Abstract
We have developed a novel linker-primer PCR assay for the detection and quantification of integrated human immunodeficiency virus type 1 (HIV) DNA. This assay reproducibly allowed the detection of 10 copies of integrated HIV DNA, in a background of 2 × 105 cell equivalents of human chromosomal DNA, without amplifying extrachromosomal HIV DNA. We have used this assay and a near-synchronous one-step T-cell infection model to investigate the kinetics of viral DNA accumulation following HIV infection. We report here that integrated HIV DNA started accumulating 1 h after the first appearance of extrachromosomal viral DNA and accounted for ∼10% of the total HIV DNA synthesized in the first round of viral replication. These results highlight the efficient nature of integrase-mediated HIV integration in infected T cells.
Integration of newly synthesized viral DNA into the host cell chromosome is common to all retroviruses and is essential for a productive human immunodeficiency virus (HIV) infection (12, 22, 28, 30). Upon reverse transcription of the viral genomic RNA, the resulting linear DNA molecule is actively transported to the nucleus within a complex of host and viral proteins known as the preintegration complex, which is thought to be the immediate precursor to the integration reaction (2, 3, 5, 13, 19, 24, 26). Analyses of the extrachromosomal and total HIV DNA forms using both Southern hybridization and PCR-based techniques have indicated that full-length linear DNA is first observed at approximately 3 to 4 h postinfection (p.i.) (1, 20, 21, 23, 25). In reports on the kinetics of HIV DNA synthesis following cell-to-cell infection, the circular forms of viral DNA were shown to first appear at 8 h p.i., with the two long-terminal-repeat (2-LTR) species constituting a minor population compared to the 1-LTR and linear forms over the course of infection (1, 25).
In contrast to investigations on both free and total viral DNA forms, little work has been performed on the accumulation of integrated DNA within infected cells following HIV infection. This has been primarily due to the lack of an appropriate assay which can selectively detect and quantify integrated viral DNA, as chromosomal DNA preparations isolated from cells infected with HIV invariably contain significant amounts of contaminating extrachromosomal HIV DNA (1, 27, 30, 34). However, two assays able to distinguish between the extrachromosomal and integrated HIV DNA have recently been described and used to quantify the amounts of integrated proviral HIV DNA in infected patients (6–10). Here we present an alternative linker-primer PCR assay (LP-PCR) developed to specifically detect and quantify integrated HIV DNA species. This assay utilizes the presence of frequently occurring NlaIII restriction enzyme recognition sites in chromosomal DNA adjacent to the integrated provirus and at known positions within the proviral sequence. Linkers are ligated to the DNA termini generated by NlaIII digestion of chromosomal DNA and serve as templates from which priming can occur in a subsequent PCR amplifying the 5′-U3 HIV region and upstream cellular DNA sequence. In conjunction with other PCR-based assays, we have used LP-PCR to study the kinetics of total, integrated, and 2-LTR HIV DNA accumulation over time following a high-multiplicity infection of HuT-78 T cells with HIVHXB2. In addition, we also present results comparing LP-PCR to a nested Alu PCR method for the quantification of integrated HIV DNA.
Establishment of LP-PCR for the detection and quantification of integrated HIV DNA.
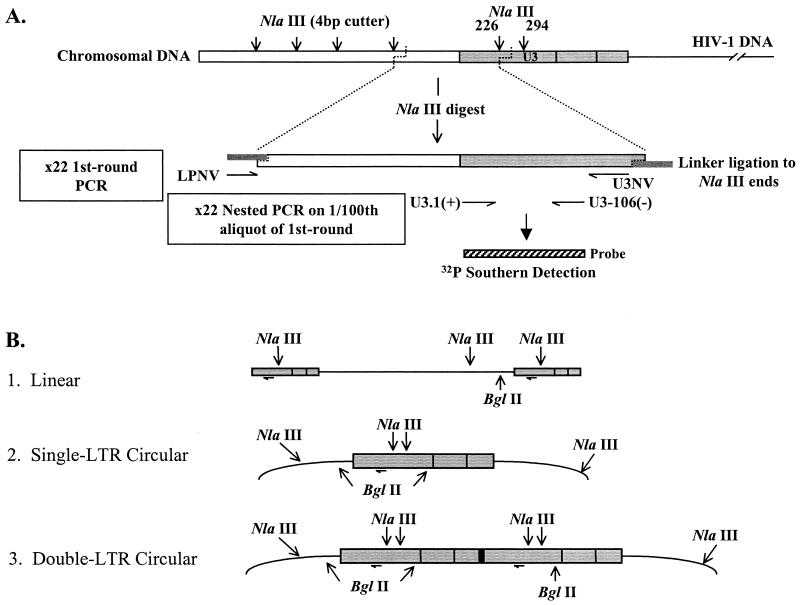
To specifically detect integrated HIV DNA in the presence of contaminating extrachromosomal viral DNA forms, we modified a previously described linker ligation PCR protocol used for sequence analysis of the human T-cell leukemia virus type 1 integration junctions (32). Briefly, chromosomal DNA was digested with the restriction enzyme NlaIII. NlaIII has a 4-bp recognition sequence and generates a 4-bp 3′ overhang to which the specifically designed oligonucleotide linker LPNV is annealed and ligated (Fig. 1A). This linker generates a region from which priming can occur in a subsequent PCR using the same linker oligonucleotide (LPNV) in conjunction with a primer (U3NV) designed to anneal within the U3 region of the HIV LTR. Since retroviral integration is random with respect to cellular sequences, LP-PCR generates a population of cellular-5′ HIV junction DNA sequences of various lengths. A nested PCR was performed to generate a product of a defined length, which was then quantified against a known set of standards (see below).
