Abstract
Twenty-four of over 24,000 patients genotyped over the past 3 years were found to have human immunodeficiency virus (HIV) isolates that possess an insert in the protease gene. In this report, we evaluated the spectrum of protease gene insertion mutations in patient isolates and analyzed the effect of these various insertion mutations on viral phenotypes. The inserts were composed of 1, 2, 5, or 6 amino acids that mapped at or between codons 35 and 38, 17 and 18, 21 and 25, or 95 and 96. Reduced susceptibility to protease inhibitors was found in isolates which possess previously reported drug resistance mutations. Fitness assays, including replication and competition experiments, showed that most of the isolates with inserts grew somewhat better than their counterparts with a deletion of the insert. These experiments demonstrate that, rarely, insertion mutations can develop in the HIV type 1 protease gene, are no more resistant than any other sequences which have similar associated resistance mutations, and can provide a borderline advantage in replication.
The low fidelity of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT), combined with the lack of an associated proofreading function (20), results in high levels of mutation and increasing genetic variation that produces quasispecies of HIV. These mechanisms make it possible for a number of mutations in the protease and RT genes of HIV to emerge and to be selected as the predominant isolates during drug treatment (3, 15, 23, 32). The resulting amino acid substitutions affect the structure of the viral enzyme, which can alter the kinetics of enzyme function or change the ability of inhibitors to access the active site (16, 18), thus providing the mutant virus a competitive advantage under drug pressure (13). Recently it has been shown that resistance to multiple nucleoside analogs can result from several insertion mutations near codon 69 of the RT gene (2, 14, 28, 31). However, all previously described mutations associated with resistance to protease inhibitors have been single codon substitutions that have resulted from 1- or 2-base point mutations in the protease gene (1, 16, 30). In this study, we characterized HIV-1 isolates possessing various amino acid insertions in the protease gene using several different methodological approaches, including drug susceptibility assays, kinetics of viral antigen production, and competitive replication assays.
Genotypes of insert-containing isolates and patterns of insertion mutations.
The patient plasma or serum specimens studied here were submitted for HIV-1 protease genotyping to Quest Diagnostics Inc., San Juan Capistrano, California; Stanford University Hospital, Stanford, Calif.; and the University of Oregon, Portland. To make an RT-PCR artifact less likely, genotyping was performed independently by population-based sequencing of plasma-derived HIV RNA at both Stanford University and Quest Diagnostics as previously described (30). In addition, genotyping was repeated over time and the presence of an insertion was confirmed in the four instances where additional patients' specimens were available. Both laboratories demonstrated the same sequence result in all cases. The patterns of insertion mutations in the protease gene showed that the nucleic acid compositions of the inserts were typically duplications of neighboring sequences (Table 1). Most of the inserts (in 19 out of 24 isolates) were composed of 1 to 5 amino acids that mapped between codons 35 and 38. Two were found to be a single-amino-acid insertion between codons 21 and 25, one was a single-amino-acid insertion positioned between codons 17 and 18, and 5- and 6-amino-acid insertions were observed at positions 95 to 96 (Table 1). Ten isolates had at least two major resistance-associated protease gene mutations (G48V, I54V, V82A, I84V, and/or L90M), with an average of 10 other amino acid changes from consensus B. The eight other isolates did not possess any major protease gene mutations but did posses an average of seven other changes compared to consensus B. Nineteen insertions appeared preferentially in the flap region (Table 1), which encompasses amino acids 33 to 62 (4, 5, 21, 29). Furthermore, the database available to us showed that 10 of 11 simian immunodeficiency virus-African green monkey isolates had two amino acid insertions in the protease gene at codons 34 to 37 (25; http://hivdb.stanford.edu/hiv/). Molecular modeling experiments have shown that the insertions cause conformational changes in the geometry of the flap region and contribute to structural alterations in more distant region of the molecule (M. A. Winters, E.-Y. Kim, S. Chou, A. Warford, R. Kagan, R. Fenwick, L. Kovari, and T. C. Merigan, Abstr. 7th Conf. Retrovir. Opportun. Infect., abstr. 723, 2000; L. Kovari, personal communication, 2000). Because the flap region does not contribute strongly to the enzyme's stability (29) and the flap region overlies the catalytic aspartate residues located in the substrate binding site (4, 5), mutation of flap residues might provide an effective means for the virus to block protease inhibitor access.
