Abstract
The spike glycoprotein E2 of Sindbis virus (SIN) is synthesized in the infected cell as a PE2 precursor protein, which matures through cleavage by a cellular furin-like protease. Previous work has shown that SIN mutants impaired in PE2 cleavage are noninfectious on BHK-21 cells, the block in infection being localized at a step after virus-receptor interaction but prior to RNA replication. Here, we studied the membrane fusion properties of SIN PE2 cleavage mutants and observed that these viruses are impaired in their ability to form an E1 homotrimer and to fuse with liposomes at a mildly acidic pH. The block in spike rearrangement and fusion could be overridden by exposure of the mutant viruses to very low pH (<4.5). Cleavage mutants with second-site resuscitating mutations in PE2 were highly infectious for BHK-21 cells. The ability of these viruses to form E1 homotrimers and to fuse at a mildly acidic pH was completely restored despite a sustained lack of PE2 cleavage.
Alphaviruses, such as Sindbis virus (SIN), Semliki Forest virus (SFV), and Venezuelan equine encephalitis virus (VEE), are enveloped viruses which contain three major structural proteins, the capsid protein, C, and two envelope glycoproteins, E2 and E1 (38). The glycoproteins are organized on the surface of the virion in 80 heterooligomeric spikes, which mediate the infectious entry of these viruses into cells. The E2 glycoprotein is primarily involved in the interaction of a virus particle with cell surface attachment receptors (37), whereas the E1 glycoprotein is necessary for the subsequent membrane fusion process (8, 40). Very recent crystallographic and cryoelectron microscopy studies have revealed that E1 in many respects resembles the flavivirus E glycoprotein. The alphavirus E1 protein appears to lie flat on the viral surface, driving lateral spike interactions, whereas E2 forms the spike protrusions (21, 30).
In the infected cell, the alphavirus structural proteins are translated as a large polyprotein. Once the C protein is cleaved off the polypeptide chain, the NH2 terminus of PE2, the precursor protein of E2 (PE2 is called p62 in SFV), serves to direct the cotranslational translocation of the remaining polyprotein to the endoplasmic reticulum (ER) (38). This polyprotein is processed by a signal peptidase within the ER, and the two envelope proteins associate to form PE2/E1 heterodimers (38). The PE2/E1 heterodimer matures further while passing through the Golgi and trans-Golgi network (TGN). In the TGN or in a post-TGN compartment, PE2 is cleaved close to its amino terminus to form E2 and a residual peptide, E3 (24). PE2 cleavage is mediated by a furin-like host protease at a consensus recognition sequence, XBXBBX, in which B is a basic and X a hydrophobic amino acid (18). Peptide E3, which contains the XBXBBX sequence, is retained on the SFV spike, but it is released in the case of SIN (24, 28).
Several investigators have shown that the efficiency of PE2 cleavage can be influenced by amino acid changes within the cleavage site (5, 13, 19, 22, 34, 39). Thus, a number of PE2 cleavage mutants of SIN, SFV, and VEE have been identified, and these viruses appear to have defects in one or more biological functions due to the presence of uncleaved PE2. For example, in SFV, cleavage of p62 was prevented by replacement of Arg by Leu at position −1 of E2 (34). This p62 cleavage mutant virus (mL) was found to be noninfectious on BHK-21 cells. The block in infection appeared to involve both the virus-receptor interaction and fusion activity of the mutant virus. Viral infectivity on BHK-21 cells was restored by in vitro cleavage of p62 with trypsin or by exposure of the virus to very low pHs (22, 34, 40). Likewise, several SIN PE2 cleavage mutants have been shown to be noninfectious. For example, replacement of the Arg or Ser residue at position 1 of E2 (the last amino acid of the XBXBBX cleavage site) by an Asn residue within the context of the infectious clones TRSB (corresponding to a laboratory-adapted strain of SIN) and TR339 (containing the consensus sequence of wild-type SIN) almost completely blocked viral infectivity (13, 14, 19, 25). SIN cleavage mutants in which part of the cleavage site was deleted were also found to be essentially noninfectious on BHK-21 cells (19).
Interestingly, it has been found that the lethality of PE2 cleavage mutations in TRSB-N (Arg-to-Asn substitution at position 1 of E2) could be reversed by resuscitating second-site mutations in PE2 (14). These investigations showed that after infection with TRSB-N virions, small plaques were occasionally formed. Sequence analysis of the purified plaques revealed that some of these viruses carried second-site mutations in PE2 but remained cleavage deficient. While these PE2 cleavage mutants with a resuscitating mutation in PE2 were found to be infectious on BHK-21 cells, the viruses appeared to have an attenuated virulence in CD-1 mice compared to the parental TRSB virus.
Recent data showed that the block in infection of PE2 cleavage-deficient SIN viruses without second-site resuscitating mutations is not at the level of the initial virus-receptor interaction (19). SIN PE2 cleavage mutants with an intact cleavage site do bind very efficiently to BHK-21 cells, even better than the parental TR339 virus. It has been shown that the presence of the basic XBXBBX sequence mediates an efficient interaction with heparan sulfate (HS), which is abundantly expressed on BHK-21 cells and thus acts as a receptor for the virus (4, 20). Furthermore, it has been shown that the presence of uncleaved PE2 in virions does not influence RNA replication or virus assembly and release (14, 19). Taken together, these data indicate that the block in infection of these SIN PE2 cleavage mutants lies downstream of the interaction of a virus particle with a cellular receptor but prior to RNA replication, suggesting that these mutant viruses are impaired in membrane fusion.
Here, we studied the fusogenic properties of PE2 cleavage mutants, based on the infectious clone TR339, using a liposomal model system. PE2 cleavage mutants in which either the Ser residue at position 1 of E2 was replaced by Asn (E2:N1) or from which the BXBB sequence within the PE2 cleavage site was deleted (FDF) were used. The results show that PE2 cleavage mutants were unable to fuse with liposomes at pH 5.0, indicating that the block in infection lies at the level of the fusion process. Incubation of PE2 cleavage mutants at low pH demonstrated that the viruses are impaired in their ability to form an E1 homotrimer, the fusion-active conformation of the viral spike protein. The block in rearrangement and fusion could be overridden by exposure of the virus to very low pHs (pH < 4.5). Furthermore, E2:N1 cleavage mutants with resuscitating mutations in PE2 were generated, analogous to those identified in TRSB-N, and found to be infectious in BHK-21 cells. These PE2 resuscitated mutant viruses do form E1 homotrimers at a physiologically relevant acidic pH despite the lack of PE2 cleavage. Accordingly, their membrane fusion activity appeared to be completely restored.
MATERIALS AND METHODS
Constructs.
