Abstract
Membrane fusion by human immunodeficiency virus type 1 (HIV-1) is promoted by the refolding of the viral envelope glycoprotein into a fusion-active conformation. The structure of the gp41 ectodomain core in its fusion-active state is a trimer of hairpins in which three antiparallel carboxyl-terminal helices pack into hydrophobic grooves on the surface of an amino-terminal trimeric coiled coil. In an effort to identify amino acid residues in these grooves that are critical for gp41 activation, we have used alanine-scanning mutagenesis to investigate the importance of individual side chains in determining the biophysical properties of the gp41 core and the membrane fusion activity of the gp120-gp41 complex. Alanine substitutions at Leu-556, Leu-565, Val-570, Gly-572, and Arg-579 positions severely impaired membrane fusion activity in envelope glycoproteins that were for the most part normally expressed. Whereas alanine mutations at Leu-565 and Val-570 destabilized the trimer-of-hairpins structure, mutations at Gly-572 and Arg-579 led to the formation of a stable gp41 core. Our results suggest that the Leu-565 and Val-570 residues are important determinants of conserved packing interactions between the amino- and carboxyl-terminal helices of gp41. We propose that the high degree of sequence conservation at Gly-572 and Arg-579 may result from selective pressures imposed by prefusogenic conformations of the HIV-1 envelope glycoprotein. Further analysis of the gp41 activation process may elucidate targets for antiviral intervention.
Infection by human immunodeficiency virus type 1 (HIV-1) is initiated by fusion of the viral and cellular membranes to allow virus entry into the cell. This process is mediated by the viral envelope glycoprotein through interaction with cellular receptors. The HIV-1 envelope glycoprotein is synthesized as a precursor polyprotein, gp160, that is proteolytically processed to generate two subunits, the surface glycoprotein gp120 and the transmembrane glycoprotein gp41 (53). These subunits remain noncovalently associated to form the oligomeric envelope glycoprotein spike on the viral membrane. gp120 is responsible for the sequential binding of the envelope glycoprotein spike to CD4 and a chemokine receptor (typically CCR5 or CXCR4) on the surface of target cells (reviewed in references 2 and 75). These events trigger gp41 to undergo conformational changes that are crucial for activation of HIV-1 membrane fusion. By analogy with the pH-induced structural changes in the hemagglutinin (HA) protein of influenza virus, the HIV-1 fusion activation process likely involves substantial conformational changes from a metastable prefusogenic state to an energetically more stable fusogenic conformation (reviewed in references 14 and 64). The control of these structural rearrangements is thought to be central to HIV-1 entry and to strategies for intervention.
The structure and mechanism of the gp41 molecule have been extensively studied. Protein dissection studies demonstrated that the N- and C-terminal heptad repeat regions of the gp41 ectodomain associate to form an α-helical trimer of antiparallel dimers (50). X-ray crystallographic analysis confirmed that this gp41 core is a trimer of hairpins (13, 68, 69). Three N-terminal helices form a trimeric coiled coil, and three C-terminal helices pack in the reverse direction into three hydrophobic grooves on the surface of the coiled coil. Recent evidence indicates that this helical-hairpin structure corresponds to the fusion-active conformation of gp41 (13, 36, 50, 68, 69). Because the membrane anchor and the fusion peptide of the gp41 ectodomain are embedded in the viral and target cell membranes, respectively, the formation of the fusogenic hairpin structure results in the colocalization of the two membranes and thus overcomes the energy barrier for membrane fusion (27, 32, 69).
Peptides corresponding to the C-terminal heptad repeat region, referred to as C peptides, can specifically inhibit viral entry into target cells at nanomolar concentrations (37, 73). One such peptide (T-20) is in clinical study and has shown antiviral activity in humans (42). T-20 binds to gp41 only after interaction of the envelope glycoprotein complex with the cellular receptors (27, 39, 56) and is thought to act in a dominant-negative manner by binding to the N-terminal coiled coil of gp41 during its conformational change to the fusogenic state (15, 50, 72). Considerations of the efficiency of C peptide competition with its cognate sequence suggest a relatively long-lived prehairpin intermediate (14, 27, 39, 56). Intermediate states of the gp41 molecule may serve as a target for the development of small-molecule HIV-1 inhibitors (22, 25).
Considerable evidence now implies that packing interactions between the central coiled-coil trimer and the C-terminal helix are important determinants of HIV-1 entry and its inhibition (35, 54, 60, 63). For example, the replacement of a conserved glutamine (Gln-652), buried in this helical interface, by the hydrophobic residue leucine increases HIV-1 infectivity and underlies an enhancement in the antiviral activity of C peptides (8, 63). Biophysical and structural characterization directly demonstrates that this substitution strengthens the interhelix interaction in the fusogenic hairpin structure by providing additional hydrophobic packing forces (63). Taken together, these results have been interpreted to indicate that the receptor-triggered conformational changes of the HIV-1 envelope glycoprotein are thermodynamically controlled and that the process of membrane apposition and lipid bilayer fusion is driven by the currency of energy released from the formation of the fusogenic gp41 core (33, 49, 51, 63, 69).
Therefore, it is of fundamental importance to understand the structural and mechanistic basis for the helical interactions in this core. We show here, using alanine-scanning mutagenesis, that amino acids Leu-556, Leu-565, Val-570, Gly-572, and Arg-579 at e and g positions of the N34 coiled coil are essential for envelope glycoprotein function. Whereas Leu-565 and Val-570 contribute to the conserved packing interactions between the N- and C-terminal helices of gp41, Gly-572 and Arg-579 appear to be critical for prefusogenic conformations of the HIV-1 envelope glycoprotein complex.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
High-fidelity XL PCR (rTth and Vent DNA polymerases; PE Applied Biosystems) and oligonucleotide primers envA and envN (29) were used to adapt the HIV-1 HXB2 rev and env genes from the plasmid pIIIenv 3-1 (65). Expression of the HXB2 envelope glycoprotein was within the context of the eukaryotic expression vector pCR-Uni 3.1 (Invitrogen) (45). Alanine mutations were introduced into the HXB2 envelope glycoprotein and the N34(L6)C28 peptide (51) using QuikChange mutagenesis (Stratagene, La Jolla, Calif.) and single-stranded mutagenesis (44), respectively. Mutations were verified by DNA sequencing.
Envelope glycoprotein expression.
Simian COS-7 cells were used for transient expression of the envelope glycoprotein (75). Typically, 3 μg of the HIV-1 env expression plasmid and 9 μl of FuGENE-6 reagent (Roche Molecular Biochemicals) were used, according to the manufacturer's instructions, to transfect 4 × 105 to 8 × 105 cells in a 6-cm culture dish. Cultures were washed 18 h later with physiological buffered saline (PBS) and refed with Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Transfection efficiencies were determined in microcultures from the transfected cell cultures. These microcultures were fixed with −20°C methanol-acetone (1:1) and stained using biotinylated anti-HIV immunoglobulin (HIVIG) from infected persons, NeutrAvidin-horseradish peroxidase (HRP) (Pierce Chemical Corp., Rockford, Ill.), and diaminobenzidine.
