Abstract
Objectives: Prostate cancer is characterized by diverse genetic mutations that influence disease progression and treatment response. This study was launched to explore the genetic basis of prostate cancer patients. Methods: We employed Next Generation Sequencing (NGS) to analyze 14 cancer-susceptible genes in prostate cancer patients. Results: Our study identified genetic mutations in BRCA1, BRCA2, TP53, and PMS2. In BRCA1 gene, we identified two pathogenic mutations, c.181T>G (p.Cys61Gly) and c.2457delC (p.Ala819fs), found in 10 patients, along with three benign mutations, c.5357T>G (p.Leu1786Arg), c.1111T>C (p.Leu371Pro), and c.1201C>G (p.Thr401Arg), present in 13, 11, and 15 patients, respectively. For the BRCA2 gene, one pathogenic mutation, c.6275_6276del (p.Val2092fs), was detected in 10 patients, and four benign mutations, c.5347A>T (p.Met1783Leu), c.5198A>G (p.Asp1733Gly), c.5158A>G (p.Thr1720Ala), and c.5117G>C (p.Gly1706Ala), were found in 17, 21, 34, and 12 patients, respectively. In the TP53 gene, we found two pathogenic mutations, c.1014_1015insT (p.Glu339Ter) and c.916C>T (p.Arg306Ter), in 10 and 11 patients, respectively, and two benign mutations, c.311T>C (p.Ser104Pro) and c.1129C>T (p.Arg377Cys), in 8 and 9 patients, respectively. Lastly, the PMS2 gene exhibited 16 benign mutations. Notably, the detected pathogenic mutations are rare in the broader Asian population according to the gnomAD database. Functional analyses using RT-qPCR and immunohistochemistry showed decreased expression of BRCA1, BRCA2, and TP53 in samples with pathogenic mutations, corroborating their impact on tumor suppressor function. Furthermore, drug sensitivity analysis revealed that BRCA1 and BRCA2 mutations are associated with increased sensitivity to a range of chemotherapeutic agents, supporting the concept of synthetic lethality. However, TP53 did not significantly impact drug sensitivity. Conclusion: This comprehensive analysis emphasizes the critical roles of BRCA1, BRCA2, TP53, and PMS2 in prostate cancer pathogenesis and highlights the importance of population-specific genetic screening.
Keywords: Prostate cancer, Next Generation Sequencing, BRCA1, BRCA2, TP53
Introduction
Prostate cancer is the second most common cancer in men worldwide and remains a leading cause of cancer-related mortality [1,2]. According to the Global Cancer Observatory (GLOBOCAN) 2023 data, prostate cancer accounts for approximately 15% of all new cancer diagnoses in men, with an estimated 1.4 million new cases and 375,000 deaths annually [3,4]. The burden of this disease is significant, particularly in developed countries where screening practices and an aging population contribute to higher incidence rates [3-6].
Despite advancements in early detection and treatment, prostate cancer exhibits a heterogeneous clinical course, ranging from indolent tumors that may never cause symptoms to aggressive forms that rapidly progress to metastatic cancer [7,8]. Understanding the genetic underpinnings of prostate cancer is crucial for improving risk stratification, prognostication, and the development of targeted therapy.
Recent studies have highlighted the importance of germline mutations in several cancer susceptibility genes in the pathogenesis of prostate cancer. Mutations in genes such as BRCA1, BRCA2, ATM, CHEK2, PALB2, RAD51C, and RAD51D, and others have been associated with increased risk and poor clinical outcome [9-13]. The role of these genes in DNA damage repair and genomic stability underscores their relevance in cancer biology. For instance, BRCA1 and BRCA2 mutations, well-known for their association with breast and ovarian cancers, are also implicated in aggressive prostate cancer phenotypes [14,15]. Similarly, mutations in mismatch repair genes like MLH1, MSH2, MSH6, and PMS2 are linked to Lynch syndrome, which includes a predisposition to prostate cancer [16-18].
Recently, next-generation sequencing (NGS) technology has emerged as a powerful tool for comprehensive genetic profiling [19]. NGS allows for the simultaneous analysis of multiple genes, offering detailed insight into the mutational landscape of cancer [20]. This approach has paved the way for personalized medicine, where genetic information guides tailored treatment strategies [21].
Our study aims to analyze genetic mutations in 14 key cancer susceptibility genes (BRCA1/2, ATM, CHEK2, PALB2, RAD51C, RAD51D, NBN, CDH1, TP53, MLH1, MSH2, MSH6, and PMS2) in prostate cancer patients using NGS. By elucidating the mutation spectrum in these genes, we seek to enhance our understanding of their role in prostate cancer and identify targets for therapeutic intervention. This research not only contributes to the growing body of knowledge on prostate cancer genetics but also has significant implications for clinical practice in the era of precision oncology.
Methods
Sample collection
We collected prostate cancer tissue samples from 50 patients diagnosed with prostate cancer who visited Nishtar Hospital, Multan, Pakistan, between 2021 and 2023 (Table 1). The inclusion criteria for patient selection encompassed individuals with histologically confirmed prostate cancer, irrespective of their cancer stage or grade. Exclusion criteria were patients who had received chemotherapy or radiotherapy prior to tissue sample collection, so as to avoid treatment-related alterations in genetic material. The tissue samples were obtained through transrectal ultrasound-guided (TRUS) biopsy or radical prostatectomy, based on the clinical requirements and treatment plans of the patients. Each sample was immediately placed in RNAlater® (Ambion, Austin, TX, USA) solution to stabilize and protect RNA and DNA until further processing. The samples were then stored at -80°C until DNA extraction could be performed. Informed consent was obtained from all participants before sample collection, in accordance with the ethical guidelines of the Helsinki Declaration. This study was approved by the Ethics Committee of Nishtar Medical University, ensuring compliance with ethical standards and patient confidentiality.
Table 1.
Overview of prostate cancer patient characteristics in the study
| Sr. no | Characteristic | Sample count (n) |
|---|---|---|
| 1 | Sex | |
| Male | 50 | |
| Female | 0 | |
| 2 | Age | |
| >60 | 15 | |
| <60 | 35 | |
| 3 | Treatment | |
| Pre-treatment | 50 | |
| Post-treatment | 0 |
DNA and RNA extraction
From the collected prostate cancer tissue samples, DNA extraction was performed using the organic method [22], ensuring high-quality and intact genomic DNA suitable for NGS. Initially, tissue samples were homogenized and lysed in a buffer containing SDS and proteinase K, followed by incubation at 56°C to ensure complete protein digestion. The lysate was then subjected to phenol-chloroform extraction to separate DNA from proteins and other cellular debris. This step involved the addition of an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), followed by centrifugation to separate the aqueous phase containing DNA. The aqueous phase was carefully transferred to a new tube, and DNA was precipitated using cold ethanol. The DNA precipitate was then washed with 70% ethanol, air-dried, and resuspended in TE buffer (Tris-EDTA) for storage.
For RNA extraction, the TRIzol® (Thermo Fisher Scientific, Waltham, MA, USA) method [23] was used, ensuring efficient isolation of high-quality total RNA. Tissue samples were homogenized in TRIzol® reagent, which facilitates the disruption of cells and the denaturation of proteins. Following homogenization, chloroform was added to the mixture, which was then vigorously shaken and centrifuged to separate the mixture into aqueous and organic phases. The aqueous phase, containing RNA, was carefully collected. To precipitate the RNA, isopropanol was added to the aqueous phase. The resulting RNA pellet was washed with 75% ethanol, air-dried, and resuspended in RNase-free water for storage.
The purity of the extracted DNA and RNA was assessed using the A260/A280 ratio, measured by a NanoDrop spectrophotometer (Thermo Fisher Scientific). A ratio of approximately 1.8 for DNA and 2.0 for RNA was considered indicative of high purity, ensuring that the nucleic acids were free from protein contamination and suitable for downstream applications. High-purity DNA and RNA were crucial for the reliability and accuracy of the NGS and other molecular analyses conducted in this study.