The construction of the cDNA clones pTR339, pE2:N1 (called p39N1 in previous articles), and pFDF has been described previously (19, 20). The p prefix indicates the cDNA form of the virus clone. The cDNA pE2:N1/E3:R25 was constructed by substitution into pE2:N1 of an AatII-to-StuI fragment from pTRSB-E3R25 (14). The cDNA clones pE2:N1/T191 and pE2:N1/G216 were constructed by substitution into pE2:N1 of a StuI-to-BssHII fragment from pTRSB-NE2T191 and pTRSB-NE2G216, respectively (14). The introduced mutations were confirmed by DNA sequence analysis at the University of North Carolina at Chapel Hill Automated Sequencing Facility with a 373A DNA sequencer with the Taq DyeDeoxy terminator cycle sequencing kits (Applied Biosystems).
Production and characterization of pyrene- and [35]methionine-labeled virus particles.
Viruses were produced on BHK-21 cells. The cells were cultured in Glasgow's modification of Eagle's minimal essential medium (Gibco-BRL, Breda, The Netherlands), supplemented with 5% foetal calf serum (FCS), 10% tryptose phosphate broth, 200 mM glutamine, 25 mM HEPES, and 7.5% sodium bicarbonate. For the production of pyrene-labeled virus particles, BHK-21 cells cultured for 48 h on medium containing 10 μg of 16-(1-pyrenyl)hexadecanoic acid (pyrene fatty acid; Molecular Probes, Eugene, Oreg.) per ml were transfected by electroporation with in vitro transcripts of linearized cDNA clones. Pyrene-labeled SIN particles released from the cells at 20 h posttransfection were harvested and purified from the medium as described before (3, 27, 36). For the production of [35S]methionine-labeled virus particles, BHK-21 cells were transfected with in vitro-transcribed viral RNA by electroporation, essentially as previously described (3, 27, 36). Briefly, at 3 to 4 h posttransfection, the culture medium was replaced by methionine-free Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 5% FCS and 200 mM glutamine. After 2 h of starvation, 200 μCi of [35S]methionine per 5 ml was added to the medium, and incubation was continued overnight. At 20 h posttransfection, [35S]methionine-labeled virus was harvested and purified from the medium as described before (3, 27, 36, 43). The purity of the virus was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Visualization of the protein bands and quantification of the PE2 content in virions were done by phosphorimaging analysis using Image Quant 3.3 software (Molecular Dynamics, Sunnyvale, Calif.). The percent PE2 in virions was determined by relating the intensity of PE2 to the total intensity of E1, PE2, and E2, corrected for the contribution of E1 on the basis of the relative number of methionine residues in the E1, E2, and PE2 proteins (19, 32). The specific infectivity (PFU per count per minute [cpm]) was calculated for all batches of [35S]methionine-labeled viruses by plaque assay on BHK-21 cells (20).
Binding assays.
Virus attachment to BHK-21 cells and to heparin- and bovine serum albumin-agarose beads (both from Sigma Chemical Co, St. Louis, Mo.) was performed essentially as described previously (19, 20). Briefly, BHK-21 cells (cultured in a 12-well plate [Costar, Cambridge, Mass.]) were washed two times with cold 5 mM HEPES–150 mM NaCl–0.1 mM EDTA (pH 7.4) (HNE) plus 1% fetal calf serum (HNE+). Subsequently, 150 μl of [35S]methionine-labeled virus particles was added to the cells (ranging from 106 to 107 cpm, approx. 109 to 1010 virus particles), and incubation was continued at 4°C for 2 h with gentle agitation. The cells were washed twice with HNE+ and trypsinized. For binding to heparin-agarose and albumin-agarose beads, 1 ml of beads was washed with HNE+ three times and resuspended in 1 ml of HNE+. Fifty microliters of beads was added to 50 μl of virus particles, similar to that in the cell-binding assay, and incubated for 2 h at 4°C with gentle agitation. Subsequently, the mixtures were washed three times with HNE+ and resuspended in 0.6% Triton (Sigma) in HNE. Radioactivity was quantified by liquid scintillation analysis.
Preparation of liposomes.
Liposomes (large unilamellar vesicles) were prepared by freeze/thaw extrusion as described before (3, 27, 36, 43). Briefly, lipid mixtures dried from chloroform-methanol were hydrated in HNE and subjected to five cycles of freezing and thawing. Subsequently, the suspension was extruded 21 times through two 0.2-μm filters (Nucleopore, Inc., Pleasanton, Calif.) in a LiposoFast miniextruder (Avestin, Ottawa, Canada). Liposomes consisted of phosphatidylcholine (PC), phosphatidylethanolamine (PE), sphingomyelin(SPM), and cholesterol (Chol) in a molar ratio of 1:1:1:1.5. The phospholipids were obtained from Avanti Polar Lipids (Alabaster, Ala.), and cholesterol was from Sigma. The phospholipid concentration of the liposomes was determined by phosphate analysis (2).
Fusion assay.
Fusion of pyrene-labeled SIN with liposomes was monitored on-line in an AB2 fluorimeter (SLM/Aminco, Urbana, Ill.) at excitation and emission wavelengths of 345 and 480 nm, respectively (3, 27, 36, 43). Briefly, pyrene-labeled SIN was mixed with liposomes in 0.665 ml of HNE at final concentrations of 0.5 μM viral and 200 μM liposomal phospholipid (corresponding to approximately 1010 virus particles and 2.5 × 1011 liposomes per 0.7 ml). The mixture was stirred continuously and maintained at 37°C. Fusion was initiated by the addition of 35 μl of 0.1 M MES (morpholinoethanesulfonic acid)–0.2 M acetic acid, pretitrated with NaOH to achieve the final desired pH. The fusion scale was calibrated so that 0% fusion corresponded to the initial pyrene-excimer fluorescence level and 100% fusion to the fluorescence value obtained after the addition of 35 μl of 0.2 M octaethylene glycol monododecyl ether (Fluka Chemie AG, Buchs, Switzerland) (3, 27, 36, 43). The extent of fusion was determined 60 s after acidification.
Analysis of conformational changes in viral spike protein.
The conformational changes occurring in the viral spike protein were examined under the same conditions as in the fusion experiments (3, 36). After the indicated pH treatment, samples were neutralized by addition of a pretitrated volume of NaOH, solubilized in SDS-PAGE sample buffer, and analyzed by SDS-PAGE. Running gels were further incubated for 30 min in 1 M sodium salicylate and dried. Visualization and quantification of the E1 trimer were done by phosphorimaging analysis, relating the intensity of the E1 trimer to the total intensity of E1, PE2, E2, and E1 trimer, as described above.
RESULTS
Characterization of PE2 cleavage mutant SIN viruses.
