Abstract
Extracellular vesicles (EVs) have garnered significant attention in biomedical applications. However, the rapid, efficient, and unbiased separation of EVs from complex biological fluids remains a challenge due to their heterogeneity and low abundance in biofluids. Herein, we report a novel approach to reconfigure and modify an artificial insertion peptide for the unbiased and rapid isolation of EVs in 20 min with ∼80% recovery in neutral conditions. Moreover, the approach demonstrates exceptional anti-interference capability and achieves a high purity of EVs comparable to standard ultracentrifugation and other methods. Importantly, the isolated EVs could be directly applied for downstream protein and nucleic acid analyses, including proteomics analysis, exome sequencing analysis, as well as the detection of both epidermal growth factor receptor (EGFR) and V-Ki-ras2 Kirsten Rat Sarcoma Viral Oncogene Homologue (KRAS) gene mutation in clinical plasma samples. Our approach offers great possibilities for utilizing EVs in liquid biopsy, as well as in various other biomedical applications.
Keywords: Extracellular vesicles, Peptide, Protein assay, DNA mutation detection, Functionalized interface
Graphical abstract
Highlights
-
•
We present a novel approach for the unbiased isolation of EVs with high purity.
-
•
An artificial insertion peptide was reconfigured and modified to achieve the purpose.
-
•
The BAIP system enables unbiased isolation of EVs with ∼80 % recovery in 20 min.
-
•
The isolated EVs can be directly utilized for various downstream protein and nucleic acid analyses.
-
•
We successfully detected gene mutations in EVs from plasma in clinical samples.
1. Introduction
Extracellular vesicles (EVs) are cell secreted vesicle substances with a phospholipid bilayer structure and sizes ranging from 30 nm to 1000 nm [1,2]. They are present in various body fluids, such as blood, urine, tear ascites and saliva. Previous studies reveal that EVs contain diverse nucleic acids (RNA and DNA), proteins, lipid and other substances inherited from their original parent cells, and participate in various physiological activities, such as intercellular communication [3,4], tissue repair and regeneration [5], and immunomodulation [[6], [7], [8]]. Additionally, numerous studies have shown that EVs play important roles in tumor progression processes, including epithelial-mesenchymal transition [9,10], angiogenesis [11,12], immune system avoidance [13], cancer migration, invasion and metastasis [14,15], and drug resistance [16,17]. Thus, EVs have been recognized as promising biomarkers in liquid biopsy, which is crucial for early cancer diagnosis and improvement of survival rate [15,18,19]. However, the detection of tumor-associated EVs encounters challenges due to their ultra-trace concentration at the early stage of cancer and the complex matrix interference in the sample [3,20,21]. Therefore, the rapid, simple and highly efficient isolation and enrichment of EVs from complex clinical samples is the first key issue for the EVs based biomedical applications [[22], [23], [24], [25]].
Several label-free methods have been developed for isolation and enrichment of EVs, including the gold standard method of ultracentrifugation (UC) [[26], [27], [28]], polymer-based precipitation [29,30] and microfluidic-based methods [[31], [32], [33]]. However, these methods often require large sample volumes, multiple tedious treatment steps, a relatively long processing time (∼4 h), as well as poor yields of EVs (5%–20%) [26]. Sometimes, they also cause significant aggregation with unacceptable purity, cumbersome chip fabrication and operation procedures, which limit their applications in clinical trials [26,[34], [35], [36], [37]]. Further, immunoaffinity approaches, such as specific antibody or aptamers-based isolation methods, have also been applied for the separation of EVs [23,38,39]. However, the EVs exhibit heterogeneity [[40], [41], [42]], which leads to a bias in the separation of EVs with high expression of specific antigen. The EVs with low or no expression of specific antigen will be missed, which could impede the efficient enrichment of tumor-associated EVs, and lead to the omission of crucial cancer-related information, as well as hinder the discovery of new biomarkers on EVs [3]. Moreover, the high cost and challenging storage of the antibodies, as well as the susceptibility to degradation of aptamers, have also hindered the efficient capture of EVs. To address the challenges, we previously used a lipid-nanoprobe (LNP), biotin-tagged 1,2-distearoyl-sn-glycero-3-phosphethanolamine-poly (ethylene glycol) (DSPE-PEG), inserted into the membranes of EVs for the separation and enrichment of EVs [43]. The results demonstrated that more than 80% of EVs were successfully recovered in the buffer solution. However, in clinical plasma samples, only ∼50% capture efficiency was achieved, which might be attributed to the interferences of complex ingredients in plasma. Recently, a study reported unbiased isolation of EVs using choline phosphate-functionalized magnetic beads [44]. The isolation mechanism relied on the coordination between choline phosphate on magnetic beads and phosphatidylcholine on EV membranes. Although this study was impressive, the process of functionalizing magnetic beads with choline phosphate was complicated. Thus, there is a demand for developing novel unbiased, rapid and highly efficient approaches to enrich EVs for diverse applications.
In previous studies, many cell penetrating peptides, such as HIV-1 TAT peptide, antennapedia peptide and other artificially designed peptides, possessed the ability to penetrate the lipid bilayer of the cells, and they have been employed for targeted drug delivery, cancer imaging and other applications [[45], [46], [47]]. Despite possessing the ability to penetrate the cell membrane, few of them have been considered for the isolation of EVs. Among them, a 36-residue polypeptide, namely pH-low insertion peptide (pHLIP), derived from the C helix of bacteriorhodopsin, was discovered by Engelman's lab [48]. The wild-type pHLIP (WT pHLIP) is capable of stably inserting into the phospholipid bilayer of cells in an α-helical conformation under an acidic microenvironment. Under neutral conditions, the WT pHLIP assumes nonhelical and peripheral conformations while retaining the ability to bind to the phospholipid bilayer. However, this binding is reversible and unstable under natural biological conditions [49]. That's the reason that the WT pHLIP is frequently utilized for targeted drug delivery owing to its acid-responsive property [49,50], but it has been rarely employed for EVs isolation and enrichment. Zong et al. [51] recently introduced a technique aimed at creating a double-switch pHLIP to target the tumor microenvironment (TME), while incorporating a pH-driven hook-like switch to prevent pHLIP dissociation during circulation. While this work could selectively isolate TME-associated EVs in vivo, it may inadvertently sacrifice some heterogeneous information from EVs.
We initially hypothesized that the pHLIP might be employed for unbiased isolation of EVs by leveraging the strategy of “old tricks catch new birds”, given the structural similarity between the EVs and cell membrane. However, it became evident that the WT pHLIP was not suitable for isolating EVs within common biological microenvironments. Therefore, it is necessary to reconfigure and modify the WT pHLIP peptide, enabling the peptide to transform into a more stable α-helical conformation under neutral conditions and successfully insert into the phospholipid bilayer of EVs. Hence, we engineered an artificial peptide for the first time to unbiasedly isolate EVs within 20 min, achieving an isolation efficiency of ∼80% and high purity. Compared to the existing EV isolation technologies, our research introduced distinct innovations in unbiased isolation of EVs via peptide engineering and the study of EV biomarkers, with a focus on improving the translatability for large-scale clinical and biomedical applications. This sets our work stand out from the current methods, positioning it as a promising avenue for advancing the field.
2. Results
2.1. Design, preparation and characterization of engineered peptide
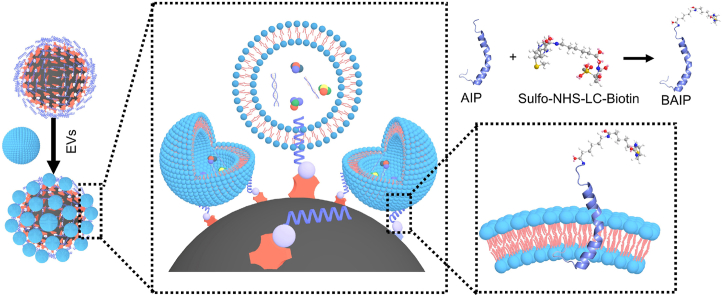
The overall workflow of this study is illustrated in Fig. 1. Our objective was to establish a novel method for unbiased isolation of EVs from biological samples and explore downstream applications, including nucleic acid and protein analysis with various technologies, to enhance translatability for clinical and biomedical purposes. In this study, we chose WT pHLIP as a basic template for reconfiguration and functionalization to isolate EVs. We selected it due to its evolution in nature, forming a transmembrane α-helix in acidic microenvironments (pH < 6.5) and spontaneously inserting into the phospholipid bilayer [48,49]. As mentioned above, we would like to reconfigure and modify the WT pHLIP in order to form α-helical conformation under neutral conditions. As is well-established, enhancing the appropriate hydrophobicity and intramolecular hydrogen bonds of a peptide could be an efficient approach to render it a more stable α-helical conformation [52,53]. We changed the N-terminal amino acid of WT pHLIP to increase the hydrophobicity and promote the formation of intramolecular hydrogen bonds, thereby facilitating formation of α-helical structure under natural pH conditions. Meanwhile, we intended to maintain the stability and unchanged nature of the remaining amino acids. This approach was driven by the fact that the middle segment of pHLIP forms an α-helix conformation, while the C-terminal of pHLIP inserts into the phospholipid bilayer (depicted as the orange and green regions in Table S2). Nevertheless, there are still millions of reconfigurations available to enhance the hydrophobicity of the pHLIP. We calculated the grand average of hydrophilicity (GRAVY) values, which reflected their relevant hydrophilic properties. The GRAVY values of 20 common amino acids and part of reconfigured peptide were shown in Table S1 and Table S2, respectively. A higher positive GRAVY value indicates a stronger hydrophobic characteristic. Thus, we meticulously selected a reconfiguration that enhanced appropriate hydrophobicity of the peptide while maintaining a balance and avoiding excessive hydrophobicity, which could lead to poor water solubility.