FIG. 1.
LP-PCR method for detection of integrated HIV DNA. (A) LP-PCR-mediated amplification of the integrated HIV DNA forms. The nested PCR product was detected using the U3-106 probe fragment (hatched box) (Table 1). (B) BglII-mediated selection against amplification of the three main extrachromosomal HIV DNA forms.
To avoid LP-PCR amplification of extrachromosomal viral DNA, the chromosomal DNA preparations were also digested with the restriction enzyme BglII. BglII has a recognition sequence of 6 bp and cleaves potential LP-PCR DNA templates within extrachromosomal HIV DNA forms, generating 4-bp 5′ overhangs to which LPNV cannot ligate (Fig. 1B). Religation of BglII fragments was inhibited by filling in two nucleotides (G and A) of the BglII site with Klenow (lacking 3′-5′ exonuclease activity [3′-5′ exo−]) polymerase before the ligation reaction was done. Due to the relative sizes of the restriction enzyme recognition sites, the chances of a BglII site occurring prior to an NlaIII site in the chromosomal sequence upstream of the 5′LTR is once every 16 integration events. Therefore, theoretically 94% of all integrated forms should be detectable by this technique.
The LP-PCR procedure was performed as follows: chromosomal DNA (isolated by the method of Hirt [17, 31]) was first digested to completion with 20 U of BglII (New England Biolabs) in 2× OPA Plus buffer (Pharmacia) for 3 h at 37°C in a final volume of 20 μl. Following this digestion, buffering conditions were then altered to final concentrations of 1× OPA Plus, 20 mM Tris-acetate (pH 7.9), 0.1 mg of bovine serum albumin (New England Biolabs) per ml, and 1 mM dithiothreitol (Boehringer Mannheim) prior to the addition of 10 U of NlaIII (New England Biolabs) and incubation at 37°C for 3 h in a final volume of 40 μl. All digestion reactions were confirmed to have proceeded to completion by both gel electrophoresis and PCR-based assays (data not shown). Two nucleotides (G and A) of the BglII overhang generated by digestion were filled in with 5 U of Klenow (3′-5′ exo−) (New England Biolabs) after modification of the buffering conditions to final concentrations of 7.5 mM dithiothreitol, 0.25 mM dGTP (Promega), and 0.25 mM dATP (Promega) and incubation at 37°C for 30 min in a final volume of 50 μl. Samples were then extracted with phenol-chloroform-isoamyl alcohol (25:24:1), ethanol precipitated in the presence of glycogen (Boehringer Mannheim), and washed in 70% ethanol prior to resuspension of the pellet in water. Linker ligation reactions in 1× ligation buffer (New England Biolabs) using 50 pmol (vast excess) of LPNV (Table 1) were heated to 60°C for 10 min and snap-cooled to minimize inter- and intramolecular ligation of NlaIII fragments, followed by the addition of 400 U of T4 DNA ligase (New England Biolabs) and incubation overnight at 16°C. First-round PCR was performed in 1× PCR Buffer II (Perkin-Elmer), 2 mM MgCl2, and 0.2 mM deoxynucleoside triphosphates (dNTPs) (Promega) using 150 pmol of LPNV, 100 pmol of U3NV (Table 1), and 5 U of AmpliTaq Gold DNA polymerase in a final volume of 100 μl. PCRs were as follows: 95°C for 12 min; 22 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min; and 72°C for 10 min. Nested PCRs were performed on 1/100 of the first-round PCR product in 1× PCR Buffer II (Perkin-Elmer), 1.5 mM MgCl2, and 0.2 mM dNTPs (Promega) using 25 pmol each of primers U3.1(+) and U3-106(−) (Table 1) and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer) in a final volume of 25 μl. PCRs were cycled as follows: 94°C for 3 min; 22 cycles of 94°C for 45 s, 58°C for 30 s, and 72°C for 45 s; and 72°C for 10 min. Amplified DNA was analyzed by subjecting 10 μl of each reaction mixture to electrophoresis through 8% polyacrylamide gels and then Southern transfer (electroblot apparatus) onto Hybond N+ nylon filters (Amersham). Following denaturation and fixation using 0.4 M NaOH, the filters were hybridized using the U3-106 probe (Table 1) in Ultrahyb solution (Ambion). Following Southern hybridization, bands were quantified using PhosphorImager ImageQuant analysis and a standard curve was generated from the simultaneous PCR of known copy numbers of standards.