In this study we investigated the membrane fusion activity of a number of SIN PE2 cleavage mutant viruses. PE2 cleavage mutations were introduced into the background of the infectious SIN clone TR339. Two mutants were chosen that have previously been shown to affect PE2 cleavage efficiency (Table 1) (19). In the mutant construct E2:N1, the Ser residue at position 1 of E2 is replaced by an Asn residue. This residue creates an N-linked glycosylation signal. Glycosylation of an Asn residue at position 1 of E2 has been proposed to interfere with furin activity (19). SDS-PAGE analysis of the E2:N1 mutant showed that, indeed, PE2 cleavage was completely inhibited. Furthermore, the virus was poorly infectious, as determined by plaque assay on BHK-21 cells. Likewise, the FDF mutant, in which the BXBB sequence was deleted, incorporated 100% uncleaved PE2 and also had a very low specific infectivity (Table 1).
TABLE 1.
Characterization of PE2 cleavage mutant SIN viruses
| Virus | Amino acids at PE2 cleavage site | Furin cleavage site | % PE2 | BHK specific infectivity (PFU/cpm) |
|---|---|---|---|---|
| TR339 | GRSKRS | Intact | 4 | 18 |
| FDF | G----S | Deleted | 100 | 0.5 |
| E2:N1 | GRSKRN | Intact | 100 | 1.5 |
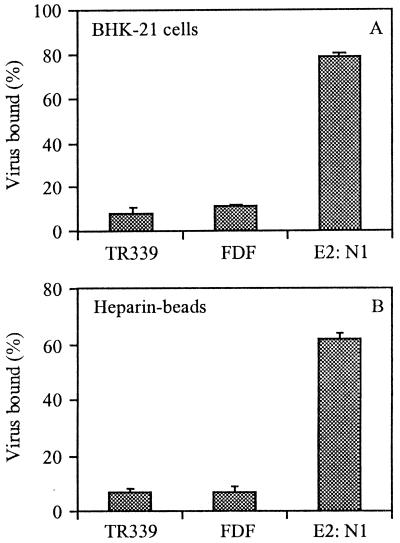
The first step in virus entry is the interaction of a virus particle with a cellular receptor. Consistent with previous reports (20), we found that TR339 virus does not bind very efficiently to BHK-21 cells (Fig. 1A). The FDF mutant bound equally poorly to BHK-21 cells. On the other hand, the E2:N1 virus did bind efficiently to BHK-21 cells. Recent data have demonstrated that SIN viruses with an intact cleavage site interact with HS, a cellular receptor which is abundantly expressed on BHK-21 cells (19). Accordingly, we found that the E2:N1 mutant bound efficiently to heparin-agarose beads (Fig. 1B). In a control in which albumin-agarose beads were used, none of the viruses bound to the beads (data not shown). Since the two mutant viruses, compared to TR339, bind similarly (FDF) or much better (E2:N1) to BHK-21 cells, it is clear that the block in infection for these mutants lies after binding of the virus particle to the cell surface, presumably at the level of the viral membrane fusion process.
FIG. 1.
Binding of PE2 cleavage-deficient SIN viruses to BHK-21 cells or heparin-agarose beads. Approximately 1010 [35S]methionine-labeled virus particles were added to the cells or beads, and binding was measured after 2 h of incubation at 4°C, as described in Materials and Methods. Each bar represents the mean of triplicate binding assays. Each set of triplicates was repeated three times. The error bars represent standard deviations. (A) Binding to BHK-21 cells. (B) Binding to heparin-agarose beads.
Fusion activity of pyrene-labeled PE2 cleavage mutant SIN viruses.
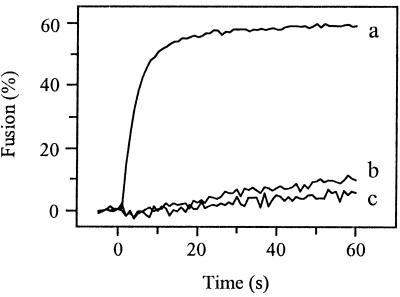
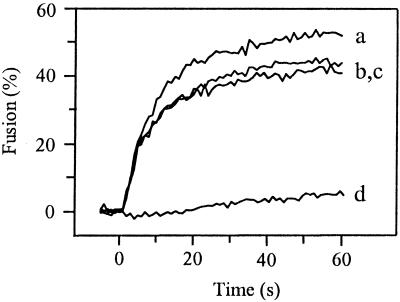
To study the fusogenic properties of the PE2 cleavage mutants, we used SIN variants biosynthetically labeled with the fluorescent probe pyrene. Because the mutant viruses are almost non-infectious, RNA transcripts derived from the cDNA were transfected directly into BHK-21 cells, cultured beforehand in the presence of pyrene fatty acid. At 20 h posttransfection the pyrene-labeled virus particles were harvested and purified from the medium (3, 27, 36). With this procedure, no additional passage of the virus is involved, minimizing the possibility of generating revertant viruses. Subsequently, fusion of pyrene-labeled SIN was measured in a liposome model system (3, 27, 36). Upon fusion of a pyrene-labeled virus particle with a target liposome, the pyrene phospholipids are diluted into the liposomal membrane, resulting in a decrease in pyrene excimer fluorescence intensity by more than an order of magnitude. Figure 2 presents the results. SIN derived from the infectious clone TR339 fused rapidly and efficiently with the liposomes at pH 5.0 (curve a), 60% of the virus fusing within 60 s after acidification. No fusion was seen at neutral pH (data not shown). In contrast to TR339, PE2 cleavage mutant E2:N1 hardly fused with the liposomes at pH 5.0 (curve b). At 60 s after acidification to pH 5.0, an extent of fusion of only 7% was observed. Like E2:N1, PE2 cleavage mutant FDF was unable to fuse at pH 5.0 (curve c).
FIG. 2.
Fusion of PE2 cleavage-deficient SIN viruses with liposomes at pH 5.0. On-line fusion experiments with pyrene-labeled viruses were performed at 37°C as described in Materials and Methods. All fusion measurements were repeated at least three times. Liposomes consisted of PC/PE/SPM/Chol (molar ratio, 1:1:1:1.5). Curve a, TR339; curve b, E2:N1; curve c, FDF.
Construction and characteristics of viable E2:N1 revertants.
Using SIN PE2 cleavage mutant TRSB-N, Heidner et al. (14) showed that after infection of cells with TRSB-N virions, small plaques were occasionally formed. These viruses were found to be infectious revertants either with a restored PE2 cleavage phenotype or with second-site mutations in PE2 along with retention of the PE2 cleavage defect.
Some of the second-site resuscitating mutations identified in TRSB-N were now introduced into the E2:N1 PE2 cleavage mutant virus, i.e., in the TR339 background. In the first mutant, E2:N1/T191, the Pro residue (CCG) at position 191 of E2 was replaced by a Thr (ACG). In the second mutant, E2:N1/G216, the Glu residue (GAA) at position 216 of E2 was replaced by a Gly (GGA). In the third mutant, E2:N1/E3:R25, the Cys residue (TGT) at a position corresponding to position 25 of E3 was replaced by an Arg (CGT). We were interested in whether the introduced mutations promoted viral viability and membrane fusion capacity, with retention of the PE2 cleavage-defective phenotype.
