Abstract
Cold temperatures negatively impact crop yield and quality, posing significant limitations to the advancement of the vegetable industry. MYB transcription factors are pivotal in enhancing plant resilience against various abiotic stresses, including low-temperature stress. Pepper (Capsicum annuum L.) is a nutrient-rich vegetable crop sensitive to low temperatures. This study aimed to determine the function of CaMYB80 in the cold stress response of pepper through virus-induced silencing. The study also conducted heterologous expression of CaMYB80 in Arabidopsis and tomato plants. The results showed that CaMYB80 could respond to low-temperature stress in pepper. CaMYB80 was localized in the nucleus and cytoplasm and exhibited transcriptional activation ability. Moreover, CaMYB80 silencing decreased cold tolerance in pepper, while its heterologous overexpression increased cold tolerance in Arabidopsis and tomato. Further analysis showed that CaMYB80 interacted with CaPOA1 (peroxidase N1-like). Similarly, the expression of CaPOA1 also responded to low-temperature stress. Overexpression of CaPOA1 enhanced freezing tolerance in Arabidopsis, while its silencing reduced cold stress tolerance in pepper. Furthermore, overexpression of CaMYB80 in Arabidopsis and tomato could increase the activity of peroxidases and the expression levels of genes in the ICE-CBF-COR (inducer of CBF expression, C-repeat binding factor, cold-responsive) regulatory network. In conclusion, our research results indicate that CaMYB80 enhances pepper cold tolerance by interacting with CaPOA1 to increase peroxidase activity and influence the expression of ICE-CBF-COR related genes.
Introduction
Low temperature stress is a prevalent abiotic factor that impacts plant growth and distribution, leading to diminished crop yield and quality [1]. Cold stress can be classified as either chilling (0–15°C) stress or freezing (<0°C) stress. Chilling stress inhibits plant growth and development, while freezing stress disrupts cell structure, leading to cell death [2]. Plants have developed sophisticated regulatory mechanisms, encompassing numerous signaling pathways, to promptly detect and efficiently counteract cold stress [3]. These pathways constitute an intricate network involving a multitude of genes encoding both structural and regulatory proteins, which directly or indirectly shield plants from the adverse effects of low temperatures [4, 5].
Transcription factors (TFs) play a crucial role as regulatory proteins, governing the expression of target genes by binding to specific cis-regulatory elements within their promoter regions [6]. Under cold stress conditions, a variety of TFs are activated, such as CBF, bHLH, MYB, and NAC families. These TFs stimulate the transcription of downstream cold-responsive (COR) genes, thereby propagating and intensifying the cold signal and initiating a cascade of physiological responses [6–8]. As one of the most extensive families of TFs, MYB TFs exert significant regulatory control over plant responses to abiotic stress by binding to MYB elements and modulating the expression of relevant genes [1]. Named after their highly conserved DNA-binding domain located at the N-terminal region, termed the MYB domain, these TFs typically contain one to four repeats of the R motif [9]. The MYB TF family can be categorized into four subfamilies based on the number of R repeats present in their sequence: 1R-MYB (R1/R2, R3-MYB), 2R-MYB (R2R3-MYB), 3R-MYB (R1R2R3-MYB), and 4R-MYB (R1/R2-MYB) [8–10].
Within these subfamilies, R2R3-MYB has garnered considerable attention owing to its multifaceted roles in plant biology. Numerous studies have elucidated the involvement of certain R2R3-MYB TFs, such as AtMYB15, AtMYB88, AtMYB44, GmMYB92, TaMYB2A, and OsMYB2, in modulating plant responses to various environmental stresses, including drought, salinity, and cold [11–16]. Additionally, research has demonstrated that DgMYB2 can enhance chrysanthemum’s cold tolerance by regulating DgGPX1 [1]. Moreover, overexpression of SlMYB102 in tomato has been shown to upregulate SlP5CS and SlAPX2 genes under low temperatures, thereby bolstering tomato plants’ cold tolerance via the CBF and proline synthesis pathways [17].
During episodes of low-temperature stress, the equilibrium of reactive oxygen species (ROS) within plants becomes disturbed, resulting in swift ROS buildup and subsequent oxidative harm [18, 19]. Peroxidases (POD, PRX, and POA) play pivotal roles as antioxidant enzymes within the ROS detoxification system. These enzymes facilitate the oxidation of various substrates using H2O2 as an electron donor, thereby curtailing ROS accumulation induced by abiotic stress and preserving membrane integrity [20, 21]. Nevertheless, the precise regulatory pathways governing the activity of antioxidant enzymes mediated by MYB transcription factors in pepper remain largely unexplored.
Pepper (Capsicum annuum L.), an important vegetable crop, is an annual or perennial herbaceous plant belonging to the Solanaceae family. It thrives under normal growth temperatures of 20–30°C and is susceptible to various environmental stresses [6]. Notably, unseasonably cold conditions during winter and spring can significantly impede the growth and maturation of pepper plants, leading to reduced productivity and economic value [22]. Therefore, investigating the response of pepper to low-temperature stress is imperative for enhancing both its quality and yield. In this study, we isolated a low-temperature-induced R2R3-MYB TF, CaMYB80, from pepper and investigated its role in regulating low-temperature tolerance through virus-induced gene silencing (VIGS). The analysis was also conducted in Arabidopsis and tomato plants. We found that the C-terminal region of CaMYB80 possesses transcription activation ability, allowing it to directly bind the CaPOA1 promoter and activate its expression to enhance plant cold tolerance. Additionally, the heterologous overexpression of CaPOA1 also improved the cold resistance of Arabidopsis. In summary, our findings prove that CaMYB80 plays a crucial role in plant cold stress by directly regulating CaPOA1 expression, thereby enhancing peroxidase activity.
Results
CaMYB80 is responsive to low-temperature stress
Pepper leaves exposed to cold conditions (0 h, 6 h, and 24 h treatment at 4°C) underwent transcriptome analysis, with data accessible under the NCBI accession number PRJNA778231. The findings revealed a notable upregulation of 30 MYB gene family members at the transcriptional level following low-temperature treatment (Fig. S1a, see online supplementary material), with statistical significance (P < 0.05). Among these genes, LOC107845708 (GenBank accession number: NC_061120) demonstrated substantial induction (log2[fold change] = 1.62 ~ 1.73) and was consequently earmarked for further investigation.
The full-length cDNA sequence of CaMYB80 comprised a 957 bp open reading frame (ORF) encoding 318 amino acids, with a calculated molecular weight of 35.16 kDa and a theoretical isoelectric point of 8.32. Analysis of the amino acid sequence revealed the presence of two SANT/MYB conserved domains (13~63, 66~114), indicating that CaMYB80 conforms to the typical structure of an R2R3-MYB transcription factor. Phylogenetic assessment demonstrated a high degree of sequence similarity between CaMYB80 and the SlMYB80 sequence from Solanum lycopersicum (Fig. 1a).
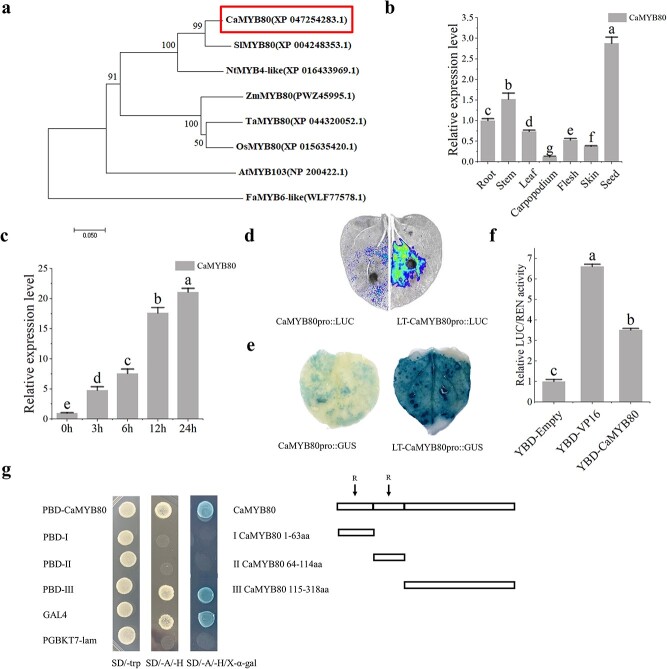
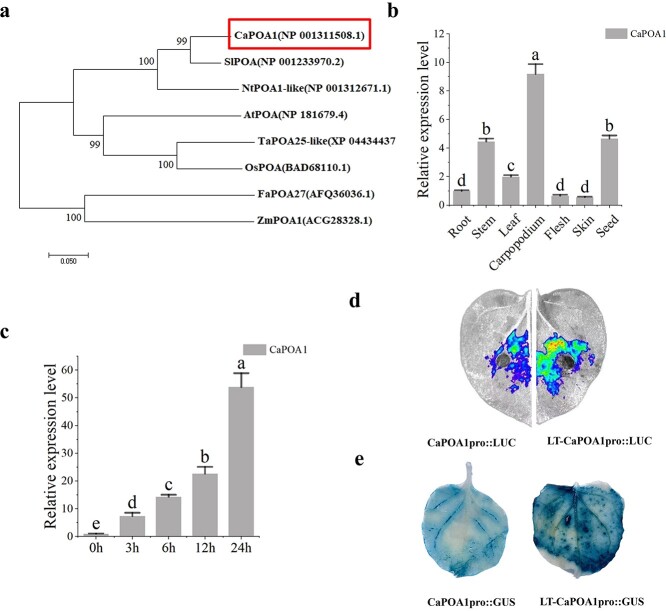
Figure 1.
