Abstract
Several herpesviruses encode Fc receptors that may play a role in preventing antibody-mediated clearance of the virus in vivo. Human cytomegalovirus (HCMV) induces an Fc-binding activity in cells upon infection, but the gene that encodes this Fc-binding protein has not been identified. Here, we demonstrate that the HCMV AD169 open reading frame TRL11 and its identical copy, IRL11, encode a type I membrane glycoprotein that possesses IgG Fc-binding capabilities.
Many herpesviruses encode proteins that interfere with the humoral and cellular immune response (18, 29). Presumably, these immune evasion gene products are important for allowing the virus to replicate in a host that already has a battery of specific antiviral defenses in place. Herpes simplex virus type 1 (HSV-1) and HSV-2, murine cytomegalovirus (MCMV) and varicella-zoster virus produce molecules that bind to the Fc portion of host immunoglobulins (6, 12, 17, 28). These virally encoded Fc receptors (v-FcRs) may prevent antiviral immunoglobulin G (IgG) from neutralizing free virus and engaging in antibody-dependent cytotoxic activity against infected cells (19). The well-characterized HSV-1 v-FcR is a heterodimer of the gE and gI glycoproteins and is able to inhibit complement activation and antibody-dependent cell-mediated cytotoxicity in in vitro experiments (8, 9). In a mouse model of HSV-1 infection, a functional v-FcR was necessary for viral evasion of antibody-mediated clearance (23). For MCMV, the role of the v-FcR has not been well defined. An MCMV strain lacking the v-FcR gene (fcr-1 or m138) replicated to low titers in mice with and without B cells (7). Thus, m138 could be important for aspects of MCMV in vivo replication that are unrelated to the binding of IgG Fc.
Human cytomegalovirus (HCMV) induces an Fc-binding activity in infected cells (3, 10, 14, 21, 25). Although there is a large amount of data regarding alphaherpesvirus-encoded Fc receptors, it is not known whether the Fc-binding molecule induced during HCMV infection is encoded by the virus or by the host. Flow cytometry has been used to demonstrate that the Fc-binding molecule in HCMV-infected cells is present at the cell surface, while immunofluorescence data indicates that Fc-binding activity can also be detected within the infected cell (10, 14, 20). HCMV-infected cells can bind IgG from several different species; they can also bind all subtypes of human IgG, but not other human Ig isotypes (1, 20, 22). Additional immunoelectron microscopy data indicates that an Fc-binding activity may be present in the tegument of HCMV virions (27). Although attempts have been made to characterize biochemically the protein or proteins that are responsible for the Fc-binding activity in infected cells, the gene that encodes the HCMV-induced FcR has not been identified (27, 30). The goal of this study was to identify and characterize the Fc-binding protein(s) induced by HCMV. We demonstrate that the HCMV open reading frame (ORF) TRL11/IRL11 encodes a glycoprotein of 34 kDa that binds to IgG Fc.
In order to identify the Fc-binding protein(s) induced by HCMV, the following approach was taken. Human foreskin fibroblasts (HFFs) (number of passages, 10 to 20) were infected with HCMV AD169 at a multiplicity of infection of 5. Infected cells were metabolically labeled with Expre35S35S protein labeling mix (NEN) for 30 min at various times postinfection (p.i.) (2). The cells were then lysed in a buffer containing: 0.5% NP-40, 150 mM NaCl, 2 mM CaCl2, 50 mM Tris-Cl (pH 7.4), 1 mM phenylmethylsulfonylfluoride, and 10 μM leupeptin, and the debris was removed by centrifugation. After preclearing of lysates with streptavidin-agarose (Pierce), human IgG Fc or a human IgG1 myeloma protein (Calbiochem) that had been biotinylated with NHS-LC-biotin (Pierce) was added at a concentration of 10 μg/ml. The biotinylated IgG proteins (Fcbiotin and IgG1biotin, respectively) and material bound to them were retrieved by the addition of streptavidin-agarose (30 μl of a 50% [vol/vol] slurry) and washed several times. Bound proteins were released by the addition of sodium dodecyl sulfate (SDS) sample buffer, and were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiography (15, 24).
