Abstract
Beyond its clinical diversity and severity, acute myeloid leukemia (AML) is known for its complex molecular background and for rewiring biological processes to aid disease onset and maintenance. FLT3 mutations are among the most recurring molecular entities that cooperatively drive AML, and their inhibition is a critical molecularly oriented therapeutic strategy. Despite being a promising avenue, it still faces challenges such as intrinsic and acquired drug resistance, which led us to investigate whether and how autophagy and inflammasome interact and whether this interaction could be leveraged to enhance FLT3 inhibition as a therapeutic strategy. We observed a strong and positive correlation between the expression of key genes associated with autophagy and the inflammasome. Gene set enrichment analysis of the FLT3-ITD samples and their ex vivo response to five different FLT3 inhibitors revealed a common molecular signature compatible with autophagy and inflammasome activation across all poor responders. Inflammasome activation was also shown to strongly increase the likelihood of a poor ex vivo response to the FLT3 inhibitors quizartinib and sorafenib. These findings reveal a distinct molecular pattern within FLT3-ITD AML samples that underscores the necessity for further exploration into how approaching these supportive parallel yet altered pathways could improve therapeutic strategies.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-74168-z.
Keywords: Acute myeloid leukemia, Autophagy, Inflammasome, FLT3-ITD inhibitors, Drug resistance
Subject terms: Targeted therapies, Haematological cancer
Introduction
Acute myeloid leukemia (AML) is a remarkably heterogeneous group of clonal hematological disorders whose pathogenesis consists of the accrual of multiple genetic and epigenetic alterations of hematopoietic stem and progenitor cells1–5. The different molecular entities that cooperatively drive leukemogenesis not only provide the hematopoietic stem and progenitor cells with malignant features such as abnormal stemness and enhanced fitness, but also yield a diverse array of molecular backgrounds with potential for molecularly oriented therapeutic strategies for the disease6–8.
Among the molecular entities that support leukemogenesis, alterations in the FMS-like tyrosine kinase III (FLT3) receptor are considerably frequent and are present in roughly one-third of all AML patients8–10. Additionally, patients harboring FLT3-internal tandem duplication (ITD), approximately one-quarter of all AML patients, were recently classified as intermediate risk according to the molecular risk stratification guidelines proposed by the European LeukemiaNet consortium10–13. Therapy strategies for intermediate-risk AML patients are limited and ill-defined due to the lack of differentially effective approaches for clinical management14; therefore, efforts to either reclassify these patients or detect vulnerabilities and biological features that could be exploited as potential avenues for therapy are a pressing matter.
In addition to genetic alterations, therapy opportunities might also lie in altered biological processes, a hallmark of AML onset and maintenance3,4,15. Biological processes such as autophagy and inflammasome are not only described to be functionally related in healthy contexts16–22, but also to be altered in AML and positively affect the biological fitness of AML cells in an isolated fashion1,23–29.
Given these previous findings, in the present study, we sought to further characterize the molecular profile of FLT3-ITD AML samples, and the relationships among the investigated biological processes, autophagy and inflammasome, and the ex vivo response to five different FLT3 small molecule inhibitors: crenolanib; gilteritinib; midostaurin, quizartinib, and sorafenib. In this way, we can investigate whether associated altered biological processes could be considered and exploited as additional targets to overcome drug resistance and improve AML therapy.
Results
Central autophagy and inflammasome gene expression are mutually and positively correlated in acute myeloid leukemia samples
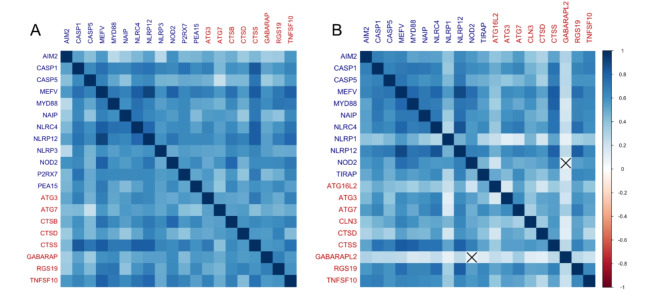
The correlation matrix is depicted considering the top 20 genes most strongly and positively correlated among all tested genes within autophagy and inflammasome genes (Supplementary Table). The TCGA acute myeloid leukemia (AML) cohort presented 12 out of 20 genes (60%) associated with inflammasome, and 8 out of 20 genes (40%) associated with autophagy (Fig. 1A). In the BeatAML 1.0 cohort, 11 out of 20 genes (55%) were associated with inflammasome genes, while 9 out of 20 genes (45%) were associated with autophagy (Fig. 1B). These correlation analyses revealed a 75% overlap between the top 20 positively correlated genes in both cohorts, nine inflammasome genes (AIM2, CASP1, CASP5, MEFV, MYD88, NAIP, NLRC4, NLRP12, and NOD2) and six autophagy genes (ATG3, ATG7, CTSD, CTSS, RGS19, and TNFSF10) (Figure S9).
Fig. 1.
