Abstract
Using the Olink Explore 1536 platform, we measured 1,463 unique proteins in 303 cerebrospinal fluid (CSF) specimens from four clinical centers contributed by uninfected controls and 12 groups of people living with HIV-1 infection representing the spectrum of progressive untreated and treated chronic infection. We present three initial analyses of these measurements: an overview of the CSF protein features of the sample; correlations of the CSF proteins with CSF HIV-1 RNA and neurofilament light chain protein (NfL) concentrations; and comparison of CSF proteins in HIV-associated dementia (HAD) and neurosymptomatic CSF escape (NSE). These reveal a complex but coherent picture of CSF protein changes with highest concentrations of many proteins during CNS injury in the HAD and NSE groups and variable protein changes across the course of systemic HIV-1 progression that included two common patterns, designated as lymphoid and myeloid patterns, related to principal involvement of their underlying inflammatory cell lineages. Antiretroviral therapy reduced CSF protein perturbations, though not always to control levels. The dataset of these CSF protein measurements, along with background clinical information, is posted online. Extended studies of this unique dataset will supplement this report to provide more detailed characterization of the dynamic impact of HIV-1 infection on the CSF proteome across the spectrum of HIV-1 infection, advancing the mechanistic understanding of HIV-1-related CNS pathobiology.
Author summary
We measured more than 1,400 proteins in 303 cerebrospinal fluid (CSF) specimens from a representative broad range of untreated and treated people living with HIV-1 infection (PLWH) along with uninfected controls who volunteered for our studies. The results defined a complex, but generally coherent set of changes in many immune-inflammatory proteins and several central nervous system injury biomarkers as systemic HIV-1 infection progressed. There was a marked increase in many of these protein biomarkers in individuals with HIV-associated dementia and neurosymptomatic CSF viral escape, two conditions distinguished by overt, and characteristically severe, neurological impairment. The resultant data and initial analyses advance the characterization and understanding of the evolution of HIV-driven neuropathogenesis. The large dataset from this study is posted online and available to other investigators for further analyses, extending the utility of these data and potentially aiding in developing future prevention, diagnosis and mitigation of the deleterious impact of HIV-1 infection on the brain.
Introduction
Exposure of the central nervous system (CNS) to HIV-1 is a nearly universal facet of untreated systemic infection that is readily documented by analysis of cerebrospinal fluid (CSF) obtained by lumbar puncture (LP) [1–11]. During the earlier phases of chronic infection, CSF viral isolates are genetically similar to plasma viruses and adapted to infect CD4+ T-lymphocytes (R5 T-cell-tropic viruses) [12]. As a consequence of the migration of infected CD4+ T-cells from blood into the CNS, where they release virus [12–15], CSF HIV-1 RNA levels are typically maintained at levels about one-tenth of those in blood [5,11,16–18]. Anatomically, HIV-1 RNA detected in CSF during this period predominantly represents a sample of virus from the leptomeninges, the site of a clinically silent aseptic meningitis with mild CSF pleocytosis [19–21]. An exception to this 1:10 ratio of CSF to plasma HIV-1 RNA concentrations develops in untreated individuals with very low CD4+ T-cell levels (< 50 cells per μL) in whom the ratio is closer to 1:100 when CSF white blood cell (WBC) counts are low, consistent with the concept that in these groups CSF virus is largely associated with CD4+ T-cell trafficking that includes infected cells and that when the number of these cells is diminished in late systemic infection, the CSF HIV-1 levels also fall [3,4,6,11,17,22–24].
The CSF:plasma HIV-1 ratio is altered in the opposite direction in patients presenting with subacute HIV-1-associated dementia (HAD) in whom high CSF HIV-1 RNA levels are nearly equal to those of plasma as a consequence of local viral replication within the CNS parenchyma in association with multinucleated-cell HIV-1 encephalitis (HIVE) [25–29]. This is generally caused by HIV-1 variants capable of infecting macrophages and related myeloid cells that express low cell-surface densities of CD4 receptors (M-tropic viruses) [30–39].
In addition to the hallmark immunosuppression that, in its severe form, underlies susceptibility to HAD and the major opportunistic complications defining the acquired immunodeficiency syndrome (AIDS), chronic HIV-1 infection is also accompanied by systemic immune activation that predisposes to an additional set of chronic co-morbidities [40–44]. CSF sampling in cohort studies shows that intrathecal immune activation and inflammation are also constant features of chronic CNS HIV-1 infection [45]. Moreover, the character of this inflammation changes over the course of untreated infection as blood CD4+ T-cells decline [45,46]. In a recent study measuring a limited panel of inflammatory biomarkers, we identified two common patterns of CSF biomarker changes as blood CD4+ T-cell declined during untreated infection in the absence of HAD. The first pattern was characterized by a rise and then a fall in lymphocyte-related inflammation as blood CD4+ T-cells decreased from >500 to below 50 cells per μL, exemplified by CSF concentration changes in CXCL10, TNF-alpha and MMP-9 [45]. We have referred to this ‘quadratic’ pattern (based on trends modeling using a quadratic equation) of CSF viral load change with CD4+ T-cell loss as the lymphoid pattern of biomarker change. The second ‘linear’ pattern (modelled with a linear trends equation), that we refer to as the myeloid pattern of CSF inflammation, was exemplified by CSF concentration changes in the macrophage inflammatory markers, CCL2 and CD163, that rose more gradually and steadily through the course of blood CD4+ T-cell decline to reach their highest levels when blood CD4+ T lymphocytes fell to <50 cells per μL in those without HAD [45]. We introduce these two terms because they serve as prototypes for similar patterns of CSF protein biomarker change noted in this analysis, as discussed later. Importantly, as previously reported [45], CSF inflammatory protein reactions are generally compartmentalized, relate to local inflammatory processes within the CNS (including the leptomeninges, perivascular tissues and CNS parenchyma), and most often evolve independently from the concentrations of the same protein biomarkers in the blood, though this is a complex issue because the cellular sources and their functional context is strongly influenced (or determined) by systemic immune status. As a result, CNS inflammatory responses may change in parallel with or diverge from their systemic counterparts but are likely largely produced within the CNS in response to local conditions. CSF in HAD patients (with the underlying multinucleated-cell form of HIVE) characteristically exhibits a broad inflammatory profile with marked elevation of the CSF biomarkers that participate in both the lymphoid and myeloid CSF patterns, augmented by additional inflammatory biomarkers that may show only limited change in the non-HAD untreated individuals across the broad range of blood CD4+ decline [45].
While CNS immune and inflammatory reactions to local infection may both aid in local viral control and facilitate viral entry, they likely also contribute to neuronal dysfunction and injury, either directly by effects on neurons or through indirect effects on uninfected myeloid cells and astrocytes by largely uncertain immunopathological pathways [47–49]. Though a number of putative mechanisms of CNS injury have been identified, the exact contributions of individual virus-encoded and inflammatory molecules to in vivo injury remain largely inferential [50–62].
In addition to revealing inflammatory changes, CSF analysis provides evidence of neuronal injury during HIV-1 infection that can now be usefully monitored by measuring neurofilament light chain protein (NfL) concentrations in either CSF or blood [63–65]. The highest CSF and plasma NfL levels in untreated people living with HIV infection (PLWH) are detected in individuals with HAD. Elevated levels are also observed in a subset of neuroasymptomatic individuals, particularly some with low CD4+ T-cell counts, indicating subclinical axonal injury. These elevated CSF and blood NfL levels can also predict the subsequent development of HAD, with examples of increased CSF levels as long as one to two years prior to overt dementia symptoms in patients developing HAD in the pre-ART era [63]. While elevations of this axonal protein are not specific to HIVE [65], in the appropriate clinical context they usefully assess ongoing disease activity, and complement clinical examinations and neuropsychological test evaluations of cognitive-motor impairment that may be confounded by the cumulative effects of inactive (legacy) HIV-1-related and non-HIV-1-related CNS injury [66–68]. Even though HAD is uncommon in treated HIV-1 infection, mild neurocognitive impairment can sometimes be found in patients on ART. In these cases, blood NfL may serve as a useful marker to help discriminate between active ongoing neuronal injury and inactive, legacy CNS injury, whether due to HIV-1 infection or to other causes.
Combination antiretroviral therapy (ART) has a major impact on CNS infection, so that effective systemic viral suppression usually also induces similar CSF HIV-1 RNA reduction to below the clinical levels of detection [16,69–71]. Indeed, even when assessed by more sensitive ‘single-copy’ assays, ART that controls plasma viremia usually suppresses CSF HIV-1 RNA to very low or undetectable levels [71,72]. Interestingly, even in the presence of systemic treatment failure, CSF HIV-1 RNA levels usually are maintained at one-tenth or, more characteristically, even lower proportions in relation to those of plasma [17,70]. Moreover, systemically effective ART can prevent development of HAD and usually has a salutary effect on newly-presenting HAD, curtailing clinical progression and inducing variable clinical improvement, particularly when adjusted to deliver an effective combination of drugs to the CNS site of infection [73,74]. Monitoring reduction of CSF NfL can also be used to confirm the effectiveness of ART in preventing and mitigating CNS injury [68].
CSF/CNS inflammation accompanying local HIV-1 infection in the meninges and brain parenchyma is also reduced by effective ART. This includes resolution of CSF pleocytosis and reduction in the levels of various inflammatory biomarkers [17]. The dynamic effect of ART on inflammatory biomarkers has been nicely shown by rapid reduction in levels of CSF neopterin, an extensively studied inflammatory biomarker in HIV-1 infection, after treatment initiation [75,76]. Similar overall therapeutic effects have been documented with other inflammatory biomarkers [46,77–82].
However, CNS inflammation may not always fully subside to normal levels after suppressive treatment. For example, CSF neopterin, immunoglobulin production and T-cell activation may persist at levels above those of HIV-1-uninfected controls despite undetectable CSF HIV-1 RNA [83–86]. Even very low levels of CSF HIV-1 RNA, below the clinical cutoff of ‘undetectable’, may be associated with higher levels of local inflammatory activity than more complete CSF viral suppression [72,76,84,87,88]. Although the factors driving persistent inflammation are not firmly established, a CNS HIV-1 reservoir may be an underlying factor [15,89–93]. Whether this persistent inflammation is harmful or protective (or both) remains to be fully defined.
There are important, more substantial exceptions to the general CSF/CNS virological treatment efficacy in the setting of systemic viral suppression. These are referred to as CSF HIV-1 escape syndromes in which CSF HIV-1 RNA concentrations exceed those of blood, with the levels of virus in the blood generally suppressed or substantially reduced by the therapy [94]. They have been classified into three distinct types: neurosymptomatic, asymptomatic, and secondary CSF escape (abbreviated here as NSE, AsE and 2ryE, respectively) [94–100]. NSE is the most important of these, since it is accompanied by neurological symptoms and signs, and in some patients by severe neurological injury [95,96,101–106]. Its neuropathology frequently exhibits a distinct variant of HIVE with prominent CD8+ T-lymphocytes [102]. Factors contributing to the development of NSE include reduced treatment adherence, drug resistance and component drugs in the ART regime with limited CNS penetration [103–105,107–110]. Fortunately, adjustments in ART, taking into account drug resistance, drug potency and CNS drug penetration, can usually halt disease progression and, to variable extent, reverse neurological deficits in many of these individuals [95,96], although for some NSE patients the course can be complicated by recrudescence [103,105,111]. In contrast, AsE, as the name implies, is not accompanied by neurological symptoms or signs, and likely is mostly transitory, perhaps akin to viral blips in plasma [112–114]. It may indeed be benign, though further studies of outcomes in these individuals are needed to more thoroughly characterize the natural history and clinical consequences of these episodes. 2ryE is provoked by other (non-HIV-1) infections within the neuraxis in which the resultant inflammatory responses activate local HIV-1 production or replication. It characteristically resolves with effective treatment of the other infection [100].
Previous CSF studies of HIV-1 infection have focused on a number of the components of local CNS inflammation in diverse cohorts and case series, often emphasizing an individual stage of infection [17,54,75,115–122]. Most have also measured a relatively small number of inflammatory molecules [62]. For this reason, a more comprehensive picture of evolving CNS inflammation in relation to the stages of systemic infection, CNS injury, and treatment, has yet to be firmly defined [22,75,76,87,123].
The aim of this study, measuring a large number of CSF proteins in parallel, was to characterize the features of CNS inflammation and tissue responses more broadly as they change over the course of chronic HIV-1 infection in both untreated and treated persons. To this end we used the Olink Explore 1536 platform to measure the concentrations of 1463 unique proteins in a total of 307 CSF samples, including 30 samples from HIV-seronegative controls and 277 samples from 272 PLWH (five individuals contributed a second CSF sample after their clinical status had changed during the cohort study in Gothenburg) representing a broad spectrum of clinical states defined by blood CD4+ T-cells, CSF HIV-1 RNA concentrations, treatment status, and clinically recognized neurological injury during chronic infection.
This report provides an introduction and initial overview of these identified CSF protein changes in relation to stages of systemic infection, local CNS infection, and CNS injury. Importantly, the online posting of the resultant dataset opens to other investigators the opportunity to extend the value of these data by exploring additional questions related to the CSF protein changes over the course of HIV-1 infection.
Materials and methods
Ethics statement
Ethical approvals were obtained from the institutional review boards of each center (Gothenburg Ethical Committee DNr 0588–01 and 060–18; UCSF IRB Protocol 10–0727; Milan, San Raffaele Scientific Institute IRB Protocol 235/2015; and Poona Hospital and Research Centre, Pune, India RECH/EC/2018/19/257). The samples from the cohort participants and the patients in the disease groups were obtained after written informed consent; if patients were unable to directly consent (some of those with HAD and NSE), this was obtained from a person with power of attorney or the equivalent.
Study design and study subjects
This was a cross-sectional, exploratory study of proteins in a convenience sample of CSF specimens obtained in research studies at four clinical centers. These specimens were derived from existing study archives and represent 11 major clinical categories, along with a small number of additional samples from uncommon clinical settings that were not included in the main analyses in this report. The majority of CSF specimens were from longstanding cohort studies at the University of Gothenburg in Gothenburg, Sweden [112] and the University of California San Francisco (UCSF) in San Francisco California, USA [17]. These cohort samples were supplemented by clinically-derived samples from ongoing HIV-1 studies in these two centers and selectively expanded by clinically-derived specimens from Milan, Italy [124] and Pune, India [125] representing the two neurological disease groups, HAD and NSE. Specimens were obtained between 1990 and 2020 within the context of research protocols. The sample groups included an uninfected control group and 12 chronically HIV-1-infected groups defined by blood CD4+ T-cell counts, neurological presentations, treatment status and viral suppression as previously described [123]. When available samples exceeded the planned number within each of these predefined groups, specimens were chosen at random, while in groups with sparse samples all available specimens were included. In brief, the following were the group criteria (the final specific individual group characteristics are described in the Results and Discussion section below).
HIV-1 uninfected control (HIV-) group
These were drawn from the Gothenburg and San Francisco populations with similar demographic and background characteristics to the PLWH at those centers. Notably, the Gothenburg specimens were all selected from a group of individuals taking pre-exposure prophylaxis (PrEP) because of HIV-1 infection risk. As with the PLWH, all of these participants volunteered for the clinical examinations, background blood tests and the study lumbar punctures (LPs). All were screened with HIV-1 serology and blood HIV-1 RNA measurements.
HIV-1-infected, untreated included 7 groups of PLWH:
Elite viral controllers (elites) were defined by positive HIV-1 serology and undetectable plasma HIV-1 RNA concentrations on repeated occasions using a standard clinical cutoff (<20 HIV-1 RNA copies per mL) in the absence of ART [23,24,126].
Five CD4-defined groups were untreated (either naïve to treatment or off ART for at least 6 months) PLWH who did not have evidence (symptoms or signs) of active or confounding neurological disease on clinical examination. Following our experience in previous studies [45,127], CSF samples were segregated into five strata based on the accompanying blood CD4+ T-cell counts: >500, 350–499, 200–349, 50–199, and <50 blood CD4+ T-lymphocytes per μL, respectively. This division was based on previous experience in analyzing CSF biomarkers with emphasis on individuals with lower blood CD4+ T-cell counts since this is the range of vulnerability to HAD. None of the PLWH exhibited symptoms or signs of active or progressive neurological impairment based on bedside clinical history and physical examinations at the time of their visit. They were all characterized as neuroasymptomatic or exhibiting only chronic, stable neurological symptoms or signs. We did not attempt to segregate those with stable asymptomatic neurocognitive impairment (ANI) or minor neurocognitive disorder (MND) discerned by formal neuropsychological testing, but rather classified the subjects solely using the broad biological and diagnostic categories provided by the CD4+ group designations.
HAD patients were PLWH who presented clinically with new symptomatic, subacute or progressive neurological disease attributed to CNS HIV-1 infection by their caring physicians based on their clinical presentation and diagnostic evaluations. Some were studied before publication of the formal Frascati criteria for HAD [128] and diagnosed with AIDS dementia complex (ADC) stages 2–4 [129] while also meeting the American Academy of Neurology criteria for HIV-1-related dementia in place at the time [130]. Retrospectively, they also met the functional criteria for the Frascati diagnosis of HAD without the requisite formal neuropsychological assessment, and therefore this term was used to encompass the full group of these patients.
HIV-1-infected, treated groups included 5 main groups of PLWH taking ART.
Two groups of treated, virally suppressed individuals were selected from the Gothenburg and San Francisco cohorts: one with either normal age-corrected CSF NfL or previously unmeasured CSF NfL (RxNFL-), and a second unusual group of special interest selected on the basis of elevated age-adjusted CSF NfL (RxNFL+) levels when measured at the time of their cohort study evaluation.
CSF escape was diagnosed in individuals taking ART with suppression of plasma HIV-1 RNA below pretreatment levels but with higher concentrations of HIV-1 RNA in CSF than plasma [94,113,131]. Three types of CSF escape were distinguished: NSE in which individuals presented clinically with new moderate to severe neurological symptoms and signs that were attributed to CNS HIV-1 infection and without alternative cause after diagnostic studies; AsE were neurologically asymptomatic individuals identified by CSF and blood HIV-1 RNA findings during participation in cohort studies; and secondary CSF escape (2ryE) which was diagnosed on the basis of CSF and plasma measurements in the context of another, non-HIV-1 nervous system infection [100].
Miscellaneous PLWH included four anecdotally interesting individuals who did not fit the above categories: one who sustained HIV-1 cure (the ‘Berlin patient’) [132], two with very early initiation of ART [133], and one ambiguous escape patient with a history of progressive multifocal leukoencephalopathy (PML). These individuals were included in the full data table (File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx accessible on Dryad, DOI: 10.5061/dryad.x3ffbg7tv) [134] but not in the analyses described in this report which emphasizes group findings.
CSF sampling
CSF from the cohort subjects was obtained according to previously described standard protocols [17,112,135,136]. Similar methods were used to process clinically-obtained samples from HAD, NSE and 2ryE groups that were preserved for study purposes. In brief, CSF, obtained by LP, was placed immediately on wet ice and subjected to low-speed centrifugation to remove cells before cell-free fluid was either submitted directly to the Clinical Laboratory for HIV-1 RNA measurement along with plasma, or aliquoted and stored within 2 hours of collection for later HIV-1 RNA measurement along with stored plasma. Stored samples were maintained at ≤-70°C until the time of biomarker assays or delayed HIV-1 RNA measurement.
Clinical evaluations and background laboratory methods
Cohort subjects underwent standardized general medical and neurological assessments at study visits as previously described [137]. Symptomatic CNS disorders (HAD, NSE, and 2ryE) were evaluated using standard clinical and neurological assessments, including neuroimaging along with CSF and blood measurements, to confirm their diagnoses and assess disease severity [137]. All cohort participants had routine clinical bedside screening for symptoms or signs of abnormalities in cognitive or motor function or evidence of CNS opportunistic infections or other conditions that might impact CSF biomarker concentrations. Individuals with CNS opportunistic infections or other conditions confounding these analyses were omitted with the exceptions of those in the 2ryE group in which these infections were part of the clinical group definition. We initially set a general target of 25 separate samples for each major clinical group. Subsequently, some groups of particular interest were augmented with additional specimens, while for other categories there were shortfalls related to limited specimen availability.
