Abstract
The pandemic outbreak caused by SARS‐CoV‐2 coronavirus brought a crucial issue in public health causing up to now more than 600 million infected people and 6.5 million deaths. Conventional diagnostic methods are based on quantitative reverse transcription polymerase chain reaction (RT‐qPCR assay) and immuno‐detection (ELISA assay). However, despite these techniques have the advantages of being standardized and consolidated, they keep some main limitations in terms of accuracy (immunoassays), time/cost consumption of analysis, the need for qualified personnel, and lab constrain (molecular assays). There is crucial the need to develop new diagnostic approaches for accurate, fast and portable viral detection and quantification. Among these, PCR‐free biosensors represent the most appealing solution since they can allow molecular detection without the complexity of the PCR. This will enable the possibility to be integrated in portable and low‐cost systems for massive and decentralized screening of SARS‐CoV‐2 in a point‐of‐care (PoC) format, pointing to achieve a performant identification and control of infection. In this review, the most recent approaches for the SARS‐CoV‐2 PCR‐free detection are reported, describing both the instrumental and methodological features, and highlighting their suitability for a PoC application.
Keywords: biosensors, CRISPR‐Cas, graphene technology, PCR‐free, point‐of‐care, SARS‐CoV‐2 PCR‐free detection
PCR‐free biosensors for SARS‐CoV‐2 detection are characterized by a high degree of technological integration and miniaturization, proposing device on which the viral sample can be directly processed and detected in a simple and fast way. This allows the viral infections analysis to be decentralized, addressing the growing need of massive outdoor screening in a PoC format.
1. Introduction
Since January 2020 the world has been forced to face SARS‐CoV‐2, a coronavirus that is responsible for the human severe respiratory syndrome COVID‐19.[ 1 , 2 , 3 ] From its first report in Wuhan, China, the SARS‐CoV‐2 contagion quickly spread worldwide becoming a pandemic, as declared by the World Health Organization (WHO), bringing up more than 600 million infected people and 6.5 million deaths.[ 4 , 5 ]
Currently, the most reliable test is based on reverse‐transcriptase polymerase chain reaction (RTPCR) that based on the recommendation of the WHO and the American Center for Disease Control (ACDC) is the unique standard for recognition of COVID‐19.[ 6 ] However, although RTPCR allows the detection of few copies of viral genome, it includes complex laboratory procedures. In the standard RT‐PCR, the viral RNA of SARS‐CoV‐2 must be reverse transcribed (RT) into complementary DNA and then amplified through PCR. This diagnostic test exhibits the highest sensitivity and limit of detection (LOD) for SARS‐CoV‐2: typically RT‐PCR carried out on nasopharyngeal samples has a LOD of ≈100 copies of viral RNA per millilitres of transport media, while the LODs of other techniques vary over 10,000‐fold.[ 7 , 8 ] However, notwithstanding the growing efforts that have been done to improve the speed of the molecular analysis integrating these processes into biochip[ 9 ] PCR‐based systems mostly need for qualified personnel and the use of costly reagents so that they can be executed only in laboratory environment. Therefore, these methods are not suitable for large‐scale diagnosis, limiting, de facto, their massive use for screening and control of COVID outbreak.
Immunological tests allow rapid diagnosis (Rapid Diagnosis Tests ‐RTD) and they are used in the frontline testing. These methods give result within 20 min but cause a high rate of false‐negative results.[ 10 ]
To control and mitigate the disease outbreaks effect it is mandatory the availability of new diagnostic tools for fast virus identification allowing the population to access to the early diagnosis, containing the dissemination. In this context, the development of point‐of‐care technologies (PoCT) able to perform molecular detection of SARS‐CoV‐2 can certainly make a revolution in the diagnostic procedure of the virus allowing the possibility to decentralize and make faster the molecular response.[ 9 , 11 , 12 , 13 ] The integration of these biosensor technology with IoT (Internet of things) enables the possibility to automatically transmit data by wireless network modalities (e.g., cellular data service, Wi‐Fi, Bluetooth) for a cloud heath database.[ 14 , 15 , 16 ]
However, the need to execute PCR amplification to reliably detect the virus make a limitation in the development of low‐cost and portable systems because of the need of thermal module and optical apparatus that cannot be easily integrated in portable systems. Therefore, PoCTs able to perform molecular analysis of SARS‐CoV‐2 RNA needs innovative biotechnologies for the genetic detection that simplify the detection avoid the many analytical steps required by the conventional PCR that increases the complexity and costs of the final system.
In this context, PCR‐free sensing strategies represent, therefore, one of the most appealing approaches to intercept the above‐reported need since these methods can avoid thermal cycling and simplify both analytical procedures and device architecture.[ 17 , 18 , 19 ]
A comprehensive review of innovative PCR‐free strategies for the PoC detection of SARS‐CoV‐2 is reported compared with the standard analytical approached involving both PCR‐based and immuno‐detection. Compared to other works in literature that are related to a general description of PoC technologies[ 20 ] and electrochemistry‐based detection,[ 21 , 22 ] this review provides an updated description of innovative strategies for the detection of SARS‐CoV‐2 focusing the innovative PCR‐free approaches. These new biosensors, most of which have not been thoroughly investigated and commercialized yet, involve a variety of revelation strategies suitable to provide fast and accurate analysis of the virus toward its early and massive screening and an active surveillance for the viral contagions containment.[ 21 , 23 , 24 ] This would avoid the collapse of the health systems that is due to the numerous assistances for severe patients.
2. SARS‐CoV‐2 Conventional Analysis
The SARS‐CoV‐2 virus, responsible for the COVID‐19 disease, belongs to the Coronaviridae family. Its 30 Kilobases (Kb) genome consists of a single strand RNA encoding for factors involved in the infection process, such as the spike protein (S), that is affine toward the angiotensin converting enzyme 2 (ACE 2) receptor exposed by host human cell, and the viral structure synthesis. From the pandemic outbreak, multiple variants of SARS‐CoV‐2 have been reported [ 25 ] that include several mutations accumulated inside the genome. This has altered the diagnosis, reduced the efficacy of therapeutic (vaccines and antivirals) and increased the disease transmissibility and severity.
Conventional technologies used for routine COVID‐19 diagnosis and infection control are mainly based on a) lateral flow immunoassays and b) molecular NAT (Nucleic Acid Tests) assays, as schematized in Figure 1 .
Figure 1.
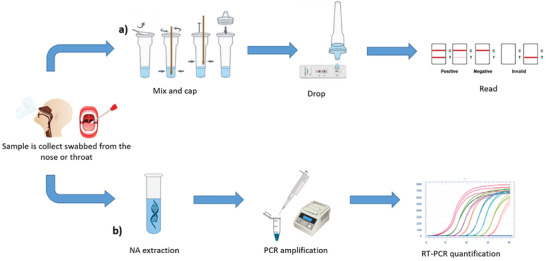
Scheme of a) immunoassay and b) NAT assay main steps.
A series of immunoassays based on the lateral flow technology (LFIA) have been proposed as rapid diagnostic tools for COVID‐19. These tests are based on immunochromatography and detected the anti‐SARS‐Cov‐2 antibodies in qualitatively o semi‐quantitatively way through optical (colorimetric/fluorescence) transduction.[ 26 , 27 ] On that, Au nanoparticles (NPs) are used as optical reporter for most of the tests.[ 28 , 29 ] An example of Au‐NPs LFIA is from Huang et al. in 2020 , reported in Figure 2a.[ 30 ] The proposed assay demonstrated a rapid and on‐site diagnosis of the IgM antibody with an ELISA indirect assay. The AuNP‐LF strips were coated with SARS‐CoV‐2 nucleoproteins that reveals within 15 min the virus by a molecular sandwich combining the Anti‐SARS‐CoV‐2 antibodies (SARS‐CoV‐2 IgMs) to the AuNPs‐conjugated antibody (AuNPs‐antihuman IgM). Chen et al.[ 31 ] improved the LFIA performances proposing a rapid and sensitive detection trough an anti‐SARS‐CoV‐2 IgG functionalized with lanthanide nanoparticles (LNPs) as fluorescent reporter (Figure 2b). The method led to the identification of SARS‐CoV‐2 IgG in human serum within 10 min, which is faster compared to the previous approach, together with a high level of reproducibility and accuracy, as confirmed by statistics, that addresses the requirements for clinical diagnostics.
Figure 2.
