Abstract
Background
Glioblastoma (GBM) is an aggressive primary brain tumor with a high recurrence rate and poor prognosis. Necroptosis, a pathological hallmark of GBM, is poorly understood in terms of its role in prognosis, tumor microenvironment (TME) alteration, and immunotherapy.
Methods & Results
We assessed the expression of 55 necroptosis-related genes in GBM and normal brain tissues. We identified necroptosis-stratified clusters using Uni-Cox and Least Absolute Shrinkage and Selection Operator (LASSO) regression to establish the 10-gene Glioblastoma Necroptosis Index (GNI). GNI demonstrated significant prognostic efficacy in the TCGA dataset (n = 160) and internal validation dataset (n = 345) and in external validation cohorts (n = 591). The GNI-high subgroup displayed a mesenchymal phenotype, lacking the IDH1 mutation, and MGMT methylation. This subgroup was characterized by significant enrichment in inflammatory and humoral immune pathways with prominent cell adhesion molecules (CD44 and ICAM1), inflammatory cytokines (TGFB1, IL1B, and IL10), and chemokines (CX3CL1, CXCL9, and CCL5). The TME in this subgroup showed elevated infiltration of M0 macrophages, neutrophils, mast cells, and regulatory T cells. GNI-related genes appeared to limit macrophage polarization, as confirmed by immunohistochemistry and flow cytometry. The top 30% high-risk score subset exhibited increased CD8 T cell infiltration and enhanced cytolytic activity. GNI showed promise in predicting responses to immunotherapy and targeted treatment.
Conclusions
Our study highlights the role of necroptosis-related genes in glioblastoma (GBM) and their effects on the tumor microenvironment and patient prognosis. TheGNI demonstrates potential as a prognostic marker and provides insights into immune characteristics and treatment responsiveness.
Keywords: Brain tumor, single-cell analysis, immunotherapy, glioma, central nervous system, inflammation
1. Introduction
Glioblastoma (GBM), classified as a grade IV astrocytoma, is a highly aggressive and frequently recurring brain tumor with a grim prognosis [1]. GBM is the most common primary malignant brain tumor, with a prevalence of 54% among gliomas and 16% among all primary brain tumors [2]. Primary GBM, which accounts for approximately 80% of GBM cases, refers to de novo development of glioblastoma. In contrast, the transformation from a lower to high-grade IV astrocytoma is characterized as secondary GBM [3]. The current mainstay of treatment involves feasible surgical resection with subsequent radiotherapy combined with concomitant and adjuvant temozolomide (TMZ) chemotherapy [4]. Untreated GBM patients typically experience a median survival of only 3 months, whereas those undergoing conventional treatment may survive for 12–15 months [5,6]. Overall, only 5% of diagnosed patients are expected to survive for five years or more [6].
The tumor microenvironment (TME) plays a crucial role in cancer development, therapeutic response, and prognosis [7]. In glioblastoma (GBM), an immunosuppressive microenvironment is evident both within the tumor and in peripheral blood, contributing to rapid disease progression and poor outcomes [8]. Various strategies such as immune checkpoint inhibitors, vaccine therapies, oncolytic viruses, and gene therapy are being explored to disrupt the immunosuppressive TME and enhance treatment specificity [9]. However, immunosuppressive stromal factors, majorly the tumor-associated macrophages (TAMs), have shown to restrict tumor penetration affecting the efficiency of ICBs and oncolytic viruses [10,11]. Hence, further research into factors that modulate the GBM TME is needed to enhance the efficacy of current treatments or independently improve patient outcomes.
There is an increasing interest in the investigations of the role of programmed cell death, such as ferroptosis, pyroptosis, and necroptosis, in the modulation of TME and therapeutic response [12,13]. The process of cell death plays a crucial role in the development and homeostasis of the body, and the ability to resist cell death has been identified as a key factor in the formation of tumors and resistance to treatment [14]. Apoptosis, a form of programmed cell death, is defective in GBM; however, numerous intratumoral necrosis foci have been identified [15,16]. Necrosis, a form of non-programmed cell death, is considered a pathological and radiological hallmark of GBM [17,18]. Historically, necrosis has been viewed as an unintentional, unregulated, passive mechanism of cell death. Nevertheless, contemporary findings have uncovered a controlled variant of necrosis known as necroptosis [19–26]. In contrast to apoptosis, which is characterized by nuclear compaction, preserved organelle integrity, cytoplasmic reduction, and membrane retention, necroptosis is characterized by the rupture of the cell membrane, leading to the efflux of cellular constituents [20]. The initiation of necroptosis can be prompted by the activation of diverse death receptors, primarily tumor necrosis factor (TNF) receptors, such as TNFR1 and FAS, and toll-like receptors, such as TLR3 and TLR4 [27–30]. Ligation of TNFR1 results in the formation of death-inducing signaling complex (DISC), a cytosolic complex that includes RIPK1 (receptor-interacting serine/threonine protein kinase 1), Fas-associated death domain (FADD), RIPK3, and caspase-8. If caspase-8 is inactivated, the interactions between RIPK1 and RIPK3 lead to the formation of the necrosome and oligomerization of mixed lineage kinase domain-like (MLKL) [27,28,31]. Activation of TLR4 by lipopolysaccharide (LPS) or damage-associated molecular patterns (DAMPs) and TLR3 by intra-endosomal double-stranded RNA (dsRNA) results in RIPK3 activation via RHIM (RIP homotypic interaction motif) engagement of TRIF (TIR-domain-containing adapter-inducing interferon-β) [29,30]. Moreover, in specific cell types, such as macrophages, necroptosis can be initiated by IFNAR1 (interferon alpha receptor 1) and IFNGR1 (interferon gamma receptor 1), primarily resulting from the prolonged activation of transcription factors, including signal transducer and activator of transcription 1 and 2 (STAT1 and STAT2), as well as interferon regulatory factor 9 (IRF9) [32–34]. ZBP1 (Z-DNA binding protein 1), an exogenous DNA sensor in the cytosol that stimulates the synthesis of type-I interferon and triggers nuclear factor kappa B (NF-κB), can independently activate RIPK3 by physically interacting with it through RHIM , irrespective of RIPK1 [35]. Membrane pores are formed by oligomerized MLKL after translocation to the cytoplasm, resulting in the release of intracellular contents, a characteristic shared by both necrosis and necroptosis, with consequences for the tumor microenvironment [20–26].
The objective of this study was to investigate the role of necroptosis in glioblastoma (GBM) by understanding how necroptosis impacts GBM prognosis and contributes to TME remodeling. We sought to identify and analyze key necroptosis-related genes and their functional implications in GBM, and to evaluate the potential of these biomarkers for predicting treatment responses and guiding therapeutic strategies. To achieve these objectives, we conducted a comprehensive literature review to identify relevant necroptosis-related genes and employed machine learning models on GBM transcriptional data to elucidate their roles in disease progression and treatment response. An overview of the workflow is shown in Figure S1.
2. Materials and methods
2.1. Public databases and retrieved datasets
For the training set, transcriptome sequencing data (human genome reference: GRCh38 [hg38]; alignment: Illumina HiSeq 2000/2500; quantification: RNA-Seq FPKM values) and clinical information for glioblastoma (GBM) samples (n = 170) and normal adjacent tissue samples (n = 5) were obtained from the TCGA Data Portal (https://portal.gdc.cancer.gov/). For the internal validation cohort, datasets for GBM patients (n = 358) were sourced from two RNA-Seq FPKM value datasets (mRNAseq_325 and mRNAseq_693; alignment: Illumina HiSeq 2,000/2,500/4,000; human genome reference: GRCh37 [hg19]) available on the Chinese Glioma Genome Atlas (CGGA) website (http://www.cgga.org.cn/). Detailed participant characteristics of both cohorts are summarized in Supplementary Table S1.
To compare tumor samples to normal brain tissues, mRNA sequencing data from 1152 normal brain tissue samples and TCGA GBM tumor samples, accessible through the University of California, Santa Cruz (UCSC) Xena website (https://xenabrowser.net/), were used. In this instance, the initial log-transformation was reversed for both the datasets. The TCGA GBM samples were log2-transformed with an offset of 1, whereas the GTEx normal brain samples were log2-transformed with an offset of 0.001. This reversal was accomplished using the following codes: 2^data and log2(data-0.001 + 1).
Furthermore, we included seven additional GBM microarray datasets for external validation: CGGA array_301 (n = 124), GSE108474 (REMBRANDT; n = 210), GSE122586 (n = 88), GSE83300 (n = 50), GSE43378 (n = 32), GSE13041 (n = 27), and GSE74187 (n = 60) (https://www.ncbi.nlm.nih.gov/geo/). The datasets GSE108474, GSE43378, and GSE13041 utilized the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array platform, known as GPL570. On the other hand, the CGGA array_301, GSE122586, GSE83300, and GSE74187 datasets are based on the Agilent-014850 Whole Human Genome Microarray 4 × 44 K G4112F platform, labeled as GPL6480.
CNV (copy number variations) data was retrieved from the Xena browser (https://xenabrowser.net/) in order to evaluate the location and frequency of aberrations in NRGs.
Additionally, we acquired the stemness score for TCGA GBM dataset from the Xena browser (https://xenabrowser.net/). The oncoplot illustrating the GBM variant classification and mutation frequency in necroptosis-related genes (NRGs) was obtained from the Gene Set Cancer Analysis database (http://bioinfo.life.hust.edu.cn/GSCA/) using the mutation module. STRING (Search Tool for the Retrieval of Interacting Genes), version 11.0 (https://string-db.org/), a protein-protein interaction (PPI) network was constructed to investigate the interaction patterns among differentially expressed genes (DEGs).
2.2. Normalization, batch effect removal, and visualization
Normalization was conducted using the limma R package, while batch effects were addressed using the Combat function from the sva package. To visualize the effects of batch correction, Principal Component Analysis (PCA) was performed. The prcomp function from the stats package was utilized to compute PCA, and scatter plots were generated using the ggscatter function from the ggpubr package.
2.3. Identification of necroptosis-related genes
A literature review was conducted to identify genes related to necroptosis utilizing databases such as PubMed, Scopus, and Web of Science to find relevant publications up to September 2023. The search strategy included keywords like ‘necroptosis’, ‘necroptosis-related genes’, ‘necroptosis pathway’, ‘RIPK1’, ‘RIPK3’, ‘MLKL’, and ‘necroptotic cell death’. Each publication was independently reviewed by two researchers (M.K. and X.H.) to ensure objectivity and minimize bias. Duplicate entries were identified and removed using reference management software (EndNote), followed by a manual check for accuracy and originality. Genes with direct role in necroptosis were identified in three categories as initiators, key mediators, and regulators. In cases of conflicting results or discrepancies between reviewers, a third senior researcher (J.L.) was consulted. Discussions were held until a consensus was reached among all authors. Cross-validation was performed by referring to well-known literature reviews on necroptosis and The Molecular Signatures Database (MSigDB), which revealed a total of 55 genes associated with necroptosis (referred to as necroptosis-related genes or NRGs) (Supplementary Table S2) [19–70].
2.4. Consensus clustering
Consensus clustering analysis, a precise unsupervised clustering technique, was employed to classify the patients according to the expression profiles of NRGs. This involved systematic exploration of the cluster matrices across a range of clustering variable values (k) from k = 2 to k = 11. The goal was to determine the optimal cluster number (k) and assess consensus stability by analyzing cumulative distribution function (CDF) plots. To perform consensus clustering, we leveraged the R package ‘ConsensuClusterPlus’ with 1000 repetitions [71]. To assess survival disparities among the identified clusters, Kaplan-Meier survival analysis was performed utilizing the R package ‘survival’.
2.5. Identification of necroptosis-related prognostic signature
To identify differentially expressed genes (DEGs) between clusters, we utilized the ‘limma’ package with criteria set at a log2 fold change (log2FC) ≥ 1 and a false discovery rate (FDR) < 0.01. The resulting DEGs were then extracted from the training dataset (TCGA) and the validation dataset (CGGA) after intersection for common genes expression that involved normalization and batch correction using the ‘limma’ and ‘sva’ packages. Following this, we conducted univariate Cox regression analysis in TCGA GBM samples to identify DEGs with prognostic significance (p < 0.05). For further refinement and construction of the prognostic signature, we employed LASSO (Least Absolute Shrinkage and Selection Operator) regression analysis through the R package ‘glmnet’ with TCGA GBM as training set and CGGA as validation set. This approach facilitates effective reduction of candidate genes [72]. The individual risk scores for each patient were computed by considering their specific gene expression levels and the corresponding coefficients using the following formula: Risk Score = (exp = gene expression, β = coefficient).
2.6. Risk model assessment
GBM samples were categorized into high- and low-glioblastoma necroptosis index (GNI) groups using the median risk score. To explore the spatial distribution and integration of risk within these groups, we utilized Principal Component Analysis (PCA) by employing the ‘prcomp’ function from the ‘stats’ R package. The prognostic and predictive significance of the risk groups were evaluated through KM (Kaplan-Meier) and Receiver Operating Characteristic (ROC) curves. These assessments used R packages, including ‘survival’, ‘survminer’, and ‘time-ROC’. Moreover, we assessed the predictive performance of the GNI by estimating the Concordance Index through the ‘concordance.index’ R package [73,74]. To ascertain the independent prognostic significance of the risk subgroups, we performed both uni- and multi-variate Cox-regression analyses. This process was repeated for validation of the CGGA cohort. Subsequently, a nomogram was developed involving the GNI and common clinicopathological characteristics identified through regression analysis to predict the GBM survival probability. Calibration curves were generated to assess the effectiveness of the nomogram. Additionally, we employed Decision Curve Analysis (DCA), a statistical approach that accounts for clinical implications, to evaluate the diagnostic and prognostic efficacy of the nomogram. This analysis was performed using the ‘ggDCA’ package [75].
2.7. Functional enrichment analysis
Differential expression analysis was conducted to identify DEGs between risk subgroups (logFC = 1 and fdr < 0.05). The resulting DEGs were then subjected to Gene Ontology (GO) enrichment analysis using the ‘clusterProfiler’ package. Gene set enrichment analysis (GSEA) was performed using hallmark gene sets (h.all.v2022.1) to detect signaling pathways operating within each subgroup (http://gsea-msigdb.org/gsea/msigdb/). The ‘gsva’ package of R was utilized to estimate the activity of the KEGG pathways in the clusters as described by Hänzelmann et al. [76].
2.8. Annotation of the tumor immune microenvironment (TIME)
Infiltration of stromal (stromal score) and immune cells (immune score), collectively termed the ESTIMATE score, and tumor purity were evaluated by running the ESTIMATE algorithm (https://sourceforge.net/projects/estimateproject/) [77]. The enrichment of immune cells and immune-related pathways was estimated via single-sample GSEA (ssGSEA) using the ‘gsva’ R package. The CIBERSORT algorithm was employed to assess the proportion of immune cells, including diverse subtypes, resulting in 22 categories (https://cibersortx.stanford.edu/) [78]. Seven types of immune checkpoint molecules and signaling pathways were classified based on previously published studies [79,80]. We employed the tracking tumor immunophenotype (TIP) website to assess the seven-step antitumor response (http://biocc.hrbmu.edu.cn/TIP/) [81]. This evaluation was based on enrichment of a carefully curated set of 178 genes using ssGSEA.
2.9. Immune response prediction and validation
The total mutation burden (TMB) data were retrieved from the Xena website (https://xenabrowser.net/) to assess the mutational load. The TIDE, which represents Tumor Immune Dysfunction and Exclusion, was used to estimate the anti-tumor and immune-evasive abilities of the risk subgroups [82]. Immunophenoscore (IPS) and ICB (immune checkpoint blockade (ICB) responses were obtained from the TCIA (https://tcia.at/home) [83]. Finally, the response to anti-PD1 and anti-CTLA4 blockade therapy was assessed using the submap algorithm [84]. Response to immunotherapy was validated by investigating GNI activity in IMvigor210 (urothelial carcinoma; n = 348; ICI = anti-PD-L1), GSE176307 (metastatic urothelial carcinoma; n = 88; anti-PD-1/PD-L1), GSE91061 (melanoma; n = 51), GSE78220 (metastatic melanoma; n = 27; ICI = anti-PD-1), and E-MTAB-3218 (array express) (clear cell renal cell carcinoma [ccRCC]; n = 58). The E-MTAB-3218 dataset utilized the GPL13070 platform, featuring the Affymetrix Human Genome U219 Array. In contrast, the RNA-sequencing datasets employed four distinct platforms: GPL24014 (Ion Torrent S5 XL; human genome reference: GRCh37 [hg19]; TPM values) for GSE176307, GPL11154 (Illumina HiSeq 2000; human genome reference: GRCh37 [hg19]; FPKM values) for GSE78220, GPL9052 (Illumina Genome Analyzer; human genome reference: GRCh37 [hg19]; FPKM values) for GSE91061, and GPL11154 (Illumina HiSeq 2500; human genome reference: GRCh38 [hg38]; TPM values) for IMvigor210. The ‘oncoPredict’ R package was utilized for predicting drug sensitivities [85].