HIV-1 RNA levels were measured in cell-free CSF and plasma at each site in local Clinical Research or Clinical Laboratories using the ultrasensitive Amplicor HIV-1 Monitor assay (versions 1.0 and 1.5; Roche Molecular Diagnostic Systems, Branchburg, NJ), Cobas TaqMan RealTime HIV-1 (version 1 or 2; Hoffmann-La Roche, Basel, Switzerland) or the Abbott RealTime HIV-1 assay (Abbot Laboratories, Abbot Park, IL, USA), depending on the site and time period of collection. All recorded viral loads reported below 20 copies per mL were standardized to an assigned ‘floor’ value of 19 copies per mL for descriptive and computational purposes. Each cohort study visit included assessments by local Clinical Laboratories using routine methods to measure CSF white blood cell counts (WBCs), blood CD4+ and (in many) CD8+ T lymphocyte counts by flow cytometry, and CSF and blood albumin to assess blood-brain barrier integrity.
CSF NfL concentrations had been measured in multiple runs in a portion of the subjects prior to this study using a sensitive sandwich enzyme-linked immunosorbent assay (NF-light ELISA kit, UmanDiagnostics AB, Umeå, Sweden) [138] performed in the Clinical Neurochemistry Laboratory at the University of Gothenburg by board-certified laboratory technicians blind to clinical data; intra-assay coefficients of variation were below 10% for all analyses. To compare these NfL values across all individuals, we calculated age-adjusted NfL values for 50 years of age and compared them to the 50 year-old upper limit of normal of 991 ng/L, as outlined by Yilmaz and colleagues [68].
Olink Explore methods
Samples from all sites were maintained at ≤-70°C, aggregated in San Francisco, shipped together to Olink Laboratory in Watertown, MA, USA in previously stored, frozen aliquoted tubes, and analyzed in a single run in November 2020. Samples were transferred using local Olink procedures. The Explore 1536 battery deployed at that time consisted of four plates: Inflammation, Cardiometabolic, Oncology and Neurology plates. The Olink immunoassays are based on the Proximity Extension Assay (PEA) technology [139], that uses a pair of oligonucleotide-labelled antibodies to bind to their respective target protein. When the two antibodies are in close proximity after binding, a new polymerase chain reaction target sequence is formed, which is then detected and quantified by Next Generations Sequencing (NGS). Output is reported in NPX units on a log2 scale of relative concentrations which are used for within-assay comparison of protein concentrations (https://olink.com/). Proteins measured using the Olink platform are identified in this study using the gene nomenclature and numbers as they appear in the UniProt database (https://www.uniprot.org/uniprotkb/) [140].
The level of detection (LOD) for each assay is listed in the data output and based on the background, estimated from the negative controls on every plate, plus three standard deviations. In the results presented below for this analysis, we include the returned values without adjustment or substitution when below the LOD. This is one of three strategies discussed by Olink and adopted for this study both for simplicity, and because it may increase the statistical power and provide a more normal distribution of the data without an increase in false positives (https://olink.com/faq/how-is-the-limit-of-detection-lod-estimated-and-handled/). Each protein measurement is comprised of four subgroups, each with a related LOD; these are listed in the full dataset now posted online (File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx. DOI: 10.5061/dryad.x3ffbg7tv) [134]. Our analysis also included measurements that were flagged in the data output file with quality control (QC) Warnings. These QC Warnings were generated if incubation Control 2 or Detection Control (corresponding to that specific sample) deviated by more than a pre-determined value (+/- 0.3) from the median value of all samples on the plate (https://olink.com/faq/what-does-it-mean-if-samples-are-flagged-in-the-qc/). Our own analysis of the effect of the QC warnings in the three proteins with quadruplicate repeats supports our decision to include these values as shown and is discussed in the context of S1 Fig.
Statistics analysis
R version 4.1.1 was used for the main data analysis. The “hclust” function in R was used to perform hierarchical clustering to identify three clusters of the proteins. The “prcomp” function in R was used to perform the principal component analysis. One-way ANOVA was used to test the differential expression of proteins between the clinical groups. Biomarker associations were analyzed across the entire sample set using Spearman’s rank correlation, graphically presented as a heat map for some. Comparison of biomarker concentrations between subject groups to address selected a priori questions used non-parametric methods including Mann-Whitney to compare two groups and Kruskal-Wallis with Dunn’s post hoc test to compare three or more groups.
Two-sample t tests were used to compare the protein expression between the HAD and NSE groups. Partial correlation was used to assess the association between protein markers and the CSF HIV-1 level while controlling for the CSF NfL level (NEFL in Olink output). Similarly, partial correlation was used to assess the association between protein markers and the CSF NEFL level while controlling for the CSF HIV-1 level. Gene signature set enrichment analysis (GSEA) was used to calculate the enrichment score of the immune related pathways [141].
The immune related pathways were defined as the child terms of the term “immune system process GO:0002376” [142]. Because the Olink Explore 1536 platform only measured a selected set of the human proteins, we only included an immune pathway if more than five of its proteins were measured by Olink assay. All statistical tests were adjusted for multiple testing using the FDR method.
Comparisons of the quadruplicate results of three proteins that were analyzed on each of the four plates and of earlier NfL measurements to the Olink NEFL results by linear regression used Prism 9.5.1 and 10.1, GraphPad Software, San Diego, California USA (www.graphpad.com). Selected comparisons of groups by ANOVA with correction for multiple comparisons and graphs of subject profiles were also produced using Prism 9.5.1 and 10.1 which was also used for redrawing some of the graphs produced by other methods.
Dryad DOI
Results and discussion
This is the initial report of a study using the Olink Explore 1536 platform to measure CSF proteins in large sample of grouped specimens representing the spectrum of untreated and treated chronic HIV-1 infection. The resultant full study dataset (File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx) is posted online at Dryad: DOI: 10.5061/dryad.x3ffbg7tv (https://datadryad.org/stash) [134].
We have divided the presentation of the study results and discussion into two principal sections. The first, Background results section, describes the clinically-defined groups in the study cohort along with selected characteristics of the Olink Explore 1536 data output that are illustrated in S1 and S2 Figs by analysis of three sets of quadruplicate-repeat measurements embedded in the Olink panel and measurements of seven CSF proteins that have served as biomarkers of different cellular components of CNS injury in previous studies. The second, Main results section, examines salient features of the full set of measured CSF proteins in three sets of analyses that focus on: 1. The overall changes in proteins across the study groups; 2. correlations of the measured CSF proteins with biomarkers of CNS infection (CSF HIV-1 RNA) and CNS injury (CSF NfL); and 3. comparison of CSF proteins in two clinical forms of HIVE: HAD and NSE.
Background analysis of the study groups and selected features of the Olink Explore platform applied to the CSF samples
Study group characteristics
This cross-sectional analysis of a convenience sample of 303 CSF samples follows a strategy used in several of our previous studies of CSF biomarkers [45,75]. It draws on the experience and archived CSF specimens of four research centers with long-term interests in HIV-1-related CNS disease. The complete set of 307 analyzed CSF samples included 30 specimens from HIV-1-seronegative controls and 277 from 272 PWH (five PLWH contributed a second CSF sample after their clinical status had changed during the cohort study in Gothenburg) representing 7 untreated and 5 ART-treated groups that are the focus of this report, along with a miscellaneous group of four samples from individuals who do not clearly fit into any of the main categories and therefore are not included in this analysis which centers on group identities and differences. The background demographic and HIV-1-related clinical laboratory characteristics of the individual study participants, along with the Olink protein measurements of the entire set of 307 CSF samples, are all included in Dryad-posted dataset (DOI: 10.5061/dryad.x3ffbg7tv) [134].
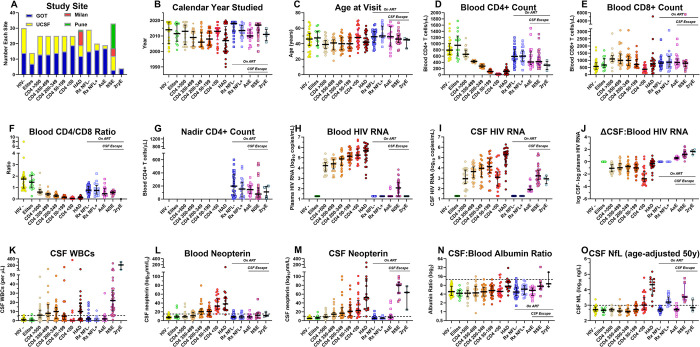
The background features of the subject groups are summarized in S1 Table and graphically detailed in Fig 1. The latter shows the individual values for each of these background variables within the group designations. The main features of these subject groups are discussed in more detail in the legend to this figure.
Fig 1. Study group background characteristics.
Except for the first panel (Fig 1A), the graphs (Fig 1B–1O) use the same format that is applied to subsequent figures showing the Olink-generated protein measurement profiles across the study groups. This includes the symbols and the median and IQR bars. The dashed horizontal lines in Fig 1K–1O indicate the mean +2 SD of the uninfected, seronegative (HIV-) control group in the measured variable as a rough estimate of their upper normal limit and as a visual guide to compare groups; this convention is also used in the Olink CSF protein measurement figures showing the group profiles in subsequent figures. A. Study sites. Most of the samples were obtained in the context of scheduled outpatient visits during long-term cohort studies in Gothenburg, Sweden (GOT) and San Francisco, California, USA (SF) that included lumbar punctures (LPs) as part of the study protocols (Fig 1A). This included CSF specimens from the HIV-, elite controllers (elites), five neuroasymptomatic CD4-defined groups, two ART-treated and virally suppressed groups (one with normal CSF NfL and one with elevated CSF NfL, designated as RxNFL+ and RxNFL-) and the asymptomatic escape (AsE) group. The HIV-associated dementia (HAD), neurosymptomatic escape (NSE) and secondary escape (2ryE) samples were obtained during clinical evaluations after informed consent. Additionally, the NSE and HAD groups were augmented by CSF specimens from Milan and Pune to attain a size comparable to the other main groups. When there was a larger number of CSF specimens available for a given group than required for the study, samples were chosen at random from archived collections (e.g., CD4-defined groups). An exception to this random selection was the choice of the HIV- group from GOT: all of these were from individuals taking pre-exposure prophylaxis (PrEP) because of their self-identified risk for HIV-1 infection. HIV- controls from SF were seronegative individuals from the same clinical site and demographic population as the people living with HIV (PLWH) included in the local cohort, but not on PrEP. The Rx NFL+ and the AsE groups were almost all from GOT related to the local interest in these conditions. The small number of secondary escape (2ryE) were also all from GOT. In general, the imbalances in the subject group sizes related to scarcity of available samples (e.g., elites and 2ryE, resulting in smaller sample sizes), or to inclusion of larger numbers of specimens to augment particularly important groups, including the HIV- group, related to its comparative utility, and HAD and NSE groups because of our interest in more severe HIV-1-related CNS injury. These imbalances, along with those of calendar years and subject ages (B and C below), emphasize that this exploratory study used a convenience sample rather than one that was balanced with respect to individual demographic variables. B. Calendar year. The dates of study varied among the groups. In part this reflected the history of our cohort studies in concert with the changes in acceptance and efficacy of treatment regimens over the collection period. These time variations impacted the length of untreated chronic infection, susceptibility to disease progression and presence of viral suppression among our study cohorts. For example, there were nearly 10 years separating the median years of the HAD group from both treatment-suppressed groups (RxNFL- and RxNFL+). The NSE group sampling also reflected later recognition and attention to this clinical entity. C. Age. The ages of the subjects were not specifically selected and consequently also varied among groups, with group medians ranging from 39 to 50 years with largely similar ranges. Overall, the median ages of the treated PLWHs were similar to those of the HIV-negative (HIV-) controls, while the untreated groups were generally younger. The balance of sex was not taken into account with specimen choices, so distribution also varied among the groups. D. Blood CD4+ T-lymphocytes. The median blood CD4+ T-cell count in the elites was above that of the uninfected controls and the other HIV-1-infected groups, including the two treatment suppressed groups. The blood CD4+ T-cell concentrations in the 5 groups defined by these counts show the stepwise decrease from >500 to <50 CD4+ T-cells/μL dictated by the study design. The HAD group had a median CD4+ T-cell count of 108 cells/μL that was similar to the 50–199 CD4+ group median of 106 cells/μL, in keeping with the role of advanced immunosuppression in the development of this subacute disorder and its underlying HIVE pathology. The CD4+ T-cell counts of the two treatment-suppressed groups were nearly equal, with both having blood CD4+ T-cell counts above those of the viremic CD4-defined groups except for the group with the highest blood CD4+ T-cell counts (CD4>500 group), indicating the partial CD4+ cell preservation or recovery with suppressive treatment. By contrast, the blood CD4+ T-cell counts in the three CSF escape groups were lower than the two treatment-suppressed groups, consistent with less robust recovery or ongoing loss during the escape episodes. However, these levels were not as low as in the untreated HAD group. The CD4+ blood counts of the 2ryE subjects were also low, indeed below the other two escape groups, likely contributing to their vulnerability to intercurrent infections (herpes zoster in three and HSV2 meningitis in the fourth) and factoring into their clinically-directed LPs. E. Blood CD8+ T lymphocytes. Blood CD8+ T-cell counts were on the high side of normal in the elites, rose as blood CD4+ cells decreased in the untreated CD4-defined groups except for those in the CD4 <50 group (median of 380 CD8+ T-cells per μL). They were relatively increased in the HAD group (median 750 CD8+ T-cells per μL) and in all the treated groups, including the two virally suppressed and the NSE and ASE groups (medians in 800s per μL). F. Blood CD4+/CD8+ T-cell ratio. The differences in the trajectories of the two T-cell subpopulations resulted in lowered ratios in all infected groups compared to the HIV- controls, reflecting variable reductions in CD4+ and elevations of CD8+ T-lymphocytes. Reductions were most severe in the CD4 -defined groups, most notably in those with CD4+ T-cell counts below 200 cells per μL and in the HAD group. These ratios were relatively increased in the four treated groups (0.47 to 0.77), though all remained below the level of the uninfected controls (median ratio of 1.75). Reduced ratios persisted in the two ART-suppressed groups (RxNFL- and RxNFL+) and the AsE and NSE groups. G. Nadir blood CD4+ T-lymphocytes. Nadir CD4+ T-cell counts are shown only for the ART-treated groups, since CD4+ T-cell values in the untreated groups at their study visits were generally at or near their nadirs. The treated groups showed varying degrees of presumed blood CD4+ T-cell recovery, higher in the two virally suppressed (Rx NFL- and NFL+) groups (medians of 590 and 600 CD4+ T-cells per μL) than in the CSF escape NSE and ASE groups (medians of 425 and 412 CD4+ T-cells per μL, respectively), the latter perhaps either contributing to development of CSF escape or, alternatively, reflecting an impact of escape on CD4+ T-cell dynamics. H. CSF HIV-1 RNA. Highest CSF HIV-1 RNA concentrations were present in the HAD group, while the CD4-defined groups showed the ‘inverted U’, or lymphoid, pattern of change with lower concentrations at the extremes (in the CD4+ >500 and CD4+ <50 groups) than in the middle ranges (CD4+ 350–499, 200–345 and 50–199 groups). We previously reported the association of this pattern with that of the CSF WBC counts as also noted in this study (see Fig 1K below), suggesting that these two findings were causally related [45]. In this study the correlation of CSF HIV-1 RNA to CSF WBC count across the CD4-defined groups was significant (Pearson correlation P = 0.006). This lymphoid pattern of infection and cell response likely underlies this same pattern in many of the CSF proteins included in the Olink Explore 1536 panel as shown in later figures. The CSF HIV-1 RNA concentrations of three escape groups were generally below those of the untreated groups except for the CD4 <50 group. CSF HIV-1 RNA concentrations were all below detection in the elites. I. Blood HIV-1 RNA. In the untreated individuals (not including the elites) there was a steady increase in blood HIV-1 RNA through the full range of CD4+ T-cell loss that reached its highest mean levels in the HAD group. Unlike with CSF, there was no decrement in the CD4 <50 group, showing a distinct difference between the CSF and the systemic blood viral dynamics. The presence of low-level blood HIV-1 RNA in the NSE group may have reflected systemic partial drug resistance in some or spillover of the ‘escaped’ CNS/CSF infection into the blood. J. CSF:Blood HIV-1 RNA differences. This panel shows the differences in viral loads between the two fluids (calculated as the log10 CSF HIV-1 RNA copies per mL–log10 plasma HIV-1 RNA copies per mL) and emphasizes the largely consistent relationship between CSF and blood HIV-1 RNA levels in untreated infection at blood CD4+ T-cell levels between 50 and >500 cells/μL in which the CSF concentrations were approximately 10-fold lower than those in blood, and thus maintaining a nearly 1:10 ratio of CSF to blood HIV-1 RNA. This may relate to the kinetics in the influx of infected CD4+ T-cells and of viral release into the CSF over this blood CD4+ T-cell range. This ratio was disrupted when blood CD4+ T-cells fell to <50 cells/μL and the CSF WBC count decreased to negligible levels; in this group the CSF:blood HIV-1 RNA ratio decreased nearly 10-fold to an overall ratio of <1:100. This underscores the importance of lymphocyte traffic and likely direct virus release by trafficking infected CD4+ T-cells in the determining the CSF HIV-1 levels in the neurologically asymptomatic individuals, particularly those with blood CD4+ T-cell counts above 50 cells per μL. This relationship changed markedly with the development of HAD and the underlying direct neuropathic parenchymal CNS infection, i.e., HIVE, in which the CSF HIV-1 RNA concentrations reached high levels, and the differences between it and the blood viral load decreased. Thus, in the HAD group local brain infection rather than hematogenous sources likely was responsible for the high levels of CSF HIV-1 RNA. The reversed CSF:blood HIV-1 RNA ratios in the three CSF escape groups were consonant with their definitions predicated on CSF > blood HIV-1 RNA concentrations that indicated the direct CNS sources of the measured CSF HIV-1 RNA in the face of systemic viral suppression. K. CSF WBC counts. The median CSF cell counts rose and then fell over the course of untreated infection in the CD4-defined groups. CSF WBC counts were highest in the 200–349 group and declined to lowest levels when blood CD4+ T-cells fell below 50 cells/μL, defining the lymphoid pattern. As discussed above, this likely was causally linked to the changes in CSF HIV-1 RNA in these groups. Both neurologically defined groups were associated with augmented local inflammation: CSF WBCs were elevated in HAD, and even higher in NSE (median counts of 10 and 22 cells per μL, respectively). In these two settings the CSF WBC counts likely involved a response to local CNS infection and injury. L. CSF neopterin. This pteridine is predominantly, though perhaps not exclusively, a macrophage-related activation marker [143]. CSF levels showed a steady median increase as CD4 cells declined, including the highest levels in the CD4+ <50 group among the CD4-defined sequence of groups, thus providing a prototypical example of the myeloid pattern of CSF biomarker change. CSF concentrations were even higher in the HAD group, and, notably, were highest in the NSE group, while suppressive treatment brought CSF neopterin concentrations back to near normal levels in the absence of symptomatic CSF escape. M. Blood neopterin. In blood, neopterin concentrations showed a similar, though more restricted elevation with CD4 decline but were notably increased in both the CD4 < 50 and HAD groups, presumably indicating systemic macrophage activation in these settings that either links these two sites of infection or marks parallel changes in myeloid cell populations in the CNS and systemically. In contrast to CSF, the blood neopterin was only mildly elevated in the NSE and 2ryE groups in keeping with the underlying compartmentalized CNS HIV-1 and intercurrent infections in these two groups in the face of systemic HIV-1 suppression. N. CSF: blood albumin ratio. This ratio provides an index of blood-brain barrier integrity, though with considerable individual and age-related variability [144]. In this sample set, only direct HIVE in the HAD group and, to a lesser degree, in the NSE group was associated with elevated median albumin ratios. Minor, though not significant, increases were present in the RxNFL+ group. One-way ANOVA with Tukey’s multiple comparison test found significant differences (P<0.05) only for HAD (CD4 >500: P = 0.0004; Elites: P = 0.0028; AsE: P = 0.0067; CD4 200–349: P = 0.0069; RxNFL-: P = 0.0116; CD4 50–199: P = 0.0202; CD4 350–499 P = 0.0228) and NSE (CD4 >500: P = 0.0023; Elites: P = 0.0075; AsE: P = 0.0173; CD4 200–349 P = 0.0211; HIV-1- P = 0.024; RxNFL- P = 0.0337; CD4 50–99 P = 0.0479). Other intergroup comparisons were not significant. Overall, the albumin ratio elevations were small, and likely to have had only limited, if any, impact on the CSF proteins measured in this panel. O. CSF NfL. Prior to this study, CSF NfL was measured using the UMAN ELISA method (UmanDiagnostics, Umeå, Sweden) in 209 of the 307 (71.3 percent) of the specimens. In this figure, the NfL values have been adjusted to age 50 years using the normative data and methods of Yilmaz and colleagues [68] to allow more direct subject comparisons and estimations of abnormal values. While these data were incomplete and superseded in this study by the Olink measurements performed on all samples, they guided our initial definition of the RxNFL+ group. The highest levels of NfL were in the HAD group, and there were elevated levels in a substantial proportion of the CD4 <50 group presumably related to subclinical neurological injury. Notably, the NfL elevation of the HAD group had nearly a 10-fold higher median value than that of the NSE group. These observations were confirmed and extended by the measurement of NfL within the Olink Explore panel as shown later in S2 Fig.