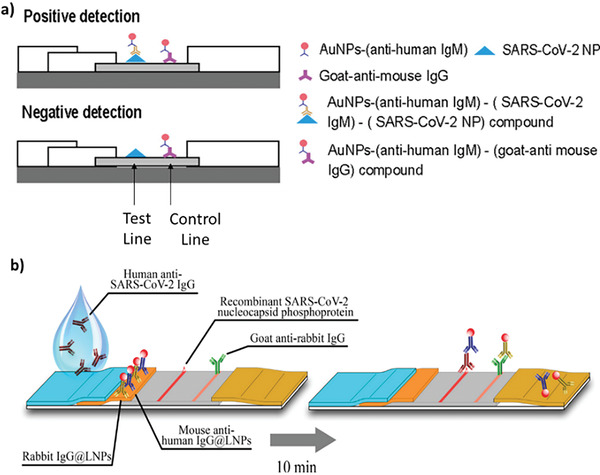
a) AuNP‐LF‐based assay for anti‐SARS‐CoV‐2 IgM chromatographic detection. Reproduced with permission.[ 30 ] Copyright 2022, American Chemical Society. b) LNP‐based LFIA for anti‐SARS‐CoV‐2 IgG fluorescent detection. Reproduced with permission.[ 31 ] Copyright 2022, American Chemical Society.
The conventional LFIA technologies allow the SARS‐CoV‐2 detection very quickly, since do not require any preliminary virus processing step, but they suffer of low levels of sensitivity and accuracy compared to the molecular methods, as it will be described below. Moreover, these methods are also limited by the stability of the antibodies and antigens used in the recognition process.
Molecular methods are based on NATs and exhibit the best sensitivity and accuracy.[ 32 ] These tests are performed through the quantitative Real‐Time RT‐PCR (RT‐qPCR) analysis that reverse‐transcribes and amplifys the ssRNA viral genome target that is detected by fluorescence recording. RT‐qPCR allows a very specific and sensitive quantification of virus and it can accurately measure the patient's viral load also during the early onset of the infection. However, PCR‐based methods need to be performed by trained personnel and in specialized laboratories and also are affected by expensive reagents and long procedures due to thermal cycling of amplification.[ 33 , 34 , 35 ] Loop‐mediated isothermal amplification (LAMP) methods have been introduced to contain these limitations. LAMP are performed at constant temperature, excluding the thermal cycling typical of the PCR reaction and simplifying the analysis.[ 36 ] Various LAMP techniques have been adapted to detect SARS‐CoV‐2 RNA[ 37 , 38 ] with sensitivities that are comparable to conventional RT‐PCR and a shorter time of analysis (<1 h). A recent work based on LAMP was developed by Ye et al. in 2022.[ 39 ] The method, called Plasmonic LAMP, includes i) an isothermally amplification of a specific SARS‐CoV‐2 genomic sequence, ii) a digestion by restriction enzymes, and iii) detection through hybridization on gold‐silver (Au‐Ag) nanoshells functionalised with oligonucleotide probes. The detection is performed by a plasmonic sensors. The plasmonic LAMP method improved the detection performances using accessible temperatures and achieving a Limit of Detection (LoD) of 10 copies per reaction, comparable with the standard PCR method (Figure 3 ).
Figure 3.
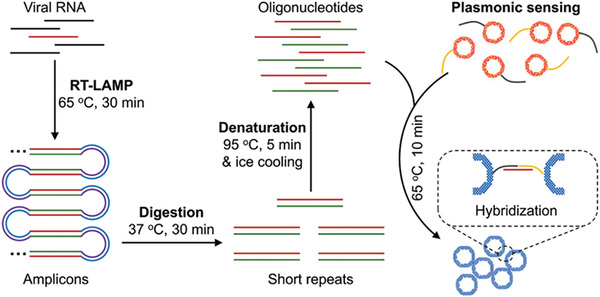
Plasmonic LAMP for SARS‐CoV‐2 RNA detection. Reproduced with permission.[ 39 ] Copyright 2022, Wiley‐VCH GmbH.
A technological advancement of PCR technology is the digital PCR that provides sensitive detection and precise quantification of nucleic acid targets minimising the bias interference. The sample is split into small aliquots (nL‐pL) that undergoes a RT‐qPCR reaction. Each aliquot is considered positive (1, fluorescent) if it contains the target, or considered negative (0) otherwise. By combining the positive and negative results and with the help of the Poisson statistic, it is possible to estimate the exact number of the nucleic acids (NA) target copies and to calculate the absolute concentration of the template in the original sample.[ 33 ] Most of the available molecular methods for SARS‐CoV‐2 are based on the digital droplet PCR (ddPCR) achieving a high sensitivity, down to 10−2 copies mL−1 of viral genome.[ 40 , 41 ]
The conventional methods above described do not fulfill the need to merge a sensitive quantification at the molecular level (10 copies of virus) together with rapid detection (within 15 min). At this regard, new approaches must be considered to create innovative methods allowing SARS‐CoV‐2 detection in PoC format, like those described below.
3. PCR‐Free Strategies for SARS‐CoV‐2 PoC Detection
The worldwide spreading of SARS‐CoV‐2 increased the demand of advanced biosensors for the COVID‐19 diagnosis and massive control in a Point‐of‐Care (PoC) format.[ 42 ] Due to the high level of integration and miniaturization,[ 43 ] the possibility of surface actuation[ 44 , 45 , 46 , 47 ] and the combination with advanced detection systems,[ 48 , 49 ] these technologies should combine the speed of analysis with the high level of accuracy and sensitivity, also, enabling the possibility for a self‐monitoring, which could exclude both laboratory training and bulky instrumentations, and a fast, integrated and decentralized analysis of the biological sample.
One of the main features that limit the conventional analytical strategies toward this PoC application is the length and complexity of the procedure and the instrumentation used for the screening. Conventional molecular tests for SARS‐CoV‐2 are based on the consolidated RT‐PCR reaction that allows to selectively and quantitatively detect the viral genome keeping the infection under control. However, this technique suffers of some limitations that mostly concern the sample collection and handling, the long‐time of analysis (given by the amplification thermal cycling) and the expensive instrumentations used, that do not match with the need of easy, fast, and portable PoC analysis.
In this scenario, the PCR‐free approach for the SARS‐CoV‐2 detection introduced a new frontier of molecular diagnostics where the viral genome is directly revealed without its amplification. To accomplish this type of alternative molecular analysis, however, is very important to guarantee the best compromise between a high‐resolution analysis, which could be able to provide the sensitive and accurate revelation of a few copies of a genetic target, and an integrable and miniaturizable dedicated measurement setup that is consistent with the idea of a PoC application. In this sense, a lot of PCR‐free approaches have been developed, so far, focusing on enzymes,[ 50 , 51 ] nucleic acids (RNA or DNA),[ 52 ] antibodies,[ 53 ] proteins[ 54 , 55 ] based recognition combined to electrochemical, electrical, optical, piezoelectric and calorimetric transduction.[ 56 , 57 , 58 ] Recent and performant PCR‐free sensors for the SARS‐CoV‐2 detection suitable for a PoC application are reported and discussed.
3.1. Electrochemical and Electrical SARS‐CoV‐2 PCR‐Free Sensors
3.1.1. Electrochemical Biosensors
Compared to other detection strategies,[ 59 ] like the optical that is affected by the issue of the luminescent probes/analytes stability[ 60 , 61 ] and the lab constraint of the measurement equipment,[ 62 , 63 ] the electrochemical (EC) transduction (such as that based on voltammetry, conductometry, impedentiometry, etc.) offer optimal performances that are perfectly suitable toward a PCR‐free approach, due to the possibility of having a high level of sensitivity and accuracy combined to a high portability and miniaturizability of the entire detection system.
Literature reports promising solutions for the electrochemical PCR‐free detection of SARS‐CoV‐2. Farzin et al.,[ 64 ] as an example, developed a voltametric PCR‐free sensor able to determine the RNA‐dependent RNA polymerase (RdRP) sequence. The technology was based on a carbon paste (CPE) electrode modified with silver ions (Ag+), hexathia‐18‐crown‐6 (HT18C6), dendrimer‐coated silicon quantum dots and PAMAM chitosan (SiQDs@PAMAM). The top of the electrode, instead, was functionalized by immobilized 5′‐amino‐oligonucletide probes. So structured, the electrode was able to sense the hybridization between probes and viral RdRP target, producing a current signal whose intensity decreased oppositely to the target concentration, with a linear response in the range of 1.0 pm –8.0 nm and a LoD of 890 copies µL−1 (0.3 pm).
Kashefi‐Kheyrabadi et al.[ 65 ] developed an electrochemical biosensor able to detect both the S and ORF1ab gene sequences of SARS‐CoV‐2 without amplification, reported in Figure 4a. This was based on a 4‐WJ hybridization system consisting in Universal DNA‐Hairpin (UDH) probes immobilized on top of screen‐printed electrodes (SPGE), modified by gold nano‐needles. When UDH probe hybridizes to 2 adapter strands (m and f), designed to specifically identify the target viral sequences, passes from the hairpin to a straight shape enabling the hybridization between adapters and target. The m adapter is labelled by a redox marker so that the S and Orf1ab genes interactions creates a current signal on top of the electrodes detected by Square‐wave Voltammetry (SWV). So composed, the biosensor was able to detected the viral genes in a one‐step and multiplex approach, achieving a LoD of 0.15 copies µL−1 (5.0 ag µL−1) and 0.21 copies µL−1 (6.8 ag µL−1) (for S and Orf1ab genes, respectively) within 1 h of analysis.