2.10. Single cell data analysis
The single-cell expression matrix of the GBM dataset (GSE148842) along with the meta-information files were downloaded from the TISCH database (http://tisch.comp-genomics.org/) [86]. The count matrices from each dataset were preprocessed following the standard pipeline in MAESTRO [87]. The RunUMAP ‘RunUMAP’ function of the ‘Seurat’ R package was used for dimensionality reduction, and the results were visualized using umap (uniform manifold approximation and projection) plots. Furthermore, the ‘AddModuleScore’ function was employed to depict the average expression of various genes (including Necroptosis mediators such as RIPK1, RIPK3, and MLKL, as well as GNI genes) within each cell type.
2.11. Cell lines and cell culture
Human glioblastoma multiforme (GBM) cellular models (U251 and LN229) were obtained from the Chinese Academy of Sciences Committee of Type Culture Collection in Shanghai, China. These cells were cultivated in a culture medium comprising DMEM supplemented with 10% fetal bovine serum (FBS) and 100 U/ml each of penicillin and streptomycin. The cells were cultured in a humidified incubator at 37 °C in a 5% CO2 atmosphere. We regularly performed authentication checks on all cell lines utilized in this investigation by assessing their morphology and conducted tests to ensure the absence of Mycoplasma contamination.
2.12. Necroptosis induction
To induce necroptosis, cells were exposed to a mixture of recombinant human TNF-α (tumor necrosis factor-alpha; 10 ng/ml) (obtained from Peprotech, New Jersey, USA), a second mitochondrial-derived activator of caspases (SMAC) mimetic BV6 (1 nM) (sourced from Selleck Chemicals, Houston, USA), and a pan-caspase inhibitor known as zVAD-FMK (40 mM) (acquired from ENZO Life Science, New York, USA). Necrostatin-1 (procured from Enzo) was introduced one hour prior to administration of the aforementioned agents to suppress necroptosis. Before collecting the culture media, the cells were rinsed twice with phosphate buffered saline (PBS), and fresh media were replaced following a 3-hour treatment with the aforementioned agents. Subsequently, cells were incubated for an additional 12 h at 37 °C. The culture media were collected and filtered using a syringe filter (22-mm) from Merck (Darmstadt, Germany). After centrifugation at 1500 rpm for 5 min, the supernatants were collected and stored at 4 °C.
2.13. Quantitative real-time PCR
Trizol Reagent (Takara, Otsu, Japan) was used to isolate and purify the complete RNA. Complementary DNA (cDNA) libraries were generated by reverse transcription of purified RNA. SYBR Green PCR Kit (Takara) was used for quantitative real-time polymerase chain reaction (qRT-PCR). mRNA expression levels were normalized against beta activity (internal control), and the relative mRNA levels were compared between the treated and control groups. The primer sequences are listed in Supplementary Table S3.
2.14. Immunohistochemistry
Sections of formalin-fixed, paraffin-embedded tumor tissues, each measuring 4 mm in thickness, were deparaffinized through sequential treatment with xylene and ethanol. Antigen retrieval was performed by microwave boiling in citrate buffer (pH 6.0), and hydrogen peroxide (0.3%) was subsequently used to block the endogenous activity of horseradish peroxidase. The sections were then washed with 10% phosphate-buffered saline (PBS) and blocked with bovine serum albumin (5%). The sections were then incubated with primary antibodies targeting specific proteins: MLKL (Proteintech, #21066-1-AP, rabbit, 1:50), Stanniocalcin-1 (STC1) (Proteintech, #20621-1-AP, rabbit, 1:200), Lysyl oxidase homolog 1(LOXL1) (Affinity, #DF14089, rabbit, 1:35), COL22A1 (Affinity, #DF14266, rabbit, 1:50), CD68 (Abcam, #ab955, rabbit, 1:1000), and CD163 (Cell Signaling Technology, #25121, Rabbit, 1:100). Incubation was carried out overnight at 4 °C. The sections were then 20-minute incubation with biotinylated secondary antibody (goat anti-rabbit IgG) at room temperature. Visualization was achieved using a 3,5-diaminobenzidine (DAB) Substrate Kit and the sections were counterstained with hematoxylin. Staining intensity was assessed using a semiquantitative scale: 0, negative; 1, weak; 2, moderate; and 3, strong. The presence of positive cells was categorized as follows: 0, < 5%; 1 5–25%, 2 26–50%, 3 51–75%, and 4, > 75%. The Immunohistochemistry (IHC) scores were obtained by multiplying the staining intensity by the frequency of positive cells. In cases of heterogeneous tissue staining, each distinct area was individually scored and these scores were aggregated to determine the final result. Prior to conducting the study, informed consent was obtained from the patients, and approval was granted by the internal review and ethics boards of the Affiliated Cancer Hospital and the Institute of Guangzhou Medical University.
2.15. Construction of overexpressed glioma cell lines
Overexpression vectors (pcDNA3.1-STC1-3xFlag-C, plvx-CMV-LOXL1(human)-EGFP-Puro, pCDH-CMV-COL22A1(human)(repeat-opt)-3 × FLAG-CopGFP-Puro, pCDNA3.1-EGFP-P2A-hMLKL-3xflag) and negative control vectors (pcDNA3.1-3xflag, pLVX-CMV-Puro, pCDH-CMV-mcs-3 × FLAG-CopGFP-Puro, PcDNA3.1(+)-EGFP) were obtained from Hanyi Biosciences (China). Then these vectors were transfected into LN229 and U251 cells with liposome 3000 transfection reagent (Thermofisher, USA), and 48 h later, qPCR was performed three times, and western blot analysis was performed to detect the upregulation of STC1, LOXL1, COL22A1 and MLKL.
2.16. Coculture assay
THP-1 monocytes were induced in RPMI-1640 medium containing 10% fetal bovine serum (FBS). M0 macrophages were differentiated from 100 ng/ml PMA (Phorbol 12-myristate 13-acetate, Sigma, Cat# P1585) for 24 h. Once differentiated (M0 macrophages), they were incubated respectively with cell supernatant of glioblastoma cell lines (LN229 or U251) transfected with over-expressive vectors (MLKL-OE, STC1-OE, LOXL1-OE, COL22A1-OE) for 48 h.
2.17. Flow cytometry
The expression of CD206 in macrophages was detected by flow cytometry. Cells were isolated with trypsin, washed and blocked with PBS + 1% BSA solution, and then incubated with CD206 (321105, BioLegend, California, USA). Then the cells were analyzed with BD FACSCantoTM II flow cytometry analyzer and FlowJo software.
2.18. Western blot analysis
Cells were first washed twice with PBS and then lysed using RIPA buffer (CST, USA) containing protease and phosphatase inhibitors (CWBIO, China). The lysate was centrifuged at 12,000 × g for 15 min at 4 °C. Protein concentration was measured using the BCA Protein Assay Kit (CWBIO, China). An aliquot of 30 µg of total protein was separated by SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad, Minneapolis, MN). The membrane was blocked with 5% BSA in Tris-buffered saline with Tween-20 (TBST) and incubated with primary antibodies overnight at 4 °C. Following this, the membrane was washed with TBST and incubated with the appropriate HRP-conjugated secondary antibody for 1 h at room temperature. Protein bands were visualized using an enhanced chemiluminescence method (Pierce Biotechnology). The primary antibodies include: Anti-MLKL (Proteintech, #21066-1-AP, rabbit, 1:3000), anti-STC1 (Proteintech, #20621-1-AP, rabbit, 1:1000), anti-LOXL1 (Affinity, #DF14089, rabbit, 1:1000), and anti-COL22A1 (Affinity, #DF14266, rabbit, 1:1000)
2.19. Statistical analysis
Non-parametric Wilcoxon Rank-Sum Test (Mann-Whitney U Test) was used to compare gene expression/enrichment scores between the groups. Non-parametric Kruskal-Wallis test was used to compare three groups. qPCR results were compared using Two-tailed unpaired T-test. Categorical variables were compared using the chi-squared test. Correlations were estimated using the Spearman/Pearson’s correlation test. The Kaplan-Meier method was used to estimate survival differences. Statistical significance was determined using the log-rank test. Cox regression hazard models were used to perform the univariate and multivariate factor analyses. Statistical analysis was performed using the statistical software R v4.0.3 (http://www.r-project.org).
3. Results
3.1. Genomic aberrations & differential expression of necroptosis-related genes (NRGs)
A comprehensive review of existing studies identified 55 necroptosis-related genes (NRGs) [19–70]. Pathway enrichment and protein-protein interaction analyses demonstrated their participation in the necroptosis pathway and their association with a shared regulatory mechanism (Figure 2A and B and Supplementary Table S4). The chromosomal locations and copy number variations (CNVs) of these NRGs are depicted in Figure S2A, with CNV frequencies remaining low, peaking at 4% (Figure S2B). Mutation frequencies (SNVs) in the TCGA GBM cohort were similarly low (Figure S2C), with EZH2, CYLD, AXL, DIABLO, ITPK1, and TYRO3 showing consistent mutation rates of 7%, primarily featuring missense mutations. RIPK3, a key necroptosis mediator, exhibited a deletion frequency of 6%. Notably, certain negative regulators of necroptosis, including MIB2 (11%), PRKN (10%), and NDRG2 (6%), displayed higher deletion frequencies. However, genetic alterations did not significantly impact prognosis (Figure S2D), suggesting infrequent genetic aberrations in NRGs among GBM patients.
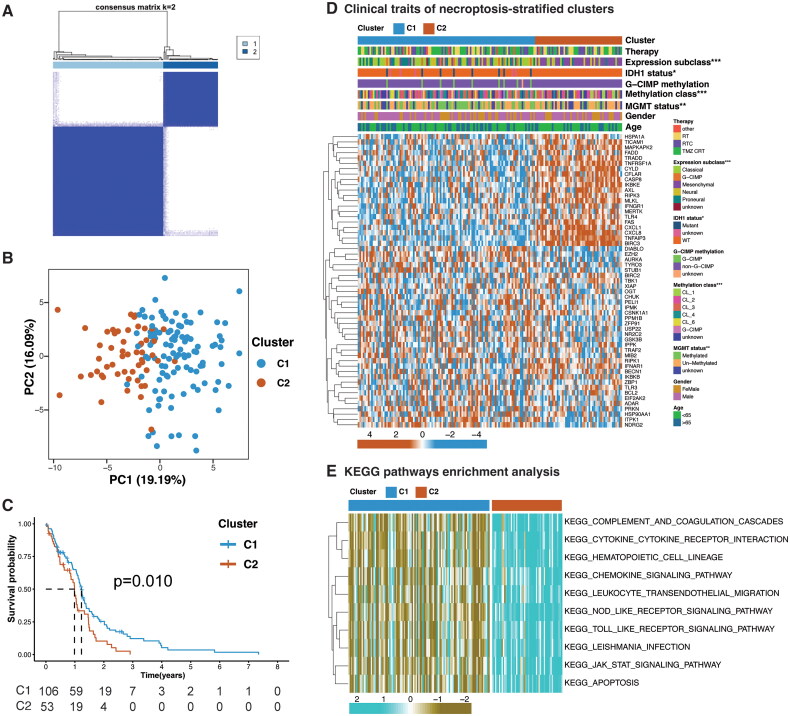
Figure 2.
Necroptosis-stratified clusters. (A) Consensus clustering of TCGA GBM samples based on expression of NRGs. (B) PCA (Principal Component analysis) plots of necroptosis-stratified clusters for TCGA cohort. (C) Overall survival difference between the clusters. (D) Heatmap illustrating association between the clusters and their clinicopathological features, and expression level of each NRG in the necroptosis-stratified clusters. Chi-square test; *p < 0.05; **p < 0.01; ***p < 0.001. (E) Top 10 Kyoto encyclopedia of genes and genomes (KEGG) enriched pathways operating between the clusters.
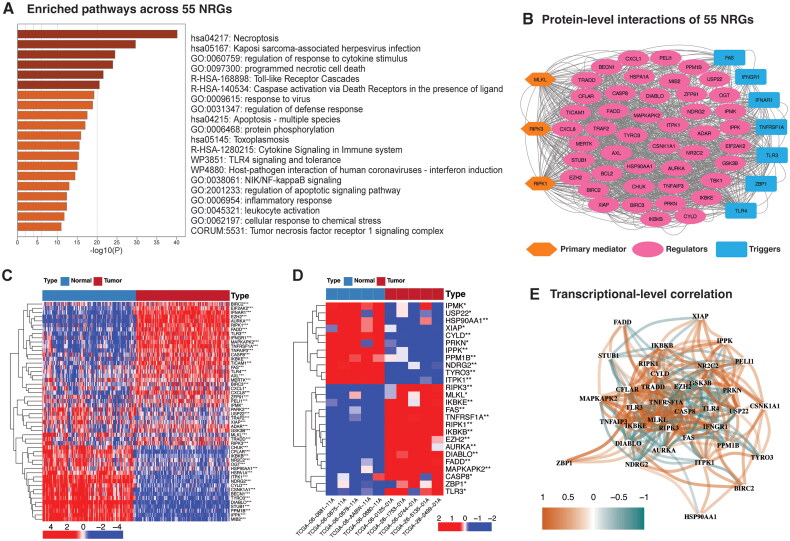
Figure 1.
Analysis of 55 necroptosis-related genes. (A) Bar graph of enriched terms (pathway or process) across 55 NRGs, colored by p-values. (B) Protein-protein interaction (PPI) network (interaction score = 0.4). (C) Heatmap illustrating the expression contrast of NRGs between TCGA GBM tumor (n = 155) and GTEx random normal brain samples (n = 155). Upregulation is represented in red, whereas downregulation is indicated in blue. Differential expression was assessed using the Wilcoxon rank-sum test with a p-value threshold of 0.05. Significance levels are denoted as follows: *p < 0.05, **p < 0.01, and ***p < 0.001. (D) Heatmap depicting the expression patterns of NRGs in TCGA GBM, comparing normal (n = 5) and randomly selected tumor samples (n = 5). Upregulation is indicated in red, whereas downregulation is shown in blue. Wilcoxon rank-sum test; *p < 0.05, **p < 0.01, and ***p < 0.001. (E) Network depicting the mRNA-level correlations among NRGs in the TCGA GBM cohort. The color intensity corresponds to the correlation strength. Pearson’s correlation test, Pearson correlation coefficient (r) cutoff of 0.2.
Next, we examined the NRGs transcriptional variation between GBM patients and normal brain tissues. Due to the limited number of normal samples in the TCGA GBM dataset (only five), we integrated GTEx-derived normal brain samples (n = 1152) with TCGA GBM primary tumor samples after correcting for batch effects (Figure S3A and B). Differential expression analysis revealed significant upregulation of necroptosis core mediators (RIPK1 and RIPK3), necroptosis-inducing receptors (TNFRSF1A, FAS, TLR3, TLR4), TAM kinases (AXL, MEERTK), and chemokine ligands (CXCL1, CXCL8) in TCGA GBM primary tumor samples (n = 155) compared to an equal number of randomly selected GTEx normal brain samples (n = 155) (Figure 1C). Conversely, a subset of NRGs, mainly negative regulators, showed downregulation. These findings were consistent when comparing TCGA tumor samples (n = 160) to normal tissues (n = 5) (Figure S3C), as well as in comparisons with five randomly selected GBM tumor samples, demonstrating a similar dysregulation pattern to that of overall TCGA GBM dataset (Figure 1D). Moreover, a significant interaction at mRNA level was also observed for the majority of the NRGs in TCGA GBM patients, as illustrated in Figure 1E. These outcomes suggest a potential dysregulation of the necroptosis pathway in GBM.