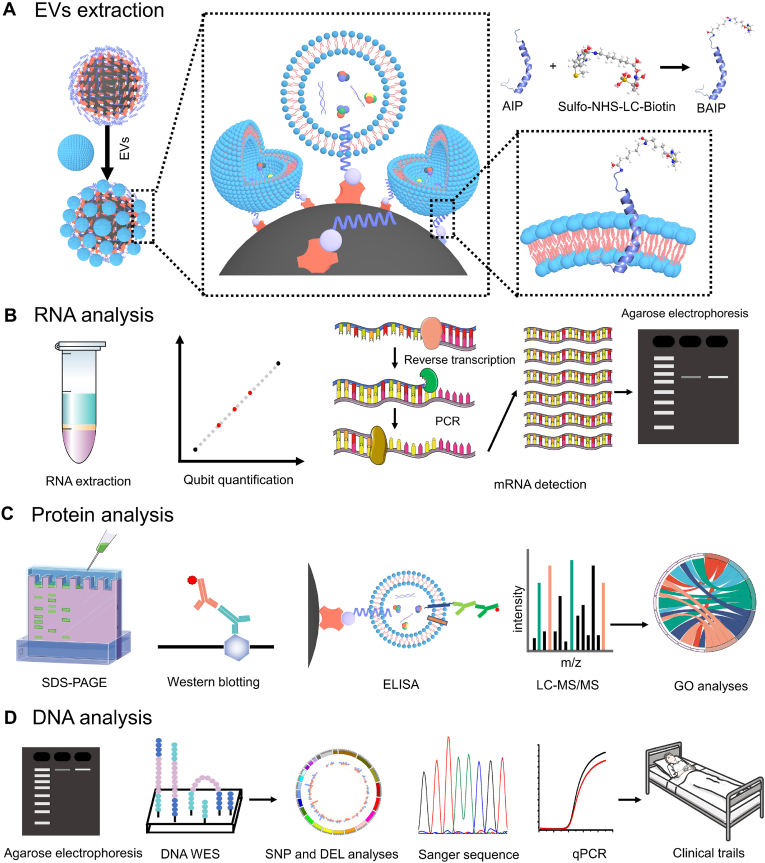
Fig. 1.
Scheme of the BAIP-based EVs enrichment and downstream analyses. (A) Scheme of capture of EVs with the BAIP-based method. (B) Scheme of the extraction of cargo RNA contents from isolated EVs and the subsequent RNA quantification, mRNA detection and RNA agarose electrophoresis characterization. (C) Scheme of the protein analysis, such as SDS-PAGE, Western blotting, enzyme-linked immunosorbent assay (ELISA) and LC-MS/MS. (D) Scheme of the DNA analysis, such as DNA electrophoresis characterization, whole exome sequencing (WES), sanger sequencing and real-time PCR (qPCR) for DNA mutation detection.
To achieve this, we reconfigured the N terminal of Cysteine (Cys) and Alanine (Ala) amino acids instead of Glycines (Gly) from the WT pHLIP and functionalized the artificial insertion peptide (AIP, Table S3) with biotin. This transformation changed the R groups of the corresponding amino acids from -H to -CH3 and -CH2SH, enhancing the hydrophobicity. We observed the GRAVY value increased from 0.158 (WT pHLIP) to 0.300 (AIP), which might be appropriate for increasing the hydrophobicity of AIP, promoting the formation of α-helix conformation. Moreover, we modified the N-terminal amino acid of AIP by introducing a biotin moiety with a relatively long alkyl chain (Fig. 2A), resulting in a little increase in hydrophobicity. Additionally, the biotin functionalized artificial insertion peptide (BAIP) facilitated the formation of intramolecular hydrogen bonds, further contributing to stabilize the α-helix structure. In order to verify above hypothesis, we used circular dichroism to measure the conformation of AIP and BAIP under neutral (pH 7.4) and weakly acidic conditions (pH 6.5). As depicted in Fig. 2B, the AIP exhibited an α-helix conformation (∼210 nm, ∼220 nm) under neutral condition, which was different from that of WT pHLIP (Fig. S1). Additionally, it also showed strong random coil structure (∼230 nm). These findings were consistent with the observations under acidic condition (Fig. S2). After modification with biotin, the α-helix conformation of AIP was maintained under both conditions. It is noteworthy that the random coil structure of BAIP was weakened than AIP (Fig. 2B and Fig. S2), which was consistent with the above hypothesis. Notwithstanding, Fig. S3 showed the successful functionalization of the AIP with biotin, and the almost identical peaks were observed for BAIP under neutral and acidic conditions, implying the BAIP might display similar structure and conformation under both conditions.
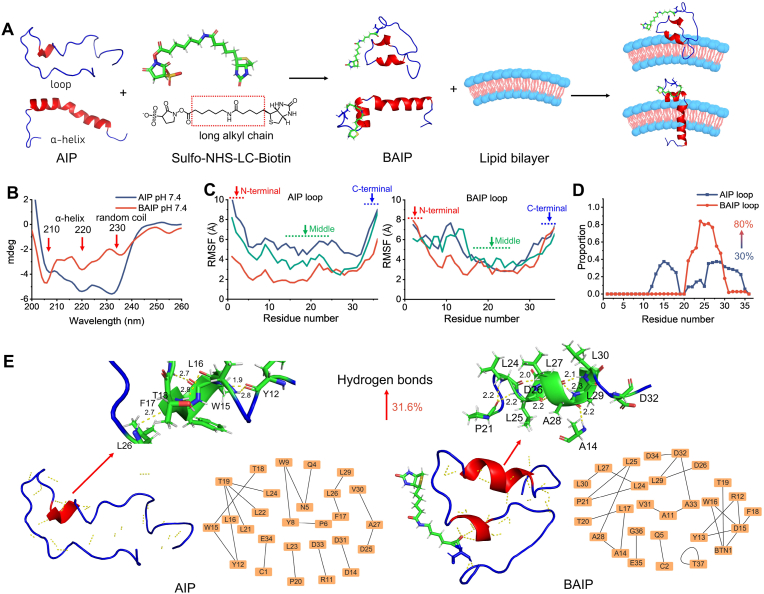
Fig. 2.
Design and engineering reconfiguration of WT pHLIP to BAIP. (A) Schematic of AIP modified with biotin and inserted into the phospholipid bilayer. (B) Circular dichroism spectra of AIP and BAIP at pH 7.4. (C) Root Mean Square Fluctuation (RMSF) of AIP and BAIP with loop conformation. The three spectra (red, blue, green) were the results of three independent simulation trajectories. RMSF refers to the structural change of an atom from its initial conformation over a period of time, reflecting the degree of freedom and flexibility of the atom. The results showed that the biotin modification had a notable effect not only on the N-terminal amino acid structure, but also on the middle and posterior amino acids of AIP when loop state was used as the initial model. (D) Proportion of each amino acid forming α-helix structure of AIP and BAIP with loop state. (E) Intramolecular hydrogen bonds of reconfigured AIP and BAIP with loop conformation analyzed by PyMOL, respectively. The enlarged hydrogen bonds diagrams were in AIP between residues L16-T18 and in BAIP between L25-L30, respectively. The unit of hydrogen bond length is Å.
We then used molecular dynamics simulations to study the changes of structure and conformation of AIP and BAIP. According to the results of circular dichroism, the AIP and BAIP exhibited α-helix and loop conformations under both conditions, which were simulated as initial states. We observed that the overall structural changes in BAIP were more pronounced compared to AIP, irrespective of whether the initial model was in α-helix or loop state (Fig. S4). This indicated that biotin modification might interact with AIP (such as hydrogen bonds), resulting in more substantial structural alterations. For more details, when α-helix state was used as the initial model for simulation, the biotin modification exhibited a more significant impact on the amino acid structure of the N-terminal of AIP. However, it had minimal effect on the middle and C-terminal portions, leading to a significantly stable structure (Fig. S5). When loop state was used as the initial model, the biotin modification had a notable effect not only on the N-terminal amino acid structure, but also on the middle and posterior amino acids of AIP. This effect was observed as reduced flexibility of amino acids, providing the evidence that the biotin modification promoted the formation of secondary structure in AIP (Fig. 2C). Furthermore, we utilized the Dictionary of Secondary Structure of Proteins (DSSP) to analyze each amino acid and statistically determine the probability of forming different secondary structures (Fig. S6). The statistical results indicated that when initial state was α-helix state, the AIP had a high probability of forming a helical secondary structure between N5-D31, which remained stable even after the biotin modification (Fig. S7, Tables S4–S5). When AIP was initially in the loop state, apart from amino acids A13-T18 and L26-E34 which had an approximately 30% probability to form helical secondary structure, the residual amino acids in AIP predominantly exhibited no secondary structure. However, the formation probability of helical secondary structure of BAIP was significantly improved, with segments L24-L27 reaching a helix probability exceeding 80% (Fig. 2D, Tables S6–S7), indicating that the modification of biotin significantly improved the formation probability of helical secondary structure.
In addition, the formation of intramolecular hydrogen bonds of AIP and BAIP was also analyzed (datasets S1). When the initial state of AIP was in a helical conformation, the modification with biotin predominantly resulted in intramolecular hydrogen bonds with N6, Q5, and W10 (Fig. S8). In the case of the initial state being a loop conformation, the intramolecular hydrogen bonds were relatively simple, with only three amino acids (L16-T18) forming a helical structure, while the remaining segments lacked any secondary structure (Fig. 2E, left). Following the biotin modification of AIP, the amino acids from D15-F18 and L25-L30 formed a helical structure, and biotin formed intramolecular hydrogen bonds with D15, W16, and T19, which contributed to the stability of the helical conformation, compared with AIP, the intramolecular hydrogen bonds of BAIP was increased by 31.6% (Fig. 2E right, Fig. S9). In summary, the AIP exhibited both α-helix and loop conformations under neutral conditions, and the biotin modification facilitated the further formation and stability of the α-helical conformation.
