region spans VP2 residues 429 to 524 (VP2:429-524) (16, 28). Polyclonal rabbit antibodies directed against this region neutralize ADV infectivity for CrFK cells and strongly react with capsids in immunoelectron microscopy (16). Infected mink also recognize a major epitope in VP2:428-446, a sequence contained within this region (16, 28).
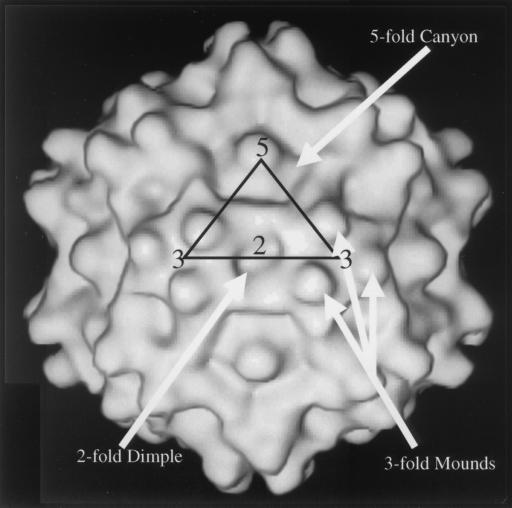
Taken together, these findings from antigenic mapping and pathogenesis studies indicate the importance of understanding the capsid structure. A three-dimensional model of the T=1 ADV capsid has been built into electron density determined to 22 Å resolution by cryoelectron microscopy and image reconstruction (Fig. 1) (41). Although the structure shares canonical features with more prototypical parvoviruses, such as canine parvovirus (CPV), feline panleukopenia virus, and minute virus of mice (5, 7, 41, 61), several prominent differences are evident (Fig. 1). Most notably, there are three knob-like mounds decorating the viral icosahedral threefold axes of symmetry and wider ridges that separate the dimple-like depression at the icosahedral twofold axes from the canyon-like depression surrounding the icosahedral fivefold axes. The differences can largely be accounted for by short stretches of amino acids inserted into the flexible loop segments of the VP2 protein molecules in ADV compared to all other parvoviruses (16, 20, 41). Several of these insertions are located in the highly immunoreactive VP2:429-524 segment. The additional peptide sequences may be potential binding sites for antibodies involved in ADV pathogenesis.
FIG. 1.
Shaded surface representation of the ADV-GVP2 three-dimensional reconstruction viewed perpendicular to a twofold icosahedral axis, obtained by the method of McKenna et al. (41). The following features are depicted on the reconstruction: twofold (“2”), threefold (“3”), and fivefold (“5”) icosahedral axes; the mounds adjacent to the threefold axis (3-fold mounds); the dimple or depression at the twofold axis (2-fold dimple); and the canyon surrounding the fivefold axis (5-fold canyon). An asymmetric icosahedral unit is superimposed on the structure as a triangle. The resolution of the image is 22 Å.
By in vitro monitoring of immune complex formation, virus neutralization in CrFK cells, and ADE in K562 cells as model systems for the involvement of antibody in ADV pathogenesis, several questions may be formulated. First, are the same antigenic determinants of ADV implicated in these phenomena? Second, at what location on the virus particle do these determinants reside? Third, do specific structural features on the ADV capsid contribute to pathogenicity and the immune response it engenders?
To address these questions, we prepared rabbit polyclonal and mouse monoclonal antibodies against several peptides contained within the VP2:429-524 portion of VP2. We have characterized these antibodies with respect to their capacity to participate in vitro in immune complex formation, to neutralize infectivity, and to mediate ADE. Our results suggested that antibodies to a short sequence in the VP2 capsid protein can be implicated in all three phenomena. Other antibodies were able to mediate ADE but failed to neutralize virus or generate immune complexes. These findings indicate that antibodies to specific capsid sequences are important in the pathogenesis of ADV infections. The observation that the same amino acid sequence can host both virus neutralization and ADE may explain the inability of capsid-based vaccines to protect against ADV infections in adult animals.
MATERIALS AND METHODS
Viruses and cells.
Crandell feline kidney cells (CrFK), K562 cells and the Spodoptera frugiperda insect cell line (Sf9) were maintained according to previously reported methods (14, 29, 41, 63). The procedure for propagating the molecularly cloned, cell culture-adapted ADV-G has also been described (14). For production of recombinant ADV capsids containing only VP2 (ADV-GVP2), Sf9 cells were infected with recombinant baculovirus in suspension cultures at a multiplicity of 1 PFU/cell for 72 h (41, 63).
Capsids were isolated as noted earlier by using chloroform extraction and 10% polyethylene glycol 8000 (PEG 8000) precipitation with slight modifications (41). Specifically, particles precipitated with PEG 8000 were dissolved in 50 mM Tris (pH 8.0)–100 mM NaCl and were passed through a 0.45-μm (pore size) filter. A commercially available cocktail of protease inhibitors in tablet form (Boehringer Mannheim Gmbh, Mannheim, Germany) was employed during isolation and storage to minimize proteolysis.
For immunoelectron microscopy, recombinant ADV-GVP2 capsids or native ADV-G virions were additionally purified over discontinuous step gradients of iodixanol (OptiPrep; Nycomed) (64). Particles were detected at an approximate density (ρ = 2.24 g/ml) similar to the density reported for recombinant adeno-associated virus particles isolated by the same procedure (64).
Peptides.
Peptides, based on the ADV-G VP2 capsid protein sequence (16, 43) and conjugated to keyhole limpet hemocyanin through an additional N-terminal cysteine, were obtained from a commercial source (Princeton Biomolecules, Columbus, Ohio) and characterized by the Structural Biology Section of the National Institute of Allergy and Infectious Diseases (NIAID) (27). Purity was estimated to be >75%. The peptide designations and amino acid sequences were as follows: VP2:428-446, (C)SNYYSDNEIEQHTAKQPL (28); VP2:455-470, (C)KIDSWEEGWPAASGTH; and VP2:487-501, (C)EQELNFPHEVLDAA. The peptides were dissolved in phosphate-buffered saline (PBS) at 0.5 mg/ml. The VP2:429-524 region is contained in a prokaryotic fusion protein (16).
Production of polyclonal and monoclonal antibodies.
Heterologous polyclonal antipeptide antisera were prepared in rabbits according to a published protocol (27).
Monoclonal antibodies against the VP2:428-446 and VP2:487-501 peptides conjugated to keyhole limpet hemocyanin were generated at the Saint Louis University Hybridoma Development Service Facility according to standard methods (27). Upon isotype testing (IsoStrip Mouse Monoclonal Antibody Isotyping Kit; Roche Diagnostics Corp., Indianapolis, Ind.), the monoclonal antibodies were found to be immunoglobulin G1 (IgG1) and failed to bind protein A (data not shown). This result is consistent with known characteristics of murine IgG1 antibodies (26). IgG concentrations were estimated by an immunoenzyme assay (Boehringer Mannheim). In addition, all of the antibodies reacted specifically in enzyme-linked immunosorbent assay (ELISA) against their cognate peptides (data not shown).