Figure 4.
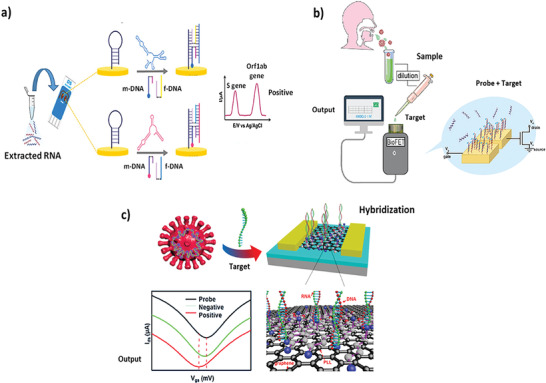
PCR‐free EC and electrical sensors for SARS‐CoV‐2 detection. a) Electrochemical sensor based on four‐way junction (4‐WJ) hybridization for the detection of SARS‐CoV‐2. Reproduced with permission.[ 65 ] Copyright 2022, Elsevier. b) EDL‐gated BioFETs to rapidly detect SARS‐CoV‐2 viral RNA in saliva. Reproduced with permission.[ 68 ] Copyright 2022, Wiley‐VCH GmbH. c) GFET biosensor for SARS‐CoV‐2 RNA detection. Reproduced with permission.[ 71 ] Copyright 2022, American Chemical Society.
Another interesting type of detection suitable for the PCR‐free sensing of SARS‐CoV‐2 detection is represented by the electrical methods based on the field effect transistor (FET). These technologies are able to directly translate interactions of the target molecules with the FET surface into readable electrical signals[ 66 ] and are potentially integrable in a PoC device due to their high sensitivity, small size, label‐free detection, although suffer of low sensitivity for direct virus detection in a physiological environment, in the so‐called Debye screening effect.[ 67 ] To overcome this limit, Paulose et al.[ 68 ] developed a saliva‐based electrical bilayer (EDL)‐gated BioFET enabling the detection of SARS‐CoV‐2 under physiological conditions. This screening tool was functionalized using oligonucleotide probes and successfully detected the viral genome up to 0.30 copies µL−1 (1 fm) of concentration in ≈1 h, reported in Figure 4b.
3.1.2. Graphene‐Based EC and Electrical Sensors
Graphene attracted great interest as alternative material of sensing surface for the PCR‐free SARS‐CoV‐2 detection,[ 69 ] due to its electrical and mechanical properties. Its excellent SVR, optical properties, thermal and electrical conductivity offer the best optimization of the sensing performances, especially, if combined to both the electrochemical and electrical detection.
In this sense, an application of graphene for PCR‐free sensing of SARS‐CoV‐2 was from Damiati et al.,[ 70 ] who developed a label free graphene‐based electrochemical sensor that can be incorporated into a flexible printed circuit board (FPCB). In the proposed strategy the graphene was used as a working electrode while a biotin‐streptavidin interaction was implied to immobilize the oligonucleotide capturing probe. Through the hybridization of the viral genome and in presence of an iron/ferrocyanide redox‐active complex, a voltametric analysis was performed for the virus detection. The careful design of the detection platform improved both selectivity and sensitivity and allowed the target quantification from 3 to 107 Copies µL−1 (100 fg mL−1 and 1 µg/ mL−1), introducing a new solution for simple and cost‐effective diagnostic test of viral infections in PoC format.
Gao et al.,[ 71 ] instead, introduced an ultrasensitive poly‐L‐lysine (PLL)‐modified graphene‐based FET (GFET) for the PCR‐free detection of SARS‐CoV‐2 genome. The PLL was used to immobilize the DNA probes, as reported in Figure 4c, and the results showed a LoD of 0.30 copies µL−1 (1 fm), with a time of analysis of 20 min. Compared to other GFET biosensors, this system gave a 113% improvement in the analytical performances, paving the basis for a new frontier of diagnostic tools suitable for the SARS‐CoV‐2 early and massive screening.
A final application of graphene was proposed by Ji et al.[ 72 ]. They presented a modified Self‐Actuated Molecular System (MECS) based on a graphene microelectrode to detect the ORF1ab genes of SARS‐CoV‐2 RNA. The MECS has a hydra‐like morphology made up of a tentacle, consisting in a flexible single‐stranded DNA with a terminal electrochemical label, and a vertical trunk, composed by a double‐stranded DNA. When the target is added, the tentacle spontaneously changes its configuration generating an electrochemical signal, revealed by DPV and SWV, by which the MECS can detect the SARS‐CoV‐2 genome within 1 minute, without any amplification, with a low LoD of 0.025 µL–1 copies, for SWV, and 0.035 µL–1 copies, for DPV.
3.1.3. Electrochemical PoC
First attempts of PCR‐free technological innovation and informatization toward a PoC application have been done by Dou et al.,[ 73 ] that proposed a universal and multiplex electrochemical wireless biosensor for the molecular detection of Sars‐CoV‐2. The WE of the sensor was functionalized with DNA tetrahedral structure probes (TSP DNA) complementary to the S gene target of the SARS‐CoV‐2 genome. The electrochemical impedance spectroscopy (EIS) was then used to measure surface steady‐state current change as function of the hybridization processes between the capture probe and the target, revealing down to 10 fm of viral RNA, with the possibility of wireless transmission to a special APP for data analysis.
A recent work by Song et al.[ 74 ] reported a diagnostic platform, shown in Figure 5 , for the on‐site detection of the Sars‐CoV‐2 N gene without amplification and with a LoD of 11.46 fm. This platform adopted a one‐pot detection protocol with an effective luminescence upconversion[ 75 ] sandwich test (PULD) and a smartphone‐controlled handheld device.
Figure 5.
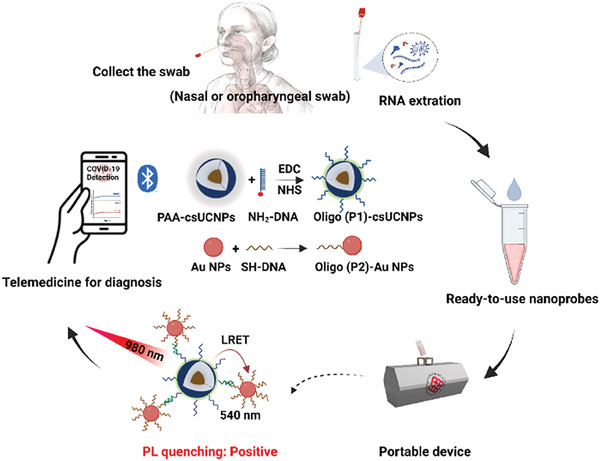
SARS‐CoV‐2 detection with smartphone‐based biosensor. The detection workflow of SARS‐CoV‐2 via PULD. Reproduced with permission.[ 74 ] Copyright 2023, Elsevier.
The Sars‐CoV‐2 N gene was captured with two core‐shell probes doped with lanthanides (csUCNP) and gold (Au NP) nanoparticles both functionalized with anchored capture probes, complementary to the N target. Each time the probes/target hybridization occurred, the csUCNPs and Au NPs were brought in proximity so that, using an excitation source at 980 nm, the fluorescence of csUCNPs could be absorbed by the Au NPs due to the luminescence resonance energy (LRET). In this way, the N gene of Sars‐CoV‐2 was quantified by measuring the variation of the fluorescence intensity and the results were shared with the patients via a bluetooth smartphone connection.
Finally Lomae et al.[ 76 ] have developed a paper‐based electrochemical sensor to detect the N gene sequence of the Sars‐Cov‐2 RNA genome. The device was functionalized with a pyrrolidinyl peptide nucleic acid (acpcPNA) used as a probe to capture the complementary RNA target. The sensor was completed with a smartphone‐assisted Sensit Smart potentiostat and a smartphone application measured the current variation given by the hybridization between the target and the acpcPNA probe. This process interfered with the redox activity of the [Fe(CN)6]3−/4− electrochemical reporter causing a decrease of the amperometric response in a dependent‐manner with the amount of Sars‐CoV‐2 genome, within a linear range of 0.1 to 200 nm and with a LoD of 1.0 pm.
3.2. Nanostructured Sensors
An important feature concerning the performances of a PCR‐free sensor in a PoC format is the surface‐to‐volume ratio (SVR). The exposure enhancement of a sensing surface can be achieved using nanostructured architectures (such as nanoparticles, nanosheet, etc.) that increases the effectiveness of affinity and interactions between the sensing surface‐active sites and the target molecules, allowing to immobilized a higher number of probes and their exposure to the target environment. This leads to an increase of sensitivity and selectivity of the PCR‐free sensor, since the probability for few copies of a molecular target to be revealed by the recognition element is maximized. Moreover, the nanostructuring of a sensing system increases the level of miniaturizability of its architecture, thus, becoming more consistent with a PoC configuration.