CaMYB80 is responsive to low-temperature stress. Values are means ± SD from three independent experiments. Values with different letters above the bars are significantly different at P < 0.05. (a) Phylogenetic tree analysis of the CaMYB80 family from pepper (Capsicum annuum), tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), corn (Zea mays), rice (Oryza sativa), arabidopsis (Arabidopsis thaliana), wheat (Triticum aestivum), and strawberry (Fragaria ananassa). (b) Relative expression levels of CaMYB80 in the roots, stems, leaves, carpopodiums, flesh, skins and seeds of wild-type (WT) pepper, as determined by RT–qPCR. (c) Relative expression levels of CaMYB80 in pepper under cold conditions (4°C). (d) Luciferase complementation imaging (LCI) of the CaMYB80 native promoter after transient expression in tobacco; the results of the control (25°C/22°C day/night) and cold (4°C for 6 h) treatment groups are compared of the CaMYB80. (e) GUS staining of the CaMYB80 native promoter after transient expression in tobacco. (f) Transcriptional transactivation in tobacco. (g) Transcriptional transactivation in yeast; the right panel shows the CaMYB80 fragment used for testing.
Expression analysis of CaMYB80 across various tissues revealed a notably high expression level in seeds, followed by the stem, while the lowest expression was observed in Carpopodiums (Fig. 1b). We also found that the expression of CaMYB80 increased continuously with the low-temperature treatment duration, reaching its highest level at 24 h (Fig. 1c). To further investigate this, we inserted the natural promoter of CaMYB80 (2000 bp) into a vector containing LUC and GUS reporters for analysis. Plant luciferase in vivo imaging and GUS staining results revealed that the low-temperature treatment boosted the activity of the CaMYB80 promoter (Fig. 1d and e). This was further confirmed by quantifying LUC and GUS enzyme activities (Fig. S1b and c).
To validate the transcriptional activity of CaMYB80, we employed a dual-luciferase reporter gene system. Our findings demonstrated that the experimental group harboring the pBD-CaMYB80 effector vector exhibited notably higher LUC/REN values compared to the negative control (pBD-empty), suggesting that CaMYB80 possesses transcriptional activation capability (Fig. 1f). To determine the transcriptional activation region of CaMYB80, we conducted yeast two-hybrid assay using the recombinant plasmids containing the corresponding fragments. It was found that the yeast cells transformed with the complete ORF sequence of CaMYB80 (1–318 aa) and the C-terminal sequence of CaMYB80 (115–318 aa) grew normally on SD/−Ade/-His medium (Fig. 1g), indicating that the C-terminus of CaMYB80 has transcriptional activation ability.
CaMYB80 silencing reduces the cold tolerance of pepper
Firstly, we explored the function of CaMYB80 through VIGS. The results showed that unlike the TRV2:00 and TRV2:CaMYB80 plants, which had green leaves, leaf whitening was observed in TRV2: CaPDS plants after 3 weeks of transient transformation, demonstrating the reliability of the experiment (Fig. S2a, see online supplementary material). Subsequently, we measured the silencing efficiency using RT-qPCR and found that the silencing efficiency was close to 70% (Fig. S2b, see online supplementary material).
Under normal temperature conditions, TRV2:CaMYB80 plants exhibited reduced growth compared to TRV2:00 plants (Fig. 2a), as evidenced by significant reductions in their total chlorophyll content, net photosynthetic rate (Pn), potential photochemical efficiency (Fv/Fm), electron transport rate (ETR), and photochemical quenching (qP) (Fig. 2b–f). After a 24-hour treatment at 4°C, TRV2:CaMYB80 plants suffered more severe photosynthetic damage compared to TRV2:00 plants, with significant decreases in their chlorophyll content, Pn, Fv/Fm, ETR, and qP. However, non-photochemical quenching (qN) showed no significant difference between the two groups (Fig. 2b–g).
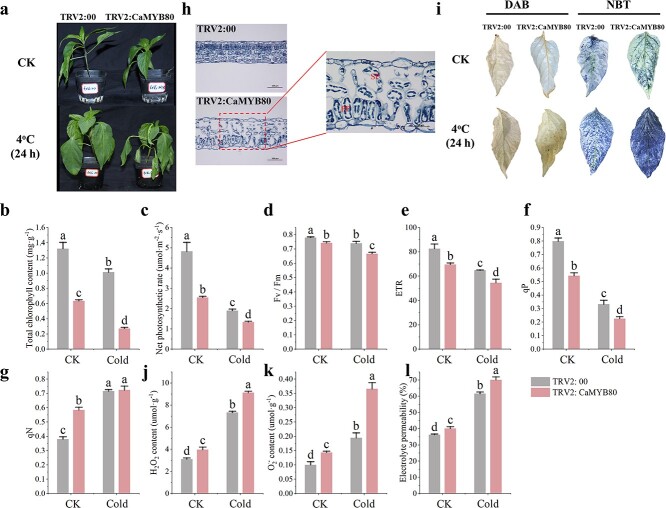
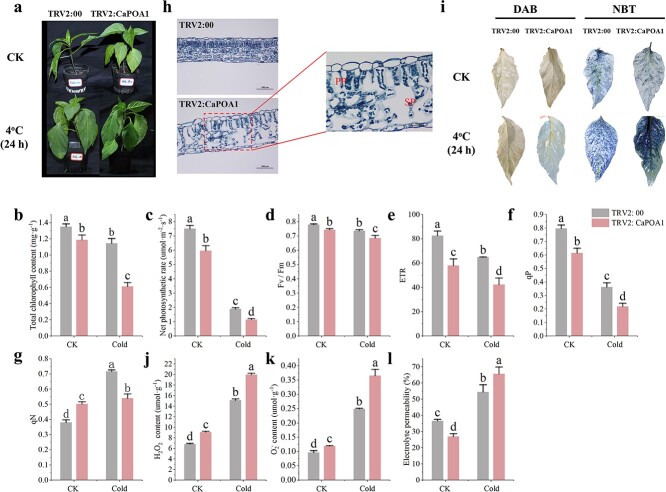
Figure 2.
CaMYB80 silencing reduces the cold tolerance of pepper. Values are means ± SD from three independent experiments. Values with different letters above the bars are significantly different at P < 0.05. (a) Comparison phenotypes between CaMYB80-silenced plants and control plants. (b) Total chlorophyll content. (c) Net photosynthetic rate. (d) Fv/Fm. (e) ETR. (f) qP. (g) qN. (h) Transverse sections of leaves of CaMYB80-silenced plants and control plants at low temperature. PP: palisade tissue; SP: spongy tissue. (i) NBT and DAB histochemical staining. (j) H2O2 content. (k) O2− content. (l) Electrolyte permeability.
Because the TRV2:CaMYB80 plants exhibited lower photosynthetic capacity, we studied their leaf anatomy. As shown in Fig. 2h, after 3 days of low-temperature treatment, TRV2:CaMYB80 leaves exhibited a more loosely arranged cell structure with disordered arrangement of fence tissues and larger and less tightly arranged intercellular spaces in sponge tissues compared to TRV2:00 plants. This indicated that TRV2:CaMYB80 plants suffered more severe leaf damage under low-temperature stress.
We utilized staining techniques with 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) to assess the accumulation of H2O2 and O2− in leaves, respectively. Compared to TRV2:00 plants, the leaves of TRV2:CaMYB80 plants exhibited a larger staining area under low-temperature stress (Fig. 2i), indicating a significant increase in the H2O2 and O2− levels (Fig. 2i–g). Additionally, there was a significant increase in electrolyte permeability of TRV2:CaMYB80 plants (Fig. 2k), suggesting increased cell membrane damage in TRV2:CaMYB80 plants under low-temperature stress.