In order to effect higher antibody concentrations, the hybridoma cell lines 282.20.1.4 (anti-VP2:428-446) and 281.12.1.7 (anti-VP2:487-501) were cultured in Integra CELLine CL 350 flasks (Integra Biosciences, Inc., Ijamsville, Md.) which yielded monoclonal antibody IgG1 concentrations in excess of 1 mg/ml.
Immune blotting.
Immune blots were performed by using lysates of ADV-G-infected CrFK cells as antigen (16). The blocked membranes were clamped into a multichannel manifold, and individual primary antibodies and developing reagents were applied to separate channels. The following slight modifications yielded very low backgrounds; blocking and dilution of primary and secondary antibodies was in PBS–0.5% Tween 20–5% nonfat dry milk. Signals from rabbit sera and mouse monoclonal antibody samples were developed by using, respectively, horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG or HRP-conjugated rabbit anti-mouse immunoglobulins.
Immune electron microscopy.
The ability of antibodies to bind native ADV-G or ADV-GVP2 capsids immobilized on Parlodion-coated grids was evaluated precisely as detailed in an earlier study (16). Antibody that decorated virus was revealed by use of goat anti-rabbit IgG or rabbit anti-mouse IgG, both conjugated to 5-nm gold particles. Reactions were graded as follows: −, negative; +, <10% of the capsids had 1 or more gold particles; ++, 30 to 70% of the capsids decorated with fewer gold particles per capsid; +++, heavy gold particle decoration of >90% of capsids.
To determine the ability of antibodies to aggregate virus particles and produce immune complexes, capsids were incubated with dilutions of serum (1/100) or monoclonal antibody (100 μg/ml) for 2 h at room temperature. Samples (5 μl) were applied to Parlodion-coated 300-mesh copper grids and allowed to adsorb for 4 h at room temperature in a humid chamber. After a wash with deionized H2O, the samples were stained with 1% ammonium molybdate. The electron microscopy methods applied here have been previously described (16).
ADE of infection.
The method for monitoring ADE was modified slightly from previous reports (29, 35). Briefly, individual wells of a 24-well culture plate were seeded with 5 × 105 K562 cells in 0.25 ml of complete medium. Equal volumes (0.4 ml) of serial 10-fold dilutions of sera or monoclonal antibodies were incubated at 37°C for 1 h with a standard dilution of ADV-G. The mixture was added to the cells and cultured for 72 h at 31.8°C. At this time, 5 × 104 cells were cytocentrifuged onto slides, acetone fixed, and stained for ADV antigens with fluorescein isothiocyanate (FITC)-conjugated IgG from ADV-infected mink. The number of positive cells was counted and compared to control cultures receiving ADV mixed with medium alone. No enhancement was observed when normal serum or an isotype-matched monoclonal antibody was used.
Virus neutralization.
The assay for antibody neutralization of ADV-G infectivity for CrFK cells was performed as detailed in earlier work (16). Any serum dilution that caused a >70% reduction in the number of ADV-positive cells was considered positive.
IFA.
Indirect immunofluorescence assay (IFA) was done on acetone-fixed cytocentrifuged samples of CrFK cells, either uninfected or infected 72 h previously with ADV-G (44). The secondary antibodies were FITC-conjugated goat anti-rabbit IgG or FITC-conjugated rabbit anti-mouse immunoglobulins. After the final wash, a drop of propidium iodide was placed on the cells for 10 s in order to delineate the nucleus. Images were captured by laser scanning confocal microscopy as reported elsewhere (44).
Structural modeling.
An isodensity map of the ADV-GVP2 capsid derived by cryoelectron microscopy, image reconstruction, and comparison with the CPV capsid atomic structure has been reported (41). In an effort to evaluate possible antibody interactions of these ADV peptide regions, Fab fragments were docked into approximate binding sites by using available lysozyme-Fab complex structures as a guide (45). Specifically, antigenic epitopes of lysozyme in the complexes were superimposed onto the ADV peptides to position the Fab fragments at allowable antigen-antibody interaction distances and orientations relative to antigenic sites (21, 56, 58). The lysozyme coordinates were then deleted from the complexes, leaving the Fab in contact with the ADV capsid.
RESULTS
Immunoblot analysis.
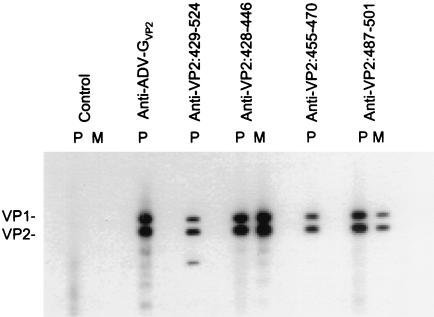
To evaluate reactivity of the antipeptide antibodies against the full-length ADV capsid proteins, antibodies were tested in immunoblot against lysates of ADV-G-infected CrFK cells (Fig. 2). All three polyclonal sera reacted against both of the overlapping capsid proteins, VP1 and VP2, as did the anticapsid serum. The monoclonal antibodies against VP2:428-446 and VP2:487-501 also recognized both capsid proteins. The polyclonal antibody against the VP2:429-524 expression protein (16) also reacted with VP1 and VP2 and, in addition, recognized a smaller protein that is likely a degradation product (3). All antibodies reacted specifically with their cognate peptides by ELISA (data not shown). Thus, immunization of rabbits and mice with the peptides induced antibodies that were reactive with denatured full-length capsid proteins.
FIG. 2.
Immunoblot with antibodies prepared against ADV VP2 peptides. The indicated rabbit polyclonal (P) or mouse monoclonal (M) antibody preparations were reacted in immunoblot against a whole-cell lysate of ADV-G-infected CrFK cells. The rabbit polyclonal sera were used at a 1/100 dilution, except for the anti-ADV-G capsid serum, which was used at 1/1,000; the mouse monoclonal antibodies were at 1 μg/ml. The positions of the ADV capsid proteins VP1 and VP2 are marked.
Immunoelectron microscopy.
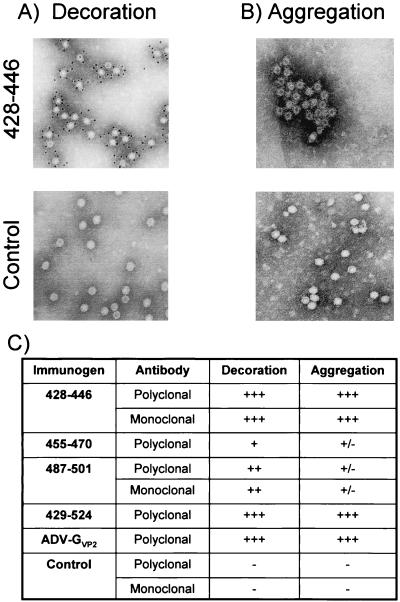
Antiviral antibody interacts with ADV in animals to form immune complexes, which are a key feature in the pathogenesis (15, 51, 53). To determine whether the antipeptide antibodies can bind and aggregate capsids, we used immunoelectron microscopy. Antibodies were assessed for their ability both to decorate immobilized capsids and to aggregate particles, the latter being a surrogate measure for immune complex formation (Fig. 3). Both the polyclonal and monoclonal anti-VP2:428-446 antibodies bound densely to immobilized capsids, as evidenced by the heavy decoration with gold-labeled secondary reagents (Fig. 3A). Furthermore, these antibodies aggregated soluble capsids to produce obvious immune complexes (Fig. 3B), in which antibody bridging was noted and the capsid margins were indistinct. Some of these complexes were very large, incorporating hundreds of particles.