Despite demographic imbalances in this convenience sample, as discussed in the legend to Fig 1, the group of CSF specimens provides a robust aggregate that encompasses the main facets of chronic, evolving HIV-1 infection in the absence and presence of treatment, and importantly includes two groups with HIV-1 encephalitis (HIVE): the untreated HAD and treated NSE groups. Here we briefly focus on selected background features that particularly bear on the main CSF protein findings and on some of the interpretive themes that follow. This includes introduction to the prototypes of the lymphoid (CSF HIV-1 RNA and CSF WBC counts) and myeloid (CSF neopterin) patterns of change across the five untreated, neurologically asymptomatic CD4-defined groups introduced earlier and defined in the MATERIALS AND METHODS section, along with the CSF NfL concentrations in a subset of the specimens that had been previously measured.
CSF HIV-1 RNA concentrations and CSF WBC counts: Lymphoid patterns of change in the CD4-defined groups
As discussed in the legend to Fig 1, The concentrations of HIV-1 RNA in the blood and CSF, and hence their interrelationships, changed with progression of untreated infection in the CD4-defined groups (Fig 1H–1J). While in the blood there was a nearly steady, stepwise increase in HIV-1 RNA concentrations as the blood CD4+ T-cell count decreased from the highest (CD4 >500) to lowest (CD4 <50) group levels, in the CSF progressive viral load increase was interrupted in the CD4 <50 group by a drop in the median HIV-1 RNA to 3.09 log10 copies per mL from a peak median of 4.14 log10 copies per mL in the CD4 50–199 group, resulting in a high CSF-blood difference in this group (2.75 log10 copies RNA per mL shown in Fig 1J). This contrasted with the more constant differences between CSF and blood HIV-1 RNA of about 1.0 log10 copies per mL in the other four CD4-defined groups. These changes in CSF HIV-1 RNA were parallel to, and likely related to, the CSF WBC counts (Fig 1K) in which there was also a decrease in the CD4 <50 group to negligible levels in comparison to the other untreated groups with higher blood CD4+ T-cell counts [17]. The low CSF WBC counts in the CD4 <50 group may have related to the limited availability of both CD4+ and CD8+ T-cells that generally comprise the majority of CSF WBCs in HIV-1 infection [83]. More particularly, the low number of CD4+ T-cells in this group likely resulted in decreased passage of HIV-1-infected cells from the blood into the leptomeninges, and, consequently, diminished viral release into the CSF spaces. The reduced presence of CSF T-cells in this group also likely impacted the levels of some of the measured proteins in this study, resulting in this same lymphoid pattern for many of the CSF proteins in the sequence of CD4-defined groups, similar to the previously reported patterns in a limited subset of inflammatory biomarkers that included CXCL10, TNF-alpha and MMP-9 as discussed above in the Introduction. In contrast, the CSF HIV-1 RNA concentrations in HAD group increased to the highest levels among the groups, to near those in the blood (CSF median of 5.27 and blood median of 5.66 log10 copies RNA per mL). This was a consequence of the HIVE underlying HAD in which local CNS HIV-1 replication within the brain likely ‘spilled over’ into the CSF [17]. The median CSF WBC count in the HAD group was 10 cells per μL, and the median WBC count was even higher in the NSE group (22 cells per μL), though in the latter group the virological impact of the pleocytosis was likely mitigated by a partial treatment effect. The higher CSF than blood HIV-1 RNA concentrations in the three CSF escape groups (on therapy with virus suppressed in the blood) defines these entities, with the AsE group’s CSF virus concentrations (median of 1.90 log10 copies per mL) more than 10-fold lower than the other two escape groups (median CSF RNA concentration in the NSE group of 3.23 and in the 2ryE of 2.94 log10 copies per mL). Though it presents an interesting comparison to NSE, the 2ryE group was small and included CSF from three individuals with herpes zoster and one with HIV-2 meningitis, so it cannot be considered as representative of the larger possibilities in this category.
The virally suppressed (RxNFL-) group was likewise typical of the effectiveness of ART in normalizing symptomatic or asymptomatic neural injury. By contrast, the RxNFL+ group was selected from our larger experience based on elevated age-adjusted CSF NfL measurement in the absence of neurological symptoms and signs. The causes of these elevations in NfL were unknown and did not appear to predict neurological deterioration in these individuals during available follow up. Among the main possibilities are that they might have reflected mild brain insults unrelated to HIV infection (e.g., trauma) or that they were due to transient subclinical HIV-1-related injury. The lack of association with concurrent elevations in CSF HIV-1 levels, or with changes in the other CSF neural or inflammatory biomarkers (as described below) likely argues against the latter and suggests they might not be related to CNS HIV-1 infection. We plan to examine these groups further in the future to address these issues.
CSF Neopterin: Myeloid pattern of change in the CD4-defined groups
Neopterin is a pteridine inflammatory mediator that is likely mainly produced by myeloid and possibly to a lesser extent by astroglial cells within the CNS [145–147]. It serves as a useful CSF inflammatory biomarker during HIV-1 infection [75]. In contrast to HIV-1 RNA and WBCs, CSF neopterin showed a steady increase with falling CD4+ T-cell counts in the untreated PLWH groups, without a subsequent decrease in the CD4 <50 group (Fig 1L). Based on this and previous observations with other myeloid-related biomarkers, including CCL2 and sCD163 [45], we have now termed this the myeloid pattern of biomarker change within the untreated groups that contrasts with the lymphoid pattern discussed above. We use these two convenient, albeit simplistic, terms in this report to designate recurring contrasting patterns of biomarker changes observed in a number of the Olink-measured CSF proteins across the CD4-defined groups as presented below. The HAD group and two of the three escape groups, NSE and 2ryE, exhibited marked elevations in CSF neopterin while median concentrations in the AsE group were near normal. Blood neopterin levels also rose in the untreated individuals; they were notably elevated in the CD4 < 50 group and also in the HAD group, consistent with systemic myeloid activation in these settings [28]. By contrast, blood neopterin levels in the three escape groups remained at or near normal despite the elevations in CSF, including in the NSE and 2ryE groups with notably high CSF values, consistent with the compartmentalized CNS infection and inflammatory responses in these groups and local production of neopterin. In AsE the median neopterin values in both fluids were at or near normal with the exceptions of three CSF samples, distinguishing these individuals from the other two escape groups (NSE and 2ry) and suggesting that AsE is a transient and less pathogenic form of CSF escape [98]. This contrasted with the parallel CSF and blood neopterin changes with CD4+ T-cell decline in the untreated individuals and with the generally low blood neopterin levels in the virally suppressed (RxNFL- and RxNFL+) groups [45].
CSF NfL in a subset of the CSF sample
Fig 1O shows the results of the CSF NfL measurements that had been assayed in a subset of the samples at the University of Gothenburg Clinical Neurochemistry Laboratory over a number of years in several different analytical runs of an enzyme-linked immunosorbent assay (ELISA) and age-adjusted to 50 years of age using a previously described method [68]. There were consistent CSF NfL elevations in the HAD and NSE groups (more than ten-fold higher in the former) and milder increases in about half of the CD4 <50 group, consistent with relatively common subclinical CNS injury in the setting of advanced immunosuppression. The general pattern of NfL change across this group showed similarity to the myeloid pattern discussed above, but a few possible differences can be noted: the elevation of NfL in the CD4 <50 group was ‘abrupt’ without a clear lead-up in the other CD4-defined groups; additionally, there was an increase in CSF NfL in the RxNFL+ group that was absent in CSF myeloid or other neural markers as discussed below. More generally, the similarities between the patterns of change of the CSF myeloid markers and CSF NfL, that defines a neuronal injury pattern in CSF, likely relates to the importance of myeloid cells in the pathogenesis of CNS injury.
Introduction to the Olink Explore output and CSF dataset with analysis of repeated measurements
Proteins measured using the Olink platform are reported in NPX units on a log2 scale of relative concentrations (https://olink.com/products/olink-explore-3072-384). Thus, the output allows comparison of the measured protein concentrations of individuals or groups only within this particular study, and would require simultaneous calibration, with assessments of known concentrations for conversion to more standard quantitative units (e.g., ng/mL). Consequently, interpretations of results for the HIV-1-infected groups depends importantly on comparisons with the HIV-uninfected controls measured within this same assay.
The Olink output measuring 1,472 proteins includes quadruplicate measurements of three proteins (CXCL7, IL6 and TNF) as outlined below. To eliminate the effect of this redundancy, we randomly selected only one set of results for each of these three proteins to include in our overall analysis. This reduced the analysis to include assays of 1,463 unique proteins applied to 303 specimens, yielding a total of 443,289 individual CSF protein measurements. However, some of the measurements fell below the LOD of the individual assays. As noted above in the MATERIALS AND METHODS section, in this report we used the measured values without any correction related to the LODs, since these low values likely had little more effect on analysis than would truncating or eliminating values below the LODs. For other investigators who may want to adjust these values differently in future studies, the four LOD values for each protein assay are included in the posting of the full data set (File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx (DOI: 10.5061/dryad.x3ffbg7tv)) [134].
Analysis of three sets of quadruplicate repeat measurements; features of Olink Explore data output
As a prelude to the main analysis, we examined the intercorrelations among the quadruplicate measurements of three CSF proteins: CXCL8, IL6 and TNF. The Olink Explore 1536 platform includes four independent measurements of these proteins, one on each of the four assay plates. Within these data we also examined the impact of Quality Control (QC) Warnings that flagged some of the measurements and the frequency of protein measurements below the LODs reported in the Olink data output file. The QC Warnings and the percentage of measurements within different strata of LODs (100; 90 - <100; 60 - <90; 40 - <60; 20 - <40; and <20 percent) are all indicated within the full dataset table (File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx (DOI: 10.5061/dryad.x3ffbg7tv)) [134].
The analyses outlined in S1A–S1C Fig show the high intercorrelations among the quadruplicate NPX values for two of the three sets of quadruplicate values and lower correlations in the third (mean R2 results of the 6 pairwise comparisons for each of the proteins were 0.9821 and 0.9508 for CXCL8 and IL6, with less robust correlation for the TNF correlations with a mean R2 = 0.7055). These correlations, along with very similar patterns of concentration changes across the subject groups (S1B Fig), were present despite differences in the absolute NPX values in some of the repeats, emphasizing the relative scale of the Olink protein measurements and the importance of a full set of embedded controls in each independent assay.
In all but two cases the measurements with the QC warnings in these assays fell within the confidence limits of the overall results. Based on this finding, and to reduce biased selection of the Olink output, we included all Olink results in the analyses that follow, including those with QC Warnings. Values below the LODs were minimal in the CXCL8 and IL6 repeated measures and did not impact correlations. However, a greater frequency of results below the LODs were noted with the TNF measurements and contributed to less robust correlations among the four repeated measures of this protein, though perhaps not providing a complete explanation. As noted above, for the analyses in this report, we did not exclude assay results below the LODs that are listed in the Dryad posted (File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx (DOI: 10.5061/dryad.x3ffbg7tv)) and discussed in the legend to S1 Fig.
Values below the LODs preferentially affected the analysis of the aviremic groups (e.g. HIV-, elites, Rx NFL- and NFL+), reducing the potential of some proteins to discriminate differences among these groups. Because of attention to the overall protein changes, and particularly to the differences among the CD4-defined groups and the HAD and NSE groups, this issue likely had relatively limited impact on the main analyses included in this report. However, it likely will be more important in future studies using these data to examine some of the proteins in these aviremic groups in which the values below the LODs will assume greater prominence.
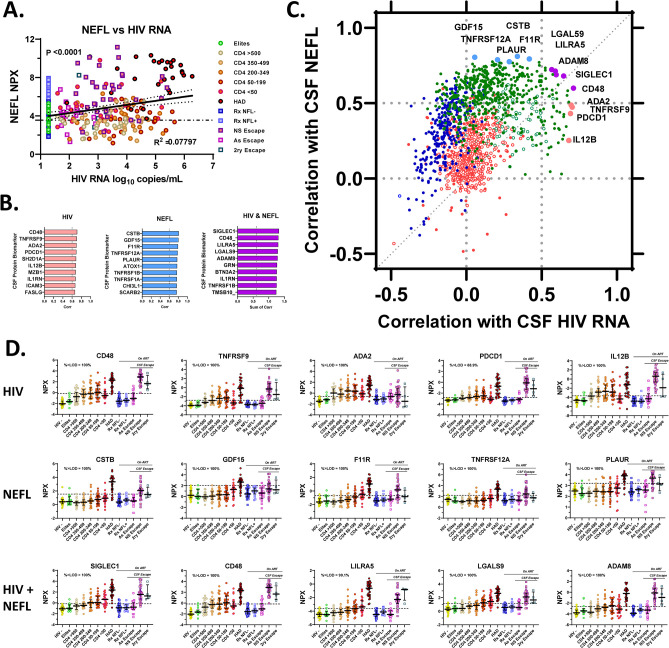
Olink CNS injury markers and the use of the Olink measurement of NEFL as a key biomarker
In an additional preliminary study, we examined the Olink measurement of NfL, that is included in the Explore 1536 platform, along with six other biomarkers of CNS injury in the panel. In several past studies we have shown that measurement of NfL in CSF (as well as in blood) serves as a valuable biomarker of active CNS injury in HIV-1 infection [66,68], just as it does in head injury, subarachnoid hemorrhage and a number of neurodegenerative conditions [65,148]. Because prior CSF NfL measurements were only available from a portion of the CSF specimens included in this study (209 of the 307, 71.3 percent), we evaluated whether the Olink measurements which are available for all specimens would be suitable for use as background variable in the main analyses of this study. [NB: In this report, we use the gene name, NEFL, when referring to the Olink measurement of the protein, and the more common abbreviation, NfL, when referring to the protein more generally and to results of measurements using other, conventional immunoassays.]
S2A Fig shows that the profile of Olink measurement of NEFL changes across the subject groups is very similar to that obtained with the NfL ELISA immunoassay performed in the subset of the sample shown earlier (comparing S2A Fig to Fig 1O). In fact, the results of the two assays are highly correlated (R2 = 0.8790) (S2B Fig). These comparisons thus support the use of the NEFL results as a principal objective marker of CNS injury in the subsequent analyses. We do not age-adjust the NEFL values in this analysis because we have not age-adjusted any of the other Olink protein results and do not have sufficient data to validate age adjustment for the Olink NEFL results. The mean NEFL values in the RxNFL+ group were higher than the RxNFL- group, indicating that the earlier immunoassay measurements in the former group were not simply laboratory errors but reflected true elevations of the CSF concentration and likely genuine axonal injury, albeit of uncertain source and meaning.
The Olink Explore 1536 platform includes several other markers that have been used in previous studies to assess CNS injury in HIV-1 infection and other neurodegenerative settings (S2C–S2H Fig). Of these, CSF CHI3L/YKL-40, TREM2, GFAP, KYNU and MAPT (all P <0.0001 with R2 = 0.6005, 0.5622, 0.4545, 0.3690 and0.3471, respectively) warrant further study as useful complements to NEFL in assessing the breadth and character of CNS injury as discussed in the legend to this figure. Of note, the elevation of CSF NEFL in the RxNFL+ group was not echoed by elevations of these other neural injury markers, adding further uncertainty to the biological meaning of the CSF concentration increases of this axonal protein in this group.
Main Olink CSF protein analyses
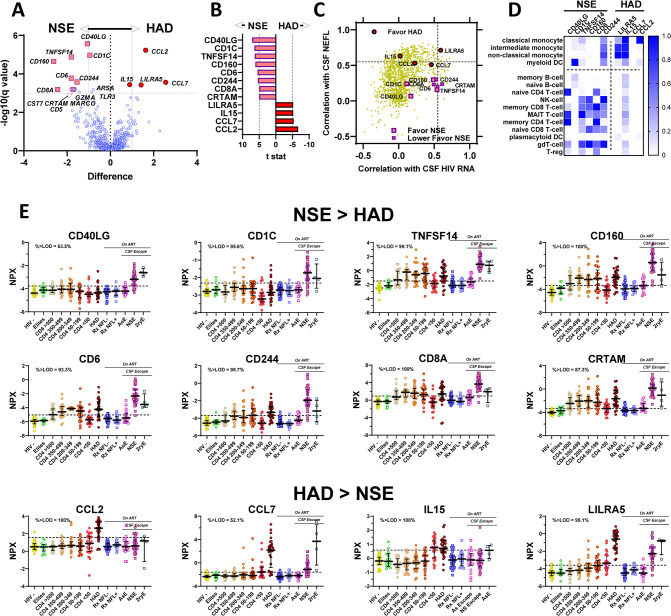
The following three main analyses explore facets of the full dataset of the measured CSF proteins as they varied over the course of systemic infection, development of HAD, successful treatment and CSF escape as represented by the specimen groups assembled for this study. The first of these analyses provides a broad introduction to the overall CSF protein changes across the study groups. The second examines the relationships of these protein changes to two major variables of CNS HIV-1 infection and its impact: CSF HIV-1 RNA concentration (as an index of CNS infection) and CSF NEFL concentration (as a measure of active CNS injury). The third compares CSF proteins in two forms of HIVE: HAD presenting in untreated individuals and NSE developing during treatment.
