Abstract
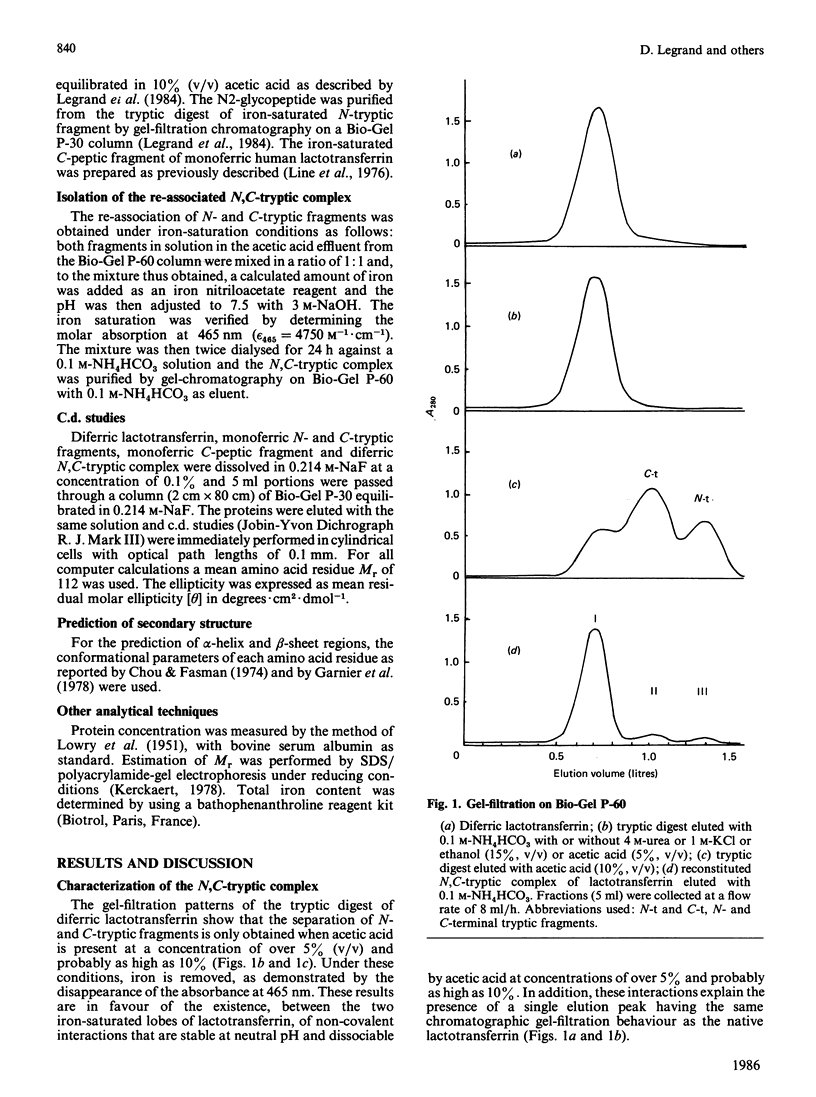
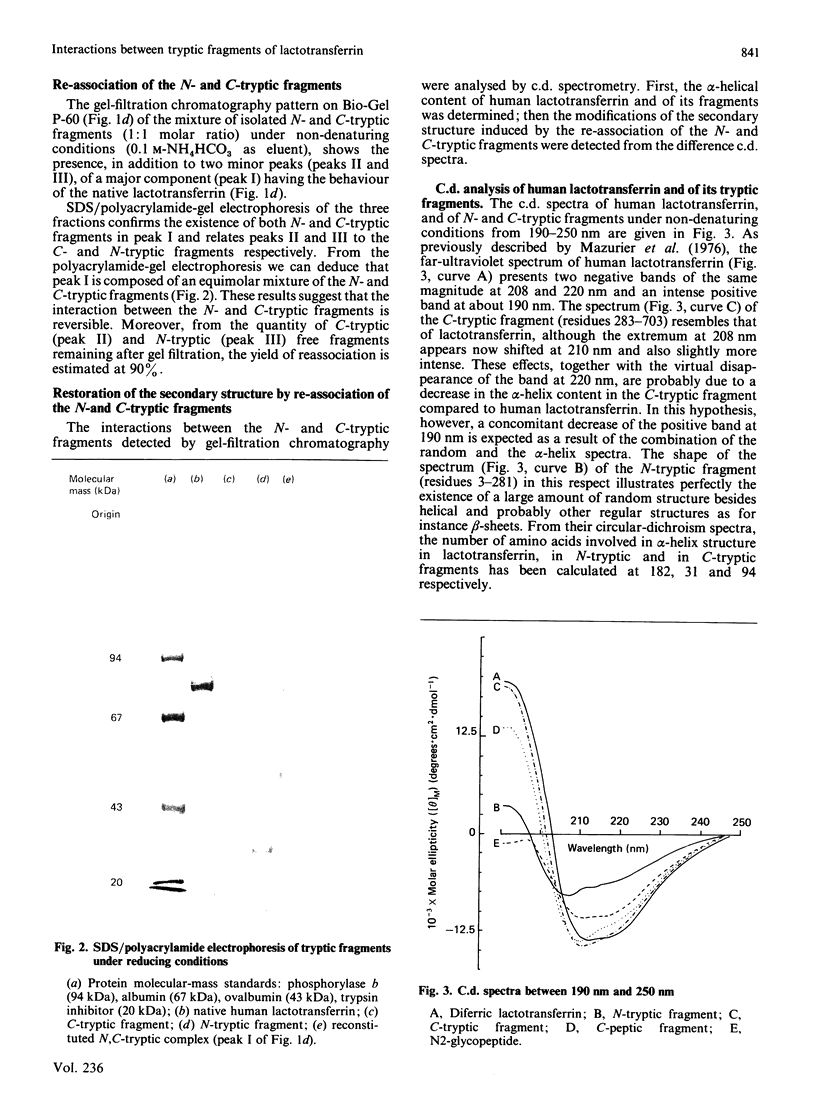
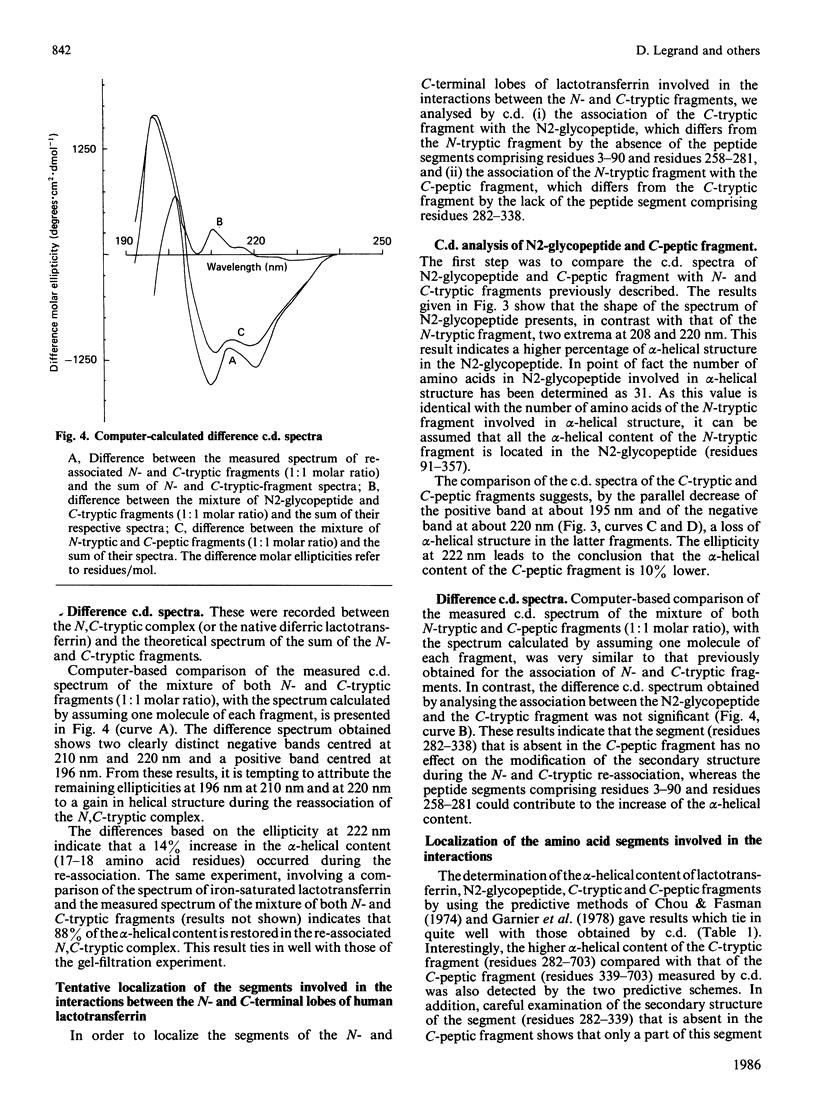
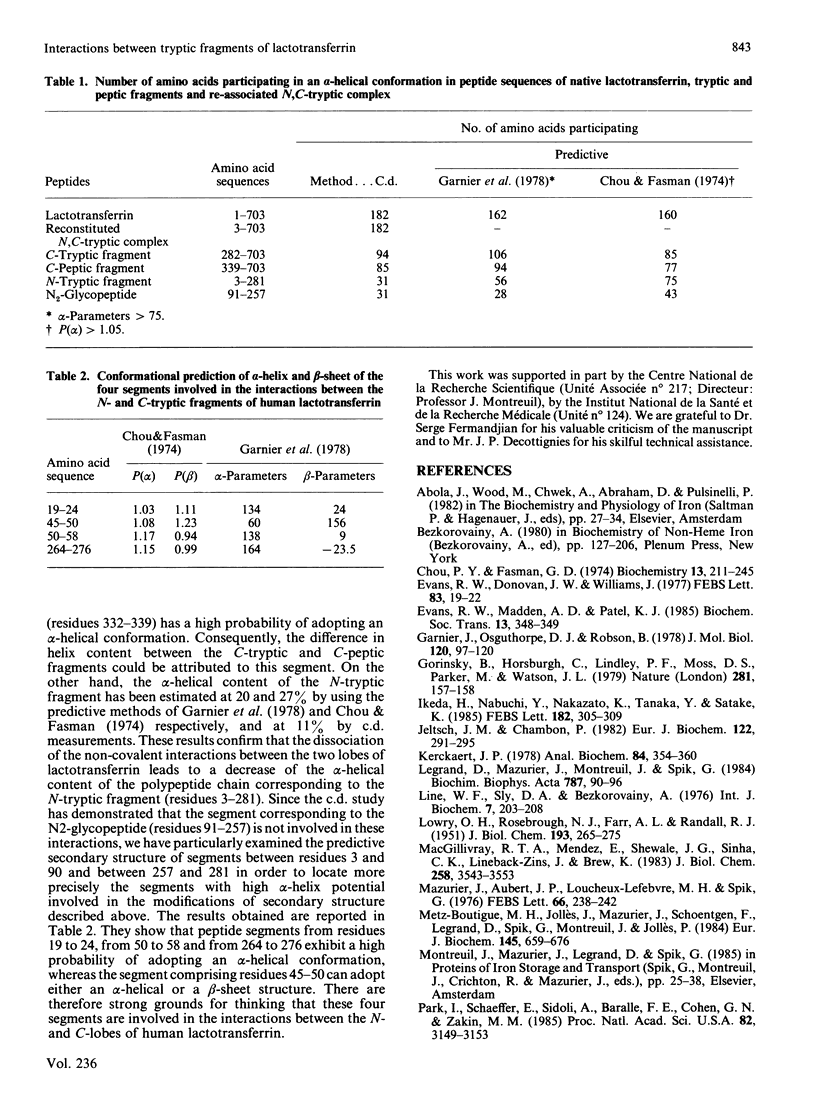
Gel filtration of a mild tryptic digest of diferric human lactotransferrin carried out in presence of 10% (v/v) acetic acid led to the isolation of two fragments, an N-terminal tryptic fragment having an Mr of 30,000 and a C-terminal tryptic fragment having an Mr of 50,000 [Legrand, Mazurier, Montreuil & Spik (1984) Biochim. Biophys. Acta 787, 90-96]. Both fragments possess a degree of organization lower than that of the native protein, as shown by the decrease of about 30% of the alpha-helical content observed by c.d. The two fragments are able to re-associate in neutral solutions, as shown by the isolation, by gel chromatography, of a re-associated 80 kDa N,C-tryptic complex having the chromatographic behaviour of the native lactotransferrin. Computer-based comparison of the measured c.d. spectrum of the mixture of N-tryptic and C-tryptic fragments (molar ratio 1:1) with the spectrum calculated by assuming one molecule of each fragment, shows that the alpha-helix content of lactotransferrin is restored. These results strongly suggest the existence of non-covalent and reversible interactions between the two