Abstract
In this report we describe the complete genome sequence of a nucleopolyhedrovirus that infects larval stages of the mosquito Culex nigripalpus (CuniNPV). The CuniNPV genome is a circular double-stranded DNA molecule of 108,252 bp and is predicted to contain 109 genes. Although 36 of these genes show homology to genes from other baculoviruses, their orientation and order exhibit little conservation relative to the genomes of lepidopteran baculoviruses. CuniNPV genes homologous to those from other baculoviruses include genes involved in early and late gene expression (lef-4, lef-5, lef-8, lef-9, vlf-1, and p47), DNA replication (lef-1, lef-2, helicase-1, and dna-pol), and structural functions (vp39, vp91, odv-ec27, odv-e56, p6.9, gp41, p74, and vp1054). Auxiliary genes include homologues of genes encoding the p35 antiapoptosis protein and a novel insulin binding-related protein. In contrast to these conserved genes, CuniNPV lacks apparent homologues of baculovirus genes essential (ie-1 and lef-3) or stimulatory (ie-2, lef-7, pe38) for DNA replication. Also, baculovirus genes essential or stimulatory for early-late (ie-1, ie-2), early (ie-0 and pe-38), and late (lef-6, lef-11, and pp31) gene transcription are not identifiable. In addition, CuniNPV lacks homologues of genes involved in the formation of virogenic stroma (pp31), nucleocapsid (orf1629, p87, and p24), envelope of occluded virions (odv-e25, odv-e66, odv-e18), and polyhedra (polyhedrin/granulin, p10, pp34, and fp25k). A homologue of gp64, a budded virus envelope fusion protein, was also absent, although a gene related to the other category of baculovirus budded virus envelope proteins, Ld130, was present. The absence of homologues of occlusion-derived virion (ODV) envelope proteins and occlusion body (OB) protein (polyhedrin) suggests that both CuniNPV ODV and OB may be structurally and compositionally different from those found in terrestrial lepidopteran hosts. The striking difference in genome organization, the low level of conservation of homologous genes, and the lack of many genes conserved in other baculoviruses suggest a large evolutionary distance between CuniNPV and lepidopteran baculoviruses.
The family Baculoviridae is a large and diverse family of occluded viruses with double-stranded DNA genomes that are pathogenic for insects, particularly of the lepidopteran, hymenopteran, and dipteran orders (23). They are divided into two genera: the nucleopolyhedroviruses (NPVs), which have large occlusion bodies (OBs) containing numerous virions (55), and the granuloviruses (GVs), which normally have single virions occluded within small granular OBs (70). Recently, the genome sequences of six NPVs and two GVs pathogenic for lepidoptera have been described (14, 34). In contrast, only limited information on the genes from dipteran and hymenopteran baculoviruses has been reported (42, 75).
Because a number of dipteran (mosquito) species are important vectors for a variety of human and veterinary diseases, extensive investigations to identify pathogens of these insects have been undertaken. The first mosquito baculovirus (AesoNPV) was isolated from Aedes sollicitans in Louisiana in 1969 (15), and over the following 20 years NPVs were isolated from 10 additional mosquito species of the genera Aedes, Anopheles, Culex, Psorophora, Uranotenia, and Wyeomyia (23). These genera are all members of the family Culicidae, which contains approximately 3,500 species. Because of the rarity of NPV isolation from mosquitoes and the difficulty of transmission in the laboratory, few cytopathological and morphological studies are available (24, 25, 66). However, the isolation and characterization of the conditions for virus transmission were recently reported for an NPV pathogenic for Culex nigripalpus (6, 42).
CuniNPV is highly pathogenic for C. nigripalpus and Culex quinquefasciatus, both of which are important vectors of St. Louis and Eastern encephalitis viruses (42), and it is responsible for epizootics in field populations of C. nigripalpus larvae. CuniNPV development is restricted to the nuclei of midgut epithelial cells in the gastric caeca and posterior stomach. As in other mosquito baculoviruses, CuniNPV has a double-stranded DNA genome that replicates in the nuclei and is packaged into rod-shaped singly enveloped nucleocapsids (42). It has two virion phenotypes, an occluded form (occlusion-derived virion [ODV]) that initiates infection in the midgut epithelial cells and a budded form (BV) that spreads the infection within the midgut. Like NPVs, the OBs are found exclusively in the nuclei of infected cells, but unlike NPVs, they are globular, not polyhedral. They are similar in size to GV OBs, but they typically contain four to eight virions, whereas each GV OB contains one or two virions. Additionally, Cuni-NPV OBs lack the envelope which surrounds the polyhedra of lepidopteran baculoviruses (42). Virion morphology and phylogenetic analysis of p74 and DNA polymerase led to the suggestion that this virus may represent a new genus, distinct from NPVs and GVs (42). In this report, we describe the complete genome sequence of CuniNPV and compare it to the genomes of other baculoviruses.
MATERIALS AND METHODS
CuniNPV DNA isolation, cloning, and sequencing.
CuniNPV was obtained from field-collected C. nigripalpus larvae isolated in 1997 in Gainesville, Fla. (6, 42), and amplified in laboratory colonies of C. nigripalpus and/or C. quinquefasciatus larvae following the procedures of Moser et al. (42). CuniNPV genomic DNA was obtained from viruses at the eighth mosquito passage. DNA was purified from OBs which were extracted from infected larvae by homogenization followed by differential centrifugation (42). Random DNA fragments of 1.5 to 6.5 kbp were obtained by incomplete enzymatic digestion with Tsp509I endonuclease (New England Biolabs, Beverly, Mass.). DNA fragments were cloned into the dephosphorylated EcoRI site of pUC19 plasmids and grown in Escherichia coli DH10B cells (GibcoBRL, Gaithersburg, Md.). pUC19 plasmids were purified using alkaline lysis according to the manufacturer's instruction (Eppendorf 5 Prime, Boulder, Colo.). DNA templates were sequenced from both ends with M13 forward and reverse primers using dideoxy chain terminator sequencing chemistries (60) and the Applied Biosystems PRISM 3700 automated DNA sequencer (PE Biosystems, Foster City, Calif.). Chromatogram traces were base-called with Phred (21, 22) and assembled with Phrap (21). Confirmatory assemblies were performed with The Institute for Genomic Research (67) and CAP3 (36) sequence assembly programs using quality files and clone length constraints (67). Quality files and default settings were used to produce a consensus sequence which was manually edited using the Consed sequence editor (29). An inconsequential degree of sequence variation was observed among sequenced clones within poly(A) stretches or short repeated motifs. This minor variation occurred in noncoding regions and did not affect sequence assembly. The final circular DNA consensus sequence represented on average eightfold redundancy at each base position and had an estimated error rate of 0.4/10 kbp.