Spearman’s correlation matrix of autophagy and inflammasome gene expression in adult AML patients from TCGA AML and BeatAML 1.0 cohorts. (A) Plot represents the TCGA AML cohort, and (B) the BeatAML 1.0 cohort data. To the right, a color scale indicates the nature and strength of the correlation according to Spearman’s rho. Each element that composes this matrix corresponds to a single correlation analysis; the diagonal of the matrix represents perfect and positive correlations, since they are being established between a gene and its own expression value. The gene symbols listed in dark blue are for inflammasome genes, while gene symbols in red are for autophagy genes. The crossed cells in the matrix represent the correlations that did not reach the pre-established statistical level of significance.
Monocyte-like cells are the main leukemic cell population as transcriptional contributor to the expression of overlapping key autophagy and inflammasome genes in AML
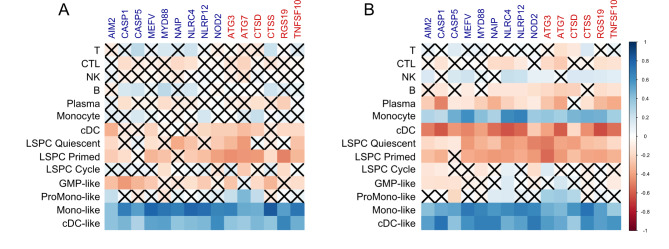
Once we observed that central autophagy and inflammasome genes are highly correlated in AML samples, we aimed to identify whether a particular component of cellularly heterogeneous samples plays a relevant role in the expression of these genes. Higher Spearman’s correlation coefficients between a particular gene’s expression and a CIBERSORTx-based cell population’s score ultimately indicate which cellular compartment contributes most to this specific gene expression within the complex sample transcriptional signal. The transcriptional deconvolution of the overlapping genes revealed that both autophagy-related and inflammasome-related genes are expressed, majorly and consistently, by monocyte-like and classical dendritic cell-like cell populations (Fig. 2A and B). In addition, in the BeatAML 1.0 cohort, even the non-malignant, regular monocytic compartment holds an important contribution to the gene expression (Fig. 2B). Also, our analysis was able to satisfactorily distinguish different cell lineages on TCGA and BeatAML 1.0 cohorts’ data according to the employment of lineage-defining genes (Figure S10). Therefore, we can safely assume an association between a cell type or a maturation stage and gene expression.
Fig. 2.
Spearman’s correlation matrix plot between autophagy and inflammasome overlapping gene expression and compartmental transcriptional contribution scores in adult AML. To the right, according to Spearman’s rho, a color scale indicates the correlation’s nature and strength. Each element that composes this matrix corresponds to a single correlation analysis. Gene symbols in dark blue are for inflammasome genes, gene symbols in red are for autophagy genes, and the black-labeled text is for cell compartments that compose a complex mixture sample. The crossed cells in the matrix represent the correlations that did not reach the pre-established statistical level of significance. The (A) plot represents the TCGA AML cohort, and (B), the BeatAML 1.0 cohort data. Abbreviations: T, T cells; CTL, cytotoxic T cells; NK, natural killer cells; B, B cells; Plasma, plasma cells; cDC, classical dendritic cells; LSPC, leukemic stem and progenitor cells, GMP, granulocyte-monocyte progenitor cells. The denomination “-like” regards malignant cell compartments that are, to a certain degree, molecularly similar to their non-malignant counterparts, albeit still being neoplastic cells.
Acute myeloid leukemia samples without FLT3-ITD from TCGA AML cohort and BeatAML 1.0 cohort exhibit enrichment of inflammatory molecular signatures
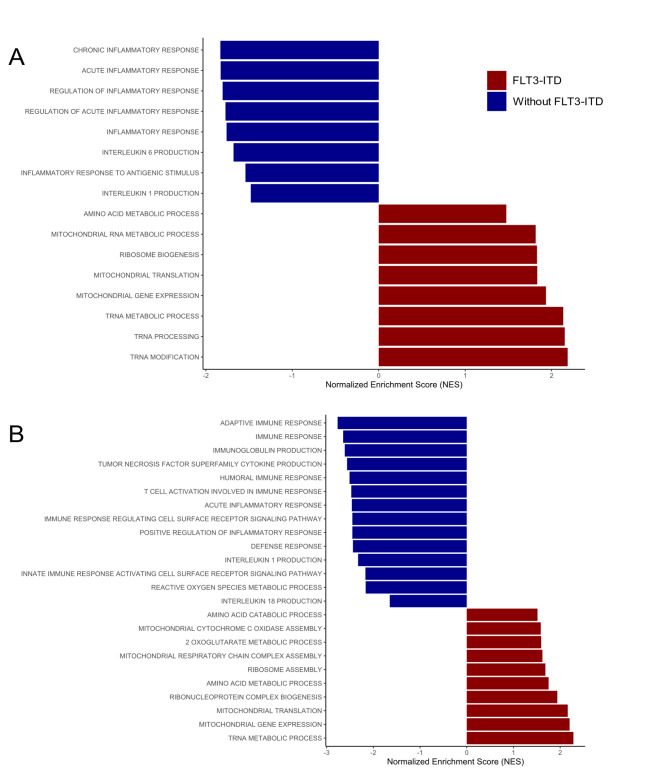
To assess how FLT3-ITD-positive samples would molecularly behave and whether these samples would inherently enrich the molecular signatures of interest, that is, gene sets compatible with autophagy activation when compared to samples without FLT3-ITD, a gene set enrichment analysis (GSEA) was performed. The GSEA has revealed that AML samples that do not bear the FLT3-ITD mutation present a predominantly pro-inflammatory molecular signature, compatible with inflammasome activation, whereas samples harboring the FLT3-ITD mutation present a molecular signature that implies increased mitochondrial activity and suggesting an oxidative metabolic state (Fig. 3).