FIG. 3.
Immunoelectron microscopy with antibodies prepared against ADV VP2 peptides. Antibody preparations were tested for the capacity to react with ADV-GVP2 capsids either in decoration or aggregation and were scored as detailed in Materials and Methods. (A and B) Decoration (A) and aggregation (B), both at +++ levels (anti-VP2:428-446 antibodies) and at negative levels (control antibodies) are depicted. (C) Tabulation of results from all sera tested. No difference was noted between rabbit polyclonal (used at a 1/100 dilution) or mouse monoclonal (used at 100 μg/ml) antibodies directed against the same peptide.
The anti-VP2:455-470 polyclonal antibody labeled <10% of the capsids, and the number of gold particles per capsid was low (Fig. 3C). Small aggregates of particles were noted when capsids were incubated in solution with the anti-VP2:455-470 antibody, although evidence of antibody bridging was not prominent (Fig. 3C).
The monoclonal and polyclonal antibodies directed against VP2:487-501 decorated ca. 40% of the virions, but the number of gold particles per capsid was less than observed with the anti-VP2:428-446 antibodies (Fig. 3C). However, when the anti-VP2:487-501 antibodies were used to aggregate capsids, only a few small clumps were noted and capsid bridging was not prominent (Fig. 3B). The polyclonal anticapsid antibody and antibody directed against VP2:429-524 strongly decorated and complexed particles to a level similar to the anti-VP2:428-446 antibodies (Fig. 3C).
The negative control polyclonal and monoclonal antibodies were unreactive (Fig. 3A and B).
Thus, antibodies directed against VP2:428-446 decorated and aggregated as efficiently as antibodies directed against the intact capsid. The high level of aggregation suggested that the anti-VP2:428-446 antibodies might be able to participate in immune complex formation in vivo.
Antibody-dependent enhancement of infection.
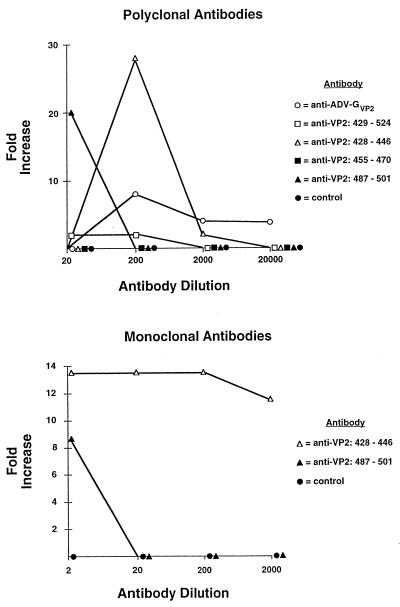
Antiviral antibody can mediate ADV infection of the monocytic cell line, K562, via the FcγRII receptor (29). This phenomenon of ADE of infection may play a role in the immunopathogenesis of ADV infections in adult mink (15, 29, 35, 36). To determine if antibodies to specific regions of the capsid could enable Fc-receptor-dependent ADE, we assayed the capacity of the antipeptide rabbit polyclonal and mouse monoclonal antibodies to mediate ADE (Fig. 4).
FIG. 4.
Antibody-dependent enhancement of ADV infection for K562 cells mediated by antibodies prepared against ADV VP2 peptides. Dilutions of the indicated rabbit polyclonal and murine monoclonal antibodies (1 mg/ml, undiluted) were incubated with ADV-G at 37°C for 1 h. The mixture was added to K562 cells. After culture for 72 h at 31.8°C, 5 × 104 cells were cytocentrifuged onto slides, acetone fixed, and stained for ADV antigens. The number of positive cells was counted and compared to control cultures receiving ADV mixed with medium alone.
Antibodies prepared against the peptide VP2:428-446 clearly enhanced ADV infection of K562 cells (Fig. 4) in a dose-dependent manner. In representative experiments, the rabbit polyclonal and mouse monoclonal antibodies increased the number of infected cells by as much as 26- and 14-fold, respectively.
Polyclonal and monoclonal antibodies prepared against the peptide VP2:487-501 also mediated ADE. However, the effect was noted only at the most concentrated dilution assayed (Fig. 4). The anti-ADV-GVP2 rabbit sera enhanced infection of K562 cells, but to a lower extent than the anti-VP2:428-446 and anti-VP2:487-501 antibodies (Fig. 4). The polyclonal antibody against VP2:455-470 showed no detectable capacity to enhance ADV infection of K562 cells (Fig. 3). Control rabbit and isotype-matched control mouse monoclonal antibody did not enhance infection (data not shown). These findings revealed that antibodies directed against at least two short VP2 sequences could mediate ADE. The fact that monoclonal antibodies were also effective indicated that reactivity with multiple epitopes was not required.
In vitro virus neutralization assay.
Although ADV is not eliminated from infected adult animals, antibody can mitigate the severity of ADV infections in mink kits and neutralize infectivity for CrFK cells in vitro (2, 11, 16, 59). Our previous studies show that in vitro neutralizing determinants are present in the segment spanning residues, i.e., VP2:429-524 (16). To extend this work, we tested the antibodies generated against smaller peptides contained within this region (Table 1). The rabbit polyclonal antibodies prepared against both capsids and VP2:429-524 and VP2:428-446 peptide neutralized ADV-G. No other polyclonal antibody had detectable neutralizing activity. The mouse monoclonal antibodies, tested either separately or as a cocktail, did not exhibit any neutralizing activity. Thus, a neutralizing determinant was located in the peptide sequence VP2:428-446.
TABLE 1.
Neutralization of ADV-G by antibodies prepared against ADV VP2 peptidesa
| Antigen | Antibody | Neutralization titer |
|---|---|---|
| VP2:428-446 | Polyclonal rabbit | 1:200 |
| Mouse monoclonal (IgG1) | Neg | |
| VP2:455-470 | Polyclonal rabbit | Neg |
| VP2:487-501 | Polyclonal rabbit | Neg |
| Mouse monoclonal (IgG1) | Neg | |
| VP2:429-524 | Polyclonal rabbit | 1:800 |
| ADV-GVP2 | Polyclonal rabbit | 1:800 |
| Control | Normal rabbit | Neg |
| Mouse monoclonal (IgG1) | Neg |
The tabulated polyclonal antibodies and monoclonal antibodies (MAbs) were assayed for the ability to neutralize ADV-G infectivity for CrFK cells as detailed in Materials and Methods. The control MAb was an isotype-matched MAb reactive with the rabies virus glycoprotein (graciously provided by Larry Ewalt).
Indirect immunofluorescence analysis.