Due to variations in the aforementioned physiological indicators induced by low-temperature stress, we conducted measurements of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), proline (Pro), soluble protein (Sp), and malondialdehyde (MDA) levels in pepper seedlings. Following a 24-hour treatment at 4°C, the SOD, POD, CAT, and Pro levels exhibited significant decreases, while the MDA level showed a significant increase in TRV2:CaMYB80 plants compared to control plants. However, there was no significant difference in the Sp level between the two groups (Fig. S2c–h, see online supplementary material).
Overall, our results demonstrate that CaMYB80 silencing reduces the ability of pepper plants to cope with ROS overproduction, resulting in increased oxidative damage.
Ectopic expression of CaMYB80 enhances Arabidopsis cold tolerance
Due to the recalcitrance of pepper in tissue culture, this experiment chose two model plants, Arabidopsis and tomato, for gene functional validation. To delve deeper into the role of CaMYB80, we first obtained T3 generation stable transgenic Arabidopsis plants. From these, we identified three transgenic Arabidopsis lines, designated OE3, OE9, and OE10, for further investigation. Relative to WT plants, these selected transgenic lines exhibited elevated expression levels of CaMYB80 (Fig. S3a, see online supplementary material). Subsequent non-acclimated (NA, −7°C) and cold-acclimated (CA, −10°C, plants exhibit stronger cold tolerance after cold acclimation, hence we chose to conduct tests at −10°C) conditions revealed that the heterologous overexpression of CaMYB80 significantly enhanced the freezing tolerance of Arabidopsis, leading to increased survival rates of transgenic Arabidopsis under both NA and CA conditions (Fig. S3c, see online supplementary material).
Compared to WT plants, the overexpression of CaMYB80 notably improved the freezing tolerance of Arabidopsis under both NA and CA conditions (Fig. 3b). Additionally, CaMYB80 overexpression increased the total chlorophyll content, as well as the SOD, POD, and CAT activities of Arabidopsis plants (Fig. 3c–f). Electrolyte permeability and MDA analysis results further indicated that OE-CaMYB80 Arabidopsis had less membrane damage than WT plants under freezing stress (Fig. 3g and h). After cold acclimation, the WT plants showed reduced membrane oxidative damage caused by freezing stress. We also verified the expression level of AtSOD, AtPRX, and AtCAT genes in Arabidopsis under NA and CA treatments. The findings indicated a significant elevation in the expression levels of AtSOD, AtPRX, and AtCAT genes in Arabidopsis under both NA and CA treatments upon overexpression of CaMYB80, aligning with the observed physiological indicators (Fig. 3i–k).
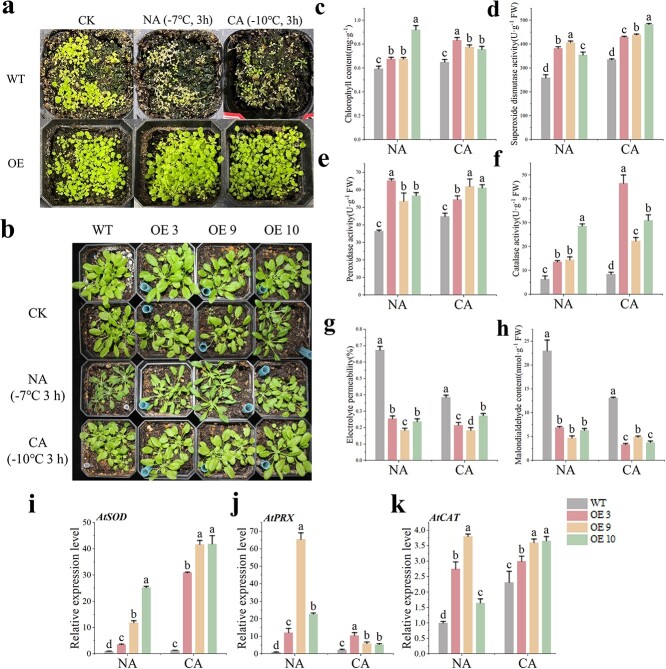
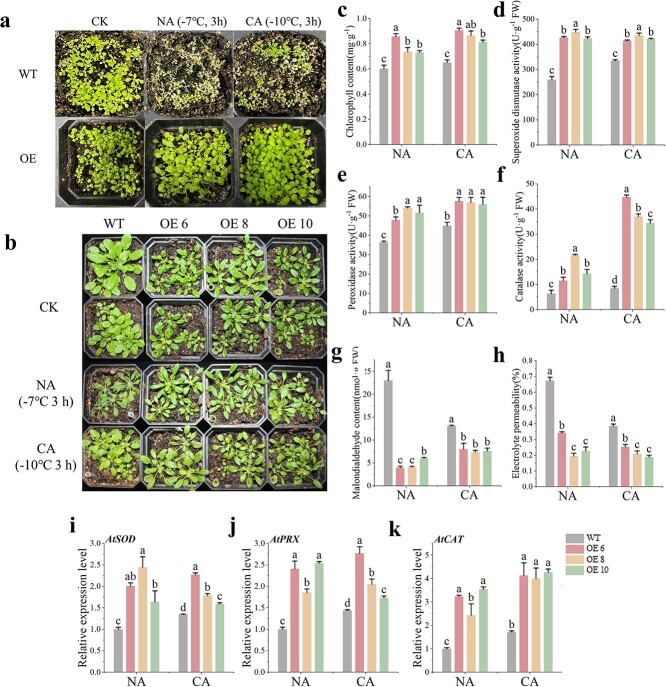
Figure 3.
Ectopic expression of CaMYB80 enhances Arabidopsis cold tolerance. Values are means ± SD from three independent experiments. Values with different letters above the bars are significantly different at P < 0.05. (a) Survival rate phenotypes of WT Arabidopsis and transgenic lines (OE3, OE9, O10). (b) Comparison phenotypes between WT Arabidopsis and transgenic lines (OE3, OE9, OE10). (c) Total chlorophyll content. (d) Superoxide dismutase activity. (e) Peroxidase activity. (f) Catalase activity. (g) Electrolyte permeability. (h) Malondialdehyde content. (i)–(k) Expression levels of AtSOD, AtPRX, and AtCAT in WT Arabidopsis and transgenic lines (OE3, OE9, OE10).
These findings indicate that CaMYB80 has the capacity to bolster the freezing tolerance of Arabidopsis by amplifying the activity of antioxidant enzymes and the expression of associated genes, consequently mitigating ROS accumulation and membrane damage induced by freezing stress. Consequently, CaMYB80 assumes a positive regulatory role in Arabidopsis’ response to freezing stress.
Ectopic expression of CaMYB80 enhances the cold tolerance of tomato
We further overexpressed CaMYB80 in tomato to obtain T1 transgenic tomato plants. We selected three transgenic tomato lines (OE1, OE4, and OE9) with high expression of CaMYB80 via PCR and RT-qPCR for further low-temperature experiments (Fig. S4a and b, see online supplementary material).
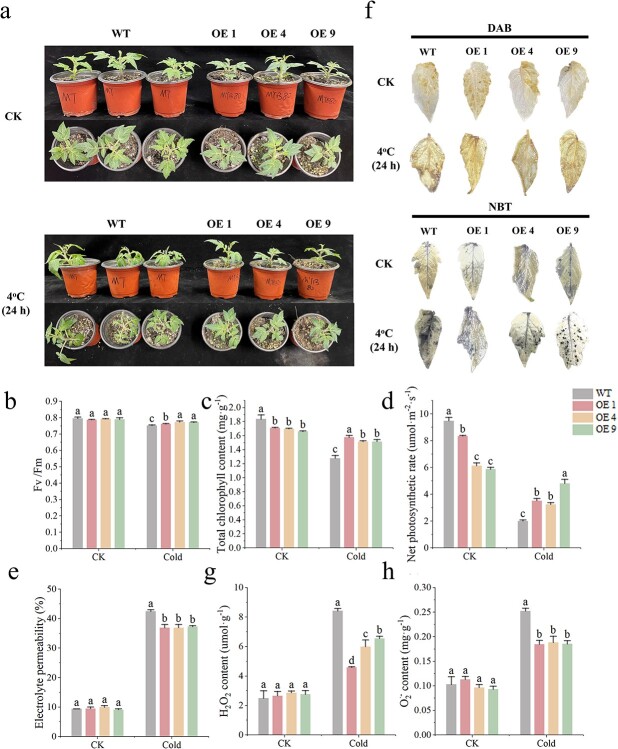
Under cold stress conditions, both WT tomato plants and three lines of tomato overexpressing CaMYB80 (OE1, OE4, and OE9) exhibited leaf wilting, albeit with more pronounced wilting observed in the WT plants (Fig. 4a). The overexpression of CaMYB80 conferred enhanced tolerance to low-temperature stress in tomato plants, mirroring the findings observed in transgenic Arabidopsis. Assessment of potential photosynthetic efficiency of PSII (Fv/Fm) revealed that, under low temperatures, CaMYB80-overexpressing tomato plants exhibited lesser inhibition of Fv/Fm (Fig. 4b), along with higher chlorophyll content and Pn levels (Fig. 4c and d) in their leaves compared to WT plants. Under normal temperature conditions, there was no notable variance in electrolyte permeability between the CaMYB80-overexpressing and WT plants. However, under low-temperature conditions, the electrolyte permeability of the WT tomato plants was significantly higher than that in the CaMYB80-overexpressing tomato plants (Fig. 4e). The changes in H2O2 and O2− contents also followed a similar pattern, with more staining observed in the leaves of the WT plants stained with DAB and NBT under low temperature compared to the CaMYB80-overexpressing plants (Fig. 4f). This was further confirmed by the quantitative analysis of H2O2 and O2− contents (Fig. 4g and h). Furthermore, the activity of ROS scavenging enzymes, as well as the levels of Pro and MDA, were measured. Compared to the WT plants, the CaMYB80-overexpressing tomato plants showed significantly increased SOD, POD, and CAT activities, enhanced proline content, and reduced accumulation of MDA under low-temperature stress (Fig. S5a–e, see online supplementary material).