2.2. Isolation and enrichment of EVs through BAIP-based system
We then performed experiments to validate the interaction between BAIP and cells. The efficiency of BAIP in inserting into the phospholipid bilayer of cells was evaluated using MCF-7 cells as model in PBS buffer at pH 7.4, while Cy3 modified streptavidin (SA-Cy3) was used to characterize the interactions. As shown in Figs. S10A–B, BAIP treated cells displayed a robust fluorescence signal on cell membranes as opposed to those untreated cells. All the cells treated with BAIP exhibited brilliant red fluorescence signals, which was further confirmed by flow cytometry analysis (Fig. S10C). These observations suggested that BAIP could proficiently insert into the phospholipid bilayer of cells under neutral condition (pH 7.4), potentially facilitating the separation and enrichment of EVs. Moreover, the binding between BAIP and cells was fast and 20 min incubation was sufficient for BAIP to insert into the cell membrane (Fig. S11). We then compared the efficiency of BAIP in inserted into the cell membrane under different pH conditions. Compared with biotin-modified WT pHLIP, which showed acid selectivity on the interaction with cell membrane (Figs. S12A–B), the BAIP exhibited the comparable efficiency under a neutral condition to that under an acidic condition (Figs. S12C–D), supporting the success reconfiguration of the WT pHLIP. Furthermore, there was no obvious decrease in cell viability even if the concentration of BAIP was as high as 100 μg/mL, demonstrating its favorable biocompatibility (Fig. S13). We investigated the ability of BAIP to insert into the phospholipid bilayer under both neutral and weakly acidic conditions, examining its efficiency across normal cells and various tumor cells. HeLa and Hepa1-6 cells were utilized to represent distinct tumor types, whereas MCF-10A, MCF-7, and MDA-MB-231 cells were employed to portray normal breast cells, non-metastatic, and highly metastatic breast cancer cells, respectively. These cells were incubated with BAIP at pH 6.5 or 7.4, followed by SA-Cy3 treatment for fluorescence imaging. As depicted in Figs. S14–S16, no substantial difference in fluorescence intensity was observed among the various types of tumor cells or normal cells.
Next, we attempted to employ the BAIP to isolate the EVs in model samples. First, the EVs secreted from MCF-7 cells were isolated through UC and identified by transmission electron microscope (TEM) (Fig. S17A). The obtained EVs displayed a saucer-like morphology, with the diameter ranging from 30 nm to 200 nm. Additionally, nanoparticle tracking analysis (NTA) results revealed that the average size of EVs was 193.8 nm, with a concentration of approximately 8.81 × 1010 particles/mL (Figs. S17B–C). The BAIP functionalized polystyrene microspheres (BAIP-PS) were then chosen to unbiasedly separate and enrich EVs. The scheme illustrated the modification of BAIP on PS (Fig. S18) and the details were presented in the supplementary information (Figs. S19–S20). We initially validated the feasibility of the BAIP-PS for unbiased isolation and enrichment of the model EVs (derived from MCF-7 cells). The SEM results clearly showed the presence of large amount of EVs (red arrows) on BAIP-PS compared to the BAIP-PS alone (Fig. 3A and Fig. S20A), supporting the capability of BAIP for isolation and enrichment of EVs. Moreover, the insertion of BAIP did not alter the size or shape of the EVs (Fig. S21), and it did not leak out any internal material of the EVs (Fig. S22). The immuno-fluorescence ELISA experiments were then conducted to prove the feasibility. CD9, a cluster-of-differentiation protein molecule that is widely and extensively expressed on the EVs, was selected as a target protein to verify the isolation of EVs by BAIP. As shown in Fig. 3B, the captured EVs were further recognized by anti-human CD9 antibody and identified by Cy3-labelled IgG. The fluorescence intensity on the BAIP-PS was significantly higher than that of the control groups, implying the successfully capture of EVs (Fig. 3C and Fig. S23).
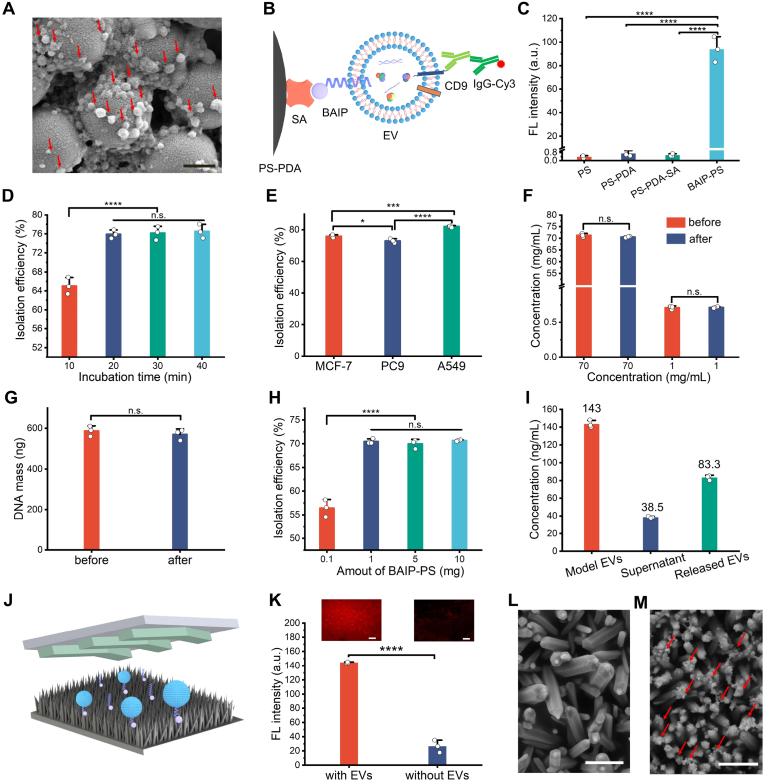
Fig. 3.
The isolation and release of EVs. (A) SEM image of EVs captured on BAIP-PS particles. Scale bar, 1 μm. (B) Diagram of immuno-fluorescence characterization of EVs captured on BAIP-PS. (C) Verification of the feasibility of EVs captured on BAIP-PS by immuno-fluorescence experiments. Data are represented as mean ± S.D. (n = 3). Statistical analysis was assessed by two-tailed Student's t-tests; ∗∗∗∗P < 0.0001. (D) Isolation efficiency versus incubation time for EVs capture. Data are represented as mean ± S.D. (n = 3). Statistical analysis was assessed by two-tailed Student's t-tests; ∗∗∗∗P < 0.0001, n.s. no significance. (E) Isolation efficiency of EVs from different cancer cells. Data are represented as mean ± S.D. (n = 3). Statistical analysis was assessed by one-way ANOVA with Tukey's method; ∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. (F–G) Anti-nonspecific adsorption of BAIP-PS. 1 mg BAIP-PS was incubated with 1 mL solution containing either 70 mg/mL BSA, 1 mg/mL BSA, or 100 μL of 5.7 ng/μL DNA (∼160 bp) at room temperature for 20 min. The adsorption of BSA on BAIP-PS was determined by the mass difference between the initial (before) and remaining (after) amounts of BSA. The measurement of adsorption of DNA on the BAIP-PS was similarly determined by mass difference. The “before” represents the mass of initial DNA without incubation with BAIP-PS, while the “after” represents the mass of the remained DNA after incubation with BAIP-PS. Data are represented as mean ± S.D. (n = 3). Statistical analysis was assessed by two-tailed Student's t-tests; n.s. no significance. (H) Isolation of EVs from plasma samples as a function of dosage of BAIP-PS. Data are represented as mean ± S.D. (n = 3). Statistical analysis was assessed by one-way ANOVA with Tukey's method; ∗∗∗∗P < 0.0001, n.s. no significance. (I) Release of EVs. The released efficiency of EVs was calculated according to Equation (2) in the supplementary information. (J) Scheme of BAIP modified ZnO microfluidic chip for EVs capture. (K) Fluorescence image of EVs captured on ZnO microfluidic chip and the corresponding fluorescence intensities. Scale bar, 50 μm. Data are represented as mean ± S.D. (n = 3). Statistical analysis was assessed by two-tailed Student's t-tests; ∗∗∗∗P < 0.0001. (L–M) SEM images of EVs captured on ZnO microfluidic chip (M) and the corresponding control group without EVs (L). Scale bar, 1 μm.
Next, the capture parameter was optimized and capture efficiency was evaluated. It should be noted that no standard method is available for EVs quantification at present. Current methods, such as NTA and protein detection, are limited by their failure to specifically identify EVs and interference from non-exosomal particles or proteins. As an alternative, the detection of RNA in EVs is considered to be a more reliable method, as RNA molecules are protected by lipid membranes, leading to enhanced stability. The isolation efficiency was determined by calculating the mass fraction of RNA extracted from the captured EVs over that obtained from the total EVs. As depicted in Fig. 3D, the isolation efficiency increased from 65.1% to 76.5% with the incubation time increasing from 10 min to 1 h for capture of EVs by BAIP-PS and reached a plateau at 20 min (P < 0.0001, two-tailed t-test). This observation was also consistent with the results obtained from the interaction of BAIP with the cells (Fig. S11), further demonstrating the fast interaction of BAIP with phospholipid bilayer. Under this condition, the isolation efficiency of EVs using PS-PDA-SA without BAIP modification was less than 2.5%.
To validate the versatile and unbiased enrichment of EVs by BAIP, we utilized the peptide to capture EVs derived from various cell types. Specifically, we selected EVs derived from MCF-7, PC9, and A549 cells, which exhibited high, medium, and low EpCAM expression levels respectively. The results revealed that the isolation efficiencies of EVs from three types of cells were 76%, 72% and 82%, respectively (Fig. 3E). These findings suggested that our proposed method appeared to be capable of capturing EVs from various cell subpopulations with less bias and regardless of the heterogeneity. We further investigated the anti-inference capability towards the protein and nucleic acid as actual plasma/serum samples contain diverse proteins and short DNA fragments that may potentially impact the capture efficiency. As shown in Fig. 3F and G, despite whether the presence of high (70 mg/mL)/low (1 mg/mL) level of proteins or DNA fragments (∼160 bp, 570 ng), the capture efficiency was negligibly (< 1%), supporting that no discernible interaction between the BAIP and interference proteins or DNA.