TABLE 1.
Protease gene inserts in primary HIV-1 strains
| Subdomain | Straina | Amino acid(s) of protease gene at codon position(s)b:
|
Resistance mutation(s) | ||||
|---|---|---|---|---|---|---|---|
| 33 | 34 | 35 through 38 | 39 | 40 | |||
| Flap | |||||||
| WT | L | E | E M N L | P | G | ||
| TTA | GAA | GAA ATG AAT TTG | CCA | GGA | |||
| Q058 | L | E | E T V L E E I N L | P | G | I54V, L90M | |
| TTA | GAA | GAA ACA GTA TTA GAA GAA ATA AAT TTG | CCA | GGA | |||
| Q650 | L | E | E T N L N L | P | G | None | |
| TTA | GAA | GAA ACG AAT TTG AAT TTG | CCA | GGA | |||
| Q552 | L | E | E T N L N L | P | G | None | |
| TTA | GAA | GAA ACA AAT TTA AAT TTG | CCA | ||||
| Q340 | L | E | D M N L N L | P | G | None | |
| TTA | GAA | GAC ATG AAT TTG AAT TTG | CCA | GGA | |||
| Q781 | L | E | D M N L N L | P | G | None | |
| TTA | GAA | GAC ATG AAT TTG AAT TTG | CCA | GGG | |||
| U099 | V | E | D T D I N L | P | G | None | |
| GTA | GAA | GAC ACA GAC ATA AAT TTG | CCA | GGA | |||
| Q288 | L | E | D T N L N L | P | G | I84V, L90M | |
| TTA | GAA | GAC ACG AAT TTG AAT TTG | CCA | GGA | |||
| Q645 | L | E | E T G L N L | P | G | L10I, I84V | |
| TTA | GAA | GAA ACG GGT TTA AAT TTG | CCA | GGA | |||
| Q164 | L | E | E V N L N L | P | G | V82A | |
| TTA | GAA | GAA GTA AAT TTA AAT TTG | CCA | GGA | |||
| Q111 | L | E | E M N L N L | P | G | L10I, M46I, I84V, L90M | |
| TTA | GAA | GAA ATG AAT TTG AAT TTG | CCA | GGA | |||
| V493 | L | E | E M D L N L | P | G | L10I, M46I, I54V, I84V | |
| TTA | GAA | GAA ATG GAT TTG AAT TTG | CCA | GGG | |||
| Q333 | L | E | E T N L N L | P | G | None | |
| TTA | GAA | GAA ACA AAT CTA AAT TTG | CCA | GGA | |||
| Q874 | L | E | D T N L N L | P | G | I84V, L90M | |
| TTA | GAG | GAC ACG AAT TTG AAT TTG | CCA | GGA | |||
| Q928 | L | E | E M N L N L | P | G | L10I, M46I, I84V, L90M | |
| TTA | GAA | GAA ATG AAT TTG AAT TTG | CCA | GGG | |||
| Q121 | L | E | E T N L N L | P | G | L10I, I84V, L90M | |
| TTA | GAA | GAA ACA AAT TTA AAT TTG | CCA | GGA | |||
| Q822 | L | E | E D L N L | P | G | None | |
| TTA | GAA | GAA GAT CTA AAT TTG | CCA | GGA | |||
| Q060 | L | E | E D I D L | P | G | L10I, G48V, V82A, L90M | |
| TTA | CAA | GAA GAC ATA GAT TTG | CCA | GGA | |||
| Q530 | L | E | E G I S L | P | G | L10I, V82A, I84V | |
| TTA | GAA | GAG GGA ATA AGT TTA | CCA | GGA | |||
| Q745 | L | E | E N I S L | P | G | None | |
| CTA | GAA | GAA AAT ATA AGT TTG | CCA | GGA | |||
| 16 | 17 | Insert | 18 | 19 | |||
| Core | |||||||
| WT | G | G | Q | L | |||
| GGG | GGG | CAA | CTA | ||||
| Q008 | G | G | R | Q | L | M46I, L90M | |
| GGG | GGG | CGG | CAA | CTA | |||
| 21 | 22 through 25 | 26 | 27 | ||||
| WT | E | A L L D | T | G | |||
| GAA | GCT CTA TTA GAT | ACA | GGA | ||||
| Q478 | E | D V L L D | T | G | M46I, I84V | ||
| GAG | GAT GTT CTA TTA GAT | ACA | GGA | ||||
| Q970 | E | A L L D H | T | G | M46I, L90M | ||
| GAA | GCC CTG CTA GAC CAC | ACA | GGA | ||||
| 94 | 95 | Insert | 96 | 97 | |||
| Dimerization | |||||||
| WT | G | C | T | L | |||
| GGT | TGC | ACT | TTA | ||||
| Q804 | G | C | T L N F P I | T | L | L10I, L90M | |
| GGT | TGC | ACT TTA AAT TTT CCC ATT | ACT | TTA | |||
| Q102 | G | C | T L N F P | T | L | L10I, I54V, I84V, L90M | |
| GGT | TGC | ACT TTA AAT TTT CCC | ACT | TTA | |||
WT, wild type.