Western blot analysis.
Two days after transfection, cell culture supernatants were harvested and filtered. gp120 shed from expressing cells was immunoprecipitated using HIVIG and protein A-Sepharose (Sigma). Cell monolayers were surface biotinylated using EZ-Link NHS-LC-biotin (Pierce Chemical) as previously described (48, 76). Cells were then lysed on ice in 50 mM Tris-HCl (pH 7.5)–150 mM NaCl–1% Triton X-100 containing 1 μg ml−1 each of aprotinin, leupeptin, and pepstatin. Biotinylated surface proteins were isolated by using NeutrAvidin-agarose (Pierce Chemical). Envelope glycoprotein was detected by Western blot analysis using the gp120-specific monoclonal antibody (MAb) Chessie B13 (kindly provided by George Lewis [1]). Western blots were visualized by chemifluorescence using ECL-Plus (Amersham Pharmacia Biotech) and quantitated by fluorescence imaging using a Fuji FLA3000G instrument.
Deglycosylation of cell surface Env.
Biotinylated envelope glycoprotein was isolated from the cell surface using NeutrAvidin-agarose and was deglycosylated using protein N-glycanase F (New England Biolabs, Beverly, Mass.). The resulting polypeptides were analyzed by Western blot using MAb Chessie 12 (kindly provided by George Lewis [1]). gp160 from a cleavage-defective envelope glycoprotein mutant (26) and soluble gp120 from a truncated envelope glycoprotein construct were deglycosylated and used as size markers.
Pulse-chase analysis.
COS-7 cells expressing envelope glycoprotein were pulse-labeled for 30 min in replicate six-well cluster dishes using 100 μCi each of [35S]cysteine and [35S]methionine in 1 ml of cysteine- and methionine-free medium. Cultures were then washed and grown in cysteine- and methionine-containing medium for the indicated times. gp120 was isolated from cell culture supernatants by immunoprecipitation. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by phosphorimaging.
Cell-cell fusion assay.
The ability of the wild-type and mutant envelope glycoproteins to mediate cell-cell fusion was determined by coculturing envelope glycoprotein-expressing COS-7 cells with U87 cells expressing CD4 and CXCR4 coreceptor (31, 45, 49). Transfected COS-7 cells were resuspended using PBS with 0.5 mM EDTA, and 5 × 103 cells were added to 96-well microcultures containing 5 × 103 U87-CD4-CXCR4 cells. In order to assess the rate and extent of syncytium formation, cocultures were fixed and immunochemically stained after 6 and 24 h. The number of envelope glycoprotein-expressing cells involved in syncytium formation and the number of nuclei contained within each syncytium were determined microscopically.
Peptide production.
Mutant N34(L6)C28 peptides were expressed in Escherichia coli BL21(DE3)/pLysS using the T7 expression system (67). Cells, freshly transformed with an appropriate plasmid, were grown to late log phase. Protein expression was induced by addition of 0.5 mM isopropylthio-β-d-galactoside. After another 3 h of growth at 37°C, the bacteria were harvested by centrifugation, and the cells were lysed by glacial acetic acid as described previously (52). Proteins were purified from the soluble fraction to homogeneity by reverse-phase high-pressure liquid chromatography (Waters) with a Vydac C18 preparative column using a water-acetonitrile gradient in the presence of 0.1% (vol/vol) trifluoroacetic acid.
CD spectroscopy.
Circular dichroism (CD) spectra were acquired on an AVIV 62DS CD spectrometer with a thermoelectric sample temperature controller. Samples for wavelength spectra were 10 μM peptide in 50 mM sodium phosphate (pH 7.0) and 150 mM NaCl. The cuvette was 0.1 cm in path length. The wavelength dependence of molar ellipticity, [θ], was monitored at 4°C as the average of five scans, using a 5-s integration time at 1.0-nm wavelength increments. Spectra were baseline corrected against the value for the cuvette with buffer alone. Fractional helix content was calculated from the CD signal by dividing the mean residue ellipticity at 222 nm by the value expected for 100% helix formation by helices of comparable size (−33,000° cm2 dmol−1) (17). Thermal stability was determined in the same buffer by measuring [θ]222 as a function of temperature. A 1.0-cm path length cell was used with continuous stirring. Thermal melts were monitored in 2°C intervals with a 2-min equilibration at the desired temperature and an integration time of 30 s. The midpoint of the thermal unfolding transition (Tm) was determined from the maximum of the first derivative, with respect to the reciprocal of the temperature, of the [θ]222 values (7). The error in estimation of Tm is ±1°C. Protein concentrations were determined by measuring absorbance at 280 nm in 6 M guanidinium chloride (23).
Sedimentation equilibrium.
Sedimentation equilibrium measurements were performed on a Beckman XL-A Optima analytical ultracentrifuge, using an An-Ti rotor and six-sectored equilibrium centrifugation centerpieces. Protein samples were dialyzed overnight against 50 mM sodium phosphate (pH 7.0)–150 mM NaCl, loaded at initial concentrations of 10, 30, and 100 μM, and analyzed at rotor speeds of 20 and 23 krpm at 20°C. Data were acquired at two wavelengths per rotor speed and processed simultaneously with a nonlinear least-squares fitting routine (38). Solvent density and protein partial specific volume were calculated according to solvent and protein composition, respectively (47). Molecular weights were all within 10% of those calculated for an ideal trimer, with no systematic deviation of the residuals.
Crystallization, data collection, and structure determination.
The G572A and R579A peptides were crystallized by hanging-drop vapor diffusion at room temperature. Initial crystallization conditions were screened by using sparse matrix crystallization kits (Crystal Screen I and II; Hampton Research, Riverside, Calif.) and then optimized. Rhombohedral crystals of G572A were grown from 10 mg ml−1 of peptide, 0.1 M sodium acetate (pH 5.2), 0.2 M ammonium sulfate, and 10% polyethylene glycol 4000. Rhombohedral crystals of R579A peptide were obtained from 20 mg ml−1 of peptide, 0.1 M sodium acetate (pH 4.5), 0.2 M ammonium sulfate, and 29% polyethylene glycol methyl ether 2000. Crystals were transferred to a cryoprotectant solution containing 15% (vol/vol) glycerol in the corresponding mother liquor and frozen in liquid nitrogen for data collection. Diffraction data on the G572A peptide trimer were collected at 95 K using an R-axis IV image plate detector mounted on a Rigaku RU200 rotating anode X-ray generator at the X-ray Crystallography Facility at the Weill Medical College of Cornell University. Data on the R579A peptide trimer were collected at 95 K using a Quantum-4 charge-coupled device-based detector at the X12B beamline of the National Synchrotron Light Source. Diffraction intensities were integrated by using Denzo and Scalepack software (59) and reduced to structural factors with the program Truncate from the CCP4 program suite (10).