TABLE 1.
Primer sequences and probes used in this studya
| Primer or probe | Sequence | Coordinates (nt) |
|---|---|---|
| Primers | ||
| PBS-659(−) | 5′-TTTCAGGTCCCTGTTCGGGCGCCAC-3′ | 659–635b |
| R7 | 5′-GGGTCTCTCTGGTTAGACC-3′ | 454–472b |
| U3NV | 5′-GGCTTCTTCTAACTTCTCTGGCTC-3′ | 179–156b |
| LPNV | 5′-TCATGATCAATGGGACGATCACATG-3′ | Same as B101c |
| U3PNV | 5′-GGTACTAGCTTGTAGCACCATCC-3′ | 151–129b |
| U3-106(−) | 5′-CCTGGCCCTGGTGTGTAGTTC-3′ | 106–86b |
| U3.1(+) | 5′-GGAAGGGCTAATTCACTCC-3′ | 2–20b |
| Alu164 | 5′-TCCCAGCTACTGGGGAGGCTGAGG-3′ | 164–187d |
| Probee | ||
| U3-106 | 2–106b |
In order to assess the sensitivity of the LP-PCR procedure, an integrated proviral DNA standard (designated HA8) was produced by mixing 5 × 105, 1 × 106, and 1 × 106 cells of the H3B (25), ACH-2 (11), and 8E5 (14) cell lines, respectively, and preparing chromosomal DNA by the method of Hirt (17, 31). These cell lines contain 2, 1, and 1 copies of the integrated HIV proviral DNA, respectively, with little or no detectable extrachromosomal forms (11, 14, 25). Rather than a single cell line, a mixture of three clonal cell lines was used as the integrated DNA standard to account for variations associated with different integration events. DNA cell equivalents were calculated based on the average signal obtained after PCR amplification of the β-globin gene (31) on six chromosomal DNA extractions from cells counted independently. The HA8 standards were used as copy number controls for quantifying total HIV DNA, integrated HIV DNA, and the β-globin content of samples. HuT-78 (15) chromosomal DNA was used as background DNA. To confirm that all four integration sites within the HA8 cell mix could be amplified by LP-PCR, the chromosomal sequence immediately upstream of the 5′ HIV LTR region of the integrated provirus(es) present in H3B, ACH-2, and 8E5 cells was obtained. In all cases, an NlaIII site preceded the BglII site in the flanking sequence (data not shown).
By comparison with the HA8 composite integrated HIV DNA standard, LP-PCR was shown to routinely detect 20 copies of integrated HIV-1 DNA in a background of 500 cell equivalents of HuT-78 chromosomal DNA (Fig. 2A). In addition, we were able to reproducibly detect 10 copies of the HA8 integrated standard in the presence of 2 × 105 cell equivalents of HuT-78 chromosomal DNA (1.2 μg) when elevated nested-PCR cycle numbers were used (data not shown). Furthermore, amplification of a construct precisely mimicking the linear viral DNA form spiked onto 1.2 μg of HuT-78 DNA routinely resulted in a signal intensity equivalent to ∼7.5% of that generated by an equivalent HIV DNA copy number of the HA8 integrated standard. This result indicated that LP-PCR was approximately 15-fold more specific for the integrated than extrachromosomal HIV DNA forms (data not shown). Sample heating and snap-cooling in the presence of a vast excess of the linker prior to the ligation reaction (to minimize intermolecular NlaIII fragment ligation), as well as the use of a hot-start PCR (to fully dissociate NlaIII fragments and inhibit linker-mediated amplification of all NlaIII fragments), were found to be critical to the success of this assay (data not shown). Furthermore, the efficiency of linker ligation to NlaIII termini was demonstrated to be approximately 100% (data not shown).