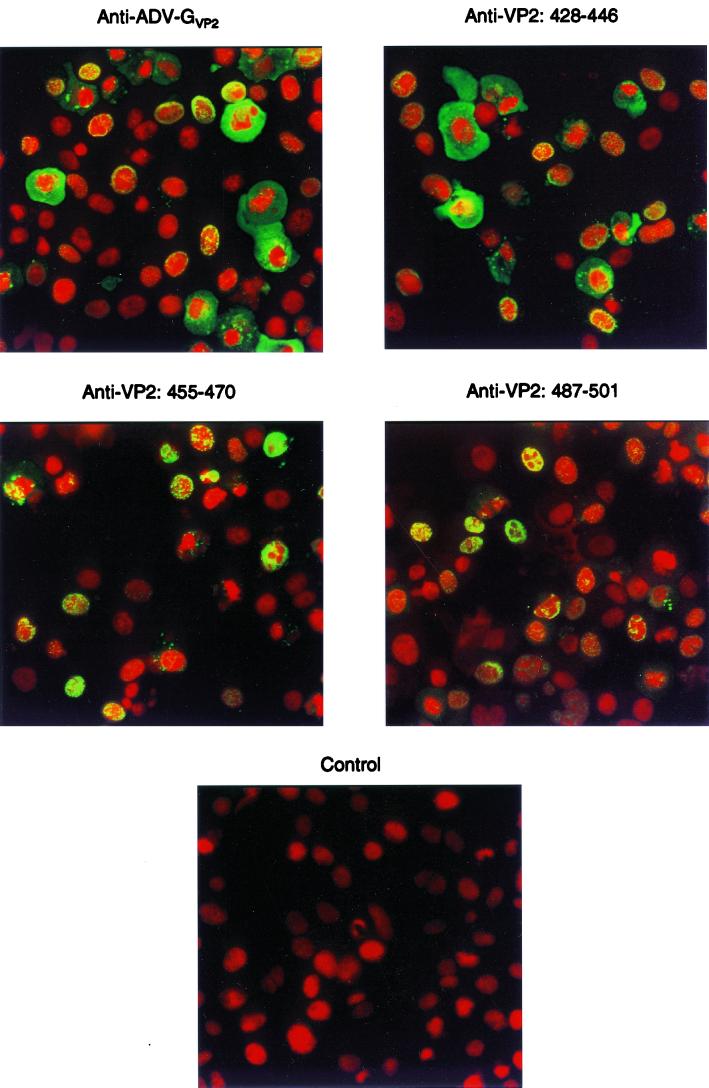
The results in the previous sections indicated that the antibodies against the ADV VP2 peptides exhibited different reactivities in several assays. We also used these antibodies in indirect immunofluorescence to determine whether they produced different staining patterns on ADV-G-infected CrFK cells and to determine whether the specific epitopes might also be differentially available for staining.
Antibody generated against capsids (anti-ADV-GVP2) (Fig. 5, upper left panel) stained viral antigens (green) in both the nucleus and the cytoplasm of infected cells. In some instances, the antigen was found in an intranuclear shell-like distribution, as previously reported (44). This was apparent because the green signal from the FITC-labeled antiviral antibodies overlapped with the nuclei that were stained red due to the propidium iodide. Antibodies directed against the peptide VP2:428-446 yielded staining patterns indistinguishable from the anticapsid antibodies (Fig. 5, upper right panel). It is likely that at least some of this signal is from assembled particles, because these antibodies strongly reacted with virus capsids in immunoelectron microscopy.
FIG. 5.
Immunofluorescent staining of ADV-G-infected CrFK cells with antibodies prepared against ADV VP2 peptides. Acetone-fixed cytocentrifuged preparations of ADV-G-infected CrFK cells were reacted with rabbit polyclonal or murine monoclonal antibodies with the indicated specificities. Staining was revealed with FITC-conjugated anti-rabbit IgG or anti-mouse IgG. Nuclei were counterstained with propidium iodide. As indicated in the text, no difference was observed between monoclonal and polyclonal antibodies with the same specificity.
A patently distinct distribution of antigen was evident with antibodies against peptides VP2:455-470 (Fig. 5, middle left panel) and VP2:487-501 (Fig. 4, middle right panel). These antibodies yielded a staining pattern that was predominantly nuclear in distribution. Since the antibody against VP2:455-470 reacted poorly with intact capsids by immunoelectron microscopy, this limited distribution of staining may indicate that the antibody only recognizes certain conformations of the peptide.
Structural considerations.
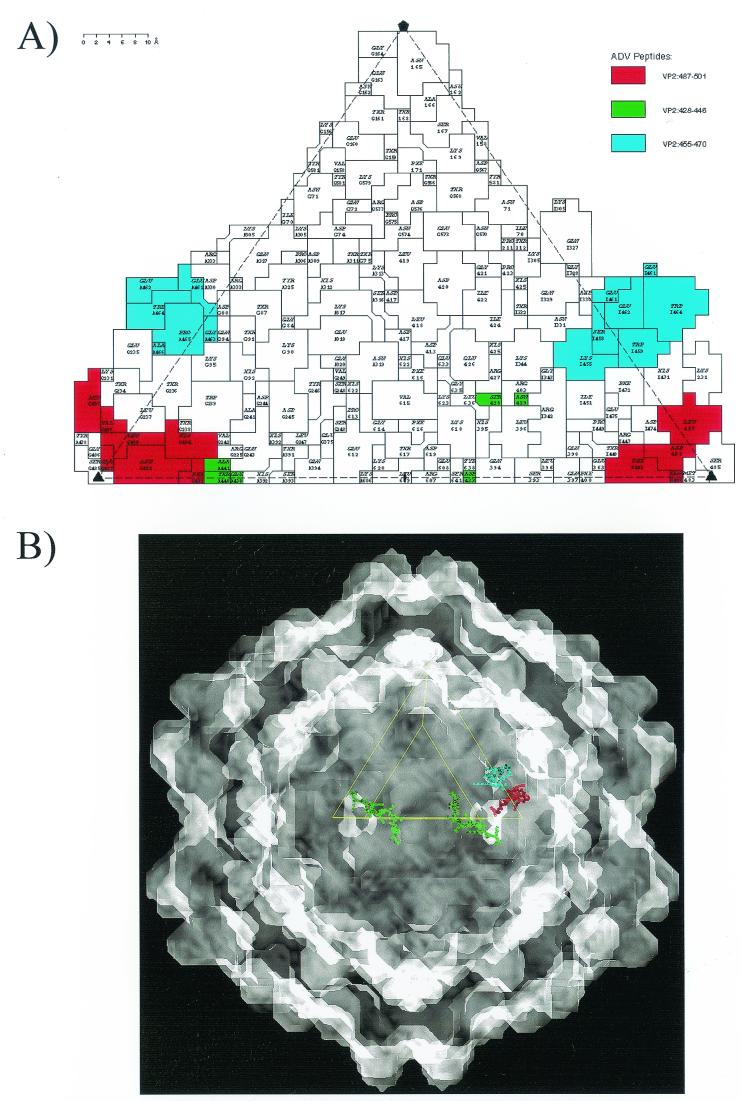
The three ADV peptide regions examined for their ability to elicit antibodies capable of immune complex formation, virus neutralization, and ADE were mapped onto a two-dimensional “roadmap” (19) representation of the ADV-GVP2 capsid surface by using an available pseudo-atomic model (41, 61) (Fig. 6A). All three peptides contain capsid surface-exposed residues (Fig. 6A). In addition, the location of the peptides was depicted on a three-dimensional reconstruction (Fig. 6B), which gives a perspective on the disposition of the peptides on the actual particle.
FIG. 6.