The structures of the G572A and R579A peptides were determined by molecular replacement with the program AMoRe (57) using the N34(L6)C28 trimer (Protein Data Bank [PDB] file name 1SZT) as a search model. The initial models were built by using conventional (2Fo − Fc)Fcalc and (Fo − Fc)Fcalc maps at 3.0 Å. Overall anisotropic B-factor and bulk solvent corrections were applied. Many cycles of torsional angle simulated annealing and grouped B-factor refinement (4) were followed by extensive rebuilding. The models were rebuilt to reflect the sequences of the G572A and R579A peptides by using the (2Fo − Fc)Fcalc and (Fo − Fc)Fcalc maps with the program O (40). The structures were refined by using positional and B-factor refinements (4). Crystallographic refinement of the structures was done with the program CNS 1.0 (5). Model geometry was analyzed by Procheck (46), with all residues occupying most-preferred regions of the Ramanchandran space. Ser-546, Gly-547, Asn-651, Gln-652, Gln-653, Glu-654, and Lys-655 of the G572A and R579A peptides were left out of the model because of the absence of interpretable electron density for these atoms. Atomic coordinates for G572A peptide (PDB file name 1I5Y) and R579A peptide (PDB file name 1I5X) have been deposited in the PDB.
RESULTS
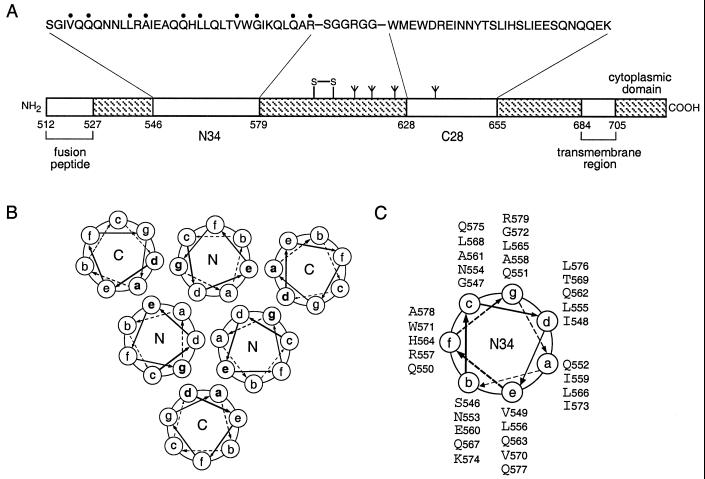
In the trimer-of-hairpins structure, residues at positions a and d of the C28 helix pack against residues at the e and g positions of the N34 coiled-coil trimer in an antiparallel orientation (Fig. 1). Sequence comparisons among HIV-1 isolates and between HIV-1 and simian immunodeficiency virus (SIV) show that the residues at these contact positions are highly conserved (43). This high degree of sequence conservation may result from selective pressure to maintain interhelical packing interactions and, hence, the structural integrity of the trimer of hairpins. To directly test this hypothesis, we changed the individual e and g residues of N34 to alanine in both the recombinant N34(L6)C28 peptide model and the intact envelope glycoprotein (Fig. 1C). Alanine-scanning mutagenesis was chosen because alanine, albeit a good helix-inducing residue, has a relatively low hydrophobicity and contributes little to protein-protein interactions (19). We reasoned that alanine replacements were likely to alter the stability of the trimer-of-hairpins structure but not to disrupt its folding per se. In this analysis, the biophysical properties of mutant N34(L6)C28 peptides were compared with the biological phenotypes of mutant envelope glycoproteins.
FIG. 1.
Core structure of the HIV-1 gp41 envelope glycoprotein. (A) Schematic representation of gp41. The important functional features of the gp41 ectodomain and the amino acid sequences of the N34 and C28 segments are shown. Amino acids mutated to alanine in this study are indicated by dots above the sequence. The naturally occurring alanine at position 558 was not altered. The disulfide bond and four potential N-glycosylation sites are depicted. The residues are numbered according to their position in gp160. The recombinant N34(L6)C28 peptide model consists of N34 and C28 plus a linker of six hydrophilic residues. (B) Cross-section of helix packing in the gp41 core. Residues at the a and d positions of the N helices form the hydrophobic interface of the trimeric coiled coil, and residues at the a and d positions of the C helices pack in an antiparallel orientation against residues at the e and g positions of adjacent N helices. (C) Helical wheel projection of the heptad repeat sequence of the N34 peptide. The view is from the NH2 terminus.
Expression of mutant envelope glycoproteins.
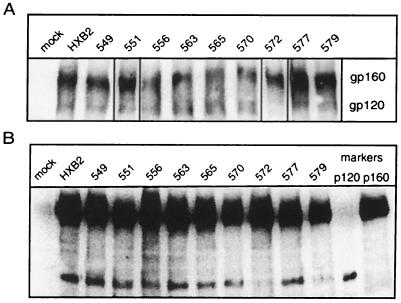
Alanine mutant envelope glycoproteins were expressed by transient transfection in COS-7 cells. As determined by Western blot analysis of cell surface-biotinylated proteins, all mutant envelope glycoproteins were expressed and transported to the cell surface with efficiencies comparable to that of the wild-type glycoprotein (Fig. 2A). The predominant species of envelope glycoprotein on the cell surface was gp160. The incomplete cleavage of the gp160 precursor is likely due to saturation of the cellular furin-like proteases that are responsible for envelope glycoprotein processing (3). gp120 represented ≈10 to 20% of the envelope glycoprotein on the cell surface. This was observed in the wild-type and in all alanine mutant proteins except G572A.
FIG. 2.
Cell surface expression of mutant envelope glycoproteins. Mutant and wild-type envelope glycoproteins were transiently expressed in COS-7 cells, and cell surface proteins were biotinylated using NHS-LC-biotin. Mutant envelope glycoprotein names are abbreviated by the position number. (A) Envelope glycoprotein was immunoprecipitated using HIVIG and detected by Western blot analysis using NeutrAvidin-HRP and ECL-Plus imaging. Representative images obtained from the analysis of comparably transfected cell cultures (40 to 60% transfection efficiency) are shown. (B) Biotinylated envelope glycoproteins were deglycosylated and detected by Western blot analysis using the gp120-specific MAb Chessie 12 (1) and ECL-Plus imaging. HXB2 gp160 and gp120 were similarly deglycosylated to provide size markers (p160, 94 kDa; and p120, 55 kDa). Using this method of analysis, we noted a consistent diminution in the ratio of gp120 to gp160. Although the basis for this apparent skewing is not known, the relative amounts of gp120 among the mutants are retained relative to the analysis of the unmodified glycoproteins. A dark image is used to emphasize deglycosylated gp120s.