3.2. Necroptosis-stratified clusters correlate with immune cell migration and poor prognosis
Two distinct necroptosis-related clusters, denoted as Cluster 1 (C1) comprising 106 samples and Cluster 2 (C2) consisting of 53 samples, were identified by analyzing the expression patterns of 55 Necroptosis-Related Genes (NRGs) within the TCGA cohort (Figure 2A). PCA showed noticeably separated necroptosis-stratified clusters, and a lower survival probability curve was demonstrated for cluster 2 (p = 0.010) (Figure 2B and C). The core necroptosis mediators (RIPK1, RIPK3, and MLKL), showed elevated expression in C2, indicating a high necroptosis index (Figure 2D). The defined clusters also showed statistically significant variation in clinical traits such as MGMT promoter methylation, methylation class, and molecular expression subtypes (Figure 2D). Cluster 2 predominantly accounted for the more malignant mesenchymal subtype and lacked the IDH1 mutation and MGMT methylation (Figure 2D and Supplementary Table S5). Moreover, the pathways associated with immune response and immune cell migration were enriched in this necroptosis-high cluster (Figure 2E and Supplementary Table S6). These outcomes suggest the association of necroptosis to immune activity and poor prognosis.
3.3. Generation of glioblastoma necroptosis index
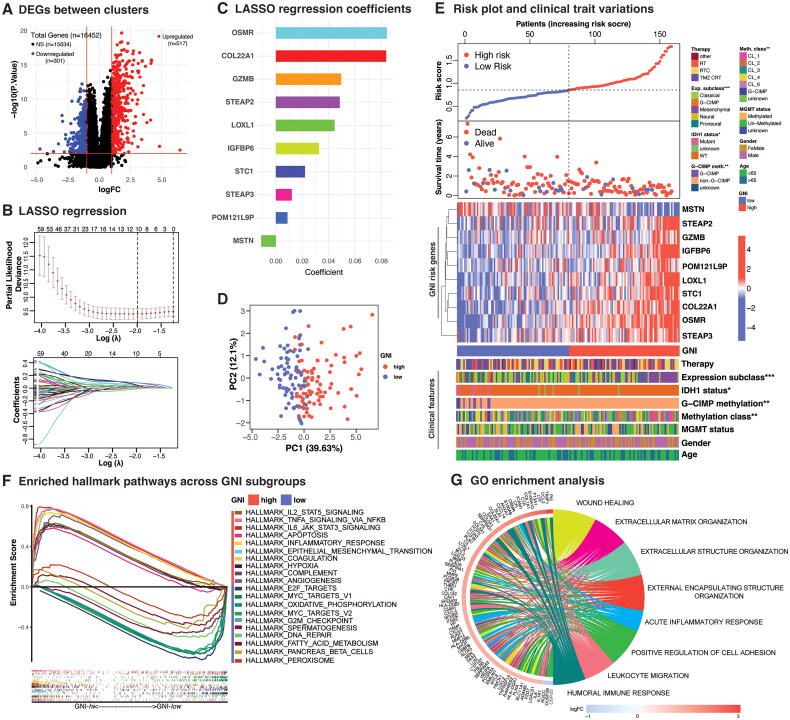
Before developing the prognostic index, the TCGA training dataset and CGGA validation dataset were intersected for common gene expression. This involved normalization and batch correction using the ‘limma’ and ‘sva’ packages (Figure S3D and E). This integration was crucial to prevent overfitting in the TCGA cohort and enhance the model’s generalizability. To develop necroptosis-related prognostic index, we first identified 818 DEGs between the two clusters by performing differential expression analysis in TCGA cohort (log2FC ≥ 1; FDR < 0.01) (Figure 3A and Supplementary Table S7). Of the 818 DEGs, 636 were shared by both cohorts. Univariate Cox regression analysis indicated prognostic relevance of 111 DEGs (p < 0.05) (Supplementary Table S8). Of the 111 survival-related genes, 102 were associated with poor prognosis, as indicated by a Hazard Ratio (HR) greater than 1. A 10-gene prognostic risk signature, referred to as the GNI, was derived using least absolute shrinkage and selection operator (LASSO) Cox regression analysis (Figure 3B and C). GBM samples were stratified into two subgroups, namely GNI-high (high-risk subgroup) and GNI-low (low-risk subgroup), based on their respective median risk scores. Principal component analysis (PCA) showed spatially separated GNI subgroups with PC1 capturing about 40% of the variation in both cohorts (Figure 3D and S3F). The distribution of patients into GNI-high and GNI-low subgroups was evenly represented, as illustrated by the plot of the risk scores (Figure 3E and S3G). Notably, patients in the GNI-high subgroup exhibited a higher frequency of deaths and shorter survival time, indicating a negative correlation compared to those in the GNI-low subgroup.
Figure 3.
Analysis of GNI signature genes’ expression, distribution, and correlation with clinical characteristics. (A) Volcano plot depicting the differentially expressed genes (DEGs) between the necroptosis-based clusters. Wilcoxon rank-sum test; DEGs were defined according to the following criteria: log 2 fold change (log2FC) = 1, and the false discover rate (FDR) < 0.01. (B) LASSO regression of the 111 OS-related genes identified via uni-cox regression analysis and cross-validation for tuning the parameter selection in the LASSO regression. (C) Bar plot depicting lasso regression coefficients. (D) Principal component analysis (PCA) plot of GNI in TCGA GBM cohort. (E) Risk plot, survival plot and heatmap displaying the expression of 10 GNI risk genes (upregulation is represented in red, while downregulation is indicated in blue) within the GNI subgroups (red: GNI-high; blue: GNI-low) and association with clinical and pathological characteristics. Chi-square test; significance: *p < 0.05; **p < 0.01; ***p < 0.001. (F) HALLMARK pathways enrichment analysis. (G) Gene Ontology (GO) terms enrichment analysis.
3.4. Characterization of GNI subgroups
The expression patterns of the ten risk genes and their association with clinical attributes for both cohorts are illustrated in heatmaps (Figure 3E and S3G). Except for MSTN, the rest of the nine risk genes were upregulated in the GNI-high subgroup. Individually, necroptosis mediators were positively correlated with GNI in the TCGA cohort (Figure. S3H–J). Major differences in clinical traits included methylation and expression classes and IDH1 mutations (Figure 3E and Supplementary Table S9). The GNI-low subgroup had a higher incidence of IDH1 mutations. The mesenchymal subtype was the predominant subtype in the GNI-high subgroup. Investigation of the CGGA cohort, which comprised a younger group (mostly <65 years), showed that MGMT methylation was significantly superior in the GNI-low subgroup (Figure S3G). In this cohort, the frequency of IDH1 mutation, 1p19q co-deletion, and female participants were prevalent in the GNI-low subgroup. Overall, similar characteristics were apparent in the GNI subgroups to that of necroptosis-stratified clusters.
Functional enrichment analysis indicated the enrichment of oncogenic and immune-related gene sets in the GNI-high subgroup, as illustrated in Figure 3F. The GNI-high subgroup is characterized by hyperactivation of IL6_JAK_STAT3 signaling, epithelial-to-mesenchymal transition (EMT), hypoxia, and angiogenesis [88,89]. Immune-related gene sets included activation of complement, coagulation, acute inflammatory response, and signaling pathways, such as TNFA and IL2_STAT5. IL2_STAT5 signaling pathway plays a dynamic role in effector and regulatory T cells (Tregs) [90]. Conversely, the GNI-low subgroup exhibited enrichment in gene sets associated with DNA repair and cell cycle, along with metabolic pathways, including oxidative phosphorylation and fatty acid metabolism. Differentially expressed genes between the GNI subgroups also endorsed the implication of inflammatory and immune-associated pathways, such as wound healing, acute inflammatory response, leukocyte migration, and humoral immune response (Figure 3G and Supplementary Tables S10, S11).
3.5. Validation of GNI genes induction during necroptosis
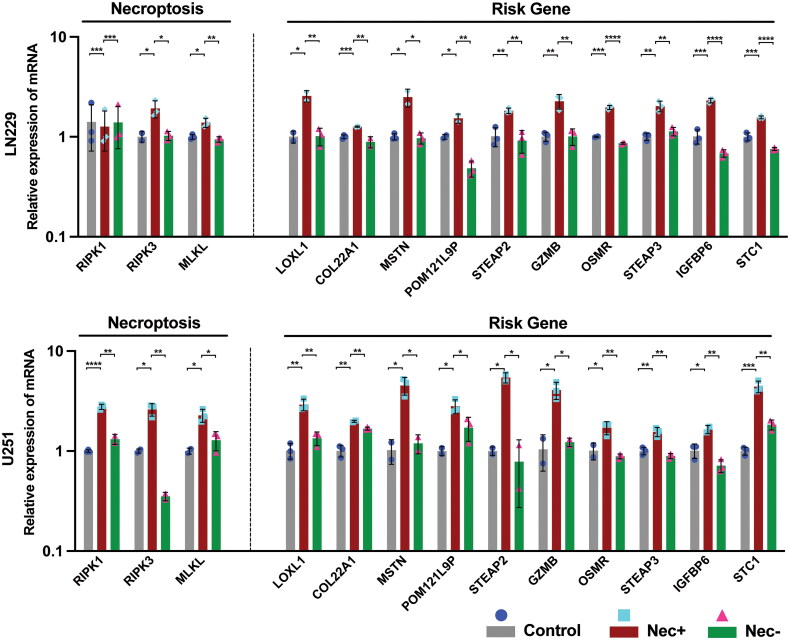
Subsequently, we proceeded to empirically verify the initiation of GNI gene activation during necroptosis induction in GBM cancer cell lines (U251 and LN229). This validation process involved utilizing the TNF-α, SMAC mimetic, and zVAD-FMK (TSZ) combination, in accordance with previous recommendations [91,92]. The validation of necroptosis induction was substantiated by the observed increase in the mRNA expression levels of necroptosis mediators such as RIPK1, RIPK3, and MLKL (Figure 4). Conversely, the introduction of the necroptosis inhibitor, necrostatin-1, led to the suppression of these genes. Furthermore, a comparable expression pattern was observed for all the GNI oncogenes.
Figure 4.
A) The relative mRNA level of necroptosis core mediators (RIPK1, RIPK3, MLKL) and GNI genes (LOXL1, COL22A1, MSTN, POM121L9P, STEAP2, GZMB, OSMR, STEAP3, IGFBP6 and STC1) in glioblastoma cells (U251 and LN229) following treatment with TNF-a, SMAC mimetic, and zVAD-FMK (TSZ) to induce necroptosis or the addition of the necroptosis inhibitor (necrostatin-1) to inhibit necroptosis. The data represent the mean ± SEM (standard error of mean) of n = 3 independent experiments (independent biological replicas) for each condition. Two-tailed unpaired T-test; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant.
3.6. GNI effectively predicts GBM prognosis
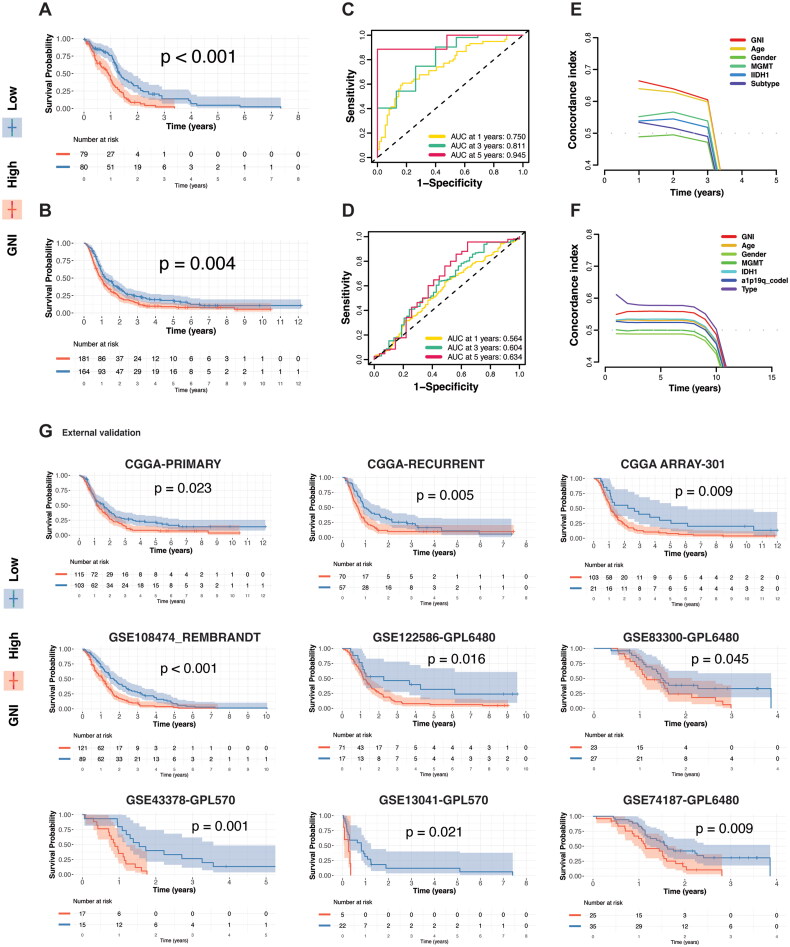
GNI could effectively identify GBM patients with the worst prognosis in both cohorts, as depicted in Figure 5A and B. The predictive performance of the prognostic model was assessed using time-dependent receiver operating characteristic (ROC) analysis. In TCGA cohort, the area under the curve (AUCs) was 0.750, 0.811, and 0.945 at 1, 3, and 5 years, respectively. Similarly, in the CGGA cohort, the AUCs were 0.564, 0.604, and 0.634 at 1, 3, and 5 years, respectively (Figure 5C and D). The concordance index identified GNI as the best-ranked model of survival in TCGA cohort (Figure 5E). In the CGGA cohort, recurrence was the best, followed by the GNI, as the CGGA cohort included patients with recurrent glioblastoma (Figure 5F). GNI could also effectively predicted the progression-free and disease-specific survival in of TCGA GBM patients (Figure S3K and L). We separately evaluated the GNI efficiency in the primary and recurrent GBM samples from the CGGA cohort, which also indicated that GNI could predict prognosis regardless of recurrence status (Figure 5G). Moreover, the ability of GNI to stratify GBM patient survival probabilities was also demonstrated in seven external datasets comprising 591 GBM patients (Figure 5G). Overall, a significant prognostic value of the GNI was evident in diverse GBM patients.
Figure 5.
GNI Predict survival probability. (A) Overall survival difference between risk subgroups in the TCGA cohort and B) CGGA cohort. (C) Receiver operating characteristic (ROC) curves and corresponding area under the curve (AUC) analyses over time, illustrating the predictive efficacy of the risk score in the TCGA cohort and D) CGGA cohort. (E) Concordance index (C-index) analyses depicting the GNI model performance in TCGA cohort and F) CGGA cohort. (G) GNI-stratified subgroups discriminating survival probabilities in the GBM cohorts.
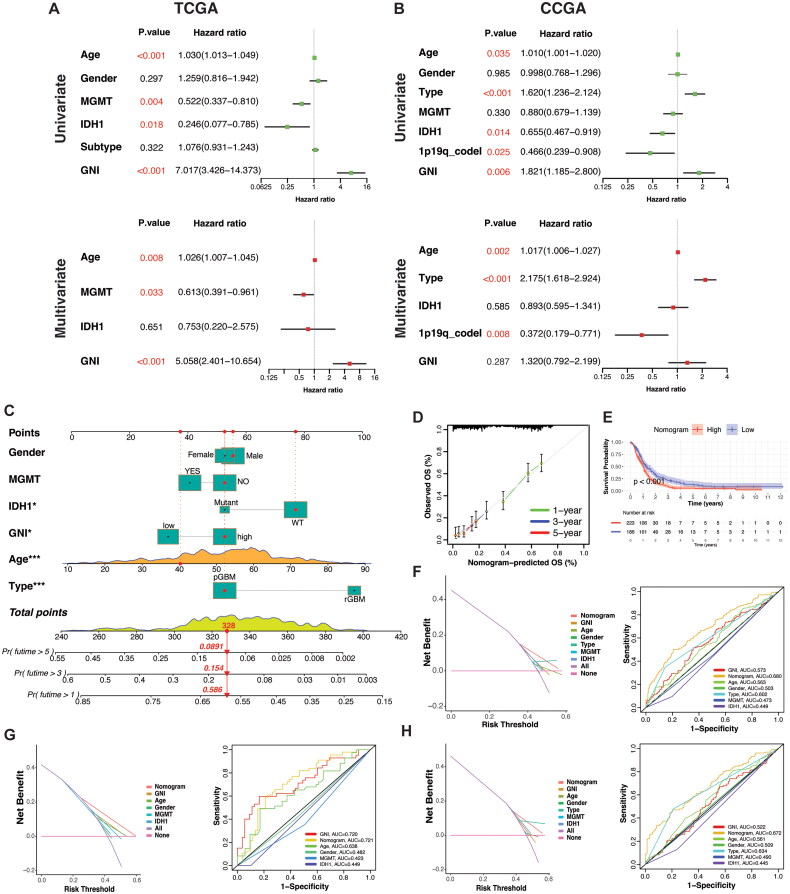
3.7. Construction of a GNI-based prognostic nomogram
Further exploration of the prognostic and predictive efficiency of GNI was conducted using uni- and multi-variate cox regression. GNI was identified as a hazardous factor in TCGA cohort at both uni- and multivariate levels (Figure 6A). However, in the CGGA cohort, recurrence was more hazardous than GNI, as previously mentioned (Figure 6B). A nomogram was developed involving the common prognostic factors between the two datasets, including age, gender, MGMT methylation, IDH1 status, type of primary disease (primary or recurrent), and GNI (Figure 6C). There was a good correspondence between the predicted probabilities based on the nomogram and the actual 1-, 3-, and 5-year OS rates of GBM patients (Figure 6D). Effective discrimination in the survival probabilities of subgroups stratified by the nomogram was demonstrated by the Kaplan-Meier survival curve (p < 0.0001) (Figure 6E). The DCA and ROC graphs illustrate the discriminative capability of the nomogram in the combined cohort and each cohort separately (Figure 6F–H).