Fig. 3.
Gene set enrichment analysis (GSEA) depicting the molecular signature of AML samples according to the presence and absence of FLT3-ITD. The positively enriched gene sets represent the FLT3-ITD group according to the normalized enrichment score (NES), since they are the reference group for the comparison. In contrast, the negatively enriched gene sets are attributed to the AML samples without FLT3-ITD. (A) TCGA AML cohort GSEA; and (B) BeatAML 1.0 cohort GSEA. Both GSEAs were based on gene sets derived from the collection of human gene sets from Gene Ontology: Biological Process (GOBP).
The molecular signatures of FLT3-ITD-positive AML patients who are poorex vivo responders to FLT3 inhibitors are compatible with autophagy and inflammasome activation
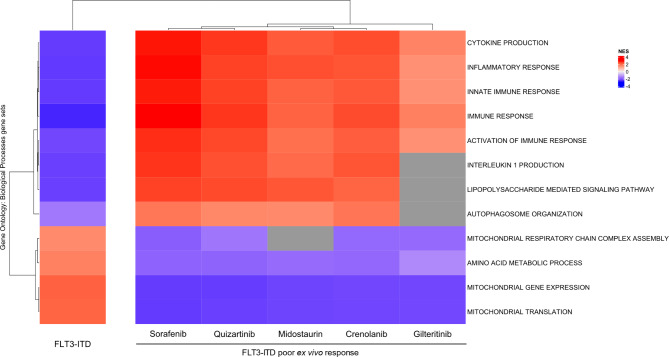
Molecular signatures such as the inflammatory response, cytokine production, innate, adaptive, cellular, and humoral immune responses, LPS-mediated signaling, and autophagy organization were consistently present as molecular patterns for the group of FLT3-ITD AML samples that are poor responders to all tested FLT3 inhibitors when compared to good responders (Fig. 4, Figure S4-S8), while the opposite behavior was observed in the FLT3-ITD samples when compared to samples without FLT3-ITD (Figs. 3 and 4).
Fig. 4.
Gene set enrichment analysis (GSEA) comparing the molecular signatures of FLT3-ITD samples with poor ex vivo response to crenolanib, gilteritinib, midostaurin, quizartinib and the molecular signature of the overall presence of FLT3-ITD mutation itself. The leftmost column represents the molecular signature attributed to FLT3-ITD samples derived from the comparison between FLT3-ITD and samples without FLT3-ITD. The remaining columns represent the molecular signature attributed to poor ex vivo responders to each of the indicated FLT3 inhibitors established from the comparison between FLT3-ITD samples that were poor and good responders according to their area under the curve (AUC) response value. The measurement of enrichment in nature and intensity was determined by the Normalized Enrichment Score (NES). The cells in shades of red are positively enriched, whilst the cells in shades of blue are negatively enriched on the indicated condition. The gray cells at the heatmap represent gene sets whose NES have not met the pre-established statistical criteria for significance. All gene sets were obtained from the Gene Ontology: Biological Process (GOBP) human gene set collection.
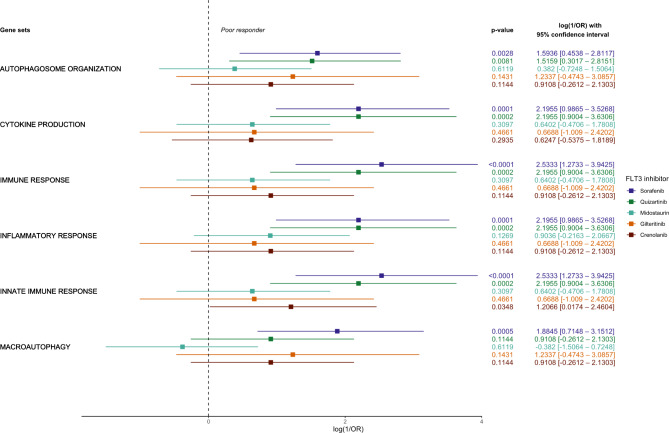
Higher inflammatory and autophagic molecular signature scores are associated with poorer response of FLT3-ITD AML patients to FLT3 inhibitors
The most recurrent inflammasome- and autophagy-related Gene Ontology: Biological Process (GOBP) collection gene sets among the poor responders (autophagosome organization, cytokine production, immune response, inflammatory response, innate immune response, and macroautophagy) were selected for ssGSEA-derived association analyses with FLT3-ITD ex vivo FLT3 inhibitors response. In contrast to conventional group-oriented GSEA, ssGSEA offers individualized enrichment scores for each sample related to a gene set in the GOBP collection. The individualized enrichment score allowed for arbitrary discretization of the samples based on the median of the enrichment score into what we denominated higher and lower signature samples. Once each sample is classified into categories of ex vivo response and molecular signature, association analysis employing Fisher’s exact test is possible. The effect size of the association was measured in odds ratio using good response as the reference group. Therefore, the reciprocal odds ratio (1/OR) was calculated to approach increases in the chance for a sample to be classified as a poor responder according to their enrichment degree for the selected gene sets.