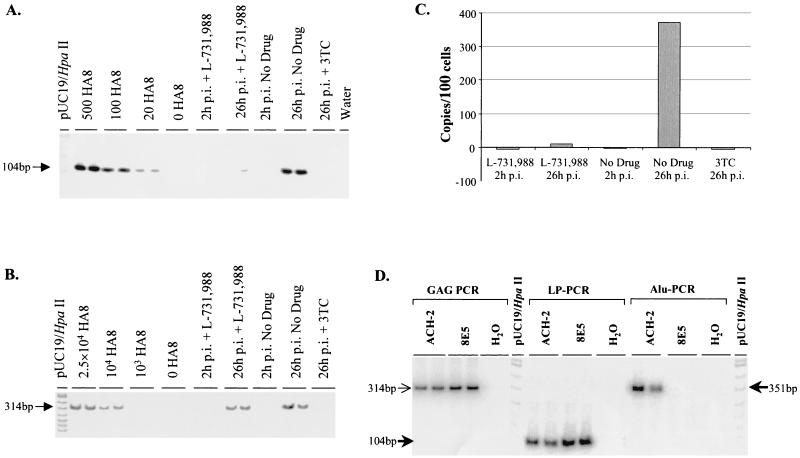
FIG. 2.
Sensitivity and specificity of LP-PCR and comparison with a nested Alu PCR protocol. (A to C) Viral DNA accumulation following cell-free infection in the presence or absence of inhibitors. HuT-78 T cells were infected using the centrifugal enhancement protocol at 0.5 TCID50 per cell and cellular DNA prepared from infected cells harvested at 26 h p.i. 3TC and L-731,988 were used as specific inhibitors of reverse transcription and integration, respectively. (A) Sensitivity of LP-PCR (as measured by amplification of the HA8 integrated HIV DNA standard) and integrated HIV DNA accumulation following infection as measured by LP-PCR performed on 100 cell equivalents of Hirt pellet (chromosomal) DNA preparations. (B) Total reverse-transcribed DNA as measured by GAG-PCR performed on combined Hirt supernatant (extrachromosomal) and Hirt pellet (chromosomal) DNA samples. (C) Graphical representation of the accumulation of integrated HIV DNA. Data were obtained by PhosphorImager analysis of the bands in panel A. (D) Comparison of PCR detection of integrated HIV DNA by LP-PCR and Alu PCR. Chromosomal DNA was isolated from ACH-2 or 8E5 cells and shown to contain equivalent amounts of total HIV DNA by GAG-PCR (314-bp band). Sizes of expected bands for LP-PCR (measuring integrated HIV DNA) are given on the left (104-bp fragment), while the expected size of the product obtained following Alu PCR (also measuring integrated HIV DNA) is indicated on the right (351-bp fragment).
To further confirm that extrachromosomal DNA forms were not detected by the LP-PCR procedure, HuT-78 cells were infected with HIVHXB2 (0.5 50% tissue culture infective dose [TCID50] per cell) in the presence or absence of the integration inhibitor L-731,988 (16) as previously described (31). The reverse transcriptase inhibitor lamivudine (3TC; final concentration, 10 μM) served as a control for inhibition of extrachromosomal HIV DNA synthesis prior to integration. Following analyses of 100 cell equivalents of cellular DNA using LP-PCR and a GAG-PCR protocol (31), strong signals corresponding to total and integrated HIV DNA were observed by 26 h p.i. in drug-free cultures, respectively (Fig. 2A and B). As expected, cultures infected in the presence of 3TC were negative for both total and integrated HIV DNA. Analysis of DNA from cells infected in the presence of L-731,988 indicated that the accumulation of integrated HIV DNA had been abolished (Fig. 2A and C), while the accumulation of extrachromosomal HIV DNA was largely unaffected (Fig. 2B). This result clearly demonstrates that LP-PCR specifically detects integrated HIV DNA.