Figure 6.
A) Uni- and multi-variate regression demonstrating prognostic significance of GNI and other factors in TCGA and B) CGGA cohorts. (C) a nomogram integrating GNI, clinical, genetic, pathological variables and overall survival probability of the combined cohort (TCGA + CGGA). (D) Correlation of actual and predicted overall survival rates in combined cohort (TCGA + CGGA). (E) survival probability of the nomogram-stratified clusters in combined cohort (TCGA + CGGA). (F) DCA (Decision curve analysis) and ROC curve plot of evaluating the nomogram in combined (TCGA + CGGA), G) TCGA, and H) CGGA cohorts.
3.8. GNI-driven immune landscape and its effects on macrophage polarization in GBM
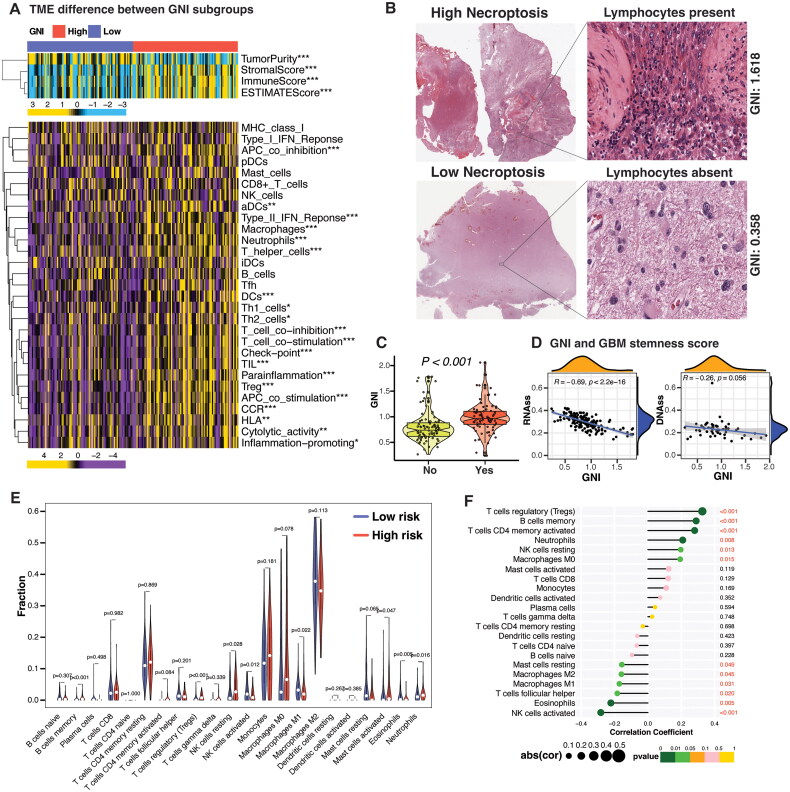
Consistent with pathway enrichment analysis, the GNI-high subgroup exhibited a significantly higher immune score compared to the GNI-low subgroup (Figure 7A). In the TCGA cohort, various immune cells, including myeloid cells and lymphocytes, were more abundant in the GNI-high subgroup. This included macrophages, neutrophils, regulatory T cells (Tregs), dendritic cells (DCs), activated dendritic cells (aDCs), and T helper cells such as Th1 and Th2 cells (Figure 7A). Notably, all 13 immune-related pathways were enriched in the GNI-high subgroup, except for the MHC class I and Type I interferon pathways (Figure 7A). Histopathological examination of TCGA GBM samples further supported the higher presence of lymphocytes in the GNI-high subgroup compared to the GNI-low subgroup (Figure 7B and C). Since, cancer stemness was negatively correlated with immune cell infiltration, we evaluated the relationship between GNI and the TCGA GBM stemness score [93]. As expected, there was a negative correlation between GNI and both RNAss and DNAss stemness scores (Figure 7D). Overall, these results suggest that a high necroptosis index indicate an elevated immune activity.
Figure 7.
TME Landscape in GNI-stratified GBM subgroups. (A) Differences in tumor microenvironment (TME) and the enrichment (ssGSEA) of immune cells (16 types) and immune-related pathways (13 types) in the TCGA GBM samples between GNI subgroups (Wilcoxon rank-sum test; *p < 0.05; **p < 0.01; ***p < 0.001). (B) Pathological HE staining (TCGA database) showing the lymphocyte infiltration between the GNI-high and GNI-low subgroups. (C) Violin plot depicting the difference in the GNI of TCGA GBM samples characterized as lymphocyte-enriched (yes) and lymphocyte-deficient (no). (D) spearman’s correlation between GNI and TCGA GBM stemness score. (E) Enrichment of immune cells in GNI subgroups as assessed by CIBERSORT algorithm (Wilcoxon rank-sum test; *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant). (F) Correlation between abundance of immune cells and GNI. Spearman’s correlation test; red indicates significance p < 0.05.
Given the crucial role of macrophages in the immunosuppressive microenvironment of GBM, we investigated the phenotype of macrophages in the GNI-high risk subgroup [94]. CIBERSORT algorithm results confirmed the infiltration of various immune cells in the GNI-high subgroup, including neutrophils, activated mast cells, monocytes, M0 macrophages, memory B cells, regulatory T cells, and activated CD4 memory T cells (Figure 7E and F). The M0 macrophage phenotype was predominant in the GNI-high risk subgroup, while the M1 and M2 phenotypes were reduced, suggesting that GNI may restrict macrophage polarization in the GBM microenvironment. Additionally, these infiltrated cells were associated with worse prognosis in the TCGA dataset (Figure S4A and Supplementary Table S12).
3.9. Validating GNI-Mediated modulation of macrophage polarization in GBM
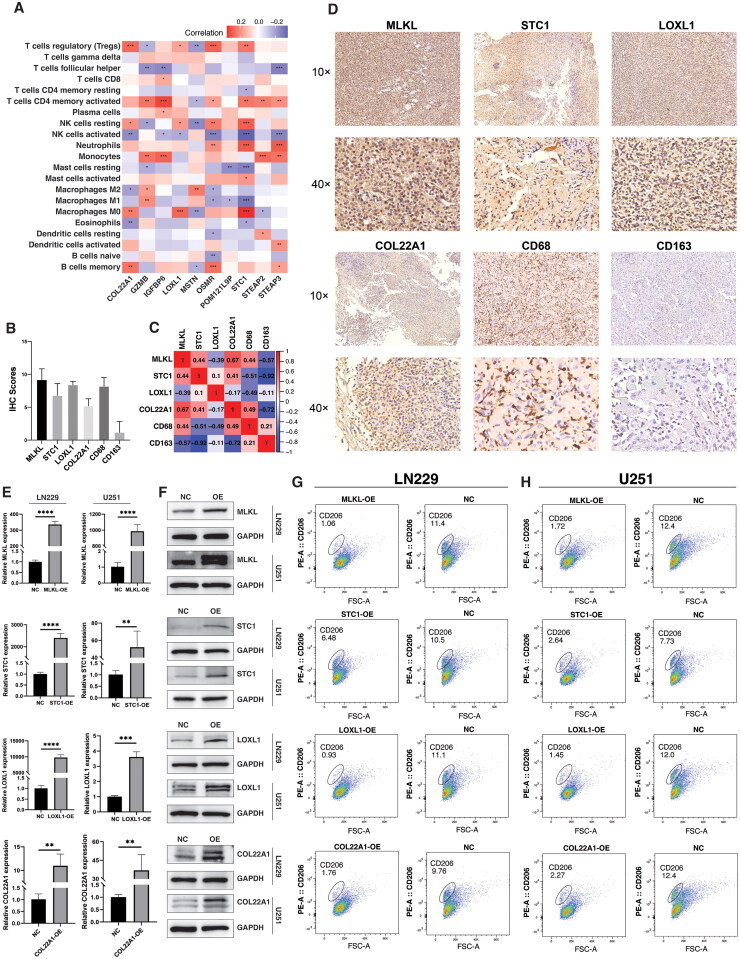
To validate this association of GNI and macrophage polarization, we first evaluated the specific GNI components related to macrophage dynamics in TCGA GBM cells. Within this framework, a cluster of GNI genes comprising COL22A1, LOXL1, OSMR, POM121L9P, and STC1 displayed a positive relationship with M0 macrophages and exhibited a negative correlation with M1 and M2 types, suggesting their potential role in preventing polarization (Figure 8A). Consequently, we opted to validate three of these genes, COL22A1, LOXL1, and STC1 using clinical samples from patients with glioblastoma. Immunohistochemical analysis of GBM tissues (n = 5) revealed a positive correlation among MLKL (a necroptosis marker), the three GNI genes, and the expression of the M0 macrophage marker (CD68) (Figure 8B–D). Conversely, the expression of CD163, a polarization marker associated with the M1/M2 macrophage phenotype, remained minimal. To further investigate the role of GNI genes (MLKL, STC1, LOXL1, and COL22A1) in modulating macrophage polarization, we conducted an indirect co-culture experiment. Glioma cell lines (LN229 and U251) transfected with overexpression vectors for the GNI genes were co-cultured with M0-like macrophages differentiated from THP-1 cells by PMA (Phorbol 12-myristate 13-acetate) (Figure 8E and F). Flow cytometry analysis revealed a considerable decrease in the proportion of CD206+ cells in the presence of GNI gene overexpression compared to the wild type (normal control), indicating that the overexpression of GNI genes reduce macrophage polarization (Figure 8G and H).
Figure 8.
GNI Genes modulates macrophage polarization. (A) Correlation between infiltration of immune cells and individual GNI genes (n = 10) in TCGA GBM cohort. (spearman’s correlation test; Statistical significance is indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001). (B) IHC quantification of expression level of MLKL, STC1, LOXL1, COL22A1, CD68 and CD163 in the clinical samples of glioblastoma. (C) Pearson’s correlation of expression level of MLKL, STC1, LOXL1, COL22A1, CD68 and CD163 in the clinical samples of glioblastoma. (D) Representative images of expression (brown, cell cytoplasmic/nucleus stain) of MLKL, STC1, LOXL1, COL22A1, CD68 and CD163 in the clinical samples of glioblastoma. (E) PCR analysis for measuring the relative mRNA expression of MLKL, STC1, LOXL1, COL22A1 in LN229 and U251 GBM over-expressive (OE) cells and normal control (NC). The data represent the mean ± SEM (standard error of mean) of n = 3 independent experiments (independent biological replicas) for each condition. Two-tailed unpaired T-test; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant. (F) Western blot analysis of protein expression of MLKL, STC1, LOXL1, COL22A1 in LN229 and U251 GBM over-expressive (OE) cells and normal control. (G) Flow cytometry plots demonstrating the difference in CD206 expression on THP-1 differentiated macrophages after coculture with LN229 and U251 GBM over-expressive (OE) cells and normal control.
3.10. Mapping necroptosis mediators and GNI genes in GBM microenvironment at single-cell resolution
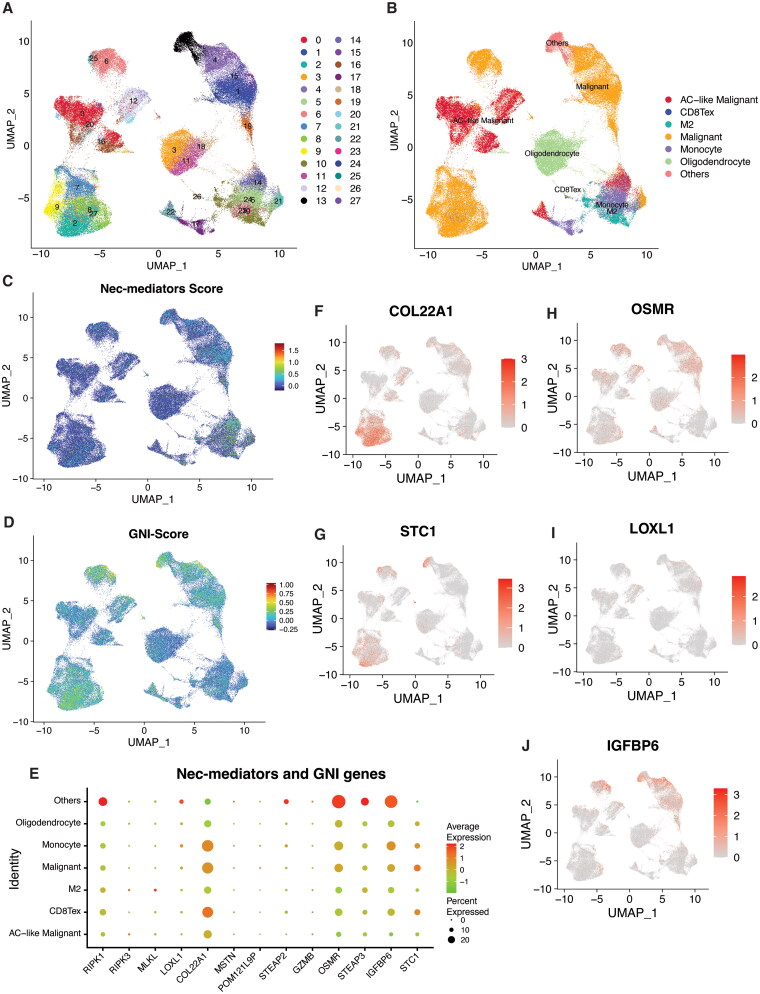
Consequently, we investigated the expression patterns of necroptosis mediators and GNI genes at the single-cell level. We accessed a preprocessed single-cell GBM dataset (GSE148842) containing normalized counts and metadata from the TISCH database [86]. This dataset encompassed seven GBM samples, totaling 111,397 single cells, following initial quality control and data standardization. A comprehensive set of 28 cell population clusters was identified, corresponding to seven distinct cell types involving both immune and cancer cells (Figure 9A and B). Utilizing the ‘Seurat’ R package, we generated UMAP visualizations of both core necroptosis regulators and GNI genes. The prevailing expression pattern of necroptosis mediators was most noticeable within immune cells (including monocytes and M2 macrophages), and to a certain extent, it was also observed in malignant cells (Figure 9C). In contrast, GNI genes were primarily expressed in the malignant cells (Figure 9D). Individually, RIPK1 was ubiquitously expressed across all cell types, whereas RIPK3 and MLKL exhibited more pronounced expression within monocytes and M2 macrophages (Figure 9E). The spectrum of GNI gene expression spanned all cell types, although discernible variations in expression patterns emerged within individual cell types. For instance, COL22A1 and STC1 were expressed in separate clusters of malignant cells, in contrast to OSMR, LOXL1, and IGFBP6, indicating potential diversity in their oncogenic roles (Figure 9F–J). Single-cell analysis indicated that necroptosis could potentially take place in myeloid cells, accompanied by the activation of GNI genes in malignant cells; conversely, the activation of GNI genes could lead to necroptosis in myeloid cells.
Figure 9.