With excellent performance towards model EVs (derived from MCF-7, PC9, A549 cells), we attempted to isolate EVs from plasma samples. In our previous study, we found that albumin could interfere with the insertion of lipid-nanoprobe (LNP, DSPE-PEG), into the membranes of EVs, leading to increase the quantity of LNP for isolation of EVs in plasma [43]. However, even with excessive lipid probes (200 nmol), we only achieved a 48.3% isolation efficiency. Doubling the amount of the LNP only slightly increased isolation efficiency to 49.5% (not statistically significant; P > 0.05, two-tailed t-test). In contrast, we found that using 1 mg of the BAIP-PS to enrich EVs in plasma resulted in an isolation efficiency of 70%, which was slightly lower than that achieved in PBS buffer (76%). The isolation efficiency of EVs in plasma remained constant at 70–72% regardless of the dosage of BAIP-PS from 1 mg to 20 mg (Fig. 3H) (not statistically significant; P > 0.05, one-way ANOVA with Tukey's method). This could be due to the limitations in reaction dynamics equilibrium between nano-sized EVs and micro-sized PS particles [54,55]. Nevertheless, our proposed BAIP based method achieved superior stability and reliability in isolation efficiency compared to that of LNP-based method, making it potentially more suitable for clinical applications.
The BAIP was also utilized to modify variety of interfaces, such as ZnO nanorods integrated microfluidic chips, PDA coated nickel foams, and SA-functionalized magnetic beads (Fig. 3J, Fig. S24 and Fig. S25A-B). The experimental results indicated that the diverse BAIP-functionalized interfaces could efficiently and unbiasedly isolate and enrich EVs (Fig. 3K–M and Fig. S25C–J). In light of the multiple applications of EVs, including drug delivery and wound healing, release of the captured EVs was equally necessary. To achieve this, we employed Sulfo-NHS-SS-Biotin, a molecule containing a disulfide bond, to modify AIP (referred to as ss-BAIP, Fig. S26). This modification facilitated the release of the captured EVs by cleaving the disulfide bond between biotin and AIP using dithiothreitol (DTT). It should be noted that the DTT treatment did not affect the protein on the surface of EVs (Fig. S27). Immuno-fluorescence analysis showed that fluorescence signal on BAIP-PS disappeared after the release of fluorescence labelled EVs (Fig. S28). The release efficiency was calculated to be 79.5% ± 2.6% (Fig. 3I), indicating that the EVs could be efficiently released and utilized in subsequent functional assays.
2.3. Detection of proteins and nucleic acids in EVs samples
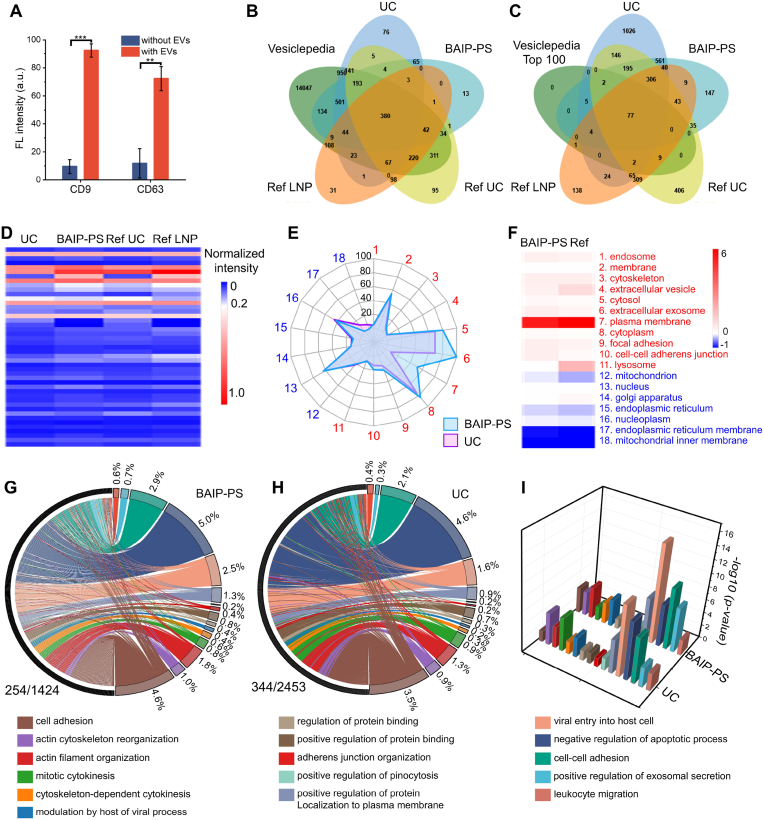
The BAIP-functionalized interfaces facilitate the isolation and enrichment of EVs, allowing for subsequent profiling of proteins and nucleic acids of EVs. We first validated the isolated EVs could be directly used for protein analysis. Initially, proteins on the captured EVs were examined and compared through SDS-PAGE. The outcome revealed numerous diffuse bands in EVs, ranging from 25 kD to 180 kD (Fig. S29A). No significant difference was observed when comparing the protein bands of the isolated EVs through BAIP-PS based approach (Lane 3) with those of the conventional UC-based method (Lane 2). Next, Western blot technique was used to identify the specific membrane proteins on the EVs, CD9 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, housekeeping protein). The results revealed comparable CD9 and GAPDH bands, as photographed in Fig. S29B, indicating that our proposed isolation method was suitable for subsequent protein analysis of EVs. These results were further proved by the immunofluorescence detection of the tetraspanin proteins, CD9 and CD63 (Fig. 4A and Fig. S29C-D).
Fig. 4.
Protein analyses of EVs. (A) The fluorescence detection of CD9 and CD63 proteins on the isolated EVs. Data are represented as mean ± S.D. (n = 3). Statistical analysis was assessed by two-tailed Student's t-tests; ∗∗P < 0.01, ∗∗∗P < 0.001. (B) Venn diagram of the overlap of all proteomic data from BAIP, UC, our previously published LNP method [43], and public EV database Vesiclepedia. (C) Comparation of top 100 proteins in public EV database Vesiclepedia. (D) Protein proportion of GO cellular components in proteomics. (E) Radar chart illustrating the protein proportion involved in the composition proteins and interference proteins of EVs from BAIP and UC-based method. (F) Comparation of the proportion of protein changes involved in composition protein and contaminating protein of our method and Ref. [43]. (G–H) Chord of GO analyses for biological processes involved in tumor progression obtained by EVs. The numerator represented the number of detected gene types that are associated with tumor progression, and the denominator represents the total number of detected gene types. (I) Correlations between the proteome of EVs and tumor regulation pathways. GO analyses for biological process were performed by DAVID database (https://david.ncifcrf.gov/home.jsp). P value for protein enrichment in annotated GO terms (compared to human genome background) was determined by the EASE Score.
Encouraged by the good performance of BAIP to isolate EVs, we used liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify potential proteins of EVs, with the comparison among those from UC (datasets S2), our LNP-based method [43] and EVs proteins listed in the publicly available database Vesiclepedia (www.microvesicles.org). The Venn diagram illustrated the overlap between proteomic data of EVs from BAIP-PS, UC and LNP-based methods, as well as Vesiclepedia (Fig. 4B-C). Approximately 93.9% of the proteins in EVs isolated by our BAIP based method was identified in 17,204 EV cargo proteins listed in Vesiclepedia, while 93.7% of the proteins in EVs isolated by UC was identified. However, in our previously reported LNP-based method, around 87.0% proteins in EVs were identified in the database. When considering the top 100 proteins in the database, our proposed method was able to identify 93 proteins in EVs, while UC and LNP-based methods found 90 and 88 proteins, respectively, indicating a slightly higher efficiency in identifying cargo proteins in EVs. Our LC-MS/MS data were also compared with the LNP-based methods on 30 key proteins in EVs (Table S8) and the results revealed that similar proteins of EVs were found in our method.
The gene ontology (GO) analysis for cellular components of the identified proteins of EVs from BAIP-PS was further conducted and compared with those from UC-based method and our LNP-based method. As depicted in Fig. 4D, the proteins of EVs obtained by BAIP-PS were comparable to those obtained by other methods. Furthermore, the changes of cellular components of the identified proteins from BAIP-PS and UC showed high consistency with those from LNP-based method in our previous report [43]. Meanwhile, recent studies have demonstrated that the protein subpopulations in EVs mainly originated from the cytoplasm, cell membrane and extracellular domain, rather than from subcellular organelles or cell nucleus [56]. These findings provided a theoretical foundation for assessing the purity of EVs based on proteomics. Hence, the proteomes of EVs from both BAIP and UC-based methods were further analyzed and the levels of purity were evaluated. The Radar chart showed the protein proportion of GO cellular components present in both the composition proteins and the interference proteins of EVs obtained from the BAIP and UC-based methods (Fig. 4E and Table S9). In the chart, the red numbers 1 to 11 represent the composition proteins of EVs, whereas the blue numbers 12 to 18 represent the interference proteins. As shown in Fig. 4E, the larger areas in the red region and smaller areas in the blue region demonstrated that our BAIP-based method identified more composition proteins of EVs and fewer interference proteins compared to UC. This result was consistent with our previous LNP-based method (Fig. S30), which also showed similar changes (verse UC method) in protein proportions of GO cellular components (Fig. 4F), as evidenced by the red and blue markers indicating composition proteins and contaminant proteins, respectively (Table S9). These findings suggested that our BAIP-based method could isolate EVs of higher purity than UC, which may facilitate downstream analysis, such as screening of new biomarkers of EVs.