Except for the innate immune response molecular signature for the FLT3-ITD samples treated ex vivo with crenolanib, no type I FLT3 inhibitors (crenolanib, gilteritinib, and midostaurin) presented association between the degree of molecular enrichment and the intrinsic ex vivo drug response to a single sample extent. Otherwise, sorafenib and quizartinib, both type II FLT3 inhibitors, were strongly associated with increased poor response likelihood and higher molecular enrichment statuses. In other words, higher enrichment scores for inflammatory increased the chances of being a poor responder to quizartinib and sorafenib in 9 to 12 times, while higher enrichment scores for autophagy-related gene sets increases the chances of being a poor responder by approximately 5 to 6.5 times, except for the macroautophagy gene set and quizartinib response which were not significantly associated (Fig. 5).
Fig. 5.
Forest plot representing the extension of the association between the enrichment score for autophagy- and inflammasome-related biological processes in single sample regimen and drug response for FLT3-ITD samples to FLT3 inhibitors. The x-axis represents the natural logarithm of the reciprocal odds ratio [log (1/OR)] from a Fisher’s exact test of discretized enrichment score (high or low enrichment), and FLT3-ITD ex vivo drug response (poor and good response). Roughly, a 2.3 log (1/OR) indicates an absolute 10-fold increase in the chance of the event occurring. In this case, the chance assessed is for poor response. To the right of the plot, the p-value, natural logarithm of the odds ratio, and 95% confidence interval are displayed. The selected gene sets stemmed from the Gene Ontology gene set collection that were overlapped among all assessed FLT3 inhibitors.
Discussion
The correlation analyses in this work suggest the existence of an alleged transcriptional balance between autophagy and the inflammasome in the context of AML. A positive mutual correlation indicates appreciable behavioral coincidence from a transcriptomic standpoint that potentially entails functional interactions. This nature of the relationship may point toward a regulatory character when considering their inherent biological features. The high overlap rate depicted among the genes in both cohorts potentially indicates that the inferred interaction is stable across the patients and may represent a much-desired conserved integration between the biological processes in a disease widely known for its heterogeneity.
Genes that encode alternative inflammasome sensor proteins such as AIM2, NAIP, NLRC4, and MEFV, were among the list of overlapping positively correlated genes between the cohorts. These inflammasome sensor proteins are also able to assemble inflammasome complexes as well as NLRP3 sensor protein and share functional endpoints such as gasdermin-D cleavage-induced pyroptosis, along with maturation and release of interleukin-1β and interleukin-1817. The caspase-1 gene, the main canonical effector protein and functional endpoint of inflammasome assembly, as well as the caspase-5 gene, responsible for alternative non-canonical interleukin-1β maturation were also overlapping inflammasome-related genes between both cohorts17. As for autophagy overlapping genes, ATG3 and ATG7 are crucial to autophagosome maturation, thus continuity of autophagic flux30,31; and it is worth noting the presence of lysosomal hydrolases cathepsin D and cathepsin S as important effectors of autophagy. The MEFV gene encodes the TRIM20 protein, which represents an important and well-described interface between autophagy and inflammasome19; this observation further endorses the mutual regulatory relationship between the processes in AML context, a feature first described in this work.
We would like to highlight the presence of the NLRP12 gene as a gene overlapped in both AML databases. Even though it shares homology to a certain extent with pro-inflammatory inflammasome sensor proteins, NLRP12 is widely described as an inflammation suppressor in activated monocytes32,33. In fact, NLRP12 mutations frequently result in protein loss-of-function, which is often associated with the onset of autoinflammatory diseases such as familial cold autoinflammatory syndrome type 234,35. NLRP12 was also described to inhibit canonical and non-canonical NF-κΒ activation pathways, which are important for NLRP3 inflammasome priming in dendritic cells and macrophages32.
Carey et al.., by immunophenotyping AML primary samples, observed that IL-1 expression originated mostly from monocytic cells24. This observation aligns with the findings in our transcriptional deconvolution analyses in which all overlapping genes related to the inflammasome were revealed to be especially expressed on samples which possessed an appreciable monocytic molecular signature, either malignant or stromal. Therefore, these cell populations seemingly play a crucial role in creating a leukemia-favorable microenvironment inflammation-wise and are the major promoters of IL-1β production. It is worth emphasizing that Carey et al.., have shown that IL-1β is an important normal CD34-positive hematopoietic cell suppressor, while enhancing AML cells growth and survival24.