To further characterize the LP-PCR procedure, a direct comparison of LP-PCR and a previously published method for the detection of integrated HIV DNA (a nested Alu PCR protocol [31]) was performed. Chromosomal preparations of the clonal cell lines ACH-2 and 8E5 (each containing one copy of integrated provirus) were shown to contain equivalent amounts of viral DNA by GAG-PCR (31) and then subjected to the LP-PCR and the nested Alu PCR procedures to detect integrated HIV DNA. While integrated DNA within the ACH-2 cell line was efficiently amplified, the nested Alu PCR method was unable to facilitate amplification of integrated DNA in the preparation of 8E5 chromosomal DNA (Fig. 2D). In contrast, the LP-PCR procedure allowed the efficient amplification of integrated DNA present in both cell lines (Fig. 2D). BLAST analyses of chromosomal sequence upstream of HIV integration sites in the 8E5 and ACH-2 cell lines revealed that in 8E5 cellular DNA, the Alu repeat element immediately upstream of the integrated DNA existed in the same orientation as the PBS-659(−) primer. Consequently, amplification between Alu elements upstream of the integration site in 8E5 cells, instead of amplification between the Alu164 and the PBS-659(−) primers, would have occurred. In contrast, the analogous Alu element in ACH-2 chromosomal DNA was present in the correct orientation for successful amplification with the PBS-659(−) primer (data not shown). We therefore propose that the nested Alu PCR technique allows amplification of only those integrants inserted at chromosomal sites immediately adjacent to an Alu element present in an orientation opposite to that of the PBS-659(−) primer. Statistically, then, the nested Alu PCR approach would be expected to successfully amplify at best half of all integrated proviral forms. Consequently, we believe that the LP-PCR assay is a potentially more appropriate protocol for detecting integrated HIV DNA. A comprehensive comparison between LP-PCR, nested Alu PCR, and an alternative assay currently being developed in our laboratory to detect integrated proviral forms based on the use of degenerate primers will be published elsewhere.
Kinetics of HIV-1 DNA integration following a one-step viral infection of HuT-78 cells.
To investigate the kinetics of viral DNA accumulation following infection, a highly synchronous one-step infection of HuT-78 cells with cell-free HIV at a multiplicity of infection of 1 TCID50 per cell was performed as previously described (31). Extensive washing of cells to remove residual noninternalized virus prior to plating minimized the chance of any additional infection events occurring after the initial infection period. Viral release into the culture supernatant following infection (as measured by P24 release using a commercial kit [NEN]) was evident by 26 h p.i., indicating that one round of replication was complete by this stage (see Fig. 4A). Consequently, the proportions of each viral DNA form assessed following infection were calculated at 26 h p.i. to ensure that the results obtained were not skewed by secondary cell-free (or cell-to-cell) infection events.
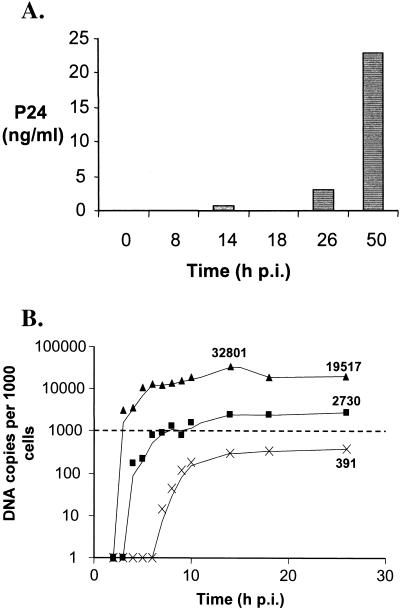
FIG. 4.
HIV replication parameters following high-multiplicity infection of HuT-78 T cells. Cells were infected with HIVHXB2 at 1 TCID50 per cell using a centrifugally enhanced protocol. (A) P24 levels in culture supernatants were measured at various time points. (B) Comparison of the total (▴), integrated (■), and 2-LTR (×) HIV DNA forms. Data were determined by PhosphorImager quantification of bands in Fig. 3. The levels of total, integrated, and 2-LTR DNA at 26 h p.i. and total HIV DNA at 14 h p.i. are shown. The dashed line indicates one DNA copy/cell.
DNA was extracted by the method of Hirt (17, 31) from infected cells harvested at various time points following infection to ensure that the bulk (>80%) of extrachromosomal forms were separated from the chromosomal DNA forms. Chromosomal DNA preparations were subjected to PCR amplification of the β-globin gene to determine the cell equivalent DNA content of each sample. Samples were volume adjusted based on the results of initial β-globin PCR quantification, and upon reanalyses, little variation between samples was observed (Fig. 3D). Extrachromosomal DNA preparations were equalized between samples based on a semiquantitative PCR assay (31) measuring the mitochondrial complement of this fraction. The mitochondrial DNA PCR results showed only minor variation between all samples, and therefore adjustment was not necessary (Fig. 3E).