Moreover, EVs may serve as an indicator for predicting tumor progression. For this purpose, the GO biological process analyses for proteins of EVs obtained through BAIP and UC-based methods were conducted. Sixteen crucial biological process terms associated with tumor progression were then particularly examined and the corresponding chords from EVs isolated by BAIP and UC-based methods were illustrated in Fig. 4G and H, respectively. After comparing the results, we found that our proposed BAIP based method could recognize 27% more of special tumor-relevant progression proteins in contrast to that of UC-based method. Meanwhile, the correlations between the proteome of EVs and tumor regulation pathways were determined by the EASE Score (P value). Notably, higher correlations in tumor regulation pathways were achieved in our BAIP based method than UC-based method, with 10 out of 16 crucial biological process terms associated with tumor progression showing either comparable or significant improvement (Fig. 4I). These findings suggested that our BAIP based method for isolating EVs resulted in a significant enrichment of tumor-related proteins which were actively involved in tumor development and progression.
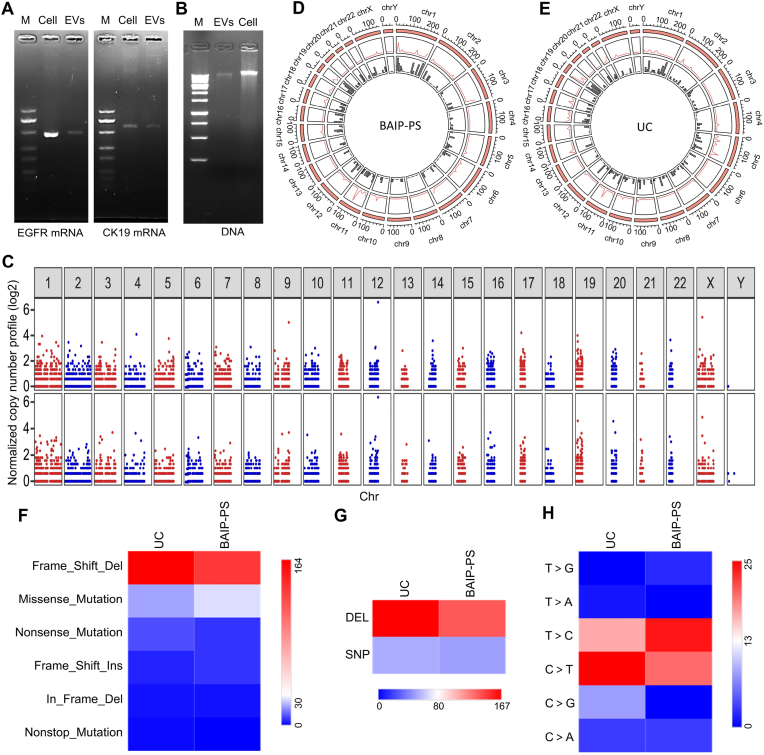
The obtained EVs by BAIP could be used not only for protein analyses, but also for nucleic acid analyses. For RNA analysis, the ability to detect mRNA in EVs was examined using EGFR and CK19 transcripts as examples. As Fig. 5A depicted, the bands of 285 bp fragments of the EGFR coding region and 346 bp fragments of the CK19 coding region were clearly observed on the 2% agarose gel, which indicated that the EVs isolated by our BAIP based method could be directly used for mRNA detection. For DNA analysis, the exosomal DNA was extracted from isolated EVs and then detected by 2% agarose gel electrophoresis. Fig. 5B showed the presence of long fragments of DNA fragments in the extracted DNA (>10 kbp), which differed from circulating cell-free DNA with a typical apoptotic DNA ladder [57]. Afterward, DNA from the isolated EVs with BAIP and model EVs (PC9 cells derived) were analyzed by WES. As shown in Fig. 5C, the DNA from isolated EVs by BAIP and UC based methods spanned all chromosomes. The DNA content of EVs isolated using BAIP closely resembled that of EVs isolated through UC, as evidenced by the copy number variation (CNV) plots for EV samples obtained from both methods (Pearson correlation coefficient was 0.92). The distribution patterns of Single Nucleotide Polymorphisms (SNP) and Deletion or Insertion (Del) densities were then carefully examined. The SNP density map unveiled a non-random distribution, showcasing distinct characteristics. Specifically, EVs isolated with BAIP displayed elevated SNP density in chr1, chr3 and chr11, while EVs isolated through UC demonstrated higher SNP density in chr3, chr11, and chr19 (Fig. 5D and E). Concurrently, mutations in the DNA from EVs isolated by both methods were meticulously annotated and summarized. The findings revealed that these mutations were classified into six types based on their impact on protein coding, with Frame_Shift_Del emerging as the most prevalent type (Fig. 5F). Moreover, an intriguing observation was found regarding the prevalence of DEL mutations over SNP mutations in isolated EVs by BAIP and UC based methods (Fig. 5G). Further categorization of SNV mutations into six types highlighted T > C and C > T as the predominant SNV mutation types in both isolated EVs (Fig. 5H).
Fig. 5.
The nucleic acid analysis of EVs. (A) Agarose gel electrophoresis for EGFR mRNA (left) and CK19 mRNA (right) detection. (M, 500 bp DNA ladder). The sequences of the fragments of EGFR and CK19 coding region were listed in Table S10. (B) Electrophoresis characterization of DNA extracted from isolated EVs and MCF-7 cells. DNA ladder: 1 kbp. (C) Plot of the exon-wide copy-number profiles of EVs isolated with UC (top) and BAIP (bottom) using Control-FREEC. (D–E) Circos plots of DNA from PC9 EVs isolated by BAIP and UC. The outer ring is chromosome length, the second layer is SNP density line chart, and the innermost layer is Indel density histogram. (F–H) Heat maps of frequency distribution with different variant classification (F), different variant types (G) and different SNV mutation types (H) from BAIP and UC based methods, respectively.
2.4. Detection of mutated DNA in EVs from plasma samples
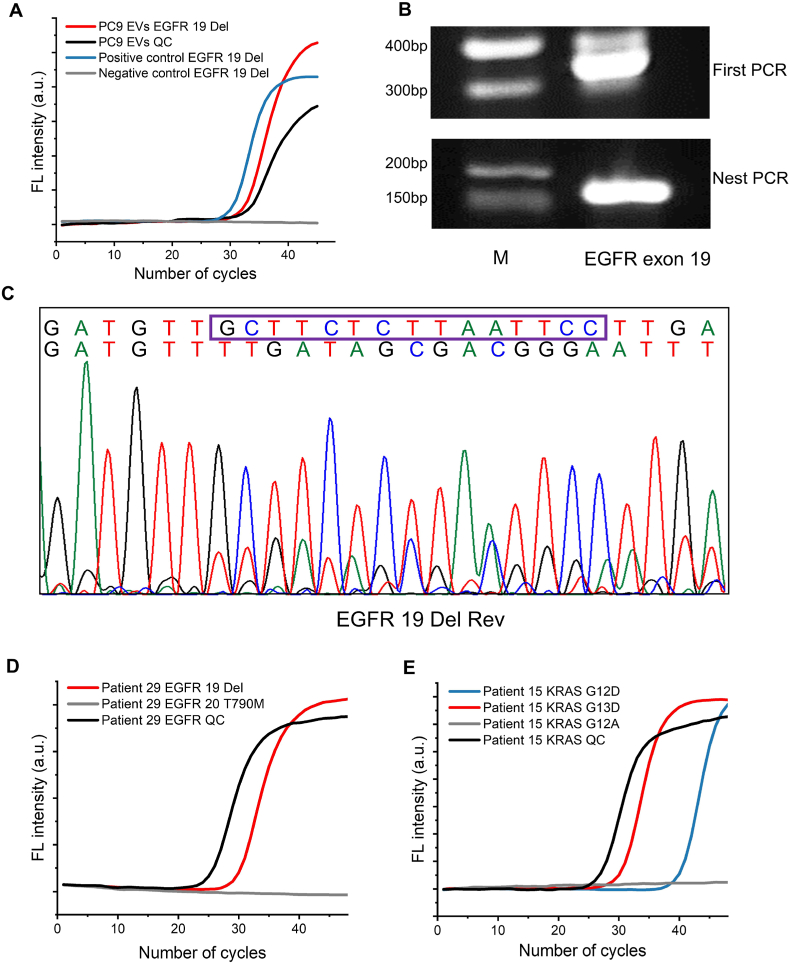
In order to promote BAIP-based method for EVs isolating to clinical trials, molecular biology methods were used to detect gene mutations in EVs directly. Firstly, 1 μL of the model EVs derived from PC9 cells was added into 99 μL of the plasma from the healthy donors to simulate clinical samples, which were used to test the performance of the gene mutation detection. qPCR assay was implemented to detect the EGFR deletion mutation in exon 19 (EGFR19Del). As shown in Fig. 6A, the EGFR19Del mutation in the extracted DNA from isolated EVs by our method was successfully identified with qPCR. Secondly, EGFR-mutant-enriched nested PCR was implemented to enhance the sensitivity for Sanger sequence analysis of EGFR19Del mutation. The desired PCR products of EGFR19Del mutation were detected on the agarose gel (Fig. 6B) and the EGFR19Del mutation was successfully identified by the Sanger sequencing analysis (Fig. 6C). These findings suggested that our BAIP-based method for EVs isolation had the potential to be employed for DNA mutation detection in clinical samples.
Fig. 6.
Detection of exosomal DNA mutations from isolated EVs. (A) Detection of EGFR19Del mutation from isolated EVs by BAIP in a simulated sample by qPCR. (B) Gel electrophoresis analysis of EGFR DNA fragments from isolated EVs after a first PCR amplification (top) and a second EGFR-mutant-enriched nested PCR (bottom). M, 500-bp DNA ladder. (C) Identification of EGFR19Del mutation in isolated EVs by Sanger sequence. (D–E) qPCR profiles for KRAS mutations or EGFR mutations in the isolated EVs by BAIP from patients 29 and 15, respectively.