Overview of the CSF protein changes across the spectrum of chronic HIV-1 infection
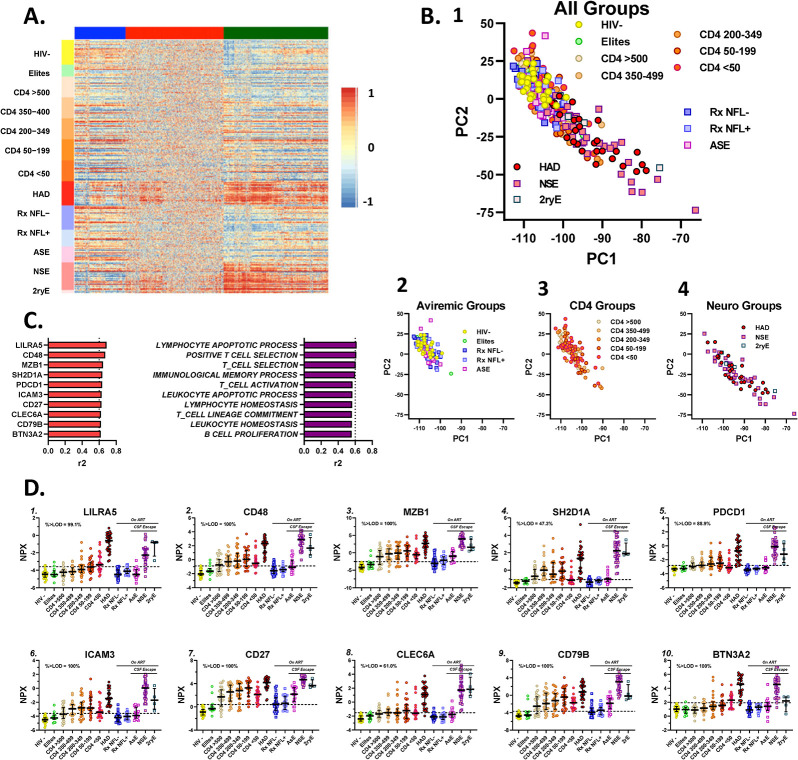
As an initial step in analyzing the broad contours of the protein changes during the course of chronic infection, we performed a hierarchical cluster analysis that empirically groups the measured proteins into three clusters with different spectra of protein changes across the full sample set. These are shown in Fig 2A as the BLUE, RED and GREEN clusters.
Fig 2. Overview of CSF proteins across subject groups.
The assembled panels depict some of the main features of the CSF protein changes across the set of subject groups. A. This hierarchical cluster analysis displays the full set of the Olink CSF protein measurements in a heatmap. The study groups (and the individual CSF samples within each group) are arranged on the vertical axis, while the proteins are grouped across the figure after segregation into three clusters designated by colors at the top within BLUE, RED and GREEN columns of the measurements of individuals proteins in each sample. The heatmap relative scale from -1 (blue) to 0 (yellow) to 1 (red) is shown to the right of the main figure. The individual proteins are color-scaled according to their relative concentrations. The most conspicuous gradations in scale are within the GREEN cluster (right column) in which many samples from the HAD group and the two symptomatic escape groups (NSE and 2ryE) stand out with higher (red) concentrations compared to the generally lower concentrations (blue and yellow) in the non-viremic individuals (HIV-, elites, and RxNFL+ and RxNFL-) with some mixed colors over this spectrum in the CD4-defined groups. By contrast, the RED cluster (middle column) showed a more even and mottled spectrum except for some of the proteins shown on the left side of this cluster in which there was red predominance in the HAD and two symptomatic escape groups similar to, but less pronounced than, in the GREEN cluster. The changes among groups in the BLUE cluster (left column) are less distinct but with some low (blue) concentrations in the CD4 <50, HAD and two symptomatic escape groups (NSE and 2ryE), and higher concentrations in the HIV- group, i.e., showing some patterns that are opposite to (or inverted in comparison to) those in the GREEN cluster. Overall, the HAD and two symptomatic escape groups accounted for the greatest concentration differences from the controls among the proteins, most prominently in the GREEN cluster. B. This PCA analysis using results of all the measured CSF proteins to assess effects of the overall CSF measurements on individual subjects and their groups provides a different perspective on the protein changes across the groups. 1. This panel includes the results from all the groups and shows a ‘gradient’ downward and to the right of the panel associated with HIV-1 systemic disease progression (as indicated by falling blood CD4+ T-cell counts) and, more particularly, with the symptomatic neurological disease groups, so that individuals with HAD and NSE reach farther in this direction. Because of the superimposition of many of the individual in this panel, three smaller panels (Fig 2B2-4) separate these groups for better visual definition of these changes. 2. the aviremic groups including the HIV-, the largely superimposed elites, treated-suppressed (RxNFL- and RxNFL+) and the ASE group are all grouped together with overlap in the upper left of the plot; 3. the five CD4-defined groups are also largely clustered in the same upper left area but with some individuals ‘moving’ down and to the right as immunosuppression advanced with lower blood CD4+ T-cell counts; 4. the HAD and two symptomatic escape groups (NSE and 2ryE) showed some overlap with the other groups but with further movement down and to the right (higher PC1 and lower PC2 values), again consistent with these neurological groups importantly ‘driving’ the main group differences in the aggregate CSF protein changes across the large cohort sample. Thus, these groups, defined clinically, are also, not surprisingly, separated in this analysis by the changes in their CSF proteomes. C. Proteins and pathways influential in distinguishing subject groups. The two bar plots show the proteins and pathways that most influenced the overall differences among the subject groups. The left bar graph lists the most influential proteins, all 10 of which are markers of inflammatory processes. The right bar graph lists the most influential defined pathways extracted from the protein changes, and again their designations emphasize that they identify inflammatory processes, including several involving T-cells. While, in part this may reflect the large number of inflammatory proteins that were included in the Explore 1536 panel, many with intersecting or overlapping functions, it also clearly shows that changes in inflammatory profiles largely determine the contours of the overall measured CSF proteome as systemic disease progressed and, particularly, as overt CNS disease with HIVE developed. D. Influential protein patterns across study subjects. These plots show the subject group concentrations of the 10 most influential proteins (from the left side of Fig 2C above) across the subject groups using the color schema introduced in Fig 1 and used throughout this report. This includes the color symbols, medians and IQR bars, and the dashed horizontal line that indicates the mean + 2 SD of the HIV- control group in this study as an estimate of the upper limit of these variables and aids as visual reference across the groups. Protein concentrations are expressed in log2 NPX units with varying scales. The plots show the prominence of the increases in the HAD and NSE groups and their dominating influence; these two groups had the highest concentrations of all 10 of these CSF proteins, with lower and more variable patterns of elevation in the CD4-defined and other groups. As annotated in the panels, of the 10 listed proteins, the measurements in six were all (100%) above the LODs for the assay. The exceptions included LILRA5, SH2D1A, PCDC1, and CLEC6A in which 99.1%, 47.3%, 88.9% and 61.0%, respectively, of the proteins were above the LODs (shown in the individual panels here as in subsequent figures of protein patterns across the subject groups). These influential proteins are all within the GREEN cluster in Fig 2A and are listed in the Glossary of Featured Proteins in Fig 2 included in S1 Appendix with brief functional descriptions extracted from UniProt database (https://www.uniprot.org/uniprotkb?query=*) [140], along with comments on selected features of the individual panels.
The initial division into three clusters in this figure provides a starting point for identifying proteins and groups of proteins with similar or contrasting overall protein profiles. As detailed in the legend to Fig 2, the widest and most distinct gradations in protein concentrations are grouped in the GREEN cluster with the conspicuously high protein concentrations (at the red and orange end of the scale) in the HAD, NSE, and 2ryE groups. These contrast with the lower concentrations (in the blue or blue mixed with yellow bands) in the aviremic HIV-, elites, and treatment suppressed (RxNFL- and RxNFL+) groups, and the more heterogeneously colored bands in the CD4+-defined groups. Similar, though less distinct and less consistent, gradations are also present on the left side of the RED cluster column, while the remaining two-thirds of the RED group show more even concentrations (and thus narrower concentration differences) through the full range of specimens. While some of these were likely proteins that were not altered over the course of infection, others are proteins with concentrations outside of the levels of measurement of the Olink assay, mainly below the LODs (see below). The BLUE cluster shows more mixed gradations, including a substantial number of proteins with seemingly ‘inverted’ concentrations—i.e. with lower concentrations in the HAD and NSE groups and in the lower CD4 defined groups than in the HIV- controls. These initial similarities and contrasts bear further examination within smaller groupings and subdivisions of these clusters in the future.
The principal component analysis (PCA) (Fig 2B1–2B4) shows the impact of the overall CSF protein changes on separating the individual participants and their groups. The virologically undetectable PLWH (Elites, Rx NFL- and NFL+, and ASE groups), along with the HIV- control group, all largely overlapped in this figure. The CD4-defined groups also overlapped with these same groups, but with some individuals with lower CD4 counts ‘moving down and to the right’ in the compound graph and again seen more clearly in isolation (Fig 2B3). Most notably, the HAD, NSE and 2ryE groups ‘migrated’ still further in this same direction with greater separation of many of the samples from the other groups (Fig 2B4) consonant with the higher concentrations of many CSF proteins in these groups. Given the conspicuous elevations of many proteins in the HAD, NSE and 2ryE groups, one might ask why the separation of subjects in this PCA is not greater. In part this may relate to inclusion of all measured proteins in the analysis, including those that show little (RED proteins in Fig 2A) or even opposite protein gradients (some of BLUE proteins in Fig 2A), that might ‘counterbalance’ or obscure the distinct group differences and reduce the separation exerted by the proteins in the GREEN group in Fig 2A).
Together the cluster analysis heat map and the PCA provide two consonant perspectives on the same CSF protein changes: the groups identified in the heat map as having the highest concentrations of many CSF proteins (the HAD, NSE and 2ryE groups) are segregated furthest from the HIV- uninfected and neurologically asymptomatic groups in the PCA analysis. Thus, these symptomatic neurological conditions have a dominating effect on the CSF concentrations of many proteins that, in turn, results in segregation of these neurological subject groups from those without neurological disease based on empirical CSF protein analysis.
Fig 2C lists the 10 proteins and 10 pathways that exhibited the most significant differences in CSF proteins across groups. Most of these are inflammation-related proteins, and the defined pathways involve lymphocyte-related functions as their names imply. These proteins are associated with T-cell, B cell and NK cell functions. Thus, although the clinical settings of two types of neurological injury had the greatest impact on the separation of the proteins, it was the inflammatory responses in CSF in these settings that dominated their impact among the measured proteins. Fig 2D shows the group profiles of the concentration changes across the subject groups of the 10 highest correlating proteins listed in Fig 2C. While the overall patterns varied, the most conspicuous common feature is the high protein levels in the HAD and NSE groups (highest for the former in some, but in this group of proteins more frequently highest in the latter). These proteins are all from the GREEN group in the cluster analysis in Fig 2A, and their profiles again emphasize that the two neurological disease groups are the important drivers of the greatest CSF protein differences in the sample set. These graphs also introduce other prominent features of the CSF protein spectra across subject-group, including the two patterns of CSF biomarker change over the course of CD4+ T-cell decline in the five untreated CD4-defined groups introduced earlier:
The lymphoid type, with highest levels in the CD4 50–199 or 200–350 groups set off by lower levels in the CD4 >500 and CD4 <50 groups (and most notably the latter) in the ‘inverted U’ pattern discussed earlier in the context of the HIV-1 RNA and WBC concentrations. This is the most frequent pattern in this group of proteins.
the myeloid type, with progressive increases leading to highest levels in the CD4 <50 group as noted earlier with CSF neopterin, is less frequent but exemplified here with the LILRA5 and BTN1A2 proteins at the upper left and lower right of this panel.
Although these ten proteins provide examples of the highest correlations, the PCA segregation of the subject group involves the entire set of measured proteins, including not only those that determined the groups separations but also those that ‘pulled’ them together because of limited protein concentrations differences. Also, while a number of these proteins share features with the lymphoid profile and two with the myeloid profile, there are also variations within these individual profiles that bear focused and more nuanced analysis in the future.
These data also illustrate the limited view provided by most previous studies, including our own, that measured only a few selected inflammatory mediators [2,45,75,82,149]. The results clearly underscore both the complexity and the broad participation of many proteins, including particularly many inflammatory proteins, in response to CSF/CNS infection and CNS injury. The findings, of course, do not distinguish which proteins contribute to CNS injury rather than simply respond to it. Nor do they distinguish which inflammatory proteins are involved in initiating and sustaining or determining the magnitude and character of the group inflammatory profile. These, and other, crucial mechanistic issues must be inferred and synthesized from the known functions of the proteins and their relative responses in the different clinical groups. In this report we focus on the empirical findings and leave a more fully synthesized mechanistic construction to future extensions of these analyses.
Of note, at least three of these proteins show mild elevation in the AsE group, indicating the low-level inflammatory responses in this setting that warrant future focused attention to this group. Further, while the HIV-, aviremic elites and the two treated groups (RxNFL- and RxNFL+) overlap in this analysis, it will be of interest to examine these groups further to identify possible distinctions among these groups to determine to what extent and how endogenously controlled (in elites) and therapeutically controlled infections may differ. We have not included these and other analyses in this introductory report, but plan to pursue these and other studies in the future.
CSF protein relations to CNS injury (CSF NEFL) and infection (CSF HIV-1 RNA)
As a step in dissecting the forces underlying the changes in CSF proteins across the subject groups, we examined the relations of CSF proteins to two major variables that can be viewed as indicators of key pathogenetic vectors involved in these changes: 1. CSF HIV-1 RNA concentrations as an index of CNS HIV-1 exposure/infection (encompassing entry, production and replication, with the latter two likely the main presumed drivers of CNS injury when located within the CNS parenchyma), and 2. CSF NEFL concentrations as a measure of the state of active CNS injury at the time of sampling. CSF HIV-1 RNA is an ambiguous indicator of CNS infection because it can reflect a variable mixture of virions derived from leptomeningeal and brain parenchymal sites of infection, depending on the clinical setting. During neuroasymptomatic infection in the CD4-defined groups (particularly those with CD4 T-cell counts >50 cells per μL), infection is largely meningeal, while HAD and NSE involve deeper parenchymal infection (HIVE). However, the contributions from these sources of origin cannot be distinguished by simple measurement of HIV-1 RNA in CSF. In Fig 3 we examine the relationships between the various CSF proteins and CSF NEFL and CSF HIV-1 RNA concentrations.
Fig 3. CSF proteins in relation to correlations of CSF NEFL and HIV-1.
This figure explores the relationships of the CSF proteins to two major variables (or pathogenic vectors) of HIV-1-related CNS disease: CSF HIV-1 RNA, an ambiguous index of CNS infection, and CSF NEFL (NfL measured by Olink in the Explore 1536 panel), an objective measurement of active CNS injury. A. Correlation of NEFL with CSF HIV-1 RNA. This plot shows a significant (P<0.0001), but relatively weak correlation (R2 = 0. 0.07797) between NEFL and CSF HIV-1 RNA by linear regression. Among the features underlying this limited correlation are the high CSF RNA values without elevation of NEFL in the untreated CD4-defined groups, the elevations of CSF NEFL in both the treated NSE group despite their relatively lower CSF HIV-1 RNA levels (related to the partial suppression from ART) and the untreated HAD patients with high levels of HIV-1 RNA. Separate analysis of the untreated and treated groups showed little improvement of the correlations (both also with P< 0.0001, but with R2 values of 0.1699 and 0.1373, respectively). Analysis of HAD patients alone showed p = 0.1709 (R2 = 0.07087). B. High correlating individual proteins. The three bar graphs show the 10 CSF proteins with: the highest correlations to 1. CSF HIV-1 and 2. CSF NEFL; and 3. the sum of the CSF HIV-1 and CSF NFL correlations. For 27 of the listed proteins, the measurements are above the LODs. Exceptions include SH2D1A in which only 47.3% of the proteins are above the LODs, and consequently this protein is omitted from further descriptions and the plotting of the proteins across subject groups below. The other two exceptions are PCDC1 (88.89% above LODs) and LILRA5 (99.1 above the LODs) which are included in the further discussion and illustration. Notably some of these proteins were also previously identified as strongly impacting the overall segregation across groups as defined earlier in Fig 2. C. Correlations of the full set of CSF proteins with CSF NEFL and HIV-1 RNA. This figure includes a total of 1662 proteins, i.e., all of the Olink-measured proteins except NEFL, plotted according to their correlations with CSF NEFL and CSF HIV-1 RNA. It provides a broad overview of the measured proteins in the context of these two important pathogenetic vectors. Each protein symbol is colored according to their grouping in the three clusters (GREEN, BLUE and RED) defined earlier in the hierarchical cluster analysis (Fig 2A) with the solid symbols showing the proteins with ≥60% of the proteins above their LODs and the open symbols showing the proteins with <60% of proteins above their LODs. Exceptions to this symbol color coding are the top five proteins identified in the three groups in Fig 3B for which the symbols have been individually enlarged, color-coded using the bar colors in Fig 3B (all were originally identified in the GREEN cluster), and labelled with their gene names. These highly correlating proteins are located at the extremes on the top, right and right upper sections of the plot (CD48 appears in both the HIV-1 and HIV-1 + NEFL groups and is colored according to the dual correlation). The dotted grid lines mark the 0 and 0.5 correlation levels for both the Y and X axes. Overall, the GREEN cluster proteins populate the highest NEFL and HIV-1 RNA correlation areas with a far larger number in the ≥0.5 of CSF NEFL correlation (Y axis) (375 proteins) than in the ≥0.5 area of CSF HIV-1 RNA correlation (X axis) (55 proteins). Twenty-two of these proteins are in the shared area in the upper right with correlations ≥0.5 for both CSF NEFL and CSF HIV-1 RNA. A minority of the GREEN proteins had <60% of their measurements above the LODs as indicated by the open green circles that exhibit NEFL correlations <-0.5 and CSF HIV-1 RNA correlations between 0.25 and 0.5. None of the 294 BLUE-clustered proteins are in the ≥0.5 HIV-1 RNA correlation area, and indeed most exhibit negative correlations with HIV-1 RNA; 52 of the BLUE-clustered proteins are in the ≥0.5 NEFL correlation area. Only a small number of the BLUE-clustered proteins show <60% of measurements above the LODs. Of the 566 RED-clustered proteins 10 are in the ≥0.5 correlations with NEFL and none correlate with HIV-1 RNA ≥0.05. These RED-clustered proteins thus are concentrated along the vertical line correlation designating a 0.0 correlation with CSF HIV-1 RNA. Additionally, many the RED-clustering proteins have <60% of proteins above the LOD (open symbols), likely contributing to their lack of correlations with either CSF NEFL or CSF HIV-1 RNA (with most grouped along the X axis at 0 correlation with CSF HIV-1 RNA). This figure with the ‘cloud’ of protein placement in relation to the two pathogenetic vectors emphasizes that the correlations of most of the Olink-measured proteins with CSF NEFL and HIV-1 RNA are largely ‘dissociated’. Only a limited number lay along the diagonal dotted line delineating equal correlations with the two major disease variables. Additionally, the much larger number of proteins correlating with NEFL at levels above 0.5 than with HIV-1 RNA, emphasizes the major influence of CNS injury on the CSF proteome. This is consistent with the strong effect of HAD and NSE on CSF proteins as can be seen by the examples below and with many of the proteins identified in other sections of the study. By contrast, high HIV-1 RNA levels in some of the CD4-defined groups with normal CSF NEFL values also contributes to this dissociation. D. Patterns of high-correlating proteins across subject groups. This Panel shows the patterns of CSF protein concentrations of the top five proteins in the three categories identified in Fig 3B. Again, the dashed horizontal lines are at the mean + 2SD of the HIV-1- group as a visual reference for comparison of group value distributions. The top row shows the five proteins with the highest correlations with HIV-1 RNA. Here the highest protein concentrations vary between the HAD and NSE groups while the CD4-defined groups exhibit the lymphoid pattern with a reduction in the protein levels in the CD4 <50 group compared to the CD4 50–199 and 200–350 groups. This is not surprising because this is the pattern of the CSF HIV-1 RNA concentrations and CSF WBC counts in these groups (see Fig 1I and 1K). The patterns in the middle row of proteins with highest correlations with NEFL exhibit two major features: highest concentrations in the HAD group and a gradual increase across the CD4-defined groups with highest values within these in the CD4 <50 group, i.e., with the myeloid pattern. This circumstantially supports the association of CNS injury with myeloid cell inflammation. The final row with highest combined HIV-1 and NEFL correlations shows a mixture of the two patterns just outlined: variability in whether HAD or NSE had highest concentrations and in the presence of either the lymphoid or myeloid patterns in the CD4-defined groups. The identified proteins in Fig 3D are listed in S1 Appendix with brief functional descriptions extracted from UniProt database (https://www.uniprot.org/uniprotkb?query=*)[140].