Even though LSCs deeply rely on mitophagy to harmonize their aberrant stemness with low internal oxidative stress, we have found the autophagy transcriptional signal to be sustained mainly by monocytic and monocyte-like cellular compartments, which suggest that such observation stems from a lineage-specific gene expression character instead of an AML-specific. Considering that LSCs are a scarce cell population within complex mixtures as the bone marrow, their cellular compartment signal would be too weak, it is a limitation that could be circumvented by sorting and enriching samples for this particular cell compartment. Also, it might be a nod to the behavior presented by stem cells of transferring damaged mitochondria, either through tunneling nanotubes or endosomes, to the monocytic compartment36. In this case, since the mitochondria do not undergo phagocytosis, they are transferred instead, which means that the damaged mitochondria are now part of the monocytic cell cytoplasm. Hence, their turnover is processed through autophagy.
All FLT3-ITD samples grouped as poor responders to the FLT3 inhibitors assessed in this study were consistently associated with a molecular signature related to autophagy and, especially, inflammasome activation. It is noteworthy that gilteritinib, midostaurin, and quizartinib, three FDA-approved FLT3 inhibitors, enriched both inflammasome- and autophagy-related gene sets among poor responders in conventional gene set enrichment analysis. Once again, this finding underscores a potential shared feature within the heterogeneous nature of the disease.
The genes MEFV and NLRP12 appeared as leading-edge genes for most inflammation regulatory gene sets enriched in poor ex vivo responders’ samples. This finding agrees with the observations that AML cells resort to differential inflammation suppression mechanisms in order to withstand the inflammatory environment at the expense of healthy hematopoietic cells37,38. Besides, autophagy, more specifically selective autophagy, is able to remove several of the endogenous inflammasome activation triggers, including the inflammasome machinery itself, and reportedly promotes greater resistance to pyroptotic cell death in AML cells while regulating internal oxidative stress16,18,39,40.
Even though all FLT3-ITD samples that were poor ex vivo responders to FLT3 inhibitors enriched inflammation-related molecular programs on GSEA, only quizartinib and sorafenib displayed strong associations from a single-sample GSEA perspective. We argue that the inhibitor’s structural nature can explain this finding. Since both sorafenib and quizartinib are type II FLT3 inhibitors, they possess a particular affinity for FLT3-ITD instead of the relative generality found within type I inhibitors as crenolanib, gilteritinib and midostaurin.
Intriguingly, the molecular pattern enriched by FLT3 inhibitors for FLT3-ITD poor ex vivo responders’ samples bear a remarkable resemblance to the molecular signature observed in AML samples without FLT3-ITD. This finding implies the presence of an entity within the FLT3-ITD samples that share molecular similarities with the samples without FLT3-ITD. This insight might provide a molecular rationale for FLT3 inhibitor’s intrinsic resistance through a molecular standpoint outwards the absence of FLT3-ITD.
Overall, we observed and proposed in a novel manner that autophagy and inflammasome do establish a transcriptional relationship in AML cohort samples. In addition to this, considering that the samples were sequenced previously to the ex vivo treatment, the enrichment of molecular signatures compatible with autophagy and inflammasome activation not only depict inherent sources of resistance to FLT3 inhibitors, but also were observed to be associated with poor response to type II FLT3 inhibitors. Nonetheless, this relationship and its practical therapeutic impacts are yet to be thoroughly investigated. Considering the inherent limitations of this work’s nature, we recognize that further experimentation is an imperative affair to assess whether this interplay is maintained on in vitro and in vivo models and whether it could be a viable potential strategy for clinical management improvement.
Methods
Transcriptomic and ex vivo response data acquisition
Mutation status and messenger RNA sequencing (RNA-seq) data from primary AML bone marrow mononuclear cells samples from The Cancer Genome Atlas (TCGA) AML cohort41 were obtained through the cBioPortal repository (cbioportal.org). The BeatAML 1.0 cohort patients’ RNA-seq, mutation status, and area under the curve (AUC) ex vivo treatment response data were obtained through the supplementary documentation in Tyner et al.42, through cBioPortal repository, and the BeatAML 1.0 associated data viewer, Vizome (vizome.org).
Correlation matrix analyses
The correlation matrices were built through Hmisc (RRID: SCR_022497) R package (Harrel, 2023; https://CRAN.R-project.org/package=Hmisc) employing Spearman’s Rank Correlation. The Hmisc’s rcorr function was employed to obtain a Spearman’s correlation coefficient matrix correlating all interest autophagy and inflammasome genes, and an overlapping matrix containing each respective analysis p-value. This feature facilitates the selection of correlations that match the established level of significance.
The autophagy and the inflammasome human genes that constituted the correlation analyses were obtained from RT2 Profiler PCR Array (QIAGEN; Hilden, NRW, Germany; catalog# 330231) product’s gene set-up. The autophagy and inflammasome genes were first thoroughly extracted and since overlapping was observed, the genes were sorted to each group according to convenience based on their reported respective involvement degree with the biological process and downstream continuity of signaling pathways. A total of 158 genes are equally distributed between autophagy (n = 79) and inflammasome (n = 79) biological processes (Supplementary Table 1). The elements of the correlation matrix were ordered according to the angular order of the matrix’s eigenvectors.