Next Generation Sequencing (NGS) analysis
For our next-generation sequencing library preparation, we employed the TruSight Cancer Sequencing Panel, a readily available targeted sequencing kit, and utilized the MiSeq platform by Illumina (San Diego, CA). All steps were carried out in strict accordance with the manufacturer’s recommended protocols. To provide a brief overview of our approach, we initiated library preparation with 50 ng of genomic DNA for each sample, employing the TruSight Rapid Capture and TruSight Cancer kits. The resultant double-stranded DNA libraries were subsequently transformed into single-stranded DNA. To target specific regions, we employed biotin-labeled probes in the first rapid capture step. Streptavidin beads were introduced to enrich the pool of mixed samples for the desired target regions. Biotinylated DNA fragments bound to the streptavidin beads were then isolated from the solution using magnetic pull-down. After this, the enriched DNA fragments were eluted from the beads and subjected to a second rapid capture step. Finally, the prepared libraries were applied to the MiSeq Flowcell for sequencing. Subsequently, the paired sequences from each sample were aligned to the human genome reference GRCh37/hg19 using BWA-MEM version 0.7.7. Duplicate sequences were identified and marked with Picard’s MarkDuplicates version 1.111 (available at https://github.com/broadinstitute/picard), and local InDel realignment was conducted using the Genome Analysis Tool Kit (GATK) version 3.1.1. The TruSight Cancer panel encompasses 94 genes associated with both common (e.g., breast, prostate) and rare cancers. Among these, we focused our analysis on 14 specific genes (BRCA1/2, ATM, CHEK2, PALB2, RAD51C, RAD51D, NBN, CDH1, TP53, MLH1, MSH2, MSH6, PMS2) using the SeqNext module within the Sequence Pilot software by JSI medical systems GmbH in Kippenheim, Germany. Our sequencing achieved a medium sequence depth of 400×, with a minimum of 30× coverage for the coding regions and the first 10 base pairs of flanking intronic regions for each gene.
The interpretation of genetic mutations followed the comprehensive guidelines set by the American College of Medical Genetics (ACMG) and the Association for Molecular Pathology (AMP). These guidelines categorize mutations into five classifications: pathogenic, likely pathogenic, variants of uncertain significance (VUS), likely benign, and benign. Furthermore, we utilized the ClinVar database to obtain valuable clinical significance data, which aggregates information on genomic variations and their health implications. By integrating these computational tools with ClinVar’s clinical data, we ensured a robust and reliable classification of mutations. This approach supported accurate clinical decision-making and enhanced our understanding of the genetic contributions to disease.
Genome aggregation database (gnomAD)
The gnomAD database is a comprehensive resource that aggregates and harmonizes exome and genome sequencing data from a wide range of large-scale sequencing projects [24]. It includes data from over 140,000 individuals, providing extensive information on human genetic variation. gnomAD is widely used for research and clinical interpretation, offering insight into population-specific allele frequencies and aiding in the identification of rare genetic variants associated with diseases. In this study, this database was utilized to analyze the frequencies of the observed mutations across prostate cancer patients from an Asian population.
cBioPortal database
The cBioPortal for Cancer Genomics is an open-access resource designed to provide visualization, analysis, and download of large-scale cancer genomics datasets [25]. It integrates data from prominent cancer studies, including The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC). cBioPortal offers tools for exploring genetic alterations, such as mutations, copy number variations, and gene expression changes, facilitating the discovery of cancer biomarkers and therapeutic targets. In this work, cBioPortal database wasused for analyzing the occurrence of observed mutations in prostate cancer patients from The Cancer Genome Atlas (TCGA) project.
RT-qPCR analysis
Using a reverse transcription kit from Promega (USA), 3 μg of RNA was reverse transcribed into cDNA in a 20 μL reaction system. Quantitative polymerase chain reaction (qPCR) was then performed using the 2× SYBR Green qPCR Master Mix (Low ROX) (Cat #: B21702, Bimake, USA). The reaction mixture, totaling 20 μL, comprised 10 μL of 2× SYBR Green qPCR Master Mix (Low ROX), 6 μL of nuclease-free water, 0.2 μM of each primer, and 2 μL of cDNA products. Relative gene expression was determined using the 2-ΔΔCT method.
The following primer sequences were used: GAPDH sense: 5’-GTCTCCTCTGACTTCAACAGCG-3’, GAPDH antisense: 5’-ACCACCCTGTTGCTGTAGCCAA-3’; BRCA1 sense: 5’-CTGAAGACTGCTCAGGGCTATC-3’, BRCA1 antisense: 5’-CTGAAGACTGCTCAGGGCTATC-3’; BRCA2 sense: 5’-GAAAATCAAGAAAAATCCTTAAAGGCT-3’, BRCA2 antisense: 5’-GTAATCGGCTCTAAAGAAACATGATG-3’; TP53 sense: 5’-CCTCAGCATCTTATCCGAGTGG-3’, TP53 antisense: 5’-TGGATGGTGGTACAGTCAGAGC-3’; PMS2 sense: 5’-TATCGGCTCTGTGTTTGGGCAG-3’, PMS2 antisense: 5’-AGCATCGGAACAGCTCAAACCG-3’.
Immunohistochemistry
A previously established protocol for immunohistochemistry was followed in this study [26]. Briefly, tissue sections underwent deparaffinization, and antigen retrieval was performed by heat treatment in an EDTA solution at pH 8.0. Protein expression levels of the mutated genes were evaluated using 4-μm-thick sections from formalin-fixed, paraffin-embedded (FFPE) specimens. Monoclonal antibodies targeting BRCA1 (Cat # TA802618AM), BRCA2 (Cat # TA802628), and TP53 (Cat # TA502870) were used, with staining conducted on the Ventana BenchMark XT staining system. A pathologist assessed tumor positivity based on the presence or absence of nuclear staining in tumor tissue, taking staining intensity into account.
Gene enrichment analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) is a bioinformatics resource that provides comprehensive functional annotation tools for researchers to understand the biological meaning behind large lists of genes [27]. DAVID integrates diverse data sources to facilitate gene enrichment analysis, allowing users to explore gene functions, biological processes, and pathways. It supports a wide range of genomic studies, enhancing the interpretation of high-throughput data and contributing to advancements in molecular biology and genetics research. In the present study, DAVID tool was used to perform gene enrichment analysis of mutated genes in prostate cancer patients.
Drug sensitivity analysis
The Gene Set Cancer Analysis (GSCA) tool, developed by Guo Lab, is a comprehensive bioinformatics platform designed to analyze gene sets in the context of cancer [28]. It integrates various data types, including genomic mutations, expression profiles, and drug responses, to provide insights into cancer biology. GSCA allows researchers to explore the relationships between gene sets and cancer phenotypes, facilitating the identification of potential biomarkers and therapeutic targets. This tool enhances our understanding of the molecular mechanisms driving cancer progression and treatment resistance. In this study, GSCA was utilized for the drug sensitivity analysis of mutated genes in prostate cancer patients.
Statistics
A Student’s t-test was employed to compare the two groups. Receiver operating curve analysis (ROC) curves were utilized to evaluate the diagnostic values of the mutated genes.