Nucleotides of each codon appear below the amino acid. Bold characters denote the presumed inserted nucleotide sequences. The insertion sequences were aligned with the consensus B sequence of HIV protease using the Stanford HIV database program (http://hivdb.stanford.edu/hiv/). The insert location was generated by the optimal sequence alignment algorithm and then manually revised. Twenty-four sequences reflecting Quest Diagnostics data as well as those of two university hospital laboratories are presented.
Recombinant viruses and drug susceptibility.
Twelve recombinant viruses of patient-derived HIV isolates were constructed by previously described homologous recombination methods (19). In brief, the purified PCR product of the protease gene was cotransfected into C8166 cells with HXB2 lacking the protease gene (pHXB2-ΔPR). Additional recombinants of the five representative isolates in which each insertion mutation was deleted by PCR-based site-directed mutagenesis were constructed (10). Primers used to remove the protease gene insert were designed after the sequences were analyzed with sequence analysis programs at the Stanford HIV database (25; http://hivdb.stanford.edu/hiv/), which is based on optimal sequence alignment of the amino acids between codons of the protease (26) and manually corrected. Recombinants were constructed for 12 of the insertion-containing isolates. In addition, five representative viral constructs lacking the insertion were created. These insertion strains were able to function as infectious clones. These results demonstrate that all the insertion-containing proteases have intact biological activities (33), a result which is not expected with a PCR artifact. In vitro susceptibility to indinavir (IDV), saquinavir (SQV), or nelfinavir (NFV) was measured for each of the 12 isolates containing the insertion and for the 5 corresponding isolates lacking the insertion using a previously described method (11). Results are expressed as mean 50% inhibitory concentrations (IC50s) of four to eight values obtained from two to four different experiments per isolate (Table 2). When we compared the susceptibilities of insertion and deletion pairs, the IC50s for three of the five insertion-containing isolates (Q781, Q822, and Q058) were similar to or higher than those for corresponding insertion-lacking isolates. The insertion in Q781 conferred reduced susceptibility to all three protease inhibitors, as demonstrated by three- to fivefold-higher IC50s than those for the corresponding constructs with the insertion deleted. The assays showed that there was no substantial difference in drug susceptibility in the insertion-containing isolates that lacked protease inhibitor resistance mutations in the protease gene (Q781, Q822, and U099) compared to that of the wild-type virus. Isolates that had major protease inhibitor resistance mutations showed a 4- to 45-fold decrease in susceptibility to protease inhibitors compared to that of the wild type. Phenotypic results suggest that previously reported drug resistance mutations seem to be primarily responsible for protease inhibitor resistance even in the presence of the insertions and that insertion mutations may not contribute directly to drug resistance.
TABLE 2.