In this sense, there are some evidences in literature reporting the use of nanostructured surface for PCR‐free sensors of SARS‐CoV‐2. Song et al.[ 77 ] proposed a SARS‐CoV‐2 sensor based on the nanowires technology. These are nanostructures having <100 nm diameter and a good conductivity and can be made from a wide variety of materials including silicon,[ 78 , 79 , 80 , 81 , 82 ] carbon and various metals. The authors developed an electrochemical biosensor based on electro‐polymerized polyaniline (PANI) nanowires for the SARS‐CoV‐2 N gene detection, reported in Figure 6a. The biosensor manages to have excellent properties thanks to the innovative biotinylated peptides designed with two anchoring branches to bind the PANI, enabling an excellent interface coverage. Subsequently, streptavidin (SA) was used to link more biotinylated oligonucleotide probes (specific for the N gene of SARS‐CoV‐2) to the peptide‐coated interface. The PCR‐free sensor was tested on serum samples and measured the SARS‐CoV‐2 genome via differential pulse voltammetry (DPV), reporting a wide linear range (10‐14 to 10‐9 m) and an exceptional LoD of 10.5 copies µL−1 (3.5 fm).
Figure 6.
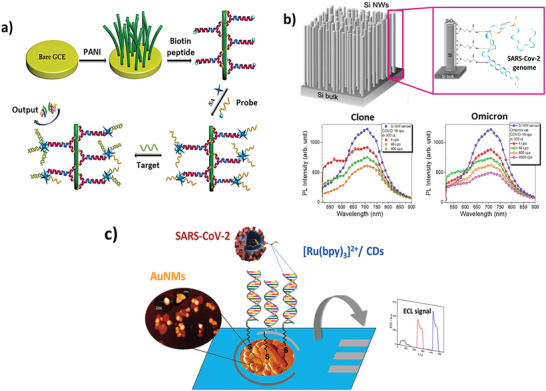
Nanostructured PCR‐free sensors. a) Antifouling electrochemical biosensor based on (PANI) nanowires for the detection of the COVID‐19 N‐gene. Reproduced with permission.[ 77 ] Copyright 2022, American Chemical Society. b) Silicon nanowires optical biosensor for the PCR‐free detection of Omicron variant of SARS‐CoV‐2. Reproduced with permission under the terms of the CC BY license.[ 83 ] Copyright 2023, the Authors. Published by Wiley‐VCH GmbH. c) Electrochemiluminescent nanostructured biosensor based on AuNMs in combination with [Ru(bpy)3]2+/CDs system for SARS‐CoV‐2 detection. Reproduced with permission.[ 85 ] Copyright 2022, Elsevier.
Leonardi et al. in 2023 proposed an optical biosensor based on silicon nanowires for the PCR‐free molecular detection of a synthetic SARS‐CoV‐2 and its real Omicron variant.[ 83 ] The nanostructuring of silicon lead to the emission of light upon the quantum confinement effect.[ 84 ] This signal was gradually quenched by the accumulation of the viral target genomes captured by oligonucleotide probes on top of the silicon nanowires, as described in Figure 6b. By measuring the intensity of this induced photo‐quenching, then, it was possible to directly reveal and quantify the viral genomes, from both a commercial SARS‐CoV‐2 clone and a swab‐isolated Omicron variant, with a LoD of 4 effective cps (red dots in Figure 6b), without any amplification and with high selectivity.
Biosensors are continuously expanding with new approaches as that of Gutiérrez‐Gálvez et al.,[ 85 ] that developed an electrochemiluminescent (ECL) nanostructured DNA biosensor for the detection of SARS‐CoV‐2. The system was composed by disposable electrodes modified with gold nanomaterials (AuNMs) and immobilized thiol‐oligonucleotide probes, reported in Figure 6c. After the hybridization of probes with specific SARS‐CoV‐2 genome sequences, the detection was carried out by using a [Ru (bpy)3]2+/CDs (carbon dots) redox‐active complex. The hybridization event generated a gap in the ECL signal which intensity was correlated to the SARS‐CoV‐2 concentration of analyzed sample. This allowed to quantify the virus with a LoD of 0.15 copies µL−1 (514 am).
3.3. NALFA‐Based Sensors
Recently, a modification of the LFA technology, widely used in the conventional immunological tests, has been developed as new PCR‐free strategy for the SARS‐CoV‐2 genome detection. This is the nucleic acid based lateral flow assay (NALFA) and allows to detect the viral RNA by combining the speed of the LFA to the sensitivity and accuracy of the molecular analysis.
Wang et al.[ 86 ] implemented a NALFA‐based biosensor for detecting SARS‐CoV‐2 in less than 1 h. The virus from the throat swab was lysed leaving the genome to hybridize to specific DNA probes. The resulting complex was, then, tagged by europium‐chelate‐based fluorescent nanoparticles (FNP) and labelled with S9.6 monoclonal antibodies. The fluorescence emission from the LFA strip was, then, revealed by hybrid capture fluorescence immunoassay (HC‐FIA), which combined the nucleic acid hybridization to the immunofluorescence analysis, excluding any extraction, reverse transcription, or amplification step. This led to the advantage for the HC‐FIA of being appliable to on‐site detection in a PoC format. Within this configuration, the biosensor achieved an accuracy of 99% on 734 samples analysed in a multi‐hospital randomized.
Also, Dighe et al.[ 87 ] designed a NALFA‐based system for SARS‐CoV‐2 detection, as described in Figure 7 . The authors proposing a system based on an RNA extraction‐free technique that utilizes a Sephadex G25 size exclusion column (NAP‐10) and biotin and 6‐carboxyfluorescein (6‐FAM) labelled oligonucleotides as highly specific probes, called ASO1 and ASO2 respectively. By using gold nanoparticles covered by cysteamine (Cyst‐AuNPs), the biosensor was able to detect the viral RNA within 30 min, achieving a Limit of Detection (LoD) of 0.02 copies µL−1.
Figure 7.
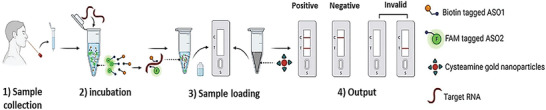
PCR‐free sensors based on NALFA technology for PoC analysis of SARS‐CoV‐2. Reproduced with permission.[ 87 ] Copyright 2022, Elsevier.
3.4. PCR‐Free Sensors Based on the RNA Sequence Direct Modification and Hybridization
The SARS‐CoV‐2 RNA identification based on its sequence modification was another diffused method that brough significant improvements of accuracy and sensitivity of PCR‐free strategies for PoC SARS‐CoV‐2 detection. One of the most representative approach was the CRISPR‐Cas technology that opened a new frontier of molecular diagnostics where the target viral genome identification and quantification is performed through a genome editing process based on the following synergic elements: CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), i.e., a short oligonucleotide sequence that act as guide during the editing process; Cas9 (CRISPR‐associated protein), i.e. an endonuclease enzyme that is able to cut specific sequences, or cleavage sites, of a target genome; and PAM (Protospace Adjacent Motif), a short oligonucleotide sequence belonging to the target genome and located close to the Cas cleavage sites. The genome editing mechanism starts when the CRISPR sequence is transcribed into small RNA fragments called guide‐RNA (or g‐RNA). The g‐RNAs, then, hybridize complementary sequences inside the target genome (i.e, the Cas cleavage sites) and, thanks to the PAM recognition, orient Cas enzyme toward its cleavage sites[ 88 ] to proceed with the final cut. If opportunely designed, the CRISPR sequence can be complementary to specific genes of SARS‐CoV‐2 genome allowing its detection by selective cut.
In 2020 Moon et al.[ 89 ] developed a SARS‐CoV‐2 colorimetric sensor based on the dCas9 enzyme, that is a mutated form of Cas lacking of the endonuclease activity and is widely used to detect pathogens sequences.[ 90 , 91 ] The method was developed on a microplate with attached the dCas9 enzyme and gRNA to specifically recognizes the target RNA. In detail, the gRNA, which contains a complementary sequence to the cleavage site of dCas9, formed a ribonucleoprotein (RNP) complex with dCas9. This complex, was incubated with viral lysate (from SARS‐CoV‐2 genome) and biotin‐PAMmer, i.e., a biotin‐protospacer adjacent motif (PAM)‐presenting oligonucleotide. After incubation, the streptavidin‐peroxidase (HRP) and 3,3′,5,5′‐tetramethylbenzidine (TMB) were added to the mix and the color change given by its oxidation allow to detect the presence of viral genome within 90 min, as schematized in Figure 8a.
Figure 8.