Location of ADV VP2 peptides on ADV capsid. VP2:428-446 (green), VP2:455-470 (blue), and VP2:487-501 (red). (A) Predicted surface-exposed residues from peptides are indicated on a roadmap representation of the surface of the ADV-GVP2 pseudo-atomic structure. The view is down a twofold icosahedral axis. (B) Predicted locations of ADV VP2 peptides are projected onto a shaded-surface representation of ADV-GVP2 viewed down a twofold icosahedral axis.
The VP2:428-446 sequence has surface-exposed residues on the walls of the icosahedral twofold depression, with the N-terminal portion adjacent to VP2:395 (Fig. 6A), a residue implicated with host range and pathogenicity (Fig. 6) (14, 31). The C-terminal portion has exposed residues at the base of the outside wall of the threefold mounds (not adjacent to the precise threefold icosahedral axis). Both the N- and the C-terminal extremes of VP2:428-446 on a single viral asymmetric surface unit, which are contributed by two twofold related peptides, are linear and surface exposed across the dimple (Fig. 6B). The VP2:455-470 and VP2:487-501 sequences have surface-exposed residues on the exterior walls and base of the “cirque” formed by the threefold mounds, respectively (Fig. 6). The VP2:455-470 peptide, forming a short helix on the top of a loop on the outer wall of a threefold mound, is adjacent to two other loops (one from the same VP2 molecule and the other from a threefold symmetry-related VP2 molecule) in the mound (Fig. 6) (41). The VP2:487-501 peptide forms a looped structure on the interior “cirque” wall (adjacent to the threefold axis), close to the base of the threefold mounds. This sequence is adjacent to VP2:229-239 from the same VP2 molecule (41).
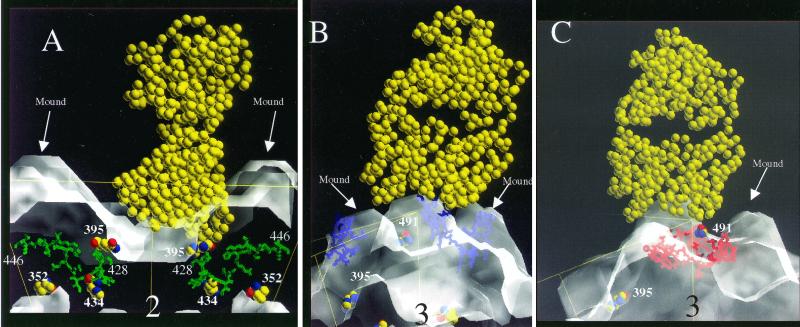
Structural data are not yet available for ADV complexed with antibodies. Thus, possible interactions of the surface-exposed regions with antibodies were modeled based on a reported lysozyme-Fab complex structure (45) (pdb accession number 3HFM) and the interactions of other small nonenveloped viruses with antibodies (6, 21, 55, 56, 57, 58, 62) (Fig. 7). For the VP2:428-446 peptide region, the N- and C-terminal residues were initially modeled separately as different epitopes (Fig. 7A). The binding of Fab fragments over the exposed amino acids at the N-terminal end would occlude a putative receptor binding site (7) and would also cover VP2:395. When viewed perpendicular to an icosahedral twofold axis, the ready accessibility of surface-exposed portions of VP2:428-446 to antibody is apparent (Fig. 7A). This suggested a possible explanation for why antibodies against this sequence are so effective in aggregating particles. In contrast, an Fab fragment docked over VP2:455-470 has relatively limited contact because the exposed residues are found at the apex of a threefold mound (Fig. 7B). Finally, we modeled the docking of an Fab over VP2:487-501. This reconstruction suggested that the Fab arm of an antibody to VP2:487-501 would bind within the “cirque” formed by the threefold mounds and that binding of more than one arm would not be allowed due to the location of this sequence (Fig. 7C).
FIG. 7.
Side-view models of ADV-GVP2 particles complexed with IgG Fab fragment (depicted as van der Waals balls). The position of the icosahedral threefold related mounds and residues involved in host range and pathogenicity (VP2:352, VP2:395, VP2:434) (14, 31) is noted. (A) Fab fragment (yellow) docked with VP2:428-446 (green) in a side view perpendicular to an icosahedral twofold axis. (B) Fab fragment (yellow) docked with VP2:455-470 (blue) in a side view perpendicular to an icosahedral threefold axis. (C) Fab fragment (yellow) docked with VP2:487-501 (red) in a side view perpendicular to an icosahedral threefold axis.
DISCUSSION
Our previous studies localized immunoreactive sites and in vitro neutralizing epitopes to a specific region of the ADV VP2 capsid protein (VP2:429-524) containing variable loops that partially comprise the surface of the virus particle (16, 41). In the present study, we analyzed antibodies against several short peptides within this region for their capacity to support immune complex formation, neutralization of ADV for CrFK cells, and Fc-receptor-mediated ADE of infection, three features implicated in ADV pathogenesis. We found that a determinant located in VP2:428-446 exhibited these three properties. Therefore, this short sequence in the capsid protein gene is involved in three key aspects of ADV pathogenesis.
Attempts to protect adult mink with capsid-based ADV vaccines have uniformly failed, even though vaccinated animals develop antiviral antibodies (1, 49, 52). Indeed, upon challenge, vaccinated animals suffer a hyperacute disease with enhanced tissue lesions (1, 52). We observed that injection of rabbits with VP2:428-446 induced both neutralizing and ADE-mediating antibody (Fig. 4; Table 1). Whereas virus neutralization by antibody may afford protection from infection, ADE facilitates the infection of macrophages that is central to the resulting immune disorder (15, 29, 35). Antibodies to VP2:428-446 have been demonstrated in ADV-infected adult mink (28) and, if these antibodies function to mediate both ADE and neutralization in vivo, any protective effect might be counteracted or supervened. Thus, our results provide possible explanations for the failure of capsid-based ADV vaccines (1, 52). As noted in the introduction, capsid-based vaccines against MEV are highly effective in mink (22, 39). In fact, peptide vaccines based on the MEV capsid sequence also induce neutralizing antibodies and confer protection (39). This indicates that a peptide vaccine can be efficacious in mink. Consequently, it will be extremely interesting to determine whether mink injected with the VP2:428-446 peptide will produce both neutralizing and ADE-mediating antibody.
Our results suggested that at least one epitope within VP2:428-446 was surface exposed and readily available for antibody binding (Fig. 6 and 7). This observation is consistent with the roadmap representation of the ADV-GVP2 pseudo-atomic structure which revealed exposed residues of this peptide on the twofold dimple walls (VP2:428-429; Fig. 6, green) and adjacent to the mounds offset from the icosahedral threefold axes (VP2:439-441) (41). Both the polyclonal and the monoclonal antibodies to VP2:428-446 aggregated virus particles (Fig. 3), but only the polyclonal antibody had neutralizing activity (Table 1). This observation supports data from CPV that virus cross-linking is not sufficient for parvovirus neutralization (6, 60, 62). Other possible mechanisms for neutralization include occlusion of the receptor attachment site (6, 38, 40, 55, 60) and stabilization of capsids against genome uncoating (13, 21, 37, 55). The modeled docking of Fab fragments over the exposed amino acids of VP2:428-446 (Fig. 7A) suggests that an antibody binding to either exposed terminus could occlude the region where a putative binding site for a CPV sialic acid receptor is located (6, 12) (Fig. 7). Thus, an antibody to the VP2:428-429 region might neutralize by impeding cell binding (Fig. 6). The same region is close to ADV VP2:395 and other residues implicated in host range and pathogenicity (Fig. 7A) (14, 31). It is currently unclear which of these mechanisms is operative for neutralization of ADV for CrFK cells by the VP2:428-446 antibodies.