DNA sequence analysis.
Genome DNA composition, structure, repeats, and restriction enzyme patterns were analyzed as previously described (1, 2, 42). Open reading frames (ORFs) longer than 30 codons with a methionine start codon (64, 65) were evaluated for coding potential using the Hexamer (ftp.sanger.ac.uk/pub/rd) and Glimmer (59) computer programs. Minor ORFs (those completely contained within larger ORFs) were excluded. bro gene family analysis was done with the MEME (19) and CAP software programs (68). Early and late promoter sequences were identified from regions located upstream (100 bp) of initiation codons as described by Kuzio et al. (39). DNA repeats were identified using the EMBOSS computer programs (Palindrome, Inverted, Tandem, Quicktandem, and Dotplot) (ftp: uk.embnet.org/pub/EMBOSS) and Blast (3). Protein homology searches were conducted using Blast (3), PsiBlast (4), FASTA (49), and HMMER (20, 63) programs with the databases PROSITE, Pfam, Prodom, Sbase, Blocks, Domo, and GenBank (11). Blast analysis was done on nonredundant databases (7270–1). GCG (19), MEMSAT (10), Psort (43), and SAPS (38) programs were used for general analysis, membrane prediction, and physical characterization of proteins.
Nucleotide sequence accession number.
The CuniNPV genome sequence has been deposited in GenBank under accession no. AF403738.
RESULTS AND DISCUSSION
Genome features.
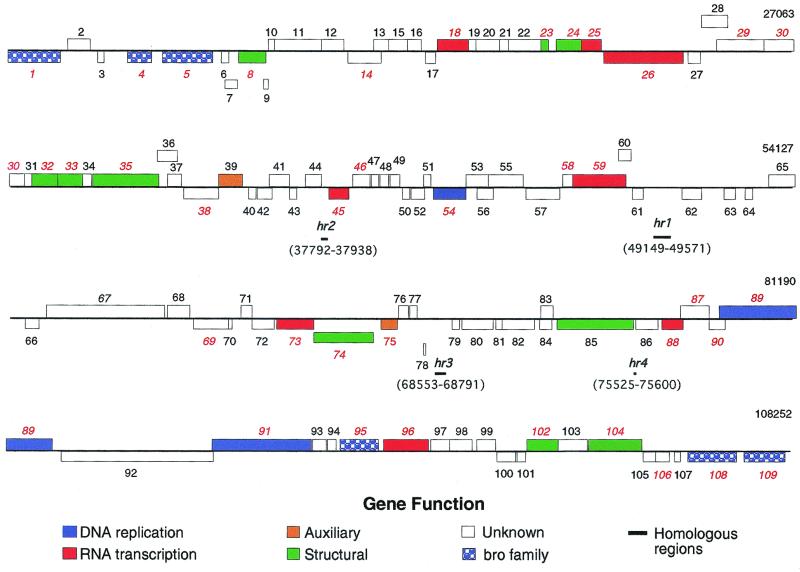
The CuniNPV genome is a circular double-stranded DNA molecule of 108,252 bp with a G+C content of 50.9%. This size is in good agreement with a previous restriction enzyme size estimate of 105 to 110 kbp (42). The CuniNPV genome contains densely arranged nonoverlapping clusters of 2 to 10 ORFs that are oriented in both directions. For descriptive purposes, we have presented the CuniNPV in a linearized form. The locations, sizes, and positions of ORFs are shown in Fig. 1 and Table 1. The CuniNPV genome contains 252 ORFs of 60 or more codons, of which 109 are likely to encode proteins (Table 1). Forty-six putative CuniNPV genes contain typical early and late baculovirus promoters upstream from the initiation codon (Table 1) (39).
FIG. 1.
Linear map of the CuniNPV genome. ORFs are numbered from left to right based on methionine initiation codon position. ORFs transcribed to the right are located above horizontal lines; ORFs transcribed to the left are below. Baculovirus homologues are indicated with red italicized numbers. Genes with related functions and members of gene families are colored according to the key.
TABLE 1.
CuniNPV ORFs
| ORF | Nucleotide position | Length (aa)a | Best match
|
ORF
|
Promoter typec | Commentsd | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accession no.b | Species | Blast score | Length (aa % identity) | AcMNPV | XcGV | |||||
| CUN001 | 1849–71 | 593 | AF162221 | Xestia c-nigrum GV | 327 | 451 (26) | 2 | 60 | bro, ATP_GTP-A motif | |
| CUN002 | 2159–2827 | 223 | L | ATP_GTP_A motif | ||||||
| CUN003 | 3366–3142 | 75 | ||||||||
| CUN004 | 5016–4147 | 290 | bro | |||||||
| CUN005 | 7085–5346 | 580 | AF271059 | Helicoverpa armigera NPV | 237 | 355 (24) | 2 | 60 | bro | |
| CUN006 | 7664–7362 | 101 | ||||||||
| CUN007 | 7986–7354 | 211 | ||||||||
| CUN008 | 8965–7979 | 329 | AF169823 | Spodoptera exigua NPV | 144 | 246 (28) | 54 | 175 | E | vp1054, virion associated, assembly |
| CUN009 | 9053–8814 | 80 | L | |||||||
| CUN010 | 9054–9257 | 68 | ||||||||
| CUN011 | 9261–10898 | 546 | L | |||||||
| CUN012 | 10919–11629 | 237 | L | |||||||
| CUN013 | 12743–13183 | 147 | ||||||||
| CUN014 | 12888–11779 | 370 | L22858 | Autographa californica NPV | 134 | 137 (26) | 92 | 101 | ||
| CUN015 | 13193–13888 | 232 | L | |||||||
| CUN016 | 13891–14277 | 129 | E | |||||||
| CUN017 | 14829–14347 | 161 | L | |||||||
| CUN018 | 14847–15920 | 358 | X71415 | Autographa california NPV | 250 | 327 (26) | 77 | 123 | L | vlf-I, very late expression factor I |
| CUN019 | 15940–16203 | 88 | L | |||||||
| CUN020 | 16203–16997 | 265 | L | |||||||
| CUN021 | 17019–17321 | 101 | ||||||||
| CUN022 | 17321–18466 | 382 | ||||||||
| CUN023 | 18476–18670 | 65 | AF271059 | Helicoverpa armigera NPV | 107 | 48 (54) | 100 | 94 | L | p6.9, SR repeat, DNA binding |
| CUN024 | 19004–19870 | 289 | P35840 | Lymantria dispar NPV | 139 | 291 (25) | 89 | 111 | L | p39-capsid, late coat protein |
| CUN025 | 19839–20513 | 225 | U75930 | Neodiprion sertifer NPV | 93 | 173 (27) | 6 | 35 | lef-2, late expression factor 2, N4_Mtase motif | |
| CUN026 | 23335–20570 | 922 | AF162221 | Xestia c-nigrum GV | 553 | 865 (26) | 50 | 148 | E | lef-8, late transcription, Rna_Pol_Beta motif |
| CUN027 | 23918–23493 | 142 | ||||||||
| CUN028 | 24026–24877 | 284 | ||||||||
| CUN029 | 24534–26102 | 523 | AF081810 | Lymantria dispar NPV | 573 | 488 (32) | 119 | 84 | L | |
| CUN030 | 26108–27529 | 474 | AF169823 | Spodoptera exigua NPV | 98 | 182 (28) | 142 | 13 | L | |
| CUN031 | 27568–27795 | 76 | L | |||||||