As shown in this figure, CSF NEFL and HIV-1 RNA concentrations are, themselves, not strongly intercorrelated across the full spectrum of study subjects (Fig 3A). Even after segregation of subsets of the groups (untreated and treated groups; and HAD and NSE groups in isolation), correlations between CSF HIV-1 RNA and NEFL remained weak. Fig 3B shows the 10 proteins that separately correlated best with CSF HIV, CSF NEFL, and with both of these variables taken together. For a broad overview of the protein relations to CNS injury and infection, we plotted all of the Olink proteins in relation to their correlations with CSF NEFL and HIV-1 RNA (Fig 3C). Proteins from the GREEN cluster dominate the higher correlations with both NEFL and HIV RNA, and a higher proportion of the proteins correlate with NEFL than with HIV-1 RNA at levels above 0.5 (designated by horizontal and vertical dotted lines). This is consonant with the protein gradients noted earlier for the GREEN cluster showing highest concentrations in the HAD and NSE subjects and the dominant effect of these two overt neurological conditions on elevated CSF protein concentrations.
By contrast, proteins from the RED cluster are mainly located in the center of Fig 3C, with HIV-1 RNA correlation values centering around zero and relatively low NEFL correlations (below 0.5) except for a few higher outliers. Many of the RED proteins returned >60% of measured values below their LODs, likely contributing to their frequent lack of correlation with either HIV-1 RNA or NEFL. Interestingly, the BLUE cluster is dominated by proteins with negative HIV-1 RNA correlations and NEFL correlations below the 0.5 correlation level, though with several showing negative correlations with CSF HIV RNA despite >0.5 correlation with NEFL, mingling with some of the GREEN cluster proteins in this location. This figure also helps to identify CSF proteins with similar functional properties (or at least correlations) and can provide a useful initial map for future studies exploring functional relationships among the CSF proteins in HIV-1-related neuropathogenesis.
In Fig 3C the five highest correlating CSF proteins identified in the three bar graphs in Fig 3B are highlighted with color-coding of their locations in the plot. All of these are inflammatory proteins, and, in a broad sense, their locations emphasize the major effect of CNS injury on CSF proteins, including particularly inflammatory proteins. However, the plot also shows that HIV-1 RNA measured in CSF is associated with protein changes that had a variable, often dissociated, relation to injury. Indeed, the highest correlations with CSF HIV-1 RNA are infrequently highly correlated with the extent of CNS injury as measured by CSF NEFL and, vice-versa, proteins correlating strongly with neuronal injury are, in general, relatively weakly correlated with HIV-1 RNA. These dissociations also provide exploratory maps for examining pathogenic relationships, including defining inflammatory increases with weak and strong relationships with CNS injury and CSF HIV-1 RNA.
Fig 3D shows the group CSF profiles for the five CSF proteins with highest correlations with HIV-1 RNA, NEFL and HIV-1 + NEFL, respectively, that were identified in Fig 3C. The patterns of change in the proteins in the first two groups show consistent differences. Those with high HIV-1 RNA correlations (top row) exhibit the lymphoid pattern across the CD4+ T-cell-defined groups while the NEFL-correlating proteins (second row) more closely followed the myeloid or perhaps neuronal pattern of change across the CD4-defined groups with a gradual or more abrupt increase to highest levels in the CD4 <50 group. There are also evident variations in these patterns among these proteins; for example, PLAUR on the far right more closely resembles the NEFL’s neuronal pattern. In the two upper rows of graphs in Fig 3D the protein levels are more often higher in the HAD group than in the NSE group, but not in all. The combined HIV-1 and NEFL examples (third row) exhibit a mixture of the patterns seen in the other two groups (e.g., CD48 with the lymphoid and LILRA5 and LGALS9 with the myeloid patterns). Of note, the RxNFL+ group does not show elevation in the inflammatory proteins that associate with NEFL, suggesting that NfL elevations in this subgroup are likely due to different processes than those involved in the two forms of HIVE. This is also in agreement with the result of the other neural injury marker measurements discussed earlier and shown in S2 Fig.
Overall, these results emphasize that the correlations of CSF proteins with CSF NEFL and HIV-1 RNA are frequently dissociated, and that, indeed, the proteins with the strongest correlations with CSF NEFL are more commonly associated with relatively weak CSF HIV-1 RNA correlations and, conversely, strong correlations with CSF HIV-1 RNA are frequently accompanied by relatively weaker correlation with CSF NEFL. Additionally, high correlations with NEFL associate with myeloid (and perhaps neuronal) patterns of change across the CD4-defined groups, while high correlations with HIV-1 RNA associate with the lymphoid pattern of change across these groups. From this, we can speculate that the proteins correlating strongly with CSF HIV-1 RNA are mainly from T-cells reacting to or even enhancing local HIV-1 production or replication, while the proteins correlating strongly with NEFL are either effecting or reacting to CNS injury and involve virus populations that evolved to grow in CNS myeloid cells, though we have not directly established these immune response-virological linkages and this first analysis does not distinguish between inflammatory biomarkers that may have different levels of importance in HAD and NSE.
CSF proteins differences between HAD and NSE
In addition to characterizing the broad changes in the CSF proteome across the defined clinical groups, this dataset can be used to explore features that distinguish individual clinical groups or sets of groups from each other. As an initial example of this application, we next compared the CSF proteins of the two groups with major HIV-1-related CNS injury: the HAD and NSE groups. Among the distinguishing background features of these two groups with HIVE are: i) the absence and presence of treatment (with full or partial plasma viral suppression); ii) consequent difference in blood CD4+ T-cell counts (median of 108 cells per μL in HAD, and 425 cells per μL in NSE); iii) difference in CSF HIV-1 RNA levels (median of 5.27 log10 copies per mL in HAD and 3.25 log10 copies per mL in NSE); iv) CSF WBC counts that were elevated in both but twice as high in NSE; and v) different CSF NEFL concentrations (higher in HAD). Previous studies also have shown that the pathology and HIV-1 genotypes/phenotypes of CSF isolates also usually differ. HAD is typically a multinucleated-cell encephalitis [129,150,151] involving myeloid cell infection with M-tropic viruses [152–154] developing in untreated individuals with advanced systemic infection, while NSE is generally a T-cell mediated disease with notable CD8+ T-cell infiltration pathologically [102] and T-tropic HIV-1 infection [155]. NSE usually occurs in the setting of limited CNS penetration of one or more antiretroviral drugs along with resistance to other drugs [94–96,101,103,104,131]. Fig 4 compares the CSF proteins of these two types of HIVE.
Fig 4. CSF proteins favoring NSE vs HAD.
This figure examines CSF protein differences between NSE and HAD. A. Comparison of CSF proteins in NSE and HAD groups. Both of these conditions are forms of HIVE with elevations of multiple CSF proteins. For the most part, the protein elevations do not differ significantly, and this volcano plot shows a high degree of protein similarity (or lack of significant differences) within the context of the full complement of measured proteins (identified by simple blue symbols) on each side of the central vertical axis. However, there are a few identified protein differences, indicated by the enlarged labelled symbols formatted using the NSE and HAD symbols. The remaining panels of the figure focus on these distinguishing proteins. B. CSF proteins distinguishing NSE and HAD subject groups. Proteins identified by t stat of ≥ +/- 5 are the same those singled out in Fig 4A volcano plot, with eight proteins favoring NSE and four favoring HAD. C. NSE and HAD distinguishing proteins in the context of their correlations with CSF NEFL and CSF HIV. This panel shows all the measured Olink proteins (except NEFL) in the context of their correlations with CSF NEFL and CSF HIV RNA using the same format as Fig 3C, but, for clarity, with a neutral yellow color for the full array of proteins rather than the GREEN, BLUE and RED designations from the hierarchical cluster analysis. The significant proteins identified in the volcano plot and t stat above (Fig 4A and 4B) are indicated by enlarged symbols. All are from the GREEN group of the cluster analysis. The proteins favoring both neurological conditions are similarly variable across a range of correlations with CSF HIV-1 RNA but located in different vertical strata with respect to NEFL correlation. Those favoring HAD are in a higher range of correlation with NEFL than those favoring NSE. This may relate, at least in part, to the generally lower levels of NEFL in NSE than HAD (Fig 2A). D. Cellular associations of proteins favoring NSE and HAD. This simple presentation draws on literature sources describing the cell associations of 10 of the proteins identified in Fig 4A and 4B, https://www.proteinatlas.org/ and [156] with the darkness of the square indicating strength of the cell relations. The proteins listed as favoring NSE are largely associated with lymphocytes (B, T and NK lineages), while those favoring HAD associate with myeloid cells, consistent with a different balance of inflammatory cells in these two forms of HIVE. E. Different patterns of high-correlating proteins in NSE and HAD across subject groups. The format of the graphs in this panel is the same as in previous figures showing Olink protein concentrations across the subject groups. The upper two rows plot the protein concentrations of the 8 proteins favoring NSE while the bottom row plots the proteins favoring HAD as identified in Fig 4A and 4B above. In the upper two rows, the median concentrations in the NSE group are greater than those of the comparable HAD group which are either normal or near the highest values among the CD4-defined groups. The CD4-defined groups exhibit the lymphoid pattern with varying levels of increase in the middle brackets. The bottom row shows the patterns of the four proteins that favor HAD. Most notably, the values of HAD group are higher than those of NSE group, and the CD4-defined group patterns are either flat or show an increase in the CD4<50 group above the groups with higher CD4+ T-cell counts, closest to the myeloid or perhaps neuronal patterns, but with clear variation in the magnitude of differences of the CD4<50 from the other CD4-defined groups. The dashed horizontal line in each panel again designates the mean +/- 2 SD of the HIV- control group for visual reference. The identified proteins in Fig 4E are listed in S1 Appendix with brief functional descriptions extracted from UniProt database (https://www.uniprot.org/uniprotkb?query=*) [140].
This figure compares the CSF proteins in NSE and HAD. While the impetus for this comparison was largely to identify protein differences, in fact, we found that the great majority of the CSF proteins were not significantly different in this analysis and that they clustered at the base of the Volcano Plot in Fig 4A. On reflection, the lack of major differences may not be surprising because both conditions are highly inflammatory types of HIVE, and whatever the root cause of virus- or inflammatory-related neuronal injury, the accompanying responses may be similar However, there are some distinctions as shown by the labelled proteins in Fig 4A and the bars in Fig 4B that identify several proteins that are more prominent in one or the other condition. Fig 4C, examining the correlations of these proteins with the two ‘pathogenic vectors’ introduced earlier shows that these proteins both stretch across the spectrum of correlations with CSF HIV RNA but appear in different strata with respect to correlation with CSF NEFL so that those favoring NSE show lower and those favoring HAD have higher correlations with this axonal injury protein. In part, at least, this likely relates in some way to the lower concentrations of CSF NEFL in the treated NSE patients, but Fig 4D using literature data suggests that these proteins relate predominantly to different cells, NSE to lymphoid and HAD to myeloid inflammatory processes. This is also consistent with the profiles of these proteins across the subject groups shown in Fig 4E in which those favoring NSE not only show higher concentrations in the NSE group than that of HAD, but also show the lymphoid pattern while those favoring HAD exhibit a myeloid or neuronal pattern across the CD4-defined groups. Thus, overall, the inflammatory components of these two types of HIVE exhibit considerable similarity, but also some discernable differences.
These initial comparisons are all consistent with the neuropathological differences between the two forms of HIVE, with HAD typically a myeloid-cell inflammatory and M-tropic virus-dominated process, and NSE mainly featuring lymphocytic infiltration and infection [102,157] in association with infection by presumed T-tropic viruses. To what extent these CSF proteins simply reflect reactions to the CNS injury or, conversely, importantly participate in driving it, remains uncertain. Are the main drivers of CNS injury in these two conditions similar but the inflammatory reactions vary because of differences in the background immune system reactive capabilities, or are the mechanisms of injury more fundamentally different?
Conclusions and future directions
This study generated an abundance of novel data related to the robust and complex changes in CSF proteins over the course of HIV-1 infection, including after treatment. The full dataset is posted so that other investigators can now join in extending the analysis of these data. This initial report is largely aimed at introducing the study and its rich dataset. It pursues an agnostic, largely empirical approach to discovering and outlining the contours of CSF protein changes over the full course of chronic HIV-1 infection. Future studies now need to use these data to examine mechanisms and unravel underlying pathogenetic issues. These might be approached from at least two directions: 1. Continued comparisons of subject groups or subgroups to characterize and understand the distinct features of each and how they differ from other groups in order to map transitions and salient differentiating features of the various stages of infection and injury; and 2. Informed analysis of the changes in particular proteins, in related proteins and in established protein networks and pathways to examine the mechanisms underlying the changing CSF proteome features at these different phases of systemic and CNS HIV-1 infection. CSF inflammation is a nearly ubiquitous feature through the course of infection, and likely a central pathogenetically facet of CNS HIV-1 infection. Its relation to both systemic infection and CNS injury are clearly key aspects of CNS infection and its consequences. Further definition and understanding of these CSF protein changes promises to provide enhanced understanding of the CNS pathobiology of HIV-1 infection and perhaps additional approaches to more precise diagnosis and to neuroprotective and neuro-mitigating therapies. This study also demonstrates the potential value of applying a similar approach, including extension to measurement of non-protein molecules, to discovery aimed at pathogenetic and diagnostic exploration of other infectious, inflammatory and degenerative neurological diseases.
Supporting information
This file contains a table that summarizes the salient background characteristics of the study groups.
(XLSX)
This appendix contains brief glossaries relating to the proteins identified in the figures.
(DOCX)
This figure examines the correlations among quadruplicate assays of three proteins and other selected features including QC warnings and LOD measurements of inflammatory molecules. The Olink Explore 1536 platform uses four individual assay plates (Cardiometabolic, Inflammation, Neurology and Oncology), each with sets of protein detection antibodies (two antibodies for each protein measured). As part of the quality control strategy, three of the proteins (CXCL8, IL6 and TNF) are measured on all four plates, i.e., in quadruplicate, using the same antibody pairs but in the context of different sets of background antibody pairs on each of these plates. S1 Fig examines the intercorrelations of the assay results and compares the patterns of study specimen profiles generated by each of the quadruplicate assays across the spectrum of the 307 study specimens (this preliminary exploration included all of the specimens, including the four miscellaneous samples omitted from the main analysis). Additionally, the plates are structured so that assay of each protein is divided into four component sets of measurements, each with a separately calculated level of detection (LOD). Examination of the three sets of quadruplicate measurements in S1 Fig provides an opportunity to concretely illustrate these methodological features which are annotated for every protein in the full data table (File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx DOI: 10.5061/dryad.x3ffbg7tv). A-C. Correlations among quadruplicate plate assay results. These panels include three montages, each with 12 graphs showing assay correlations among the quadruplicate assays with identification of the QC Warnings related to individual measurements and notations of assay LOCs. The format for the first set (CXCL8) is outlined in more detail that can then be applied to the other two sets of quadruplicates (IL6 and TNF), while the LOD notations are more prominent in the second two sets, and particularly in the third (TNF). A. Correlation of CSF CXCL8 measurements in quadruplicate assays. This panel shows the correlations of the results of four assays (one from each plate) of the CSF CXCL8 concentrations in NPX units. The assay result numbers and plate names in the panels without graphs refer to the protein results graphed on the X axis of the panels in that row, and also to the Y axis of the panels in the column, e.g., measurement 20153 was obtained on the Cardiometabolic plate and these results were used in the X axis of the other graphs in the (top) row and the Y axis of the graphs in the (left) column. The panels show both the close correlations of the results across the quadruplicate measurement and the variability of absolute results among the plates and indeed of the scales of results among plates. The graphing of the points with QC warnings shown in orange relate to the measurement shown in the column of the panel and to the protein on the Y axis (for example, the QC Warnings for the Cardiometabolic plate warnings for its CXCL8 assay are included in the three graphs below this label). With respect to the correlations, there are actually only 6 sets of correlations, but the other 6 panels show graphs of the same variable pairs mirrored across the diagonal of the panel names. These graphs show the QC Warnings of different assays (those of the columns). The six linear correlations performed across these results are all very tight with an overall mean R2 = 0.9821 +/- SD 0.0059. The “*” on the upper left box (20153 assay of CXCL8 on the Cardiometabolic plate) indicates that the results for the CXCL8 measurements from this plate were selected at random prior to analysis and used in the analyses presented in this report (and highlighted by gold-yellow background in File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx available on-line Dryad (DOI: 10.5061/dryad.x3ffbg7t). All of the QC Warning samples are within the body of other assay results. The panels do not show the LOD levels for these assays since all are well below the range of all the measurements. B. Correlation of CSF IL6 measurements in quadruplicate assays. This panel, showing the correlations among the four CSF IL6 assays (S1B Fig) is arranged in the same format as the CXCL8 panel discussed above and uses the same labelling rules. Again, the correlations among the 6 assays are high with a mean R2 mean of 0.9508 +/- SD 0.0142. The measurements with QC warnings are also again all well within the body of the overall data. The graphs also include the LODs for each of the four assays plotted on both the X and Y axes. These intrude on some of the lowest IL6 values but likely have little impact on the overall interpretation of the correlations or differences among the groups (see S1D Fig below). C. Correlation of CSF TNF measurements in quadruplicate assays. This panel is also arranged in the same format as the CXCL8 and IL6 results discussed above. However, the TNF measurement correlations among the quadruplicates are distinctly lower: the mean R2 was 0.7055. Additionally, two of the measurements with QC Warnings are outliers; this is most easily visualized in the two graphs in the Oncology plate results column (middle two subpanels on the right). Finally, the LOD plots show that a substantial number of the values are below the four LODs. The impact of this is also seen more clearly in S1D below. D. Subject group profiles of the three sets of quadruplicate CSF protein measurements across the study subject groups. This group of graphs displays the results of each of the 3 quadruplicate assays (CXCL8, IL6 and TNF in the top, middle and bottom rows of graphs, respectively) for all the subjects in this analysis. They are derived from the same measurements as the above panels but displayed according to the subject groups using the same format (including color coding of symbols and median +/- SEM error bars) for other group profiles in this report, as outlined in Fig 1. In each of these panels the horizontal dashed line is set at the mean + 2SD of the HIV negative controls for visual reference across the 13 groups while the four dotted lines indicate the four LOD values for the individual assay. The patterns of CSF protein changes across the groups are very similar for each of the quadruplicate measurements of the individual proteins (CXCL8, IL6 and TNF), including in relation to the HIV- control values, changes in medians across the CD4-defined groups as blood CD4 cells fell, elevations of HAD and NSE relative to each other (for example: modest elevations of CXCL8 with median values of NSE > HAD; elevations of IL6 in HAD with relatively low values for NSE; and nearly equal elevations of both HAD and NSE for TNF). While for each of the four replicates the relative changes in the subject groups are largely the same, there are also notable differences across some of the assays in the absolute NPX values and ranges. For example, the NPX ranges and values in the four panels of CXCL8, particularly between the right and left panels (Cardiology and Oncology results), differed. This is less marked for the IL6 repeats, although there are notable the differences in the HIV- medians in the left two panels of TNF results (Cardiology and Inflammation). For TNF, there also are differences in the mean values among the four measurements. These comparisons thus clearly illustrate that the NPX measurements report relative concentrations within each assay that can differ among the quadruplicates, yet show similar relative changes with respect to group comparisons. The figure also shows the four LOD values for each of the assays with dotted horizontal lines. In the case of CXCL8 (top row of four graphs), these are all well below all the sample values; thus, results of this assay are not impacted by low values below the LODs. For IL6 (middle row), a few of the lowest values are near or below the LODs, but this likely had little influence on the patterns of change or correlations with other biomarkers. By contrast for TNF (lowest row) the LODs are at or above the medians of the HIV- controls and several of the other groups with 25.7 to 69.2 percent of measurement results above the LODs for this protein. While the median HAD, NSE and 2ryE groups clearly show elevated TNF values in each assay, the lower values for most of the other groups that are near or below the LODs clearly interferes with interpretation and comparison of the overall pattern of change for TNF.