First, PE2 cleavage of each of the mutants was determined by SDS-PAGE analysis of [35S]methionine-labeled protein from purified particles (Table 2). In agreement with earlier observations on the corresponding TRSB-N revertants (14), all of the second-site mutant viruses retained the defect in PE2 cleavage. On the other hand, the specific infectivities of the mutants were increased by a factor of about 150 to 200 compared to the parental E2:N1 virus (Table 2). Thus, all three resuscitating mutations restored virus viability while retaining the PE2 cleavage-deficient phenotype. It is noteworthy that the specific infectivities of the resuscitated mutants (Table 2) were significantly higher than the specific infectivity of the TR339 virus (Table 1).
TABLE 2.
Characterization of resuscitated PE2 mutant SIN viruses
| Virus | Substitution mutation | % PE2 | BHK specific infectivity (PFU/cpm) |
|---|---|---|---|
| E2:N1 | None | 100 | 1.5 |
| E2:N1/T191 | E2 position 191 Pro → Thr | 100 | 230 |
| E2:N1/G216 | E2 position 216 Glu → Gly | 100 | 136 |
| E2:N1 E3:R25 | E3 position 25 Cys → Arg | 100 | 160 |
To determine whether the high specific infectivity of the resuscitated mutants was due to an improved binding of these viruses to BHK-21 cells, cell-binding assays were performed. Figure 3A shows the results. As indicated above (Fig. 1), the E2:N1 virus binds efficiently to BHK-21 cells. Likewise, the resuscitated mutants E2:N1/T191, E2:N1/G216, and E2:N1/E3:R25 also bound efficiently to BHK-21 cells. Clearly, all PE2 cleavage-deficient mutants retaining the XBXBBX sequence bind better to BHK-21 cells than TR339, explaining the higher specific infectivity of the resuscitated mutants.
FIG. 3.
Binding of PE2 cleavage-deficient second-site resuscitated SIN viruses to BHK-21 cells or heparin-agarose beads. Binding was measured after 2 h of incubation at 4°C as described in the legend to Fig. 1. Bars: a, E2:N1; b, E2:N1/T191; c, E2:N1/G216; d, E2:N1/E3:R25. (A) Binding to BHK-21 cells. (B) Binding to heparin-agarose beads.
Since the resuscitated mutants are still PE2 cleavage deficient, retaining the XBXBBX furin cleavage site, it is likely that these viruses bind to HS on BHK-21 cells (19). To investigate this directly, we determined the binding capacity of the resuscitated mutants for heparin- versus albumin-agarose beads. All viruses appeared to bind efficiently to the heparin beads (Fig. 3B). None of the viruses bound to albumin beads (data not shown). PE2 cleavage-deficient resuscitated viruses are therefore infectious and have a high specific infectivity on BHK-21 cells, presumably due to an interaction of the uncleaved PE2 with HS on the cell surface. The high infectivity of the resuscitated mutant viruses compared to the low infectivity of the parent E2:N1 virus suggests that the second-site mutations somehow restore the fusion capacity of these viruses. This question was addressed subsequently.
Fusion activity of pyrene-labeled PE2 cleavage mutant SIN viruses carrying a resuscitating mutation in PE2.
Figure 4 presents the fusion kinetics of pyrene-labeled PE2 resuscitated cleavage mutant SIN viruses with liposomes at pH 5.0. The E2:N1/T191 mutant fused efficiently with liposomes, the extent of fusion being about 55% within 60 s (curve a). The E2:N1/G216 and E2:N1/E3:R25 mutants also fused efficiently with liposomes at pH 5.0 (curves b and c). Again, the E2:N1 virus was unable to fuse at pH 5.0 (curve d). Thus, it appears that as a result of a single amino acid substitution, the noninfectious PE2 cleavage-deficient E2:N1 virus regains infectivity due to restoration of its membrane fusion competence despite retention of the uncleaved PE2 phenotype.
FIG. 4.
Fusion of PE2 cleavage-deficient SIN viruses with liposomes. On-line fusion experiments were performed at 37°C as described in the legend to Fig. 2. Curve a, E2:N1/T191; curve b, E2:N1/G216; curve c, E2:N1/E3:R25; curve d, E2:N1.
PE2 cleavage mutant SIN viruses show an acidic pH shift in activation of membrane fusion.
Previous work, based on SFV, showed that the block in infection of the p62 cleavage mutant mL (Arg-to-Leu substitution at position −1 of E2) could be overridden by exposure of the virus to very low pH (34). Therefore, we were interested in whether incubation at a very low pH could activate fusion activity of PE2 cleavage mutant SIN.
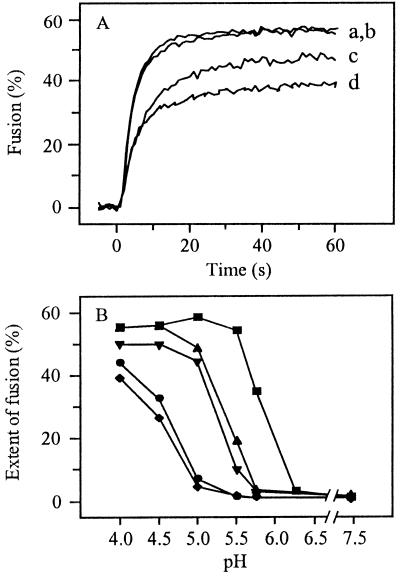
Figure 5A presents the fusion kinetics of several pyrene-labeled PE2 cleavage mutants at pH 4.0 in the liposome model system. As expected, the resuscitated mutant E2:N1/T191 (curve a) fused efficiently, with fusion kinetics identical to those of the TR339 virus (curve b). Importantly, however, under these extreme pH conditions the noninfectious E2:N1 and FDF mutants were able to fuse as well (curves c and d). As a control, the different pyrene-labeled SIN viruses were incubated at pH 4.0 in the absence of liposomes. In all cases, the fluorescence intensity remained constant, demonstrating that under these extreme pH conditions, the decrease in pyrene excimer fluorescence intensity observed in the presence of liposomes was due to dilution of the fluorescent probe from the viral membrane into the liposomal membrane (data not shown). These results indicate that the PE2 cleavage mutants FDF and E2:N1 are fusion competent, but only when the viruses are exposed to an unphysiologically low pH.
FIG. 5.
Low-pH-dependent fusion of PE2 cleavage-deficient SIN viruses with liposomes. On-line fusion experiments were performed as described in the legend to Fig. 2. (A) Time course of fusion at pH 5.0. Curve a, E2:N1/T191; curve b, TR339; curve c, E2:N1; curve d, FDF. (B) The extent of fusion at different pHs was determined 60 s after acidification. ■, TR339; ▴, E2:N1/T191; ▾, E2:N1/G216; ●, E2:N1; ⧫, FDF.