| CUN032 | 27856–28659 | 268 | AF081810 | Lymantria dispar NPV | 85 | 250 (22) | 144 | 112 | odv-ec27, occlusion-derived envelope | |
| CUN033 | 28666–29523 | 286 | AF169823 | Spodoptera exigua NPV | 64 | 186 (20) | 80 | 121 | L | gp41, tegument protein |
| CUN034 | 29526–29849 | 108 | L | |||||||
| CUN035 | 29909–32131 | 741 | AF081810 | Lymantria dispar NPV | 639 | 510 (31) | 83 | 118 | vp91-capsid, virion protein | |
| CUN036 | 32145–32780 | 212 | L | |||||||
| CUN037 | 32501–32929 | 143 | ||||||||
| CUN038 | 34228–33020 | 403 | AF081810 | Lymantria dispar NPV | 992 | 371 (52) | 22 | 45 | ||
| CUN039 | 34250–34963 | 238 | AF236641 | Spodoptera frugiperda (armyworm) | 145 | 215 (24) | L | |||
| CUN040 | 35503–35207 | 99 | ||||||||
| CUN041 | 36007–36579 | 191 | ||||||||
| CUN042 | 36062–35547 | 172 | ||||||||
| CUN043 | 36937–36656 | 94 | ||||||||
| CUN044 | 37270–37791 | 174 | ||||||||
| CUN045 | 38765–38061 | 235 | AF271059 | Helicoverpa armigera NPV | 180 | 149 (32) | 14 | 82 | lef-1, late expression factor 1 | |
| CUN046 | 38897–39505 | 203 | AF169823 | Spodoptera exigua NPV | 319 | 203 (31) | 115 | 32 | L | |
| CUN047 | 39510–39752 | 81 | ||||||||
| CUN048 | 39764–40114 | 117 | L | |||||||
| CUN049 | 40171–40491 | 107 | L | |||||||
| CUN050 | 40861–40538 | 108 | ||||||||
| CUN051 | 41329–41586 | 86 | ||||||||
| CUN052 | 41384–40854 | 177 | ||||||||
| CUN053 | 42768–43583 | 272 | ||||||||
| CUN054 | 42769–41669 | 367 | AF271059 | Helicoverpa armigera NPV | 245 | 295 (28) | 133 | 145 | alk-exo, DNA processing, exonuclease | |
| CUN055 | 43559–44716 | 386 | L | |||||||
| CUN056 | 43726–43193 | 178 | ||||||||
| CUN057 | 46055–44826 | 410 | E | |||||||
| CUN058 | 46089–46499 | 137 | AF037358 | Epiphyas postvittana NPV | 98 | 109 (26) | 68 | 135 | ||
| CUN059 | 46471–48240 | 590 | L33180 | Bombyx mori NPV | 303 | 388 (23) | 62 | 139 | lef-9, transcription, RNA polymerase motif | |
| CUN060 | 48054–48443 | 130 | ||||||||
| CUN061 | 48917–48477 | 147 | ||||||||
| CUN062 | 50968–50222 | 249 | ||||||||
| CUN063 | 52057–51665 | 131 | ||||||||
| CUN064 | 52686–52345 | 114 | L | |||||||
| CUN065 | 53089–54120 | 344 | E | |||||||
| CUN066 | 55021–54539 | 161 | ||||||||
| CUN067 | 55312–59382 | 1357 | L | |||||||
| CUN068 | 59508–60233 | 242 | E | |||||||
| CUN069 | 61562–60336 | 409 | AF162221 | Xestia c-nigrum GV | 93 | 273 (25) | 109 | 53 | ||
| CUN070 | 61746–61549 | 66 | L | |||||||
| CUN071 | 62028–62390 | 121 | E | |||||||
| CUN072 | 63185–62421 | 255 | E | |||||||
| CUN073 | 64553–63237 | 439 | AF081810 | Lymantria dispar NPV | 171 | 241 (25) | 40 | 78 | p47, transcription regulator | |
| CUN074 | 66537–64495 | 681 | AF271059 | Helicoverpa armigera NPV | 1018 | 634 (35) | 138 | 77 | L | p74, envelope protein |
| CUN075 | 67409–66858 | 184 | L22858 | Autographa californica NPV | 55 | 172 (18) | 135 | p35, apoptosis inhibitor | ||
| CUN076 | 67480–67809 | 110 | ||||||||
| CUN077 | 67883–68092 | 70 | ||||||||
| CUN078 | 68478–68173 | 102 | ||||||||
| CUN079 | 69621–69310 | 104 | ||||||||
| CUN080 | 70706–69624 | 361 | ||||||||
| CUN081 | 71035–70820 | 72 | ||||||||
| CUN082 | 72206–71031 | 392 | ||||||||
| CUN083 | 72412–72774 | 121 | ||||||||
| CUN084 | 72722–72348 | 125 | E | |||||||
| CUN085 | 75563–72918 | 882 | Occlusion body | |||||||
| CUN086 | 76454–75669 | 262 | E | ATP_GTP_A motif | ||||||
| CUN087 | 77217–78125 | 303 | U75930 | Orgyia pseudotsugata NPV | 268 | 288 (34) | 98 | 96 | L | |
| CUN088 | 77267–76470 | 266 | AF121349 | Neodiprion sertifer NPV | 96 | 200 (23) | 99 | 95 | lef-5, late expression factor | |
| CUN089 | 78649–82644 | 1332 | AF127530 | Choristoneura fumiferana NPV | 219 | 855 (20) | 95 | 98 | L | helicase-1, DNA replication |
| CUN090 | 78756–78151 | 202 | L22858 | Autographa california NPV | 216 | 169 (29) | 96 | 97 | L | |
| CUN091 | 88220–91633 | 1138 | AF215639 | Spodoptera littoralis NPV | 473 | 636 (27) | 65 | 132 | dna-pol, DNA replication, polymerase | |
| CUN092 | 88221–82915 | 1769 | L | |||||||
| CUN093 | 91721–92149 | 143 | ||||||||
| CUN094 | 92213–92479 | 89 | ||||||||
| CUN095 | 92641–93981 | 447 | AF162221 | Xestia c-nigrum GV | 89 | 89 (31) | 2 | 60 | bro | |
| CUN096 | 94194–95684 | 497 | AF162221 | Xestia c-nigrum GV | 230 | 493 (25) | 90 | 110 | lef-4, transcription | |
| CUN097 | 95822–96598 | 259 | L | |||||||
| CUN098 | 96469–97212 | 248 | L | |||||||
| CUN099 | 97438–98010 | 191 | ||||||||
| CUN100 | 98824–98075 | 250 | ||||||||
| CUN101 | 99108–98743 | 122 | ||||||||
| CUN102 | 99162–100244 | 361 | Q83953 | Orgyia pseudotsugata NPV | 318 | 302 (33) | 148 | 15 | L | odv-e56, envelope |
| CUN103 | 100301–101215 | 305 | E/L | |||||||
| CUN104 | 101319–103079 | 587 | AE002638 | Drosophila melanogaster | 184 | 250 (24) | 23 | 27 | L | Ld130, envelope fusion protein |
| CUN105 | 103598–103164 | 145 | L | |||||||
| CUN106 | 104155–103598 | 186 | AF162221 | Xestia c-nigrum GV | 111 | 139 (23) | 81 | 120 | ||
| CUN107 | 104512–104261 | 84 | ||||||||