(PDF)
The figure shows the results of seven Olink Explore 1536 protein measurements that have been applied to assess CNS injury in other studies. In S2A and S2B-H Fig the format is the same as that for other CSF biomarker changes across the groups, including the horizontal dashed line that indicates the mean + 2 SD of the HIV- controls as a visual reference for comparison across group (or– 2 SD in the case of APP in S2D Fig in which the concentration changes in the PWH were negative). The percentage of measurement above the LODs for each of these is also listed, with all in a range that allows interpretation of the values across the subject groups. Following the nomenclature used in the Olink Explore 1536 platform, the proteins are designated by their gene names. This includes the Olink measurement of NfL. In this paper we use the abbreviation, NEFL, for the Olink results while using NfL as the abbreviation for general references to this protein and to the results of other assays, Including the UMAN ELISA used for the earlier measurements performed in the University of Gothenburg Laboratory of Neurochemistry that are presented in Fig 1O. The following sections briefly discuss each panel in the figure. A. The NEFL measurements generated in the Olink Explore 1536 exhibit a pattern of concentration changes very similar to that seen with the UMAN ELISA results presented earlier in Fig 1O. Greatest elevations are present in the HAD group, minor increases in the CD4 <50 group, and substantial elevations in the NSE group (though lower than in HAD). The Rx NFL+ group shows clear elevation with a median near the level of the HIV- mean + 2 SD dashed line. This pattern might be considered as the prototypic neuronal pattern of change across the sample group. It is similar to the myeloid pattern, perhaps distinguished by the flat concentration levels in the CD4-defined groups with blood CD4+ T cells above 50 per μL, along with elevation in the NSE. Whether the general elevation in the Rx NfL+ group should be considered a part of this pattern or is peculiar to NfL alone requires further understanding of this group. Clearly minor distinctions from the myeloid pattern require further analysis and validation. B. Correlation of CSF NEFL (Olink) with prior NfL measurements without age adjustment. The regression shows the high correlation (R2 = 0.8790) between the Olink NEFL and the prior Uman ELISA assay results in the subset of the subjects presented in Fig 1O but without age adjustment. In this comparison the high correlation is noted despite the earlier measurements having been performed in multiple separate laboratory runs over several years. As in S1 Fig comparing analysis of the quadruplicate repeated assays, the NEFL measurements with QC Warnings, shown by orange symbols, fall within the central body of the results along the regression line, further supporting the inclusion of measurements with QC Warnings in our analysis. C. MAPT (P10636). In this figure, only the HAD group exhibits a clear median increase with about half of the results above the 2 SD level of the HIV- control mean. The median of the NSE group is similar to that of the control group with a tail of about 25% of the samples falling above the 95% confidence limit. Hence, this protein appears to be a less sensitive marker of neural injury in HIV-1 infection. D. APP (P05067). Measurement of this protein in the panel shows only a modest decrease in median level in the HAD group and a seeming lower median in the 2ryE group. Thus, both MAPT and APP CSF measurements, which serve as indicators of Alzheimer’s disease pathology, are not sensitive to HIV-1-related CNS injury. E. GFAP (P14136). CSF GFAP is normal in most of the groups except for HAD and to a lesser extent in NSE with about 50% and <50% concentrations above the controls +2 SD line, respectively. Either GFAP is a relatively insensitive biomarker of astrocytic reactions in this setting, or astrocytic changes are a less prominent aspect of HIV-induced neuropathology than neuronal injury (as documented by CSF NEFL changes). F. CHI3L1/YKL-40 (P36222). CHI3L1/YKL-40 concentrations in CSF are distinctly elevated in HAD and NSE with an overall pattern similar to that of NEFL, but with an earlier median increase in the CD4-defined groups. Incidentally, the value in the Rx NFL+ group is similar to that of the Rx NFL- group, which might imply that if injury is present in the Rx NFL+ group, it is circumscribed, affecting axons but sparing other cellular elements. The small 2ryE group values are also elevated, presumably related to strong glial reaction in these individuals with a myeloid pattern in the CD4-defined groups. Overall, CHI3L1/YKL-4 appears to be a sensitive biomarker of HIV-induced pathology. G. TREM2 (Q9NZC2). In this study, there was an increase in TREM2 mean values in the lower CD4-defined groups and highest levels in the HAD group, but largely normal levels in NSE except for some elevated outliers possibly indicating a difference in their pathological substrates. This is consistent with its putative association with microglial activation and a myeloid predominance in HAD but not in NSE. The gradual increase in its concentration with falling CD4+ T cells, including the CD4 <50 group, also shows the myeloid pattern. This differences from CHI3L1/YKL-4, perhaps most notably the absence of elevation in the NSE group suggests that these two markers detect different cell changes, with YKL-40 importantly encompassing astroglial changes present in both HAD and NSE, while TREM2 largely indicates myeloid activation that is more prominent in HAD than in NSE. H. KYNU (Q16719). (Kynureninase). There was a prominent increase in HAD and lesser increase in NSE with little change from control in the other groups. The pattern in CD4-defined groups was perhaps ambiguous, but the overall median values of these groups were close to that of the HIV- controls. KYNY might be a useful marker of severe CNS injury. The highest correlations (P <0.0001) of these markers with NEFL included: CHI3L1 (R2 = 0.6005), TREM2 (R2 = 0.5622), GFAP (R2 = 0.4545) and MAPT (R2 = 0.3471).
(PDF)
Data Availability
The full dataset for this study is posted online to Dryad, 10.5061/dryad.x3ffbg7tv.
Funding Statement
This study was supported principally by NIH/NINDS research grants R01 NS094067 (RWP) and 3R01 NS094067-05S1 (RWP) and by the Swedish state under an agreement between the Swedish government and the county councils (ALF agreement ALFGBG-965885) (MG). Previous research grants supporting CSF specimen collections and background data assays included: P01 MH094177 (RS), P01 DA026134 (RWP Project PI), R01 MH081772 (SSS), R01 NS043103 (RWP), R01 NS37660 (RWP), R21 MH096619 (RWP); R01 MH062701 (RWP), R01 MH096619 (RWP), R21-MH083520 (RWP); Sahlgrenska Academy at University of Gothenburg (project ALFGBG-11067) (MG); and the Swedish Research Council (project 2007-7092.3) (MG). The funders had no role in the study design, data collection, decision to publish, or publication of the manuscript.
References
- 1.Davis LE, Hjelle BL, Miller VE, Palmer DL, Llewellyn AL, Merlin TL, et al. Early viral brain invasion in iatrogenic human immunodeficiency virus infection. Neurology. 1992;42(9):1736–9. Epub 1992/09/01. doi: 10.1212/wnl.42.9.1736 . [DOI] [PubMed] [Google Scholar]
- 2.Gisslen M, Chiodi F, Fuchs D, Norkrans G, Svennerholm B, Wachter H, et al. Markers of immune stimulation in the cerebrospinal fluid during HIV infection: a longitudinal study. Scand J Infect Dis. 1994;26(5):523–33. Epub 1994/01/01. doi: 10.3109/00365549409011810 . [DOI] [PubMed] [Google Scholar]
- 3.Ellis RJ, Hsia K, Spector SA, Nelson JA, Heaton RK, Wallace MR, et al. Cerebrospinal fluid human immunodeficiency virus type 1 RNA levels are elevated in neurocognitively impaired individuals with acquired immunodeficiency syndrome. HIV Neurobehavioral Research Center Group. Ann Neurol. 1997;42(5):679–88. Epub 1997/12/10. doi: 10.1002/ana.410420503 . [DOI] [PubMed] [Google Scholar]
- 4.McArthur JC, McClernon DR, Cronin MF, Nance-Sproson TE, Saah AJ, St Clair M, et al. Relationship between human immunodeficiency virus-associated dementia and viral load in cerebrospinal fluid and brain. Ann Neurol. 1997;42(5):689–98. Epub 1997/12/10. doi: 10.1002/ana.410420504 . [DOI] [PubMed] [Google Scholar]
- 5.Gisslen M, Hagberg L, Fuchs D, Norkrans G, Svennerholm B. Cerebrospinal fluid viral load in HIV-1-infected patients without antiretroviral treatment: a longitudinal study. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17(4):291–5. Epub 1998/04/03. doi: 10.1097/00042560-199804010-00001 . [DOI] [PubMed] [Google Scholar]
- 6.Gisslen M, Fuchs D, Svennerholm B, Hagberg L. Cerebrospinal fluid viral load, intrathecal immunoactivation, and cerebrospinal fluid monocytic cell count in HIV-1 infection. J Acquir Immune Defic Syndr. 1999;21(4):271–6. Epub 1999/07/31. doi: 10.1097/00126334-199908010-00003 . [DOI] [PubMed] [Google Scholar]
- 7.Ellis RJ, Gamst AC, Capparelli E, Spector SA, Hsia K, Wolfson T, et al. Cerebrospinal fluid HIV RNA originates from both local CNS and systemic sources. Neurology. 2000;54(4):927–36. Epub 2000/02/26. doi: 10.1212/wnl.54.4.927 . [DOI] [PubMed] [Google Scholar]
- 8.Spudich S, Gisslen M, Hagberg L, Lee E, Liegler T, Brew B, et al. Central nervous system immune activation characterizes primary human immunodeficiency virus 1 infection even in participants with minimal cerebrospinal fluid viral burden. J Infect Dis. 2011;204(5):753–60. Epub 2011/08/17. doi: 10.1093/infdis/jir387 ; PubMed Central PMCID: PMC3156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valcour V, Chalermchai T, Sailasuta N, Marovich M, Lerdlum S, Suttichom D, et al. Central nervous system viral invasion and inflammation during acute HIV infection. J Infect Dis. 2012;206(2):275–82. Epub 2012/05/04. doi: 10.1093/infdis/jis326 ; PubMed Central PMCID: PMC3490695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peluso MJ, Meyerhoff DJ, Price RW, Peterson J, Lee E, Young AC, et al. Cerebrospinal fluid and neuroimaging biomarker abnormalities suggest early neurological injury in a subset of individuals during primary HIV infection. J Infect Dis. 2013;207(11):1703–12. doi: 10.1093/infdis/jit088 ; PubMed Central PMCID: PMC3636785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulfhammer G, Eden A, Antinori A, Brew BJ, Calcagno A, Cinque P, et al. Cerebrospinal Fluid Viral Load Across the Spectrum of Untreated Human Immunodeficiency Virus Type 1 (HIV-1) Infection: A Cross-Sectional Multicenter Study. Clin Infect Dis. 2022;75(3):493–502. Epub 2021/11/09. doi: 10.1093/cid/ciab943 ; PubMed Central PMCID: PMC9427147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturdevant CB, Joseph SB, Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalized replication of R5 T cell-tropic HIV-1 in the central nervous system early in the course of infection. PLoS Pathog. 2015;11(3):e1004720. Epub 2015/03/27. doi: 10.1371/journal.ppat.1004720 ; PubMed Central PMCID: PMC4374811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J Virol. 2010;84(5):2395–407. Epub 2009/12/18. doi: 10.1128/JVI.01863-09 ; PubMed Central PMCID: PMC2820937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kincer LP, Schnell G, Swanstrom R, Miller MB, Spudich S, Eron JJ, et al. HIV-1 is Transported into the Central Nervous System by Trafficking Infected Cells. Pathog Immun. 2022;7(2):131–42. Epub 2023/03/04. doi: 10.20411/pai.v7i2.524 ; PubMed Central PMCID: PMC9973728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kincer LP, Joseph SB, Gilleece MM, Hauser BM, Sizemore S, Zhou S, et al. Rebound HIV-1 in cerebrospinal fluid after antiviral therapy interruption is mainly clonally amplified R5 T cell-tropic virus. Nat Microbiol. 2023;8(2):260–71. Epub 2023/01/31. doi: 10.1038/s41564-022-01306-6 ; PubMed Central PMCID: PMC10201410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gisslen M, Svennerholm B, Norkrans G, Franzen C, Sall C, Svensson R, et al. Cerebrospinal fluid and plasma viral load in HIV-1-infected patients with various anti-retroviral treatment regimens. Scand J Infect Dis. 2000;32(4):365–9. Epub 2000/08/26. doi: 10.1080/003655400750044926 . [DOI] [PubMed] [Google Scholar]
- 17.Spudich SS, Nilsson AC, Lollo ND, Liegler TJ, Petropoulos CJ, Deeks SG, et al. Cerebrospinal fluid HIV infection and pleocytosis: relation to systemic infection and antiretroviral treatment. BMC Infect Dis. 2005;5:98. Epub 2005/11/04. doi: 10.1186/1471-2334-5-98 ; PubMed Central PMCID: PMC1299327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price RW, Spudich SS, Peterson J, Joseph S, Fuchs D, Zetterberg H, et al. Evolving character of chronic central nervous system HIV infection. Semin Neurol. 2014;34(1):7–13. Epub 2014/04/10. doi: 10.1055/s-0034-1372337 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell JE. The neuropathology of adult HIV infection. Rev Neurol (Paris). 1998;154(12):816–29. Epub 1999/02/05. . [PubMed] [Google Scholar]
- 20.Petito CK, Chen H, Mastri AR, Torres-Munoz J, Roberts B, Wood C. HIV infection of choroid plexus in AIDS and asymptomatic HIV-infected patients suggests that the choroid plexus may be a reservoir of productive infection. J Neurovirol. 1999;5(6):670–7. Epub 1999/12/22. doi: 10.3109/13550289909021295 . [DOI] [PubMed] [Google Scholar]
- 21.Burkala EJ, He J, West JT, Wood C, Petito CK. Compartmentalization of HIV-1 in the central nervous system: role of the choroid plexus. Aids. 2005;19(7):675–84. Epub 2005/04/12. doi: 10.1097/01.aids.0000166090.31693.aa . [DOI] [PubMed] [Google Scholar]
- 22.Price RW, Spudich S. Antiretroviral therapy and central nervous system HIV type 1 infection. J Infect Dis. 2008;197 Suppl 3:S294–306. Epub 2008/06/14. doi: 10.1086/533419 ; PubMed Central PMCID: PMC2628635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Probasco JC, Deeks SG, Lee E, Hoh R, Hunt PW, Liegler T, et al. Cerebrospinal fluid in HIV-1 systemic viral controllers: absence of HIV-1 RNA and intrathecal inflammation. Aids. 2010;24(7):1001–5. Epub 2010/03/20. doi: 10.1097/QAD.0b013e328331e15b . [DOI] [PubMed] [Google Scholar]
- 24.Dahl V, Peterson J, Spudich S, Lee E, Shacklett BL, Price RW, et al. Single-copy assay quantification of HIV-1 RNA in paired cerebrospinal fluid and plasma samples from elite controllers. Aids. 2013;27(7):1145–9. Epub 2012/12/06. doi: 10.1097/QAD.0b013e32835cf235 ; PubMed Central PMCID: PMC4004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snider WD, Simpson DM, Nielsen S, Gold JW, Metroka CE, Posner JB. Neurological complications of acquired immune deficiency syndrome: analysis of 50 patients. Ann Neurol. 1983;14(4):403–18. Epub 1983/10/01. doi: 10.1002/ana.410140404 . [DOI] [PubMed] [Google Scholar]
- 26.Navia BA, Cho ES, Petito CK, Price RW. The AIDS dementia complex: II. Neuropathology. Ann Neurol. 1986;19(6):525–35. Epub 1986/06/01. doi: 10.1002/ana.410190603 . [DOI] [PubMed] [Google Scholar]
- 27.Budka H. Neuropathology of human immunodeficiency virus infection. Brain Pathol. 1991;1(3):163–75. Epub 1991/04/01. doi: 10.1111/j.1750-3639.1991.tb00656.x . [DOI] [PubMed] [Google Scholar]
- 28.Rosenblum MK. Infection of the central nervous system by the human immunodeficiency virus type 1. Morphology and relation to syndromes of progressive encephalopathy and myelopathy in patients with AIDS. Pathol Annu. 1990;25 Pt 1:117–69. Epub 1990/01/01. . [PubMed] [Google Scholar]
- 29.Masliah E, Achim CL, Ge N, De Teresa R, Wiley CA. Cellular neuropathology in HIV encephalitis. Res Publ Assoc Res Nerv Ment Dis. 1994;72:119–31. Epub 1994/01/01. . [PubMed] [Google Scholar]
- 30.Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, Chen IS. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236(4803):819–22. Epub 1987/05/15. doi: 10.1126/science.3646751 . [DOI] [PubMed] [Google Scholar]
- 31.Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, et al. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol. 1999;73(12):9741–55. Epub 1999/11/13. doi: 10.1128/JVI.73.12.9741-9755.1999 ; PubMed Central PMCID: PMC113021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, et al. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J Virol. 2001;75(21):10073–89. Epub 2001/10/03. doi: 10.1128/JVI.75.21.10073-10089.2001 ; PubMed Central PMCID: PMC114582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, et al. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol. 2002;76(12):6277–92. Epub 2002/05/22. doi: 10.1128/jvi.76.12.6277-6292.2002 ; PubMed Central PMCID: PMC136234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, et al. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol. 2004;78(13):6915–26. Epub 2004/06/15. doi: 10.1128/JVI.78.13.6915-6926.2004 ; PubMed Central PMCID: PMC421670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, et al. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci U S A. 2006;103(41):15160–5. Epub 2006/10/04. doi: 10.1073/pnas.0605513103 ; PubMed Central PMCID: PMC1586182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F. HIV-1 tropism for the central nervous system: Brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology. 2006;346(1):169–79. Epub 2005/11/29. doi: 10.1016/j.virol.2005.10.031 . [DOI] [PubMed] [Google Scholar]
- 37.Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog. 2011;7(10):e1002286. Epub 2011/10/19. doi: 10.1371/journal.ppat.1002286 ; PubMed Central PMCID: PMC3188520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burdo TH, Lackner A, Williams KC. Monocyte/macrophages and their role in HIV neuropathogenesis. Immunol Rev. 2013;254(1):102–13. Epub 2013/06/19. doi: 10.1111/imr.12068 ; PubMed Central PMCID: PMC3704190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, Hoxie JA, et al. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. J Virol. 2014;88(4):1858–69. Epub 2013/12/07. doi: 10.1128/JVI.02477-13 ; PubMed Central PMCID: PMC3911544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neaton JD, Neuhaus J, Emery S. Soluble biomarkers and morbidity and mortality among people infected with HIV: summary of published reports from 1997 to 2010. Curr Opin HIV AIDS. 2010;5(6):480–90. Epub 2010/10/28. doi: 10.1097/COH.0b013e32833ed75d ; PubMed Central PMCID: PMC3079321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunt PW. HIV and inflammation: mechanisms and consequences. Curr HIV/AIDS Rep. 2012;9(2):139–47. Epub 2012/04/25. doi: 10.1007/s11904-012-0118-8 . [DOI] [PubMed] [Google Scholar]