Figure 5B shows the detailed pH dependence of fusion of SIN PE2 cleavage mutants with liposomes. For TR339, optimal fusion was observed at pH 5.0 (squares). The threshold for fusion was pH 6.25, similar to that of the laboratory-adapted SIN strain AR339 (36). The PE2 cleavage mutants FDF (diamonds) and E2:N1 (circles) showed a clear-cut downward shift in the pH dependence of fusion, with a threshold at pH 5.0. Only at very low pHs was fusion of these viruses fast and extensive. Interestingly, an intermediate pH dependence was observed for the resuscitated mutant viruses E2:N1/T191 (upward triangles) and E2:N1/G216 (downward triangles). The other resuscitated mutant virus, E2:N1/E3:R25, showed a pH dependence comparable to that of the E2:N1/T191 and E2:N1/G216 mutants (data not shown). The threshold for fusion was very close to pH 5.75 for all cleavage-deficient SIN viruses with a resuscitating mutation in PE2. In summary, SIN PE2 cleavage mutants exhibit an acidic pH shift in their ability to fuse with liposomes, and resuscitating mutations in PE2 partly reverse this pH shift.
Rearrangement of spike heterodimer is impaired in PE2 cleavage mutants.
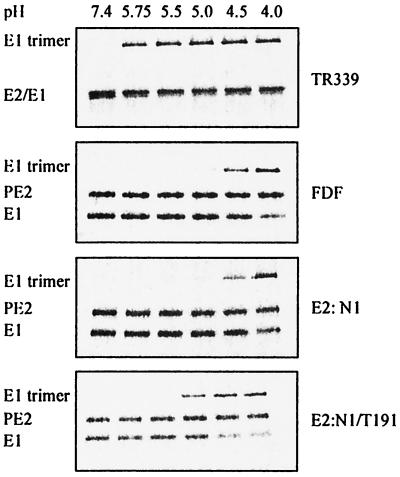
Earlier studies on SFV and SIN have demonstrated that under low-pH conditions, the E2/E1 heterodimer dissociates and a trypsin-resistant E1 homotrimer is formed. (3, 16, 29, 36, 40, 41). It is believed that the E1 homotrimer is the fusion-active conformation of the viral spike protein (16, 40). To determine whether the PE2 cleavage mutants were blocked in their ability to rearrange to the fusion-active conformation, the conformational changes of the viral spike proteins were studied. [35S]methionine-labeled virus was incubated at low pH in the presence of liposomes. At 60 s after acidification, the mixture was neutralized and the appearance of the E1 homotrimer was analyzed by SDS-PAGE.
Figure 6 shows the results. The spike protein of TR339 had already rearranged to form an E1 homotrimer at pH 5.75. Under optimal conditions for fusion (pH 5.0), 76% of the TR339 E1 protein was converted to the trimeric configuration. The PE2 cleavage mutants FDF and E2:N1 showed a large pH shift in the formation of the E1 trimer. Clearly, with these viruses, at pHs of 5.0 or above, no significant trimer formation occurred. Only when these mutants were incubated at a very low pH did the E1 spike protein rearrange to its trimeric configuration. By contrast, the resuscitated cleavage mutant E2:N1/T191 did convert to an E1 homotrimer at pH 5.0. The extent of E1 trimerization increased by incubation at pHs lower than pH 5.0. Similar results were obtained with the other resuscitated mutants, E2:N1/G216 and E2:N1/E3:R25 (data not shown). These results suggest that the PE2/E1 heterodimer of the PE2 cleavage mutants FDF and E2:N1 is stable at pH 5.0, rearranging to the fusion-active state only at very low, unphysiological, pHs. On the other hand, the PE2/E1 heterodimer of the resuscitated viruses is less stable than that of the parental E2:N1 virus and rearranges to a fusion-active conformation at a physiological mildly acidic pH (pH 5.0).
FIG. 6.
E1 trimerization of PE2 cleavage-deficient SIN viruses at 37°C and the indicated pHs. Approximately 1010 [35S]methionine-labeled SIN particles were incubated with an excess of liposomes consisting of PC/PE/SPM/Chol (molar ratio, 1:1:1:1.5). After 60 s, samples were neutralized and analyzed for the appearance of E1 trimers by SDS-PAGE, as described in Materials and Methods.
Figure 7 presents a comparison of E1 trimerization and fusion for the PE2 cleavage mutant SIN viruses at different pHs. Clearly, there is a distinct correlation between the appearance of the E1 homotrimer and the ability of the virus to fuse with liposomes. Under optimal conditions for fusion, the extent of E1 trimerization and fusion of TR339 at 60 s after acidification were 76 and 59%, respectively. At pH 5.75, 64% of the E1 glycoprotein had already rearranged to the fusion-active conformation, and half-maximal fusion was observed. On the other hand, the PE2 cleavage mutant FDF, at pH 5.0, showed extents of E1 trimerization and fusion of only 5 and 3%, respectively. At pH 4.0, the FDF mutant did become fusogenic, 70% of the E1 protein converting to a homotrimer and 40% of the virus particles fusing with liposomes under these conditions. Likewise, the PE2 cleavage mutant E2:N1 showed little E1 trimerization and fusion at pH 5.0 (6 and 8%, respectively), whereas at pH 4.0 the extents of E1 homotrimer formation and fusion for this virus were 70 and 43%, respectively.
FIG. 7.
Relative pH dependence of E1 trimerization and fusion for several PE2 cleavage-deficient SIN viruses. The extents of E1 trimerization (solid bars) and fusion (open bars) are shown at 60 s postacidification to the indicated pHs. To compare E1 trimerization and fusion, the final extents of these processes at pH 4.0 were set to 100%. (A) TR339 (absolute extents: 76% E1 trimerization, 55% fusion). (B) FDF (absolute extents: 70% E1 trimerization, 40% fusion). (C) E2:N1 (absolute extents: 70% E1 trimerization, 43% fusion). (D) E2:N1/T191 (absolute extents: 70% E1 trimerization, 56% fusion). E1 trimerization was determined as described in the legend to Fig. 6. Fusion was measured as described in the legend to Fig. 2.
At pH 5.0, the resuscitated mutant E2:N1/T191 showed extents of E1 trimerization and fusion of 38 and 49%, respectively. At a pH lower than 5.0, the extent of E1 trimerization and fusion increased further. At pH 5.75, only a very small amount (2%) of the E1 glycoprotein was converted to a homotrimer, and accordingly, fusion was negligible. Similar results were obtained for the other resuscitated mutants, E2:N1/G216 and E2:N1/E3:R25 (data not shown). The pH threshold of fusion for TR339 is pH 6.2 (Fig. 5B), which is 0.5 pH unit higher than that of the resuscitated viruses. The resuscitated mutants clearly showed a pH shift in E1 trimer formation and fusion compared to TR339. Altogether, the lower stability of the PE2/E1 heterodimer of the resuscitated mutants compared to the parental E2:N1 mutant enables the virus to become fusion active under physiological pH conditions, consistent with the restored infectivity of these viruses in BHK-21 cells.