| CUN108 | 106474–104672 | 601 | AF162221 | Xestia c-nigrum GV | 354 | 449 (27) | 2 | 60 | bro, ATP_GTP_A motif | |
| CUN109 | 108129–106660 | 490 | AF162221 | Xestia c-nigrum GV | 138 | 198 (23) | 2 | 60 | bro, ATP_GTP_A motif | |
aa, amino acids.
Accession numbers are from the GenBank or SwissProt databases.
Putative promoter. E, early; L, late.
Function was deduced from the degree of amino acid similarity to known genes or by the presence of PROSITE signatures.
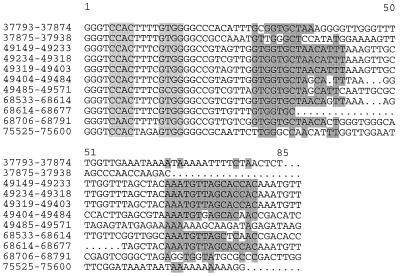
The CuniNPV genome contains four putative homologous regions (hrs) composed of 64- to 85-bp repeats located in intergenic regions which contain inverted repeats but lack sequence similarity to hrs from other baculoviruses (Fig. 2). Five copies of an imperfect 85-bp repeat motif (hr1) are located at positions 49149 to 49571. Imperfect and incomplete related sequences are also present at nucleotide positions 37792 to 37938 (hr2), 68533 to 68791 (hr3), and 75525 to 75600 (hr4). Homologous regions have been identified in all baculovirus genomes sequenced. These are composed of sets of closely related repeated sequences present at several locations throughout the genome. Evidence suggests that hrs may function as viral origins of replication (48) and enhancers of early gene expression (30). All previously sequenced baculovirus genomes contain 4 (Plutella xylostella GV) to 13 (Lymantria dispar MNPV [LdMNPV]) hrs located in multiple genomic locations (32, 39). They are composed of direct DNA repeats and imperfect DNA palindromic sequences. In general, hrs share significant intragenomic sequence homology, although they may show very low homology between viruses (32, 33, 48, 53).
FIG. 2.
CuniNPV homologous regions. Shaded areas indicate inverted repeats. Genomic nucleotide positions are on the left. Nucleotide positions in repeats are at the top. Dots represent gaps introduced by the multiple alignment program Pileup (19).
Gene categories.
Gene order and orientation are not conserved between the CuniNPV genome and genomes of baculoviruses infecting lepidopteran species (34). Only 36 of the 109 putative CuniNPV genes demonstrate clear homology to genes from other baculoviruses, implying that the core of conserved baculovirus genes may be smaller than the previously suggested 65 genes (14) or that there has been such extensive evolution that gene relatedness cannot be determined. Seventy-two CuniNPV ORFs show no homology to any other known baculovirus ORFs. In addition, the amino acid conservation between CuniNPV and lepidopteran baculovirus ORFs is low (18 to 54%), with an average of 28% identity to Autographa californica NPV (AcMNPV) (34). This is in contrast with homologues of Xestia c-nigrum and P. xylostella GVs, which show about 33% amino acid identity with the corresponding ORFs in AcMNPV (32, 33).
Early transcription.
CuniNPV lacks homologues of the lepidopteran baculovirus early transcription factors and regulators ie-0, ie-1, ie-2, and pe38 (34). Promoters of early baculovirus genes are transcribed by a combination of host RNA polymerase II and virally encoded transcription factors (26). The absence of homologues of these genes may indicate that CuniNPV is dependent on host factors for early transcription or that unique CuniNPV genes are involved in these functions. Alternatively, conservation of certain CuniNPV genes may be extremely low, thus making identification of homologues of other baculoviruses impossible.
DNA replication.
CUN045 (lef-1), CUNO25 (lef-2), CUN089 (helicase-1), CUN091 (dna-pol), CUN054 (alk-exo), and CUN018 (vlf-1) are homologues of genes likely involved in DNA replication (40). Significant amino acid changes in these homologues and the absence of a number of essential and nonessential DNA replication genes distinguish CuniNPV from lepidopteran baculoviruses.
In addition to DNA polymerase (CUN091), which was described previously (42), other CuniNPV genes implicated in DNA replication show significant differences from their baculovirus homologues. CUN045 (lef-1) has a 58-amino-acid deletion at the amino terminus and a 31-amino-acid insertion in the central part of the ORF in comparison to its closest homologue (Table 1) (5). CUN025 (lef-2) contains an N-4 cytosine-specific methylase Prosite signature (PS00093) at its amino-terminal end which is absent in other baculovirus LEF-2 proteins. In prokaryotes, N-4 cytosine-specific DNA methylases (EC 2.1.1.113) methylate the amino group at the C-4 position of cytosines in DNA, thus protecting DNA from cleavage by type II restriction enzymes. An N-4 cytosine-specific methylase signature motif may be the remnant of an ancestral restriction modification mechanism or a may indicate a dual function for the lef-2 homologue. Active restriction modification systems have been found in large DNA viruses of the chlorella algae but not in baculoviruses (51). CUN054 (alk-exo) lacks 68 amino acids at the carboxy terminus in comparison to its closest homologue (Table 1) (40).