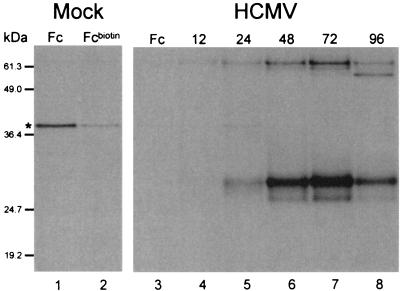
A protein of approximately 34 kDa was immunoprecipitated by Fcbiotin specifically in AD169-infected cells (Fig. 1A, lanes 5 to 8). The Fc-binding protein was detected as early as 12 h p.i. (evident in longer exposures of the autoradiogram shown in Fig. 1A), and expression levels were highest at 72 h p.i. An additional species of approximately 63 kDa was also retrieved from infected cell lysates. The heterogeneous migration pattern of the 34-kDa species suggested that it may be a glycoprotein. Indeed, digestion with PNGaseF (New England Biolabs) reduced the molecular mass of the 34 kDa protein to approximately 24 kDa (Fig. 2B, lanes 1 and 3), consistent with the presence of at least 3 N-linked glycans and a core polypeptide molecular mass of 24 kDa. The size of the 63 kDa protein was reduced to 33 kDa upon PNGaseF digestion, consistent with the presence of approximately 10 N-linked glycans. We conclude that HCMV infection induces the expression of an Fc-binding glycoprotein with a molecular mass of 34 kDa and the expression of an additional, highly glycosylated, Fc-binding protein of 63 kDa. Both the 34-kDa and the 63-kDa glycoproteins were also retrieved using IgG1biotin, indicating that both glycoproteins are capable of binding to the Fc portion of whole IgG (data not shown).
FIG. 1.
Infection of HFFs with HCMV AD169 induces the expression of IgG Fc-binding proteins. Cells were pulse-labeled and immunoprecipitations were performed. Lane 1, material immunoprecipitated with IgG Fc (Fc) from a lysate of mock-infected cells; lane 2, material immunoprecipitated with biotinylated IgG Fc (Fcbiotin) from a lysate of mock-infected cells; lane 3, material immunoprecipitated from a lysate of HCMV-infected cells 48 h p.i. using IgG Fc; lanes 4, 5, 6, 7, and 8, material immunoprecipitated at, respectively, 12, 24, 48, 72, and 96 h p.i. from lysates for HCMV-infected cells using Fcbiotin. The asterisk indicates a polypeptide retrieved nonspecifically and in variable amounts when using Fcbiotin. Positions of prestained molecular size standards (Gibco BRL) shown in this figure are meant to represent approximate molecular sizes only. More accurate estimates of the molecular sizes of the proteins of interest (described in the text) were determined using other molecular size standards (data not shown).
FIG. 2.
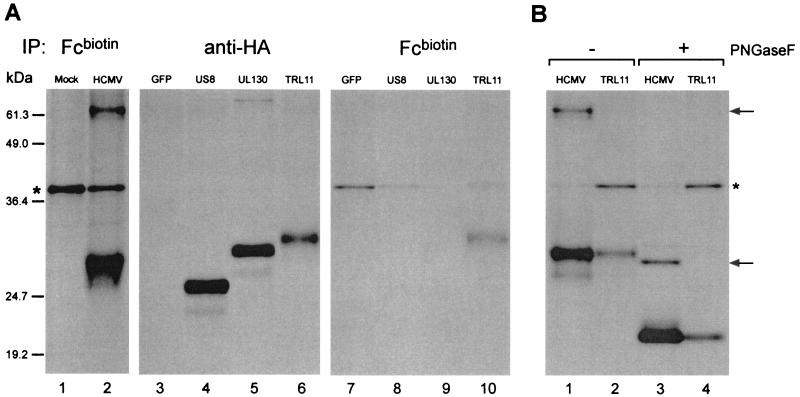
HCMV AD169 ORF TRL11/IRL11 encodes the 34-kDa Fc-binding protein present in HCMV-infected cells. (A) Results of immunoprecipitation experiments with tagged constructs. For lanes 1 and 2, HFFs were pulse-labeled and immunopreciptiations were performed with Fcbiotin. For lanes 3 to 10, 293 cells were pulse-labeled, and immunoprecipitations were performed from equal amounts of lysate using either an anti-HA antibody (lanes 3 to 6) or Fcbiotin (lanes 7 to 10). Lane 1, mock-infected HFFs; lane 2, HCMV-infected HFFs (48 h p.i.); lanes 3 and 7, 293 cells transfected with pcDNA3.1 construct containing untagged GFP (negative control); lanes 4 and 8, 293 cells transfected with pcDNA3.1 construct containing US8-HA (US8); lanes 5 and 9, 293 cells transfected with pcDNA3.1 construct containing UL130-HA (UL130); lanes 6 and 10, 293 cells transfected with pcDNA3.1 construct containing TR11/IRL11-HA (TRL11). (B) Results of immunoprecipitation experiments with untagged construct. Cells were pulse-labeled, and immunoprecipitations were performed with Fcbiotin. Half of the bound material was treated with PNGase F. Lane 1, HCMV-infected HFFs (48 p.i.) not treated; lane 2, 293 cells transfected with a construct containing untagged TR11/IRL11 and not treated; lane 3, HCMV-infected HFFs (48 h p.i.) treated with PNGaseF; lane 4, 293 cells transfected with a construct containing untagged TR11/IRL11 and treated with PNGaseF. The asterisks indicate a polypeptide retrieved nonspecifically and in variable amounts when using Fcbiotin. The arrows at the right indicate the positions of the 63-kDa and the deglycosylated 33-kDa forms of the additional Fc-binding protein present in HCMV-infected cells. The positions of prestained molecular size standards (Gibco BRL) shown in this figure are meant to represent approximate molecular sizes only. More accurate estimates of the molecular masses of the proteins of interest (described in the text) were determined using other molecular size standards (data not shown).