DISCUSSION
In this paper, we provide evidence that the block in viral infectivity of SIN PE2 cleavage mutant viruses on BHK-21 cells lies at the level of the membrane fusion activity of the viral envelope glycoprotein (Fig. 2, 6, and 7). Earlier studies on similar SIN PE2 cleavage mutants had shown that viral infection in vertebrate cells was not blocked at the level of virus-receptor interaction, RNA replication, or virus assembly (14, 19). Our present results show that SIN PE2 cleavage mutants are impaired in their fusion activity because of the inability of the immature PE2/E1 heterodimer to rearrange to an E1 homotrimer at a physiological acidic pH. The E1 trimer has been shown to represent the fusion-active conformation of the alphavirus spike (3, 11, 16, 36, 40). The lethality of SIN PE2 cleavage defect could be overridden by second-site resuscitating mutations in PE2 (Table 2), which destabilize the PE2/E1 heterodimer interaction. Thus, the spike regains the ability to form a fusion-competent E1 trimer at a physiologically relevant acidic pH (Fig. 4 to 7).
Among members of the alphavirus genus, PE2 cleavage has been associated with different biological functions of the viruses involved. Recent work has shown that SIN PE2 cleavage mutants with an intact cleavage site (XBXBBX) interact electrostatically with HS, a glycosaminoglycan which is abundantly expressed on BHK-21 cells (19). These findings are in agreement with our results, showing that the E2:N1 mutant (with an intact cleavage site) binds efficiently to BHK-21 cells and heparin beads compared to the PE2 cleavage mutant FDF (with a deleted cleavage site) (Fig. 1). PE2 cleavage mutant E2:N1 binds much more efficiently to BHK-21 cells than the parental SIN TR339, confirming that the TR339 virus does not interact with HS (20). For SFV, p62 cleavage mutants have been generated by mutation of the cleavage site from RHRR to RHRL (mL mutant) (22, 34) or SHQL (1, 39). In contrast to our PE2 cleavage-deficient SIN viruses, these SFV mutants show a large reduction in binding capacity to BHK-21 cells compared to wild-type SFV. However, in the case of SFV, the cleavage site was disrupted to create the p62 cleavage-deficient viruses. Therefore, it is difficult to interpret these results in the context of our present data on SIN mutants which are known to bind to cells through the furin cleavage site. Despite the efficient binding of the E2:N1 PE2 cleavage mutant to BHK-21 cells, the virus was still found to be noninfectious, due to its inability to fuse (Table 1, Fig. 2). However, after introduction of resuscitating mutations in PE2, E2:N1 regained fusion competence and infectivity on BHK-21 cells, as discussed in more detail below. Interestingly, the specific infectivity of these viruses on BHK-21 cells was found to be very high, even 10-fold higher than that of the parental TR339 (Table 2). Our results demonstrate that the resuscitated PE2 cleavage mutants are more infectious than TR339, very likely as a result of the more efficient interaction of these viruses with HS (Fig. 3).
The inability of cleavage mutants E2:N1 and FDF to fuse at a physiologically relevant acidic pH appears to be caused by a downward shift in the pH dependence of the viral membrane fusion reaction (Fig. 5B). Within the overall process of low-pH-dependent alphavirus membrane fusion, the E2/E1 heterodimer first dissociates, followed by formation of a trypsin-resistant E1 homotrimer (3, 11, 16, 40). In the present study, it became clear that the PE2/E1 heterodimer of the cleavage mutant viruses is too stable to rearrange to the fusion-active E1 homotrimeric conformation at a physiologically relevant acidic pH (Fig. 6 and 7). However, when the viruses were incubated at pH 4.0, the spike did form an E1 homotrimer, resulting in expression of full membrane fusion activity (Fig. 6 and 7). In conclusion, the low specific infectivity of SIN PE2 cleavage mutants is the result of an impairment of the ability of the viral spike to rearrange to the fusion-active conformation at a mildly acidic pH due to the high stability of the immature PE2/E1 heterodimer. This is in agreement with observations on the SFV p62 cleavage-deficient mL mutant, showing that viral infectivity on BHK-21 cells was restored by exposure of the virus to pH 4.5 (34).
Several investigators have identified second-site resuscitating mutations in the E3, E2, and E1 glycoproteins which promote infectivity of PE2 cleavage-deficient alphaviruses (5, 14, 39). Some of these resuscitating mutations, found with the TRSB-N SIN clone (14), were now introduced in the E2:N1 virus to investigate the role of these mutations in viral infectivity versus the stability of the spike heterodimer. These E2:N1 mutants with resuscitating mutations in PE2 were indeed infectious on BHK-21 cells (Table 2), and at the same time exhibited a restored membrane fusion capacity at a physiologically relevant acidic pH (Fig. 4 and 5). In this respect it is important to note that viruses which do cleave PE2 and also interact efficiently with the cell attachment receptor HS are more infectious on BHK-21 cells than the resuscitated PE2 cleavage-deficient viruses (20). This indicates that SIN cleavage mutants with resuscitating mutations in PE2 have an intermediate infectivity on BHK-21 cells, which in fact correlates precisely with the intermediate pH dependence of their membrane fusion activity with liposomes (Fig. 5B). For example, the TR339 virus, with a PE2 cleavage-deficient phenotype, is already fully fusion competent at pH 5.5, whereas the resuscitated PE2 cleavage mutant viruses express full membrane fusion activity only at pHs of 5.0 or below. Analysis of the conformational changes occurring in the viral spike protein showed that the PE2/E1 spikes of the resuscitated mutants were able to form an E1 homotrimer at a physiological acidic pH, in contrast to the heterodimer of the E2:N1 virus (Fig. 6). In conclusion, the presence of the resuscitating mutations in PE2 destabilizes the PE2/E1 heterodimer so, that the viral spike protein regains the ability to undergo conformational changes, with formation of a fusion-active E1 trimer at a physiologically relevant pH despite retention of the uncleaved PE2 phenotype.