Each aliquot of 500 μL genuine plasma samples collected from 31 patients and 14 healthy donors was directly processed with BAIP-PS and the DNA was extracted from the isolated EVs to examine the EGFR and KRAS mutations via qPCR. Meanwhile, the mutation detection results of qPCR in patients' samples were compared with those obtained from next generation sequencing (NGS) or qPCR measurements in corresponding tissue samples. The whole results are listed in Table 1 and Table S11. The mutation analyses revealed the presence of KRAS G12A, KRAS G12V, KRAS G12D and KRAS G13D point mutations, EGFR19Del mutations, EGFR18G719C mutations in patients 3, 4, 7, 9, 15, 18, 24, 25 and 29, as well as WT KRAS in patients 17, 18, 19, 21, 26, 27 and WT EGFR in patients 2, 30 and 31, which matched well with the results of corresponding tissue samples of patients (Fig. 6D–E and Fig. S31). These mutations have been previously linked to the development and progression of various types of cancer, making them potentially valuable for guiding accurate personalized medical treatment. Moreover, we identified EGFR19Del mutations in patients 6, 11, 12, 17 and 20 (Fig. S32). However, these mutations were not verified through NGS or qPCR analysis of their corresponding tissue samples, due to the limited quantity of samples. Despite of our efforts, we failed to detect some mutations in patients, such as 1, 8, 16, 19 and 28, while the NGS or qPCR analysis of tissue identified them, presumably due to the low abundance of cancer-relevant EVs in the plasma samples or due to the alterations in mutation status between the original tissue biopsy and blood collection. Notably, the EGFR19Del mutations in patient 9, 22, 25 and 27 and EGFR20T790M mutations in patient 18 and 27 were detected. Moreover, KRAS G12D point mutations were detected in patients 15 and 24, and KRAS G12S point mutations were found in patients 22 and 24 (Fig. S33). However, no EGFR or KRAS mutations were detected in the tissue through NGS or qPCR analysis. This highlights that our method has the ability to detect mutations in EVs from plasma when the patient has low abundance of mutations in tissue samples. In addition, all KRAS and EGFR alleles in healthy donors were detected as wild type following EVs isolation via the BAIP based method. Nonetheless, the identification of EGFR and KRAS mutations in some patients underscores the potential utility of our assay for mutation analyses in clinical settings.
Table 1.
KRAS and EGFR mutations in samples from patients.
| Patient number | Sex | Age | Tissue source | KRAS mutation |
EGFR mutation |
||
|---|---|---|---|---|---|---|---|
| EVs | Tissue | EVs | Tissue | ||||
| 1 | M | 63 | Lung | NA | WT | WT | exon 21 L858R |
| 2 | M | 67 | Lung | WT | G12R | WT | WT |
| 3 | F | 76 | Colon | G12A | G12A | NA | NA |
| 4 | M | 52 | Colon | G12V | G12V | NA | NA |
| 5 | M | 74 | Lung | NA | WT | WT | exon 21 L858R |
| 6 | F | 67 | Liver | NA | NA | exon 19 Del, exon 20 Ins | NA |
| 7 | M | 75 | liver | G12D | G12D | NA | NA |
| 8 | F | 68 | Rectum | WT | G12D | NA | NA |
| 9 | M | 56 | Lung | NA | WT | exon 18 G719X, exon 19 Del | exon 18 G719C |
| 10 | M | 77 | Lung | NA | WT | WT | exon 21 L858R |
| 11 | M | 44 | Liver | NA | NA | exon 19 Del | NA |
| 12 | F | 59 | Liver | NA | NA | exon 19 Del | NA |
| 13 | F | 82 | Lung | NA | WT | WT | exon 21 L858R |
| 14 | M | 48 | Lung | NA | WT | WT | exon 21 L858R |
| 15 | M | 59 | Colon | G12D, G13D | G13D | NA | NA |
| 16 | F | 83 | Lung | NA | WT | WT | exon 21 L858R |
| 17 | M | 73 | Colon | WT | WT | exon 19 Del | NA |
| 18 | M | 57 | Lung | WT | WT | exon 19 Del, exon 20 T790M | exon 19 Del |
| 19 | M | 76 | Lung | WT | WT | WT | exon 20 T790M |
| 20 | M | 63 | Colon | WT | NA | exon 19 Del | NA |
| 21 | F | 62 | Rectum | WT | WT | NA | NA |
| 22 | M | 45 | Lung | G12S | WT | exon 19 Del | WT |
| 23 | M | 79 | Colon | WT | G12V | NA | NA |
| 24 | M | 50 | Lung | G12S, G12D | WT | exon 19 Del | exon 19 Del |
| 25 | M | 66 | Colon | G12A | G12A | exon 19 Del | WT |
| 26 | M | 61 | Liver | WT | WT | NA | NA |
| 27 | F | 59 | Lung | WT | WT | exon 19 Del, exon 20 T790M | WT |
| 28 | F | 59 | Lung | NA | WT | WT | exon 21 L858R |
| 29 | M | 73 | Lung | NA | NA | exon 19 Del | exon 19 Del |
| 30 | M | 49 | Lung | NA | NA | WT | WT |
| 31 | M | 53 | Lung | NA | WT | WT | WT |
Bold text indicated that the mutation of EVs was consistent with that of tissue. NA, not available. WT, wild type. Del, deletion.
3. Discussion
The efficient separation and enrichment of EVs with high purify from body fluids is crucial for various biomedical applications [27,58,59]. However, the EVs display heterogenous properties in terms of their membrane proteins and nucleic acids. Thus, conventional approaches for EVs isolation and detection, such as those relying on specific antibodies or aptamers-based techniques, frequently encounter biases and fall short in capturing disease-relevant EVs. In this study, we provided a novel strategy to reconfigure and modify the membrane-penetrating peptide to achieve rapid (20 min), efficient (∼80%), cost-effective and unbiased isolation and enrichment of EVs with high purity. We changed the amino acid residue at the N-terminal of pH-sensitive WT pHLIP and modify it with biotin. The modification appropriately increased the hydrophobicity and facilitated the formation of stable intramolecular hydrogen bonds of the peptide, both of which promoted the generation of the α-helical conformation. Thus-modified peptide, BAIP, could insert into the phospholipid bilayer to isolate EVs beyond the pH limitation.
Our BAIP-based isolation approach for EVs offers several advantages. (1) The meticulously-engineered peptide provided a size- and antigen-independent isolation approach, enabling the unbiased isolation of EVs. This would effectively address the challenge of missing EVs with low antigen expression due to the heterogeneity. (2) The BAIP-PS system can achieve a ∼80% recovery and significantly reduce the isolation time of EVs to 20 min. In comparison to existing methods (Table S12), our BAIP-based method demonstrates significantly faster capture speed and more consistent capture efficiency between EVs in PBS buffer and in plasma. (3) The BAIP system exhibits exceptional anti-interference capability against non-target nucleic acids, proteins and other substances in plasma. The experiments confirmed that our BAIP-based method achieved a consistent capture efficiency of EVs in both buffer and clinical plasma samples. This capture efficiency is significantly higher than that of our previously-reported method, which utilized a lipid-nanoprobe but only achieved ∼50% capture efficiency in plasma samples [43]. (4) The purity of EVs by our method was higher than that of the unbiased UC. In addition, our high quality and purified EVs can be used for downstream analysis, such as proteomics analysis and gene sequencing analysis. For the clinical trials, we successfully used the BAIP system in combination with qPCR to detect EGFR and KRAS gene mutations in EVs from clinical samples. (5) The versatility of BAIP allows for its conjugation with various bioinspired interfaces, enabling widespread applications in EVs isolation. (6) Our approach displayed a low cost of ∼7.3 $ to process each clinical sample (Table S13), which was beneficial for the clinical translation. (7) Our BAIP system offers simplicity in operation and eliminates the need for expensive and bulky equipment, which may extend the potential applications in resource-limited areas.
Despite the encouraging findings of our study, there are some limitations and areas that can be improved in future research. Firstly, there are millions of possible reconfigurations available to modify the peptide. However, we have only investigated one specific reconfiguration. It is possible that other reconfigurations might behave more efficiently in capturing the EVs in our ongoing studies. Anyhow, we have provided a new strategy for the peptide reconfiguration, particularly aiming at the EVs isolation, which could potentially be extended to modify the other peptides besides the WT pHLIP. Secondly, although our approach demonstrated some positive clinical trial outcomes, it is essential to conduct large-scale cohort studies to gather an extensive amount of clinical data. This will enable us to establish the relationship between the data and the corresponding cancer type or its origin, to extend the method for real clinical applications. Thirdly, our unbiased approach for isolating and enriching EVs may play a vital role in identifying new biomarkers for cancer diagnostics using omics-based technology in clinical settings, holding great promise in enhancing the accuracy and effectiveness of EV-based cancer diagnosis.
4. Conclusion
In summary, we provided a novel strategy to reconfigure and modify the membrane-penetrating peptide to achieve rapid, cost-effective and unbiased isolation and enrichment of EVs by inserting the modified peptide into phospholipid bilayer of EVs. We changed the amino acid residue at the N-terminal of pH-sensitive WT pHLIP and modify it with biotin. The modification appropriately increased the hydrophobicity and facilitated the formation of stable intramolecular hydrogen bonds of the peptide, both of which promoted the generation of the α-helical conformation. Thus, the resulting peptide, BAIP, could insert into the phospholipid bilayer for the isolation of EVs. The simulation results and capture experiments confirmed the rapid (20 min), highly efficient (∼80% recovery) and unbiased enrichment of the EVs by the BAIP-based method, effectively overcoming the pH limitations. Our unbiased approach for isolating and enriching EVs may play a vital role in identifying new biomarkers for cancer diagnostics using omics-based technology in clinical settings, holding great promise in enhancing the accuracy and effectiveness of EV-based cancer diagnosis.