Results
Mutations landscape across prostate cancer patients
We analyzed genetic mutations in 14 cancer-susceptible genes in ovarian cancer patients through NGS technology. Our results revealed mutations in four genes: BRCA1, BRCA2, TP53, and PMS2. In the BRCA1 gene, we identified two pathogenic mutations, c.181T>G (p.Cys61Gly) and c.2457delC (p.Ala819fs), found in 10 patients (Table 2 and Figure 1), along with three benign mutations, c.5357T>G (p.Leu1786Arg), c.1111T>C (p.Leu371Pro), and c.1201C>G (p.Thr401Arg), present in 13, 11, and 15 patients, respectively (Table 2 and Figure 1). For the BRCA2 gene, one pathogenic mutation, c.6275_6276del (p.Val2092fs), was detected in 10 patients, and four benign mutations, c.5347A>T (p.Met1783Leu), c.5198A>G (p.Asp1733Gly), c.5158A>G (p.Thr1720Ala), and c.5117G>C (p.Gly1706Ala), were found in 17, 21, 34, and 12 patients, respectively (Table 2 and Figure 1). In the TP53 gene, we found two pathogenic mutations, c.1014_1015insT (p.Glu339Ter) and c.916C>T (p.Arg306Ter), in 10 and 11 patients, respectively, and two benign mutations, c.311T>C (p.Ser104Pro) and c.1129C>T (p.Arg377Cys), in 8 and 9 patients, respectively (Table 2 and Figure 1). Lastly, the PMS2 gene exhibited multiple benign mutations. Notably, c.1532C>T (p.Thr511Met) was found in 5 patients, while c.1531A>G (p.Thr511Ala) appeared in 11 patients (Table 2 and Figure 1). Additional mutations included c.1452G>A (p.Lys484Lys) in 12 patients, c.2002A>G (p.Asn668Ser) in 11 patients, and c.2184G>A (p.Lys728Lys) in 3 patients. The c.2244T>C (p.Glu748Glu) mutation was identified in 11 patients, c.2522A>G (p.Ile841Val) in 1 patient, and c.2565G>A (p.Leu855Leu) in 5 patients. Furthermore, c.2634T>C (p.Ser878Ser) was found in 7 patients, c.2724C>T (p.Pro908Pro) in 4 patients, and c.2748C>T (p.Ala916Ala) in 13 patients. Other mutations such as c.2907T>C (p.Tyr969Tyr), c.3003G>A (p.Gln1001Gln), and c.3144T>C (p.Ser1048Ser) were present in 21, 12, and 23 patients, respectively. The mutation c.3216C>T (p.Asp1072Asp) appeared in 2 patients, c.3276G>A (p.Leu1092Leu) in 1 patient, and c.3420T>C (p.Val1140Val) in 6 patients. Lastly, c.3579A>G (p.Arg1193Arg) was found in 11 patients, c.3720C>T (p.Pro1240Pro) in 14 patients, c.3801G>A (p.Glu1267Glu) in 16 patients, and c.3900A>G (p.Ile1300Ile) in 12 patients (Table 2 and Figure 1). Overall, our findings reveal a diverse array of mutations in the PMS2 gene among prostate cancer patients.
Table 2.
Frequency and classification of mutations detected in the BRCA1, BRCA2, TP53, and PMS2 genes among prostate cancer patients
| Sr. no | Gene | NM. DNA | Protein | Nature | No. patients |
|---|---|---|---|---|---|
| 1 | BRCA1 | NM_007294.4 | p.Cys61Gly | Pathogenic | 10 |
| c.181T>G | |||||
| 2 | NM_007294.4 | p.Ala819fs | Pathogenic | 10 | |
| c.2457delC | |||||
| 3 | NM_007294.4 | p.Leu1786Arg | Benign | 13 | |
| c.5357T>G | |||||
| 4 | NM_007294.4 | p.Leu371Pro | Benign | 11 | |
| c.1111T>C | |||||
| 5 | NM_007294.4 | p.Thr401Arg | Benign | 15 | |
| c.1201C>G | |||||
| 6 | BRCA2 | NM_000059.4 | p.Val2092fs | Pathogenic | 10 |
| c.6275_6276del | |||||
| 7 | NM_007294.4 | p.Met1783Leu | Benign | 17 | |
| c.5347A>T | |||||
| 8 | NM_007294.4 | p.Asp1733Gly | Benign | 21 | |
| c.5198A>G | |||||
| 9 | NM_007294.4 | p.Thr1720Ala | Benign | 34 | |
| c.5158A>G | |||||
| 10 | NM_007294.4 | p.Gly1706Ala | Benign | 12 | |
| c.5117G>C | |||||
| 11 | TP53 | NM_000546.6 | p.Glu339Ter | Pathogenic | 10 |
| c.1014_1015insT | |||||
| 12 | NM_000546.6 | p.Arg306Ter | Pathogenic | 10 | |
| c.916C>T | |||||
| 13 | NM_000546.6 | p.Ser104Pro | Benign | 8 | |
| c.311T>C | |||||
| 14 | NM_000546.6 | p.Arg377Cys | Benign | 9 | |
| c.1129C>T | |||||
| 15 | PMS2 | NM_000535.7 | p.Thr511Met | Benign | 5 |
| c.1532C>T | |||||
| 16 | NM_000535.7 | p.Thr511Ala | Benign | 11 | |
| c.1531A>G | |||||
| 17 | NM_000535.7 | p.Lys484Lys | Benign | 12 | |
| c.1452G>A | |||||
| 18 | NM_000535.7 | p.Asn668Ser | Benign | 11 | |
| c.2002A>G | |||||
| 19 | NM_000535.7 | p.Lys728Lys | Benign | 3 | |
| c.2184G>A | |||||
| 20 | NM_000535.7 | p.Glu748Glu | Benign | 11 | |
| c.2244T>C | |||||
| 21 | NM_000535.7 | p.Ile841Va | Benign | 1 | |
| c.2522A>G | |||||
| 22 | NM_000535.7 | p.Leu855Leu | Benign | 5 | |
| c.2565G>A | |||||
| 23 | NM_000535.7 | p.Ser878Ser | Benign | 7 | |
| c.2634T>C | |||||
| 24 | NM_000535.7 | p.Pro908Pro | Benign | 4 | |
| c.2724C>T | |||||
| 25 | NM_000535.7 | p.Ala916Ala | Benign | 13 | |
| c.2748C>T | |||||
| 26 | NM_000535.7 | p.Tyr969Tyr | Benign | 21 | |
| c.2907T>C | |||||
| 27 | NM_000535.7 | p.Gln1001Gln | Benign | 12 | |
| c.3003G>A | |||||
| 28 | NM_000535.7 | p.Ser1048Ser | Benign | 23 | |
| c.3144T>C | |||||
| 29 | NM_000535.7 | p.Asp1072Asp | Benign | 2 | |
| c.3216C>T | |||||
| 30 | NM_000535.7 | p.Leu1092Leu | Benign | 1 | |
| c.3276G>A | |||||
| 31 | NM_000535.7 | p.Val1140Val | Benign | 6 | |
| c.3420T>C | |||||
| 32 | NM_000535.7 | p.Arg1193Arg | Benign | 11 | |
| c.3579A>G | |||||
| 33 | NM_000535.7 | p.Pro1240Pro | Benign | 14 | |
| c.3720C>T | |||||
| 34 | NM_000535.7 | p.Glu1267Glu | Benign | 16 | |
| c.3801G>A | |||||
| 35 | NM_000535.7 | p.Ile1300Ile | Benign | 12 | |
| c.3900A>G |
Figure 1.
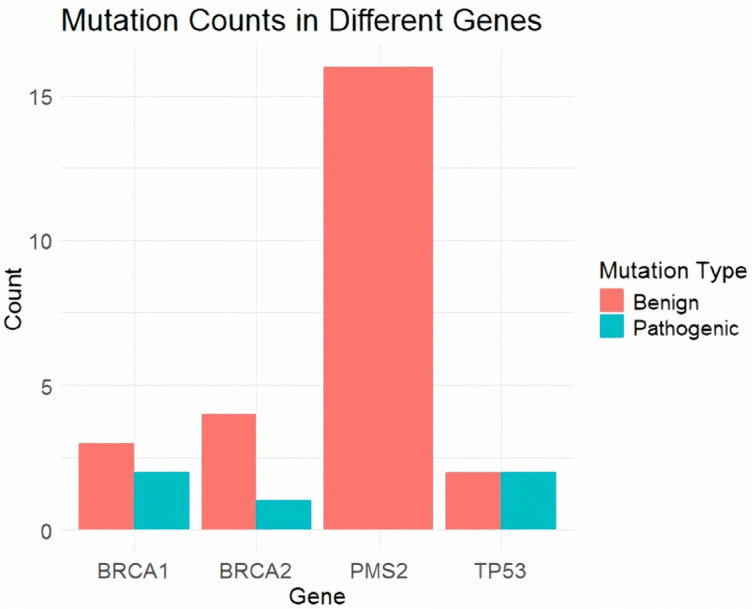
Total mutation count for each gene. This bar chart illustrates the total mutation count for each gene analyzed in the study, specifically BRCA1, BRCA2, TP53, and PMS2. The mutations are categorized into benign and pathogenic, with benign mutations represented in blue and pathogenic mutations in red. BRCA1 shows a total of five mutations, three benign and two pathogenic. BRCA2 has the highest number of mutations, totaling five, with four benign and one pathogenic. TP53 also has an equal number of benign and pathogenic mutations, each accounting for two out of the four total mutations. PMS2 shows sixteen benign mutations without any pathogenic mutations. This visualization underscores the distribution and frequency of benign versus pathogenic mutations across the different genes studied in prostate cancer patients.