CUN089 (helicase-1) has statistically significant amino acid identity (P = 10−13) to the helicase-1 gene of Choristoneura fumiferana NPV (Table 1) and contains regions of similarity to baculovirus helicase domains I to IV (35). Interestingly, one of the most conserved regions corresponds to a helicase-1 domain involved in host range specificity (17). In comparison to the helicase-1 of other baculoviruses, CUN089 has low amino acid identity (Table 1) and lacks domain VI, the nucleotide binding motif, and the leucine zipper DNA binding motif. It also has 97 additional amino acids in the amino-terminal region.
CuniNPV lacks recognizable homologues of lef-3, which has the properties of a single-stranded-DNA binding protein and is involved in the transport of helicase into the nucleus (71). It should be noted, however, that lef-3 and the early and late gene activator ie-1 are very poorly conserved in baculoviruses (32, 33, 34). Other baculovirus genes apparently lacking are present only in some baculoviruses and include dbp (DNA binding protein), lef-7, pcna (proliferating cell nuclear antigen), DNA ligase, rnr1 and rnr2 (large and small subunits of ribonucleotide reductase), and dUTPase (34, 50).
Late and very late transcription.
CUN096 (lef-4), CUN026 (lef-8), CUN059 (lef-9), and CUN073 (p47) are homologues of genes (Table 1) that encode a multisubunit RNA polymerase involved in late transcription (31, 41). In addition, CUN088 (lef-5) has been implicated in late gene transcription (52). CUN018 is similar to very late transcription factor 1 (vlf-1), the major transactivator of very late gene expression (41, 72–74).
In addition, a number of genes have been implicated in transcription by means of a transfection-based transient gene expression assay (52). Although CuniNPV contains the minimal complex necessary for late polymerase activity (lef-4, lef-8, lef-9, and p47) (31, 41), it lacks homologues of other lepidopteran baculovirus expression factors which are required for optimum levels of late transcription (lef-6, lef-10, lef-11, lef-12, and pp31 [39k]) (Table 2). Some of the transcription factors absent in CuniNPV are species specific (lef-7, lef-10, lef-12, and host cell specific factor 1), but others are conserved in all completely sequenced lepidopteran baculoviruses (lef-6, lef-11, pk-1, and pp31 [39k]) (34, 41).
TABLE 2.
Comparison between CuniNPV and conserved lepidopteran genesa
| Gene function | Gene(s) present in CuniNPV | Genes absent in CuniNPV |
|---|---|---|
| Transcription | p47, lef-8, lef-9, vlf-1, lef-4, lef-5 | lef-6, pp31 (39k), lef-11 |
| Replication | lef-2, lef-1, dna-pol, helicase-1 | lef-3, ie-1 |
| Structural proteins | vp1054, gp41, vp39, vp91 p6.9, p74, odv-ec27, odv-e56 | polyhedrin/granulin, pk-1, p10, odv-e18, odv-e25, odv-e66 |
| Host metabolism | p35 | fgf, iap, sod, ubiquitin |
| Other | alk-exo | dbp, fp, ie-0 |
| Unknown (AcMNPV ORF) | 023, 068, 081, 096, 098, 109, 115 | 013, 022, 029, 038, 053, 066, 071, 075, 076, 078, 082, 092, 093, 101, 102, 103, 106, 107, 110, 119, 139, 142, 145, 146, 150 |
Conserved genes correspond to the genomes of seven completely sequenced baculoviruses (34).
Virion structural proteins.
CuniNPV contains only 8 of 17 structural protein genes conserved among lepidopteran baculoviruses (vp39, vp91, vp1054, odv-ec27, odv-e56, p6.9, gp41, and p74) (Table 1). Eleven structural genes are absent or not identifiable. These include orf1629, p87, gp64, p24, odv-e18, odv-e25, odv-e66, p10, pp34, polyhedrin/granulin, and fp25k (34).
Nucleocapsid proteins.
CUN024 (vp39), CUN035 (vp91), and CUN008 (vp1054) are homologues of lepidopteran baculovirus genes encoding capsid-associated proteins. CUN024 contains codons for only two of the eight cysteine residues that are conserved in the vp39 capsid protein. CUN035 resembles the gene encoding a protein found in both the capsid and envelope of ODVs (58). CUN008 (vp1054) resembles the gene for a virion-associated protein that functions in nucleocapsid formation (27). Homologues of the capsid-associated proteins p87, p24, and orf1629, an essential protein associated with the basal structure of the capsid, are absent or not identifiable in CuniNPV (27).
BV proteins.
CuniNPV lacks a homologue of AcMNPV gp64, but it does encode a homologue (CUN104) of the LdMNPV envelope protein Ld130. The CUN104 product also demonstrates relatedness to Se8, the envelope fusion protein of Spodoptera exigua NPV (SeMNPV) (37), and CG4715, a Drosophila gene product related to Ld130 (56) (Table 1). Lepidopteran baculoviruses can be divided into two groups based on the envelope fusion proteins of their budded viruses (47). AcMNPV and its close relatives utilize gp64, which is a low-pH-activated envelope fusion protein (9). In contrast, many other diverse baculoviruses, including both GVs and NPVs, lack homologues of gp64 but encode proteins related to Ld130, the envelope fusion protein of LdMNPV.
Proteins associated with the occluded virus.
CUN032, CUN074, and CUN102 are homologues of the genes for three proteins associated with the occluded viral envelope, and CUN033 is the homologue of a tegument protein. CUN032 is the homologue of odv-ec27, which encodes a protein present in ODV nucleocapsids and envelopes and may function as a cyclin (7). CUN074 is similar to the gene for p74, an ODV protein required for oral infectivity. CUN102 is the homologue of odv-e56, which encodes a protein associated with both virus-induced intranuclear vesicles and envelopes. CUN033 is the homologue of the gene for the ODV tegument protein gp41 (27). gp41 is required for egress of nucleocapsids from the nucleus in the pathway of budded virus synthesis (44).