Figure 4.
Ectopic expression of CaMYB80 enhances the cold tolerance of tomato. (a) Comparison of phenotypes between WT tomatoes and transgenic tomatoes (OE1, OE4, OE9). (b) Fv/Fm. (c) Total chlorophyll content. (d) Net photosynthetic rate. (e) Electrolyte permeability. (f) NBT and DAB histochemical staining. (g) H2O2 content. (h) O2− content. Values are means ± SD from three independent experiments. Values with different letters above the bars are significantly different at P < 0.05.
CaPOA1 is responsive to low-temperature stress
The complete cDNA sequence of CaPOA1 (peroxidase N1-like, LOC107840027) contained an ORF spanning 996 base pairs, which encoded a protein of 332 amino acids. The protein exhibited a molecular weight of 36.07 kDa and a theoretical isoelectric point of 6.88. Phylogenetic analysis indicated a close relationship between CaPOA1 and SlPOA (S. lycopersicum) (Fig. 5a).
Figure 5.
CaPOA1 is responsive to low-temperature stress. Values are means ± SD from three independent experiments. Values with different letters above the bars are significantly different at P < 0.05. (a) Phylogenetic tree analysis of the CaPOA1 family from (Capsicum annuum), tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), corn (Zea mays), rice (Oryza sativa), arabidopsis (Arabidopsis thaliana), wheat (Triticum aestivum), and strawberry (Fragaria ananassa). (b) Relative expression levels of CaPOA1 in the roots, stems, leaves, carpopodiums, flesh, skins and seeds of WT pepper, as determined by RT–qPCR. (c) Relative expression levels of CaPOA1 in WT pepper under cold conditions (4°C). (d) Luciferase complementation imaging (LCI) of the CaPOA1 native promoter after transient expression in tobacco. (e) GUS staining of the CaPOA1 native promoter after transient expression in tobacco. The results of the control (25°C/22°C day/night) and cold (4°C for 6 h) treatment groups are compared of the CaPOA1.
Analysis of CaPOA1 expression across various tissues revealed its highest expression in the Carpopodium, followed by the stem, while the lowest expression was observed in the skin (Fig. 5b). Additionally, we noted a steady increase in CaPOA1 expression with prolonged low-temperature treatment, peaking at 24 hours (Fig. 5c). To investigate further, we inserted the native promoter of CaPOA1 (2000 bp) into a vector containing LUC and GUS reporter for detection. Through in vivo imaging and GUS staining, we found that low-temperature stress enhanced the activity of the CaPOA1 promoter (Fig. 5d and e). We validated these results through LUC reporter gene assays and GUS enzyme activity tests (Fig. S6a and b, see online supplementary material).
CaMYB80 binds to the promoter of CaPOA1
The reduction of ROS is associated with various antioxidant enzymes. We further analyzed all cis-element analysis in the promoter regions of CaSOD, CaPOA1, CaCAT, CaAPX1, and CaDHAR2. Based on their functional associations, we categorized them into four categories (Fig. S7, see online supplementary material): biotic and abiotic stress-related elements (LTR, TC-rich repeats, and MBS), hormone-responsive elements (ABRE, TGACG-motif, P-box, TGA-element, CGTCA-motif, and TCA-element), light-responsive elements (MRE, G-box, GT1-motif, GA-motif, ACE, AE-Box, GATA-motif, I-box, and TCCC-motif), and MYB-response elements (MYB). Our results indicate that the promoters of the CaSOD, CaPOA1, and CaCAT contain MYB elements, but only the natural promoter of CaPOA1 (186–200 bp) includes a binding site for CaMYB80 (AATAATTAGGTAACC). Therefore, we speculate that CaMYB80 can directly regulate the expression of CaPOA1.
We first determined the subcellular localization of CaMYB80 and CaPOA1 by co-expressing GFP-CaMYB80 and GFP-CaPOA1 fusion proteins with cytoplasmic and nuclear markers in tobacco leaves. Confocal microscopy observation showed that GFP-CaMYB80 and GFP-CaPOA1 were localized in the nucleus and cytoplasm (Fig. 6a).
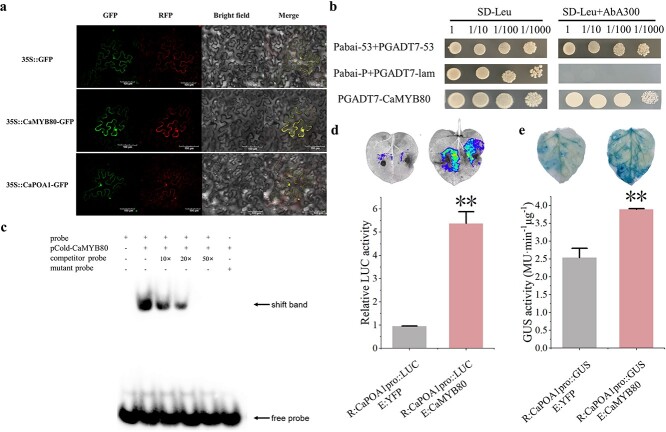
Figure 6.
CaMYB80 binds to the promoter of CaPOA1. (a) Subcellular localization; scale bars = 10 μm. (b) Results of the Y1H experiment. (c) EMSA, from left to right: 6-FAM labeled prope; CaMYB80 and 6-FAM labeled probe; CaMYB80, 6-FAM labeled probe and 10× unlabeled probe; CaMYB80, 6-FAM labeled probe and 20× unlabeled probe; CaMYB80, 6-FAM labeled probe and 50× unlabeled probe; CaMYB80 and 6-FAM labeled mutant probe. (d) LUC analysis results. (e) GUS enzyme activity results. Asterisks indicate significant difference (*P < 0.05; **P < 0.01).
To test the hypothesis of direct regulation of CaPOA1 expression by CaMYB80, we conducted yeast one-hybrid experiments. The results demonstrated normal growth of yeast cells co-expressing prey and bait plasmids on selective media, indicating the ability of CaMYB80 to bind to the MYB binding site in the CaPOA1 promoter (Fig. 6b). Furthermore, validation through EMSA assay confirmed the binding capability of CaMYB80 to the MYB binding site within the CaPOA1 promoter (Fig. 6c).
We further verified the Y1H and EMSA results by performing dual luciferase and GUS reporter assays. The results showed that the LUC and GUS activities were significantly higher in tobacco leaves co-infiltrated with 35S:CaMYB80 and CaPOA1pro: LUC/GUS than those infiltrated with 35S: empty and CaPOA1pro: LUC/GUS. The plant luciferase in vivo imaging and GUS staining results were consistent with the LUC/GUS activity results (Fig. 6d and e). These results indicate that CaMYB80 specifically binds the CaPOA1 promoter and activates its transcription.
CaPOA1 silencing reduces the cold tolerance of pepper
We validated the cold resistance function of CaPOA1 using VIGS and conducted RT-qPCR to determine the silencing efficiency of CaPOA1, which was found to be close to 76.9% (Fig. S8a, see online supplementary material). Under normal temperature conditions, there was no significant difference in the phenotype of TRV2:CaPOA1 and TRV2:00 plants. However, after 24 h of treatment at 4°C, the leaves of TRV2:CaPOA1 plants showed more severe wilting (Fig. 7a), as demonstrated by a significant decrease in the total chlorophyll, Pn, Fv/Fm, ETR, and qP contents, and a significant increase in qN (Fig. 7b–g). Further leaf anatomy study revealed that TRV2:CaPOA1 plants exhibited a looser cell structure, sparse palisade tissue, and larger gaps between sponge tissue after three days of cold treatment (Fig. 7h).
Figure 7.