- 42.Baker JV, Sharma S, Grund B, Rupert A, Metcalf JA, Schechter M, et al. Systemic Inflammation, Coagulation, and Clinical Risk in the START Trial. Open Forum Infect Dis. 2017;4(4):ofx262. Epub 2018/01/09. doi: 10.1093/ofid/ofx262 ; PubMed Central PMCID: PMC5751061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hart BB, Nordell AD, Okulicz JF, Palfreeman A, Horban A, Kedem E, et al. Inflammation-Related Morbidity and Mortality Among HIV-Positive Adults: How Extensive Is It? J Acquir Immune Defic Syndr. 2018;77(1):1–7. Epub 2017/10/11. doi: 10.1097/QAI.0000000000001554 ; PubMed Central PMCID: PMC5720921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnittman SR, Deitchman AN, Beck-Engeser G, Ahn H, York VA, Hartig H, et al. Abnormal Levels of Some Biomarkers of Immune Activation Despite Very Early Treatment of Human Immunodeficiency Virus. J Infect Dis. 2021;223(9):1621–30. Epub 2020/09/12. doi: 10.1093/infdis/jiaa580 ; PubMed Central PMCID: PMC8136975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gisslen M, Keating SM, Spudich S, Arechiga V, Stephenson S, Zetterberg H, et al. Compartmentalization of cerebrospinal fluid inflammation across the spectrum of untreated HIV-1 infection, central nervous system injury and viral suppression. PLoS One. 2021;16(5):e0250987. Epub 2021/05/14. doi: 10.1371/journal.pone.0250987 ; PubMed Central PMCID: PMC8118251 paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gisslen M, Heslegrave A, Veleva E, Yilmaz A, Andersson LM, Hagberg L, et al. CSF concentrations of soluble TREM2 as a marker of microglial activation in HIV-1 infection. Neurol Neuroimmunol Neuroinflamm. 2019;6(1):e512. Epub 2018/12/21. doi: 10.1212/NXI.0000000000000512 ; PubMed Central PMCID: PMC6278890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hategan A, Bianchet MA, Steiner J, Karnaukhova E, Masliah E, Fields A, et al. HIV Tat protein and amyloid-beta peptide form multifibrillar structures that cause neurotoxicity. Nat Struct Mol Biol. 2017;24(4):379–86. Epub 2017/02/22. doi: 10.1038/nsmb.3379 ; PubMed Central PMCID: PMC5383535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hategan A, Masliah E, Nath A. HIV and Alzheimer’s disease: complex interactions of HIV-Tat with amyloid beta peptide and Tau protein. J Neurovirol. 2019;25(5):648–60. Epub 2019/04/25. doi: 10.1007/s13365-019-00736-z ; PubMed Central PMCID: PMC9056081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson TP, Nath A. Biotypes of HIV-associated neurocognitive disorders based on viral and immune pathogenesis. Curr Opin Infect Dis. 2022;35(3):223–30. Epub 2022/06/07. doi: 10.1097/QCO.0000000000000825 ; PubMed Central PMCID: PMC9179892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong S, Banks WA. Role of the immune system in HIV-associated neuroinflammation and neurocognitive implications. Brain, behavior, and immunity. 2015;45:1–12. Epub 2014/12/03. doi: 10.1016/j.bbi.2014.10.008 ; PubMed Central PMCID: PMC4342286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McArthur JC, Johnson TP. Chronic inflammation mediates brain injury in HIV infection: relevance for cure strategies. Current opinion in neurology. 2020;33(3):397–404. Epub 2020/03/27. doi: 10.1097/WCO.0000000000000807 ; PubMed Central PMCID: PMC7331798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spudich S, Gonzalez-Scarano F. HIV-1-Related Central Nervous System Disease: Current Issues in Pathogenesis, Diagnosis, and Treatment. Cold Spring Harbor perspectives in medicine. 2012;2(6):a007120. Epub 2012/06/08. doi: 10.1101/cshperspect.a007120 ; PubMed Central PMCID: PMC3367536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cassol E, Misra V, Morgello S, Gabuzda D. Applications and limitations of inflammatory biomarkers for studies on neurocognitive impairment in HIV infection. J Neuroimmune Pharmacol. 2013;8(5):1087–97. Epub 2013/11/22. doi: 10.1007/s11481-013-9512-2 ; PubMed Central PMCID: PMC3889222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamat A, Lyons JL, Misra V, Uno H, Morgello S, Singer EJ, et al. Monocyte activation markers in cerebrospinal fluid associated with impaired neurocognitive testing in advanced HIV infection. J Acquir Immune Defic Syndr. 2012;60(3):234–43. Epub 2012/05/10. doi: 10.1097/QAI.0b013e318256f3bc ; PubMed Central PMCID: PMC3383928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burdo TH, Weiffenbach A, Woods SP, Letendre S, Ellis RJ, Williams KC. Elevated sCD163 is a marker of neurocognitive impairment in HIV infection. Aids. 2013. Epub 2013/02/26. doi: 10.1097/QAD.0b013e32836010bd . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joseph SB, Gianella S, Burdo TH, Cinque P, Gisslen M, Letendre S, et al. Biotypes of Central Nervous System Complications in People With Human Immunodeficiency Virus: Virology, Immunology, and Neuropathology. J Infect Dis. 2023;227(Suppl 1):S3–S15. Epub 2023/03/18. doi: 10.1093/infdis/jiac370 ; PubMed Central PMCID: PMC10022721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irollo E, Luchetta J, Ho C, Nash B, Meucci O. Mechanisms of neuronal dysfunction in HIV-associated neurocognitive disorders. Cell Mol Life Sci. 2021;78(9):4283–303. Epub 2021/02/16. doi: 10.1007/s00018-021-03785-y ; PubMed Central PMCID: PMC8164580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu S, Xing Y, Wang J, Pan R, Li G, Tang H, et al. The Dual Role of HIV-1 gp120 V3 Loop-Induced Autophagy in the Survival and Apoptosis of the Primary Rat Hippocampal Neurons. Neurochemical research. 2019;44(7):1636–52. Epub 2019/04/22. doi: 10.1007/s11064-019-02788-3 . [DOI] [PubMed] [Google Scholar]
- 59.Rozzi SJ, Avdoshina V, Fields JA, Mocchetti I. Human immunodeficiency virus Tat impairs mitochondrial fission in neurons. Cell Death Discov. 2018;4:8. Epub 2018/03/14. doi: 10.1038/s41420-017-0013-6 ; PubMed Central PMCID: PMC5841280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh H, Ojeda-Juarez D, Maung R, Shah R, Roberts AJ, Kaul M. A pivotal role for Interferon-alpha receptor-1 in neuronal injury induced by HIV-1. Journal of neuroinflammation. 2020;17(1):226. Epub 2020/07/31. doi: 10.1186/s12974-020-01894-2 ; PubMed Central PMCID: PMC7388458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellis RJ, Marquine MJ, Kaul M, Fields JA, Schlachetzki JCM. Mechanisms underlying HIV-associated cognitive impairment and emerging therapies for its management. Nature reviews Neurology. 2023. Epub 2023/10/11. doi: 10.1038/s41582-023-00879-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams ME, Naude PJW. The relationship between HIV-1 neuroinflammation, neurocognitive impairment and encephalitis pathology: A systematic review of studies investigating post-mortem brain tissue. Reviews in medical virology. 2024;34(1):e2519. Epub 2024/01/29. doi: 10.1002/rmv.2519 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gisslen M, Hagberg L, Brew BJ, Cinque P, Price RW, Rosengren L. Elevated cerebrospinal fluid neurofilament light protein concentrations predict the development of AIDS dementia complex. J Infect Dis. 2007;195(12):1774–8. Epub 2007/05/12. doi: 10.1086/518043 . [DOI] [PubMed] [Google Scholar]
- 64.Jessen Krut J, Mellberg T, Price RW, Hagberg L, Fuchs D, Rosengren L, et al. Biomarker evidence of axonal injury in neuroasymptomatic HIV-1 patients. PLoS One. 2014;9(2):e88591. Epub 2014/02/14. doi: 10.1371/journal.pone.0088591 ; PubMed Central PMCID: PMC3921217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bridel C, van Wieringen WN, Zetterberg H, Tijms BM, Teunissen CE, and the NFLG, et al. Diagnostic Value of Cerebrospinal Fluid Neurofilament Light Protein in Neurology: A Systematic Review and Meta-analysis. JAMA neurology. 2019;76(9):1035–48. Epub 2019/06/18. doi: 10.1001/jamaneurol.2019.1534 ; PubMed Central PMCID: PMC6580449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gisslen M, Price RW, Andreasson U, Norgren N, Nilsson S, Hagberg L, et al. Plasma Concentration of the Neurofilament Light Protein (NFL) is a Biomarker of CNS Injury in HIV Infection: A Cross-Sectional Study. EBioMedicine. 2016;3:135–40. doi: 10.1016/j.ebiom.2015.11.036 ; PubMed Central PMCID: PMC4739412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zetterberg H. Neurofilament Light: A Dynamic Cross-Disease Fluid Biomarker for Neurodegeneration. Neuron. 2016;91(1):1–3. Epub 2016/07/09. doi: 10.1016/j.neuron.2016.06.030 . [DOI] [PubMed] [Google Scholar]
- 68.Yilmaz A, Blennow K, Hagberg L, Nilsson S, Price RW, Schouten J, et al. Neurofilament light chain protein as a marker of neuronal injury: review of its use in HIV-1 infection and reference values for HIV-negative controls. Expert Rev Mol Diagn. 2017;17(8):761–70. doi: 10.1080/14737159.2017.1341313 . [DOI] [PubMed] [Google Scholar]
- 69.Mellgren A, Antinori A, Cinque P, Price RW, Eggers C, Hagberg L, et al. Cerebrospinal fluid HIV-1 infection usually responds well to antiretroviral treatment. Antivir Ther. 2005;10(6):701–7. Epub 2005/10/13. . [PubMed] [Google Scholar]
- 70.Spudich S, Lollo N, Liegler T, Deeks SG, Price RW. Treatment benefit on cerebrospinal fluid HIV-1 levels in the setting of systemic virological suppression and failure. J Infect Dis. 2006;194(12):1686–96. Epub 2006/11/17. doi: 10.1086/508750 . [DOI] [PubMed] [Google Scholar]
- 71.Yilmaz A, Svennerholm B, Hagberg L, Gisslen M. Cerebrospinal fluid viral loads reach less than 2 copies/ml in HIV-1-infected patients with effective antiretroviral therapy. Antivir Ther. 2006;11(7):833–7. Epub 2007/02/17. . [PubMed] [Google Scholar]
- 72.Dahl V, Peterson J, Fuchs D, Gisslen M, Palmer S, Price RW. Low levels of HIV-1 RNA detected in the cerebrospinal fluid after up to 10 years of suppressive therapy are associated with local immune activation. Aids. 2014;28(15):2251–8. Epub 2014/07/16. doi: 10.1097/QAD.0000000000000400 ; PubMed Central PMCID: PMC4492794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Letendre S, Marquie-Beck J, Capparelli E, Best B, Clifford D, Collier AC, et al. Validation of the CNS Penetration-Effectiveness rank for quantifying antiretroviral penetration into the central nervous system. Arch Neurol. 2008;65(1):65–70. Epub 2008/01/16. doi: 10.1001/archneurol.2007.31 ; PubMed Central PMCID: PMC2763187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Letendre SL, McCutchan JA, Childers ME, Woods SP, Lazzaretto D, Heaton RK, et al. Enhancing antiretroviral therapy for human immunodeficiency virus cognitive disorders. Ann Neurol. 2004;56(3):416–23. Epub 2004/09/07. doi: 10.1002/ana.20198 . [DOI] [PubMed] [Google Scholar]
- 75.Hagberg L, Cinque P, Gisslen M, Brew BJ, Spudich S, Bestetti A, et al. Cerebrospinal fluid neopterin: an informative biomarker of central nervous system immune activation in HIV-1 infection. AIDS Res Ther. 2010;7:15. Epub 2010/06/08. doi: 10.1186/1742-6405-7-15 ; PubMed Central PMCID: PMC2890504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yilmaz A, Yiannoutsos CT, Fuchs D, Price RW, Crozier K, Hagberg L, et al. Cerebrospinal fluid neopterin decay characteristics after initiation of antiretroviral therapy. Journal of neuroinflammation. 2013;10:62. Epub 2013/05/15. doi: 10.1186/1742-2094-10-62 ; PubMed Central PMCID: PMC3657550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hagberg L, Fuchs D, Rosengren L, Gisslen M. Intrathecal immune activation is associated with cerebrospinal fluid markers of neuronal destruction in AIDS patients. J Neuroimmunol. 2000;102(1):51–5. Epub 2000/01/08. doi: 10.1016/s0165-5728(99)00150-2 . [DOI] [PubMed] [Google Scholar]
- 78.Cinque P, Bestetti A, Marenzi R, Sala S, Gisslen M, Hagberg L, et al. Cerebrospinal fluid interferon-gamma-inducible protein 10 (IP-10, CXCL10) in HIV-1 infection. J Neuroimmunol. 2005;168(1–2):154–63. Epub 2005/08/11. doi: 10.1016/j.jneuroim.2005.07.002 . [DOI] [PubMed] [Google Scholar]
- 79.Peluso MJ, Thanh C, Prator CA, Hogan LE, Arechiga VM, Stephenson S, et al. Cerebrospinal fluid soluble CD30 elevation despite suppressive antiretroviral therapy in individuals living with HIV-1. J Virus Erad. 2020;6(1):19–26. Epub 2020/03/17. ; PubMed Central PMCID: PMC7043898. [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang W, Luo Z, Stephenson S, Li H, Di Germanio C, Norris PJ, et al. Cerebrospinal fluid and plasma lipopolysaccharide (LPS) levels in HIV-1 infection and associations with inflammation, blood-brain barrier permeability and neuronal injury. J Infect Dis. 2020. Epub 2020/12/16. doi: 10.1093/infdis/jiaa765 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Premeaux TA, D’Antoni ML, Abdel-Mohsen M, Pillai SK, Kallianpur KJ, Nakamoto BK, et al. Elevated cerebrospinal fluid Galectin-9 is associated with central nervous system immune activation and poor cognitive performance in older HIV-infected individuals. J Neurovirol. 2019;25(2):150–61. Epub 2018/11/28. doi: 10.1007/s13365-018-0696-3 ; PubMed Central PMCID: PMC6506351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hermansson L, Yilmaz A, Axelsson M, Blennow K, Fuchs D, Hagberg L, et al. Cerebrospinal fluid levels of glial marker YKL-40 strongly associated with axonal injury in HIV infection. Journal of neuroinflammation. 2019;16(1):16. Epub 2019/01/27. doi: 10.1186/s12974-019-1404-9 ; PubMed Central PMCID: PMC6345016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ho EL, Ronquillo R, Altmeppen H, Spudich SS, Price RW, Sinclair E. Cellular Composition of Cerebrospinal Fluid in HIV-1 Infected and Uninfected Subjects. PLoS One. 2013;8(6):e66188. Epub 2013/07/05. doi: 10.1371/journal.pone.0066188 ; PubMed Central PMCID: PMC3688831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ulfhammer G, Eden A, Mellgren A, Fuchs D, Zetterberg H, Hagberg L, et al. Persistent central nervous system immune activation following more than 10 years of effective HIV antiretroviral treatment. Aids. 2018;32(15):2171–8. Epub 2018/07/14. doi: 10.1097/QAD.0000000000001950 . [DOI] [PubMed] [Google Scholar]
- 85.Abdulle S, Hagberg L, Svennerholm B, Fuchs D, Gisslen M. Continuing intrathecal immunoactivation despite two years of effective antiretroviral therapy against HIV-1 infection. Aids. 2002;16(16):2145–9. Epub 2002/11/01. doi: 10.1097/00002030-200211080-00006 . [DOI] [PubMed] [Google Scholar]
- 86.Hawes IA, Alvarenga BD, Browne W, Wapniarski A, Dandekar R, Bartley CM, et al. Viral co-infection, autoimmunity, and CSF HIV antibody profiles in HIV central nervous system escape. J Neuroimmunol. 2023;381:578141. Epub 2023/07/08. doi: 10.1016/j.jneuroim.2023.578141 . [DOI] [PubMed] [Google Scholar]
- 87.Yilmaz A, Price RW, Spudich S, Fuchs D, Hagberg L, Gisslen M. Persistent intrathecal immune activation in HIV-1-infected individuals on antiretroviral therapy. J Acquir Immune Defic Syndr. 2008;47(2):168–73. Epub 2007/11/01. doi: 10.1097/QAI.0b013e31815ace97 ; PubMed Central PMCID: PMC2628632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eden A, Rydberg F, Yilmaz A, Hagberg L, Gostner J, Nilsson S, et al. Residual Central Nervous System Immune Activation Is Not Prevented by Antiretroviral Therapy Initiated During Early Chronic HIV Infection. Open Forum Infect Dis. 2023;10(2):ofad064. Epub 2023/03/03. doi: 10.1093/ofid/ofad064 ; PubMed Central PMCID: PMC9969734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burbelo PD, Bayat A, Rhodes CS, Hoh R, Martin JN, Fromentin R, et al. HIV antibody characterization as a method to quantify reservoir size during curative interventions. J Infect Dis. 2014;209(10):1613–7. Epub 2013/11/30. doi: 10.1093/infdis/jit667 ; PubMed Central PMCID: PMC3997576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gisslen M, Hunt PW. Antiretroviral Treatment of Acute HIV Infection Normalizes Levels of Cerebrospinal Fluid Markers of Central Nervous System (CNS) Inflammation: A Consequence of a Reduced CNS Reservoir? J Infect Dis. 2019;220(12):1867–9. Epub 2019/01/23. doi: 10.1093/infdis/jiz031 ; PubMed Central PMCID: PMC6833976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Veenhuis RT, Clements JE, Gama L. HIV Eradication Strategies: Implications for the Central Nervous System. Curr HIV/AIDS Rep. 2019;16(1):96–104. Epub 2019/02/09. doi: 10.1007/s11904-019-00428-7 ; PubMed Central PMCID: PMC6494469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spudich S, Robertson KR, Bosch RJ, Gandhi RT, Cyktor JC, Mar H, et al. Persistent HIV-infected cells in cerebrospinal fluid are associated with poorer neurocognitive performance. J Clin Invest. 2019;129(8):3339–46. Epub 2019/07/16. doi: 10.1172/JCI127413 ; PubMed Central PMCID: PMC6668666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Farhadian SF, Lindenbaum O, Zhao J, Corley MJ, Im Y, Walsh H, et al. HIV viral transcription and immune perturbations in the CNS of people with HIV despite ART. JCI Insight. 2022;7(13). Epub 2022/07/09. doi: 10.1172/jci.insight.160267 ; PubMed Central PMCID: PMC9310520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Winston A, Antinori A, Cinque P, Fox HS, Gisslen M, Henrich TJ, et al. Defining cerebrospinal fluid HIV RNA escape: editorial review AIDS. Aids. 2019;33 Suppl 2:S107–S11. Epub 2019/12/04. doi: 10.1097/QAD.0000000000002252 . [DOI] [PubMed] [Google Scholar]
- 95.Canestri A, Lescure FX, Jaureguiberry S, Moulignier A, Amiel C, Marcelin AG, et al. Discordance between cerebral spinal fluid and plasma HIV replication in patients with neurological symptoms who are receiving suppressive antiretroviral therapy. Clin Infect Dis. 2010;50(5):773–8. Epub 2010/01/27. doi: 10.1086/650538 . [DOI] [PubMed] [Google Scholar]