Using the size of the polypeptide backbone and the estimated number of glycosylation sites observed for the 34 kDa Fc-binding protein, we compiled a list of HCMV AD169 ORFs that fit these properties (Table 1). We chose to investigate viral genes because several herpesviruses encode Fc receptors and HCMV encodes a variety of other proteins involved in immune evasion (19, 29). An HCMV strain (RV798) that carries a deletion encompassing US2-11, a region encoding several gene products that interfere with major histocompatibility complex (MHC) class I processing, was used to rule out several candidate ORFs in that region of the HCMV genome (data not shown) (13). Those ORFs most consistent with the observed properties of the 34 kDa Fc-binding protein were amplified from AD169 genomic DNA by PCR with specific primers that incorporated an influenza virus hemagglutinin (HA) epitope tag at the C terminus and were cloned into the pcDNA3.1 mammalian expression vector (Invitrogen) by standard procedures (26). Sequence analysis was used to verify that no mutations had arisen as a consequence of PCR amplification. We note that the AD169 genome contains a second, identical copy of TRL11, designated IRL11 (5).
TABLE 1.
Characteristics of HCMV ORFs with properties similar to that of the 34-kDa IgG Fc-binding proteina
| HCMV ORF | No. of amino acids | Predicted size (kDa) | No. of N-glycosylation sites |
|---|---|---|---|
| TRL11/IRL11 | 234 | 26.6 | 3 |
| UL1 | 224 | 25.5 | 9 |
| UL7 | 222 | 24.3 | 11 |
| UL9 | 228 | 26.8 | 5 |
| UL16 | 230 | 26.1 | 8 |
| UL40 | 221 | 24.3 | 3 |
| UL118 | 209 | 24.5 | 8 |
| UL130 | 214 | 24.6 | 3 |
| US8 | 227 | 26.6 | 2 |
| US9 | 247 | 28.0 | 2 |
Data on HCMV ORFs are from reference 5.
The various ORF-HA constructs were transfected into 293 cells using LipofectAMINE (Life Technologies) according to the manufacturer's specifications. One day after transfection, cells were pulse-labeled for 15 min and lysed as described above. Equal amounts of radiolabeled lysate were subjected to immunoprecipitation using Fcbiotin or the anti-HA monoclonal antibody 12CA5. A construct expressing untagged green fluorescent protein (GFP) was used as a negative control for immunoprecipitations from 293 transfectant lysates (Fig. 2A, lanes 3 and 7). Cells transfected with HA-tagged versions of US8 (an HCMV-encoded type I membrane glycoprotein) or with the candidate ORF UL130 produced proteins reactive with the anti-HA antibody but were not immunoprecipitated by Fcbiotin (Fig. 2A, lanes 4 and 5 and lanes 8 and 9). However, cells transfected with the tagged version of TRL11/IRL11 produced a protein that was retrieved by anti-HA and Fcbiotin (Fig. 2A, lanes 6 and 10). The difference in size between the 34 kDa Fc-binding protein from HCMV-infected cells and the tagged version of gpTRL11/IRL11 is likely due to the additional amino acids of the HA tag. The difference in the amount of material present in the anti-HA and Fcbiotin immunoprecipitates likely reflects a difference in the affinities of the 12CA5 antibody for the HA tag and of the Fcbiotin for the gpTRL11/IRL11 molecule.