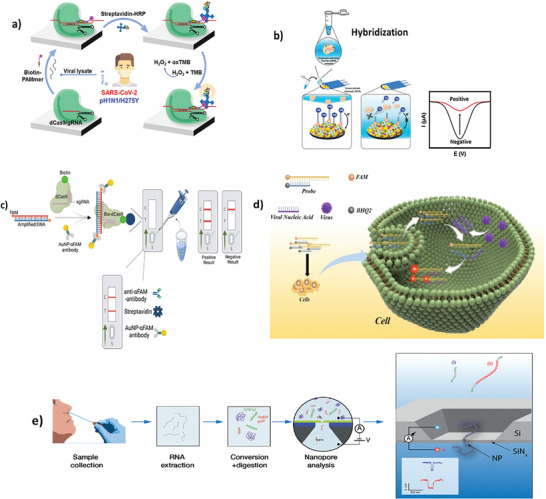
SARS‐CoV‐2 PCR‐free detection based on CRISPR‐Cas system. a) Colorimetric detection of SARS‐CoV‐2 and drug‐resistant pH1N1 using CRISPR/dCas9. Reproduced with permission.[ 89 ] Copyright 2022, American Chemical Society. b) Electrochemical biosensor for detection of SARS‐CoV‐2 RNA via CRISPR/Cas13a trans‐cleavage reaction. Reproduced with permission.[ 92 ] Copyright 2022, Elsevier. c) Bio‐SCAN platform for Rapid, Specific, and Sensitive Detection of SARS‐CoV‐2. Reproduced with permission.[ 93 ] Copyright 2022, American Chemical Society. d) PCR‑free nucleic acid testing method for RNA viruses based on linear molecular beacon probes. Reproduced with permission.[ 94 ] Copyright 2023, Springer Nature. e) PCR‐and label‐free solid‐state nanopore sensor for the quantification of SARS‐CoV‐2 RNA. Reproduced with permission.[ 95 ] Copyright 2023, Royal Society of Chemistry.
Heo et al.[ 92 ] reported a CRISPR‐Cas13a‐based electrochemical biosensor for the rapid, sensitive, and amplification‐free detection of SARS‐CoV‐2. The biosensor, shown in Figure 8b, was built by depositing nanocomposite (NC) and gold nanoflower (AuNF) on the surface of screen‐printed carbon electrodes (SPCE) to improve the conductivity, thus, the detection performances. So structured, the CRISPR‐Cas13a complex‐assisted electrochemical sensor detected the ORF and S genes of the viral genome in a linear range of 10−1–105 fg mL−1 and LoDs of 0.001 and 0.002 Copies µL−1 (4.4 × 10‐2 and 8.1 × 10‐2 fg mL−1), respectively. This was the first case in the literature of so sensitive SARS‐CoV‐2 quantification. Moreover, the platform is suitable for all‐in‐one cartridge integration and PoC applications.
Another example is the Bio‐SCAN platform from Ali et al.[ 93 ] which combines the CRISPR‐Cas method with the LFA technique, as reported in Figure 8c. Authors exploited the NAT mediated by Cas9 with appropriate modifications used to simplify the analysis. The fragment of the target genome was amplified by RPA at 42 °C with custom primers and labelled with FAM (DNA‐FAM). The extracted RNA samples are recognized by an RNP complex labelled with biotin and dCas9 (sgRNA‐bio‐dCas9). The sgRNA‐bio‐dCas9 complex recognized the DNA‐FAM target forming FAM‐DNA‐bio‐dCas9‐sgRNA and was applied to the LFA strip. This accumulated anti‐FAM antibodies labelled with gold nanoparticles (AuNP‐αFAM antibody) and gave a visual detection of the target nucleic acid on the test line. The Bio‐SCAN platform was able to detect the SARS‐CoV‐2 genome in less than 1 h from sample collection to analysis and complies with many PoC criteria such as portability, fast analysis times, selectivity and easy to use. Bio‐SCAN results were 100% in accordance to the RT‐qPCR results, used as comparison; moreover, the platform is appliable in the detection of SARS‐CoV‐2 alternative variants by, simply, customizing the sequences of FAM‐conjugated primers.
A recent work from Du et al. reported a sensing technology based on the selective recognition of the N gene of SARS‐CoV‐2 genome by a linear molecular beacon fluorescent probe.[ 94 ] The probe, a double‐stranded DNA modified with a quencher and a reporter, was able to hybridize with the target viral N gene sequence causing the release of the quencher modified strand and inducing the ‘‘Signal on’’ state of fluorescence. The signal intensity increased in a dose‐dependent manner using various SARS‐CoV‐2 genome concentrations from 0.75 to 100 nm. This PCR‐free detection was confirmed also in macrophages used as cell model to simulate the SARS‐CoV‐2 infection and the possibility of an in vitro diagnostics, as shown in Figure 8d.
Another interesting type of PCR‐free identification of SARS‐CoV‐2 genome based on its sequence modification and reading has been proposed by van Kooten et al. in 2022.[ 95 ] They developed a PCR‐and label‐free solid‐state nanopore sensor, reported in Figure 8e, for the quantification of SARS‐CoV‐2 RNA in clinical nasal swab samples. By using a reverse transcription of the RdRP gene of the extracted viral genome and its subsequent enzymatic digestion, the device was able to directly identify a SARS‐Cov‐2 target gene in a single‐molecule counting and a length‐based approach performed by a microfluidic silicon chip having an electroactive 4 nm diameter nanopore where the digested cDNA target is translocated and its nucleotide composition is electrically revealed by an ion current profiling. This allows to operate a sequencing‐like detection of the marker gene, increasing the level of sensitivity and accuracy of the PCR‐free analysis. Table 1 reports a comparison of the most representative PCR‐free sensors above described. It can be noticed that most of these biosensors are tested using artificial samples simulating clinical uses (mock samples). The input samples are very variegated (sputum, swab, serum etc etc) and the tested targets are also various from SARS‐Cov‐2 whole genome to shorten specific genes. Based on these variabilities, the corresponding sensing performances are wide and they range between some thousands of cps to few cps per reaction (105 – 2 cps/reaction). Respect to the real clinical samples, there are two examples[ 65 , 87 ] that prove the possibility for real use in qualitative test (yes/no answers), while only one study[ 83 ] demonstrated a quantitative detection using extracted real SARS‐Cov‐2 samples with a LoD of 4 cps/reaction, overcoming the PCR LoD that corresponds to ≈10 cps/reaction. Based on the above considerations, it is evident that, although these technology are very promising, however further extensive clinical validations are necessary to be used in commercial assays.
Table 1.
PCR‐free sensors for SARS‐CoV‐2 detection
| Sensor type | Sample type | Target | Transduction method | LoD | LoD [cps/µL] | LoD [cps /reaction] | NOTE | Ref |
|---|---|---|---|---|---|---|---|---|
| EC | Sputum | RdRP sequence | CV | 0.3 pm * | 3 × 104 ** | 6 × 105 ** | Tested on mock samples | [64] |
| EC | Respiratory clinical sample | S and ORF1ab gene | SWV | 5.0 ag µL−1 * | 3 × 105 ** | n.a. | Verified on clinical samples for YES/NO test | [65] |
| Electrical | RNA sequence suspended in Saliva | SARS‐CoV‐2 RNA | FET | 1 fm * | 102 ** | 7 × 103 ** | Tested on mock samples | [68] |
| Graphene‐based EC | Synthetic SARS‐CoV‐2 Orf in Saline‐sodium citrate buffer | SARS‐CoV‐2 RNA | Voltammetry | 100 fg mL−1 * | 6.5 ** | 130** | Tested on analytical samples | [70] |
| Graphene‐based Electrical | Throat swab samples | SARS‐CoV‐2 RNA | G‐FET | 1 fm * | 102 ** | 7 × 103 ** | Tested on mock samples | [71] |
| Nanostructured | Human serum | N Covid‐19 gene | DPV | 3.5 fm * | 3.5 × 102 ** | n.a. | Tested on mock samples | [77] |
| Nanostructured | Human saliva | SARS‐CoV‐2 RNA | Optical | 17 zm ** | 0.01** | 4* | Verified on extracted clinical samples | [83] |
| Nanostructured | SARS‐CoV‐2 suspended in human serum | SARS‐CoV‐2 RNA | ECL | 514 am * | 5.14 × 102 ** | 5.14 × 104 ** | Tested on mock samples | [85] |
| LFA‐based | Nasal/nasopharyngeal swab | SARS‐CoV‐2 RNA | Colorimetric | / | 0.02* | 2** | Verified on clinical samples for YES/NO test | [87] |
| CRISPR‐Cas‐based | Saliva | ORF and S genes | DPV | 3 fg mL−1 * | 0.2 ** | n.a. | Tested on mock samples | [92] |
Value from the original paper.
Calculated from data reported in the original paper.
n.a = Not available.