Frequencies of the pathogenic mutations in Asian prostate cancer patients
NGS analysis revealed several pathogenic mutations: in the BRCA1 gene, c.181T>G (p.Cys61Gly) and c.2457delC (p.Ala819fs) were found in 10 patients; in the BRCA2 gene, c.6275_6276del (p.Val2092fs) was detected in 10 patients; and in the TP53 gene, c.1014_1015insT (p.Glu339Ter) and c.916C>T (p.Arg306Ter) were found in 10 patients. Given the clinical significance of pathogenic mutations, we further analyzed their frequency in the Asian population using the gnomAD database. Our investigation revealed that these pathogenic mutations have a frequency of 0 in the Asian population according to gnomAD. This finding suggests that these mutations are rare in our Pakistani population, highlighting their potential significance and the necessity for targeted genetic screening within this demographic to identify at-risk individuals and tailor personalized treatment strategies.
Analysis of the pathogenic mutations in The Cancer Genome Atlas (TCGA)
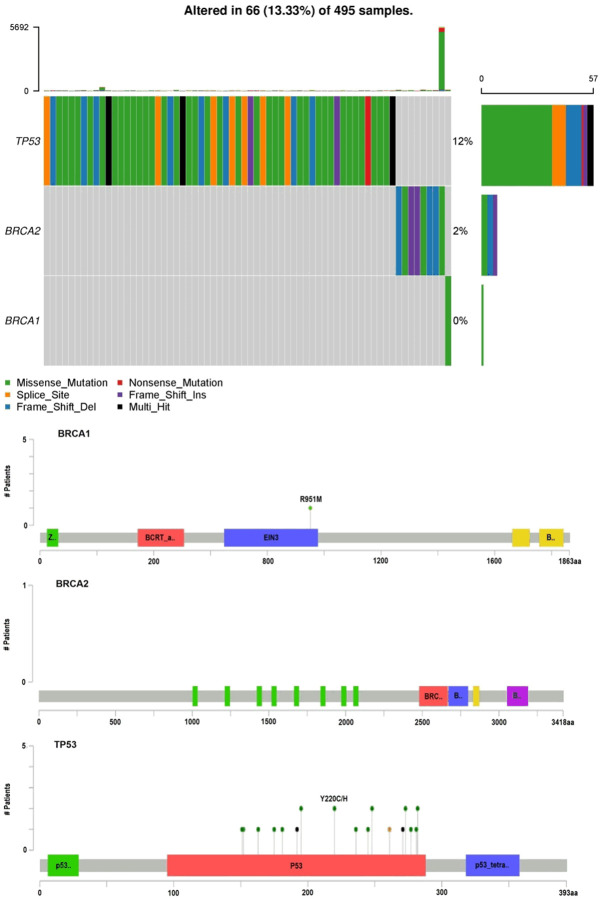
In this part of our study, we thoroughly investigated the presence of identified pathogenic mutations in the BRCA1 gene (c.181T>G, p.Cys61Gly, and c.2457delC, p.Ala819fs), the BRCA2 gene (c.6275_6276del, p.Val2092fs), and the TP53 gene (c.1014_1015insT, p.Glu339Ter, and c.916C>T, p.Arg306Ter) within the TCGA prostate cancer patient cohort, utilizing the cBioPortal database. Our detailed analysis confirmed that these specific pathogenic mutations were absent in the TCGA prostate cancer patients, as they were not detected in the TCGA dataset (Figure 2). This significant finding highlights the exceptional rarity of the pathogenic mutations identified in our study.
Figure 2.
Mutation analysis of BRCA1, BRCA2, and TP53 in The Cancer Genome Atlas (TCGA) prostate cancer samples. The figure provides a comprehensive overview of mutations in the TP53, BRCA2, and BRCA1 genes across 495 prostate cancer samples, highlighting the percentage of samples altered. The top panel presents a heatmap illustrating the distribution of different mutation types, including missense mutations (green), nonsense mutations (red), splice site mutations (purple), frame shift insertions (pink), frame shift deletions (blue), and multi-hit events (black). The bottom panel maps the specific mutation sites on the BRCA1, BRCA2, and TP53 protein domains, with green dots representing the positions and number of patients affected. Key functional domains are indicated on the protein maps, including the BRCT domain in BRCA1, the BRC repeats in BRCA2, and the P53 and P53_tetra domains in TP53.
Functional consequence analyses of the observed pathogenic mutations
RT-qPCR analysis
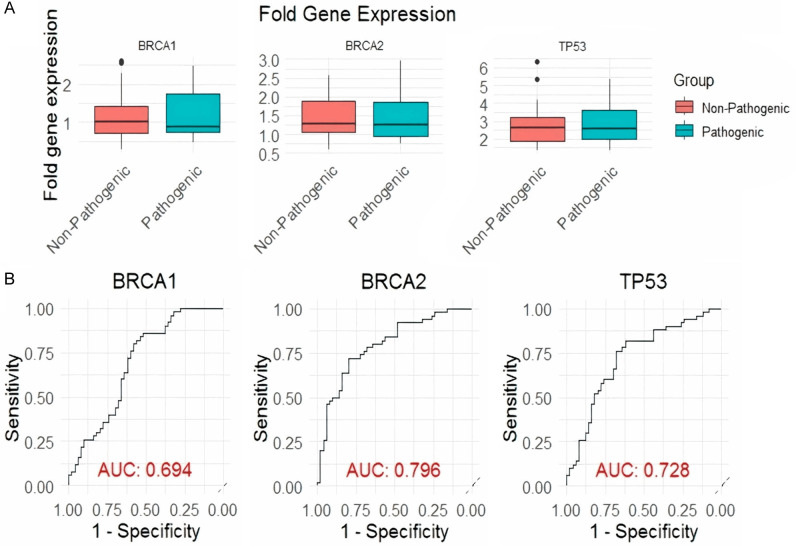
We categorized prostate cancer tissue samples into two distinct groups. The first group consisted of 10 prostate cancer samples with documented pathogenic mutations in the BRCA1, BRCA2, and TP53 genes (Pathogenic group), while the second group included 40 prostate cancer samples without such pathogenic mutations (Non-pathogenic group). We then performed an expression analysis of BRCA1, BRCA2, and TP53 genes in these two cohorts using RT-qPCR. The box plots in Figure 3A reveal that all three genes (BRCA1, BRCA2, and TP53) show decreased expression in the Pathogenic group, indicating an association with pathogenic mutations. The ROC curves and AUC values illustrate the discriminatory power of these genes: BRCA2 has the highest AUC (0.796), suggesting it is the most effective in distinguishing between the groups, followed by TP53 (AUC 0.728) and BRCA1 (AUC 0.694) (Figure 3B). These results imply that while all three genes are down-regulated in the presence of pathogenic mutations, BRCA2 is the most promising biomarker for identifying such mutations in prostate cancer samples.
Figure 3.