In order to provide additional evidence that TRL11/IRL11 encodes the 34 kDa Fc-binding protein seen in HCMV-infected cells, an untagged version of TRL11/IRL11 was amplified by PCR and cloned into pcDNA3.1. Transfection of the untagged TRL11/IRL11 construct into 293 cells followed by pulse-labeling and immunoprecipitation using Fcbiotin yielded a protein that comigrated with the 34-kDa Fc-binding protein from HCMV-infected cells in SDS-PAGE (Fig. 2B, lanes 1 and 2), and comigration was also observed after digestion with PNGaseF (Fig. 2B, lanes 3 and 4). Cells transfected with the untagged TRL11/IRL11 construct also produced a protein that bound to IgG1biotin, indicating that gpTRL11/IRL11 can interact with the Fc region of whole IgG (data not shown). We conclude that the HCMV ORF TRL11/IRL11 encodes an Fc-binding protein present in infected cells. Importantly, gpTRL11/IRL11 is capable of binding to IgG Fc in the absence of any other viral protein (Fig. 2A and B). However, these data do not formally rule out the possibility that gpTRL11/IRL11 may form a complex with another viral or human protein to form a functional Fc receptor that may have a specificity or affinity different from that of gpTRL11/IRL11 alone.
The ORFs that encode the Fc-binding protein, TRL11 and IRL11, are identical in sequence to one another and are located, respectively, in the terminal repeat long (TRL) and internal repeat long (IRL) regions of the AD169 genome (Fig. 3). The 234-aa protein encoded by TRL11/IRL11 is a predicted type I membrane glycoprotein with three N-glycosylation sites and has a predicted cytoplasmic tail of 31 aa. We note the presence of a consensus motif for engagement of the endocytic machinery (DXXXLL) located in the predicted cytoplasmic tail (Fig. 3) (11, 16). The amino acid sequence of TRL11/IRL11 does not show homology to any of the known herpesvirus v-FcRs, suggesting that different herpesviruses have evolved the ability to bind to host IgG Fc at multiple times in the past. The protein sequence of TRL11/IRL11 shows limited similarity to the UL153 ORF of the Towne strain of HCMV and is a member of the RL11 gene family of AD169, which comprises a set of ORFs that are moderately similar (4, 5). However, TRL11/IRL11 does not bear any obvious homology to other proteins presently in the databases.
FIG. 3.
Locations of TRL11 and IRL11 in the AD169 genome and corresponding amino acid sequence. At the top, a schematic of the HCMV AD169 genome showing the position of the TRL11 and IRL11 ORFs in the TRL and IRL regions, respectively, is shown. The arrows denote the respective coding sequence orientations. At the bottom, the deduced amino acid sequence of the TRL11/IRL11 ORF is shown. The putative signal peptide is in lowercase, and the putative transmembrane domain is underlined. N-glycosylation sites are underlined and in bold and the internalization motif (DXXXLL) is highlighted in bold.
The identification of gpTRL11/IRL11 as an HCMV-encoded IgG Fc-binding protein adds to the lengthy list of HCMV gene products that interact with molecules important for immune system function (29). With the availability of a gene that encodes an HCMV IgG Fc-binding protein, it will now be possible to characterize its interaction with IgG Fc and its potential role as a v-FcR that may be involved in evasion of the antibody response by HCMV.
Acknowledgments
This work was supported by the National Institutes of Health (R37-AI33456 and P01-AI42257). B.N.L. is a Howard Hughes Medical Institute predoctoral fellow, and R.S.T. is a Novartis Fellow of the Life Sciences Research Foundation.
REFERENCES
- 1.Antonsson A, Johansson P J. Binding of human and animal immunoglobulins to the IgG Fc receptor induced by human cytomegalovirus. J Gen Virol. 2001;82:1137–1145. doi: 10.1099/0022-1317-82-5-1137. [DOI] [PubMed] [Google Scholar]
- 2.Beersma M F, Bijlmakers M J, Ploegh H L. Human cytomegalovirus down-regulates HLA class I expression by reducing the stability of class I H chains. J Immunol. 1993;151:4455–4464. [PubMed] [Google Scholar]
- 3.Beersma M F C. Ph.D thesis. Amsterdam, The Netherlands: University of Amsterdam; 1993. [Google Scholar]
- 4.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell. Hutchison T C A, III, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 6.Costa J, Yee C, Nakamura Y, Rabson A. Characteristics of the Fc receptor induced by herpes simplex virus. Intervirology. 1978;10:32–39. doi: 10.1159/000148965. [DOI] [PubMed] [Google Scholar]