In order to quantitate the relative amounts of cell surface gp120 among the mutants, we deglycosylated the cell surface glycoproteins to generate discrete polypeptides (Fig. 2B). Steady-state levels of gp120 in the alanine mutants V549A, Q551A, Q563A, L565A, V570A, and Q577A were similar to that in the wild-type glycoprotein. We conclude that alanine substitutions at Val-549, Gln-551, Gln-563, Leu-565, Val-570, and Gln-577 did not affect envelope glycoprotein transport to the cell surface, gp160 cleavage, or gp120-gp41 association. By contrast, the G572A mutant glycoprotein exhibited a 10-fold reduction in the relative amount of cell surface gp120. Lesser reductions were also noted in the L556A and R579A mutants (two- and fourfold, respectively).
To examine whether the marked decrease in the steady-state level of gp120 in the G572A mutant was due to a reduction in the efficiency of gp160 cleavage, we analyzed the rate of appearance of gp120 in the cell culture medium. Given the high rate of spontaneous gp120 shedding from the envelope glycoprotein complex (55), we reasoned that this assay would provide a sensitive measure for gp160 cleavage efficiency. If proteolytic cleavage were reduced by the mutation, the amount of shed gp120 would likewise be reduced. Figure 3 shows the time course from a pulse-chase labeling experiment. The rate and/or extent of gp120 shedding in G572A was not decreased relative to that of the wild-type glycoprotein. We conclude that the reduction in gp120 accumulation in G572A is not due to a defect in proteolytic cleavage. By extension, the reduction in the steady-state level of gp120 in the G572A mutant may be caused by an increase in gp120 shedding; the Gly-572-to-Ala mutation may affect the stability of the gp120-gp41 association.
FIG. 3.
Pulse-chase analysis of gp120 shedding by mutant envelope glycoproteins. Cells expressing mutant (L556A, G572A, Q577A, or R579A) and wild-type envelope glycoproteins were pulse-labeled for 30 min using [35S]cysteine and [35S]methionine. Comparable amounts of radioactivity were incorporated in envelope glycoproteins by all cultures (1.2 × 104 to 2.2 × 104 phosphostimulated luminescence units in gp160). Culture supernatants were harvested at 1, 3, 6, and 20 h, and gp120 was immunoprecipitated using HIVIG. Proteins were resolved by SDS-polyacrylamide gel electrophoresis, and radioactivity was visualized by phosphorimaging.
In the pulse-chase labeling experiments (Fig. 3), the L556A and R579A mutants exhibited a slight reduction in the rate and/or extent of gp120 shedding relative to the wild-type glycoprotein. The significance of this finding is unclear, as a similar decrease was observed in Q577A, a mutant in which the gp120 accumulation on the cell surface was unaltered. The mechanism for the two- to fourfold reduction in the steady-state levels of gp120 in L556A and R579A remains undefined.
Taken together, our results indicate that the biosynthesis and expression of the V549A, Q551A, Q563A, L565A, V570A, and Q577A envelope glycoproteins were unaffected by alanine substitutions (Table 1). On the other hand, G572A, L556A, and R579A displayed reduced steady-state levels of gp120 on the cell surface.
TABLE 1.
Expression and fusion activity of mutant envelope glycoproteins
| Glycoprotein | Cell surface expressiona | Cell surface gp120/gp160b ratio | gp120 sheddingc | Fusion activityd |
|---|---|---|---|---|
| Wild type | +++ | +++ | +++ | +++ |
| V549A | +++ | +++ | +++ | +++ |
| Q551A | +++ | +++ | +++ | +++ |
| L556A | +++ | ++ | ++ | ± |
| Q563A | +++ | +++ | +++ | +++ |
| L565A | +++ | +++ | +++ | ± |
| V570A | +++ | +++ | +++ | ± |
| G572A | +++ | + | +++ | − |
| Q577A | +++ | +++ | ++ | +++ |
| R579A | +++ | ++ | ++ | ± |
+++, comparable to wild-type envelope glycoprotein.
+++, comparable to wild-type envelope glycoprotein; ++, 2- to 4-fold reduction; +, 10-fold reduction.
+++, comparable to wild-type envelope glycoprotein, as assessed by Western blot analysis and/or by pulse-chase measurements; ++, slight reduction in gp120 shedding.
+++, comparable to wild-type envelope glycoprotein; ±, severe (≥100-fold) reduction in fusion ability; −, complete absence of fusion. Intermediate levels of impairment were not represented in these mutations.
Fusogenic potential of mutant envelope glycoproteins.
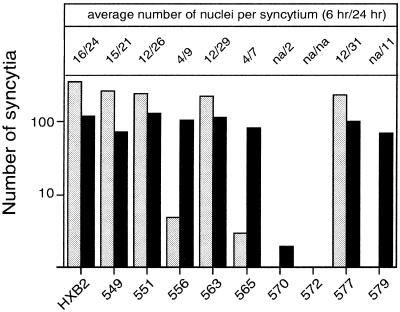
The ability of the alanine mutant envelope glycoproteins to mediate cell-cell fusion was assessed by coculturing transfected COS-7 cells with a fusion partner, U87 cells expressing CD4 and CXCR4. In this assay, syncytium formation proceeds rapidly, and the maximum number of syncytia is typically attained by 6 h of coculture. Thereafter, neighboring syncytia begin to fuse, resulting in a decreased number of syncytia and an increased number of nuclei per syncytium. By these criteria, V549A, Q551A, Q563A, and Q577A were indistinguishable from the wild-type envelope glycoprotein in fusogenic potential (Fig. 4). In contrast, V570A, G572A, and R579A failed to produce syncytia within 6 h of coculture, and L556A and L565A yielded 100-fold fewer syncytia than the wild-type glycoprotein. By 20 h of coculture, R579A, L556A, and L565A were able to generate similar numbers of syncytia as the wild-type envelope glycoprotein, but these syncytia were small and similar to those seen at 6 h with the wild-type glycoprotein. The V570A mutant produced only a few binucleated cells after 20 h of coculture, and G572A was entirely defective.
FIG. 4.
Cell-cell fusion activity of mutant envelope glycoproteins. Cells expressing mutant and wild-type envelope glycoproteins were cocultured with U87-CD4-CXCR4 cells for either 6 or 24 h. Cultures were fixed with −20°C methanol-acetone (1:1), and envelope glycoprotein-expressing cells were visualized by immunochemical staining. The number of envelope glycoprotein-expressing cells involved in syncytium formation and the average number of nuclei contained within each syncytium were determined. Mutant envelope glycoprotein labels are abbreviated by the position number. The graph indicates the number of syncytia at 6 and 24 h (gray and black bars, respectively). The average size of the syncytia (number of nuclei per syncytium at 6 and 24 h) is indicated at the top. na, not applicable. Results depicted are from one experiment and are representative of relative differences among mutants. Note that the number of syncytia is biphasic with time and typically decreases at longer times as syncytia merge.
In summary, V570A, G572A, R579A, L556A, and L565A were severely debilitated in their ability to mediate membrane fusion (Table 1). Because the alanine substitutions in these mutants did not grossly affect envelope glycoprotein biosynthesis, our results raise the possibility that the alanine mutations may perturb the interhelical packing interactions in the gp41 core structure, thereby modulating the membrane fusion activity of the envelope glycoprotein complex.