Differential gene expression and ROC analysis for pathogenic vs. non-pathogenic groups of prostate cancer samples. A. Box plots showing the fold gene expression levels of BRCA1, BRCA2, and TP53 in non-pathogenic (red) and pathogenic (blue) groups. Each plot displays the distribution of gene expression levels with median values indicated by the central line and interquartile ranges represented by the boxes. B. Receiver Operating Characteristic (ROC) curves for BRCA1, BRCA2, and TP53, evaluating their ability to distinguish between pathogenic and non-pathogenic groups. Sensitivity (true positive rate) is plotted against 1-specificity (false positive rate) for each gene. The Area Under the Curve (AUC) values are provided for each ROC curve, with BRCA1 showing an AUC of 0.694, BRCA2 an AUC of 0.796, and TP53 an AUC of 0.728, indicating the discriminative power of each gene in identifying pathogenic cases. A P<0.05 was used as the cut-off criterion.
Immunohistochemistry
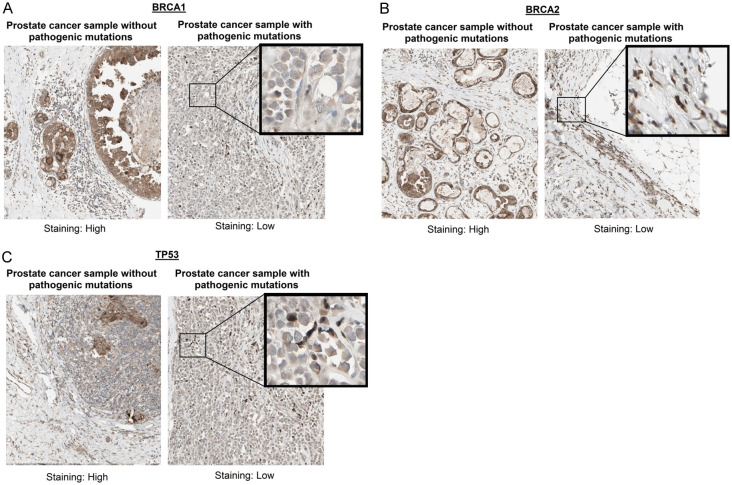
In our study, we conducted IHC analysis of BRCA1, BRCA2, and TP53 proteins in two representative prostate cancer tissue samples. These samples were carefully chosen to represent two distinct genetic profiles: one with pathogenic mutations in the BRCA1, BRCA2, and TP53 genes, and the other without mutations in these genes. The immunohistochemical staining results for BRCA1, BRCA2, and TP53 in prostate cancer tissue samples show a clear distinction between samples without pathogenic mutations and those with pathogenic mutations. For all three genes, samples without pathogenic mutations exhibited high staining intensity, indicating higher protein expression levels (Figure 4). In contrast, samples with pathogenic mutations demonstrated low staining intensity, reflecting reduced protein expression (Figure 4). This pattern suggests that the presence of pathogenic mutations in BRCA1, BRCA2, and TP53 genes is associated with decreased expression of these tumor suppressor proteins, aligning with the gene expression data and highlighting the potential impact of these mutations on protein expression in prostate cancer.
Figure 4.
Immunohistochemical staining of BRCA1, BRCA2, and TP53 in pathogenic vs. non-pathogenic prostate cancer samples. A. Immunohistochemical staining results for BRCA1 between samples with and without pathogenic mutations. B. Immunohistochemical staining results for BRCA2 between samples with and without pathogenic mutations. C. Immunohistochemical staining results for TP53 between samples with and without pathogenic mutations.
Gene enrichment analysis
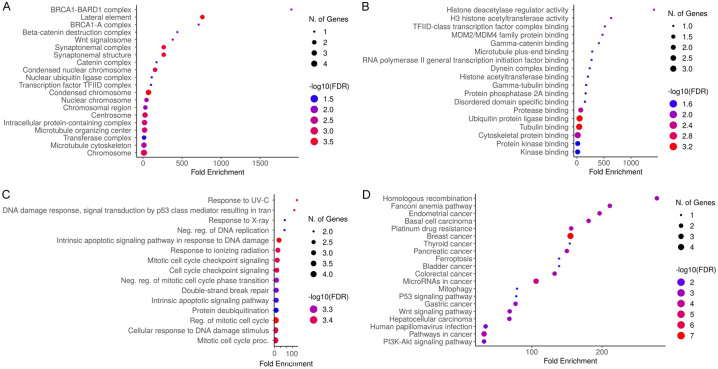
DAVID tool was used to perform gene enrichment analysis of the mutated genes (BRCA1, BRCA2, TP53, and PMS2). Figure 5 presents a comprehensive analysis of the functional enrichment, highlighting significant biological processes (BP), molecular functions (MF), cellular components (CC), and pathways. In CC analysis (Figure 5A), highly enriched components include “condensed chromosome, microtubule organizing center, centrosome, synaptonemal complex, and BRCA1-related complexes”, indicating a strong association with chromosome structure and DNA repair mechanisms. Figure 5B focuses on MF, revealing significant enrichment in “H3 histone acetyltransferase activity, various protein binding activities, and histone deacetylase regulator activity”, underscoring the importance of epigenetic regulation and protein interactions in these mutations. Figure 5C examines BP, showing enrichment in “DNA damage response, regulation of DNA replication, mitotic cell cycle checkpoint signaling, and apoptotic signaling pathways”, which are critical for maintaining genomic stability and cellular response to DNA damage. Finally, Figure 5D illustrates pathway enrichment, with significant involvement in cancer-related pathways such as “PI3K-Akt signaling, P53 signaling, and the Fanconi anemia pathway”, emphasizing their roles in cell growth, survival, DNA repair, and apoptosis. Overall, these results highlight the pivotal roles of these mutated genes in crucial cellular functions and pathways that contribute to prostate cancer progression and response to treatment.
Figure 5.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of BRCA1, BRCA2, and TP53 Genes via Database for Annotation, Visualization, and Integrated Discovery (DAVID). A. BRCA1, BRCA2, and TP53 genes-related Cellular Component (CC) terms. B. BRCA1, BRCA2, and TP53 genes-related Molecular Function (MF) terms. C. BRCA1, BRCA2, and TP53 genes-related Biological Process (BP) terms. D. BRCA1, BRCA2, and TP53 genes-related KEGG pathway terms. A p-value of less than 0.05 was used as the cut-off criterion for significance.
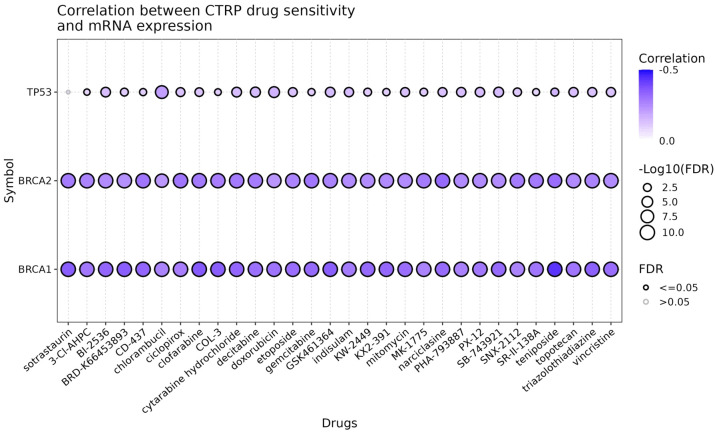
Drug sensitivity analysis
Next, the drug sensitivity analysis of BRCA1, BRCA2, and TP53 was performed using GSCA database. The results show that TP53 does not significantly affect drug sensitivity, as evidenced by the predominantly small and grey circles (Figure 6). In contrast, both BRCA1 and BRCA2 exhibit significant negative correlations with all the drugs tested, indicated by the large, dark blue circles with black borders (Figure 6). This suggests that lower expression levels of BRCA1 and BRCA2 are associated with increased sensitivity to a wide range of drugs, including sotrastaurin, 3-Cl-AHPC, BI-2536, BRD-K65433903, CD-437, chlorambucil, ciclopirox, clofarabine, COL-2, cytarabine hydrochloride, decitabine, doxorubicin, etoposide, gemcitabine, GSK-461364, indisulam, KW-2449, mitomycin C, MK-1775, narcidase, PHA-793887, PI-103, SB-743921, SNX-2112, teniposide, topotecan, triazolothiadiazine, and vincristine (Figure 6).