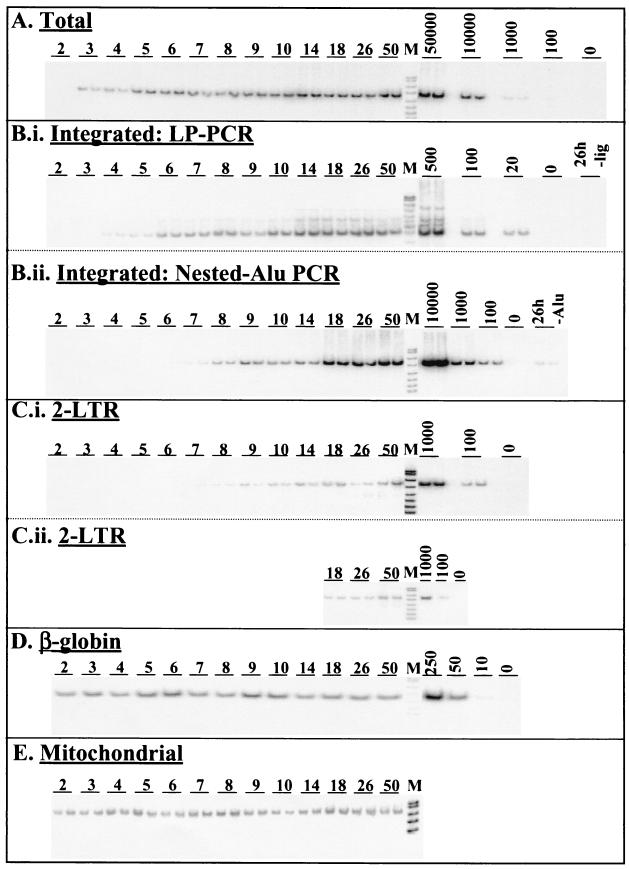
FIG. 3.
Accumulation kinetics of total, integrated, and 2-LTR viral DNA forms following high-multiplicity infection of HuT-78 T cells. Infections were performed using 1 TCID50 of HIVHXB2 per cell with centrifugal enhancement. All PCRs were confirmed to amplify DNA in a linear fashion by quantification of standards (A to D) or dilution sets (E) (dilutions not shown). DNA markers (pUC19/HpaII) are indicated (M). (A) Total HIV DNA forms as measured by GAG-PCR using 500 cell equivalents of total DNA (combined Hirt supernatant and Hirt pellet). (B.i) Integrated HIV DNA levels as measured by LP-PCR on 100 cell equivalents of chromosomal DNA (Hirt pellet). (B.ii) Integrated HIV DNA levels as measured by the modified nested Alu PCR method performed on 1,000 cell equivalents of chromosomal DNA. (C.i) 2-LTR HIV DNA levels as measured by 2-LTR PCR on 500 cell equivalents of total DNA. (C.ii) Reanalysis of later time points for the 2-LTR DNA forms using 1,000 cell equivalents of total DNA. (D) β-Globin levels assayed by PCR on 50 cell equivalents of chromosomal DNA. Standards represent amplification of various amounts (based on cell counts) of HA8 chromosomal DNA. (E) Mitochondrial DNA levels assayed by PCR on 50 cell equivalents of Hirt supernatant (extrachromosomal) fraction.
The total viral DNA complement was measured by mixing Hirt supernatant and Hirt pellet DNA fractions from the same time points and analyzing the pooled samples by PCR for the presence of GAG DNA sequences that are synthesized in the mid-late stages of the HIV reverse transcription process (20). PCRs were performed on 500 cell equivalents of DNA as described previously (31). The results (Fig. 3A) indicated that near full-length viral DNA was detected approximately 3 h after infection, which is in close agreement with previous studies (1, 21, 25). PhosphorImager analysis of bands showed that total HIV DNA had peaked at a level of approximately 30 copies/cell at 14 h p.i. and declined to levels of approximately 20 copies per cell by 26 h p.i. (Fig. 4B). The reduction in the total viral DNA complement after 14 h p.i. (∼40%) can be attributed to the degradation of extrachromosomal HIV DNA within the cellular environment. This result is consistent with a previous report showing that significant proportions of reverse-transcribed viral DNA degrade within the intracellular environment following cell-to-cell infection (1). Most viral DNA had been reverse transcribed by 5 h p.i. (Fig. 4B), providing further evidence that this one-step HIV infection was nearly synchronous. Consistent with the results of P24 release following infection (Fig. 4A), the GAG signal at 50 h p.i. was higher than at 26 h p.i., implying that some degree of either second-round cell-free or cell-to-cell infection (superinfection) may have occurred during this time.