5. Materials and methods
Study design
This study aimed to develop a novel method for the unbiased isolation of EVs in biological samples and the investigation of EV biomarkers, with the goal of enhancing transformation for large-scale clinical and biomedical applications. With the inspiration of the natural WT pHLIP for the drug delivery, we supposed to reconfigure and modify the peptide for the unbiased isolation of EVs, independent of the pH limitation. We reconfigured the WT pHLIP to a novel AIP and functionalized it with biotin to obtain the meticulously engineered BAIP. Using molecular dynamics simulations and circular dichroism, we investigated the conformation of AIP and BAIP. The BAIP-based method was assessed and optimized to isolate EVs in physical environment. To verify the universality of BAIP, we modified BAIP on different substrates to isolate EVs from neutral environment. The purity of the isolated EVs was investigated by proteomics, and the suitability of the isolated EVs for downstream molecular analysis was investigated by proteomics and WES. For liquid biopsy, we used the BAIP coping with qPCR to detect the gene mutations in EVs (EGFR and KRAS) from the clinical samples, the results were compared with the clinical results. The AIP was synthesized and purified by Motif Biotech (Motif Biotechnology Co., Ltd, Suzhou, China). It was characterized by high-performance liquid chromatography and mass spectrometry. The details of experimental methods can be found in Supplementary Information.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The blood samples of cancer patients and healthy person were obtained from Renmin Hospital of Wuhan University and Tongji Hospital of Huazhong University of Science and Technology (HUST). The study was reviewed and approved by the Institutional Review Board (IRB) of Tongji Hospital of HUST ([2021] IEC(A146). Written consent was obtained from either the participant or next of kin.
CRediT authorship contribution statement
Le Wang: Writing – original draft, Methodology, Investigation, Data curation, Conceptualization. Zhou Gong: Formal analysis. Ming Wang: Resources, Methodology, Formal analysis. Yi-Zhong Liang: Investigation. Jing Zhao: Resources. Qi Xie: Formal analysis. Xiao-Wei Wu: Investigation. Qin-Ying Li: Writing – review & editing. Cong Zhang: Investigation. Li-Yun Ma: Writing – review & editing. Si-Yang Zheng: Investigation. Ming Jiang: Writing – review & editing. Xu Yu: Writing – review & editing, Writing – original draft, Supervision, Project administration, Funding acquisition, Conceptualization. Li Xu: Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant No. 21804105, 22174049, 31971155), the Program for HUST Academic Frontier Youth Team (Grant No. 2019QYTD09), the Natural Science Foundation of Hubei Province of China (No.2021CFB335), the Fundamental Research Funds for the Central Universities in China (Grant Nos. 2172019KFYRCPY112, 2172020kfyXJJS082), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (Grant No. 2020329) and the Fundamental Research Funds for the Central Universities, HUST: Grant Nos. 2023JYCXJJ060. The authors thank the Analytical and Testing Center of HUST for the material characterization.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.09.023.
Contributor Information
Ming Wang, Email: morgan@whu.edu.cn.
Xu Yu, Email: xuyu@hust.edu.cn.
Li Xu, Email: xulpharm@mails.tjmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Shao H., Im H., Castro C.M., Breakefield X., Weissleder R., Lee H. New technologies for analysis of extracellular vesicles. Chem. Rev. 2018;118:1917–1950. doi: 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Pan M.M., Xu L., Yu X., Zheng S.Y. Recent advances of emerging microfluidic chips for exosome mediated cancer diagnosis. Smart Materials in Medicine. 2021;2:158. doi: 10.1016/j.smaim.2021.06.001. 17. [DOI] [Google Scholar]
- 3.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367 doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi R., Zhan A., Li X., Kong B., Liang G. Biomimetic extracellular vesicles for the tumor targeted treatment. Eng. Regen. 2023;4:427–437. doi: 10.1016/j.engreg.2023.08.002. [DOI] [Google Scholar]
- 5.Su N., Hao Y., Wang F., Hou W., Chen H., Luo Y. Mesenchymal stromal exosome-functionalized scaffolds induce innate and adaptive immunomodulatory responses toward tissue repair. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marar C., Starich B., Wirtz D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021;22:560–570. doi: 10.1038/s41590-021-00899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan L., Wang L., Wang X., Zhang H. Exosomes-based particles as inhalable COVID-19 vaccines. Biomed. Technol. 2023;4:24–27. doi: 10.1016/j.bmt.2023.01.003. [DOI] [Google Scholar]
- 8.Yu W.-W., Wan Q.-Q., Wei Y., Li Y.-T., Li Q.-H., Ye T., Xu K.-H., Song J.-H., Lei C., Wan M. -Chen, Jiao K., Tay F.R., Niu L.-N. Engineered extracellular vesicles: regulating the crosstalk between the skeleton and immune system. Eng. Regen. 2022;3:270–282. doi: 10.1016/j.engreg.2022.06.004. [DOI] [Google Scholar]
- 9.Syn N., Wang L., Sethi G., Thiery J.P., Goh B.C. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol. Sci. 2016;37:606–617. doi: 10.1016/j.tips.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 10.He X., Ma Y., Wen Y., Zhang R., Zhao D., Wang G., Wang W., Huang Z., Guo G., Zhang X., Lin H., Zhang L. Tumor-derived apoptotic extracellular vesicle-mediated intercellular communication promotes metastasis and stemness of lung adenocarcinoma. Bioact. Mater. 2024;36:238–255. doi: 10.1016/j.bioactmat.2024.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todorova D., Simoncini S., Lacroix R., Sabatier F., Dignat-George F. Extracellular vesicles in angiogenesis. Circ. Res. 2017;120:1658–1673. doi: 10.1161/circresaha.117.309681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei X., Zhang D., Zhu Y. Exosomes: toward a potential application in bladder cancer diagnosis and treatment. Smart Med. 2024;3 doi: 10.1002/SMMD.20230027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buzas E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2023;23:236–250. doi: 10.1038/s41577-022-00763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paskeh M.D.A., Entezari M., Mirzaei S., Zabolian A., Saleki H., Naghdi M.J., Sabet S., Khoshbakht M.A., Hashemi M., Hushmandi K., Sethi G., Zarrabi Z., Kumar A.P., Tan S.C., Papadakis M., Alexiou A., Islam M.A., Mostafavi E., Ashrafizadeh M. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J. Hematol. Oncol. 2022;15:83. doi: 10.1186/s13045-022-01305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian F., Zhang S., Liu C., Han Z., Liu Y., Deng J., Li Y., Wu X., Cai L., Qin L., Chen Q., Yuan Y., Liu Y., Cong Y., Ding B., Jiang Z., Sun J. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat. Commun. 2021;12:2536. doi: 10.1038/s41467-021-22913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao H., Chung J., Lee K., Balaj L., Min C., Carter B.S., Hochberg F.H., Breakefield X.O., Lee H., Weissleder R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu C., Chen Q., Wu T., Du X., Dong Y., Peng Z., Xue W., Sunkara V., Cho Y.-K., Dong L. The role of extracellular vesicles in the treatment of prostate cancer. Small. 2024 doi: 10.1002/smll.202311071. [DOI] [PubMed] [Google Scholar]
- 18.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N., Reissfelder C., Pilarsky C., Fraga M.F., Piwnica-Worms D., Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun N., Lee Y.-T., Zhang R.Y., Kao R., Teng P.-C., Yang Y., Yang P., Wang J.J., Smalley M., Chen P.-J., Kim M., Chou S.-J., Bao L., Wang J., Zhang X., Qi D., Palomique J., Nissen N., Han S.-H.B., Sadeghi S., Finn R.S., Saab S., Busuttil R.W., Markovic D., Elashoff D., Yu H.-H., Li H., Heaney A.P., Posadas E., You S., Yang J.D., Pei R., Agopian V.G., Tseng H.-R., Zhu Y. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat. Commun. 2020;11:4489. doi: 10.1038/s41467-020-18311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P., Zhou X., He M., Shang Y., Tetlow A.L., Godwin A.K., Zeng Y. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 2019;3:438–451. doi: 10.1038/s41551-019-0356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu Q., Gao J., Zhao K., Chen X., Lin X., Huang C., An Y., Xiao X., Wu Q., Cui L., Zhang P., Wu L., Yang C. Fluid nanoporous microinterface enables multiscale-enhanced affinity interaction for tumor-derived extracellular vesicle detection. Proc. Natl. Acad. Sci. 2022;119 doi: 10.1073/pnas.2213236119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C., Zhao J., Tian F., Chang J., Zhang W., Sun J. λ-DNA- and aptamer-mediated sorting and analysis of extracellular vesicles. J. Am. Chem. Soc. 2019;141:3817–3821. doi: 10.1021/jacs.9b00007. [DOI] [PubMed] [Google Scholar]
- 23.Zhang P., Wu X., Gardashova G., Yang Y., Zhang Y., Xu L., Zeng Y. Molecular and functional extracellular vesicle analysis using nanopatterned microchips monitors tumor progression and metastasis. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aaz2878. eaaz2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y., Zhu Q., Cheng L., Wang Y., Li M., Yang Q., Hu L., Lou D., Li J., Dong X., Lee L.P., Liu F. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods. 2021;18:212–218. doi: 10.1038/s41592-020-01034-x. [DOI] [PubMed] [Google Scholar]