Figure 6.
Correlation between CTRP Drug Sensitivity and mRNA Expression of BRCA1, BRCA2, and TP53. The dot plot illustrates the correlation between drug sensitivity (measured in the CTRP database) and mRNA expression levels of three genes: BRCA1, BRCA2, and TP53. Each row corresponds to a different gene, and each column represents a specific drug. The color intensity of the dots indicates the strength of the negative correlation, with darker shades of blue representing stronger negative correlations. The size of the dots reflects the significance of the correlation, as measured by the -log10(FDR) value. Larger dots indicate more significant correlations (lower FDR values), and a filled dot signifies an FDR of ≤0.05, whereas an open dot indicates an FDR >0.05. The drugs are listed along the x-axis and include a variety of chemotherapeutic agents and other compounds.
Discussion
Prostate cancer is a heterogeneous disease characterized by a variety of histologic subtypes and genetic alterations [29,30]. It can be one of the most lethal malignancies due to its often late diagnosis and complex biology [30]. Genetic mutations, particularly in genes involved in DNA repair mechanisms such as BRCA1, BRCA2, and TP53, play a significant role in the pathogenesis of prostate cancer [31-33]. Understanding the landscape of these mutations can aid in developing targeted therapies and improving patient outcome.
Our study provides a comprehensive analysis of the mutation landscape in prostate cancer, focusing on 14 cancer-susceptible genes (BRCA1/2, ATM, CHEK2, PALB2, RAD51C, RAD51D, NBN, CDH1, TP53, MLH1, MSH2, MSH6, and PMS2) using NGS technology. This approach has revealed a diverse array of mutations in BRCA1, BRCA2, TP53, and PMS2 genes. We identified pathogenic mutations in BRCA1 (c.181T>G, p.Cys61Gly; c.2457delC, p.Ala819fs) and BRCA2 (c.6275_6276del, p.Val2092fs) in Pakistani cohort of prostate cancer patients. These mutations are known to be associated with increased cancer risk due to their critical roles in homologous recombination repair of DNA double-strand breaks [34,35]. Earlier studies have reported similar pathogenic variants in BRCA1 and BRCA2 among different cancer types, including breast and ovarian cancers, highlighting their universal significance in cancer biology [36-38]. However, our findings indicate that these specific mutations are rare in the broader Asian population, as evidenced by their absence in the gnomAD database, which emphasizes the importance of population-specific genetic screening.
The detection of pathogenic TP53 mutations (c.1014_1015insT, p.Glu339Ter; c.916C>T, p.Arg306Ter) aligns with the well-documented role of TP53 as a tumor suppressor gene frequently mutated in various cancers [39,40]. Our results are consistent with existing literature that underscores the prevalence and significance of TP53 mutations in cancer progression and prognosis [41,42]. Similarly, the identified benign mutations in PMS2 (c.1532C>T, p.Thr511Met; c.1531A>G, p.Thr511Ala) add to the growing body of evidence that variations in mismatch repair genes, while not always pathogenic, can contribute to cancer susceptibility and genetic diversity within populations [43,44].
Our functional analyses using RT-qPCR and immunohistochemistry (IHC) demonstrated decreased expression of BRCA1, BRCA2, and TP53 in samples harboring pathogenic mutations. This reduction in gene and protein expression is indicative of the functional impact these mutations have on the tumor suppressor capabilities of these genes. Our findings are in line with previous studies that have shown similar patterns of down-regulation in BRCA1 and BRCA2-mutated breast and ovarian cancer [45,46]. Additionally, the ROC curve analysis highlighted BRCA2 as the most effective biomarker for distinguishing between pathogenic and non-pathogenic groups, suggesting its potential utility in clinical diagnostics.
Gene enrichment analysis revealed significant associations with key biological processes, molecular functions, and cellular components involved in DNA repair, cell cycle regulation, and apoptotic pathways. These results corroborate earlier research emphasizing the critical roles of BRCA1, BRCA2, TP53, and PMS2 in maintaining genomic stability and cellular homeostasis [47,48]. The enrichment in cancer-related pathways such as PI3K-Akt signaling and p53 signaling further highlights the relevance of these genes in oncogenesis and therapy response [49].
Our drug sensitivity analysis indicated that BRCA1 and BRCA2 mutations are associated with increased sensitivity to a range of chemotherapeutic agents. This finding supports the concept of synthetic lethality, where BRCA1/2-deficient cells are more susceptible to DNA-damaging agents and PARP inhibitors [50]. In contrast, TP53 did not show a significant impact on drug sensitivity, which may reflect the complexity of p53-mediated drug responses and the influence of other genetic and epigenetic factors [51].
This study offers several significant merits, including the comprehensive analysis of mutations in 14 cancer-susceptible genes using NGS technology, which provides a detailed landscape of genetic alterations in prostate cancer patients. The functional consequence analyses through RT-qPCR and immunohistochemistry add a robust layer of validation to the genetic findings, linking mutations to changes in gene and protein expression. The study’s focus on population-specific mutations, particularly in a Pakistani cohort, addresses a critical gap in existing literature and underscores the need for tailored genetic screening strategies. Additionally, the integration of gene enrichment and drug sensitivity analyses provides a holistic view of the biological and therapeutic implications of the identified mutations. However, the study also has limitations. The relatively small sample size limits the generalizability of the findings, and the absence of functional validation in cellular or animal models restricts the ability to draw definitive conclusions about the biological impact of the mutations. Furthermore, the lack of longitudinal data prevents assessment of the mutations’ prognostic significance and their potential role in disease progression and treatment response over time. Despite these limitations, the study makes a valuable contribution to the understanding of genetic mutations in prostate cancer and highlights avenues for future research.
Conclusion
Our study provides valuable insight into the mutational landscape of key cancer-susceptible genes in prostate cancer. The identified pathogenic mutations in BRCA1, BRCA2, and TP53, along with their functional consequences and implications for drug sensitivity, underscore the importance of targeted genetic screening and personalized treatment strategies. Our findings contribute to the growing understanding of the genetic underpinnings of prostate cancer and highlight the need for further research to explore the therapeutic potential of these insights.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP2024R470), King Saud University, Riyadh, Saud Arabia.
Disclosure of conflict of interest
None.