Deconvolution analysis
The bulk RNA-seq data of the BeatAML 1.0 and TCGA AML cohorts were submitted to a digital cytometry tool known as CIBERSORTx43 (https://cibersortx.stanford.edu/). The CIBERSORTx promotes deconvolution of the transcriptome signal from a previous cell mixture. In the current context, bone marrow mononuclear cells transcriptome signal was deconvoluted into the different cellular compartments that constitutes it and assigned a compartment score for each type of cell population assessed according to a reference single-cell RNA sequencing (scRNA-seq) data, in this case van Galen et al.. leukemia and healthy scRNAseq bone marrow primary samples44. The compartment score indicates which cell type is predominant within a sample, therefore its cellular composition regarding abundance and how much of the total transcriptome signal corresponds to a particular cell type. CIBERSORTx-derived deconvolution data from both BeatAML 1.0 and TCGA AML cohorts is available in Zeng et al.. supplementary material45.
Gene set enrichment analysis
FLT3-ITD-positive AML patients from BeatAML 1.0 cohort were arbitrarily dichotomized according to their median ex vivo treatment response to FLT3 inhibitors: midostaurin, quizartinib, gilteritinib, crenolanib, and sorafenib into good responders when bellow the AUC median and poor responder otherwise. The statistical characterization of each response data group to each FLT3 inhibitors is provided as supplementary material. These groups were then associated with their RNA-seq data and underwent differential gene expression.
The gene set enrichment analysis (RRID: SCR_003199) was performed from the gene expression logarithm fold change (logFC)-based pre-ranking. LogFC values were obtained from differential gene expression analysis using the Galaxy (RRID: SCR_006281) platform and under the limma-voom algorithm tool46 (usegalaxy.org). The pre-ranked genes served as the input for the fgsea (RRID: SCR_020938) Bioconductor R package. The gene sets are obtainable from the Molecular Signature Database (MSigDB)47 (https://www.gsea-msigdb.org/).
The pre-ranked genes enrichment was submitted to 10,000 permutations under weighted enrichment statistics. The level of significance was pre-established at 5% and adjusted to a false discovery rate of 25%. This work has employed the Gene Ontology: Biological Process (GOBP; 7751 gene sets) collection of human gene sets that were loaded using the qusage (RRID: SCR_024255) Bioconductor R package. The GOBP gene sets were selected and plotted by convenience according to the statistical significance criteria along with the false discovery rate using the R 4.3.1 software package ggplot2 (RRID: SCR_014601).
Single-sample gene set enrichment analysis
The ssGSEA was performed in R 4.3.1 employing Pranali’s script (https://rpubs.com/pranali018/SSGSEA) according to the methodology described by Barbie et al.48. Enrichment score was plotted according to the regrouped samples original category in order to support the GSEA findings. The ssGSEA enrichment scores (ES) respective to each molecular program found in GSEA output were dichotomized into a binary categorical variable comprising high and low signatures according to the ES median. The association between ex vivo response – poor and good response – and the enrichment scores – high and low signature – was inquired through Fisher’s exact test inbuilt R 4.3.1 function.
Statistical analyses
The data were compared according to the fulfillment of the assumptions each statistical inference model inherently requires. The normality of distribution on the data linear regression model residuals was inquired using a quantile-quantile plot (Figure S2), supported by data residuals density plot (Figure S1) and the Shapiro-Wilk model. Data scedasticity, was assessed by the modified robust Brown-Forsythe Levene-type test for absolute deviations from the median when data residuals for two or more groups that are not normally distributed, F-test when the data residuals of two groups are normally distributed, and Bartlett’s test when comparing three or more groups that are normally distributed (Supplementary Table 2).
Considering two homoscedastic groups whose data residuals are normally distributed, Student’s T-test was the more fitting statistical model to perform the comparative analysis, while when the groups were heteroscedastic Welch’s T-test was the model of preference. In case of non-normally distributed data residuals, the comparison between two groups was performed using Wilcoxon rank sum test regardless of scedasticity (Supplementary Table 2).
Contingency tables were analyzed by Fisher’s exact test and the effect size was measured according to the odds ratio (OR) and a 95% confidence interval. Since the good responder groups are the reference groups for comparison, the natural logarithm of the reciprocal OR (1/OR) was plotted instead for a more informative reading of the data.
All the statistical analyses were performed using R programming language version 4.3.1 (R Core Team (2022) - https://www.R-project.org/) (RRID: SCR_001905) and RStudio Integrated Development Environment (IDE) version 2023.03.0 + 386 (RStudio Team (2020) – http://www.rstudio.com/) (RRID: SCR_000432). The established level of significance (α) was 5% for all the analyses.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the São Paulo Research Foundation (FAPESP) (grants #2013/08135-2 [Research, Innovation and Dissemination Centers – Cell Therapy Center], #2022/03871-1, and #2021/11112-0), the National Council for Scientific and Technological Development (CNPq) (465539/2014-9 [National Institute of Science and Technology in Stem Cell and Cell Therapy in Cancer], 409401/2021-8, and 309614/2022-8), and the Coordination for the Improvement of Higher Education Personnel (CAPES) (88887.650052/2021-00), funding code #001.