The monoclonal antibody to VP2:428-446 cross-linked particles and formed immune complexes (Fig. 3). This mandated that the epitope be located in a position such that a single IgG molecule could bridge two particles. By using previously determined allowable antigen-antibody interaction distances and orientations relative to antigenic sites (45, 21, 56, 58), we modeled potential interactions between an antibody molecule and the surface-exposed portions of VP2:428-446 (Fig. 7). The results suggested that the epitope should be located near the icosahedral twofold axes where the particle has the smallest cross-sectional diameter (41). Furthermore, binding of the Fab arms from a single monoclonal antibody to two virus particles should be readily accommodated (21, 55–58), resulting in particle bridging. A direct evaluation of this prediction can be done by analyzing cryoelectron microscopy-based reconstructions of particles complexed with Fab fragments. These experiments are currently under way.
Structural mapping of antigens for CPV and several other parvoviruses places major neutralizing domains adjacent to the icosahedral threefold axes and on the shoulder of the wall between the fivefold canyon-like region and the icosahedral twofold dimple (6, 38, 60, 62). The mounds on the ADV capsid, which contain the VP2:455-470 and VP2:487-501 peptides, would alter the general capsid topography where major neutralizing epitopes of CPV map (6, 41). It is possible that these mounds might disrupt potential major neutralizing domains and direct the antibody response toward targets that mediate ADE and immune complex formation. This might contribute significantly to the distinct immunopathogenesis of ADV infections.
The failure of the antibody to VP2:455-470 to aggregate, neutralize, or mediate ADE was at first somewhat surprising, because the peptide is mostly surface exposed near the apex of the mounds (Fig. 6 and 7B). The reasons for this are unclear at present; however, upon inspection of the ADV-GVP2 pseudo-atomic model, the region around VP2:455-470 has significant secondary structure (41). If VP2:455-470 is part of a complex conformational structure, it may not be recognized by the antipeptide antibody in all its configurations. This might also explain the limited reactivity of this antibody in our IFA studies (Fig. 5).
The limited ability of antibodies to VP2:487-501 to aggregate particles and mediate ADE is consistent with the surface exposure of the linear peptide residues (Fig. 6). The low degree of particle aggregation and gold particle decoration (Fig. 3) may be explained by the observation that only a single Fab arm can be accommodated in the “cirque-like” gap between the threefold mounds where three VP2:487-501 epitopes would reside (41) (Fig. 7C). Thus, the expected occupancy would only be one-third of that predicted for VP2:428-446 where each of the 60 epitopes on the capsid is available.
The pathogenicity and host range of ADV are controlled by coordinate interactions among a small number of capsid amino acids (14, 31, 43). One of the residues controlling host range (VP2:434) is located in the VP2:428-446 peptide. Other involved amino acids also are found close to this site (Fig. 6 and 7). The fact that these residues cluster suggests that this small part of the capsid is important not only in the immunopathogenesis of ADV infections but also in viral host range and pathogenicity.
In summary, we have implicated short peptide sequences in key aspects of ADV pathogenesis and in the adverse response of mink to capsid-based vaccines (1, 52). In addition, unique features of the ADV capsid may also play a role in the unusual nature of infections with this virus. Clearly, our studies do not address the entire capsid protein sequence or potential conformational epitopes (43, 54). Consequently, the results leave open the possibility that complex functional classes of epitopes exist on the ADV capsid: some mediating only neutralization, others involved with ADE and immune complex formation, and others having mixed characteristics. It is possible that nonneutralizing epitopes might dominate by inducing antibodies that interfere with neutralization and accentuate disease (25, 32).
ACKNOWLEDGMENTS
We acknowledge the services and advice of Peter Yaciuk of the Hybridoma Development Service (St. Louis University School of Medicine) for monoclonal antibody production and Jan Lukszo (Peptide Synthesis and Analysis Unit, NIAID) for peptide synthesis and analysis. Thomas Kindt, DIR, NIAID, graciously provided funds for the monoclonal antibody production. Gary Hettrick assisted in preparation of the figures, and members of the Laboratory of Persistent Viral Diseases provided critical input.
REFERENCES
- 1.Aasted B, Alexandersen S, Christensen J. Vaccination with Aleutian mink disease parvovirus (AMDV) capsid proteins enhances disease, while vaccination with the major non-structural AMDV protein causes partial protection from disease. Vaccine. 1998;16:1158–1165. doi: 10.1016/s0264-410x(98)80114-x. [DOI] [PubMed] [Google Scholar]
- 2.Aasted B, Alexandersen S, Hansen M. Treatment of neonatally Aleutian disease virus (ADV) infected mink with gammaglobulin containing antibodies to ADV reduces the death rate of mink kits. Acta Vet Scand. 1988;29:323–330. doi: 10.1186/BF03548625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aasted B, Race R E, Bloom M E. Aleutian disease virus, a parvovirus, is proteolytically degraded during in vivo infection in mink. J Virol. 1984;51:7–13. doi: 10.1128/jvi.51.1.7-13.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aasted B, Tierney G S, Bloom M E. Analysis of the quantity of antiviral antibodies from mink infected with different Aleutian disease virus strains. Scand J Immunol. 1984;19:395–402. doi: 10.1111/j.1365-3083.1984.tb00947.x. [DOI] [PubMed] [Google Scholar]
- 5.Agbandje-McKenna M, Llamas-Saiz A L, Wang F, Tattersall P, Rossmann M G. Functional implications of the structure of the murine parvovirus minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 6.Agbandje M, Parrish C R, Rossmann M G. The recognition of parvovirus capsids by antibodies. Semin Virol. 1995;6:219–231. [Google Scholar]
- 7.Agbandje M, Parrish C R, Rossmann M G. The structure of parvoviruses. Semin Virol. 1995;6:299–309. [Google Scholar]