Susceptibilities of patient HIV-1 recombinant isolates to protease inhibitors
| Isolate | Insertion | Major protease inhibitor mutation(s) | Recombinantb | IC50 (μM)a
|
||
|---|---|---|---|---|---|---|
| IDV | SQV | NFV | ||||
| NL4-3 | Wild type | 0.02 ± 0.02 | 0.01 ± 0.00 | 0.02 ± 0.01 | ||
| Q058 | 35TVLEE | I54V, L90M | Insertion | 0.52 ± 0.04 (22.2) | 0.16 ± 0.07 (16.5) | 0.13 ± 0.03 (5.7) |
| Deletion | 0.09 ± 0.08 (3.9) | 0.10 ± 0.10 (10.2) | 0.15 ± 0.02 (6.8) | |||
| Q781 | 36NL | None | Insertion | 0.05 ± 0.01 (2.0) | 0.03 ± 0.03 (2.8) | 0.03 ± 0.04 (1.5) |
| Deletion | 0.02 ± 0.00 (0.7) | 0.00 ± 0.00 (0.5) | 0.01 ± 0.01 (0.5) | |||
| U099 | 35TD | None | Insertion | 0.02 ± 0.02 (0.7) | 0.00 ± 0.00 (0.2) | 0.04 ± 0.02 (1.6) |
| Deletion | 0.03 ± 0.02 (1.2) | 0.00 ± 0.00 (0.5) | 0.02 ± 0.03 (1.0) | |||
| Q164 | 36NL | V82A | Insertion | 0.06 ± 0.02 (2.6) | 0.01 ± 0.02 (1.4) | 0.04 ± 0.02 (1.6) |
| Deletion | 0.02 ± 0.00 (0.7) | 0.02 ± 0.01 (1.6) | 0.06 ± 0.06 (2.9) | |||
| Q822 | 37D | None | Insertion | 0.01 ± 0.02 (0.6) | 0.04 ± 0.04 (3.8) | 0.04 ± 0.02 (1.9) |
| Deletion | 0.04 ± 0.00 (1.6) | 0.01 ± 0.01 (0.8) | 0.01 ± 0.00 (0.4) | |||
| Q650 | 36NL | None | Insertion | 0.01 ± 0.00 (0.2) | 0.01 ± 0.00 (0.9) | 0.01 ± 0.00 (0.3) |
| Q552 | 36NL | None | Insertion | 0.01 ± 0.01 (0.4) | 0.01 ± 0.00 (0.6) | 0.01 ± 0.00 (0.4) |
| Q645 | 36GL | L10I, I84V | Insertion | 0.02 ± 0.00 (0.8) | 0.03 ± 0.01 (3.5) | 0.02 ± 0.00 (0.9) |
| Q288 | 35TN | I84V, L90M | Insertion | 0.03 ± 0.01 (1.2) | 0.07 ± 0.06 (7.3) | 0.10 ± 0.07 (4.5) |
| V493 | 36DL | L10I, M46I, I54V, I84V | Insertion | 0.27 ± 0.33 (6.0) | 1.21 ± 1.64 (45.2) | 0.30 ± 0.38 (13.6) |
| Q008 | 17R | L46I, L90M | Insertion | 0.11 ± 0.12 (4.6) | 0.09 ± 0.03 (9.3) | 0.28 ± 0.06 (8.2) |
| Q804 | 95TLNFPI | L10I, L90M | Insertion | 0.16 ± 0.12 (6.8) | 0.06 ± 0.05 (6.2) | 0.12 ± 0.09 (5.2) |
IC50s are the means of results of three to four tests. Recombinant isolates were prepared and tested as described in the text. Values in parentheses are fold changes in IC50s relative to that of the wild type.
“Wild type” represents a recombinant virus containing an NL4-3 PR gene.
Replication kinetics and competition studies.