Single-cell transcriptomic analysis of necroptosis mediators and GNI genes in glioblastoma. A, B) UMAP plots displaying main clusters and cell-types in single-cell gastric cancer dataset (GSE148842), colored by cluster (A) and (B) cell type. C, D) UMAP plots depicting the expression scores of (C) necroptosis mediators (RIPK1, RIPK3, MLKL) and (D) GNI genes, colored by expression level. (E) The bubble plot depicting the expression levels of necroptosis mediators (RIPK1, RIPK3, MLKL) and GNI genes across cell types. F-I) UMAP plot exhibiting expression of selected GNI genes in single cell GBM dataset.
3.11. Immunomodulators in GNI-defined GBM subgroups and anti-tumor immune activity in GBM
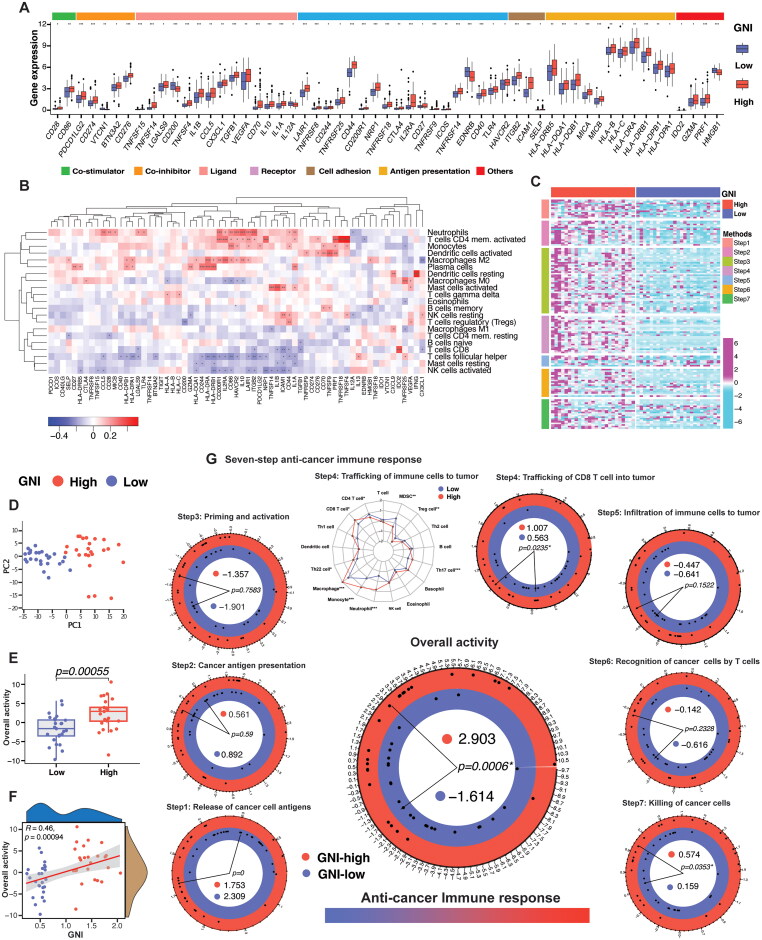
Immunomodulators (IMs) are central to the immune evasion strategy of cancer cells and are critical for cancer immunotherapy. The IMs associated with cell adhesion and migration, such as CD44, ICAM1, SELP, and ITGB2, were elevated (Figure 10A). In addition, several chemokines (CX3CL1, CXCL9, and CCL5) and cytokines (TGFB1, IL1B, and IL10) were upregulated in the GNI-high subgroup and positively correlated with monocytes, macrophages, and neutrophils, among others (Figure 10B). These cytokines are central to the immunosuppressive roles of CD4 + T cells (Th1, Th2, and Treg) and myeloid-derived suppressor cells [95,96]. MHC signaling was elevated in the GNI-high subgroup. MHC-I molecules were positively associated with the majority of immune cells and were associated with better prognosis, for example, HLA-A, HLA-C, and MICB (Figure 10B, S4B, and Supplementary Table S13). MHC-II molecules were also upregulated in the GNI-high subgroup and were mainly correlated with plasma cells, M2 macrophages, monocytes, activated CD4 memory T cells, and neutrophils (Figure 10A and B). MHC-II signaling was associated with the worst prognosis (Figure S4B). The costimulatory molecule CD27 (also a marker of memory B cells) and its ligand CD70 were upregulated and showed a positive correlation with naïve and memory B cells (Figure 10A and B). Additionally, CD27-CD70 signaling also plays a critical role in the regulation of T-cell responses [97]. Other TNFRs, such as TNFRSF4 (OX40, CD134), TNFRSF9 (4-1BB, CD137), TNFRSF8 (CD30), TNFRSF14 (HVEM), and TNFRSF18 (GITR), were also elevated in the GNI-high subgroup, which is expressed by T cells and influences T cell responses directly or indirectly by inducing inflammation or activating APCs [98,99]. The GNI-high subgroup showed upregulation of several immune inhibitors, such as PD-1, PD-L1, CTLA-4, CD274, CD200R1, TIGIT, VEGFA, and IDO, indicating T-cell exhaustion (Figure 10A) [100–107].
Figure 10.
Immunomodulators (IMs) And anti-cancer immune response in GNI subgroups. (A) Differential upregulation of IMs in GNI subgroups. (Wilcoxon rank-sum test; *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant). (B) Heatmap of correlation between immune cells and IMs. *p < 0.05; **p < 0.01; ***p < 0.001. (C) Heatmap of the seven-step anti-cancer immune response signature gene expression (178 genes) between top 15% GNI-high (n = 25) and GNI-low (n = 25). Purple and cyan represent upregulation & downregulation respectively. (D) Principal component analysis (PCA) plots of GNI subgroups for TCGA cohort (top 30%) based on overall activity. (E) Boxplot of overall activity difference between GNI subgroups. Wilcoxon rank-sum test. (F) Pearson’s correlation between GNI and overall activity. (G) Difference in the individual seven-step activity performance and overall activity between the GNI subgroups. Wilcoxon rank-sum test; p < 0.05 was considered significant.
The trafficking of immune cells and the activity of immunomodulators have been incorporated to estimate the tumor immunophenotype, which is characterized by a seven-step process and indicates the status of anti-cancer immune response [81]. The seven-step response was significantly higher in the top 15% of GNI-high subgroup compared to top 15% of low-GNI subgroup, as illustrated in Figure 10C–G. Significant trafficking of CD8 T cells, killing of the cancer cells, and overall anti-cancer immune response activity in the GNI-high subgroup was revealed (Figure 10C–G). These outcomes further endorse the earlier observations of GNI association immune activity in GBM.
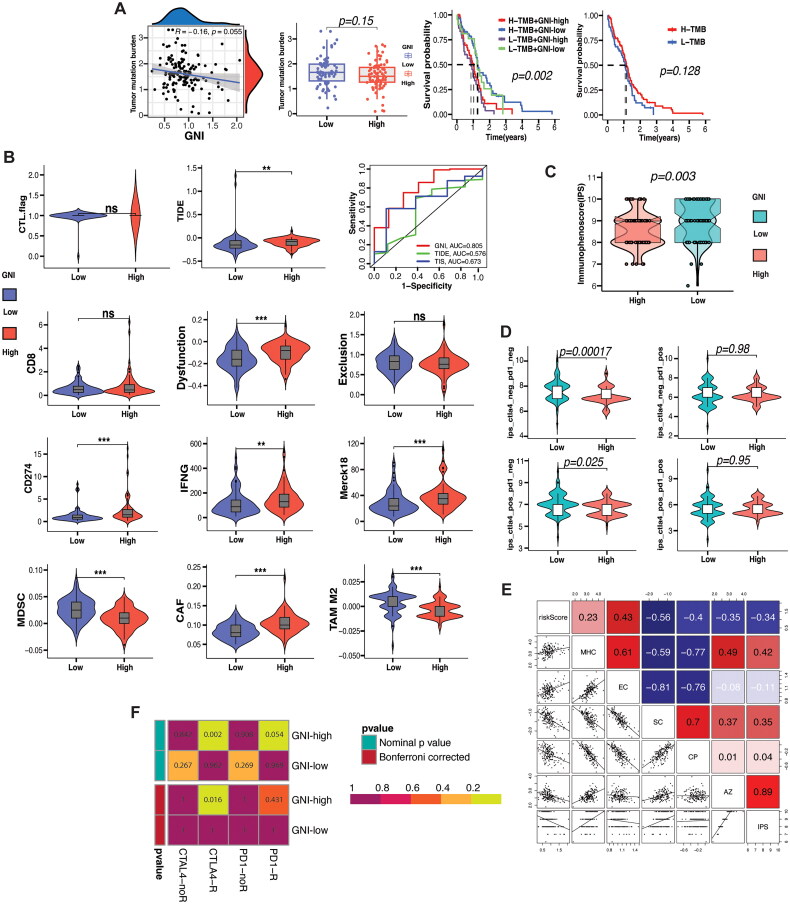
3.12. GNI’s potential to predict ICI response
Immune response prediction based on biomarkers has been mainly evaluated in melanoma, lung cancer, and bladder cancer, which have shown a good response to immunotherapy in the form of immune checkpoint blockade (ICB) such as CTLA-4 and PD-1/PD-L1 blockade [108]. Tumor mutation burden has been proven to be a valuable biomarker at different cutoff values for predicting response to ICB [109]. We estimated the TMB level, which indicated an inverse relationship with the GNI (Figure 11A). Interestingly, high TMB was associated with improved survival, as shown in Figure 11A. Tumor Immune Dysfunction and Exclusion (TIDE) is a computational framework that models immune evasion strategies by cancers in terms of T-cell dysfunction (high CTL infiltration) or exclusion (low CTL infiltration), and predicts the tumor response to ICI therapy [82]. The GNI-high subgroup had a high T cell dysfunction score, as indicators such as CD276, IFN, and Merck were higher in this subgroup (Figure 11B). However, the exclusion score showed no difference even though the TAM2 and MDSCs scores were high in the GNI-low subgroup, indicating that the immune evasion strategy operating in the GNI-high subgroup was via T cell dysfunction. According to the TIDE score, ICI response was better in the GNI-low subgroup. Nonetheless, T-cell dysfunction was the main immune evasion mechanism employed by the GNI-high subgroup. We further evaluated the prediction efficiency of another ICI response indicator, immunophenoscore (IPS), which is based on the activity of four factors: MHC signaling, tumor suppressor cells (MSDC and TAM2), effector cells (T cells), and immunomodulators (immune checkpoints) [83]. The GNI-low subgroup achieved a low score, indicating a better response to ICI therapy (Figure 11C). When limiting the IPS to CTLA-4 and PD-1 scores, a response difference was observed when both CTTLA-4 and PD-1 were negative or CTLA-4 positive plus PD-1 negative, favoring the GNI-low subgroup (Figure 11D). There was no difference in the IPS score when PD-1 was positive, regardless of the status of CTLA-4 (Figure 11D). In accordance with the TIDE outcome, the IPS also indicated a positive correlation between GNI and MHC and effector cells (Figure 11E). However, immune checkpoint molecules are positively associated with suppressor cells rather than effector cells, which corresponds to the unique microenvironment in GBM. Finally, we employed the submap algorithm to infer the response to ICI checkpoint inhibitors (anti-CTLA-4 and PD-1) based on response data from a melanoma cohort treated with ICI immunotherapy [110]. Interestingly, the outcome indicated that the GNI-high subgroup would respond to anti-CTLA-4 blockade and also showed a trend toward a better response to anti-PD-1 blockade (Figure 11F).
Figure 11.
Immunotherapy response prediction. (A) Correlation between GNI and tumor mutational burden (TMB) and prognostic impact of TMB in GBM and GNI subgroups. (B) Comparison of TIDE (tumor immune dysfunction and exclusion) algorithm results (including markers: CTL, CD8, CD276, IFNG, MERCK18, MDSCs, CAFs, TAM2; and scores: T cell dysfunction score, T cell exclusion score and TIDE score) between the GNI subgroups. Wilcoxon rank-sum test; *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant. (C) Immunophenoscore (IPS) difference between GNI subgroups predicting immunotherapy response. Wilcoxon rank-sum test (D) IPS difference between GNI subgroups predicting anti-CTLA-4 and/or anti-PD-1 immunotherapy response. Wilcoxon rank-sum test. (E) Correlation between GNI and IPS and its components of IPS including MHC, effector cells (EC), suppressor cells (SC), checkpoint protein (CP) and sum of weighted averaged Z scores of the four components (AZ). (F) probability of immunotherapy (anti-CTLA-4 and/or anti-PD-1) response of GNI subgroups estimated by submap algorithm.
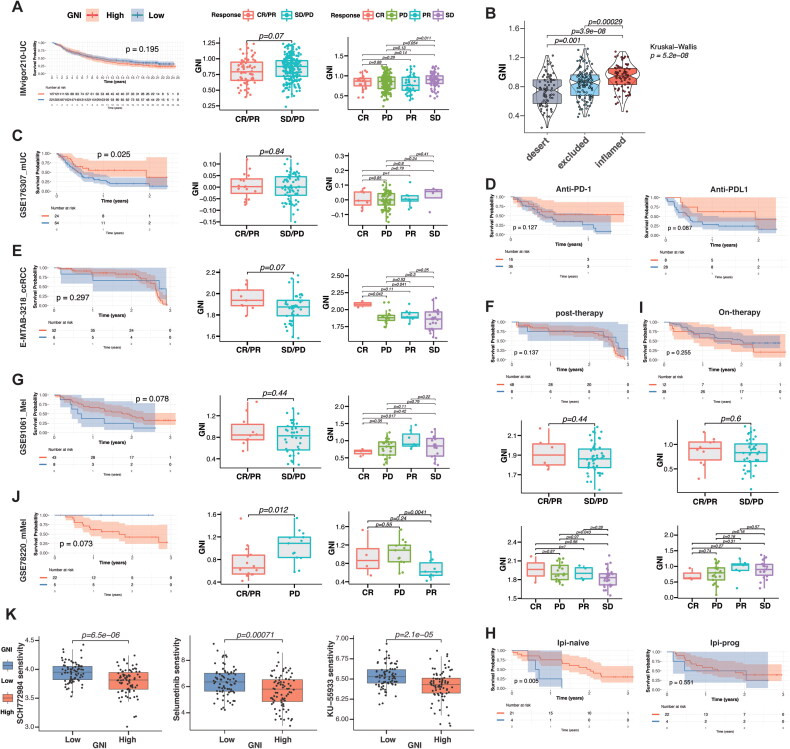
To validate whether GNI could predict the response to ICI immunotherapy, we investigated its activity in ICI-treated cohorts, including IMvigor210 (urothelial carcinoma, n = 348; ICI, anti-PD-L1), GSE176307 (metastatic urothelial carcinoma, n = 88; anti-PD-1/PD-L1), E-MTAB-3218 (array express) (ccRCC, n = 58), GSE91061 (melanoma, n = 51), and GSE78220 (metastatic melanoma, n = 27; ICI = anti-PD-1). Surprisingly, the integration of these patients into high- and low-risk subgroups based on GNI demonstrated promise for ICI responses (Figure 12). In the IMvigor210 cohort comprising urothelial carcinoma patients treated with pembrolizumab, there was no survival advantage for either GNI subgroup, and the overall response tended to favor the GNI-low subgroup (Figure 12A). Nonetheless, stable disease was significantly higher than disease progression (p = 0.054), and GNI was significantly progressively correlated with the immune microenvironment from desert to inflamed (Figure 12B). Moreover, a significantly better prognosis was demonstrated in patients with metastatic urothelial carcinoma receiving pembrolizumab and/or atezolizumab (Figure 12C). In this cohort, initial treatment with an anti-PD-L1 agent (atezolizumab) resulted in slightly better survival compared to pembrolizumab (Figure 12D). Clear cell renal cell carcinoma (ccRCC) cohort receiving anti-CTLA-4 and anti-PD-1agents also showed a promising response (CR/PR vs SD/PD: p = 0.07; CR vs SD: p = 0.041; CR vs PD: p = 0.042) (Figure 12E). Integration of risk subgroup based on post-therapy RNA sequencing showed a positive correlation with progression (PD vs. SD: p = 0.043), indicating a change in the activity of GNI after immunotherapy initiation (Figure 12F). The GNI-based subgrouping of melanoma patients also showed a trend toward better survival (p = 0.078) (Figure 12G). A rather significant improvement was demonstrated in ipilimumab-naïve patients receiving anti-PD-1 therapy compared to patients receiving anti-PD-1 therapy after progression on CTLA-4 blockade (Figure 12H). On-therapy RNA sequencing failed to show such a response (Figure 12I). GNI failed to show any difference in response in the cohort of patients with metastatic melanoma (Figure 12J). Finally, drug sensitivity analysis indicated that the GNI-high subgroup was only sensitive to three targeted drugs (out of 198 drugs), suggesting a strong resistance profile (Figure 12K). In conclusion, these analyses indicate that GNI-high is resistant to the majority of molecular targeted drugs but may respond to immune checkpoint blockade.