CaPOA1 silencing reduces the cold tolerance of pepper. Values are means ± SD from three independent experiments. Values with different letters above the bars are significantly different at P < 0.05. (a) Comparison phenotypes between CaPOA1-silenced plants and control plants. (b) Total chlorophyll content. (c) Net photosynthetic rate. (d) Fv/Fm. (e) ETR. (f) qP. (g) qN. (h) Transverse sections of leaves of CaPOA1-silenced plants and control plants at low temperature. PP: palisade tissue; SP: spongy tissue. (i) NBT and DAB histochemical staining. (j) H2O2 content. (k) O2− content. (l) Electrolyte permeability.
The histological staining results showed that compared to TRV2:00 plants, TRV2:CaPOA1 plants had more spots on the leaves under normal and low-temperature conditions (Fig. 7i). We quantitatively analysed the levels of H2O2 and O2− (Fig. 7j and k), and the results were consistent with the histological staining results. Moreover, the electrolyte permeability of TRV2:CaPOA1 plants was significantly lower than control plants under normal temperature but significantly higher under low-temperature conditions (Fig. 7i).
Additionally, we conducted measurements of SOD, POD, and CAT activities, along with Pro, Sp, and MDA levels in pepper leaves. The findings revealed markedly decreased levels of SOD, POD, Sp, and Pro, accompanied by a notable increase in MDA level in CaPOA1-silenced plants compared to the control plants. However, there was no significant disparity in CAT activity between the two groups (Fig. S8b–g, see online supplementary material). These outcomes suggest that silencing of CaPOA1 diminishes the cold tolerance of pepper plants, rendering them more vulnerable to oxidative harm.
Ectopic expression of CaPOA1 enhances Arabidopsis cold tolerance
To further investigate the function of CaPOA1, we produced transgenic Arabidopsis plants overexpressing CaPOA1 in the T3 generation. PCR and RT-qPCR were employed to select three transgenic lines (OE6, OE8, and OE10) exhibiting high CaPOA1 expression levels for freezing tolerance assessment (Fig. S9a and b, see online supplementary material). Remarkably, compared to WT plants, these transgenic Arabidopsis lines displayed notably enhanced survival rates under both NA and CA conditions (Fig. S9c, see online supplementary material).
Overexpression of CaPOA1 significantly enhanced the freezing tolerance (Fig. 8a and b) and increased total chlorophyll content and the SOD, POD, and CAT activities (Fig. 8c–f) of Arabidopsis under NA and CA conditions plants. Further analysis of the MDA content and electrolyte permeability indicated that the membrane damage was less severe in the transgenic Arabidopsis plants overexpressing CaPOA1 than in the WT plants under cold stress (Fig. 8g and h). We also found that the expression levels of AtSOD, AtPRX, and AtCAT genes in Arabidopsis under NA and CA treatments were consistent with the physiological indexes (Fig. 8i–k). These results suggest that CaPOA1 improves Arabidopsis’ ability to withstand freezing temperatures by enhancing the activities of antioxidant enzymes and the expression of related genes, thereby positively regulating the plant’s response to low-temperature stress.
Figure 8.
Ectopic expression of CaPOA1 enhances Arabidopsis cold tolerance. Values are means ± SD from three independent experiments. Values with different letters above the bars are significantly different at P < 0.05. (a) Survival rate test phenotypes of WT Arabidopsis and transgenic lines (OE6, OE8, OE10). (b) Comparison phenotypes between WT Arabidopsis and transgenic lines (OE6, OE8, OE10). (c) Total chlorophyll content. (d) Superoxide dismutase activity. (e) Peroxidase activity. (f) Catalase activity. (g) Electrolyte permeability. (h) Malondialdehyde content. (i)–(k) Expression levels of AtSOD, AtPRX, and AtCAT in WT Arabidopsis and transgenic lines (OE6, OE8, OE10).
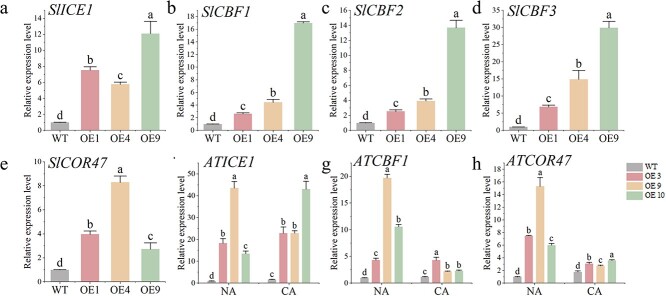
CaMYB80 promotes the expression of the CBF regulatory network-related genes
To explore the potential regulatory role of CaMYB80 on genes within the CBF regulatory network, we assessed the expression of ICE, CBF, and COR47 genes in both WT and CaMYB80-overexpressing tomato and Arabidopsis plants under NA and CA conditions using RT-qPCR. Notably, the expression levels of cold-responsive genes SlICE1, SlCBF1, SlCBF2, SlCBF3, and SlCOR47 were markedly elevated in CaMYB80-OE tomato plants (Fig. 9a–e). Similarly, in CaMYB80-OE Arabidopsis lines, the expression levels of ATICE1, ATCBF1, and ATCOR47 were significantly augmented under both NA and CA treatments (Fig. 9f–h). These findings suggest that CaMYB80 modulates the expression of genes within the CBF regulatory network in response to low-temperature stress, consequently enhancing the plants’ cold tolerance.
Figure 9.
CaMYB80 promotes the expression of the CBF regulatory network-related genes. (a)–(e) Expression levels of SlICE1, SlCBF1, SlCBF2, SlCBF3, and SlCOR47 in WT tomatoes and transgenic tomatoes (OE1, OE4, OE9) at low temperature. (f)–(h) Expression levels of AtICE1, AtCBF1, and AtCOR47 in WT Arabidopsis and transgenic lines (OE3, OE9, OE10). Values are means ± SD from three independent experiments. Values with different letters above the bars are significantly different at P < 0.05.
Discussion
As a warmth-loving vegetable, pepper experiences reduced yield and quality and even plant death under low-temperature stress [22]. Therefore, exploring the cold tolerance mechanism of pepper is of great significance for improving its quality and yield. In our investigation, we identified an R2R3-type MYB TF, CaMYB80. Phylogenetic analysis demonstrated that CaMYB80 shares significant homology with other plant R2R3 MYB proteins, particularly showing close relatedness to tomato SlMYB80 (Fig. 1a). Previous research has linked the SlMYB80 gene with cold tolerance in tomato plants [23]. In this article, we reported that CaMYB80 is a cold-responsive TF that enhances plant cold tolerance. We discovered that CaMYB80 interacts with the promoter of CaPOA1, increasing CaPOA1 transcript levels and thereby positively regulating cold tolerance in peppers. These findings provide important insights for the molecular breeding of cold-resistant pepper varieties.
Previous studies have shown that MYB TFs play a crucial role in regulating plant cold resistance [1, 11]. These proteins can regulate plant cold tolerance by regulating the ROS scavenging system. Overexpression of the R2R3-MYB gene LeAN2 could increase antioxidant enzyme activity and enhance the low-temperature stress tolerance of tomato [24]. Additionally, overexpressing the MYB-related grape TF, AQUILO, in Arabidopsis could increase SOD and POD activity and upregulate the expression of their encoding genes [25]. In this experiment, CaMYB80-silenced plants showed a more sensitive phenotype to low temperature (Fig. 2a), while CaMYB80-overexpressing (OE) Arabidopsis and tomato plants showed increased tolerance to low temperature (Figs 3b and 4a). Damaged Photosystem II (PSII) and reduced chlorophyll content are common in stress-sensitive plants, low-temperature stress can reduce the ability of PSII to utilize light energy, leading to photoinhibition, while also causing significant damage to membrane structures [26]. Under cold stress, the Pn and chlorophyll contents of CaMYB80-silenced plants were significantly lower than those of the control plants (Fig. 2b and c). Simultaneously, there is more damage to leaf structure, resulting in disordered arrangement of cellular tissues (Fig. 2h). However, the chlorophyll content of CaMYB80-OE plants was higher (Fig. 3c). ROS are the products of plant metabolism and have extremely high activity and toxicity [26]. The ROS accumulation was significantly reduced in CaMYB80-OE tomato plants (Fig. 4f) but significantly increased in CaMYB80-silenced plants compared to the control plants (Fig. 2k and l). This finding was confirmed by tissue staining, as CaMYB80-silenced plants had deeper DAB and NBT staining under low-temperature stress (Fig. 2i). In plants, MDA content and electrolyte permeability are indicators of ROS-mediated cell membrane damage [27, 28]. In chrysanthemum, the MDA content and relative electrolyte permeability in the DgMYB2-OE lines were the lowest, followed by the WT and DgMYB2-RNAi lines [1]. In our study, the electrolyte permeability and MDA levels of CaMYB80-OE plants were significantly reduced after low-temperature stress compared to the WT plants (Fig. 3g and h). These findings indicate that during cold stress, CaMYB80 can maintain leaf photosynthetic capacity, elevate antioxidant enzyme activity for ROS scavenging, uphold the equilibrium of ROS within cells, and thereby enhance plant cold resistance. These results provide evidence for the positive role of CaMYB80 in response to low-temperature stress.