Biophysical analysis of mutant N34(L6)C29 peptides.
Variants of the recombinant N34(L6)C28 peptide with single alanine substitutions at the e and g positions of the N34 coiled coil were produced by bacterial expression and purified by reverse-phase high-performance liquid chromatography. Sedimentation equilibrium measurements indicated that each peptide sedimented as a discrete trimer over a total peptide concentration of 10 to 100 μM at neutral pH (Fig. 5A; Table 2). The helical content and stability of mutant N34(L6)C28 complexes were quantitated by CD. Each peptide was >90% helical at 4°C at 10 μM peptide (Table 2), and each exhibited a cooperative thermal unfolding transition (Fig. 5B; Table 2). Under these conditions, the midpoints of the thermal transition (Tm) for N34(L6)C28 (wild type), V549A, Q551A, L556A, Q563A, L565A, V570A, G572A, Q577A and R579A were 70, 73, 74, 64, 68, 50, 56, 82, 76, and 72°C, respectively (Fig. 5B; Table 2). Therefore, alanine mutations at the e and g residues of the N34 helix caused only changes in thermal stability, without significantly influencing the overall topology. In general, introduction of alanine for bulky hydrophobic residues markedly destabilized the trimer-of-hairpins structure; the decrease in thermal stability, ΔTm, between the wild-type and mutant peptides was 6, 20, and 14°C for Leu-556, Leu-565, and Val-570, respectively. On the other hand, the Gly-572-to-Ala substitution led to appreciable stabilization of the trimer of hairpins, reinforcing the observation that glycine in α-helices is generally destabilizing (reviewed in reference 41).
FIG. 5.
Biophysical characterization of mutant N34(L6)C28 peptides. (A) The L556A (open rhombs), L565A (open circles), and G572A (open triangles) peptides form three-stranded bundles. Sedimentation equilibrium data (20 krpm) were collected at 20°C at pH 7.0 for L556A (30 μM), L565A (100 μM), and G572A (10 μM). The natural logarithm of the absorbance at 280, 236, and 230 nm is plotted against the square of the radial position. For an ideal single-species system, this plot is linear, with the slope proportional to the molecular weight of the molecule. (B) Thermal melts monitored by CD at 222 nm for L556A (open rhombs), L565A (open circles), V570A (solid circles), G572A (open triangles), and R579A (solid triangles) peptides at 10 μM at pH 7.0.
TABLE 2.
Alanine mutants of the HIV-1 gp41 ectodomain core form trimer-of-hairpins structures
| Mutant | −[θ]222 (degrees cm2 dmol−1) | Tm (°C) | Molecular mass (kDa) |
|---|---|---|---|
| N34(L6)C28 | 31,300 | 70 | 24.4 |
| V549A | 31,100 | 73 | 23.9 |
| Q551A | 30,800 | 74 | 23.7 |
| L556A | 30,500 | 64 | 23.2 |
| Q563A | 31,200 | 68 | 24.2 |
| L565A | 31,500 | 50 | 22.9 |
| V570A | 29,900 | 56 | 23.3 |
| G572A | 31,400 | 82 | 23.5 |
| Q577A | 31,000 | 76 | 23.6 |
| R579A | 31,900 | 72 | 24.1 |
These results indicate that the destabilization of the gp41 ectodomain core by alanine mutations at Leu-565 and Val-570 may underlie the fusion-defective phenotype of the mutant envelope glycoproteins (Tables 1 and 2). In contrast, alanine mutations at Gly-572 and Arg-579 stabilized the gp41 core structure but severely impaired membrane fusion activity. This effect on fusogenicity may reflect the reduced level of mature gp120 in these mutants. The L556A mutant had an intermediate phenotype, where both the thermal stability of the gp41 core and the level of cell surface gp120 in the envelope glycoprotein complex were slightly reduced. Interestingly, the Gly-572 residue is involved in the formation of hydrophobic cavities on the surface of the N34 trimer that are crucial for interhelical packing interactions (13, 35, 60). The Arg-579 side chain, on the other hand, does not directly participate in the helical interactions (13, 68, 69).
Crystal structures of G572A and R579A peptides.
The unexpected observation that alanine mutations at Gly-572 and Arg-579 disrupt envelope glycoprotein function while maintaining the thermal stability of the gp41 ectodomain core led us to examine the atomic detail of the respective trimer-of-dimer structures. The G572A and R579A peptides were crystallized, and their X-ray structures were determined to 2.1 and 1.8 Å, respectively, using molecular replacement methods. The crystals of both G572A and R579A belong to the space group R3 and contain the trimer formed around the crystallographic threefold axis. The electron density maps of G572A and R579A reveal the positions of all of the amino acid residues except a few disordered side chains at the helix termini (see Materials and Methods). Simulated-annealing omit maps were calculated to confirm the assignment of the side chains. The structure of the the G572A peptide was refined to a conventional R-factor of 20.6%, with a free R-factor of 23.4% over a resolution range of 50.0 to 2.1 Å, and the final model consists of 61 amino acid residues and incorporates 38 water molecules and a sulfate ion. The structure of the R579A peptide was refined against 50.0 to 1.8 Å resolution data to yield a conventional R-factor of 17.9%, with a free R-factor of 19.8%, and the final model includes 61 residues, 83 water molecules, and a sulfate ion. Table 3 summarizes the data collection and refinement statistics for these two structures.
TABLE 3.
X-ray data collection and refinement statistics
| Statistic | G572A | R579A |
|---|---|---|
| Unit cell dimensions | ||
| a, b, c (Å) | 52.06, 52.06, 59.74 | 51.25, 51.25, 59.87 |
| α, β, γ (degrees) | 90, 90, 120 | 90, 90, 120 |
| Space group | R3 | R3 |
| Data processing | ||
| Resolution (Å) | 50–2.1 | 50–1.8 |
| Measured reflections | 23,645 | 40,929 |
| Unique reflections | 3,509 | 5,440 |
| Completeness (%) | 99.5 | 99.6 |
| Rmerge (%)a | 2.9 | 3.5 |
| Refinement | ||
| Resolution (Å) | 50–2.1 | 50–1.8 |
| Reflections in working set | 3,153 | 4,860 |
| Reflections in test set | 356 | 580 |
| Protein nonhydrogen atoms | 486 | 490 |
| Water molecules | 38 | 83 |
| Sulfate ion | 1 | 1 |
| Rfree (%)b | 23.4 | 22.2 |
| Rcryst (%)b | 20.6 | 17.9 |
| Average B-factor (Å) | 37.8 | 19.8 |
| Rms deviations from ideality | ||
| Bond lengths (Å) | 0.004 | 0.008 |
| Bond angles (degrees) | 0.7 | 1.0 |
| Torsion angles (degrees) | 15.0 | 14.8 |
Rmerge = Σ|I − 〈I〉|/ΣI, where I is the intensity of an individual measurement and 〈I〉 is the average intensity from multiply recorded reflections.