4. Future Perspectives
The proposed sensing strategies (Table 1) represent promising solutions for fast and reliable detection of COVID‐19 and data collection in a PoC format, with the aim to contribute to an efficient patient management in terms of fast diagnosis and prompt answer to the treatment, and to address the current and future outbreaks of coronavirus infection.
In contrast to the conventional amplification‐based methods, the reported PCR‐free sensors proved to be able to perform the direct detection of the SARS‐CoV‐2 genome without its amplification, that is particularly challenging since the starting viral RNA concentration in an early or suspected infected human sample is usually very low. However, in order to be suitable for a PoC application, the PCR‐free approach should be, furtherly, implemented.
All sensors, in fact, lacked of an on‐board integration of a specific chemistry and module by which performing the SARS‐CoV‐2 RNA purification that, so far, was always executed outside of the sensor.[ 77 , 78 , 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 , 87 , 88 , 89 , 90 , 91 , 92 , 93 , 94 , 95 , 96 , 97 , 98 ] In this sense, literature reports some solutions based on miniaturized microfluidic and silicon‐based modules[ 96 , 97 ] or high performances lysis buffers combined to heat inactivation process[ 98 ] that could be suitable for the integrated RNA/DNA extraction in a PoC format. This type of integration would increase the speed and automation of the entire molecular analysis toward a self‐patient screening approach. Moreover, the integrated genome extraction would reduce the cost of analysis, that are, mostly, affected by the consumption of a lot of commercial RNA purification kits.
Another perspective, instead, concerns the informatic implementation, as schematized in Figure 9 . A PCR‐free sensor (schemed in Figure 9a,b) should be opportunely designed to easily communicate with dedicated clouds where to, fast, store a high amount of analytical data acquired by the sensor (Figure 9c). The cloud repository (Figure 9d) would be useful in the way it could innovate the way to control of the infection data. Encrypted Wi‐Fi modules and protocols, for example, could be used to connect the sensor with the decentralised environments such as hospitalized recovery structures and Public Health surveillance worldwide centres. Then, it would be possible to, quickly, register the infection, discriminating between asymptomatic and pauci‐symptomatic subjects, and proceed with the patient isolation and the tracking of its contagion history. This could provide a massive global screening and efficient control of the COVID‐19 pandemic.
Figure 9.
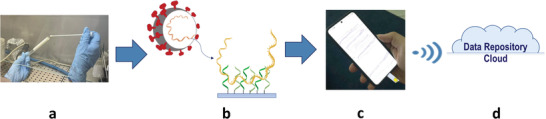
PoC Molecular Analysis of SARS‐CoV‐2: a) swab collection; b) PCR‐free virus genome analysis; c) integrated and miniaturized system for data acquisition; d) remote data repository on cloud for large‐scale screening.
5. Conclusion
COVID‐19 pandemic introduced the need for new accurate and efficient diagnostic tools for the SARS‐CoV‐2 detection. Many efforts, nowadays, have been spent toward the development of innovative biosensing systems that could quickly and precisely screen the virus in a PoC format, solving the main drawbacks related to the conventional diagnostic techniques. Commercial immunological tests, in fact, allow the fast detection of SARS‐CoV‐2 but lack in sensitivity, thus, limiting the screening of the infection only to its advanced stages. Conventional PCR‐based methods for SARS‐CoV‐2 molecular detection, instead, offer a highly sensitive and specific diagnosis but have some limitations. They are, in fact, intrinsically quite laborious, involving several analytical steps for the sample detection, so that result constrained to specialized and centralized laboratories and expert personnel. Moreover, PCR‐based methods need long time to perform the analysis (especially due to the thermal cycling), with a total time from specimen collection to test results that can add up to days, and imply a certain cost consumption, with ≈30−80 € per sample analysis.
In this scenario, literature reported the PCR‐free biosensors as innovative solutions to face the growing spread of SARS‐CoV‐2 and COIVD‐19 pandemic. These technologies have been developed to overcome the drawbacks of conventional molecular methods and opened a new frontier of the infectious disease analysis proposing fast, sensitive, accurate and portable analytical strategies for the SARS‐CoV‐2 detection.
PCR‐free technologies are designed to perform the direct detection of SARS‐CoV‐2 RNA without the PCR amplification, thus, optimizing time, costs, and level of decentralization of the molecular analysis. In order to achieve the best condition for the direct identification and quantification of the viral genome, in the last two years, a lot of PCR‐free strategies have been proposed.
The EC and FET‐based PCR‐free sensors, especially if combined to the properties of the graphene, reported the most performant detection approach since exclude the optical labelling of target, that usually affect the stability and sensitivity of analysis, and can be easily shaped for miniaturized sensing applications, according to the PoC skills.
The nanostructured PCR‐free sensors, instead, proposed improved SVR of the sensing surface able to increase the number of immobilized probes to be exposed to the direct hybridization of the genetic target, thus, enhancing the level of sensitivity and accuracy.
The NALFA‐based sensors combined the analytical speed typical of the LFA technology with the high sensitivity of the NA analysis. By using a chromatographic approach applied to the SARS‐CoV‐2 genome detection, these devices allowed to perform fast and highly sensitive viral screening with LoD below 1 copies µL−1.
Lastly, the sensors based on the RNA sequence direct modification and hybridization increased the level of accuracy and sensitivity of the PCR‐free analysis, as the genome editing of the CRISPR‐Cas technology that introduced the possibility to directly identify few copies of SARS‐CoV‐2 RNA through its enzyme‐based recognition.
Taken together, these proposed PCR‐free sensing strategies and their potential implementations, in terms of integrated genetic extraction and informatic configuration, pave the basis for a new frontier of fast, precise, massive, and early screening tools, suitable for the PoC decentralized and low‐resource settings analysis, which could improve the quality of SARS‐CoV‐2 infection diagnosis and the efficiency of viral pandemic control.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
The research was supported by the co‐financing of the European Union ‐FSE‐REACT‐EU, PON Research and Innovation 2014‐2020 DM1062 / 2021 (CUP J45F21001750007), and the European Union's Horizon Europe EIC Pathfinder Open programme “ECLIPSE project” (Grant Agreement Nr. 101046787).
Open access Funding provided by University of Messina within the CRUI‐CARE Agreement.
Biographies
Emanuele Luigi Sciuto obtained his Ph.D. in materials science and nanotechnologies at the University of Catania, with an industrial doctoral scholarship at STMicroelectronics. He is the winner of the 3rd Ph.D. Thesis National Award Contest “Dario Scapaticci” organized by MESAP industrial pole of Torino. Now, he is a researcher in chemistry at the University of Messina and his research is focused on the development of advanced nanotechnologies and biointerfaces for biosensing applications in biomedicine and environmental monitoring. Currently, he is working on the development and characterization of nanostructured materials for innovative genetic “point‐of‐care” sensing applications.

Sabrina Conoci is full professor of general and inorganic chemistry at the University of University of Messina and University of Bologna. She is also Rector Delegate to Technology Transfer; Scientific Advisor of STMicroelectronics in the advanced research on innovative sensors; member of the Strategic and Advisory Council of the Graphene Flaghship. She was principal investigator and work packages leader of several Italian and European research projects. Her research focuses on the development of multifunctional nanostructured systems, advanced bio‐nanotechnologies and innovative materials and lab‐on‐chip for medical point‐of‐care devices.

Calorenni P., Leonardi A. A., Sciuto E. L., Rizzo M. G., Faro M. J. L., Fazio B., Irrera A., Conoci S., PCR‐Free Innovative Strategies for SARS‐CoV‐2 Detection. Adv. Healthcare Mater. 2023, 12, 2300512. 10.1002/adhm.202300512
Contributor Information
Emanuele L. Sciuto, Email: emanueleluigi.sciuto@unime.it.
Sabrina Conoci, Email: sabrina.conoci@unime.it.
References
- 1. Gorbalenya A. E., Baker S. C., Baric R. S., de Groot R. J., Drosten C., Gulyaeva A. A., Haagmans B. L., Lauber C., Leontovich A. M., Neuman B. W., Penzar D., Perlman S., Poon L. L. M., Samborskiy D. V., Sidorov I. A., Sola I., Ziebuhr J., Nat. Microbiol. 2020, 5, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu F., Zhao S., Yu B., Chen Y.‐M., Wang W., Song Z.‐G., Hu Y., Tao Z.‐W., Tian J.‐H., Pei Y.‐Y., Yuan M.‐L., Zhang Y.‐L., Dai F.‐H., Liu Y., Wang Q.‐M., Zheng J.‐J., Xu L., Holmes E. C., Zhang Y.‐Z., Nature 2020, 579, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou P., Yang X.‐L., Wang X.‐G., Hu B., Zhang L., Zhang W., Si H.‐R., Zhu Y., Li B., Huang C.‐L., Chen H.‐D., Chen J., Luo Y., Guo H., Jiang R.‐D., Liu M.‐Q., Chen Y., Shen X.‐R., Wang X., Zheng X.‐S., Zhao K., Chen Q.‐J., Deng F., Liu L.‐L., Yan B., Zhan F.‐X., Wang Y.‐Y., Xiao G.‐F., Shi Z.‐L., Nature 2020, 579, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.“Coronavirus Disease (COVID‐19) Situation Reports,” can be found under https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019/situation‐reports, (accessed: January 2023).