The absence of several baculovirus ODV gene homologues suggests involvement of another set of viral or perhaps cellular proteins in CuniNPV ODV assembly. odv-e25, odv-e66, and odv-e18 are absent in CuniNPV. In lepidopteran baculoviruses there are four highly conserved ODV envelope proteins (ODV-E18, ODV-E25, ODV-E56, and ODV-E66). ODV-E25 and ODV-E66 are found in the intranuclear microvesicles that are believed to be viral envelope precursors and are necessary for the morphogenesis of preoccluded virions. In addition, CuniNPV lacks a p24 homologue (AcMNPV ORF 128), a protein associated with ODV and BV. Lepidopteran ODVs enter the brush border microvilli of midgut epithelial cells by fusion of the viral envelope with the cellular plasma membrane (69). The lack of ODV proteins suggests that other CuniNPV proteins may perform these cell entry functions.
Proteins involved with virion occlusion.
CuniNPV OBs are globular, lack a polyhedron envelope and calyx structure, and typically contain about four individually enveloped virions (42). In addition, although they appear to be composed of a peptide similar in size to lepidopteran polyhedrin or granulin (about 30 kDa), amino-terminal amino acid sequence analysis did not reveal homology to other baculovirus OB proteins (42). However, the sequence did match positions 693 to 709 of CUN085, an ORF with no homology to any other known baculovirus gene. The large size of the CUN085 product (882 amino acids) suggests that it is cleaved to produce components of CuniNPV OBs. In addition, no homologues of polyhedrin or granulin genes were identified in the genome sequence.
CuniNPV also lack homologues of the polyhedron-associated proteins p10, fp25K (a conserved protein involved in polyhedra formation), and pp34, the polyhedron envelope and calyx protein (27, 34). The lack of a pp34 homologue is reflected in the absence of a polyhedron envelope and calyx surrounding the OB. The small size of the CuniNPV OBs, the limited number of virions occluded, and the lack of an envelope and calyx structure are reminiscent of the structure of the OBs of GVs.
The absence of a p10 gene in the CuniNPV genome is supported by observed morphological differences from cells infected with lepidopteran NPVs (42). In infected lepidopteran cells, p10 is expressed as an abundant protein (69) that is associated with nuclear and cytoplasmic fibrillar bodies during terminal stages of infection. Fibrillar material produced during CuniNPV infection, however, does not resemble fibrillar bodies associated with lepidopteran baculovirus infections. Most notably, the cytoplasmic features of CuniNPV-infected cells are microtubule bundles and irregular cisternae of smooth endoplasmic reticulum.
bro gene family.
The CuniNPV genome contains six baculovirus repeated ORFs (bro ORFs) (CUN001, CUN004, CUN005, CUN095, CUN108, and CUN109). CuniNPV bro ORFs are variable in length (290 to 601 amino acids) and similarity (23 to 31% amino acid identities) to other baculovirus bro ORFs (Table 1). The closest baculovirus homologues are the genes for X. c-nigrum GV ORF60 and ORF131, followed by LdMNPV group III bro d, c, and i (39). All six CuniNPV bro ORFs contain the sequence for the 41-amino-acid motif common to all bro proteins (39) and two additional amino acid motifs conserved in group I and III bro proteins. Baculovirus repeated ORFs are present in 1 to 17 copies in most NPVs and GVs. Although their function is unknown, some bro proteins have been shown to bind DNA and may be involved in viral DNA replication (76).
Genes that may inhibit apoptosis.
CuniNPV contains one gene (CUN075) with homology to the baculovirus antiapoptosis p35 gene (16), but it lacks homologues of the iap (inhibitor of apoptosis) family of genes (18). Although up to four copies of iap genes may be present in baculovirus genomes (39), CuniNPV lacks members of this gene family. However, CUN075 demonstrates homology to p35 genes, suggesting that it may be utilized to block apoptosis. The lack of the sequence for the 110-amino-acid carboxy-terminal region, which in AcMNPV p35 mediates antiapoptotic activity (8, 13), in CUN075 is surprising and suggests a different mode of action for this gene.
Auxiliary genes.
Auxiliary genes are nonessential for viral replication in cell culture, although they likely provide selective advantages in insects. CuniNPV lacks homologues of at least 14 baculovirus auxiliary genes, including those encoding superoxide dismutase, ubiquitin, inhibitor of apoptosis, protein kinase 1, and viral enhancing factors (45).
CUN039 shows relatedness to genes for insulin binding proteins of Spodoptera frugiperda and may belong to the auxiliary gene category. It is also related to a Caenorhabditis elegans gene of unknown function and Drosophila IMP-L2 (28, 46, 61). Insulin and related peptides are very important hormones for the regulation of growth and metabolism. The insulin-related peptide binding protein secreted from S. frugiperda cells is composed of two immunoglobulin-like C2 domains, binds human insulin, and inhibits insulin signaling through the insulin receptor (61). The closest homologue of the S. frugiperda binding protein is the essential protein IMP-L2, found in Drosophila melanogaster. IMP-L2 also binds insulin and related peptides (28, 46, 61) and is implicated in neural and ectodermal development (28, 46). CUN039 may have a similar role in modifying host metabolism and/or development.
Phylogeny of CuniNPV.
Although clearly related to lepidopteran baculoviruses, the data from our sequence analyses indicate a number of major differences. These include low levels of identity between homologous ORFs, lack of conservation in gene order, absence of many genes present in all lepidopteran baculovirus genomes, and the lack of a homologue to polyhedrin or granulin, which are two of the most conserved baculovirus gene products (Table 2). These differences resemble those observed among viral subfamilies (1, 2) and suggest that the evolutionary distance between CuniNPV and lepidopteran baculoviruses is greater than the distance separating GVs from NPVs. Our previous phylogenetic analyses using DNA polymerase and p74 sequences suggested that CuniNPV is a member of a baculovirus lineage distinct from lepidopteran NPVs and GVs (43). Phylogenetic trees and distance analysis of highly conserved CuniNPV genes (CUN038 and CUN074) reveals a rate of amino acid change similar to that of Nudiviruses, a group that has recently been excluded from the Baculoviridae (data not shown).
It has been suggested that some viruses (62), including baculoviruses (54, 57), may coevolve with their hosts in a process called host-dependent evolution. Molecular evidence indicates that the ancestral Lepidoptera and Diptera separated about 280 million years ago (12). The extent of the differences between the lepidopteran baculoviruses and CuniNPV may reflect this ancient separation.
ACKNOWLEDGMENTS
We thank G. F. Rohrmann for his many helpful comments and suggestions on the manuscript and A. Ciupryk and A. Zsak for excellent technical assistance.