Integrated HIV DNA levels were measured using both the LP-PCR procedure (Fig. 3, panel B.i) and a modified version of the previously published nested Alu PCR protocol (31) in chromosomal DNA preparations of infected cells harvested at each time-point (Fig. 3, panel B.ii). Integrated DNA was first detected by LP-PCR (performed on 100 cell equivalents of chromosomal DNA) at 4 h p.i. (that is, 1 h after the first appearance of newly synthesized viral DNA) (Fig. 3, compare panels A and B.i) and the level reached approximately three copies/cell by 26 h p.i. In contrast, when the nested Alu PCR method was performed (on 1,000 cell equivalents of DNA), integrated provirus was first detected considerably later and repeatedly displayed lower levels (Fig. 3, panel B.ii) across all time points tested. Control experiments involving first-round amplification of 26-h-p.i. samples performed in the absence of ligation (LP-PCR) or the Alu164 primer (nested Alu PCR) (Table 1) resulted in bands of very low intensity (Fig. 3, panels B.i and B.ii). This indicated that the signals obtained when amplification was performed in the presence of both ligation (LP-PCR) and the Alu164 primer (nested Alu PCR) indeed resulted from the specific amplification of integrated HIV DNA and not the nested amplification of input target sequences. The discrepancy between integration levels observed when either the LP-PCR or the nested Alu PCR protocol was used was expected and was presumed to result from the ability of the nested Alu PCR approach to detect only a small proportion of integration events (Fig. 2D). Therefore, integrated HIV DNA, at the end of the first round of HIV replication (i.e., 26 h p.i.), was found to account for approximately 10% of the total HIV DNA synthesized at the peak of viral DNA accumulation (i.e., 14 h p.i.) (Fig. 4B).
We also monitored accumulation of the 2-LTR viral DNA forms using a specific PCR amplification protocol with primers flanking the dual-repeat cassette within the circular form. To allow quantification of 2-LTR viral DNA levels, a control construct was generated by PCR amplification of Hirt supernatant samples taken from a cell-free infection of HuT-78 cells at 26 h p.i. using primers R7 and U3NV (Table 1). The 2-LTR control construct was precisely quantified (based on LTR copy number) by comparative PCR amplification against the HA8 standard mix using primers U3.1(+) and U3-106(−) (Table 1) in appropriate amounts of background DNA (data not shown). Quantification of 2-LTR circular DNA following infection was achieved by performing PCR on 500 cell equivalents of total DNA in 1× PCR Buffer II (Perkin-Elmer), 1.5 mM MgCl2, and 0.2 mM dNTPs using 25 pmol each of primers R7 and U3PNV and 1.5 U of AmpliTaq Gold DNA polymerase. Reactions were cycled as follows: 94°C for 12 min; 26 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 45 s; and 72°C for 10 min. The initial results (Fig. 3, panel C.i) clearly showed continuing 2-LTR DNA accumulation from 7 h p.i. onwards, which is in close agreement with previous studies (21). An unexpectedly low value was obtained for the 26-h-p.i. sample. Since this time point was to be used for our endpoint analysis, we reanalyzed this sample and the adjacent time points using 1,000 cell equivalents of DNA (Fig. 3, panel C.ii). Taken together, these results showed that the 2-LTR viral DNA form is a minor species compared to the integrated DNA form, with levels of ∼0.4 copy/cell (representing ∼1% of the total viral DNA species) at 26 h p.i. (Fig. 4B).
In our infection model, nearly full-length DNA species were first detected at 3 h p.i., with the appearance of integrated forms at 4 h p.i. Integrated HIV DNA as measured by LP-PCR was found to comprise approximately 10% of the total viral DNA synthesized following one round of infection (Fig. 4B). While we believe such levels to represent an efficient process, care should be taken when the integration efficiencies observed in cell-free T-cell infection systems are used for predicting the integration efficiencies in vivo. Our infection model involves the use of actively growing T cells and a high multiplicity of infection. Consequently, this model gave rise to a large number of viral DNA molecules, which might be expected to compete for cellular factors involved in a variety of early events in HIV replication, including integration. It is possible that in HIV-infected patients, where these factors might not be as limiting, the efficiency of HIV integration would be higher as measured by the amounts of integrated viral DNA expressed as a percentage of the total viral DNA. Furthermore, major differences exist in vivo with respect to not only the various activation states of T cells but also the cell type(s) infected. Cells of the macrophage/monocyte lineage are generally considered to be nondividing cells, and the early events in infection of these cells differ markedly from those observed in proliferating T cells (29, 33). Thus, the kinetics of integration within the monocyte/macrophage cell lineage should be considered in a separate study.
In conclusion, we have established a system in which the amounts of integration can be measured over the course of an infection with HIV-1. We have also demonstrated that HIV-1 integration is a rapid and relatively efficient process under one-step infection conditions and defined the levels of total, integrated, and 2-LTR HIV DNA forms during the course of infection. While this article was being revised, a short report was published that supports the conclusions of this work by demonstrating similar frequencies of proviral integration into host chromosomal DNA following a one-step, cell-free infection model (4). Studies are now under way to investigate the mechanisms of integration and the efficiency with which potential integrase inhibitors affect the accumulation of integrated proviral DNA in infected cells under similar infection conditions.