- 96.Peluso MJ, Ferretti F, Peterson J, Lee E, Fuchs D, Boschini A, et al. Cerebrospinal fluid HIV escape associated with progressive neurologic dysfunction in patients on antiretroviral therapy with well controlled plasma viral load. Aids. 2012;26(14):1765–74. Epub 2012/05/23. doi: 10.1097/QAD.0b013e328355e6b2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eden A, Fuchs D, Hagberg L, Nilsson S, Spudich S, Svennerholm B, et al. HIV-1 viral escape in cerebrospinal fluid of subjects on suppressive antiretroviral treatment. J Infect Dis. 2010;202(12):1819–25. Epub 2010/11/06. doi: 10.1086/657342 ; PubMed Central PMCID: PMC3052942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eden A, Nilsson S, Hagberg L, Fuchs D, Zetterberg H, Svennerholm B, et al. Asymptomatic Cerebrospinal Fluid HIV-1 Viral Blips and Viral Escape During Antiretroviral Therapy: A Longitudinal Study. J Infect Dis. 2016;214(12):1822–5. Epub 2016/11/02. doi: 10.1093/infdis/jiw454 . [DOI] [PubMed] [Google Scholar]
- 99.Joseph SB, Kincer LP, Bowman NM, Evans C, Vinikoor MJ, Lippincott CK, et al. Human Immunodeficiency Virus Type 1 RNA Detected in the Central Nervous System (CNS) After Years of Suppressive Antiretroviral Therapy Can Originate from a Replicating CNS Reservoir or Clonally Expanded Cells. Clin Infect Dis. 2019;69(8):1345–52. Epub 2018/12/19. doi: 10.1093/cid/ciy1066 ; PubMed Central PMCID: PMC6938202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hagberg L, Price RW, Zetterberg H, Fuchs D, Gisslen M. Herpes zoster in HIV-1 infection: The role of CSF pleocytosis in secondary CSF escape and discordance. PLoS One. 2020;15(7):e0236162. Epub 2020/07/23. doi: 10.1371/journal.pone.0236162 ; PubMed Central PMCID: PMC7375594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lescure FX, Moulignier A, Savatovsky J, Amiel C, Carcelain G, Molina JM, et al. CD8 Encephalitis in HIV-Infected Patients Receiving cART: A Treatable Entity. Clin Infect Dis. 2013. Epub 2013/03/22. doi: 10.1093/cid/cit175 . [DOI] [PubMed] [Google Scholar]
- 102.Gray F, Lescure FX, Adle-Biassette H, Polivka FM, Gallien S, Pialoux G, et al. Encephalitis with infiltration by CD8+ Lymphocytes in HIV Patients receiving Combination Antiretroviral Treatment. Brain Pathol. 2013. Epub 2013/01/26. doi: 10.1111/bpa.12038 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ferretti F, De Zan V, Gerevini S, Turrini F, Boeri E, Gianotti N, et al. Relapse of Symptomatic Cerebrospinal Fluid HIV Escape. Curr HIV/AIDS Rep. 2020;17(5):522–8. Epub 2020/09/03. doi: 10.1007/s11904-020-00526-x . [DOI] [PubMed] [Google Scholar]
- 104.Mastrangelo A, Turrini F, de Zan V, Caccia R, Gerevini S, Cinque P. Symptomatic cerebrospinal fluid escape. Aids. 2019;33 Suppl 2:S159–S69. Epub 2019/12/04. doi: 10.1097/QAD.0000000000002266 . [DOI] [PubMed] [Google Scholar]
- 105.Chan TY, De Zan V, Gregg A, Alagaratnam J, Gerevini S, Antinori A, et al. The symptomatology of cerebrospinal fluid HIV RNA escape: a large case-series. Aids. 2021;35(14):2341–6. Epub 2021/06/16. doi: 10.1097/QAD.0000000000002992 . [DOI] [PubMed] [Google Scholar]
- 106.Sachdeva RK, Naidu G, Chauhan P, Kharbanda S, Kaur J, Joseph P, et al. CSF viral escape on HAART: Analysis from single tertiary care centre. AIDS Res Hum Retroviruses. 2024. Epub 2024/02/17. doi: 10.1089/AID.2022.0187 . [DOI] [PubMed] [Google Scholar]
- 107.Ferretti F, Gianotti N, Lazzarin A, Cinque P. Central nervous system HIV infection in "less-drug regimen" antiretroviral therapy simplification strategies. Semin Neurol. 2014;34(1):78–88. Epub 2014/04/10. doi: 10.1055/s-0034-1372345 . [DOI] [PubMed] [Google Scholar]
- 108.Bingham R, Ahmed N, Rangi P, Johnson M, Tyrer M, Green J. HIV encephalitis despite suppressed viraemia: a case of compartmentalized viral escape. Int J STD AIDS. 2011;22(10):608–9. Epub 2011/10/15. doi: 10.1258/ijsa.2011.010507 . [DOI] [PubMed] [Google Scholar]
- 109.Webb AJ, Borrelli EP, Vyas A, Taylor LE, Buchanan AL. The effect of antiretroviral therapy with high central nervous system penetration on HIV-related cognitive impairment: a systematic review and meta-analysis. AIDS care. 2023;35(11):1635–46. Epub 2022/07/20. doi: 10.1080/09540121.2022.2098231 ; PubMed Central PMCID: PMC9845421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mukerji SS, Misra V, Lorenz DR, Uno H, Morgello S, Franklin D, et al. Impact of Antiretroviral Regimens on Cerebrospinal Fluid Viral Escape in a Prospective Multicohort Study of Antiretroviral Therapy-Experienced Human Immunodeficiency Virus-1-Infected Adults in the United States. Clin Infect Dis. 2018;67(8):1182–90. Epub 2018/04/05. doi: 10.1093/cid/ciy267 ; PubMed Central PMCID: PMC6160603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kang J, Wang Z, Zhou Y, Wang W, Wen Y. Learning from cerebrospinal fluid drug-resistant HIV escape-associated encephalitis: a case report. Virol J. 2023;20(1):292. Epub 2023/12/11. doi: 10.1186/s12985-023-02255-0 ; PubMed Central PMCID: PMC10712177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hagberg L, Gisslen M. Cohort profile: a longitudinal study of HIV infection in the central nervous system with focus on cerebrospinal fluid—the Gothenburg HIV CSF Study Cohort. BMJ Open. 2023;13(5):e070693. Epub 2023/05/18. doi: 10.1136/bmjopen-2022-070693 ; PubMed Central PMCID: PMC10193099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joseph J, Cinque P, Colosi D, Dravid A, Ene L, Fox H, et al. Highlights of the Global HIV-1 CSF Escape Consortium Meeting, 9 June 2016, Bethesda, MD, USA. J Virus Erad. 2016;2(4):243–50. Epub 2016/10/27. ; PubMed Central PMCID: PMC5075354. [PMC free article] [PubMed] [Google Scholar]
- 114.Joseph SB, Trunfio M, Kincer LP, Calcagno A, Price RW. What can characterization of cerebrospinal fluid escape populations teach us about viral reservoirs in the central nervous system? Aids. 2019;33 Suppl 2:S171–S9. Epub 2019/12/04. doi: 10.1097/QAD.0000000000002253 . [DOI] [PubMed] [Google Scholar]
- 115.McArthur JC, Nance-Sproson TE, Griffin DE, Hoover D, Selnes OA, Miller EN, et al. The diagnostic utility of elevation in cerebrospinal fluid beta 2-microglobulin in HIV-1 dementia. Multicenter AIDS Cohort Study. Neurology. 1992;42(9):1707–12. doi: 10.1212/wnl.42.9.1707 . [DOI] [PubMed] [Google Scholar]
- 116.Kelder W, McArthur JC, Nance-Sproson T, McClernon D, Griffin DE. Beta-chemokines MCP-1 and RANTES are selectively increased in cerebrospinal fluid of patients with human immunodeficiency virus-associated dementia. Ann Neurol. 1998;44(5):831–5. Epub 1998/11/18. doi: 10.1002/ana.410440521 . [DOI] [PubMed] [Google Scholar]
- 117.Cinque P, Vago L, Mengozzi M, Torri V, Ceresa D, Vicenzi E, et al. Elevated cerebrospinal fluid levels of monocyte chemotactic protein-1 correlate with HIV-1 encephalitis and local viral replication. Aids. 1998;12(11):1327–32. Epub 1998/08/26. doi: 10.1097/00002030-199811000-00014 . [DOI] [PubMed] [Google Scholar]
- 118.Conant K, McArthur JC, Griffin DE, Sjulson L, Wahl LM, Irani DN. Cerebrospinal fluid levels of MMP-2, 7, and 9 are elevated in association with human immunodeficiency virus dementia. Ann Neurol. 1999;46(3):391–8. doi: . [DOI] [PubMed] [Google Scholar]
- 119.Sabri F, De Milito A, Pirskanen R, Elovaara I, Hagberg L, Cinque P, et al. Elevated levels of soluble Fas and Fas ligand in cerebrospinal fluid of patients with AIDS dementia complex. J Neuroimmunol. 2001;114(1–2):197–206. Epub 2001/03/10. doi: 10.1016/s0165-5728(00)00424-0 . [DOI] [PubMed] [Google Scholar]
- 120.Price RW, Epstein LG, Becker JT, Cinque P, Gisslen M, Pulliam L, et al. Biomarkers of HIV-1 CNS infection and injury. Neurology. 2007;69(18):1781–8. Epub 2007/10/31. doi: 10.1212/01.wnl.0000278457.55877.eb . [DOI] [PubMed] [Google Scholar]
- 121.Burlacu R, Umlauf A, Marcotte TD, Soontornniyomkij B, Diaconu CC, Bulacu-Talnariu A, et al. Plasma CXCL10 correlates with HAND in HIV-infected women. J Neurovirol. 2020;26(1):23–31. Epub 2019/08/16. doi: 10.1007/s13365-019-00785-4 ; PubMed Central PMCID: PMC7018596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saloner R, Cherner M, Grelotti DJ, Paolillo EW, Moore DJ, Heaton RK, et al. Lower CSF homovanillic acid relates to higher burden of neuroinflammation and depression in people with HIV disease. Brain, behavior, and immunity. 2020;90:353–63. Epub 2020/09/24. doi: 10.1016/j.bbi.2020.09.012 ; PubMed Central PMCID: PMC7544671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peterson J, Gisslen M, Zetterberg H, Fuchs D, Shacklett BL, Hagberg L, et al. Cerebrospinal fluid (CSF) neuronal biomarkers across the spectrum of HIV infection: hierarchy of injury and detection. PLoS One. 2014;9(12):e116081. Epub 2014/12/30. doi: 10.1371/journal.pone.0116081 ; PubMed Central PMCID: PMC4277428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cinque P, Vago L, Ceresa D, Mainini F, Terreni MR, Vagani A, et al. Cerebrospinal fluid HIV-1 RNA levels: correlation with HIV encephalitis. Aids. 1998;12(4):389–94. Epub 1998/03/31. doi: 10.1097/00002030-199804000-00007 . [DOI] [PubMed] [Google Scholar]
- 125.Dravid AN, Natrajan K, Kulkarni MM, Saraf CK, Mahajan US, Kore SD, et al. Discordant CSF/plasma HIV-1 RNA in individuals on virologically suppressive antiretroviral therapy in Western India. Medicine (Baltimore). 2018;97(8):e9969. Epub 2018/02/22. doi: 10.1097/MD.0000000000009969 ; PubMed Central PMCID: PMC5841989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197(1):126–33. Epub 2008/01/04. doi: 10.1086/524143 ; PubMed Central PMCID: PMC3466592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Price RW, Peterson J, Fuchs D, Angel TE, Zetterberg H, Hagberg L, et al. Approach to cerebrospinal fluid (CSF) biomarker discovery and evaluation in HIV infection. J Neuroimmune Pharmacol. 2013;8(5):1147–58. Epub 2013/08/15. doi: 10.1007/s11481-013-9491-3 ; PubMed Central PMCID: PMC3889225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, et al. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69(18):1789–99. Epub 2007/10/05. doi: 10.1212/01.WNL.0000287431.88658.8b . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Price RW, Brew B, Sidtis J, Rosenblum M, Scheck AC, Cleary P. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science. 1988;239(4840):586–92. Epub 1988/02/05. doi: 10.1126/science.3277272 . [DOI] [PubMed] [Google Scholar]
- 130.Nomenclature and research case definitions for neurologic manifestations of human immunodeficiency virus-type 1 (HIV-1) infection. Report of a Working Group of the American Academy of Neurology AIDS Task Force. Neurology. 1991;41(6):778–85. Epub 1991/06/01. doi: 10.1212/wnl.41.6.778 . [DOI] [PubMed] [Google Scholar]
- 131.Ferretti F, Gisslen M, Cinque P, Price RW. Cerebrospinal Fluid HIV Escape from Antiretroviral Therapy. Curr HIV/AIDS Rep. 2015;12(2):280–8. doi: 10.1007/s11904-015-0267-7 . [DOI] [PubMed] [Google Scholar]
- 132.Yukl SA, Boritz E, Busch M, Bentsen C, Chun TW, Douek D, et al. Challenges in detecting HIV persistence during potentially curative interventions: a study of the Berlin patient. PLoS Pathog. 2013;9(5):e1003347. Epub 2013/05/15. doi: 10.1371/journal.ppat.1003347 ; PubMed Central PMCID: PMC3649997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Henrich TJ, Hatano H, Bacon O, Hogan LE, Rutishauser R, Hill A, et al. HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: An observational study. PLoS Med. 2017;14(11):e1002417. Epub 2017/11/08. doi: 10.1371/journal.pmed.1002417 ; PubMed Central PMCID: PMC5675377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Price RG M; Hu Z; Cinque P; Dravid A; Hagberg L; Yilmaz A; Zetterberg H; Dietmar F6; Gostner J; Blennow K; Spudich S. File 1 Dataset of CSF proteins in chronic HIV _infection_rev2024-09-10.xlsx; doi: 10.5061/dryad.x3ffbg7tv [DOI] [Google Scholar]
- 135.Andersson LM, Svennerholm B, Hagberg L, Gisslen M. Higher HIV-1 RNA cutoff level required in cerebrospinal fluid than in blood to predict positive HIV-1 isolation. J Med Virol. 2000;62(1):9–13. Epub 2000/08/10. doi: . [DOI] [PubMed] [Google Scholar]
- 136.Price RW, Paxinos EE, Grant RM, Drews B, Nilsson A, Hoh R, et al. Cerebrospinal fluid response to structured treatment interruption after virological failure. Aids. 2001;15(10):1251–9. doi: 10.1097/00002030-200107060-00006 . [DOI] [PubMed] [Google Scholar]
- 137.Price RW, Sidtis JJ. Evaluation of the AIDS dementia complex in clinical trials. J Acquir Immune Defic Syndr. 1990;3 Suppl 2:S51–60. Epub 1990/01/01. . [PubMed] [Google Scholar]
- 138.Gaetani L, Hoglund K, Parnetti L, Pujol-Calderon F, Becker B, Eusebi P, et al. A new enzyme-linked immunosorbent assay for neurofilament light in cerebrospinal fluid: analytical validation and clinical evaluation. Alzheimer’s research & therapy. 2018;10(1):8. Epub 2018/01/27. doi: 10.1186/s13195-018-0339-1 ; PubMed Central PMCID: PMC6389166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9(4):e95192. Epub 2014/04/24. doi: 10.1371/journal.pone.0095192 ; PubMed Central PMCID: PMC3995906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.UniProt C. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023;51(D1):D523–D31. Epub 2022/11/22. doi: 10.1093/nar/gkac1052 ; PubMed Central PMCID: PMC9825514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. Epub 2005/10/04. doi: 10.1073/pnas.0506580102 ; PubMed Central PMCID: PMC1239896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.The Gene Ontology C. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D8. Epub 2018/11/06. doi: 10.1093/nar/gky1055 ; PubMed Central PMCID: PMC6323945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fuchs D, Chiodi F, Albert J, Asjo B, Hagberg L, Hausen A, et al. Neopterin concentrations in cerebrospinal fluid and serum of individuals infected with HIV-1. Aids. 1989;3(5):285–8. doi: 10.1097/00002030-198905000-00006 . [DOI] [PubMed] [Google Scholar]
- 144.Blennow K, Fredman P, Wallin A, Gottfries CG, Karlsson I, Langstrom G, et al. Protein analysis in cerebrospinal fluid. II. Reference values derived from healthy individuals 18–88 years of age. Eur Neurol. 1993;33(2):129–33. Epub 1993/01/01. doi: 10.1159/000116919 . [DOI] [PubMed] [Google Scholar]
- 145.Huber C, Batchelor JR, Fuchs D, Hausen A, Lang A, Niederwieser D, et al. Immune response-associated production of neopterin. Release from macrophages primarily under control of interferon-gamma. J Exp Med. 1984;160(1):310–6. Epub 1984/07/01. doi: 10.1084/jem.160.1.310 ; PubMed Central PMCID: PMC2187425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fuchs D, Spira TJ, Hausen A, Reibnegger G, Werner ER, Felmayer GW, et al. Neopterin as a predictive marker for disease progression in human immunodeficiency virus type 1 infection. Clin Chem. 1989;35(8):1746–9. Epub 1989/08/01. . [PubMed] [Google Scholar]
- 147.Cano OD, Neurauter G, Fuchs D, Shearer GM, Boasso A. Differential effect of type I and type II interferons on neopterin production and amino acid metabolism in human astrocyte-derived cells. Neuroscience letters. 2008;438(1):22–5. Epub 2008/05/07. doi: 10.1016/j.neulet.2008.04.046 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Skillback T, Farahmand B, Bartlett JW, Rosen C, Mattsson N, Nagga K, et al. CSF neurofilament light differs in neurodegenerative diseases and predicts severity and survival. Neurology. 2014;83(21):1945–53. Epub 2014/10/24. doi: 10.1212/WNL.0000000000001015 . [DOI] [PubMed] [Google Scholar]
- 149.Jespersen S, Pedersen KK, Anesten B, Zetterberg H, Fuchs D, Gisslen M, et al. Soluble CD14 in cerebrospinal fluid is associated with markers of inflammation and axonal damage in untreated HIV-infected patients: a retrospective cross-sectional study. BMC Infect Dis. 2016;16(1):176. doi: 10.1186/s12879-016-1510-6 ; PubMed Central PMCID: PMC4839160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Navia BA, Jordan BD, Price RW. The AIDS dementia complex: I. Clinical features. Ann Neurol. 1986;19(6):517–24. Epub 1986/06/01. doi: 10.1002/ana.410190602 . [DOI] [PubMed] [Google Scholar]
- 151.Brew BJ, Rosenblum M, Cronin K, Price RW. AIDS dementia complex and HIV-1 brain infection: clinical-virological correlations. Ann Neurol. 1995;38(4):563–70. Epub 1995/10/01. doi: 10.1002/ana.410380404 . [DOI] [PubMed] [Google Scholar]
- 152.Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R. HIV-1 target cells in the CNS. J Neurovirol. 2015;21(3):276–89. Epub 2014/09/23. doi: 10.1007/s13365-014-0287-x ; PubMed Central PMCID: PMC4366351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sturdevant CB, Dow A, Jabara CB, Joseph SB, Schnell G, Takamune N, et al. Central nervous system compartmentalization of HIV-1 subtype C variants early and late in infection in young children. PLoS Pathog. 2012;8(12):e1003094. Epub 2013/01/10. doi: 10.1371/journal.ppat.1003094 ; PubMed Central PMCID: PMC3531524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Woodburn BM, Kanchi K, Zhou S, Colaianni N, Joseph SB, Swanstrom R. Characterization of Macrophage-Tropic HIV-1 Infection of Central Nervous System Cells and the Influence of Inflammation. J Virol. 2022;96(17):e0095722. Epub 2022/08/18. doi: 10.1128/jvi.00957-22 ; PubMed Central PMCID: PMC9472603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lustig G, Cele S, Karim F, Derache A, Ngoepe A, Khan K, et al. T cell derived HIV-1 is present in the CSF in the face of suppressive antiretroviral therapy. PLoS Pathog. 2021;17(9):e1009871. Epub 2021/09/24. doi: 10.1371/journal.ppat.1009871 ; PubMed Central PMCID: PMC8509856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. 2019;366(6472). Epub 2019/12/21. doi: 10.1126/science.aax9198 . [DOI] [PubMed] [Google Scholar]
- 157.Lucas SB, Wong KT, Nightingale S, Miller RF. HIV-Associated CD8 Encephalitis: A UK Case Series and Review of Histopathologically Confirmed Cases. Front Neurol. 2021;12:628296. Epub 2021/04/20. doi: 10.3389/fneur.2021.628296 ; PubMed Central PMCID: PMC8047670. [DOI] [PMC free article] [PubMed] [Google Scholar]