Rcryst = Σ|Fobs − Fcalc|ΣFobs, where Fobs and Fcalc are observed and calculated structural factors, respectively. No ς-cutoff was applied. Rfree is calculated for a set of reflections that were excluded from refinement.
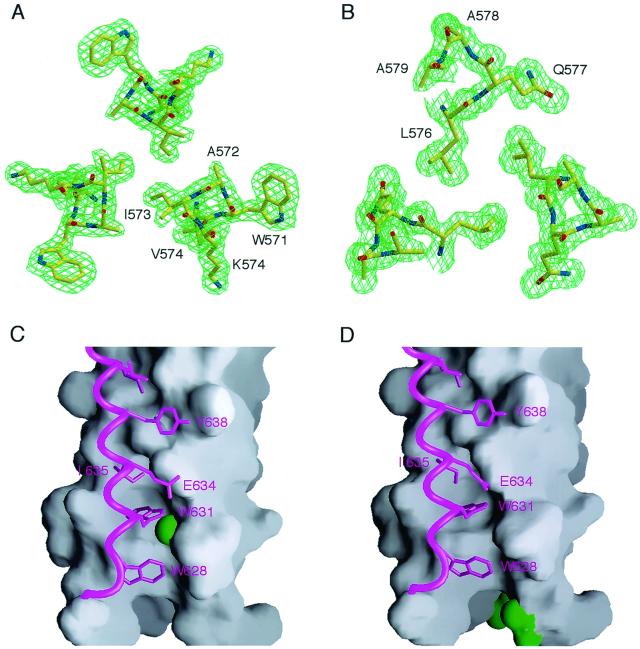
As expected, the overall folds of G572A and R579A are very similar to those of the wild-type molecule. In all cases, the interior N34 coiled coil of three parallel helices, wrapped in a gradual left-handed superhelix, is surrounded by a sheath of antiparallel C28 helices (Fig. 6). At the center of the trimer of hairpins, the N34 three-stranded coiled coil displays typical acute knobs-into-holes packing, where the a and d side chains (knobs) fit into the spaces (holes) between four residues on the neighboring helices (Fig. 7A and B) (18, 30). Residues at the e and g positions of the N34 helices lie on the outside of the trimeric coiled coil and form three hydrophobic grooves on the surface of this coiled coil. The Gly-572 and Arg-579 residues replaced in G572A and R579A, respectively, are both in g positions of N34. The root-mean-square (rms) deviations between all Cα atoms of the central N34 coiled coil in the wild-type and mutant molecules are 0.27 Å for G572A and 0.30 Å for R579A. The C28 helices in G572A and R579A can also be superimposed upon the wild-type counterpart with rms deviations of 0.40 and 0.35 Å, respectively. Thus, the fusion-defective Gly-572-to-Ala and Arg-579-to-Ala mutations do not significantly alter the trimer-of-hairpins structure.
FIG. 6.
Overall views of the mutant G572A and R579A peptide six-helix bundles. (A) Side view. The amino termini of N34 and the carboxyl termini of C28 are at the top of the figure. Helices of G572A (yellow) and R579A (pink) peptides were used for the superposition. (B) View from the top, looking down the threefold axis of the trimer of hairpins. Figures were generated with the program Setor (24).
FIG. 7.
Conserved interhelical interactions in the crystal structures of the G572A and R579A trimers. The final 2Fo − Fc electron density maps of the G572A and R579A peptides are shown with the refined model superimposed (panels A and B, respectively). Oxygen, nitrogen, and carbon atoms are colored red, blue, and yellow, respectively. The maps are contoured at 1.0 standard deviation above the average density. Interactions of the C28 helix with a deep cavity on the surface of the N34 coiled coil of the G572A and R579A peptides are shown (panels C and D, respectively). The C28 helices, represented as ribbons, are drawn against a surface representation of the N34 coiled coil. The side chains of the mutated residues are colored green. Figures were generated with the programs Setor (24) and Grasp (58).
On the surface of the N34 trimeric coiled coil, there are three prominent, symmetry-related cavities (≈400 Å3) that each accommodate three hydrophobic residues from the abutting C28 helix: Trp-628, Trp-631, and Ile-635 (13, 68, 69). These conserved coiled-coil cavities have been shown to be critical for HIV-1 entry and its inhibition and provide a potential antiviral drug target (12, 13, 22, 25, 35, 66). The Gly-572 residue, which is involved in forming this cavity, is invariant in all 251 fully sequenced M group HIV-1 isolates (43). The substituted Ala-572 side chains point into the triangular interhelical space between two N34 helices and a buttressing C28 helix (Fig. 7C). The favorable van der Waals interactions between the Ala-572 and Trp-631 side chains can strengthen interhelical packing and stabilize the trimer-of-hairpins structure, as suggested by the large increase in Tm of G572A relative to that of N34(L6)C28. In addition, the Gly-572-to-Ala substitution also leads to local structural rearrangements: the side chains of Trp-628 and Ile-635 deviated substantially. Overall, the methyl groups of the Ala-572 residues in G572A pack efficiently into the hydrophobic cavity of N34 and make a good C28 interaction (Fig. 7C).
The conserved grooves along the N34 coiled-coil surface are lined with predominantly hydrophobic residues (Fig. 1C). Interestingly, the Arg-579 residue at a g position is completely conserved in 245 of the 251 sequenced HIV-1 strains. Of the remaining six isolates with different residues at this position, three possess a conservative lysine substitution (43). In gp41 core crystal structures (13, 68, 69), the Arg-579 side chain is poorly ordered, as suggested by very high individual atomic B factors. While electron density for Ala-579 in the R579A peptide is unambiguous, this substituted residue contributes little to the interhelical packing interactions. It would therefore appear that the slight stabilization of the trimer-of-hairpins structure associated with the alanine substitution at Arg-579 reflects the favorable helix propensity of alanine relative to arginine in the wild-type molecule.
DISCUSSION
The HIV-1 envelope glycoprotein complex controls the key process of viral entry. The current model of HIV-1 membrane fusion implies that, as the trimer of hairpins is assembled, a ready source of energy can be made available for overcoming the activation energy needed for lipid bilayer fusion. Because the native and prehairpin intermediate conformations of gp41 are not known, the basic property of gp41 activation during HIV-1 membrane fusion is not well understood. Nevertheless, as has been established for the case of influenza virus HA protein (6, 9, 74), it is widely believed that the native gp41 structure is substantially different from the fusogenic trimer-of-hairpins structure. In the simplest model, the N-terminal heptad repeat region of gp41 exists in an non-coiled-coil conformation in the native state but forms a coiled-coil trimer in the prehairpin intermediate and fusogenic hairpin conformations (16, 21, 34, 51, 70, 73). The N-terminal heptad repeat sequence is one of the most highly conserved regions within the primate immunodeficiency virus envelope glycoproteins (11, 20, 28). Residues at the a and d positions form a hydrophobic interface at the interior of the N-terminal trimeric coiled coil (62). There is only one nonconservative substitution at an a position between HIV-1 and SIV (Ile-573 in HIV-1 and Thr in the corresponding position of SIV). Mutations at this position affect the folding and stability of the gp41 ectodomain core and, coordinately, the ability of the mutant envelope glycoproteins to mediate membrane fusion (21, 49, 51). Formation of the N-terminal three-stranded coiled coil appears to occur as an early event in the gp41 refolding process.