Author contributions
B.G.S.M. executed formal data analysis and interpretation, original manuscript writing, prepared Figs. 1, 2, 3, 4 and 5, prepared supplementary material, and contributed to conceptualization. M.A.M. contributed to conceptualization. D.A.P.M contributed to data analysis and interpretation. J.A.M.N and F.T contributed with conceptualization, formal analysis, supervision, and manuscript writing. All authors reviewed the manuscript and approved its final version for submission.
Data availability
The RNA sequencing and ex vivo drug screening datasets analysed during the current study are publicly available in the cBioPortal and Vizome repositories, respectively at https://www.cbioportal.org/ and http://www.vizome.org/. Deconvolution analysis for both datasets is available at A. G. X. Zeng et al., “A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia,” Nat Med, vol. 28, no. 6, pp. 1212–1223, Jun. 2022, doi: 10.1038/s41591-022-01819-x supplementary material.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Culp-Hill, R., D’Alessandro, A. & Pietras, E. M. Extinguishing the embers: Targeting AML metabolism. Trends Mol Med27(4), 332–344. 10.1016/j.molmed.2020.10.001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Kouchkovsky, I. & Abdul-Hay, M. ‘Acute myeloid leukemia: A comprehensive review and 2016 update’. Blood Cancer Journal6(7), e441–e441. 10.1038/bcj.2016.50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khwaja, A. et al. Acute myeloid leukaemia. Nat Rev Dis Primers2(1), 16010. 10.1038/nrdp.2016.10 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Luciano, M., Krenn, P. W. & Horejs-Hoeck, J. The cytokine network in acute myeloid leukemia. Front Immunol13, 1000996. 10.3389/fimmu.2022.1000996 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prada-Arismendy, J., Arroyave, J. C. & Röthlisberger, S. Molecular biomarkers in acute myeloid leukemia. Blood Rev31(1), 63–76. 10.1016/j.blre.2016.08.005 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Bazinet, A. & Assouline, S. A review of FDA-approved acute myeloid leukemia therapies beyond ‘7 + 3’. Expert Rev Hematol14(2), 185–197. 10.1080/17474086.2021.1875814 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Hindley, A., Catherwood, M. A., McMullin, M. F. & Mills, K. I. Significance of NPM1 gene mutations in AML. Int J Mol Sci22(18), 10040. 10.3390/ijms221810040 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kayser, S. & Levis, M. J. The clinical impact of the molecular landscape of acute myeloid leukemia. Haematologica108(2), 308–320. 10.3324/haematol.2022.280801 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padmakumar, D. et al. A concise review on the molecular genetics of acute myeloid leukemia. Leuk Res111, 106727. 10.1016/j.leukres.2021.106727 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Marando, L. & Huntly, B. J. P. Molecular landscape of acute myeloid Leukemia: Prognostic and therapeutic implications. Curr Oncol Rep22(6), 61. 10.1007/s11912-020-00918-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daver, N., Schlenk, R. F., Russell, N. H. & Levis, M. J. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia33(2), 299–312. 10.1038/s41375-018-0357-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, H. Emerging agents and regimens for AML. J Hematol Oncol14(1), 49. 10.1186/s13045-021-01062-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Döhner, H. et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood140(12), 1345–1377. 10.1182/blood.2022016867 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Awada, H. et al. A focus on intermediate-risk acute myeloid leukemia: Sub-classification updates and therapeutic challenges. Cancers (Basel)14(17), 4166. 10.3390/cancers14174166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allert, C., Müller-Tidow, C. & Blank, M. F. The relevance of the hematopoietic niche for therapy resistance in acute myeloid leukemia. Int J Cancer10.1002/ijc.34684 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Biasizzo, M. & Kopitar-Jerala, N. Interplay between NLRP3 inflammasome and autophagy. Front Immunol11, 591803. 10.3389/fimmu.2020.591803 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanson, K. V., Deng, M. & Ting, J.P.-Y. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat Rev Immunol19(8), 477–489. 10.1038/s41577-019-0165-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu, K. K. L. & Cheng, K. K. Y. A new role of the early endosome in restricting NLRP3 inflammasome via mitophagy. Autophagy18(6), 1475–1477. 10.1080/15548627.2022.2040314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bueno, E., Wyatt, S., Duttenhefner, R., Asa, D., Dasanna, S., and Sinha, S. C. “Chapter 21 - Autophagy as an integral immune system component,” in Autophagy in Health and Disease (Second Edition), B. A. Rothermel and A. Diwan, Eds., Academic Press, 2022, pp. 303–320. 10.1016/B978-0-12-822003-0.00011-5.