Figure 12.
Validation of immunotherapy response. (A) The Kaplan-Meier curves of difference in survival probability between the GNI-stratified subgroups and boxplots of GNI variation in responsiveness to immune checkpoint blockade of IMvigor210, (C) GSE176307, (E) E-MTAB-3218, (G) GSE91061, and (J) GSE78220 cohorts. The scattered dots represent the GNI of the two subgroups. (B) Kruskal–wallis test measures the difference in the GNI among TME immunophenotypes. (D) The Kaplan-Meier curves of difference in survival probability between the GNI-stratified subgroups of GSE176307 cohort based on type of immune checkpoint blockade (anti-PD1/anti-PD-LL1). (F) the Kaplan-Meier curve of difference in survival probability between the GNI-stratified subgroups and boxplots of GNI variation in responsiveness to immune checkpoint blockade of E-MTAB-3218 cohort based on post-therapy RNA sequencing data. (H) The Kaplan-Meier curves of difference in survival probability between the GNI-stratified subgroups of GSE91061 cohort based on treatment type (ipilimumab-naïve or ipilimumab-progressed. (I) The Kaplan-Meier curve of difference in survival probability between the GNI-stratified subgroups and boxplots of GNI variation in responsiveness to immune checkpoint blockade of GSE91061 cohort based on on-therapy RNA sequencing data. (K) Drug sensitivity analysis of GNI subgroups. Wilcoxon rank-sum test.
4. Discussion
Necroptosis, a recently identified form of programmed cell death, plays a dual role in cancer and is involved in both tumor suppression and progression [111]. In various cancer types, downregulation of necroptosis mediators (such as RIPK1, RIPK3, and MLKL) is linked to tumor grade and prognosis [111–119]. Intriguingly, two-thirds of the 60 cancer cell lines exhibited RIPK3 absence, which could be restored by treatment with the hypomethylating agent, decitabine [119]. Collectively, these investigations suggest the potential anti-cancer effects of necroptosis. However, there are also pro-cancer effects associated with necroptosis, including the participation of necroptosis-related inflammation in stimulating processes such as angiogenesis, cancer cell proliferation, metastasis, and T cell death [113,114]. In agreement with our study findings, necroptosis mediators were found to be overexpressed in glioblastoma (GBM), correlating with prognosis and hinting at a carcinogenic effect. The subgroup with high GNI gene expression displayed enrichment of processes and signaling pathways conducive to cancer progression, such as hypoxia, angiogenesis, epithelial-mesenchymal transition (EMT), and the IL6_JAK_STAT3 pathway.
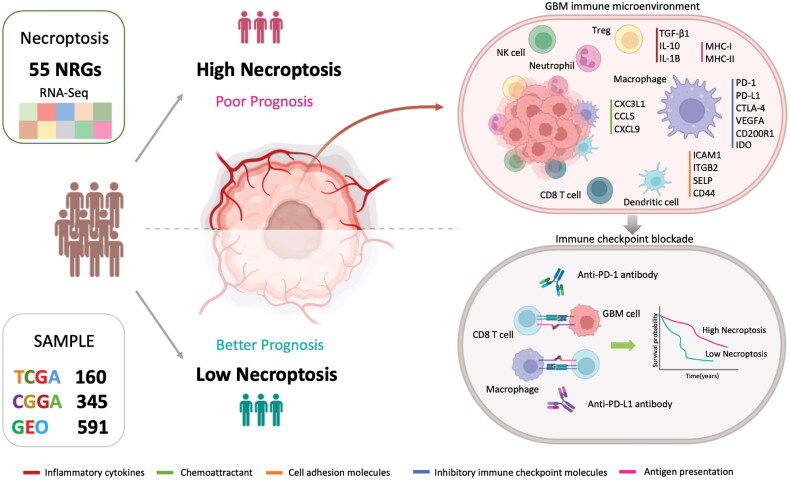
The TME framework displayed similarities reminiscent of the pattern observed in chronic inflammation, including the presence of infiltrating inflammatory cells, such as macrophages, neutrophils, and mast cells [120]. M0 macrophages were the predominant phenotype of infiltrating macrophages in the GNI-high GBM TME. The reduced presence of M1 macrophages, and to a lesser extent M2 macrophages, within the GNI-high subgroup may underscore their reported susceptibility to necroptosis compared to the M0 phenotype [121]. Notably, GNI genes, such as STC1, COL22A1, and LOXL1, also appear to impede the polarization of M0 macrophages to the M1 and M2 phenotypes. Moreover, inflammation triggered by necroptosis was associated with the activation of anti-cancer immune responses, fostering dendritic cells (DCs) via antigen presentation and stimulating T cells, which was also observed in our study [118,122,123]. The inflammation-driven immune response was associated with the subsequent development of an immunosuppressive TME, as indicated by the enrichment of inflammatory/immune pathways, abundance of immune cells, and upregulation of immune checkpoint molecules, as highlighted in Figure 13.
Figure 13.
Exploration of 55 necroptosis-related genes in diverse GBM populations revealed a high-risk subgroup with an immunosuppressive immunophenotype and poor prognosis, which was best described by a 10-gene signature termed the glioblastoma necroptosis index (GNI). The GNI signature predicts the immune response to immune checkpoint inhibition therapy in several cancers.
Individual characterization of GNI oncogenes could reflect how each member contributes to the GNI phenotype. Lysyl oxidase-like 1 (LOXL1) belongs to the LOX family of copper-dependent ε-amine lysine oxidases, which are involved in elastic fiber synthesis and homeostasis, and are implicated in fibrous disease and cancer [124]. In cancer, their role has been identified in the construction of the tumor microenvironment by covalently cross-linking collagen and elastin in the extracellular matrix (ECM), which was also reflected in our study by its positive correlation with the stromal score [125]. LOXL1 was initially identified as a tumor suppressor [126–128]; however, it has been increasingly associated with cancer progression, angiogenesis, and therapy resistance [129–131]. Similarly, the interaction between OSMR (expressed in fibroblasts) and its cognate ligand, OSM, which is usually expressed by macrophages, is reported to drive the inflammatory environment and tumor growth [132]. The outcomes of our study indicate that OSM-OSMR signaling may also operate in the GNI-high subgroup, as OSMR was significantly correlated with both stromal and immune scores. IGFBP6 has also been reported to play a role in fibrosis and chemoresistance in glioblastoma through paracrine IGF2/IGF-1R signaling [133,134]. Stanniocalcin-1 (STC1) has recently been identified as a tumor oncogene that is induced by oxidative stress, resulting in poor prognosis by downregulating ERK1/2 signaling [135]. Hypoxia-induced STC1 also promotes tumor growth and metastasis in ccRCC and breast cancer [136,137]. Moreover, fibroblast-derived STC1 restricts the differentiation of tumor-associated macrophages in lung adenocarcinoma [138]. STEAP2 and STEAP3 are ferrireductases and cupric reductases that stimulate the cellular uptake of both iron and copper; as such, these proteins may participate in ferroptosis and cuproptosis, thereby initiating these processes upon necroptosis induction [139]. Alternatively, neutrophil-induced ferroptosis has been shown to promote tumor necrosis in glioblastoma [140]. Hence, further exploration of individual oncogenes may unravel various necroptosis-initiating events and how necroptosis may affect the induction of other pathways contributing to cancer growth.
Non-neoplastic cells constitute a substantial proportion of the GBM microenvironment, including both tissue-resident and infiltrative cells [141]. Microglia, a tissue-resident cell type, and bone marrow-derived macrophages (BMDMs) make up to one-third of the tumor mass and greatly contribute to tumor progression and the immunosuppressive TME [142,143]. BMDMs are recruited specifically by tumor cells regardless of irradiation and BBB damage, and remain a distinct population despite acquiring microglial features [144–146]. Correspondingly, the GNI-high subgroup was enriched in M0 macrophages. Glioblastoma-associated macrophages (GAMs) and tumor cells play a critical role in the development of an immunosuppressive TME by secreting high levels of anti-inflammatory factors, such as IL-10, IL-4, IL-1B, MIF, TGF-β1, and PGE2 [145–148]. Several of these factors (TGF-β1, IL-10, and IL-1B) positively correlated with the fraction of various types of immune cells, indicating their role in the overall immunosuppressive TME. In particular, M0 macrophages, which showed a positive correlation with GNI, were positively associated with clinically relevant inhibitory checkpoint molecules, such as CTLA-4, PD-L1, and IDO. PD-L1 ligand expression in tumors and GAMs is reported to suppress T-cell function and proliferation and may influence Treg proliferation [105]. Interestingly, M0 macrophages and Tregs were positively correlated, which might explain this effect. IDO receptors expressed by M0 macrophages could also induce Tregs and suppress CD8+ T cells, which showed a strong association with IDO2 [100,101,149]. IDO upregulation is associated with the worst prognosis, and its genetic ablation in orthotopic and transgenic mouse glioma models has led to spontaneous rejection of brain tumors [150]. Hence, the IDO blockade may serve as a potential therapeutic target in the GNI-high subgroup. CD200R1 upregulation in the GNI-high subgroup represents another potential inhibitory immune checkpoint worthy of further investigation. Although CD200 was significantly higher in the GNI-low subgroup, CD200R1 has been shown to operate regardless of CD200 expression in suppressing the anti-tumor response [106]. Interestingly, CD200R1 was mainly correlated with neutrophils, activated CD4 memory T cells, and monocytes, and further investigation is needed to explore this connection.
Several predictive biomarkers of response to immune checkpoint blockade have been identified in several cancers, including tumor mutation and neoantigen burden, mismatch repair (MMR) deficiency, CD8+ T cell infiltration, and PD-Ll expression [151]. However, their predictive ability for GBM is unclear because of the limited number of clinical studies. The tumor mutation load (TML) in glioma is very low compared to that in high TML malignancies, and only a small fraction of GBM patients (3.5%) present with high TML. High TML in patients exhibits positively associated with MMR mutations and a lack of enriched influx of CD8+ T cells, PD-1+ T cells, or tumor-expressed PD-L1 [152]. Consistently, GNI showed a negative association with TMB and a positive correlation with CD8+ T cell density and PD-L1 expression. In contrast to other cancers, a higher TMB showed a trend toward better survival in GBM. All these indicators and the outcome of our study suggest that TMB may not be an ideal biomarker of ICB in gliomas. Newer machine learning models to predict ICB response have also been developed, such as TIDE and IPS, which are mainly based on the TME characteristics of melanoma in terms of immune cells (anti-cancer and immunosuppressive cells) and immune checkpoints [82,83]. These prediction models failed to predict the predictive ability of the GNI. Failure was anticipated, as these algorithms bank on TME characteristics of melanoma and non-small cell lung cancer, which vary greatly from GBM. Nonetheless, the submap algorithm was validated in our study to predict the response to ICB in GBM, which indicated a positive response to anti-CTLA-4 and anti-PD-1 blockade therapy. A promising response to immunotherapy was observed in the GNI-high subgroup. Moreover, the GNI-high subgroup was highly resistant to molecular targeted drugs, except Selumetinib, SCH772984 (a highly selective and ATP-competitive ERK inhibitor), and KU-55933 (ATM kinase inhibitor). KU-55933 has been demonstrated to increase TMZ responsiveness of TMZ-sensitive GBM cells and abrogate radioresistance in glioblastoma stem-like cells [153,154]. This also suggests that the GNI-high subgroup may have demonstrated greater resistance to therapy and explains the lack of association between GNI and the type of therapy, as most patients received both treatments. Selumetinib, a mitogen-activated protein kinase 1 and 2 (MEK1/2) inhibitor, has recently been approved for the treatment of tumors associated with neurofibromatosis, and its exploration in GBM is warranted [155]. A rationale for the use of ERK inhibition (SCH772984) in GBM treatment has also been developed [156].
Glioblastoma (GBM) is well-known for its immunosuppressive tumor microenvironment (TME) and resistance to immune checkpoint blockade (ICB) therapies [8–10]. Vaccine and virus-based immunotherapies offer promising alternatives by targeting tumor-associated antigens (TAAs), which enhances specificity for cancerous tissue while reducing damage to normal brain tissue [157,158]. These therapies are crucial for transforming the ‘cold’ TME into a ‘hot’ TME, as they can induce lytic cell death in tumor cells [159]. Recent research has shown that combining oncolytic viruses with bortezomib, a proteasome inhibitor, can trigger apoptosis and necroptosis, leading to greater therapeutic efficacy [160]. This suggests that necroptosis may be a key mechanism for converting a cold TME into a hot one, as observed in our study. We found that patients in the top 30% of the highest necroptosis index exhibited increased CD8 T cell infiltration and enhanced cytolytic activity.
There have few previously published studies on the role of necroptosis in the glioma patients using bioinformatic analysis. Wu et al. identified 614 necroptosis-related genes using GeneCards (https://www.genecards.org/) as opposed to our study which only included NRGs (n = 55) reported in published medical literature for direct involvement in necroptosis [161]. Wan et al.’s study developed a necroptosis-related risk signature comprising 18 genes including the main NRGs, similar to the study of Wu, et al. but our signature highlighted genes differentially expressed between necroptosis-stratified clusters, underscoring the functional implications of necroptosis in glioblastoma [162]. In contrast, Li et al. extended their analysis to various cell death pathways beyond necroptosis and did not provide empirical validation [163]. Thus, our study offers unique insights into necroptosis in GBM by detailing its functional implications and providing empirical evidence of its role in remodeling the GBM tumor microenvironment. Nonetheless, major portion of our results were also based on the transcriptomic analysis, which has its own limitations. It doesn’t not account for post-translational modifications or protein activity, which can be crucial in understanding the functional role of genes in cancer progression and treatment response. Both RIPK3 and MLKL, known mediators of necroptosis, also exhibit necroptosis-independent functions [164,165]. Specifically, RIPK3 has been linked to pathogen control and immune responses during chronic viral infections, potentially through dysregulated type 1 interferon signaling [164]. Similarly, MLKL, in association with RBM6, regulates endothelial cell adhesion molecule expression, thereby influencing EC-leukocyte interactions independent of its necroptosis-executing role [165]. Therefore, the potential impact of these necroptosis-independent functions on our study outcomes cannot be disregarded, and caution is needed when interpreting the results. Furthermore, our study lacks detailed mechanistic insights into how these GNI genes contribute to progression of glioblastoma and TME remodeling. Hence, further in-depth functional studies are necessary to clarify the exact mechanisms by which GNI genes influence GBM development and progression.
5. Conclusions
Necroptosis modulation, as evident by the variation in the expression of 55 necroptosis-related genes, in glioblastoma appears to shape its immune microenvironment and prognosis. The glioblastoma-necroptosis index (GNI) developed in this study demonstrated high predictive and prognostic efficiency in GBM and could be a useful biomarker. Leveraging these insights could potentially guide the development of targeted therapies aimed at manipulating the immune response and improving clinical outcomes for GBM patients. However, further investigations and translational studies are warranted to fully harness the potential of the GNI signature and its associated immunomodulation for effective GBM treatment strategies.
Supplementary Material
Funding Statement
National Nature Science Foundation of China (82102974, 82203966, 82373516, 82173452), Natural Science Foundation of Guangdong Province, China (No.2024A1515010830), Guangdong Basic and Applied Basic Research Foundation, China (2020A1515111074), China Postdoctoral Science Foundation (2020M672595), Plan on enhancing scientific research in GMU (GMUCR2024-01019), Project of Health Commission of Guangdong Province of China (Grant number: A2022355), Natural Science Foundation of Fujian Province, China (No.2020J011014), and Science and Technology Program of Guangzhou, China (202201011048).