The N-terminal coiled-coil surface contains three hydrophobic grooves that are the sites for the C-terminal helix interactions (13, 68, 69). The e and g residues lining these grooves pack against residues at the a and d positions of the C-terminal helices. The high sequence conservation at these contact positions has led to the proposal that interactions between the N- and C-terminal helices are important for the resolution of the prehairpin intermediate into the hairpin structure during membrane fusion (13, 68, 69). Indeed, mutagenesis studies demonstrate that mutations at these interhelical interface positions often abolish infectivity and membrane fusion (70, 71). This proposal is also consistent with a large body of data on the inhibition of HIV-1 infection and syncytium formation by derivatives of the peptides that make up the gp41 core (12, 35, 49, 61). In the present study, we have investigated the role of individual side chains at the nine e and g positions of the N34 coiled coil in conferring structural specificity and conformational stability to the gp41 core and in determining the fusion potential of the envelope glycoprotein complex.
Our alanine-scanning mutagenesis results show that the Leu-556, Leu-565, Val-570, Gly-572, and Arg-579 residues play a critical role in envelope glycoprotein function, while the Val-549, Gln-551, Gln-563, and Gln-577 side chains per se are not essential for this membrane fusion activity. These findings with regard to L556A, Q563A, V570A, G572A, and Q577A agree with those reported previously by Weiss and colleagues (70, 71). Our biophysical analysis reveals that alanine mutations in the fusion-defective L565A and V570A envelope glycoproteins destabilize the trimer of hairpins by a Tm shift of ≈14 to 20°C. In contrast, the Tms of the four mutant gp41 cores carrying the fusion-competent alanine substitutions are comparable to or even higher than that of the wild-type molecule. In general, the stability of the trimer-of-hairpins structure modulates the membrane fusion properties of the gp120-gp41 complex. Interestingly, the fusion-defective Gly-572-to-Ala and Arg-579-to-Ala mutations lead instead to enhanced stabilization of the gp41 core structure.
The conformational stability of the gp41 core structure is thought to play a critical role in the refolding of the envelope glycoprotein into a fusion-active conformation (33, 34, 49, 51). Although melting temperature has been useful as a qualitative guide to thermodynamic stability, it appears that the dependence of HIV-1 fusion potential on the stability of the trimer of hairpins can vary. In contrast to our previous observation that the Ile-573-to-Thr substitution decreases the thermal stability of the gp41 core by 25°C relative to the wild-type molecule and has only moderate effects on membrane fusion activity (49), we show here that a destabilization of the trimer of hairpins by a Tm shift of 14 to 20°C can markedly reduce the fusion potential of the envelope glycoprotein. It is possible that the a position Ile-573-to-Thr mutation affects formation of the N-terminal coiled coil of gp41 after interaction of the envelope glycoprotein complex with cellular receptors, while the alanine mutations at the e and g positions studied here act on the association of the C-terminal helices with the N-terminal coiled coil during the transition from the prehairpin intermediate to the trimer-of-hairpins structure.
The crystal structures of the wild-type gp41 core and the G572A and R579A mutants can be superimposed, with rms deviations of 0.39 and 0.40 Å, respectively. From the structures we can see that each substituted alanine side chain is readily accommodated in the trimer-of-hairpins structure by using different sets of atoms. Our data suggest that local structural perturbations in the gp41 core are unlikely to be a major cause of the fusion-defective phenotype of the G572A and R579A glycoproteins. Instead, both mutant proteins exhibit a reduction in the steady-state level of gp120 on the cell surface. In contrast to an earlier interpretation of the G572A phenotype (70), our results suggest that this reduction is not due to a deficiency in gp160 cleavage but may reflect an increase in gp120 shedding from the envelope glycoprotein complex. We propose that alanine mutations at Gly-572 and Arg-579 affect the association between the gp120 and gp41 subunits in the native envelope glycoprotein. This proposal is consistent with previous disulfide cross-linking studies that demonstrated the physical proximity of the Thr-605 residue of the gp41 ectodomain and the C-terminal region of gp120 (3). Therefore, we suggest that Gly-572 and Arg-579 are required for maintaining the structural integrity of the native gp120-gp41 complex. The defect in membrane fusion may reflect this instability and the resultant reduction in gp120 on the cell surface. Further studies of the G572A and R579A mutant glycoproteins may open new perspectives in the search for effective approaches to stabilizing the native envelope glycoprotein complex for vaccine development.
Several approaches towards antiviral intervention have recently converged to highlight the critical importance of transiently exposed conformations of the HIV-1 envelope glycoprotein. Synthetic peptides targeting the prehairpin intermediate of gp41 are now in clinical study (42). Small-molecule inhibitors of the formation of the trimer-of-hairpins structure have also been developed (22, 25, 66). Mutations in the envelope glycoprotein that specifically arrest the fusion process at critical steps may allow the accumulation of intermediates for vaccine development. Further analysis of protein structure and conformation will also provide insights into the complex biology of HIV-1 entry.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI48385 (M.L.) and AI44669 (J.H.N.). J.H.N. is grateful to the J. B. Pendleton Charitable Trustfor enabling the purchase of key instrumentation.
We thank Meg Trahey, Scott Larson, and Amanda Wilhelm for technical help in this project and George Lewis (Institute of Human Virology, University of Maryland) for providing Chessie B13 and 12 monoclonal antibodies.