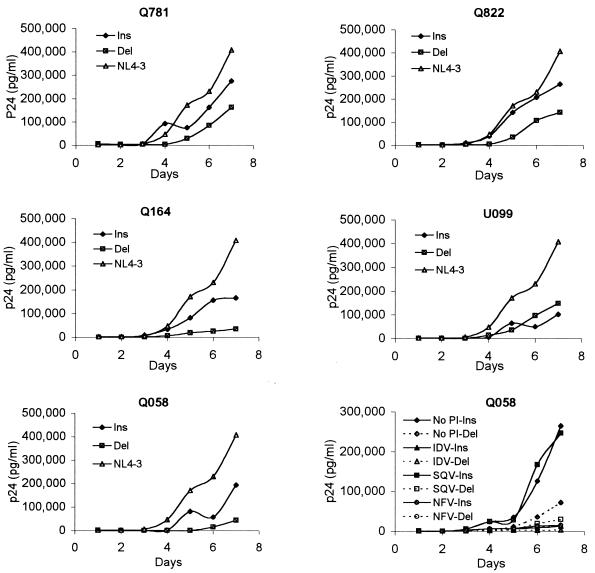
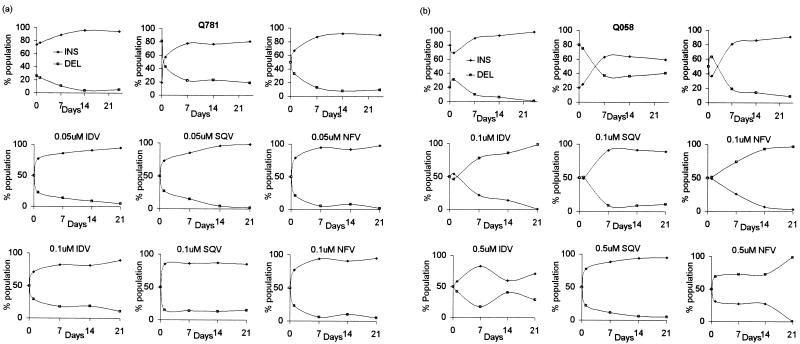
A replication kinetics assay was carried out by modification of previously described methods (27). Five thousand 50% tissue culture infective doses (TCID50) (9) of each virus was used to infect 5 × 106 phytohemagglutinin (PHA)-stimulated peripheral blood mononuclear cells (PBMCs) (multiplicity of infection, 0.001) in both replication and competition experiments. p24 antigen production was measured for 7 days, and at least three independent assays were performed with five different isolates (Fig. 1). The Q058 insertion-deletion pair was selected to serve as a related protease inhibitor-resistant control. In the absence of drug, all isolates showed lower replication rates than that of wild-type NL4-3. Although three of five isolates lacking the insertion (Q781, Q822, and U099) do not have major protease inhibitor resistance mutations, there were several differences in their genotypes. Previous studies showed that several protease mutations confer reduced enzyme activity due to either an inability to refold and autoprocess or an intrinsic lack of protease activity (18, 22, 24, 29). It is possible that point mutations or insertions in the protease gene might have caused an impairment of protease function, and the insertion and point mutations were likely selected for in order to restore protease activity and viral replicative fitness. Four of the five isolates tested (Q781, Q822, Q164, and Q058) showed that the insertion-containing isolates grew better than their corresponding insertion/lacking isolates (Fig. 1). Tests in the presence of the IC50s of IDV, SQV, and NFV revealed that Q058 isolates exhibited markedly different replication profiles (Fig. 1). To evaluate the fitness of an isolate containing the insert relative to that of its counterpart lacking the insert, we tested Q058 and Q781 isolate pairs as the most protease inhibitor resistant and a protease inhibitor sensitive insertion isolates, respectively (Fig. 2). In competition studies, the relative fitness of an insertion/insertion-less pair has been evaluated by allowing the two virus populations to compete with each other until one isolate becomes dominant (7, 13). To ensure that an increase in the proportion of one isolate suggests a relatively better replicative capacity than that of its counterpart, the isolate pairs were administered at three different ratios: 80 and 20%, 50 and 50%, and 20 and 80%, based on TCID50. The cells were fed with medium containing 1% interleukin-2 twice per week and with PHA-stimulated PBMCs 7 and 14 days after infection. RNAs were extracted from each of the mixtures of virus on day 0, and RT-PCR and sequencing verified the insertion-containing/insertion-lacking isolate ratio of the mixture. At days 1, 7, 14, and 21, chromosomal DNA from infected cells was purified and the HIV-1 protease coding region was amplified as described above. The proportion of insertion-containing and insertion-lacking isolates was determined with relative peak heights in electropherograms. In order to determine the impact of an insertion mutation on fitness under protease inhibitor pressure, two different concentrations of drugs were used as a selective condition based on the minimum and maximum IC50s. In the absence of protease inhibitors, insertion-containing isolates outgrew their corresponding insertion-lacking isolates by day 7, regardless of starting concentrations of viruses (Fig. 2). The Q781 pair showed that the insertion-containing isolate outgrew the insertion-lacking isolate regardless of the presence of drugs (Fig. 2a). In the presence of a protease inhibitor, the domination occurred earlier, indicating that the replication rates of the insertion isolates are more affected by drug pressure. In the presence of both concentrations of SQV, the Q058 insertion-containing isolate outgrew its insertion-lacking control (Fig. 2b). When competition cultures were done with IDV present, Q058 failed to outgrow its insertless partner at low drug concentrations but grew quickly at high concentrations. This result for low concentrations contrasts with the drug susceptibility results presented above.