- 7.Crnkovic-Mertens I, Messerle M, Milotic I, Szepan U, Kucic N, Krmpotic A, Jonjic S, Koszinowski U H. Virus attenuation after deletion of the cytomegalovirus Fc receptor gene is not due to antibody control. J Virol. 1998;72:1377–1382. doi: 10.1128/jvi.72.2.1377-1382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubin G, Socolof E, Frank I, Friedman H M. Herpes simplex virus type 1 Fc receptor protects infected cells from antibody-dependent cellular cytotoxicity. J Virol. 1991;65:7046–7050. doi: 10.1128/jvi.65.12.7046-7050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank I, Friedman H M. A novel function of the herpes simplex virus type 1 Fc receptor: participation in bipolar bridging of antiviral immunoglobulin G. J Virol. 1989;63:4479–4488. doi: 10.1128/jvi.63.11.4479-4488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa T, Hornberger E, Sakuma S, Plotkin S A. Demonstration of immunoglobulin G receptors induced by human cytomegalovirus. J Clin Microbiol. 1975;2:332–336. doi: 10.1128/jcm.2.4.332-336.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heilker R, Spiess M, Crottet P. Recognition of sorting signals by clathrin adaptors. Bioessays. 1999;21:558–567. doi: 10.1002/(SICI)1521-1878(199907)21:7<558::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones T R, Hanson L K, Sun L, Slater J S, Stenberg R M, Campbell A E. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller R, Peitchel R, Goldman J N, Goldman M. An IgG-Fc receptor induced in cytomegalovirus-infected human fibroblasts. J Immunol. 1976;116:772–777. [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Letourneur F, Klausner R D. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- 17.Litwin V, Sandor M, Grose C. Cell surface expression of the varicella-zoster virus glycoproteins and Fc receptor. Virology. 1990;178:263–272. doi: 10.1016/0042-6822(90)90402-d. [DOI] [PubMed] [Google Scholar]
- 18.Loenen W A, Bruggeman C A, Wiertz E J. Immune evasion by human cytomegalovirus: lessons in immunology and cell biology. Semin Immunol. 2001;13:41–49. doi: 10.1006/smim.2001.0294. [DOI] [PubMed] [Google Scholar]
- 19.Lubinski J, Nagashunmugam T, Friedman H M. Viral interference with antibody and complement. Semin Cell Dev Biol. 1998;9:329–337. doi: 10.1006/scdb.1998.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacCormac L P, Grundy J E. Human cytomegalovirus induces an Fc gamma receptor (Fc gammaR) in endothelial cells and fibroblasts that is distinct from the human cellular Fc gammaRs. J Infect Dis. 1996;174:1151–1161. doi: 10.1093/infdis/174.6.1151. [DOI] [PubMed] [Google Scholar]
- 21.Mackowiak P A, Marling-Cason M. Immunoreactivity of cytomegalovirus-induced Fc receptors. Microbiol Immunol. 1987;31:427–434. doi: 10.1111/j.1348-0421.1987.tb03105.x. [DOI] [PubMed] [Google Scholar]
- 22.Murayama T, Natsuume-Sakai S, Shimokawa K, Furukawa T. Fc receptor(s) induced by human cytomegalovirus bind differentially with human immunoglobulin G subclasses. J Gen Virol. 1986;67:1475–1478. doi: 10.1099/0022-1317-67-7-1475. [DOI] [PubMed] [Google Scholar]
- 23.Nagashunmugam T, Lubinski J, Wang L, Goldstein L T, Weeks B S, Sundaresan P, Kang E H, Dubin G, Friedman H M. In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J Virol. 1998;72:5351–5359. doi: 10.1128/jvi.72.7.5351-5359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ploegh H L. One-dimensional isoelectric focusing of proteins in slab gels. In: Coligan J E, Dunn B M, Ploegh H L, Speicher D W, Wingfield P T, editors. Current protocols in protein science. Vol. 1. New York, N.Y: John Wiley and Sons, Inc.; 1995. pp. 10.12.11–10.12.18. [DOI] [PubMed] [Google Scholar]
- 25.Rahman A A, Teschner M, Sethi K K, Brandis H. Appearance of IgG (Fc) receptor(s) on cultured human fibroblasts infected with human cytomegalovirus. J Immunol. 1976;117:253–258. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Stannard L M, Hardie D R. An Fc receptor for human immunoglobulin G is located within the tegument of human cytomegalovirus. J Virol. 1991;65:3411–3415. doi: 10.1128/jvi.65.6.3411-3415.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thale R, Lucin P, Schneider K, Eggers M, Koszinowski U H. Identification and expression of a murine cytomegalovirus early gene coding for an Fc receptor. J Virol. 1994;68:7757–7765. doi: 10.1128/jvi.68.12.7757-7765.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tortorella D, Gewurz B E, Furman M H, Schust D J, Ploegh H L. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 30.Xu B, Murayama T, Ishida K, Furukawa T. Characterization of IgG Fc receptors induced by human cytomegalovirus. J Gen Virol. 1989;70:893–900. doi: 10.1099/0022-1317-70-4-893. [DOI] [PubMed] [Google Scholar]