REFERENCES
- 1.Abacioglu Y H, Fouts T R, Laman J D, Claassen E, Pincus S H, Moore J P, Roby C A, Kamin-Lewis R, Lewis G K. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retrovir. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 2.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Binley J M, Sanders R W, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Maddon P J, Olson W C, Moore J P. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brünger A T. XPLOR version 3.1: a system for X-ray crystallography and NMR. New Haven, Conn: Yale University Press; 1992. [Google Scholar]
- 5.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, Rice L M, Simonson T, Warren G L. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 6.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Cantor C, Schimmel P. Biophysical chemistry, part III. W. H. New York, N.Y: Freeman and Company; 1980. [Google Scholar]
- 8.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 10.CCP4. Collaborative computational project number 4. Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- 11.Chambers P, Pringle C R, Easton A J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 12.Chan D C, Chutkowski C T, Kim P S. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 14.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen C H, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S S-L, Lee C-N, Lee W-R, McIntosh K, Lee T-H. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane protein. J Virol. 1993;67:3615–3619. doi: 10.1128/jvi.67.6.3615-3619.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y-H, Yang J T, Chau K H. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 18.Crick F H C. The packing of α-helices: simple coiled coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 19.Cunningham B C, Wells J A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 20.Delwart E J, Mosialos G, Gilmore T. Retroviral envelope glycoproteins contain a “leucine zipper”-like repeat. AIDS Res Hum Retrovir. 1990;6:703–706. doi: 10.1089/aid.1990.6.703. [DOI] [PubMed] [Google Scholar]
- 21.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckert D M, Malashkevich V N, Hong L H, Carr P A, Kim P S. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99:103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 23.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 24.Evans S V. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 25.Ferrer M, Kapoor T M, Strassmaier T, Weissenhorn W, Skehel J J, Oprian D, Schreiber S L, Wiley D C, Harrison S C. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of nonnatural binding elements. Nat Struct Biol. 1999;6:953–960. doi: 10.1038/13324. [DOI] [PubMed] [Google Scholar]
- 26.Freed E O, Myers D J, Risser R. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J Virol. 1989;63:4670–4675. doi: 10.1128/jvi.63.11.4670-4675.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 28.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retrovir. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 29.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp P M, Shaw G M, Hahn B H WHO and NIAID Networks for HIV Isolation and Characterization. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harbury P B, Kim P S, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 31.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughson F M. Enveloped viruses: a common mode of membrane fusion? Curr Biol. 1997;7:R565–R569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- 33.Jelesarov I, Lu M. Thermodynamics of trimer-of-hairpins formation by the SIV gp41 envelope protein. J Mol Biol. 2001;307:637–656. doi: 10.1006/jmbi.2001.4469. [DOI] [PubMed] [Google Scholar]
- 34.Ji H, Bracken C, Lu M. Buried polar interactions and conformational stability in the simian immunodeficiency virus (SIV) gp41 core. Biochemistry. 2000;39:676–685. doi: 10.1021/bi991893e. [DOI] [PubMed] [Google Scholar]
- 35.Ji H, Shu W, Burling F T, Jiang S, Lu M. Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion. J Virol. 1999;73:8578–8586. doi: 10.1128/jvi.73.10.8578-8586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang S, Lin K, Lu M. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1998;72:10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang S, Lin K, Strick N, Neurath A R. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 38.Johnson M L, Correia J J, Yphantis D A, Halvorson H R. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones P L, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and coreceptors. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 40.Jones T A, Zou J W, Cowan S W, Kjelgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;D47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 41.Kallenbach N R, Lyu P, Zhou H. CD spectroscopy and the helix-coil transition in peptides and polypeptides. In: Fasman G D, editor. Circular dichroism and the conformational analysis of biomolecules. New York, N.Y: Plenum Press; 1996. pp. 201–259. [Google Scholar]
- 42.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 43.Kuiken C, Foley B, Hahn B, Marx P, McCutchan F, Mellors J W, Mullins J, Wolinsky S, Korber B. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 2000. [Google Scholar]
- 44.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 45.LaCasse R A, Follis K E, Moudgil T, Trahey M, Binley J M, Planelles V, Zolla-Pazner S, Nunberg J H. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J Virol. 1998;72:2491–2495. doi: 10.1128/jvi.72.3.2491-2495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laskowski R A, MacArthur M V, Moss D D, Thornton J M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 47.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S E, Rowe A J, Horton J C, editors. Analytical ultracentrifugation in biochemistry and polymer science. Cambridge, England: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 48.Lisanti M P, Sargiacomo M, Graeve L, Saltiel A R, Rodriguez-Boulan E. Polarized apical distribution of glycosyl-phosphatidylinositol-anchored proteins in a renal epithelial cell line. Proc Natl Acad Sci USA. 1988;85:9557–9561. doi: 10.1073/pnas.85.24.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Shu W, Fagan M B, Nunberg J H, Lu M. Structural and functional analysis of the HIV-1 gp41 core containing an Ile573 to Thr substitution: implications for membrane fusion. Biochemistry. 2001;40:2797–2807. doi: 10.1021/bi0024759. [DOI] [PubMed] [Google Scholar]
- 50.Lu M, Blacklow S C, Kim P S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 51.Lu M, Ji H, Shen S. Subdomain folding and biological activity of the core structure from human immunodeficiency virus type 1 gp41: implications for viral membrane fusion. J Virol. 1999;73:4433–4438. doi: 10.1128/jvi.73.5.4433-4438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu M, Kim P S. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J Biomol Struct Dyn. 1997;15:465–471. doi: 10.1080/07391102.1997.10508958. [DOI] [PubMed] [Google Scholar]
- 53.Luciw P A. Human immunodeficiency viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melinick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1881–1952. [Google Scholar]
- 54.Malashkevich V N, Chan D C, Chutkowski C T, Kim P S. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc Natl Acad Sci USA. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKeating J A, McKnight A, Moore J P. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J Virol. 1991;65:852–860. doi: 10.1128/jvi.65.2.852-860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 58.Nichols A, Sharp K, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 59.Otwinowski Z, Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 60.Rimsky L T, Shugars D C, Matthews T J. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Root M J, Kay M S, Kim P S. Protein design of an HIV-1 entry inhibitor. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 62.Shu W, Ji H, Lu M. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry. 1999;38:5378–5385. doi: 10.1021/bi990199w. [DOI] [PubMed] [Google Scholar]
- 63.Shu W, Liu J, Ji H, Radigen L, Jiang S, Lu M. Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides. Biochemistry. 2000;39:1634–1642. doi: 10.1021/bi9921687. [DOI] [PubMed] [Google Scholar]
- 64.Skehel J J, Wiley D C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 65.Sodroski J, Goh W C, Rosen C, Haseltine W A. Role of the HTLVIII/LAV envelope in syncytium formation and cytopathicity. Nature. 1986;322:470–474. doi: 10.1038/322470a0. [DOI] [PubMed] [Google Scholar]
- 66.Sodroski J G. HIV-1 entry inhibitors in the side pocket. Cell. 1999;99:243–246. doi: 10.1016/s0092-8674(00)81655-4. [DOI] [PubMed] [Google Scholar]
- 67.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 68.Tan K, Liu J, Wang J, Shen D, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 70.Weng Y, Weiss C D. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1998;72:9676–9682. doi: 10.1128/jvi.72.12.9676-9682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weng Y, Yang Z, Weiss C D. Structure-function studies of the self-assembly domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 2000;74:5368–5372. doi: 10.1128/jvi.74.11.5368-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wild C, Greenwell T, Shugars D, Rimsky-Clarke L, Matthews T. The inhibitory activity of an HIV type 1 peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res Hum Retrovir. 1995;11:323–325. doi: 10.1089/aid.1995.11.323. [DOI] [PubMed] [Google Scholar]
- 73.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Peptides corresponding to a predictive a-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson I A, Skehel J J, Wilson D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–375. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 75.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 76.York J, Follis K E, Trahey M, Nyambi P N, Zolla-Pazner S, Nunberg J H. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J Virol. 2001;75:2741–2752. doi: 10.1128/JVI.75.6.2741-2752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]