FIG. 1.
Replication kinetics of HIV-1 recombinant isolates. One thousand TCID50 of each virus was used per 106 PHA-stimulated PBMCs. Virus production was monitored every day by p24 antigen assay. Culture supernatants were collected every day until day 7, and p24 antigen production was monitored by enzyme-linked immunosorbent assay. Data are the means of results of three different tests. To serve as a related protease inhibitor-resistant control, Q058 insertion (Ins) and Q058 deletion (Del) isolates were cultured with the IC50 of each protease inhibitor (PI) and p24 values were measured daily for 7 days.
FIG. 2.
Competitive HIV-1 replication assay of insertion (INS) and deletion (DEL) isolate pairs of Q781 with and without the insertion (a) and Q058 with and without the insertion (b). Data were generated based on relative peak heights in electropherograms produced from DNA sequencing of the HIV-1 genome. In the absence of protease inhibitors, insertion and deletion pairs were combined at three different ratios, 80:20, 50:50, and 20:80 based on TCID50. In the presence of protease inhibitions, insertion and deletion isolates were coinfected at the same ratios and cultured in the presence of two different concentrations of three protease inhibitors (IDV, SQV, and NFV).
Of more than 24,000 patients genotyped by Quest Diagnostics over the past 3 years, 22 individuals (0.09%) were found to have HIV isolates that possess an insertion in the protease gene. The prevalence of isolates containing protease insertion mutations is substantially lower than the occurrence of other types of protease mutations and 10-fold lower than the occurrence of RT insertions in the same group of patients (31). The lower prevalence of insert-containing isolates suggests that a unique set of host conditions and virus characteristics may be required for the insert-containing isolates to occur and/or emerge under drug selection pressure. The inserts may have been selected during protease inhibitor therapy, as available pretherapy samples did not show evidence of the insert in one case (U099). The patient had been treated with stavudine, lamivudine, and IDV for 8 months and then treated with zidovudine, lamivudine, and NFV. The codon 35TD insertion was absent until 2 years of treatment had passed (Winters et al., Abstr. 7th Conf. Retrovir. Opportun. Infect.; S. Chou, personal communication, 1999). A recent study from another group reported a patient who developed an 18HL insertion. That patient also had a history of IDV and NFV treatment, and the 18HL insertion appeared following the first year of IDV treatment (17). Protease gene inserts have not been found so far in HIV-1 and HIV-2 genotypes from protease inhibitor-naïve patients at the Stanford HIV database (25) and other available databases. These patients' histories suggest that insertions, like point mutations, may be selected in vivo during protease inhibitor therapy but much more infrequently.
There are several theories about how these insertions could have been generated. Relatively to the strand transfer mechanism, hairpin structures and local sequence context can cause RT to pause during replication, leading to higher rates of mutation in specific areas (8, 12, 34). During reverse transcription, the finger domain in HIV-1 RT (p66) is in intimate contact with its template up to 6 nucleotide positions ahead of the catalytic site, and the effect on pausing of the RNA secondary structure ahead of the enzyme might be offset 5′ on the template by approximately 6 nucleotides (8). Investigation suggests that hairpin loops are common features of the protease RNA secondary structure, especially in the region encompassing bases 87 through 99, which correspond to codons 29 to 33 (data not shown). Given that 18 of the 22 isolates in this study possessed one, two, or five amino acid insertions a few bases upstream from this region, it is likely that the area of insertion, codons 35 through 38, is affected by this process. Further studies of the secondary structure of protease RNA may offer more insight regarding the possible mechanisms of the insertion patterns in HIV-1 protease (8).