Authors contributions
M. K., XT.H., and XX.Y. contributed equally to this work. M. K. wrote the manuscript. M. K., XT.H., and XX.Y. contributed significantly to the investigation, data analysis, and methodology. DH.Z., BY. W., AA. X., R. L., AB. Ren., CC. C., JJ. S., Z.R., YW. Y., and J. L. made significant contributions to the conception, formal analysis, funding, resources, supervision, validation, visualization, supervision, and final approval of the manuscript. All authors have read and approved the final and revised versions of the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are openly available in ‘The Cancer Genome Atlas’ [https://portal.gdc.cancer.gov/] and ‘Chinese Glioma Genome Atlas’ [http://www.cgga.org.cn/]. The raw data files supporting the findings from our own experiments in this study are available from the corresponding author, Jie Lin, upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the Internal Review and Ethics Boards of the Affiliated Cancer Hospital and Institute of Guangzhou Medical University [Approval number: GYZL-ZN-2023(030)]. The patients provided written informed consent to participate in the study. The study adhered to the ethical principles outlined in the Helsinki Declaration.
References
- 1.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohgaki H, Kleihues P.. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170(5):1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Koshy M, Villano JL, Dolecek TA, et al. Improved survival time trends for glioblastoma using the SEER 17 population-based registries. J Neurooncol. 2012;107(1):207–212. doi: 10.1007/s11060-011-0738-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malmström A, Grønberg BH, Marosi C, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. doi: 10.1016/S1470-2045(12)70265-6. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Shao X, Zhang Y, et al. Role of tumor microenvironment in cancer progression and therapeutic strategy. Cancer Med. 2023;12(10):11149–11165. doi: 10.1002/cam4.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quail DF, Joyce JA.. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucke-Wold B, Rangwala BS, Shafique MA, et al. Focus on current and emerging treatment options for glioma: a comprehensive review. World J Clin Oncol. 2024;15(4):482–495. doi: 10.5306/wjco.v15.i4.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomaszewski WH, Waibl-Polania J, Chakraborty M, et al. Neuronal CaMKK2 promotes immunosuppression and checkpoint blockade resistance in glioblastoma. Nat Commun. 2022;13(1):6483. doi: 10.1038/s41467-022-34175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulci G, Dmitrieva N, Gianni D, et al. Depletion of peripheral macrophages and brain microglia increases brain tumor titers of oncolytic viruses. Cancer Res. 2007;67(19):9398–9406. doi: 10.1158/0008-5472.CAN-07-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Xiong A, Liu J, et al. PANoptosis: bridging apoptosis, pyroptosis, and necroptosis in cancer progression and treatment. Cancer Gene Ther. 2024;31(7):970–983. doi: 10.1038/s41417-024-00765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Hua P, He C, et al. Non-apoptotic cell death-based cancer therapy: molecular mechanism, pharmacological modulators, and nanomedicine. Acta Pharm Sin B. 2022;12(9):3567–3593. doi: 10.1016/j.apsb.2022.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carafa V, Altucci L.. Deregulation of cell death in cancer: recent highlights. Cancers (Basel). 2020;12(12):3517. doi: 10.3390/cancers12123517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krakstad C, Chekenya M.. Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Mol Cancer. 2010;9(1):135. doi: 10.1186/1476-4598-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melo-Lima S, Lopes MC, Mollinedo F.. ERK1/2 acts as a switch between necrotic and apoptotic cell death in ether phospholipid edelfosine-treated glioblastoma cells. Pharmacol Res. 2015;95–96:2–11. doi: 10.1016/j.phrs.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Brat DJ, Castellano-Sanchez AA, Hunter SB, et al. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 2004;64(3):920–927. doi: 10.1158/0008-5472.can-03-2073. [DOI] [PubMed] [Google Scholar]
- 18.Raza SM, Lang FF, Aggarwal BB, et al. Necrosis and glioblastoma: a friend or a foe? A review and a hypothesis. Neurosurgery. 2002;51(1):2–13. discussion -3. doi: 10.1097/00006123-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanden Berghe T, Vanlangenakker N, Parthoens E, et al. Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features. Cell Death Differ. 2010;17(6):922–930. doi: 10.1038/cdd.2009.184. [DOI] [PubMed] [Google Scholar]
- 21.Dhuriya YK, Sharma D.. Necroptosis: a regulated inflammatory mode of cell death. J Neuroinflammation. 2018;15(1):199. doi: 10.1186/s12974-018-1235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi ME, Price DR, Ryter SW, et al. Necroptosis: a crucial pathogenic mediator of human disease. JCI Insight. 2019;4(15):e128834. doi: 10.1172/jci.insight.128834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Kos R, Garssen J, et al. Molecular insights into the mechanism of necroptosis: the necrosome as a potential therapeutic target. Cells. 2019;8(12):1486. doi: 10.3390/cells8121486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grootjans S, Vanden Berghe T, Vandenabeele P.. Initiation and execution mechanisms of necroptosis: an overview. Cell Death Differ. 2017;24(7):1184–1195. doi: 10.1038/cdd.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo J, Nam YW, Kim S, et al. Necroptosis molecular mechanisms: recent findings regarding novel necroptosis regulators. Exp Mol Med. 2021;53(6):1007–1017. doi: 10.1038/s12276-021-00634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galluzzi L, Kepp O, Chan FK, et al. Necroptosis: mechanisms and relevance to disease. Annu Rev Pathol. 2017;12(1):103–130. doi: 10.1146/annurev-pathol-052016-100247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandenabeele P, Declercq W, Van Herreweghe F, et al. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci Signal. 2010;3(115):re4. doi: 10.1126/scisignal.3115re4. [DOI] [PubMed] [Google Scholar]
- 28.Vercammen D, Brouckaert G, Denecker G, et al. Dual signaling of the Fas receptor: initiation of both apoptotic and necrotic cell death pathways. J Exp Med. 1998;188(5):919–930. doi: 10.1084/jem.188.5.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser WJ, Sridharan H, Huang C, et al. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J Biol Chem. 2013;288(43):31268–31279. doi: 10.1074/jbc.M113.462341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He S, Liang Y, Shao F, et al. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc Natl Acad Sci USA. 2011;108(50):20054–20059. doi: 10.1073/pnas.1116302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez DA, Weinlich R, Brown S, et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 2016;23(1):76–88. doi: 10.1038/cdd.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson N, McComb S, Mulligan R, et al. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol. 2012;13(10):954–962. doi: 10.1038/ni.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thapa RJ, Nogusa S, Chen P, et al. Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc Natl Acad Sci USA. 2013;110(33):E3109–E3118. doi: 10.1073/pnas.1301218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McComb S, Cessford E, Alturki NA, et al. Type-I interferon signaling through ISGF3 complex is required for sustained Rip3 activation and necroptosis in macrophages. Proc Natl Acad Sci U S A. 2014;111(31):E3206–E3213. doi: 10.1073/pnas.1407068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malireddi RKS, Kesavardhana S, Kanneganti T-D.. ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis). Front Cell Infect Microbiol. 2019;9:406. doi: 10.3389/fcimb.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy JM, Czabotar PE, Hildebrand JM, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39(3):443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 37.Najafov A, Mookhtiar AK, Luu HS, et al. TAM kinases promote necroptosis by regulating oligomerization of MLKL. Mol Cell. 2019;75(3):457–468.e4. doi: 10.1016/j.molcel.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Zhu K, Liang W, Ma Z, et al. Necroptosis promotes cell-autonomous activation of proinflammatory cytokine gene expression. Cell Death Dis. 2018;9(5):500. doi: 10.1038/s41419-018-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McComb S, Cheung HH, Korneluk RG, et al. cIAP1 and cIAP2 limit macrophage necroptosis by inhibiting Rip1 and Rip3 activation. Cell Death Differ. 2012;19(11):1791–1801. doi: 10.1038/cdd.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaCasse EC, Mahoney DJ, Cheung HH, et al. IAP-targeted therapies for cancer. Oncogene. 2008;27(48):6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 41.Dovey CM, Diep J, Clarke BP, et al. MLKL requires the inositol phosphate code to execute necroptosis. Mol Cell. 2018;70(5):936–948.e7. doi: 10.1016/j.molcel.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNamara DE, Dovey CM, Hale AT, et al. Direct activation of human MLKL by a select repertoire of inositol phosphate metabolites. Cell Chem Biol. 2019;26(6):863–877.e7. doi: 10.1016/j.chembiol.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roedig J, Kowald L, Juretschke T, et al. USP22 controls necroptosis by regulating receptor-interacting protein kinase 3 ubiquitination. EMBO Rep. 2021;22(2):e50163. doi: 10.15252/embr.202050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciotti S, Iuliano L, Cefalù S, et al. GSK3β is a key regulator of the ROS-dependent necrotic death induced by the quinone DMNQ. Cell Death Dis. 2020;11(1):2. doi: 10.1038/s41419-019-2202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Gong W, Wang H, et al. O-GlcNAc transferase suppresses inflammation and necroptosis by targeting receptor-interacting serine/threonine-protein kinase 3. Immunity. 2019;50(3):576–590.e6. doi: 10.1016/j.immuni.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi C-S, Kehrl JH.. Bcl-2 regulates pyroptosis and necroptosis by targeting BH3-like domains in GSDMD and MLKL. Cell Death Discov. 2019;5(1):151.Shi C-S, Kehrl JH.. Bcl-2 regulates pyroptosis and necroptosis by targeting BH3-like domains in GSDMD and MLKL. Cell Death Discov. 2019;5(1): 151. doi: 10.1038/s41420-019-0230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong Y, Zhang ZH, Wang JY, et al. Zinc finger protein 91 mediates necroptosis by initiating RIPK1-RIPK3-MLKL signal transduction in response to TNF receptor 1 ligation. Toxicol Lett. 2021;356:75–88. doi: 10.1016/j.toxlet.2021.12.015. [DOI] [PubMed] [Google Scholar]
- 48.Li D, Xu T, Cao Y, et al. A cytosolic heat shock protein 90 and cochaperone CDC37 complex is required for RIP3 activation during necroptosis. Proc Natl Acad Sci U S A. 2015;112(16):5017–5022. doi: 10.1073/pnas.1505244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston AN, Ma Y, Liu H, et al. Necroptosis-blocking compound NBC1 targets heat shock protein 70 to inhibit MLKL polymerization and necroptosis. Proc Natl Acad Sci U S A. 2020;117(12):6521–6530. doi: 10.1073/pnas.1916503117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanna-Addams S, Liu S, Liu H, et al. CK1α, CK1δ, and CK1ε are necrosome components which phosphorylate serine 227 of human RIPK3 to activate necroptosis. Proc Natl Acad Sci U S A. 2020;117(4):1962–1970. doi: 10.1073/pnas.1917112117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakabayashi O, Takahashi H, Moriwaki K, et al. MIND bomb 2 prevents RIPK1 kinase activity-dependent and -independent apoptosis through ubiquitylation of cFLIPL. Commun Biol. 2021;4(1):80. doi: 10.1038/s42003-020-01603-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Chang X, Feng J, et al. TRADD mediates RIPK1-independent necroptosis induced by tumor necrosis factor. Front Cell Dev Biol. 2019;7:393. doi: 10.3389/fcell.2019.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaiser WJ, Upton JW, Long AB, et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471(7338):368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oberst A, Dillon CP, Weinlich R, et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471(7338):363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dondelinger Y, Jouan-Lanhouet S, Divert T, et al. NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol Cell. 2015;60(1):63–76. doi: 10.1016/j.molcel.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 56.Lafont E, Draber P, Rieser E, et al. TBK1 and IKKε prevent TNF-induced cell death by RIPK1 phosphorylation. Nat Cell Biol. 2018;20(12):1389–1399. doi: 10.1038/s41556-018-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seo J, Lee E-W, Sung H, et al. CHIP controls necroptosis through ubiquitylation- and lysosome-dependent degradation of RIPK3. Nat Cell Biol. 2016;18(3):291–302. doi: 10.1038/ncb3314. [DOI] [PubMed] [Google Scholar]
- 58.Onizawa M, Oshima S, Schulze-Topphoff U, et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol. 2015;16(6):618–627. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen W, Wu J, Li L, et al. Ppm1b negatively regulates necroptosis through dephosphorylating Rip3. Nat Cell Biol. 2015;17(4):434–444. doi: 10.1038/ncb3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie Y, Zhu S, Zhong M, et al. Inhibition of aurora kinase A induces necroptosis in pancreatic carcinoma. Gastroenterology. 2017;153(5):1429–1443. doi: 10.1053/j.gastro.2017.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seo J, Seong D, Nam YW, et al. Beclin 1 functions as a negative modulator of MLKL oligomerisation by integrating into the necrosome complex. Cell Death Differ. 2020;27(11):3065–3081. doi: 10.1038/s41418-020-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hitomi J, Christofferson DE, Ng A, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135(7):1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petersen SL, Chen TT, Lawrence DA, et al. TRAF2 is a biologically important necroptosis suppressor. Cell Death Differ. 2015;22(11):1846–1857. doi: 10.1038/cdd.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lou X, Zhu H, Ning L, et al. EZH2 regulates intestinal inflammation and necroptosis through the JNK signaling pathway in intestinal epithelial cells. Dig Dis Sci. 2019;64(12):3518–3527. doi: 10.1007/s10620-019-05705-4. [DOI] [PubMed] [Google Scholar]
- 65.Zhu J, Yang L-K, Wang Q-H, et al. NDRG2 attenuates ischemia-induced astrocyte necroptosis via the repression of RIPK1. Mol Med Rep. 2020;22(4):3103–3110. doi: 10.3892/mmr.2020.11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karki R, Sundaram B, Sharma BR, et al. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep. 2021;37(3):109858. doi: 10.1016/j.celrep.2021.109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu D, Jin T, Zhu H, et al. TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell. 2018;174(6):1477–1491.e19. doi: 10.1016/j.cell.2018.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dondelinger Y, Delanghe T, Rojas-Rivera D, et al. MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat Cell Biol. 2017;19(10):1237–1247. doi: 10.1038/ncb3608. [DOI] [PubMed] [Google Scholar]
- 69.Cao K, Tait SWG.. Parkin inhibits necroptosis to prevent cancer. Nat Cell Biol. 2019;21(8):915–916. doi: 10.1038/s41556-019-0350-1. [DOI] [PubMed] [Google Scholar]
- 70.Wang H, Meng H, Li X, et al. PELI1 functions as a dual modulator of necroptosis and apoptosis by regulating ubiquitination of RIPK1 and mRNA levels of c-FLIP. Proc Natl Acad Sci U S A. 2017;114(45):11944–11949. doi: 10.1073/pnas.1715742114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilkerson MD, Hayes DN.. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishwaran H, Gerds TA, Kogalur UB, et al. Random survival forests for competing risks. Biostatistics. 2014;15(4):757–773. doi: 10.1093/biostatistics/kxu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrell FE, JrLee KL, Mark DB.. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statist Med. 1996;15(4):361–387. doi: . [DOI] [PubMed] [Google Scholar]
- 74.Pencina MJ, D’Agostino RB.. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23(13):2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 75.Vickers AJ, Elkin EB.. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hänzelmann S, Castelo R, Guinney J.. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4(1):2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang M, Wang X, Chen X, et al. Novel immune-related gene signature for risk stratification and prognosis of survival in lower-grade glioma. Front Genet. 2020;11:363. doi: 10.3389/fgene.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu L, Deng C, Pang B, et al. TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res. 2018;78(23):6575–6580. doi: 10.1158/0008-5472.CAN-18-0689. [DOI] [PubMed] [Google Scholar]