- 25.Tang J., Jia X., Li Q., Cui Z., Liang A., Ke B., Yang D., Yao C. A DNA-based Hydrogel for exosome separation and biomedical applications. Proc. Natl. Acad. Sci. 2023;120 doi: 10.1073/pnas.2303822120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S., Yu Z., Chen D., Wang Z., Miao J., Li Q., Zhang D., Song J., Cui D. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small. 2020;16 doi: 10.1002/smll.201903916. [DOI] [PubMed] [Google Scholar]
- 27.Yang B., Chen Y., Shi J. Exosome biochemistry and advanced nanotechnology for next-generation theranostic platforms. Adv. Mater. 2019;31 doi: 10.1002/adma.201802896. [DOI] [PubMed] [Google Scholar]
- 28.Xia B., Hu R., Chen J., Shan S., Xu F., Zhang G., Zhou Z., Fan Y., Hu Z., Liang X. Oral administration properties evaluation of three milk-derived extracellular vesicles based on ultracentrifugation extraction methods. Adv. Healthc. Mater. 2024 doi: 10.1002/adhm.202401370. [DOI] [PubMed] [Google Scholar]
- 29.Coumans F.A.W., Brisson A.R., Buzas E.I., Dignat-George F., Drees E.E.E., El-Andaloussi S., Emanueli C., Gasecka A., Hendrix A., Hill A.F., Lacroix R., Lee Y., Leeuwen T.G., Mackman N., Mäger I., Nolan J.P., Pol E., Pegtel D.M., Sahoo S., Siljander P.R.M., Sturk G., Wever O., Nieuwland R. Methodological guidelines to study extracellular vesicles. Circ. Res. 2017;120:1632–1648. doi: 10.1161/circresaha.117.309417. [DOI] [PubMed] [Google Scholar]
- 30.Martínez-Greene J.A., Hernández-Ortega K., Quiroz-Baez R., Resendis-Antonio O., Pichardo-Casas I., Sinclair D.A., Budnik B., Hidalgo-Miranda A., Uribe-Querol E., Ramos-Godínez M.d.P., Martínez-Martínez E. Quantitative proteomic analysis of extracellular vesicle subgroups isolated by an optimized method combining polymer-based precipitation and size exclusion chromatography. J. Extracell. Vesicles. 2021;10 doi: 10.1002/jev2.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu M., Ouyang Y., Wang Z., Zhang R., Huang P.-H., Chen C., Li H., Li P., Quinn D., Dao M., Suresh S., Sadovsky Y. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. 2017;114:10584–10589. doi: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibsen S.D., Wright J., Lewis J.M., Kim S., Ko S.-Y., Ong J., Manouchehri S., Vyas A., Akers J., Chen C.C., Carter B.S., Esener S.C., Heller M.J. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano. 2017;11:6641–6651. doi: 10.1021/acsnano.7b00549. [DOI] [PubMed] [Google Scholar]
- 33.Wunsch B.H., Smith J.T., Gifford S.M., Wang C., Brink M., Bruce R.L., Austin R.H., Stolovitzky G., Astier Y. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat. Nanotechnol. 2016;11:936–940. doi: 10.1038/nnano.2016.134. [DOI] [PubMed] [Google Scholar]
- 34.Guo S.C., Tao S.C., Dawn H. Microfluidics-based on-a-Chip systems for isolating and analysing extracellular vesicles. J. Extracell. Vesicles. 2018;7 doi: 10.1080/20013078.2018.1508271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin B., Lei Y., Wang J., Zhu L., Wu Y., Zhang H., Wu L., Zhang P., Yang C. Microfluidic-based exosome analysis for liquid biopsy. Small Methods. 2021;5 doi: 10.1002/smtd.202001131. [DOI] [PubMed] [Google Scholar]
- 36.Yasui T., Yanagida T., Ito S., Konakade Y., Takeshita D., Naganawa T., Nagashima K., Shimada T., Kaji N., Nakamura Y., Thiodorus I.A., He Y., Rahong S., Kanai M., Yukawa H., Ochiya T., Kawai T., Baba Y. Unveiling massive numbers of cancer-related urinary-MicroRNA candidates via nanowires. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1701133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Liu C., Cheng Y., Li Y., Deng J., Bai L., Qin L., Mei H., Zeng M., Tian F., Zhang S., Sun J. Cascaded microfluidic circuits for pulsatile filtration of extracellular vesicles from whole blood for early cancer diagnosis. Sci. Adv. 2023;9 doi: 10.1126/sciadv.ade2819. eade2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu Q., Shu Y., Chen Y., Huang Z., Yao Z., Chen X., Lin F., Feng J., Huang C., Wang H., Ding H., Yang C., Wu L. A fluid multivalent magnetic interface for high-performance isolation and proteomic profiling of tumor-derived extracellular vesicles. Angew. Chem. Int. Ed. 2023;62 doi: 10.1002/anie.202215337. [DOI] [PubMed] [Google Scholar]
- 39.Li W., Wang H., Zhao Z., Gao H., Liu C., Zhu L., Wang C. Yanlian yang, emerging nanotechnologies for liquid biopsy: the detection of circulating tumor cells and extracellular vesicles. Adv. Mater. 2019;31 doi: 10.1002/adma.201805344. [DOI] [PubMed] [Google Scholar]
- 40.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu D., Yan J., Shen X., Sun Y., Thulin M., Cai Y., Wik L., Shen Q., Oelrich J., Qian X., Dubois K.L., Ronquist K.G., Nilsson M., Landegren U., Kamali-Moghaddam M. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat. Commun. 2019;10:3854. doi: 10.1038/s41467-019-11486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang G., Lin G., Zhu Y., Duan W., Jin D. Emerging technologies for profiling extracellular vesicle heterogeneity. Lab Chip. 2020;20:2423–2437. doi: 10.1039/d0lc00431f. [DOI] [PubMed] [Google Scholar]
- 43.Wan Y., Cheng G., Liu X., Hao S.-J., Nisic M., Zhu C.-D., Xia Y.-Q., Li W.-Q., Wang Z.-G., Zhang W.-L., Rice S.J., Sebastian A., Albert I., Belani C.P., Zheng S.-Y. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nat. Biomed. Eng. 2017;1:58. doi: 10.1038/s41551-017-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q., Zhang Z., Wang F., Wang X., Zhan S., Yang X., Xu C., Liu D. Reversible zwitterionic coordination enables rapid, high-yield, and high-purity isolation of extracellular vesicles from biofluids. Sci. Adv. 2023;9 doi: 10.1126/sciadv.adf4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tietz O., Cortezon-Tamarit F., Chalk R., Able S., Vallis K.A. Tricyclic cell-penetrating peptides for efficient delivery of functional antibodies into cancer cells. Nat. Chem. 2022;14:284–293. doi: 10.1038/s41557-021-00866-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zorko M., Jones S., Langel U. Cell-penetrating peptides in protein mimicry and cancer therapeutics. Adv. Drug Deliv. Rev. 2022;180 doi: 10.1016/j.addr.2021.114044. [DOI] [PubMed] [Google Scholar]
- 47.Saha A., Mandal S., Arafiles J.V.V., Gómez-González J., Hackenberger C.P.R., Brik A. Structure-uptake relationship study of DABCYL derivatives linked to cyclic cell-penetrating peptides for live-cell delivery of synthetic proteins. Angew. Chem. Int. Ed. 2022;61 doi: 10.1002/anie.202207551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunt J.F., Rath P., Rothschild K.J., Engelman D.M. Spontaneous, pH-dependent membrane insertion of a transbilayer α-helix. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 49.Cheng C.J., Bahal R., Babar I.A., Pincus Z., Barrera F., Liu C., Svoronos A., Braddock D.T., Glazer P.M., Engelman D.M., Saltzman W.M., Slack F.J. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu G., Li F., Zhang F., Huang L.-L., Zhang L., Lv Y., Wei W., Xie H.-Y. Amplifying nanoparticle targeting performance to tumor via diels-alder cycloaddition. Adv. Funct. Mater. 2018;28 doi: 10.1002/adfm.201707596. [DOI] [Google Scholar]
- 51.Zong Z., Liu X., Ye Z., Liu D. A double-switch pHLIP system enables selective enrichment of circulating tumor microenvironment-derived extracellular vesicles. Proc. Natl. Acad. Sci. 2023;120 doi: 10.1073/pnas.2214912120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi M., Jiang Z., Xiao Y., Song Y., Tang R., Zhang L., Huang J., Tian Y., Zhou S. Stapling of short cell-penetrating peptides for enhanced tumor cell-and-tissue dual-penetration. Chem. Commun. 2022;58:2299–2302. doi: 10.1039/d1cc06595e. [DOI] [PubMed] [Google Scholar]
- 53.Lu H., Wang J., Bai Y., Lang J.W., Liu S., Lin Y., Cheng J. Ionic polypeptides with unusual helical stability. Nat. Commun. 2011;2:206. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]
- 54.Wu J., Lin Z., Zou Z., Liang S., Wu M., Hu T.Y., Zhang Y. Identifying the phenotypes of tumor-derived extracellular vesicles using size-coded affinity microbeads. J. Am. Chem. Soc. 2022;144:23483–23491. doi: 10.1021/jacs.2c10042. [DOI] [PubMed] [Google Scholar]
- 55.Lin Z., Zhang J., Zou Z., Lu G., Wu M., Niu L., Zhang Y. A dual-encoded bead-based immunoassay with tunable detection range for COVID-19 serum evaluation. Angew. Chem. Int. Ed. 2022;61 doi: 10.1002/anie.202203706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi D., Go G., Kim D.-K., Lee J., Park S.-M., Vizio D.D., Gho Y.S. Quantitative proteomic analysis of trypsin-treated extracellular vesicles to identify the real-vesicular proteins. J. Extracell. Vesicles. 2020;9 doi: 10.1080/20013078.2020.1757209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gormally E., Caboux E., Vineis P., Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat. Res. 2007;635:105–117. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 58.Wang W., Luo J., Wang S. Recent progress in isolation and detection of extracellular vesicles for cancer diagnostics. Adv. Healthc. Mater. 2018;7 doi: 10.1002/adhm.201800484. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y., Wong C.Y., Lim C.Z.J., Chen Q., Yu Z., Natalia A., Wang Z., Pang Q.Y., Lim S.W., Loh T.P., Ang B.T., Tang C., Shao H. Multiplexed RNA profiling by regenerative catalysis enables blood-based subtyping of brain tumors. Nat. Commun. 2023;14:4278. doi: 10.1038/s41467-023-39844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.