HIV-1 replicating in vivo may find multiple molecular pathways to increase its fitness. Despite the low prevalence of insertions in the protease gene of HIV-1, the results presented in this report demonstrate that insertions are acquired in vivo and likely confer an advantage in terms of fitness. Further studies are needed to characterize the factors that cause the selection and the biochemical properties of these insert-containing proteases. A recent report indicating that an insert-containing virus can be transmitted between patients (6) suggested that such strains will be encountered in the future and may be important if they acquire drug resistance or in vivo replicative advantages.
Acknowledgments
We thank S. Chou, University of Oregon, Portland, for kindly giving us one of the insertion isolates and that patient's treatment history. L. C. Kovari, Wayne State University, Detroit, Mich., helped us with his thoughts about structural analysis and other related topics. We thank R. Lobato and R. Shafer, Stanford University, Stanford, Calif., for helpful comments and criticism of the manuscript.
This research was supported by a National Foundation for Cancer Research grant to T. C. Merigan for a project titled “Drug resistance in infection with HIV” and by the Korea Science and Engineering Foundation.
REFERENCES
- 1.Boden D, Markowitz M. Resistance to human immunodeficiency virus type I protease inhibitors. Antimicrob Agents Chemother. 1998;42:2775–2783. doi: 10.1128/aac.42.11.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briones C, Mas A, Gomez-Mariano G, Altisent C, Menendez-Arias L, Soriano V, Domingo E. Dynamics of dominance of dipeptide insertion in reverse transcriptase of HIV-1 from patients subjected to prolonged therapy. Virus Res. 2000;66:13–26. doi: 10.1016/s0168-1702(99)00120-3. [DOI] [PubMed] [Google Scholar]
- 3.D'Aquila R T, Johnson V A, Welles S L, Japour A J, Kuritzkes D R, DeGruttola V, Reichelderfer P S, Coombs R W, Crumpacker C S, Kahn J O, Richman D D. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. Ann Intern Med. 1995;122:401–408. doi: 10.7326/0003-4819-122-6-199503150-00001. [DOI] [PubMed] [Google Scholar]
- 4.Ehnlund E, Bjorling E. Fine characterization of the antigenic site within the flap region in the protease protein of HIV-1. Arch Virol. 2000;145:365–369. doi: 10.1007/s007050050028. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald P M, Springer J P. Structure and function of retroviral proteases. Annu Rev Biophys Biophys Chem. 1991;20:299–320. doi: 10.1146/annurev.bb.20.060191.001503. [DOI] [PubMed] [Google Scholar]
- 6.Grant R M, Kahn J O, Wrin T, Drews B, Javier J, Webb M, Petropolous C J, Hecht F M. HIV-1 with an insertion in protease is drug-susceptible, replication-competent and transmissible. Antivir Ther. 2001;6(Suppl. 1):44. [Google Scholar]
- 7.Harrigan P R, Bloor S, Larder B A. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998;72:3773–3778. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison G P, Mayo M S, Hunter E, Lever A M L. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 1998;26:3433–3442. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J-M, Lane J, Black R J, Reichelderfer P S, D'Aquila R T, Crumpacker C S the RV-43 Study Group; the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahn S M, Jiang W, Borner C, O'Driscoll K, Weinstein I B. Construction of defined deletion mutants by thermal cycled fusion: applications to protein kinase C. Am J Methods Cell Mol Biol. 1990;2:27–30. [Google Scholar]
- 11.Kim E Y, Vrang L, Öberg B, Merigan T C. Anti-HIV type 1 activity of 3′-fluoro-3′-deoxythymidine for several different multidrug-resistant mutants. AIDS Res Hum Retrovir. 2001;17:401–407. doi: 10.1089/088922201750102445. [DOI] [PubMed] [Google Scholar]
- 12.Kim J K, Palaniappan C P, Wu W, Fay P J, Bambara R A. Evidence for unique mechanism of strand transfer from the transactivation response region of HIV-1. J Biol Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 13.Kosalaraksa P, Kavlick M F, Maroun V, Le R, Mitsuya H. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an in vitro comparative HIV-1 replication assay. J Virol. 1999;73:5356–5363. doi: 10.1128/jvi.73.7.5356-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larder B A, Bloor S, Kemp S D, Hertogs K, Desmet R L, Miller V, Sturmer M, Staszewski S, Ren J, Stammers D K, Stuart D I, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin P F, Samanta H, Rose R E, Patick A K, Trimble J, Bechtold C M, Revie D R, Khan N C, Federici M E, Li H, et al. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy. J Infect Dis. 1994;170:1157–1164. doi: 10.1093/infdis/170.5.1157. [DOI] [PubMed] [Google Scholar]