The maturation of the alphavirus spike heterodimer through cleavage of the PE2 precursor of E2 has a distinct function in the viral life cycle. During transport of the heterodimer from the ER to the surface of the infected cell, the uncleaved PE2 protein is presumed to function as a chaperone, protecting the spike from premature destabilization within the acidic TGN (9). Subsequently, after cleavage of PE2 in a post-TGN compartment by a furin-like protease, the mature viral spike protein is primed for expression of membrane fusion activity when exposed to a mildly acidic pH within the endosomal compartment of a new target cell. The results of this study indicate that while cleavage maturation is a common phenomenon in viral assembly, it is not absolutely required for viral infection. SIN PE2 cleavage mutants with resuscitating mutations in PE2 were found to be infectious on BHK-21 cells despite retention of the uncleaved PE2 phenotype. Apparently, the resuscitating mutations in PE2 destabilize the PE2/E1 heterodimer to such an extent that the spike has the capacity to rearrange to the fusion-active conformation at the lumenal pH of endosomes, while at the same time the heterodimer is stable enough to survive the mildly acidic lumen of the TGN (pH ∼6.0 [35]) during transport to the cell surface.
Interestingly, another alphavirus PE2 cleavage mutant called S12 (Ser-to-Asn substitution at position 1 of E2), derived from SAAR86, was found to be fully infectious on BHK-21 cells (31, 33). In the light of our present results, we would argue that the PE2/E1 heterodimer of the S12 mutant is less stable than that of the E2:N1 virus, despite the fact that both viruses carry the same amino acid substitution at E2 position 1. Differences in the genetic background of S12 and E2:N1 outside the PE2 cleavage region could account for the different spike stability, in agreement with our present observation that distant mutations in the E2:N1 mutant have the capacity to restore viral infectivity by destabilization of the spike heterodimer. We postulate that the S12 spike heterodimer has a very specific pH dependence, so that it survives the TGN during maturation but does become fusion competent at acidic pH in endosomes. The stability of the spike heterodimer and the fusion competence at low pHs of SAAR86 and S12 remain to be determined.
The results in Fig. 6 and 7 show that there is a clear correlation between expression of low-pH-dependent membrane fusion activity of SIN and formation of the E1 homotrimeric conformation. However, close scrutiny of the data reveals that mutant SIN viruses with a relatively low pH threshold for fusion require a lower extent of trimer formation to reach a certain level of fusion than the TR339 virus with a high pH threshold for fusion. This suggests that there are different levels of pH control of viral fusion activation and that, besides E1 trimerization, other pH-dependent conformational alterations are involved. In previous studies on SFV (3) and SIN (36), we have shown that, kinetically, fusion in the liposome model system occurs with a distinct delay after virus-liposome binding and E1 trimerization. This lag phase prior to the onset of fusion may well involve the rearrangement of several E1 trimers into a fusion complex. Interestingly, the duration of the lag phase decreases with decreasing pH (3, 36). Thus, our present observation suggesting that, at a relatively high pH, a relatively high extent of E1 trimerization is required for fusion may well reflect the comparatively long lag phase between E1 trimerization and the onset of fusion under these conditions.
The present results suggest that there is a direct correlation between the infectivity of SIN on BHK-21 cells and low-pH-dependent membrane fusion activity of the virus with liposomes. Not only is SIN-liposome fusion clearly dependent on low pH, it also appears that subtle shifts in the pH dependence of fusion have a profound effect on viral infectivity. This result provides additional support for the notion that exposure to a mildly acidic pH is an obligatory step in the infectious entry of SIN into its host cell. This conclusion is in agreement with recent observations of Glomb-Reinmund and Kielian (12) and DeTulleo and Kirchausen (6), indicating that SIN enters cells by receptor-mediated endocytosis and fusion from within acidic endosomes. On the other hand, very recently Hernandez et al. (15) provided evidence, based on investigation of mosquito cell infection by SIN, suggesting that exposure to an acidic pH may not be an obligatory step in alphavirus cell entry. Clearly, it cannot be excluded that alphaviruses use different routes of cell entry in mosquito and vertebrate cells. Also, our observations do not rule out the possibility that interaction of SIN with cell surface components induces conformational alterations priming the spike to subsequently rearrange to its fusion-active structure (7, 26). Nevertheless, from our present results it would appear that low pH is a crucial factor in alphavirus spike heterodimer destabilization and thus in the activation of viral membrane fusion activity. This conclusion is completely consistent with recent structural data on the SIN spike (30). Furthermore, SIN closely resembles SFV in terms of both spike organization (21, 3) and membrane fusion characteristics (3, 27, 36). It has been clearly demonstrated that SFV infects cells via receptor-mediated endocytosis and fusion from within acidic endosomes (6, 10, 12, 17, 23, 42).
ACKNOWLEDGEMENTS
This work was supported by the U.S. National Institutes of Health (grants 2R01HL16660-27 and R01AI22186-14), by The Netherlands Organization for Scientific Research (NWO) under the auspices of the Foundation for Chemical Research (CW), and by the Royal Netherlands Academy of Arts and Sciences (KNAW) (travel grant to J.M.S.).
REFERENCES
- 1.Berglund P, Sjoberg M, Garoff H, Atkins G J, Sheahan B J, Liljeström P. Semliki Forest virus expression system: production of conditionally infectious recombinant particles. Nat Biotechnol. 1993;11:916–920. doi: 10.1038/nbt0893-916. [DOI] [PubMed] [Google Scholar]
- 2.Böttcher C J F, van Gent C M, Fries C. A rapid and sensitive sub-micro phosphorus determination. Anal Chim Acta. 1961;24:203–204. [Google Scholar]
- 3.Bron R, Wahlberg J M, Garoff H, Wilschut J. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 1993;12:693–701. doi: 10.1002/j.1460-2075.1993.tb05703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrnes A P, Griffin D E. Binding of Sindbis virus to cell surface heparan sulfate. J Virol. 1998;72:7349–7356. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis N L, Brown K W, Greenwald G F, Zajac A J, Zacny V L, Smith J F, Johnston R E. Attenuated mutants of Venezuelan equine encephalitis virus containing lethal mutations in the PE2 cleavage signal combined with a second-site suppressor mutation in E1. Virology. 1995;212:102–110. doi: 10.1006/viro.1995.1458. [DOI] [PubMed] [Google Scholar]