Studies have revealed that MYB TFs predominantly regulate gene expression by binding to the promoter regions of target genes, thus either facilitating or inhibiting their expression [10, 29]. In chrysanthemums, the downstream target gene DgGPX1 of DgMYB2 has been identified, and overexpression of DgMYB2 can enhance plant cold tolerance by increasing the activity of glutathione peroxidase. [1]. Similarly, MdMYB23 has been found to directly bind to the promoter regions of MdCBF1 and MdCBF2, thereby enhancing apple cold resistance by activating the expression levels of downstream COR genes [30]. In pears, PbrMYB5 directly activates the expression of PbrDHAR2, consequently enhancing cold tolerance by promoting the synthesis of ascorbic acid (AsA) [31] (Fig. 1g). Our research indicates that the C-terminal of CaMYB80 possesses transcriptional activation activity (Fig. 1g). This attribute may be linked to the characteristic features of MYB transcription factors, where the DNA-binding domain located at the N-terminus distinguishes various types of MYB TFs, while the C-terminus is responsible for mediating transcriptional activity [32]. Through analyses involving Y1H, LUC/REN, GUS activity, and EMSA, our investigation revealed that CaMYB80 exhibits direct binding to the promoter region of the CaPOA1 gene, and positively regulates the expression of CaPOA1 (Fig. 6b–e). Additional evidence suggests that POD, an antioxidant enzyme widely present in plants, works synergistically with SOD and CAT to remove excess free radicals under stress conditions, thereby enhancing plant stress resilience [33]. In Arabidopsis, elevating the expression of AtPrx3 resulted in heightened tolerance to dehydration and salt stress, whereas suppressing AtPrx3 led to the manifestation of dehydration and salt-sensitive traits [34]. Introducing CaPOD2 from pepper into Arabidopsis enhances the plant’s resistance to pathogens [35]. Moreover, heterologous expression of OsPrx114 in rice also enhanced resistance to pathogens by inducing pathogen-related genes [36]. Our results indicated that CaPOA1 plays a crucial role in clearing excess ROS. Under low-temperature conditions, transgenic Arabidopsis plants overexpressing CaPOA1 had lower ROS levels, while CaPOA1-silenced plants had higher levels of ROS compared to WT plants. Due to the beneficial role of CaPOA1 under low temperatures, we hypothesize that CaMYB80 can confer cold tolerance. The primary mechanism by which CaMYB80 enhances plant cold resistance is by enhancing the expression of CaPOA1 associated with scavenging ROS. In this study, SOD, POD, and CAT activities were lower in CaMYB80-silenced and CaPOA1-silenced plants compared to control plants (Figs S2c–e and S8b–d, see online supplementary material), indicating that CaMYB80 and CaPOA1 silencing reduces ROS levels. Conversely, plants overexpressing CaMYB80 and CaPOA1 exhibited higher SOD, POD, and CAT activities than control plants (Figs 3d–f and 8d–f; Fig. S5a–c, see online supplementary material). Furthermore, the expression of antioxidant-related genes (AtSOD, AtPRX, and AtCAT) significantly increased in CaMYB80-OE plants and CaPOA1-OE plants (Figs 3i–k and 8i–k). These results support the notion that CaMYB80 enhances plant cold tolerance by increasing the ability of CaPOA1 to scavenge ROS molecules (Fig. 10).
Figure 10.
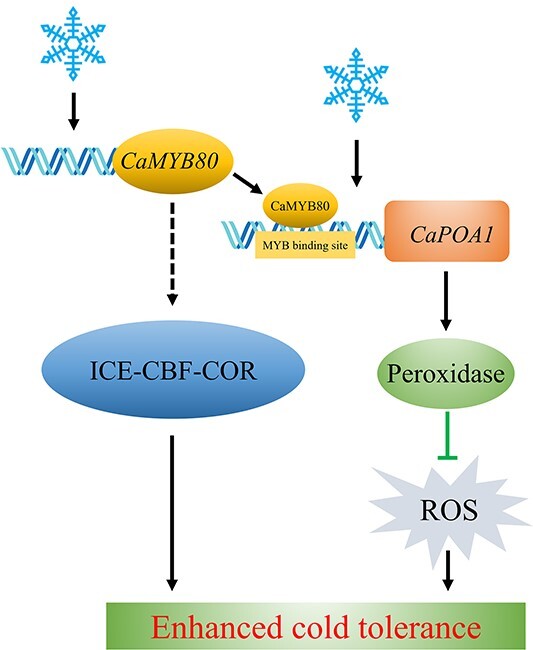
Proposed model through which CaMYB80 enhanced the cold tolerance of pepper by directly targeting CaPOA1.
Furthermore, previous research indicated that stress-related genes regulate stress responses in transgenic plants [37]. To date, the most well-studied low-temperature regulation network is the ICE-CBF-COR (inducer of CBF expression, C-repeat binding factor, cold-responsive) transcriptional model [38]. Under low temperatures, the dehydration-responsive element-binding 1 (DREB1/CBFs) genes are rapidly induced, followed by activation of downstream target COR genes, resulting in the accumulation of protective substances that promote cold acclimation and freezing tolerance, thereby enhancing their cold tolerance [39, 40]. ICE1 can bind the MYC binding site in the CBF1–3 promoter, thereby promoting CBF expression under cold stress and positively regulating plant response to low temperatures [41]. In AtICE1 mutant plants, AtCBF expression is suppressed under cold stress, resulting in reduced cold tolerance in Arabidopsis [42]. In this study, overexpression of CaMYB80 significantly increased the expression levels of ICE1, CBFs, and COR47 in tomatoes and Arabidopsis (Fig. 9a–h). These results suggest that CaMYB80 may enhance plant cold tolerance by regulating the expression of CBF network genes (Fig. 10).
Conclusion
Our study presents the inaugural framework elucidating the role of CaMYB80 in modulating cold tolerance in pepper. The research unveils that (i) CaMYB80 is directly activated by low temperatures and enhances pepper cold tolerance by upregulating the expression of cold stress-responsive genes (ICE-CBF-COR); (ii) CaMYB80 interacts directly with CaPOA1, thereby decreasing ROS accumulation through the promotion of CaPOA1 expression; and (iii) CaPOA1 positively regulates cold tolerance in peppers by enhancing ROS scavenging capacity to reduce leaf oxidative damage, thereby maintaining plant tolerance to low-temperature stress. These findings indicate that CaMYB80 potentially enhances pepper cold tolerance by impacting the CBF pathway and bolstering ROS scavenging capacity.
Materials and methods
Plant materials and growth conditions
The experimental materials, including Arabidopsis ecotype Columbia-0, wild-type MicroTOM tomato, tobacco (Nicotiana benthamiana), and pepper variety ‘Ganzi’, were obtained from the Vegetable Germplasm Innovation Laboratory of Sichuan Agricultural University. These plants were cultivated in a climate-controlled growth chamber set to a temperature of 25°C during the day and 20°C at night, with a photoperiod of 16 hours of light followed by 8 hours of darkness and maintained at 70% relative humidity.
Phylogenetic analyses
Target protein sequences (MYB80 and POA1) were obtained through BLAST (Basic Local Alignment Search Tool) on the NCBI website (https://www.ncbi.nlm.nih.gov/). The Neighbor-Joining (NJ) method was used to construct the phylogenetic trees of MYB80 and POA1 in MEGA X software.
Virus-induced gene silencing of CaMYB80 and CaPOA1
To silence CaMYB80 and CaPOA1, we inserted a 300 bp ORF of each gene into the TRV2 vector, resulting in the generation of recombinant vectors TRV2-CaMYB80 and TRV2-CaPOA1. These vectors, along with TRV2: 00, TRV2: CaPDS, and TRV1 vectors, were transformed into Agrobacterium tumefaciens strain GV3101. Subsequently, the TRV2: CaMYB80, TRV2: CaPOA1, TRV2: CaPDS, and TRV2: 00 vectors were mixed equally and infiltrated into the cotyledons of 2-week-old pepper plants. Following two days of incubation in darkness, the plants were transferred to normal growth conditions, following the protocol described by Zhang et al. [3]. After 21 days, the efficiency of CaMYB80 and CaPOA1 silencing was assessed using RT-qPCR.