- 8.Alexandersen S, Bloom M E, Perryman S. Detailed transcription map of Aleutian mink disease parvovirus. J Virol. 1988;62:3684–3694. doi: 10.1128/jvi.62.10.3684-3694.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandersen S, Bloom M E, Wolfinbarger J, Race R E. In situ molecular hybridization for detection of Aleutian mink disease parvovirus DNA by using strand-specific probes: identification of target cells for viral replication in cell cultures and in mink kits with virus-induced interstitial pneumonia. J Virol. 1987;61:2407–2419. doi: 10.1128/jvi.61.8.2407-2419.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandersen S, Larsen S, Cohn A, Uttenthal A, Race R E, Aasted B, Hansen M, Bloom M E. Passive transfer of antiviral antibodies restricts replication of Aleutian mink disease parvovirus in vivo. J Virol. 1988;63:9–17. doi: 10.1128/jvi.63.1.9-17.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandersen S, Storgaard T, Kamstrup N, Aasted B, Porter D D. Pathogenesis of Aleutian mink disease parvovirus infection: effects of suppression of antibody response on viral mRNA levels and on development of acute disease. J Virol. 1994;68:738–749. doi: 10.1128/jvi.68.2.738-749.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbis D P, Chang S-F, Parrish C R. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology. 1992;191:301–308. doi: 10.1016/0042-6822(92)90192-r. [DOI] [PubMed] [Google Scholar]
- 13.Belnap D M, Filman D J, Trus B L, Cheng N, Booy F P, Conway J F, Curry S, Hiremath C N, Tsang S K, Steven A C, Hogle J M. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloom M E, Fox J M, Berry B D, Oie K L, Wolfinbarger J B. Construction of pathogenic molecular clones of Aleutian mink disease parvovirus that replicate both in vivo and in vitro. Virology. 1998;251:288–296. doi: 10.1006/viro.1998.9426. [DOI] [PubMed] [Google Scholar]
- 15.Bloom M E, Kanno H, Mori S, Wolfinbarger J B. Aleutian mink disease: puzzles and paradigms. Infect Agents Dis. 1994;3:279–301. [PubMed] [Google Scholar]
- 16.Bloom M E, Martin D A, Oie K L, Huhtanen M E, Costello F, Wolfinbarger J B, Hayes S F, Agbandje-McKenna M. Expression of Aleutian mink disease parvovirus capsid proteins in defined segments: localization of immunoreactive sites and neutralizing epitopes to specific regions. J Virol. 1997;71:705–714. doi: 10.1128/jvi.71.1.705-714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloom M E, Race R E, Hadlow W J, Chesebro B. Aleutian disease of mink: the antibody response of sapphire and pastel mink to Aleutian disease virus. J Immunol. 1975;115:1034–1037. [PubMed] [Google Scholar]
- 18.Bloom M E, Race R E, Wolfinbarger J B. Characterization of Aleutian disease virus as a parvovirus. J Virol. 1980;35:836–843. doi: 10.1128/jvi.35.3.836-843.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman M S. Mapping the surface properties of macromolecules. Protein Sci. 1993;2:459–469. doi: 10.1002/pro.5560020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman M S, Rossmann M G. Structure, sequence and function correlates among parvoviruses. Virology. 1993;194:491–508. doi: 10.1006/viro.1993.1288. [DOI] [PubMed] [Google Scholar]
- 21.Che Z, Olson N H, Leippe D, Lee W-M, Mosser A G, Rueckert R R, Baker T S, Smith T J. Antibody-mediated neutralization of human rhinovirus 14 explored by means of cryoelectron microscopy and X-ray crystallography of virus-Fab complexes. J Virol. 1998;72:4610–4622. doi: 10.1128/jvi.72.6.4610-4622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen J, Alexandersen S, Bloch B, Aasted B, Uttenthal A. Production of mink enteritis parvovirus empty capsids by expression in a baculovirus vector system: a recombinant vaccine for mink enteritis parvovirus in mink. J Gen Virol. 1994;75:149–155. doi: 10.1099/0022-1317-75-1-149. [DOI] [PubMed] [Google Scholar]
- 23.Christensen J, Storgaard T, Bloch B, Alexandersen S, Aasted B. Expression of Aleutian mink disease parvovirus proteins in a baculovirus vector system. J Virol. 1993;67:229–238. doi: 10.1128/jvi.67.1.229-238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemens D L, Wolfinbarger J B, Mori S, Berry B D, Hayes S F, Bloom M E. Expression of Aleutian mink disease parvovirus capsid proteins by a recombinant vaccinia virus: self-assembly of capsid proteins into particles. J Virol. 1992;66:3077–3085. doi: 10.1128/jvi.66.5.3077-3085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleveland S M, Buratti E, Jones T D, North P, Baralle F, McLain L, McInerney T, Durrani Z, Dimmock N J. Immunogenic and antigenic dominance of a nonneutralizing epitope over a highly conserved neutralizing epitope in the gp41 envelope glycoprotein of human immunodeficiency virus type 1: its deletion leads to a strong neutralizing response. Virology. 2000;266:66–78. doi: 10.1006/viro.1999.0041. [DOI] [PubMed] [Google Scholar]
- 26.Coe J E, Coe P R, Ross M J. Staphylococcal protein A purification of rodent IgG1 and IgG2 with particular emphasis on Syrian hamsters. Mol Immunol. 1981;18:1007–1012. doi: 10.1016/0161-5890(81)90119-x. [DOI] [PubMed] [Google Scholar]
- 27.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W. Current protocols in immunology. New York, N.Y: John C. Wiley & Sons, Inc.; 2000. [Google Scholar]
- 28.Costello F, Steenfos N, Jensen K T, Christensen J, Gottschalk E, Holm A, Aasted B. Epitope mapping of Aleutian mink disease parvovirus virion protein VP1 and VP2. Scand J Immunol. 1999;49:347–354. doi: 10.1046/j.1365-3083.1999.00499.x. [DOI] [PubMed] [Google Scholar]
- 29.Dworak L J, Wolfinbarger J B, Bloom M E. Aleutian mink disease parvovirus infection of K562 cells is antibody-dependent and is mediated via an Fc(gamma)RII receptor. Arch Virol. 1997;142:363–373. doi: 10.1007/s007050050082. [DOI] [PubMed] [Google Scholar]
- 30.Eklund C M, Hadlow W J, Kennedy R C, Boyle C C, Jackson T A. Aleutian disease of mink: properties of the etiologic agent and the host responses. J Infect Dis. 1968;118:510–516. doi: 10.1093/infdis/118.5.510. [DOI] [PubMed] [Google Scholar]
- 31.Fox J M, Stevenson M A, Bloom M E. Replication of Aleutian mink disease parvovirus in vivo is influenced by residues in the VP2 protein. J Virol. 1999;73:3835–3842. doi: 10.1128/jvi.73.10.8713-8719.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrity R R, Rimmelzwaan G, Minassian A, Tsai W P, Lin G, de Jong J J, Goudsmit J, Nara P L. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J Immunol. 1997;159:279–289. [PubMed] [Google Scholar]
- 33.Hadlow W J, Race R E, Kennedy R C. Comparative pathogenicity of four strains of Aleutian disease virus for pastel and sapphire mink. Infect Immun. 1983;41:1016–1023. doi: 10.1128/iai.41.3.1016-1023.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanno H, Wolfinbarger J B, Bloom M E. Identification of Aleutian mink disease parvovirus transcripts in macrophages of infected adult mink. J Virol. 1992;66:5305–5312. doi: 10.1128/jvi.66.9.5305-5312.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanno H, Wolfinbarger J B, Bloom M E. Aleutian mink disease parvovirus infection of mink macrophages and human macrophage cell line U937: demonstration of antibody-dependent enhancement of infection. J Virol. 1993;67:7017–7024. doi: 10.1128/jvi.67.12.7017-7024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanno H, Wolfinbarger J B, Bloom M E. Aleutian mink disease parvovirus infection of mink peritoneal macrophages and human macrophage cell lines. J Virol. 1993;67:2075–2082. doi: 10.1128/jvi.67.4.2075-2082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolatkar P R, Bella J, Olson N H, Bator C M, Baker T S, Rossmann M G. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langeveld J P M, Casal J I, Vela C, Dalsgaard K, Smale S H, Pujik W C, Meloen R H. B-cell epitopes of canine parvovirus: distribution on the primary structure and exposure on the viral surface. J Virol. 1993;67:765–772. doi: 10.1128/jvi.67.2.765-772.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langeveld J P M, Kamstrup N, Uttenthal A, Strandbygaard B, Vela C, Dalsgaard K, Beekman N J C M, Meloen R H, Casal J I. Full protection in mink against mink enteritis virus with new generation canine parvovirus vaccines based on synthetic peptide or recombinant protein. Vaccine. 1995;13:1033–1037. doi: 10.1016/0264-410x(95)00021-r. [DOI] [PubMed] [Google Scholar]