- 5. Fakhruddin B. (S. H. M.)., Blanchard K., Ragupathy D., Prog. Disaster Sci. 2020, 7, 100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tahamtan A., Ardebili A., Expert Rev. Mol. Diagn. 2020, 20, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sohni Y., Lab. Med. 2020, 52, 107. [Google Scholar]
- 8. Fung B., Gopez A., Servellita V., Arevalo S., Ho C., Deucher A., Thornborrow E., Chiu C., Miller S., J. Clin. Microbiol. 2020, 58, 01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rizzo M. G., Carnazza S., De Plano L. M., Franco D., Nicolò M. S., Zammuto V., Petralia S., Calabrese G., Gugliandolo C., Conoci S., Guglielmino S. P. P., Sens. Actuators, B 2021, 329, 129227. [Google Scholar]
- 10. Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S., Bruno R., Mojoli F., Baldanti F., Members of the San Matteo Pavia COVID‐19 Task Force , J. Med. Virol. 2020, 92, 1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao B., Wang K., Xu H., Qin Q., Yang J., Zheng W., Jin Q., Cui D., Sens. Actuators, A 2020, 312, 112130. [Google Scholar]
- 12. Petralia S., Conoci S., ACS Sens. 2017, 2, 876. [DOI] [PubMed] [Google Scholar]
- 13. Nikolaou P., Sciuto E. L., Zanut A., Petralia S., Valenti G., Paolucci F., Prodi L., Conoci S., Biosens. Bioelectron. 2022, 209, 114165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arumugam S., Colburn D. A. M., Sia S. K., Adv. Mater. Technol. 2020, 5, 1900720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui Y., Sensors 2017, 17, 2289.28991220 [Google Scholar]
- 16. Choudhuri A., Chatterjee J. M., Garg S., in Internet of Things in Biomedical Engineering (Eds: Balas V. E., Son L. H., Jha S., Khari M., Kumar R.), Academic Press, Cambridge: 2019, pp. 131–160. [Google Scholar]
- 17. Petralia S., Sciuto E. L., Di Pietro M. L., Zimbone M., Grimaldi M. G., Conoci S., Analyst 2017, 142, 2090. [DOI] [PubMed] [Google Scholar]
- 18. Giamblanco N., Conoci S., Russo D., Marletta G., RSC Adv. 2015, 5, 38152. [Google Scholar]
- 19. Leonardi A. A., Lo Faro M. J., Petralia S., Fazio B., Musumeci P., Conoci S., Irrera A., Priolo F., ACS Sens. 2018, 3, 1690. [DOI] [PubMed] [Google Scholar]
- 20. Suleman S., Shukla S. K., Malhotra N., Bukkitgar S. D., Shetti N. P., Pilloton R., Narang J., Nee Tan Y., Aminabhavi T. M., Chem. Eng. J. 2021, 414, 128759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campos‐Ferreira D., Visani V., Córdula C., Nascimento G. A., Montenegro L. M. L., Schindler H. C., Cavalcanti I. M. F., Biochem. Eng. J. 2021, 176, 108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamidi‐Asl E., Heidari‐Khoshkelat L., Bakhsh Raoof J., Richard T. P., Farhad S., Ghani M., Microchem. J. 2022, 178, 107322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petersen E., Petrosillo N., Koopmans M., Beeching N., Di Caro A., Gkrania‐Klotsas E., Kantele A., Kohlmann R., Koopmans M., Lim P.‐L., Markotic A., López‐Vélez R., Poirel L., Rossen J. W. A., Stienstra Y., Storgaard M., Clin. Microbiol. Infect. 2018, 24, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Girum T., Lentiro K., Geremew M., Migora B., Shewamare S., Trop. Med. Health 2020, 48, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.“Tracking SARS‐CoV‐2 variants ,” can be found under https://www.who.int/activities/tracking‐SARS‐CoV‐2‐variants, (accessed: January 2023).
- 26. Cheng C.‐M., Sci. Rep. 2020, 10, 14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koczula K. M., Gallotta A., Essays Biochem. 2016, 60, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y., Liu X., Wang L., Yang H., Zhang X., Zhu C., Wang W., Yan L., Li B., Sci. Rep. 2020, 10, 9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsai T.‐T., Huang T.‐H., Chen C.‐A., Ho N. Y.‐J., Chou Y.‐J., Chen C.‐F., Sci. Rep. 2018, 8, 17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang C., Wen T., Shi F.‐J., Zeng X.‐Y., Jiao Y.‐J., ACS Omega 2020, 5, 12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G., Anal. Chem. 2020, 92, 7226. [DOI] [PubMed] [Google Scholar]
- 32. CDC , https://www.cdc.gov/coronavirus/2019‐ncov/symptoms‐testing/testing.html (accessed: January 2023).
- 33. Sciuto E. L., Leonardi A. A., Calabrese G., Luca G. D., Coniglio M. A., Irrera A., Conoci S., Biomolecules 2021, 11, 1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Almassian D. R., Cockrell L. M., Nelson W. M., Chem. Soc. Rev. 2013, 42, 8769. [DOI] [PubMed] [Google Scholar]
- 35. Jin Y.‐H., Cai L., Cheng Z.‐S., Cheng H., Deng T., Fan Y.‐P., Fang C., Huang D., Huang L.‐Q., Huang Q., Han Y., Hu B., Hu F., Li B.‐H., Li Y.‐R., Liang K., Lin L.‐K., Luo L.‐S., Ma J., Ma L.‐L., Peng Z.‐Y., Pan Y.‐B., Pan Z.‐Y., Ren X.‐Q., Sun H.‐M., Wang Y., Wang Y.‐Y., Weng H., Wei C.‐J., Wu D.‐F., et al., Mil. Med. Res. 2020, 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Notomi T., Nucleic Acids Res. 2000, 28, 63e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R. O., Wei H., Tanner N. A., 2020, 2020.02.26.20028373.