- 20. Evans, T. D., Sergin, I., Zhang, X., and Razani, B. “Target acquired: Selective autophagy in cardiometabolic disease,” Sci Signal, 10(468), p. eaag2298 (2017). 10.1126/scisignal.aag2298. [DOI] [PMC free article] [PubMed]
- 21.Lara-Reyna, S. et al. Inflammasome activation: from molecular mechanisms to autoinflammation. Clin Transl Immunology11(7), e1404. 10.1002/cti2.1404 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, L. & Xia, H. TRIM proteins in inflammation: From expression to emerging regulatory mechanisms. Inflammation44(3), 811–820. 10.1007/s10753-020-01394-8 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Zhong, C. et al. NLRP3 inflammasome promotes the progression of acute myeloid leukemia via IL-1β pathway. Front Immunol12, 661939. 10.3389/fimmu.2021.661939 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carey, A. et al. Identification of interleukin-1 by functional screening as a key mediator of cellular expansion and disease progression in acute myeloid leukemia. Cell Rep18(13), 3204–3218. 10.1016/j.celrep.2017.03.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Boer, B. et al. The IL1-IL1RAP axis plays an important role in the inflammatory leukemic niche that favors acute myeloid leukemia proliferation over normal hematopoiesis. Haematologica106(12), 3067–3078. 10.3324/haematol.2020.254987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei, S. et al. AMPK/FIS1-mediated mitophagy is required for self-renewal of human AML stem cells. Cell Stem Cell23(1), 86–100. 10.1016/j.stem.2018.05.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saulle, E., Spinello, I., Quaranta, M. T. & Labbaye, C. Advances in understanding the links between metabolism and autophagy in acute myeloid leukemia: From biology to therapeutic targeting. Cells12(11), 1553. 10.3390/cells12111553 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shevyrev, D., Tereshchenko, V., Berezina, T. N. & Rybtsov, S. Hematopoietic stem cells and the immune system in development and aging. Int J Mol Sci24(6), 5862. 10.3390/ijms24065862 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joffre, C., Ducau, C., Poillet-Perez, L., Courdy, C., and Mansat-De Mas, V. Autophagy a close relative of AML biology. Biology (Basel), 10(6), 552 (2021). 10.3390/biology10060552. [DOI] [PMC free article] [PubMed]
- 30.Hansen, M., Rubinsztein, D. C. & Walker, D. W. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol19(9), 579–593. 10.1038/s41580-018-0033-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto, H., Zhang, S. & Mizushima, N. Autophagy genes in biology and disease. Nat Rev Genet24(6), 382–400. 10.1038/s41576-022-00562-w (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arthur, J. C., Lich, J. D., Aziz, R. K., Kotb, M. & Ting, J.P.-Y. Heat shock protein 90 associates with monarch-1 and regulates its ability to promote degradation of NF-kappaB-inducing kinase. J Immunol179(9), 6291–6296. 10.4049/jimmunol.179.9.6291 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Normand, S. et al. Proteasomal degradation of NOD2 by NLRP12 in monocytes promotes bacterial tolerance and colonization by enteropathogens. Nat Commun9(1), 5338. 10.1038/s41467-018-07750-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pati, S., Sarkar, S., Das, E., Sherpa, N., Kanti Das, M., and Datta, S. Frightening fever: Familial cold autoinflammatory syndrome 2 (FCAS-2) with macrophage activation syndrome (MAS). Indian J Pediatr89(10), 1055 (2022). 10.1007/s12098-022-04322-w. [DOI] [PMC free article] [PubMed]
- 35.Wang, H.-F. NLRP12-associated systemic autoinflammatory diseases in children. Pediatr Rheumatol Online J20(1), 9. 10.1186/s12969-022-00669-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarmah, D. et al. Mitochondrial dysfunction in stroke: Implications of stem cell therapy. Transl Stroke Res10.1007/s12975-018-0642-y (2018). [DOI] [PubMed] [Google Scholar]
- 37.Avagyan, S. et al. Resistance to inflammation underlies enhanced fitness in clonal hematopoiesis. Science374(6568), 768–772. 10.1126/science.aba9304 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Hormaechea-Agulla, D. et al. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNγ signaling. Cell Stem Cell28(8), 1428–1442. 10.1016/j.stem.2021.03.002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vargas, J. N. S., Hamasaki, M., Kawabata, T., Youle, R. J. & Yoshimori, T. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol24(3), 167–185. 10.1038/s41580-022-00542-2 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Cao, W., Li, J., Yang, K. & Cao, D. An overview of autophagy: Mechanism, regulation and research progress. Bull Cancer108(3), 304–322. 10.1016/j.bulcan.2020.11.004 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Ley, T. J. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med368(22), 2059–2074. 10.1056/NEJMoa1301689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyner, J. W. et al. Functional genomic landscape of acute myeloid leukaemia. Nature562(7728), 526–531. 10.1038/s41586-018-0623-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol37(7), 773–782. 10.1038/s41587-019-0114-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Galen, P. et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell176(6), 1265–1281. 10.1016/j.cell.2019.01.031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng, A. G. X. et al. A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia. Nat Med28(6), 1212–1223. 10.1038/s41591-022-01819-x (2022). [DOI] [PubMed] [Google Scholar]
- 46.Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res44(W1), W3–W10. 10.1093/nar/gkw343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A102(43), 15545–15550. 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature462(7269), 108–112. 10.1038/nature08460 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing and ex vivo drug screening datasets analysed during the current study are publicly available in the cBioPortal and Vizome repositories, respectively at https://www.cbioportal.org/ and http://www.vizome.org/. Deconvolution analysis for both datasets is available at A. G. X. Zeng et al., “A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia,” Nat Med, vol. 28, no. 6, pp. 1212–1223, Jun. 2022, doi: 10.1038/s41591-022-01819-x supplementary material.