Subcellular localization
The coding sequences (CDS) of the CaMYB80 and CaPOA1 genes were amplified using primers designed to exclude stop codons. High-fidelity polymerase chain reaction (PCR) was employed, utilizing Takara high-fidelity enzyme (Takara, Shiga, Japan) for amplification, and the resulting gene sequences were cloned into the PBI221-GFP vector. The GFP-CaMYB80 and GFP-CaPOA1 plasmids, along with the PBI221-GFP empty vector, were individually introduced into A. tumefaciens strain GV3101. Bacterial cell suspensions containing different plasmids were mixed at equal ratios of 1:1:1 and infiltrated into tobacco leaves (OD600 = 0.6). The infiltrated plants were maintained in a growth chamber for 2–3 days before being examined and photographed under a laser confocal microscope (FV3000, Olympus, Tokyo, Japan).
Arabidopsis and tomato transformation
To establish Arabidopsis plants with CaMYB80 and CaPOA1 overexpression, we inserted the CDS of CaMYB80 and CaPOA1 into the PBI121 vector under the control of the 35S promoter. These constructs, named 35S:CaMYB80 and 35S:CaPOA1, were then introduced into A. tumefaciens strain GV3101. Arabidopsis genetic transformation was achieved using the floral dip method, and the resulting T3 plants were selected for subsequent investigations.
For tomato, two-week-old plants underwent transformation via Agrobacterium-mediated leaf infiltration. The infiltrated leaves were utilized to obtain callus tissues, which were cultured on Murashige and Skoog (MS) solid medium supplemented with 50 mg/L kanamycin to encourage elongation and rooting. This process yielded the T0 generation of transgenic tomato plants, which underwent screening to obtain seeds for the T1 generation plants. The transgenic status of both Arabidopsis and tomato plants was confirmed through PCR and RT-qPCR analyses.
Cold stress tolerance assay
To analyse the cold stress tolerance of the T3 transgenic Arabidopsis and wild-type (WT) Arabidopsis, we performed low-temperature treatment experiments on the WT and transgenic plants expressing CaMYB80 and CaPOA1. The treatments were as follows: (i) CK: normal temperature conditions; (ii) NA: −7°C (3 h) + 4°C (24 h); (iii) CA: 4°C (3 d) + −10°C (3 h) + 4°C (24 h). Following the low-temperature treatment, certain plants were relocated to normal temperature conditions for a 3-day recovery period, and their survival rate was determined. Additionally, to assess the cold tolerance of pepper and tomato plants, experiments involving low-temperature treatments were conducted on pepper plants with silenced CaMYB80 and CaPOA1 genes, CaMYB80-transgenic tomato plants, and WT plants. The treatments were: (i) CK: normal temperature conditions; (ii) 4°C (24 h). After low-temperature stress, samples were taken and stored at −80°C after treatment with liquid nitrogen for later use. Each treatment was set up for three biological replicates.
Measurement of physiological indicators
The determination details of chlorophyll content, electrolyte permeability, SOD, POD, CAT, MDA, hydrogen peroxide (H2O2), superoxide anion (O2−), and Pro were referenced from previous studies [43].Chlorophyll fluorescence parameters were recorded with a PAM2500 chlorophyll fluorometer (Walz, Nuremberg, Germany), and photosynthetic parameters were measured using a LI-6400 portable photosynthesis system (LI-COR Inc., Lincoln, NE, USA).
Plant leaf staining for visualization of H2O2 and O2− contents was performed using diaminobenzidine (DAB) and nitroblue tetrazolium (NBT), respectively. Leaves were immersed in DAB solution (1 mg/ml DAB +20 mM Na2HPO4, pH 3.8) and NBT solution (0.5 mg/ml NBT), then kept in darkness for 6 hours. Subsequently, photographs were taken (DM2500, Leica, Wetzlar, Germany) after alcohol decolorization.
RNA extraction and real-time quantitative PCR
Total RNA was extracted from pepper, Arabidopsis, and tomato leaves using the Plant RNA Extraction Kit (Takara, Shiga, Japan). Subsequently, first-strand cDNA synthesis was performed utilizing the PrimeScript kit (Takara, Shiga, Japan), followed by RT-qPCR analysis conducted on an iCycleriQ™ machine (Bio-Rad, Hercules, CA, USA). To determine the relative expression levels of the target genes, normalization was carried out against the geometric mean of Caactin2, Atactin2, and SIUBI3 expression levels using the delta–delta Ct (2−ΔΔCT) method. Details of the primers employed for the analysis can be found in Table S1 (see online supplementary material).
Low-temperature response of CaMYB80 and CaPOA1 promoters
To explore the response of CaMYB80 and CaPOA1 to low-temperature stress, we constructed CaMYB80pro-LUC, CaMYB80pro-GUS, CaPOA1pro-LUC, and CaPOA1pro-GUS vectors, which were then introduced into A. tumefaciens strain GV3101 for co-infiltration into tobacco leaves. The activities of LUC enzyme and GUS enzyme were determined according to Dual-Luciferase Reporter Gene Assay Kit (Beyotime Biotechnology, Shanghai, China) and GUS gene quantitative detection Kit (Coolaber, Beijing, China), respectively.
cis-element analysis
To understand the expression regulation mechanism of CaSOD, CaPOA1, CaAPX1, CaCAT, and CaDHHR2, sequences upstream of the start codon within a 2000 bp range were extracted, and PlantCARE was used for cis-element prediction, with visualization using TBtools.
Transcriptional activation analysis of CaMYB80
A reporter gene construct was engineered by incorporating 5 × GAL4 sequences and 1 × TATA sequence upstream of the LUC gene within the pGreenII0800-LUC vector. Subsequently, the CaMYB80 ORF was inserted into an enhanced pSuper1300-YFP vector containing gal4bd, resulting in the creation of the recombinant plasmid pBD-CaMYB80. For comparative analysis, pBD-VP16 served as a positive control, while the improved pBD-Empty vector acted as a negative control. Following transformation, the recombinant plasmids were introduced into A. tumefaciens strain GV3101. Co-infiltration of the transformed bacterial solution containing 5 × UAS-TATA-LUC with the bacterial suspensions harboring pBD-CaMYB80, pBD-VP16, and pBD-Empty constructs was conducted in tobacco leaves. The LUC/REN ratio was assessed after 2 days of incubation.
The CDS of CaMYB80, along with three sequence fragments (1–63 aa, 64–114 aa, 115–318 aa), were individually inserted into the yeast expression vector pGBKT7. These constructs were subsequently introduced into the yeast strain Y2H and plated onto selective media with or without X-α-gal under double dropout (SD/−Ade/-His) conditions. The plates were then inverted and incubated at 30°C for 3 days, after which colonies were examined and photographed.
Y1H, dual-LUC, GUS activity, and EMSA assays
The CDS of CaMYB80 was inserted into the pGADT7 vector, while the anticipated element upstream of the CaPOA1 promoter was linked to the pAbAi vector for yeast one-hybrid (Y1H) screening. The self-activation of pAbAi-CaPOA1pro using adding aureobasidin A (AbA) (Coolaber, Beijing, China) to SD/-Ura dropout media and the appropriate AbA concentration was screened. Subsequently, pGADT7-CaMYB80 was transformed into competent yeast cells prepared with pAbAi-CaPOA1pro on SD/−Leu dropout media containing AbA (300 ng/mL). After incubation at 28°C in the dark for 2–3 days, the growth of each group was assessed to determine whether there was binding between CaPOA1:pro and CaMYB80.
To further explore the interaction between CaMYB80 and the promoter cis-elements of CaPOA1, we conducted a luciferase reporter gene assay. The 2000 bp promoter region of CaPOA1 was cloned into the pGreenII0800-LUC vector to create the reporter construct. CaMYB80 was then integrated into the pSuper1300-YFP vector via homologous recombination to generate the effector construct. Subsequently, both the reporter and effector constructs were co-infiltrated into tobacco leaves. Following a 2-day incubation period, in vivo imaging was conducted for fluorescence detection, and the LUC/REN ratio was subsequently determined.
GUS activity analysis was conducted by cloning the CaMYB80 gene into the pSuper1300-YFP vector and transforming the vector into A. tumefaciens strain GV3101 as the effector and cloning the 2000 bp promoter region of CaPOA1 into the pCAMBIA2300-MCS-GUS vector as the reporter. The reporter and effector were co-infiltrated into tobacco leaves, and after 2 days of incubation, GUS staining and GUS enzyme activity assay were performed on the leaves.
Electrophoresis mobility shift assay (EMSA) was performed according to the method by Yang et al. [1]. Briefly, the CDS of CaMYB80 was cloned into the pCold I vector for HIS-tag fusion, and the HIS-CaMYB80 protein was expressed and purified from BL21 (DE3) cells. The obtained protein was subjected to gel EMSA using the EMSA Chemiluminescent Kit (Thermo Fisher Scientific, 20 148, Waltham, MA, USA). A 5′ biotin-labeled promoter region of the CaPOA1 (AATAATTAGGTAACC) was used as the probe for the analysis.