lobes of lactotransferrin. In addition it was demonstrated that short peptide segments (residues 19-24, 45-58 and 264-276) are involved in the secondary-structure modifications referred to above.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Evans R. W., Donovan J. W., Williams J. Calorimetric studies on the binding of iron and aluminium to the amino- and carboxyl-terminal fragments of hen ovotransferrin. FEBS Lett. 1977 Nov 1;83(1):19–22. doi: 10.1016/0014-5793(77)80632-7. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gorinsky B., Horsburgh C., Lindley P. F., Moss D. S., Parkar M., Watson J. L. Evidence for the bilobal nature of diferric rabbit plasma transferrin. Nature. 1979 Sep 13;281(5727):157–158. doi: 10.1038/281157a0. [DOI] [PubMed] [Google Scholar]
- Jeltsch J. M., Chambon P. The complete nucleotide sequence of the chicken ovotransferrin mRNA. Eur J Biochem. 1982 Feb;122(2):291–295. doi: 10.1111/j.1432-1033.1982.tb05879.x. [DOI] [PubMed] [Google Scholar]
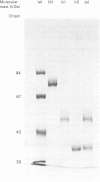
- Kerckaert J. P. Highly simplified analytical or preparative slab gel electrophoresis. Anal Biochem. 1978 Feb;84(2):354–360. doi: 10.1016/0003-2697(78)90052-0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Legrand D., Mazurier J., Metz-Boutigue M. H., Jolles J., Jolles P., Montreuil J., Spik G. Characterization and localization of an iron-binding 18-kDa glycopeptide isolated from the N-terminal half of human lactotransferrin. Biochim Biophys Acta. 1984 May 31;787(1):90–96. doi: 10.1016/0167-4838(84)90111-0. [DOI] [PubMed] [Google Scholar]
- MacGillivray R. T., Mendez E., Shewale J. G., Sinha S. K., Lineback-Zins J., Brew K. The primary structure of human serum transferrin. The structures of seven cyanogen bromide fragments and the assembly of the complete structure. J Biol Chem. 1983 Mar 25;258(6):3543–3553. [PubMed] [Google Scholar]
- Mazurier J., Aubert J. P., Loucheux-Lefevre M. H. Comparative circular dichroism studies of iron-free and iron-saturated forms of human serotransferrin and lactortransferrin. FEBS Lett. 1976 Jul 15;66(2):238–242. doi: 10.1016/0014-5793(76)80512-1. [DOI] [PubMed] [Google Scholar]
- Metz-Boutigue M. H., Jollès J., Mazurier J., Schoentgen F., Legrand D., Spik G., Montreuil J., Jollès P. Human lactotransferrin: amino acid sequence and structural comparisons with other transferrins. Eur J Biochem. 1984 Dec 17;145(3):659–676. doi: 10.1111/j.1432-1033.1984.tb08607.x. [DOI] [PubMed] [Google Scholar]