- 38. Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.‐H., Yin X., Clin. Chem. 2020, 66, 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye H., Nowak C., Liu Y., Li Y., Zhang T., Bleris L., Qin Z., Small 2022, 18, 2107832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hindson C. M., Chevillet J. R., Briggs H. A., Gallichotte E. N., Ruf I. K., Hindson B. J., Vessella R. L., Tewari M., Nat. Methods 2013, 10, 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M., Zhao Y., Zhang X., Zhu T., Peng T., Xie J., Gao Y., Wang D., Dai X., Fang X., Talanta 2021, 224, 121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vandenberg O., Martiny D., Rochas O., Van Belkum A., Kozlakidis Z., Nat. Rev. Microbiol. 2021, 19, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sciuto E. L., Petralia S., Calabrese G., Conoci S., Biotechnol. Bioeng. 2020, 117, 1554. [DOI] [PubMed] [Google Scholar]
- 44. Sciuto E. L., Bongiorno C., Scandurra A., Petralia S., Cosentino T., Conoci S., Sinatra F., Libertino S., Chemosensors 2018, 6, 59. [Google Scholar]
- 45. Petralia S., Cosentino T., Sinatra F., Favetta M., Fiorenza P., Bongiorno C., Sciuto E. L., Conoci S., Libertino S., Sens. Actuators, B 2017, 252, 492. [Google Scholar]
- 46. Sortino S., Petralia S., Condorelli G. G., Conoci S., Condorelli G., Langmuir 2003, 19, 536. [Google Scholar]
- 47. Callari F. L., Petralia S., Conoci S., Sortino S., New J. Chem. 2008, 32, 1899. [Google Scholar]
- 48. Favetta M., Valletta A., Fortunato G., Castagna M. E., Conoci S., Sciuto E. L., Cosentino T., Sinatra F., Libertino S., Sens. Biosensing Res. 2015, 6, 72. [Google Scholar]
- 49. Santangelo M. F., Sciuto E. L., Busacca A. C., Petralia S., Conoci S., Libertino S., Sens. Biosensing Res. 2015, 6, 95. [Google Scholar]
- 50. Hanpanich O., Saito K., Shimada N., Maruyama A., Biosens. Bioelectron. 2020, 165, 112383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hasanzadeh M., Nahar A. S., Hassanpour S., Shadjou N., Mokhtarzadeh A., Mohammadi J., Enzyme Microb. Technol. 2017, 105, 64. [DOI] [PubMed] [Google Scholar]
- 52. Chen Y., Qian C., Liu C., Shen H., Wang Z., Ping J., Wu J., Chen H., Biosens. Bioelectron. 2020, 153, 112049. [DOI] [PubMed] [Google Scholar]
- 53. Cui F., Zhou H. S, Biosens. Bioelectron. 2020, 165, 112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mo G. C. H., Posner C., Rodriguez E. A., Sun T., Zhang J., Nat. Commun. 2020, 11, 1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S., Sensors 2020, 20, 3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thevenot D. R., Tóth K., Durst R. A., Wilson G. S., Pure Appl. Chem. 1999, 71, 2333. [Google Scholar]
- 57. Shen Y., Anwar T. B., Mulchandani A., Sens. Actuators Rep. 2021, 3, 100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Campàs M., Katakis I., TrAC, Trends Anal. Chem. 2004, 23, 49. [Google Scholar]
- 59. Cho I.‐H., Kim D. H., Park S., Biomater. Res. 2020, 24, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sciuto E. L., Coniglio M. A., Corso D., Van Der Meer J. R., Acerbi F., Gola A., Libertino S., Water (Switzerland) 2019, 11, 1986. [Google Scholar]
- 61. Sciuto E. L., Santangelo M. F., Villaggio G., Sinatra F., Bongiorno C., Nicotra G., Libertino S., Sens. Biosensing Res. 2015, 6, 67. [Google Scholar]
- 62. Santangelo M. F., Sanfilippo D., Fallica G., Busacca A. C., Pagano R., Sciuto E. L., Lombardo S., Libertino S., presented at 2014 Fotonica AEIT Italian Conf. on Photonics Technologies , Naples, Italy, May 2014. [Google Scholar]
- 63. Santangelo M. F., Pagano R., Lombardo S., Sciuto E. L., Sanfilippo D., Fallica G., Sinatra F., Libertino S., presented at Proc. SPIE , San Francisco, CA, March 2014. [Google Scholar]
- 64. Farzin L., Sadjadi S., Sheini A., Mohagheghpour E., Microchim. Acta 2021, 188, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kashefi‐Kheyrabadi L., Nguyen H. V., Go A., Baek C., Jang N., Lee J. M., Cho N.‐H., Min J., Lee M.‐H., Biosens. Bioelectron. 2022, 195, 113649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu S., Guo X., NPG Asia Mater. 2012, 4, 23. [Google Scholar]
- 67. Chu C.‐H., Sarangadharan I., Regmi A., Chen Y.‐W., Hsu C.‐P., Chang W.‐H., Lee G.‐Y., Chyi J.‐I., Chen C.‐C., Shiesh S.‐C., Lee G.‐B., Wang Y.‐L., Sci. Rep. 2017, 7, 5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Paulose A. K., Huang C.‐C., Chen P.‐H, Tripathi A., Chen P.‐H., Huang Y.‐S., Wang Y.‐L., Adv. Mater. Technol. 2022, 7, 2100842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang X., Cui H., Gui Y., Sensors 2017, 17, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Damiati S., Sopstad S., Peacock M., Akhtar A. S., Pinto I., Soares R. R. G., Russom A., IEEE Sens. J. 2021, 21, 13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gao J., Wang C., Wang C., Chu Y., Wang S., Sun M. Y., Ji H., Gao Y., Wang Y., Han Y., Song F., Liu H., Zhang Y., Han L., Anal. Chem. 2022, 94, 1626. [DOI] [PubMed] [Google Scholar]
- 72. Ji D., Guo M., Wu Y., Liu W., Luo S., Wang X., Kang H., Chen Y., Dai C., Kong D., Ma H., Liu Y., Wei D., J. Am. Chem. Soc. 2022, 144, 13526. [DOI] [PubMed] [Google Scholar]
- 73. Dou Y., Su J., Chen S., Li T., Wang L., Ding X., Song S., Fan C., Chem. Commun. 2022, 58, 6108. [DOI] [PubMed] [Google Scholar]
- 74. Song M., Wong M.‐C., Li L., Guo F., Liu Y., Ma Y., Lao X., Wang P., Chen H., Yang M., Hao J., Biosens. Bioelectron. 2023, 222, 114987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yao J., Huang C., Liu C., Yang M., Talanta 2020, 208, 120157. [DOI] [PubMed] [Google Scholar]
- 76. Lomae A., Preechakasedkit P., Hanpanich O., Ozer T., Henry C. S., Maruyama A., Pasomsub E., Phuphuakrat A., Rengpipat S., Vilaivan T., Chailapakul O., Ruecha N., Ngamrojanavanich N., Talanta 2023, 253, 123992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Song Z., Ma Y., Chen M., Ambrosi A., Ding C., Luo X., Anal. Chem. 2021, 93, 5963. [DOI] [PubMed] [Google Scholar]
- 78. Leonardi A. A., Lo Faro M. J., Di Franco C., Palazzo G., D'Andrea C., Morganti D., Manoli K., Musumeci P., Fazio B., Lanza M., Torsi L., Priolo F., Irrera A., J. Mater. Sci.: Mater. Electron. 2020, 31, 10. [Google Scholar]
- 79. Leonardi A. A., Lo Faro M. J., Irrera A., Nanomaterials 2020, 10, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Leonardi A. A., Faro M. J. L., Irrera A., Nanomaterials 2021, 11, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Leonardi A. A., Lo Faro M. J., Irrera A., Anal. Chim. Acta 2021, 1160, 338393. [DOI] [PubMed] [Google Scholar]
- 82. Lo Faro M. J., Leonardi A. A., Priolo F., Fazio B., Miritello M., Irrera A., Sci. Rep. 2020, 10, 12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Leonardi A. A., Sciuto E. L., Lo Faro M. J., Fazio B., Rizzo M. G., Calabrese G., Francioso L., Picca R., Nastasi F., Mancuso G., Spinella C., Knoll W., Irrera A., Conoci S., Nano Select 2023, 4, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mohammad N. S., J. Phys.: Condens. Matter 2014, 26, 423202. [DOI] [PubMed] [Google Scholar]
- 85. Gutiérrez‐Gálvez L., Del Caño R., Menéndez‐Luque I., García‐Nieto D., Rodríguez‐Peña M., Luna M., Pineda T., Pariente F., García‐Mendiola T., Lorenzo E., Talanta 2022, 240, 123203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang D., He S., Wang X., Yan Y., Liu J., Wu S., Liu S., Lei Y., Chen M., Li L., Zhang J., Zhang L., Hu X., Zheng X., Bai J., Zhang Y., Zhang Y., Song M., Tang Y., Nat. Biomed. Eng. 2020, 4, 1150. [DOI] [PubMed] [Google Scholar]
- 87. Dighe K., Moitra P., Alafeef M., Gunaseelan N., Pan D., Biosens. Bioelectron. 2022, 200, 113900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li Y., Li S., Wang J., Liu G., Trends Biotechnol. 2019, 37, 730. [DOI] [PubMed] [Google Scholar]
- 89. Moon J., Kwon H.‐J., Yong D., Lee I.‐C., Kim H., Kang H., Lim E.‐K., Lee K.‐S., Jung J., Park H. G., Kang T., ACS Sens. 2020, 5, 4017. [DOI] [PubMed] [Google Scholar]
- 90. Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A., Nature 2014, 507, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jiao C., Sharma S., Dugar G., Peeck N. L., Bischler T., Wimmer F., Yu Y., Barquist L., Schoen C., Kurzai O., Sharma C. M., Beisel C. L., Science 2021, 372, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Heo W., Lee K., Park S., Hyun K.‐A., Jung H.‐I., Biosens. Bioelectron. 2022, 201, 113960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ali Z., Sánchez E., Tehseen M., Mahas A., Marsic T., Aman R., Sivakrishna Rao G., Alhamlan F. S., Alsanea M. S., Al‐Qahtani A. A., Hamdan S., Mahfouz M., ACS Synth. Biol. 2022, 11, 406. [DOI] [PubMed] [Google Scholar]
- 94. Du F., Zhang W., Yao H., Xia Y., Zhang X., Yang P., Ning P., J. Nanobiotechnol. 2022, 20, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Van Kooten X. F., Rozevsky Y., Marom Y., Ben Sadeh E., Meller A., Nanoscale 2022, 14, 4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Petralia S., Sciuto E. L., Conoci S., Analyst 2017, 142, 140. [DOI] [PubMed] [Google Scholar]
- 97. Shaw K. J., Hughes E. M., Dyer C. E., Greenman J., Haswell S. J., Lab. Invest. 2013, 93, 961. [DOI] [PubMed] [Google Scholar]
- 98. Azmi I., Faizan M. I., Kumar R., Raj Yadav S., Chaudhary N., Kumar Singh D., Butola R., Ganotra A., Datt Joshi G., Deep Jhingan G., Iqbal J., Joshi M. C., Ahmad T., Front. Cell Infect. Microbiol. 2021, 11, 632646. [DOI] [PMC free article] [PubMed] [Google Scholar]