References
- 1.Bergengren O, Pekala KR, Matsoukas K, Fainberg J, Mungovan SF, Bratt O, Bray F, Brawley O, Luckenbaugh AN, Mucci L, Morgan TM, Carlsson SV. 2022 update on prostate cancer epidemiology and risk factors-a systematic review. Eur Urol. 2023;84:191–206. doi: 10.1016/j.eururo.2023.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang W, Cao G, Wu F, Wang Y, Liu Z, Hu H, Xu K. Global burden of prostate cancer and association with socioeconomic status, 1990-2019: a systematic analysis from the global burden of disease study. J Epidemiol Glob Health. 2023;13:407–421. doi: 10.1007/s44197-023-00103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simbaña-Rivera K, Torres-Roman JS, Challapa-Mamani MR, Guerrero J, De la Cruz-Ku G, Ybaseta-Medina J, Martinez-Herrera JF. Regional disparities of prostate cancer mortality in Ecuador: an examination of trends and correlates from 2004 to 2019. BMC Public Health. 2023;23:992. doi: 10.1186/s12889-023-15941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adus-Salam A, Jimoh M, Ehiedu CG. Sexual characteristics of patients with prostate cancer seen for radiation treatment. Ecancermedicalscience. 2023;17:1577. doi: 10.3332/ecancer.2023.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hameed Y, Usman M, Liang S, Ejaz S. Novel diagnostic and prognostic biomarkers of colorectal cancer: capable to overcome the heterogeneity-specific barrier and valid for global applications. PLoS One. 2021;16:e0256020. doi: 10.1371/journal.pone.0256020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hameed Y, Usman M, Ahmad M. Does mouse mammary tumor-like virus cause human breast cancer? Applying Bradford Hill criteria postulates. Bull Natl Res Cent. 2020;44:1–13. [Google Scholar]
- 7.Madu CO, Lu Y. Novel diagnostic biomarkers for prostate cancer. J Cancer. 2010;1:150–77. doi: 10.7150/jca.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usman M, Okla MK, Asif HM, AbdElgayed G, Muccee F, Ghazanfar S, Ahmad M, Iqbal MJ, Sahar AM, Khaliq G, Shoaib R, Zaheer H, Hameed Y. A pan-cancer analysis of GINS complex subunit 4 to identify its potential role as a biomarker in multiple human cancers. Am J Cancer Res. 2022;12:986–1008. [PMC free article] [PubMed] [Google Scholar]
- 9.Graffeo R, Rana HQ, Conforti F, Bonanni B, Cardoso MJ, Paluch-Shimon S, Pagani O, Goldhirsch A, Partridge AH, Lambertini M, Garber JE. Moderate penetrance genes complicate genetic testing for breast cancer diagnosis: ATM, CHEK2, BARD1 and RAD51D. Breast. 2022;65:32–40. doi: 10.1016/j.breast.2022.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manchanda R, Patel S, Gordeev VS, Antoniou AC, Smith S, Lee A, Hopper JL, MacInnis RJ, Turnbull C, Ramus SJ, Gayther SA, Pharoah PDP, Menon U, Jacobs I, Legood R. Cost-effectiveness of population-based BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2 mutation testing in unselected general population women. J Natl Cancer Inst. 2018;110:714–725. doi: 10.1093/jnci/djx265. [DOI] [PubMed] [Google Scholar]
- 11.Lee A, Mavaddat N, Cunningham A, Carver T, Ficorella L, Archer S, Walter FM, Tischkowitz M, Roberts J, Usher-Smith J, Simard J, Schmidt MK, Devilee P, Zadnik V, Jürgens H, Mouret-Fourme E, De Pauw A, Rookus M, Mooij TM, Pharoah PP, Easton DF, Antoniou AC. Enhancing the BOADICEA cancer risk prediction model to incorporate new data on RAD51C, RAD51D, BARD1 updates to tumour pathology and cancer incidence. J Med Genet. 2022;59:1206–1218. doi: 10.1136/jmedgenet-2022-108471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullah L, Hameed Y, Ejaz S, Raashid A, Iqbal J, Ullah I, Ejaz SA. Detection of novel infiltrating ductal carcinoma-associated BReast CAncer gene 2 mutations which alter the deoxyribonucleic acid-binding ability of BReast CAncer gene 2 protein. J Cancer Res Ther. 2020;16:1402–1407. doi: 10.4103/jcrt.JCRT_861_19. [DOI] [PubMed] [Google Scholar]
- 13.Hameed Y, Ejaz S. TP53 lacks tetramerization and N-terminal domains due to novel inactivating mutations detected in leukemia patients. J Cancer Res Ther. 2021;17:931–937. doi: 10.4103/jcrt.JCRT_536_19. [DOI] [PubMed] [Google Scholar]
- 14.Oh M, Alkhushaym N, Fallatah S, Althagafi A, Aljadeed R, Alsowaida Y, Jeter J, Martin JR, Babiker HM, McBride A, Abraham I. The association of BRCA1 and BRCA2 mutations with prostate cancer risk, frequency, and mortality: a meta-analysis. Prostate. 2019;79:880–895. doi: 10.1002/pros.23795. [DOI] [PubMed] [Google Scholar]
- 15.Sundararajan S, Ahmed A, Goodman OB Jr. The relevance of BRCA genetics to prostate cancer pathogenesis and treatment. Clin Adv Hematol Oncol. 2011;9:748–755. [PubMed] [Google Scholar]
- 16.Dominguez-Valentin M, Sampson JR, Seppälä TT, Ten Broeke SW, Plazzer JP, Nakken S, Engel C, Aretz S, Jenkins MA, Sunde L, Bernstein I, Capella G, Balaguer F, Thomas H, Evans DG, Burn J, Greenblatt M, Hovig E, de Vos Tot Nederveen Cappel WH, Sijmons RH, Bertario L, Tibiletti MG, Cavestro GM, Lindblom A, Della Valle A, Lopez-Köstner F, Gluck N, Katz LH, Heinimann K, Vaccaro CA, Büttner R, Görgens H, Holinski-Feder E, Morak M, Holzapfel S, Hüneburg R, Knebel Doeberitz MV, Loeffler M, Rahner N, Schackert HK, Steinke-Lange V, Schmiegel W, Vangala D, Pylvänäinen K, Renkonen-Sinisalo L, Hopper JL, Win AK, Haile RW, Lindor NM, Gallinger S, Le Marchand L, Newcomb PA, Figueiredo JC, Thibodeau SN, Wadt K, Therkildsen C, Okkels H, Ketabi Z, Moreira L, Sánchez A, Serra-Burriel M, Pineda M, Navarro M, Blanco I, Green K, Lalloo F, Crosbie EJ, Hill J, Denton OG, Frayling IM, Rødland EA, Vasen H, Mints M, Neffa F, Esperon P, Alvarez K, Kariv R, Rosner G, Pinero TA, Gonzalez ML, Kalfayan P, Tjandra D, Winship IM, Macrae F, Möslein G, Mecklin JP, Nielsen M, Møller P. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med. 2020;22:15–25. doi: 10.1038/s41436-019-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiwari AK, Roy HK, Lynch HT. Lynch syndrome in the 21st century: clinical perspectives. QJM. 2016;109:151–158. doi: 10.1093/qjmed/hcv137. [DOI] [PubMed] [Google Scholar]
- 18.Cerretelli G, Ager A, Arends MJ, Frayling IM. Molecular pathology of Lynch syndrome. J Pathol. 2020;250:518–531. doi: 10.1002/path.5422. [DOI] [PubMed] [Google Scholar]
- 19.Satam H, Joshi K, Mangrolia U, Waghoo S, Zaidi G, Rawool S, Thakare RP, Banday S, Mishra AK, Das G, Malonia SK. Next-generation sequencing technology: current trends and advancements. Biology (Basel) 2023;12:997. doi: 10.3390/biology12070997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morganti S, Tarantino P, Ferraro E, D’Amico P, Viale G, Trapani D, Duso BA, Curigliano G. Complexity of genome sequencing and reporting: next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit Rev Oncol Hematol. 2019;133:171–182. doi: 10.1016/j.critrevonc.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Barros-Silva D, Marques CJ, Henrique R, Jerónimo C. Profiling DNA methylation based on next-generation sequencing approaches: new insights and clinical applications. Genes (Basel) 2018;9:429. doi: 10.3390/genes9090429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato Y, Sugie R, Tsuchiya B, Kameya T, Natori M, Mukai K. Comparison of the DNA extraction methods for polymerase chain reaction amplification from formalin-fixed and paraffin-embedded tissues. Diagn Mol Pathol. 2001;10:265–271. doi: 10.1097/00019606-200112000-00009. [DOI] [PubMed] [Google Scholar]