- 16.Mahalingam B, Louis J M, Reed C C, Adomat J M, Krouse J, Wang Y-F, Harrison R W, Weber I T. Structural and kinetic analysis of drug resistance of HIV-1 protease. Eur J Biochem. 1999;263:238–245. doi: 10.1046/j.1432-1327.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- 17.Marchadier E, Domingo C, Escaut L, Vittecoq D, Liotier J Y, Beyou A, Dussaix E. New insertion in HIV-1 protease gene associated with indinavir resistance. Antivir Ther. 2000;5(Suppl. 3):44. [Google Scholar]
- 18.Martineze-Picado J, Savara A V, Sutton L, D'Aquinla R T. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maschera B, Furfine E, Blair E D. Analysis of resistance to human immunodeficiency virus type 1 protease inhibitors by using matched bacterial expression and proviral infection vectors. J Virol. 1995;69:5431–5436. doi: 10.1128/jvi.69.9.5431-5436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 21.Rose R B, Craik C S, Stroud R M. Domain flexibility in retroviral proteases: structural implications for drug resistant mutations. Biochemistry. 1998;37:2607–2621. doi: 10.1021/bi9716074. [DOI] [PubMed] [Google Scholar]
- 22.Rose R E, Gong Y-F, Greytok J A, Bechtold C M, Terry B J, Robinson B S, Alam M, Colonno R J, Lin P-F. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc Natl Acad Sci USA. 1996;93:1648–1653. doi: 10.1073/pnas.93.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schapiro J M, Winters M A, Lawrence J, Merigan T C. Clinical cross-resistance between the HIV-1 protease inhibitors saquinavir and indinavir and correlations with genotypic mutations. AIDS. 1999;13:359–365. doi: 10.1097/00002030-199902250-00008. [DOI] [PubMed] [Google Scholar]
- 24.Schock H B, Garsky V M, Kuo L C. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 25.Shafer R W, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shafer W R, Jung D R, Betts B J. Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nat Med. 2000;6:1290–1292. doi: 10.1038/81407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma P L, Crumpacker C S. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J Virol. 1999;73:8448–8456. doi: 10.1128/jvi.73.10.8448-8456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamlet C, Yahi N, Tourres C, Colson P, Quinson A-M, Poizot-Martin I, Dhiver C, Fantini J. Multidrug resistance genotypes (insertions in β3-β4 finger subdomain and MDR mutations) of HIV-1 reverse transcriptase from extensively treated patients: incidence and association with other resistance mutations. Virology. 2000;270:310–316. doi: 10.1006/viro.2000.0261. [DOI] [PubMed] [Google Scholar]
- 29.Wallqvist A, Smythers G W, Covell D G. A cooperative folding unit in HIV-1 protease. Implications for protein stability and occurrence of drug-induced mutations. Protein Eng. 1998;11:999–1005. doi: 10.1093/protein/11.11.999. [DOI] [PubMed] [Google Scholar]
- 30.Winters M A, Schapiro J M, Lawrence J, Merigan T C. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J Virol. 1998;72:5303–5306. doi: 10.1128/jvi.72.6.5303-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winters M A, Coolley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winters M A, Shafer R W, Jellinger R A, Mamtora G, Gingeras T, Merigan T C. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob Agents Chemother. 1997;41:757–762. doi: 10.1128/aac.41.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrobel J A, Conrad M J, Bloedon E, Swanstrom R, Hutchison C A., III Analysis of HIV type 1 reverse transcriptase: comparing sequences of viral isolates with mutational data. AIDS Res Hum Retrovir. 2000;16:2049–2054. doi: 10.1089/088922200750054783. [DOI] [PubMed] [Google Scholar]
- 34.Wu W, Blumberg B M, Fay P J, Bambara R A. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J Biol Chem. 1995;270:325–332. doi: 10.1074/jbc.270.1.325. [DOI] [PubMed] [Google Scholar]