- Park I., Schaeffer E., Sidoli A., Baralle F. E., Cohen G. N., Zakin M. M. Organization of the human transferrin gene: direct evidence that it originated by gene duplication. Proc Natl Acad Sci U S A. 1985 May;82(10):3149–3153. doi: 10.1073/pnas.82.10.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spik G., Strecker G., Fournet B., Bouquelet S., Montreuil J., Dorland L., van Halbeek H., Vliegenthart J. F. Primary structure of the glycans from human lactotransferrin. Eur J Biochem. 1982 Jan;121(2):413–419. doi: 10.1111/j.1432-1033.1982.tb05803.x. [DOI] [PubMed] [Google Scholar]
- Uzan G., Frain M., Park I., Besmond C., Maessen G., Trépat J. S., Zakin M. M., Kahn A. Molecular cloning and sequence analysis of cDNA for human transferrin. Biochem Biophys Res Commun. 1984 Feb 29;119(1):273–281. doi: 10.1016/0006-291x(84)91648-6. [DOI] [PubMed] [Google Scholar]
- Williams J., Elleman T. C., Kingston I. B., Wilkins A. G., Kuhn K. A. The primary structure of hen ovotransferrin. Eur J Biochem. 1982 Feb;122(2):297–303. doi: 10.1111/j.1432-1033.1982.tb05880.x. [DOI] [PubMed] [Google Scholar]
- Yang F., Lum J. B., McGill J. R., Moore C. M., Naylor S. L., van Bragt P. H., Baldwin W. D., Bowman B. H. Human transferrin: cDNA characterization and chromosomal localization. Proc Natl Acad Sci U S A. 1984 May;81(9):2752–2756. doi: 10.1073/pnas.81.9.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]