- 23.Brown RAM, Epis MR, Horsham JL, Kabir TD, Richardson KL, Leedman PJ. Total RNA extraction from tissues for microRNA and target gene expression analysis: not all kits are created equal. BMC Biotechnol. 2018;18:16. doi: 10.1186/s12896-018-0421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudmundsson S, Singer-Berk M, Watts NA, Phu W, Goodrich JK, Solomonson M Genome Aggregation Database Consortium. Rehm HL, MacArthur DG, O’Donnell-Luria A. Variant interpretation using population databases: lessons from gnomAD. Hum Mutat. 2022;43:1012–1030. doi: 10.1002/humu.24309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Liu X, Yue M, Liu Z, Zhang Y, Ma Y, Luo J, Li W, Bai J, Yao H, Chen Y, Li X, Feng D, Song X. Identification of hub genes in bladder cancer based on weighted gene co-expression network analysis from TCGA database. Cancer Rep (Hoboken) 2022;5:e1557. doi: 10.1002/cnr2.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;9:P3. [PubMed] [Google Scholar]
- 28.Liu CJ, Hu FF, Xie GY, Miao YR, Li XW, Zeng Y, Guo AY. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief Bioinform. 2023;24:bbac558. doi: 10.1093/bib/bbac558. [DOI] [PubMed] [Google Scholar]
- 29.Kossaï M, Leary A, Scoazec JY, Genestie C. Ovarian cancer: a heterogeneous disease. Pathobiology. 2018;85:41–49. doi: 10.1159/000479006. [DOI] [PubMed] [Google Scholar]
- 30.Prat J. Ovarian carcinomas: five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012;460:237–249. doi: 10.1007/s00428-012-1203-5. [DOI] [PubMed] [Google Scholar]
- 31.Shah S, Rachmat R, Enyioma S, Ghose A, Revythis A, Boussios S. BRCA mutations in prostate cancer: assessment, implications and treatment considerations. Int J Mol Sci. 2021;22:12628. doi: 10.3390/ijms222312628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vietri MT, D’Elia G, Caliendo G, Resse M, Casamassimi A, Passariello L, Albanese L, Cioffi M, Molinari AM. Hereditary prostate cancer: genes related, target therapy and prevention. Int J Mol Sci. 2021;22:3753. doi: 10.3390/ijms22073753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boussios S, Rassy E, Moschetta M, Ghose A, Adeleke S, Sanchez E, Sheriff M, Chargari C, Pavlidis N. BRCA mutations in ovarian and prostate cancer: bench to bedside. Cancers (Basel) 2022;14:3888. doi: 10.3390/cancers14163888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pohlreich P, Zikan M, Stribrna J, Kleibl Z, Janatova M, Kotlas J, Zidovska J, Novotny J, Petruzelka L, Szabo C, Matous B. High proportion of recurrent germline mutations in the BRCA1 gene in breast and ovarian cancer patients from the Prague area. Breast Cancer Res. 2005;7:R728–36. doi: 10.1186/bcr1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laraqui A, Uhrhammer N, Lahlou-Amine I, El Rhaffouli H, El Baghdadi J, Dehayni M, Moussaoui RD, Ichou M, Sbitti Y, Al Bouzidi A, Amzazi S, Bignon YJ. Mutation screening of the BRCA1 gene in early onset and familial breast/ovarian cancer in Moroccan population. Int J Med Sci. 2013;10:60–67. doi: 10.7150/ijms.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardoso FC, Goncalves S, Mele PG, Liria NC, Sganga L, Diaz Perez I, Podesta EJ, Solano AR. BRCA1 and BRCA2 mutations and clinical interpretation in 398 ovarian cancer patients: comparison with breast cancer variants in a similar population. Hum Genomics. 2018;12:39. doi: 10.1186/s40246-018-0171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fanale D, Pivetti A, Cancelliere D, Spera A, Bono M, Fiorino A, Pedone E, Barraco N, Brando C, Perez A, Guarneri MF, Russo TDB, Vieni S, Guarneri G, Russo A, Bazan V. BRCA1/2 variants of unknown significance in hereditary breast and ovarian cancer (HBOC) syndrome: looking for the hidden meaning. Crit Rev Oncol Hematol. 2022;172:103626. doi: 10.1016/j.critrevonc.2022.103626. [DOI] [PubMed] [Google Scholar]
- 38.Witjes VM, van Bommel MHD, Ligtenberg MJL, Vos JR, Mourits MJE, Ausems MGEM, de Hullu JA, Bosse T, Hoogerbrugge N. Probability of detecting germline BRCA1/2 pathogenic variants in histological subtypes of ovarian carcinoma. A meta-analysis. Gynecol Oncol. 2022;164:221–230. doi: 10.1016/j.ygyno.2021.10.072. [DOI] [PubMed] [Google Scholar]
- 39.Aubrey BJ, Strasser A, Kelly GL. Tumor-suppressor functions of the TP53 pathway. Cold Spring Harb Perspect Med. 2016;6:a026062. doi: 10.1101/cshperspect.a026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soussi T. The p53 tumor suppressor gene: from molecular biology to clinical investigation. Ann N Y Acad Sci. 2000;910:121–139. doi: 10.1111/j.1749-6632.2000.tb06705.x. [DOI] [PubMed] [Google Scholar]
- 41.Li VD, Li KH, Li JT. TP53 mutations as potential prognostic markers for specific cancers: analysis of data from The Cancer Genome Atlas and the International Agency for Research on Cancer TP53 Database. J Cancer Res Clin Oncol. 2019;145:625–636. doi: 10.1007/s00432-018-2817-z. [DOI] [PubMed] [Google Scholar]
- 42.Goel S, Hall J, Pradhan K, Hirsch C, Przychodzen B, Shastri A, Mantzaris I, Janakiram M, Battini R, Kornblum N, Derman O, Gritsman K, Al-Hafidh J, Wang Y, Halmos B, Steidl U, Maciejewski JP, Braunschweig I, Verma A. High prevalence and allele burden-independent prognostic importance of p53 mutations in an inner-city MDS/AML cohort. Leukemia. 2016;30:1793–1795. doi: 10.1038/leu.2016.74. [DOI] [PubMed] [Google Scholar]
- 43.Baretti M, Le DT. DNA mismatch repair in cancer. Pharmacol Ther. 2018;189:45–62. doi: 10.1016/j.pharmthera.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell RJ, Farrington SM, Dunlop MG, Campbell H. Mismatch repair genes hMLH1 and hMSH2 and colorectal cancer: a HuGE review. Am J Epidemiol. 2002;156:885–902. doi: 10.1093/aje/kwf139. [DOI] [PubMed] [Google Scholar]
- 45.Voutsadakis IA, Stravodimou A. Homologous recombination defects and mutations in DNA damage response (DDR) genes besides BRCA1 and BRCA2 as breast cancer biomarkers for PARP inhibitors and other DDR targeting therapies. Anticancer Res. 2023;43:967–981. doi: 10.21873/anticanres.16241. [DOI] [PubMed] [Google Scholar]
- 46.Lin K, Baritaki S, Vivarelli S, Falzone L, Scalisi A, Libra M, Bonavida B. The breast cancer protooncogenes HER2, BRCA1 and BRCA2 and their regulation by the iNOS/NOS2 axis. Antioxidants (Basel) 2022;11:1195. doi: 10.3390/antiox11061195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomasova K, Cumova A, Seborova K, Horak J, Koucka K, Vodickova L, Vaclavikova R, Vodicka P. DNA repair and ovarian carcinogenesis: impact on risk, prognosis and therapy outcome. Cancers (Basel) 2020;12:1713. doi: 10.3390/cancers12071713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Figueroa BAC, Guzmán BIL, Rodríguez RR, del Campo FDJM, García JIP, García EMI. Role of tumor suppressor genes in carcinogenesis: a narrative review. Int J Med Sci Clin Res Stud. 2023;3:2707–2716. [Google Scholar]
- 49.Rascio F, Spadaccino F, Rocchetti MT, Castellano G, Stallone G, Netti GS, Ranieri E. The pathogenic role of PI3K/AKT pathway in cancer onset and drug resistance: an updated review. Cancers (Basel) 2021;13:3949. doi: 10.3390/cancers13163949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konecny GE, Kristeleit RS. PARP inhibitors for BRCA1/2-mutated and sporadic ovarian cancer: current practice and future directions. Br J Cancer. 2016;115:1157–1173. doi: 10.1038/bjc.2016.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiewe T, Haran TE. How mutations shape p53 interactions with the genome to promote tumorigenesis and drug resistance. Drug Resist Updat. 2018;38:27–43. doi: 10.1016/j.drup.2018.05.001. [DOI] [PubMed] [Google Scholar]