- 82.Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 84.Hoshida Y, Brunet JP, Tamayo P, et al. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maeser D, Gruener RF, Huang RS.. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22(6):bbab260. doi: 10.1093/bib/bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun D, Wang J, Han Y, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49(D1):D1420–D1430. d30. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang C, Sun D, Huang X, et al. Integrative analyses of single- cell transcriptome and regulome using MAESTRO. Genome Biol. 2020;21(1):198. doi: 10.1186/s13059-020-02116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson DE, O’Keefe RA, Grandis JR.. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Papale M, Buccarelli M, Mollinari C, et al. Hypoxia, inflammation and necrosis as determinants of glioblastoma cancer stem cells progression. Int J Mol Sci. 2020;21(8):2660. doi: 10.3390/ijms21082660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones DM, Read KA, Oestreich KJ.. Dynamic roles for IL-2–STAT5 signaling in effector and regulatory CD4 + T cell populations. J Immunol. 2020;205(7):1721–1730. doi: 10.4049/jimmunol.2000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cai Z, Zhang A, Choksi S, et al. Activation of cell- surface proteases promotes necroptosis, inflammation and cell migration. Cell Res. 2016;26(8):886–900. doi: 10.1038/cr.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hannes S, Abhari BA, Fulda S.. Smac mimetic triggers necroptosis in pancreatic carcinoma cells when caspase activation is blocked. Cancer Lett. 2016;380(1):31–38. doi: 10.1016/j.canlet.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 93.Miranda A, Hamilton PT, Zhang AW, et al. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci U S A. 2019;116(18):9020–9029. doi: 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Locarno CV, Simonelli M, Carenza C, et al. Role of myeloid cells in the immunosuppressive microenvironment in gliomas. Immunobiology. 2020;225(1):151853. doi: 10.1016/j.imbio.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 95.Cecere TE, Todd SM, Leroith T.. Regulatory T cells in arterivirus and coronavirus infections: do they protect against disease or enhance it? Viruses. 2012;4(5):833–846. doi: 10.3390/v4050833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goh CC, Roggerson KM, Lee HC, et al. Hepatitis C virus-induced myeloid-derived suppressor cells suppress NK cell IFN-γ production by altering cellular metabolism via arginase-1. J Immunol. 2016;196(5):2283–2292. doi: 10.4049/jimmunol.1501881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van de Ven K, Borst J.. Targeting the T-cell co-stimulatory CD27/CD70 pathway in cancer immunotherapy: rationale and potential. Immunotherapy. 2015;7(6):655–667. doi: 10.2217/imt.15.32. [DOI] [PubMed] [Google Scholar]
- 98.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3(9):745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 99.Watts TH. TNF/TNFR family members in costimulation of T cell responses. Annu Rev Immunol. 2005;23(1):23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 100.Brown NF, Carter TJ, Ottaviani D, et al. Harnessing the immune system in glioblastoma. Br J Cancer. 2018;119(10):1171–1181. doi: 10.1038/s41416-018-0258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Woroniecka KI, Rhodin KE, Chongsathidkiet P, et al. T-cell dysfunction in glioblastoma: applying a new framework. Clin Cancer Res. 2018;24(16):3792–3802. doi: 10.1158/1078-0432.CCR-18-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu L, Xiao H, Xu M, et al. Glioma-derived T cell immunoglobulin- and mucin domain-containing molecule-4 (TIM4) contributes to tumor tolerance. J Biol Chem. 2011;286(42):36694–36699. doi: 10.1074/jbc.M111.292540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Han S, Zhang C, Li Q, et al. Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br J Cancer. 2014;110(10):2560–2568. doi: 10.1038/bjc.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nishikawa H, Sakaguchi S.. Regulatory T cells in tumor immunity. Int J Cancer. 2010;127(4):759–767. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- 105.DiDomenico J, Lamano JB, Oyon D, et al. The immune checkpoint protein PD-L1 induces and maintains regulatory T cells in glioblastoma. Oncoimmunology. 2018;7(7):e1448329. doi: 10.1080/2162402X.2018.1448329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rygiel TP, Karnam G, Goverse G, et al. CD200-CD200R signaling suppresses anti-tumor responses independently of CD200 expression on the tumor. Oncogene. 2012;31(24):2979–2988. doi: 10.1038/onc.2011.477. [DOI] [PubMed] [Google Scholar]
- 107.Gajewski TF, Schreiber H, Fu YX.. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ottaviano M, De Placido S, Ascierto PA.. Recent success and limitations of immune checkpoint inhibitors for cancer: a lesson from melanoma. Virchows Arch. 2019;474(4):421–432. doi: 10.1007/s00428-019-02538-4. [DOI] [PubMed] [Google Scholar]
- 109.Sha D, Jin Z, Budczies J, et al. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 2020;10(12):1808–1825. doi: 10.1158/2159-8290.CD-20-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roh W, Chen P-L, Reuben A, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9(379):eaah3560. doi: 10.1126/scitranslmed.aah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Najafov A, Chen H, Yuan J.. Necroptosis and cancer. Trends Cancer. 2017;3(4):294–301. doi: 10.1016/j.trecan.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Philipp S, Sosna J, Adam D.. Cancer and necroptosis: friend or foe? Cell Mol Life Sci. 2016;73(11-12):2183–2193. doi: 10.1007/s00018-016-2193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gong Y, Fan Z, Luo G, et al. The role of necroptosis in cancer biology and therapy. Mol Cancer. 2019;18(1):100. doi: 10.1186/s12943-019-1029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moriwaki K, Bertin J, Gough PJ, et al. Differential roles of RIPK1 and RIPK3 in TNF-induced necroptosis and chemotherapeutic agent-induced cell death. Cell Death Dis. 2015;6(2):e1636–e1636. doi: 10.1038/cddis.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Feng X, Song Q, Yu A, et al. Receptor-interacting protein kinase 3 is a predictor of survival and plays a tumor suppressive role in colorectal cancer. Neoplasma. 2015;62(4):592–601. doi: 10.4149/neo_2015_071. [DOI] [PubMed] [Google Scholar]
- 116.Ando Y, Ohuchida K, Otsubo Y, et al. Necroptosis in pancreatic cancer promotes cancer cell migration and invasion by release of CXCL5. PLoS One. 2020;15(1):e0228015. doi: 10.1371/journal.pone.0228015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Geserick P, Wang J, Schilling R, et al. Absence of RIPK3 predicts necroptosis resistance in malignant melanoma. Cell Death Dis. 2015;6(9):e1884–e1884. doi: 10.1038/cddis.2015.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schmidt SV, Seibert S, Walch-Rückheim B, et al. RIPK3 expression in cervical cancer cells is required for PolyIC-induced necroptosis, IL-1α release, and efficient paracrine dendritic cell activation. Oncotarget. 2015;6(11):8635–8647. doi: 10.18632/oncotarget.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koo G-B, Morgan MJ, Lee D-G, et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015;25(6):707–725. doi: 10.1038/cr.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao H, Wu L, Yan G, et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct Target Ther. 2021;6(1):263. doi: 10.1038/s41392-021-00658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hao Q, Idell S, Tang H.. M1 macrophages are more susceptible to necroptosis. J Cell Immunol. 2021;3(2):97–102. doi: 10.33696/immunology.3.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yatim N, Jusforgues-Saklani H, Orozco S, et al. RIPK1 and NF-κB signaling in dying cells determines cross-priming of CD8+ T cells. Science. 2015;350(6258):328–334. doi: 10.1126/science.aad0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Snyder AG, Hubbard NW, Messmer MN, et al. Intratumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci Immunol. 2019;4(36):eaaw2004. doi: 10.1126/sciimmunol.aaw2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yuan R, Li Y, Yang B, et al. LOXL1 exerts oncogenesis and stimulates angiogenesis through the LOXL1-FBLN5/αvβ3 integrin/FAK-MAPK axis in ICC. Mol Ther Nucleic Acids. 2021;23:797–810. doi: 10.1016/j.omtn.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barker HE, Cox TR, Erler JT.. The rationale for targeting the LOX family in cancer. Nat Rev Cancer. 2012;12(8):540–552. doi: 10.1038/nrc3319. [DOI] [PubMed] [Google Scholar]
- 126.Contente S, Kenyon K, Sriraman P, et al. Epigenetic inhibition of lysyl oxidase transcription after transformation by ras oncogene. Mol Cell Biochem. 1999;194(1–2):79–91. doi: 10.1023/a:1006913122261. [DOI] [PubMed] [Google Scholar]
- 127.Csiszar K, Fong SFT, Ujfalusi A, et al. Somatic mutations of the lysyl oxidase gene on chromosome 5q23.1 in colorectal tumors. Int J Cancer. 2002;97(5):636–642. doi: 10.1002/ijc.10035. [DOI] [PubMed] [Google Scholar]
- 128.Wu G, Guo Z, Chang X, et al. LOXL1 and LOXL4 are epigenetically silenced and can inhibit ras/extracellular signal-regulated kinase signaling pathway in human bladder cancer. Cancer Res. 2007;67(9):4123–4129. doi: 10.1158/0008-5472.CAN-07-0012. [DOI] [PubMed] [Google Scholar]
- 129.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 130.Le Calvé B, Griveau A, Vindrieux D, et al. Lysyl oxidase family activity promotes resistance of pancreatic ductal adenocarcinoma to chemotherapy by limiting the intratumoral anticancer drug distribution. Oncotarget. 2016;7(22):32100–32112. doi: 10.18632/oncotarget.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zeltz C, Pasko E, Cox TR, et al. LOXL1 is regulated by integrin α11 and promotes non-small cell lung cancer tumorigenicity. Cancers (Basel). 2019;11(5):705. doi: 10.3390/cancers11050705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee BY, Hogg EKJ, Below CR, et al. Heterocellular OSM-OSMR signalling reprograms fibroblasts to promote pancreatic cancer growth and metastasis. Nat Commun. 2021;12(1):7336. doi: 10.1038/s41467-021-27607-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Oliva CR, Halloran B, Hjelmeland AB, et al. IGFBP6 controls the expansion of chemoresistant glioblastoma through paracrine IGF2/IGF-1R signaling. Cell Commun Signal. 2018;16(1):61. doi: 10.1186/s12964-018-0273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liso A, Venuto S, Coda AR, et al. IGFBP-6: at the crossroads of immunity, tissue repair and fibrosis. Int J Mol Sci. 2022;23(8):4358. doi: 10.3390/ijms23084358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nguyen A, Chang ACM, Reddel RR.. Stanniocalcin-1 acts in a negative feedback loop in the prosurvival ERK1/2 signaling pathway during oxidative stress. Oncogene. 2009;28(18):1982–1992. doi: 10.1038/onc.2009.65. [DOI] [PubMed] [Google Scholar]
- 136.Ma X, Gu L, Li H, et al. Hypoxia-induced overexpression of stanniocalcin-1 is associated with the metastasis of early stage clear cell renal cell carcinoma. J Transl Med. 2015;13(1):56. doi: 10.1186/s12967-015-0421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chang AC-M, Doherty J, Huschtscha LI, et al. STC1 expression is associated with tumor growth and metastasis in breast cancer. Clin Exp Metastasis. 2015;32(1):15–27. doi: 10.1007/s10585-014-9687-9. [DOI] [PubMed] [Google Scholar]
- 138.Kamata T, So TY, Ahmed Q, et al. Fibroblast-derived STC-1 modulates tumor-associated macrophages and lung adenocarcinoma development. Cell Rep. 2020;31(12):107802. doi: 10.1016/j.celrep.2020.107802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ohgami RS, Campagna DR, McDonald A, et al. The Steap proteins are metalloreductases. Blood. 2006;108(4):1388–1394. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yee PP, Wei Y, Kim S-Y, et al. Neutrophil-induced ferroptosis promotes tumor necrosis in glioblastoma progression. Nat Commun. 2020;11(1):5424. doi: 10.1038/s41467-020-19193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Haddad-Tóvolli R, Dragano NRV, Ramalho AFS, et al. Development and function of the blood-brain barrier in the context of metabolic control. Front Neurosci. 2017;11:224. doi: 10.3389/fnins.2017.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Graeber MB, Scheithauer BW, Kreutzberg GW.. Microglia in brain tumors. Glia. 2002;40(2):252–259. doi: 10.1002/glia.10147. [DOI] [PubMed] [Google Scholar]
- 143.Hambardzumyan D, Gutmann DH, Kettenmann H.. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bowman RL, Klemm F, Akkari L, et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 2016;17(9):2445–2459. doi: 10.1016/j.celrep.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen Z, Feng X, Herting CJ, et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 2017;77(9):2266–2278. doi: 10.1158/0008-5472.CAN-16-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cronk JC, Filiano AJ, Louveau A, et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J Exp Med. 2018;215(6):1627–1647. doi: 10.1084/jem.20180247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Roesch S, Rapp C, Dettling S, et al. When immune cells turn bad-tumor-associated microglia/macrophages in glioma. Int J Mol Sci. 2018;19(2):436. doi: 10.3390/ijms19020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wei J, Gabrusiewicz K, Heimberger A.. The controversial role of microglia in malignant gliomas. Clin Dev Immunol. 2013;2013:285246–285212. doi: 10.1155/2013/285246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sordillo PP, Sordillo LA, Helson L.. The kynurenine pathway: a primary resistance mechanism in patients with glioblastoma. Anticancer Res. 2017;37(5):2159–2171. doi: 10.21873/anticanres.11551. [DOI] [PubMed] [Google Scholar]
- 150.Hanihara M, Kawataki T, Oh-Oka K, et al. Synergistic antitumor effect with indoleamine 2,3-dioxygenase inhibition and temozolomide in a murine glioma model. J Neurosurg. 2016;124(6):1594–1601. doi: 10.3171/2015.5.JNS141901. [DOI] [PubMed] [Google Scholar]
- 151.Gibney GT, Weiner LM, Atkins MB.. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17(12):e542–e51. doi: 10.1016/S1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hodges TR, Ott M, Xiu J, et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol. 2017;19(8):1047–1057. doi: 10.1093/neuonc/nox026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nadkarni A, Shrivastav M, Mladek AC, et al. ATM inhibitor KU-55933 increases the TMZ responsiveness of only inherently TMZ sensitive GBM cells. J Neurooncol. 2012;110(3):349–357. doi: 10.1007/s11060-012-0979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Carruthers R, Ahmed SU, Strathdee K, et al. Abrogation of radioresistance in glioblastoma stem-like cells by inhibition of ATM kinase. Mol Oncol. 2015;9(1):192–203. doi: 10.1016/j.molonc.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Markham A, Keam SJ.. Selumetinib: first Approval. Drugs. 2020;80(9):931–937. doi: 10.1007/s40265-020-01331-x. [DOI] [PubMed] [Google Scholar]
- 156.Hannen R, Hauswald M, Bartsch JW.. A rationale for targeting extracellular regulated kinases ERK1 and ERK2 in glioblastoma. J Neuropathol Exp Neurol. 2017;76(10):838–847. doi: 10.1093/jnen/nlx076. [DOI] [PubMed] [Google Scholar]
- 157.Reddy R, Yan SC, Hasanpour Segherlou Z, et al. Oncolytic viral therapy: a review and promising future directions. J Neurosurg. 2024;140(2):319–327. doi: 10.3171/2023.6.JNS23243. [DOI] [PubMed] [Google Scholar]
- 158.Zhang Q, Liu F.. Advances and potential pitfalls of oncolytic viruses expressing immunomodulatory transgene therapy for malignant gliomas. Cell Death Dis. 2020;11(6):485. doi: 10.1038/s41419-020-2696-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lovatt C, Parker AL.. Oncolytic viruses and immune checkpoint inhibitors: the “hot” new power couple. Cancers (Basel). 2023;15(16):4178. doi: 10.3390/cancers15164178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee D, de los Reyes VAA, Kim Y.. Optimal strategies of oncolytic virus-bortezomib therapy via the apoptotic, necroptotic, and oncolysis signaling network. Math Biosci Eng. 2024;21(3):3876–3909. doi: 10.3934/mbe.2024173. [DOI] [PubMed] [Google Scholar]
- 161.Wu Y, Huang Y, Zhou C, et al. A novel necroptosis-related prognostic signature of glioblastoma based on transcriptomics analysis and single cell sequencing analysis. Brain Sci. 2022;12(8):988. doi: 10.3390/brainsci12080988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wan S, Moure UAE, Liu R, et al. Combined bulk RNA-seq and single-cell RNA-seq identifies a necroptosis-related prognostic signature associated with inhibitory immune microenvironment in glioma. Front Immunol. 2022;13:1013094. doi: 10.3389/fimmu.2022.1013094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li J, Song Z, Chen Z, et al. Association between diverse cell death patterns related gene signature and prognosis, drug sensitivity, and immune microenvironment in glioblastoma. J Mol Neurosci. 2024;74(1):10. doi: 10.1007/s12031-023-02181-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Preston SP, Allison CC, Schaefer J, et al. A necroptosis-independent function of RIPK3 promotes immune dysfunction and prevents control of chronic LCMV infection. Cell Death Dis. 2023;14(2):123. doi: 10.1038/s41419-023-05635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dai J, Zhang C, Guo L, et al. A necroptotic-independent function of MLKL in regulating endothelial cell adhesion molecule expression. Cell Death Dis. 2020;11(4):282. doi: 10.1038/s41419-020-2483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are openly available in ‘The Cancer Genome Atlas’ [https://portal.gdc.cancer.gov/] and ‘Chinese Glioma Genome Atlas’ [http://www.cgga.org.cn/]. The raw data files supporting the findings from our own experiments in this study are available from the corresponding author, Jie Lin, upon reasonable request.