- 6.DeTulleo L, Kirchhausen T. The clathrin endocytic pathway in viral infection. EMBO J. 1998;17:4585–4593. doi: 10.1093/emboj/17.16.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn D C, Meyer W J, Mackenzie J M, Johnston R E. A conformational change in Sindbis virus glycoproteins E1 and E2 is detected at the plasma membrane as a consequence of early virus-cell interaction. J Virol. 1990;64:3643–3653. doi: 10.1128/jvi.64.8.3643-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garoff H, Frischauf A M, Simons K, Lehrach H, Delius H. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature. 1980;288:286–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- 9.Garoff H, Hewson R, Opstelten D E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garoff H, Wilschut J, Liljeström P, Wahlberg J M, Bron R, Suomalainen M, Smyth J, Salminen A, Barth B U, Zhao H, Forsell K, Ekstrom M. Assembly and entry mechanisms of Semliki Forest virus. Arch Virol. 1994;9(Suppl):329–338. doi: 10.1007/978-3-7091-9326-6_33. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons D L, Ahn A, Chatterjee P K, Kielian M. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J Virol. 2000;74:7772–7780. doi: 10.1128/jvi.74.17.7772-7780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glomb-Reinmund S, Kielian M. The role of low pH and disulfide shuffling in the entry and fusion of Semliki Forest virus and Sindbis virus. Virology. 1998;248:372–381. doi: 10.1006/viro.1998.9275. [DOI] [PubMed] [Google Scholar]
- 13.Heidner H W, Johnston R E. The amino-terminal residue of Sindbis virus glycoprotein E2 influences virus maturation, specific infectivity for BHK cells, and virulence for mice. J Virol. 1994;68:8064–8070. doi: 10.1128/jvi.68.12.8064-8070.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidner H W, McKnight K L, Davis N L, Johnston R E. Lethality of PE2 incorporation into Sindbis virus can be suppressed by second-site mutations in E3 and E2. J Virol. 1994;68:2683–2692. doi: 10.1128/jvi.68.4.2683-2692.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez R, Luo T, Brown D T. Exposure to low pH is not required for penetration of mosquito cells by Sindbis virus. J Virol. 2001;75:2010–2013. doi: 10.1128/JVI.75.4.2010-2013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Justman J, Klimjack M R, Kielian M. Role of spike protein conformational changes in fusion of Semliki Forest virus. J Virol. 1993;67:7597–7607. doi: 10.1128/jvi.67.12.7597-7607.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kielian M, Helenius A. Role of cholesterol in fusion of Semliki Forest virus with membranes. J Virol. 1984;52:281–283. doi: 10.1128/jvi.52.1.281-283.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klenk H D, Garten W. Activation cleavage of viral spike proteins by host proteases. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 241–280. [Google Scholar]
- 19.Klimstra W B, Heidner H W, Johnston R E. The furin protease cleavage recognition sequence of Sindbis virus PE2 can mediate virion attachment to cell surface heparan sulfate. J Virol. 1999;73:6299–6306. doi: 10.1128/jvi.73.8.6299-6306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klimstra W B, Ryman K D, Johnston R E. Adaptation of Sindbis virus to BHK-21 cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lescar J, Roussel A, Wien M W, Navaza J, Fuller S D, Wengler G, Wengler G, Rey F A. The fusion glycoprotein shell of Semliki Forest virus: An Icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 22.Lobigs M, Garoff H. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the enveloppe glycoprotein p62. J Virol. 1990;64:1233–1240. doi: 10.1128/jvi.64.3.1233-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsh M, Bolzau E, Helenius A. Penetration of Semliki Forest virus from acidic prelysosomal vacuoles. Cell. 1983;32:931–940. doi: 10.1016/0092-8674(83)90078-8. [DOI] [PubMed] [Google Scholar]
- 24.Mayne J T, Rice C M, Strauss E G, Hunkapiller M W, Strauss J H. Biochemical studies of the maturation of the small Sindbis virus glycoprotein E3. Virology. 1984;134:338–357. doi: 10.1016/0042-6822(84)90302-7. [DOI] [PubMed] [Google Scholar]
- 25.McKnight K L, Simpson D A, Lin S C, Knott T A, Polo J M, Pence D F, Johannesen D B, Heidner H W, Davis N L, Johnston R E. Deduced consensus sequence of Sindbis virus strain AR339: mutations contained in laboratory strain will affect cell culture and in vivo phenotypes. J Virol. 1996;70:1981–1989. doi: 10.1128/jvi.70.3.1981-1989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer W J, Gidwitz S, Ayers V K, Schoepp R J, Johnston R E. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J Virol. 1992;66:3504–3513. doi: 10.1128/jvi.66.6.3504-3513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieva J L, Bron R, Corver J, Wilschut J. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 1994;13:2797–2804. doi: 10.1002/j.1460-2075.1994.tb06573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paredes A M, Heidner H W, Thuman-commike P, Venkararam Prasad B V, Johnston R E, Chiu W. Structural localization of the E3 glycoprotein in attenuated Sindbis virus mutants. J Virol. 1998;72:1534–1541. doi: 10.1128/jvi.72.2.1534-1541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J Cell Biol. 1991;112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pletnev S V, Zhang W, Mukhopadhyay S, Fisher B R, Hernandez R, Brown D T, Baker T S, Rossmann M G, Kuhn R J. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell. 2001;105:127–136. doi: 10.1016/s0092-8674(01)00302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Presley J F, Polo J M, Johnston R E, Brown D T. Proteolytic processing of the Sindbis virus membrane protein precursor PE2 is nonessential for growth in vertebrate cells but is required for efficient growth in invertebrate cells. J Virol. 1991;65:1905–1909. doi: 10.1128/jvi.65.4.1905-1909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice C M, Strauss J H. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc Natl Acad Sci USA. 1981;78:2062–2066. doi: 10.1073/pnas.78.4.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russel D L, Dalrymple J M, Johnston R E. Sindbis virus mutations which coordinately affect glycoprotein processing, penetration, and virulence in mice. J Virol. 1989;63:1619–1629. doi: 10.1128/jvi.63.4.1619-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salminen A, Wahlberg J M, Lobigs M, Liljeström P, Garoff H. Membrane fusion process of Semliki Forest virus. II. Cleavage dependent reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seksek O, Biwersi J, Verkman A S. Direct measurement of trans-Golgi pH in living cells and regulation by second messengers. J Biol Chem. 1995;270:4967–4970. doi: 10.1074/jbc.270.10.4967. [DOI] [PubMed] [Google Scholar]
- 36.Smit J M, Bittman R, Wilschut J. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J Virol. 1999;73:8476–8484. doi: 10.1128/jvi.73.10.8476-8484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith T J, Cheng R H, Olson N H, Peterson P, Chase E, Kuhn R J, Baker T S. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc Natl Acad Sci USA. 1995;92:10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tubulekas I, Liljeström P. Suppressors of cleavage site mutations in the p62 envelope protein of Semliki Forest virus reveal dynamics in spike structure and function. J Virol. 1998;72:2825–2831. doi: 10.1128/jvi.72.4.2825-2831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahlberg J M, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol. 1992;66:7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahlberg J M, Garoff H. Membrane fusion process of Semliki Forest virus. I. Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol. 1992;116:339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White J, Helenius A. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc Natl Acad Sci USA. 1980;77:3273–3277. doi: 10.1073/pnas.77.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilschut J, Corver J, Nieva J L, Bron R, Moesby L, Reddy K C, Bittman R. Fusion of Semliki Forest virus with cholesterol-containing liposomes at low pH: a specific requirement for sphingolipids. Mol Membr Biol. 1995;12:143–149. doi: 10.3109/09687689509038510. [DOI] [PubMed] [Google Scholar]