REFERENCES
- 1.Afonso C A, Tulman E R, Lu Z, Oma E, Kutish G F, Rock D L. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso C L, Tulman E R, Lu Z, Zsak L, Kutish G F, Rock D L. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett J W, Lauzon H A, Mercuri P S, Krell P J, Sohi S S, Arif B M. The putative LEF-1 proteins from two distinct Choristoneura fumiferana multiple nucleopolyhedroviruses share domain homology to eukaryotic primases. Virus Genes. 1996;13:229–237. doi: 10.1007/BF00366983. [DOI] [PubMed] [Google Scholar]
- 6.Becnel J J, White S E, Moser B A, Fukuda T, Rotstein M J, Undeen A H, Cockburn A. Epizootiology and transmission of a newly discovered baculovirus from the mosquitoes Culex nigripalpus and C. quinquefasciatus. J Gen Virol. 2001;82:275–282. doi: 10.1099/0022-1317-82-2-275. [DOI] [PubMed] [Google Scholar]
- 7.Belyavskyi M, Braunagel S C, Summers M D. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proc Natl Acad Sci USA. 1998;95:11205–11210. doi: 10.1073/pnas.95.19.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertin J, Mendrysa S M, LaCount D J, Gaur S, Krebs J F, Armstrong R C, Tomaselli K J, Friesen P D. Apoptotic suppression by baculovirus P35 involves cleavage by and inhibition of a virus-induced CED-3/ICE-like protease. J Virol. 1996;70:6251–6259. doi: 10.1128/jvi.70.9.6251-6259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blissard G W, Wenz J R. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brendel V, Bucher P, Nourbakhsh I R, Blaisdell B E, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burks C. Molecular biology database list. Nucleic Acids Res. 1999;27:1–9. doi: 10.1093/nar/27.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burmester T, Massey H C, Jr, Zakharkin S, Benes H. The evolution of hexamerins and the phylogeny of insects. J Mol Evol. 1998;47:93–108. doi: 10.1007/pl00006366. [DOI] [PubMed] [Google Scholar]
- 13.Cartier J L, Hershberger P A, Friesen P D. Suppression of apoptosis in insect cells stably transfected with baculovirus p35: dominant interference by N-terminal sequences p35(1–76) J Virol. 1994;68:7728–7737. doi: 10.1128/jvi.68.12.7728-7737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Ijkel W F, Tarchini R, Sun X, Sandbrink H, Wang H, Peters S, Zuidema D, Lankhorst R K, Vlak J M, Hu Z. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J Gen Virol. 2001;82:241–257. doi: 10.1099/0022-1317-82-1-241. [DOI] [PubMed] [Google Scholar]
- 15.Clark T B, Chapman H C, Fukuda T. Nuclear-polyhedrosis and cytoplasmic-polyhedrosis virus infections in Louisiana mosquitoes. J Invertebr Pathol. 1969;14:284–286. doi: 10.1016/0022-2011(69)90120-7. [DOI] [PubMed] [Google Scholar]
- 16.Clem R J, Fechheimer M, Miller L K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 17.Croizier G, Croizier L, Argaud O, Poudevigne D. Extension of Autographa californica nuclear polyhedrosis virus host range by interspecific replacement of a short DNA sequence in the p143 helicase gene. Proc Natl Acad Sci USA. 1994;91:48–52. doi: 10.1073/pnas.91.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crook N E, Clem R J, Miller L K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eddy S R, Mitchison G, Durbin R. Maximum discrimination hidden Markov models of sequence consensus. J Comput Biol. 1995;2:9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- 21.Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 22.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 23.Federici B A. Viral pathogens of mosquito larvae. Bull Am Mosquito Control Assoc. 1985;6:62–74. [Google Scholar]
- 24.Federici B A. Mosquito baculovirus: sequence of morphogenesis and ultrastructure of the virion. Virology. 1980;100:1–9. doi: 10.1016/0042-6822(80)90546-2. [DOI] [PubMed] [Google Scholar]
- 25.Federici B A, Lowe R E. Studies on the pathology of a baculovirus in Aedes triseriatus. J Invertebr Pathol. 1972;20:14–21. doi: 10.1016/0022-2011(72)90075-4. [DOI] [PubMed] [Google Scholar]
- 26.Friesen P D. Regulation of baculovirus early gene expression. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 141–170. [Google Scholar]
- 27.Funk C J, Braunagel S C, Rohrmann G F. Baculovirus structure. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 7–32. [Google Scholar]
- 28.Garbe J C, Yang E, Fristrom J W. IMP-L2: an essential secreted immunoglobulin family member implicated in neural and ectodermal development in Drosophila. Development. 1993;119:1237–1250. doi: 10.1242/dev.119.4.1237. [DOI] [PubMed] [Google Scholar]
- 29.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 30.Guarino L, Summers M D. Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J Virol. 1986;60:215–223. doi: 10.1128/jvi.60.1.215-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guarino L A, Xu B, Jin J, Dong W. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol. 1998;72:7985–7991. doi: 10.1128/jvi.72.10.7985-7991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashimoto Y, Hayakawa T, Ueno Y, Fujita T, Sano Y, Matsumoto T. Sequence analysis of the Plutella xylostella granulovirus genome. Virology. 2000;275:358–372. doi: 10.1006/viro.2000.0530. [DOI] [PubMed] [Google Scholar]
- 33.Hayakawa T, Ko R, Okano K, Seong S-I, Goto C, Maeda S. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 34.Hayakawa T, Rohrmann G F, Hashimoto Y. Patterns of genome organization and content in lepidopteran baculoviruses. Virology. 2000;278:1–12. doi: 10.1006/viro.2000.0668. [DOI] [PubMed] [Google Scholar]
- 35.Heldens J G M, Liu Y, Zuidema D, Goldbach R W, Vlak J M. Characterization of a putative Spodoptera exigua multicapsid nucleopolyhedrovirus helicase gene. J Gen Virol. 1997;78:3101–3114. doi: 10.1099/0022-1317-78-12-3101. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.IJkel W F J, Westenberg M, Goldbach R W, Blissard G W, Vlak J M, Zuidema D. A novel baculovirus fusion envelope protein with a proprotein convertase cleavage site. Virology. 2000;275:30–41. doi: 10.1006/viro.2000.0483. [DOI] [PubMed] [Google Scholar]
- 38.Jones D T, Taylor W R, Thornton J M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 39.Kuzio J, Pearson M N, Harwood S H, Funk C J, Evans J T, Slavicek J M, Rohrmann G F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 40.Lu A, Krell P J, Vlak J M, Rohrmann G F. Baculovirus DNA replication. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 171–191. [Google Scholar]