Acknowledgments
We thank Linda Mundy and Helen Hocking for preparing the viral stocks and Melissa Egberton and Steven Young (Merck and Co.) for the sample of L-731,988 used in this study.
This work was supported by the Australian Commonwealth AIDS Research Grant Programme.
REFERENCES
- 1.Barbosa P, Charneau P, Dumey N, Clavel F. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res Hum Retrovir. 1994;10:53–59. doi: 10.1089/aid.1994.10.53. [DOI] [PubMed] [Google Scholar]
- 2.Bukrinsky M I, Sharova N, Dempsey M P, Stanwick T L, Bukrinskaya A G, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler S L, Hansen M S, Bushman F D. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun T W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y H, Brookmeyer R, Zeiger M A, Barditch-Crovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 7.Chun T W, Engel D, Berrey M M, Shea T, Corey L, Fauci A S. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun T W, Fauci A S. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc Natl Acad Sci USA. 1999;96:10958–10961. doi: 10.1073/pnas.96.20.10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun T W, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano R F. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 10.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clouse K A, Powell D, Washington I, Poli G, Strebel K, Farrar W, Barstad P, Kovacs J, Fauci A S, Folks T M. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J Immunol. 1989;142:431–438. [PubMed] [Google Scholar]
- 12.Englund G, Theodore T S, Freed E O, Engleman A, Martin M A. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J Virol. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 14.Folks T M, Powell D, Lightfoote M, Koenig S, Fauci A S, Benn S, Rabson A, Daugherty D, Gendelman H E, Hoggan M D. Biological and biochemical characterization of a cloned Leu-3− cell surviving infection with the acquired immune deficiency syndrome retrovirus. J Exp Med. 1986;164:280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazdar A F, Carney D N, Bunn P A, Russell E K, Jaffe E S, Schechter G P, Guccion J G. Mitogen requirements for the in vitro propagation of cutaneous T-cell lymphomas. Blood. 1980;55:409–417. [PubMed] [Google Scholar]
- 16.Hazuda D J, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler J A, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller M D. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 17.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 18.Jurka J, Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci USA. 1988;85:4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retrovir. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 20.Karageorgos L, Li P, Burrell C J. Stepwise analysis of reverse transcription in a cell-to-cell human immunodeficiency virus infection model: kinetics and implications. J Gen Virol. 1995;76:1675–1686. doi: 10.1099/0022-1317-76-7-1675. [DOI] [PubMed] [Google Scholar]
- 21.Kim S Y, Byrn R, Groopman J, Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaFemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Simm M, Potash M J, Volsky D J. Human immunodeficiency virus type 1 DNA synthesis, integration, and efficient viral replication in growth-arrested T cells. J Virol. 1993;67:3969–3977. doi: 10.1128/jvi.67.7.3969-3977.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Yoder K, Hansen M S, Olvera J, Miller M D, Bushman F D. Retroviral cDNA integration: stimulation by HMG I family proteins. J Virol. 2000;74:10965–10974. doi: 10.1128/jvi.74.23.10965-10974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P, Burrell C J. Synthesis of human immunodeficiency virus DNA in a cell-to-cell transmission model. AIDS Res Hum Retrovir. 1992;8:253–259. doi: 10.1089/aid.1992.8.253. [DOI] [PubMed] [Google Scholar]
- 26.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauza C D, Galindo J. Persistent human immunodeficiency virus type 1 infection of monoblastoid cells leads to accumulation of self-integrated viral DNA and to production of defective virions. J Virol. 1989;63:3700–3707. doi: 10.1128/jvi.63.9.3700-3707.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai H, Kawamura M, Sakuragi J, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonza S, Maerz A, Deacon N, Meanger J, Mills J, Crowe S. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J Virol. 1996;70:3863–3869. doi: 10.1128/jvi.70.6.3863-3869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandegraaff N, Kumar R, Hocking H, Burke T, Mills J, Rhodes D, Burrell C J, Li P. Specific inhibition of human immunodeficiency virus type 1 (HIV-1) integration in cell culture: putative inhibitors of HIV-1 integrase. Antimicrob Agents Chemother. 2001;45:2510–2516. doi: 10.1128/AAC.45.9.2510-2516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wattel E, Vartanian J P, Pannetier C, Wain-Hobson S. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J Virol. 1995;69:2863–2868. doi: 10.1128/jvi.69.5.2863-2868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg J B, Matthews T J, Cullen B R, Malim M H. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med. 1991;174:1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanussi S, Bortolin M T, Giacca M, De Paoli P. Quantitative assessment of integrated and episomal HIV DNA. AIDS. 2000;16:931–933. [PubMed] [Google Scholar]