Statistical analysis
The data were organized using Excel 2019 software (Microsoft, Redmond, WA, USA), SPSS23.0 (IBM, New York, NY, USA) as used for statistical analysis at P < 0.05 with Tukey’s test. The graphs were plotted using Orign 2019b (Electronic Arts Inc, Redwood, CA, USA) and the data was presented as the mean ± standard deviation, Values with different letters above the bars are significantly different at P < 0.05.
Acknowledgements
This work was supported by breeding research in vegetables (2021YFYZ0022).
Author contributions
J.X.: Writing – review & editing, Writing – original draft, Formal analysis, Data curation. D.W.: Data curation. L.L.: Data curation. M.X.: Data curation. Y.T.: Investigation. Y.-S.L.: Investigation. B.S.: Data curation. Z.H.: Methodology. Y.Z.: Methodology. H.L.: Conceptualization, Funding acquisition, Writing – review & editing.
Supplementary Material
Contributor Information
Jiachang Xiao, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Dong Wang, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Le Liang, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Minghui Xie, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Yi Tang, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Yun-Song Lai, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Bo Sun, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Zhi Huang, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Yangxia Zheng, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Huanxiu Li, College of Horticulture, Sichuan Agricultural University, Chengdu 611130, China.
Data availability
The data are presented within the paper and supplementary files.
Conflict of interest statement
The authors declare no conflict of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
References
- 1. Yang X, Luo Y, Bai H. et al. DgMYB2 improves cold resistance in chrysanthemum by directly targeting DgGPX1. Hortic Res. 2022;9:uhab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X, Ding Y, Li Z. et al. PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev Cell. 2019;51:222–235.e5 [DOI] [PubMed] [Google Scholar]
- 3. Zhang L, Jiang X, Liu Q. et al. The HY5 and MYB15 transcription factors positively regulate cold tolerance in tomato via the CBF pathway. Plant Cell Environ. 2020;43:2712–26 [DOI] [PubMed] [Google Scholar]
- 4. Yan Y, Mintao S, Si M. et al. Mechanism of CsGPA1 in regulating cold tolerance of cucumber. Hortic Res. 2022;9:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ding Y, Yang S. Surviving and thriving: How plants perceive and respond to temperature stress. Dev Cell. 2022;57:947–58 [DOI] [PubMed] [Google Scholar]
- 6. Wang Z, Zhang Y, Hu H. et al. CabHLH79 acts upstream of CaNAC035 to regulate cold stress in pepper. Int J Mol Sci. 2022;23:2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yi S, Ding Y, Yang S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018;23:623–37 [DOI] [PubMed] [Google Scholar]
- 8. Feller A, Machemer K, Braun EL. et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116 [DOI] [PubMed] [Google Scholar]
- 9. Tombuloglu H. Genome-wide identification and expression analysis of R2R3, 3R- and 4R-MYB transcription factors during lignin biosynthesis in flax (Linum usitatissimum). Genomics. 2020;112:782–95 [DOI] [PubMed] [Google Scholar]
- 10. Dubos C, Stracke R, Grotewold E. et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–81 [DOI] [PubMed] [Google Scholar]
- 11. Agarwal M, Hao Y, Kapoor A. et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281:37636–45 [DOI] [PubMed] [Google Scholar]
- 12. Ding Z, Li S, An X. et al. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics. 2009;36:17–29 [DOI] [PubMed] [Google Scholar]
- 13. Jung C, Seo JS, Han SW. et al. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liao Y, Zou HF, Wang HW. et al. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008;18:1047–60 [DOI] [PubMed] [Google Scholar]
- 15. Mao X, Jia D, Li A. et al. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct Integr Genomics. 2011;11:445–65 [DOI] [PubMed] [Google Scholar]
- 16. Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63:2541–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang M, Hao J, Chen X. et al. SlMYB102 expression enhances low-temperature stress resistance in tomato plants. PeerJ. 2020;8:e10059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahmad P, Jaleel CA, Salem MA. et al. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit Rev Biotechnol. 2010;30:161–75 [DOI] [PubMed] [Google Scholar]
- 19. Alkadi H. A review on free radicals and antioxidants. Infect Disord Drug Targets. 2020;20:16–26 [DOI] [PubMed] [Google Scholar]
- 20. Raggi S, Ferrarini A, Delledonne M. et al. The arabidopsis class III peroxidase AtPRX71 negatively regulates growth under physiological conditions and in response to cell wall damage. Plant Physiol. 2015;169:2513–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shigeto J, Tsutsumi Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016;209:1395–402 [DOI] [PubMed] [Google Scholar]
- 22. Zhang H, Pei Y, Zhu F. et al. CaSnRK2.4-mediated phosphorylation of CaNAC035 regulates abscisic acid synthesis in pepper (Capsicum annuum L.) responding to cold stress. Plant J. 2023;117:1377–91 [DOI] [PubMed] [Google Scholar]
- 23. Chen X, Duan X, Wang S. et al. Virus-induced gene silencing (VIGS) for functional analysis of MYB80 gene involved in Solanum lycopersicum cold tolerance. Protoplasma. 2019;256:409–18 [DOI] [PubMed] [Google Scholar]
- 24. Meng X, Wang JR, Wang GD. et al. An R2R3-MYB gene, LeAN2, positively regulated the thermo-tolerance in transgenic tomato. J Plant Physiol. 2015;175:1–8 [DOI] [PubMed] [Google Scholar]
- 25. Sun X, Matus JT, Wong DCJ. et al. The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J Exp Bot. 2018;69:1749–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khorobrykh S, Havurinne V, Mattila H. et al. Oxygen and ROS in Photosynthesis. Plants (Basel). 2020;9:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gómez R, Vicino P, Carrillo N. et al. Manipulation of oxidative stress responses as a strategy to generate stress-tolerant crops. From damage to signaling to tolerance. Crit Rev Biotechnol. 2019;39:693–708 [DOI] [PubMed] [Google Scholar]
- 28. Ozturk M, Turkyilmaz Unal B, García-Caparrós P. et al. Osmoregulation and its actions during the drought stress in plants. Physiol Plant. 2021;172:1321–35 [DOI] [PubMed] [Google Scholar]
- 29. Ma D, Constabel CP. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019;24:275–89 [DOI] [PubMed] [Google Scholar]
- 30. An JP, Li R, Qu FJ. et al. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018;96:562–77 [DOI] [PubMed] [Google Scholar]
- 31. Xing C, Liu Y, Zhao L. et al. A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ. 2019;42:832–45 [DOI] [PubMed] [Google Scholar]
- 32. Luo Y, Wang Y, Li X. et al. Transcription factor DgMYB recruits H3K4me3 methylase to DgPEROXIDASE to enhance chrysanthemum cold tolerance. Plant Physiol. 2024;194:1104–19 [DOI] [PubMed] [Google Scholar]
- 33. Dunand C, Meyer DM, Crèvecoeur M. et al. Expression of a peroxidase gene in zucchini in relation with hypocotyl growth. Plant Physiol Biochem. 2003;41:805–11 [Google Scholar]
- 34. Llorente F, López-Cobollo RM, Catalá R. et al. A novel cold-inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. Plant J. 2002;32:13–24 [DOI] [PubMed] [Google Scholar]
- 35. Choi HW, Kim YJ, Lee SC. et al. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 2007;145:890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wally O, Punja ZK. Enhanced disease resistance in transgenic carrot (Daucus carota L.) plants over-expressing a rice cationic peroxidase. Planta. 2010;232:1229–39 [DOI] [PubMed] [Google Scholar]
- 37. Nguyen KH, Mostofa MG, Li W. et al. The soybean transcription factor GmNAC085 enhances drought tolerance in Arabidopsis. Environ Exp Bot. 2018;151:12–20 [Google Scholar]
- 38. Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–51 [DOI] [PubMed] [Google Scholar]
- 39. Park S, Lee CM, Doherty CJ. et al. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015;82:193–207 [DOI] [PubMed] [Google Scholar]
- 40. Vogel JT, Zarka DG, Van Buskirk HA. et al. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211 [DOI] [PubMed] [Google Scholar]
- 41. Zarka DG, Vogel JT, Cook D. et al. Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 2003;133:910–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim YS, Lee M, Lee JH. et al. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol Biol. 2015;89:187–201 [DOI] [PubMed] [Google Scholar]
- 43. Xiao J, He M, Chen P. et al. Proanthocyanidins delay the senescence of young asparagus stems by regulating antioxidant capacity and synthesis of phytochemicals. Food Chem X. 2024;21:101222 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are presented within the paper and supplementary files.