- 41.Lu A, Miller L K. Regulation of baculovirus late and very late gene expression. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 193–216. [Google Scholar]
- 42.Moser B A, Becnel J J, White S E, Afonso C L, Kutish G F, Shanker S, Almira E. Morphological and molecular evidence that Culex nigripalpus baculovirus is an unusual member of the family Baculoviridae. J Gen Virol. 2001;82:283–297. doi: 10.1099/0022-1317-82-2-283. [DOI] [PubMed] [Google Scholar]
- 43.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 44.Olszewski J, Miller L K. A role for baculovirus GP41 in budded virus production. Virology. 1997;233:292–301. doi: 10.1006/viro.1997.8612. [DOI] [PubMed] [Google Scholar]
- 45.O'Reilly D R. Auxiliary genes of baculoviruses. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 267–300. [Google Scholar]
- 46.Osterbur D L, Fristrom D K, Natzle J E, Tojo S J, Fristrom J W. Genes expressed during imaginal disc morphogenesis: IMP-L2, a gene expressed during imaginal disc and imaginal histoblast morphogenesis. Dev Biol. 1988;129:439–448. doi: 10.1016/0012-1606(88)90391-0. [DOI] [PubMed] [Google Scholar]
- 47.Pearson M N, Groten C, Rohrmann G F. Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J Virol. 2000;74:6126–6131. doi: 10.1128/jvi.74.13.6126-6131.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson M N, Rohrmann G F. Lymantria dispar nuclear polyhedrosis virus homologous regions: characterization of their ability to function as replication origins. J Virol. 1995;69:213–321. doi: 10.1128/jvi.69.1.213-221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 50.Possee R D, Rohrmann G F. Baculovirus genome organization and evolution. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 109–140. [Google Scholar]
- 51.Que Q, Zhang Y, Nelson M, Ropp S, Burbank D E, Van Etten J L. Chlorella virus SC-1A encodes at least five functional and one nonfunctional DNA methyltransferases. Gene. 1997;190:237–244. doi: 10.1016/s0378-1119(96)00862-1. [DOI] [PubMed] [Google Scholar]
- 52.Rapp J C, Wilson J A, Miller L K. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J Virol. 1998;72:10197–10206. doi: 10.1128/jvi.72.12.10197-10206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasmussen C, Leisy D J, Ho P S, Rohrmann G F. Structure-function analysis of the Autographa californica multinucleocapsid nuclear polyhedrosis virus homologous region palindromes. Virology. 1996;224:235–245. doi: 10.1006/viro.1996.0525. [DOI] [PubMed] [Google Scholar]
- 54.Rohrmann G F. Evolution of occluded baculoviruses. Vol. 1. Boca Raton, Fla: CRC Press; 1986. [Google Scholar]
- 55.Rohrmann G F. Nucleopolyhedrovirus. 2nd ed. London, United Kingdom: Academic Press; 1999. [Google Scholar]
- 56.Rohrmann G F, Karplus P A. Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC Evol Biol. 2001;1:1–9. doi: 10.1186/1471-2148-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohrmann G F, Pearson M N, Bailey T J, Becker R R, Beaudreau G S. N-terminal polyhedrin sequences and occluded baculovirus evolution. J Mol Evol. 1981;17:329–333. doi: 10.1007/BF01734354. [DOI] [PubMed] [Google Scholar]
- 58.Russell R L, Rohrmann G F. Characterization of P91, a protein associated with virions of an Orgyia pseudotsugata baculovirus. Virology. 1997;233:210–223. doi: 10.1006/viro.1997.8599. [DOI] [PubMed] [Google Scholar]
- 59.Salzberg S L, Delcher A L, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sloth Andersen A, Hertz Hansen P, Schaffer L, Kristensen C. A new secreted insect protein belonging to the immunoglobulin superfamily binds insulin and related peptides and inhibits their activities. J Biol Chem. 2000;275:16948–16953. doi: 10.1074/jbc.M001578200. [DOI] [PubMed] [Google Scholar]
- 62.Soeda E, Maruyama T, Arrand J, Griffin B. Host-dependent evolution of three papova viruses. Nature. 1980;285:165–167. doi: 10.1038/285165a0. [DOI] [PubMed] [Google Scholar]
- 63.Sonnhammer E L L, Eddy S R, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staden A, McLachlan A D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982;10:141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982;10:2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stiles B, Dunn P E, Paschke J D. Histopathology of a nuclear polyhedrosis infection in Aedes epactius with observations in four additional mosquito species. J Invertebr Pathol. 1983;41:191–202. doi: 10.1016/0022-2011(83)90219-7. [DOI] [PubMed] [Google Scholar]
- 67.Sutton G G, White O, Adams M D, Kerlavage A R. TIGR assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Technol. 1995;1:9–19. [Google Scholar]
- 68.Tatusov R L, Altschul S F, Koonin E V. Detection of conserved segments in proteins: iterative scanning of sequence databases with alignment blocks. Proc Natl Acad Sci USA. 1994;91:12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams G V, Faulkner P. Cytological changes and viral morphogenesis during baculovirus infection. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 61–107. [Google Scholar]
- 70.Winstanley D, O'Reilly D. Granuloviruses. 2nd ed. London, United Kingdom: Academic Press; 1999. [Google Scholar]
- 71.Wu Y, Carstens E B. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology. 1998;247:32–40. doi: 10.1006/viro.1998.9235. [DOI] [PubMed] [Google Scholar]
- 72.Yang S, Miller L K. Activation of baculovirus very late promoters by interaction with very late factor 1. J Virol. 1999;73:3404–3409. doi: 10.1128/jvi.73.4.3404-3409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang S, Miller L K. Control of baculovirus polyhedrin gene expression by very late factor 1. Virology. 1998;248:131–138. doi: 10.1006/viro.1998.9272. [DOI] [PubMed] [Google Scholar]
- 74.Yang S, Miller L K. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology. 1998;245:99–109. doi: 10.1006/viro.1998.9152. [DOI] [PubMed] [Google Scholar]
- 75.Zanotto P M, Kessing B D, Maruniak J E. Phylogenetic interrelationships among baculoviruses: evolutionary rates and host associations. J Invertebr Pathol. 1993;62:147–164. doi: 10.1006/jipa.1993.1090. [DOI] [PubMed] [Google Scholar]
- 76.Zemskov E A, Kang W, Maeda S. Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. J Virol. 2000;74:6784–6789. doi: 10.1128/jvi.74.15.6784-6789.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]