- 40.Lopez de Turiso J A, Cortes E, Ranz A I, Garcia J, Sanz A, Vela C, Casal J I. Fine mapping of canine parvovirus B cell epitopes. J Gen Virol. 1991;72:2445–2456. doi: 10.1099/0022-1317-72-10-2445. [DOI] [PubMed] [Google Scholar]
- 41.McKenna R, Olson N H, Chipman P R, Baker T S, Booth T F, Christensen J, Aasted B, Fox J M, Bloom M E, Agbandje-McKenna M. The three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J Virol. 1999;73:6882–6891. doi: 10.1128/jvi.73.8.6882-6891.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mori S, Wolfinbarger J B, Miyazawa M, Bloom M E. Replication of Aleutian mink disease parvovirus in lymphoid tissues of adult mink: involvement of follicular dendritic cells and macrophages. J Virol. 1991;65:952–956. doi: 10.1128/jvi.65.2.952-956.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oie K L, Durrant G, Wolfinbarger J B, Martin D, Costello F, Perryman S, Hadlow W J, Bloom M E. The relationship between capsid protein (VP2) sequence and pathogenicity of Aleutian mink disease parvovirus (ADV): a possible role for raccoons in the transmission of ADV infections. J Virol. 1996;70:852–861. doi: 10.1128/jvi.70.2.852-861.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oleksiewicz M B, Costello F, Huhtanen M, Wolfinbarger J B, Alexandersen S, Bloom M E. Subcellular localization of Aleutian mink disease parvovirus proteins and DNA during permissive infection of Crandell feline kidney cells. J Virol. 1996;70:3242–3247. doi: 10.1128/jvi.70.5.3242-3247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Padlan E A, Silverton E W, Sheriff S, Cohen G H, Smith-Gill S J, Davies D R. Structure of an antibody-antigen complex:crystal structure of the HyHEL-10 Fab-lysozyme complex. Proc Natl Acad Sci USA. 1989;86:5938–5942. doi: 10.1073/pnas.86.15.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parrish C R. Pathogenesis of feline panleukopenia and canine parvovirus. Bailliere Clin Hematol. 1995;8:57–71. doi: 10.1016/S0950-3536(05)80232-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parrish C R, Carmichael L E, Antczak D F. Antigenic relationships between canine parvovirus type 2, feline panleukopenia virus, and mink enteritis virus using conventional antisera and monoclonal antibodies. Arch Virol. 1982;72:267–278. doi: 10.1007/BF01315223. [DOI] [PubMed] [Google Scholar]
- 48.Parrish C R, Leathers C W, Pearson R, Gorham J R. Comparisons of feline panleukopenia virus, canine parvovirus, raccoon parvovirus, and mink enteritis virus and their pathogenicity for mink and ferrets. Am J Vet Res. 1987;48:1429–1435. [PubMed] [Google Scholar]
- 49.Porter D D. Aleutian disease: a persistent parvovirus infection of mink with a maximal but ineffective host immune response. Prog Med Virol. 1986;33:42–60. [PubMed] [Google Scholar]
- 50.Porter D D, Larsen A E. Aleutian disease of mink: infectious virus-antibody complexes in the serum. Proc Soc Exp Biol Med. 1967;126:680–682. [Google Scholar]
- 51.Porter D D, Larsen A E, Porter H G. The pathogenesis of Aleutian disease of mink. I. In vivo viral replication and the host antibody response to viral antigen. J Exp Med. 1969;130:575–589. doi: 10.1084/jem.130.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porter D D, Larsen A E, Porter H G. The pathogenesis of Aleutian disease of mink. II. Enhancement of tissue lesions following the administration of a killed virus vaccine or passive antibody. J Immunol. 1972;109:1–7. [PubMed] [Google Scholar]
- 53.Porter D D, Larsen A E, Porter H G. The pathogenesis of Aleutian disease of mink. III. Immune complex arteritis. Am J Pathol. 1983;71:331–344. [PMC free article] [PubMed] [Google Scholar]
- 54.Race R E, Chesebro B, Bloom M E, Aasted B, Wolfinbarger J. Monoclonal antibodies against Aleutian disease virus distinguish virus strains and differentiate sites of virus replication from sites of viral antigen sequestration. J Virol. 1986;57:285–293. doi: 10.1128/jvi.57.1.285-293.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith T J, Baker T. Picornaviruses: epitopes, canyons, and pockets. Adv Virus Res. 1999;52:1–23. doi: 10.1016/s0065-3527(08)60297-3. [DOI] [PubMed] [Google Scholar]
- 56.Smith T J, Chase E S, Schmidt T J, Olson N H, Baker T S. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature. 1996;383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith T J, Olson N H, Cheng R H, Chase E S, Baker T S. Structure of a human rhinovirus-bivalently bound antibody complex: implications for viral neutralization and antibody flexibility. Proc Natl Acad Sci USA. 1993;90:7015–7018. doi: 10.1073/pnas.90.15.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith T J, Olson N H, Cheng R H, Liu H, Chase E S, Lee W-M, Leippe D, Mosser A G, Rueckert R R, Baker T S. Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody. J Virol. 1993;67:1148–1158. doi: 10.1128/jvi.67.3.1148-1158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stolze B, Kaaden O-R. Apparent lack of neutralizing antibodies in Aleutian disease is due to masking of antigenic sites by phospholipids. Virology. 1987;158:174–180. doi: 10.1016/0042-6822(87)90251-0. [DOI] [PubMed] [Google Scholar]
- 60.Strassheim M L, Gruenberg A, Veijalainen P M L, Sgro J-Y, Parrish C R. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology. 1994;198:175–184. doi: 10.1006/viro.1994.1020. [DOI] [PubMed] [Google Scholar]
- 61.Tsao J, Chapman M S, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith T J, Rossmann M G, Compans R W, Parrish C R. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 62.Wikoff W R, Wang G, Parrish C R, Cheng R H, Strassheim M L, Baker T S, Rossmann M G. The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment. Structure. 1994;2:595–607. doi: 10.1016/s0969-2126(00)00062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu W-H, Bloom M E, Berry B D, McGinley M J, Platt K B. Expression of Aleutian mink disease parvovirus capsid proteins in a baculovirus expression system for potential diagnostic use. J Vet Diagn Investig. 1994;6:23–29. doi: 10.1177/104063879400600105. [DOI] [PubMed] [Google Scholar]
- 64.Zolotukhin S, Byrne B, Mason E, Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski R, Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]